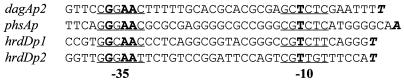Abstract
The sigE gene of Streptomyces coelicolor A3(2) encodes an RNA polymerase sigma factor belonging to the extracytoplasmic function (ECF) subfamily. Constructed sigE deletion and disruption mutants were more sensitive than the parent to muramidases such as hen egg white lysozyme and to the CwlA amidase from Bacillus subtilis. This correlated with an altered muropeptide profile, as determined by reverse-phase high-performance liquid chromatography analysis of lytic digests of purified peptidoglycan. The sigE mutants required high levels of magnesium for normal growth and sporulation, overproducing the antibiotic actinorhodin and forming crenellated colonies in its absence. Together, these data suggest that sigE is required for normal cell wall structure. The role of ςE was further investigated by analyzing the expression of hrdD, which is partially sigE dependent. The hrdD gene, which encodes the ςHrdD subunit of RNA polymerase, is transcribed from two promoters, hrdDp1 and hrdDp2, both similar to promoters recognized by other ECF sigma factors. The activities of hrdDp1 and hrdDp2 were reduced 20- and 3-fold, respectively, in sigE mutants, although only hrdDp1 was recognized by EςE in vitro. Growth on media deficient in magnesium caused the induction of both hrdD promoters in a sigE-dependent manner.
The ςE subunit of Streptomyces coelicolor RNA polymerase was originally identified by its ability to direct transcription in vitro from dagAp2, one of four promoters of the dagA gene, which encodes an extracellular agarase (8, 9). Cloning of the ςE gene (sigE) led to the discovery of a distinct subfamily of sigma factors, named the extracytoplasmic function (ECF) subfamily, which are structurally distinct from other members of the ς70 family (39). ECF sigma factors have now been described in a wide range of gram-positive and gram-negative bacteria and, where studied, have been shown to be involved in regulating a variety of functions, typically concerned with the extracytoplasmic environment (44). Well-studied examples include ςE of Escherichia coli, required for transcription of genes involved in the turnover and correct folding of periplasmic proteins (13, 54, 56); ςFecI, which responds to extracellular iron(III) dicitrate and directs the transcription of genes required for its uptake in E. coli (2); ςCarQ, required for carotenoid biosynthesis in Myxococcus xanthus (22); and ςAlgU, which regulates the production of extracellular alginate in Pseudomonas aeruginosa (16, 24).
Members of the ECF subfamily have a number of common features which distinguish them from other members of the ς70 family. First, of the four conserved regions in ς70-related sigma factors (38, 39), region 3 and much of region 1 are usually absent, resulting in the typically small size of ECF sigma factors (usually 20 to 30 kDa). Second, promoters regulated by ECF sigma factors are strikingly similar, especially in the −35 region, where a GAAC motif is conserved (39, 44). Third, their activity is often regulated by specific anti-sigma factors encoded by downstream genes. Examples are RseA in the case of E. coli ςE (14, 45) and CarR in the case of ςCarQ (22).
One of the most striking aspects of ECF sigma factors is that relatively few have been identified by traditional genetic means, although very large numbers of ECF sigma factor genes are now being uncovered in a variety of bacteria through genome sequencing. For example, in Bacillus subtilis there are seven ECF sigma factor genes (37), none of which was discovered genetically. This seems to imply either that they are functionally redundant or that they control the expression of genes not relevant to normal laboratory culture conditions. Recently, it was shown that there is indeed some redundancy among the ECF sigma factors of B. subtilis. A sigX mutant of B. subtilis has slightly increased sensitivity to heat and oxidative stress but has no other obvious phenotype (30). ςX contributes to the transcription from at least seven promoters, four of which are also recognized by one or more different ECF sigma factors in vivo (29). The other three promoters are completely dependent on ςX in vivo, but a second, ςX-independent promoter also contributes to the expression of the respective genes. Therefore, each of the seven known members of the ςX regulon is transcribed by more than one RNA polymerase holoenzyme, meaning that disruption of sigX will not abolish the expression of any of these genes. This may explain, at least in part, the subtlety of the sigX mutant phenotype (29).
To date, the biological role of ςE in Streptomyces has been investigated only in the actinomycin producer S. antibioticus. An S. antibioticus sigE null mutant was deficient in actinomycin production, although direct targets for ςE have not been identified (34). Here we investigate the biological role of ςE in S. coelicolor. We show that constructed sigE null mutants have an altered cell wall structure, increased sensitivity to cell wall-lytic enzymes, and a distinct peptidoglycan muropeptide profile. We also show that sigE mutants require high levels of magnesium for normal growth and sporulation, and we identify a ςE-dependent promoter that is induced when cultures are grown under conditions of magnesium deficiency.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. coelicolor A3(2) M600 (11) and its derivatives were cultivated on MM, R2, R2YE (28), MS agar (mannitol plus soy flour) (27), SMMS agar (19), and NMMP liquid media (28), essentially as described previously (28). Unmethylated DNA for introduction into S. coelicolor was isolated from E. coli ET12567 (dam dcm hsdS) (40). Plasmids are described in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pIJ2925 | Cloning vector based on pUC18 (bla lacZa) | 33 |
| pMT3000 | pIJ2925 with modified polycloning site (bla lacZa) | 50 |
| pDH5 | E. coli phagemid cloning vector (bla tsr lacZa) | 25 |
| pSET151 | E. coli conjugative plasmid (bla tsr lacZa oriT) | 5 |
| pSET152 | E. coli/Streptomyces conjugative plasmid containing attP and integration functions of φC31 [aac(3)IV lacZa oriT] | 5 |
| pET11c | T7 expression vector (bla) | Novagen |
| pUZ8002 | RK2 derivative with defective oriT (aph) | 62 |
| pIJ2020 | The multicopy vector pIJ486 carrying dagA on a 5-kb BamHI fragment | 7 |
| pIJ2027 | The promoter region of dagA cloned in pUC18 | 7 |
| pIJ2036 | 1.9-kb SmaI fragment containing hrdD cloned in pIJ2925 | 7 |
| pIJ5950 | 2.05-kb PvuII fragment containing sigE cloned in pIJ2925 | This work |
| pIJ5951 | ΔsigE derivative of pIJ5950 | This work |
| pIJ5952 | ΔsigE cloned in pDH5 | This work |
| pIJ5954 | sigE::hyg derivative of pIJ5950 | This work |
| pIJ5855 | sigE::hyg cloned in pSET151 | This work |
| pIJ2076 | 0.87-kb HindIII-XhoI fragment of pIJ5950 replaced with two complementary oligonucleotides generating an NdeI site at the initiation codon of sigE | This work |
| pIJ2078 | sigE-containing NdeI-BglII fragment cloned in pET11c | This work |
Overproduction of ςE.
A 2.05-kb PvuII fragment carrying sigE was cloned into SmaI-cut pIJ2925 (33) such that, in the resulting plasmid, pIJ5950, the HindIII site in the polylinker was upstream of sigE. The 870 bp of DNA between the HindIII site and a unique XhoI site 10 bp downstream of the sigE ATG start codon was replaced with two complementary oligonucleotides (5′-AGCTTCCATATGGGTGAAGTTC-3′ and 5′-TCGAGAACTTCACCCATATGGA-3′) that introduced an NdeI site overlapping the ATG start codon and also replaced the second, third, and fourth codons with synonymous codons commonly associated with genes expressed at high levels in E. coli. The cassette replacement was verified by sequencing the resulting plasmid (pIJ2076), and sigE was excised as a 1.2-kb NdeI-BglII fragment and cloned into the expression vector pET11c (Novagen), which had been cut with NdeI and BamHI, to generate pIJ2078.
pIJ2078 was introduced into E. coli BL21λDE3(pLysS) (59), and sigE expression was induced in exponentially growing cells (optical density at 600 nm, 0.5) by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). ςE was recovered from inclusion bodies essentially as described previously (48). Inclusion bodies were solubilized with 0.25 (wt/vol) Sarkosyl (N-lauroylsarcosine), followed by extensive dialysis to remove Sarkosyl and to allow ςE to refold. ςE was further purified by Mono-Q anion-exchange column chromatography.
Approximately 10 μg of purified ςE was subjected to sequential Edman degradation in order to determine the sequence of the 1st 10 N-terminal residues. The result—GEVLEFEEYV—showed complete agreement with that predicted from the DNA sequence of sigE (39) and showed that the N-terminal N-formylmethionine had been removed, as it is in S. coelicolor (39).
In vitro transcription.
Runoff transcription assays were performed with [α-32P]CTP (600 Ci mmol−1; Dupont-NEN) as described by Buttner et al. (8). Typical reaction mixtures contained 1.25 pmol of E. coli core RNA polymerase (Epicentre Technologies, Madison, Wis.) and 12.5 pmol of ςE. Transcription from the hrdD promoter region was assayed by using two fragments isolated from pIJ2036: a 490-bp HindIII-NarI fragment and a 630-bp HindIII-SstI fragment. Transcripts were analyzed on 6% polyacrylamide-7 M urea gels with heat-denatured 32P-labelled HpaII digests of pBR322 as size standards. S. coelicolor RNA polymerase holoenzyme, purified from YEME-grown cultures, was a gift from T. Fujii and E. Takano.
Construction of sigE mutants of S. coelicolor A3(2).
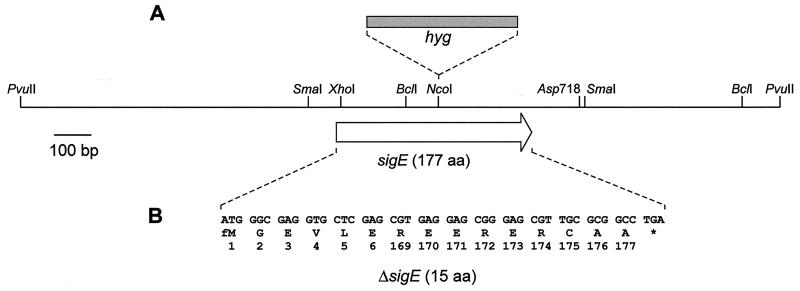
A PCR-based approach was used to generate an internal in-frame deletion in sigE. By using pIJ5950 as a template, DNA downstream from sigE was amplified with a primer complementary to the C-terminal region of sigE which incorporated an XhoI site at the 5′ end (MP1; 5′-CGCTCGAGCGTGAGGAGCGG-3′) and a primer complementary to a sequence just downstream from the Asp718 site (KB5; 5′-GCCGGTACCCCCGGTCC-3′) (see Fig. 1). The resulting 150-bp product was digested with XhoI and Asp718, then inserted into XhoI-Asp718-digested pMT3000 (50) and checked by DNA sequencing. The XhoI-Asp718 fragment was reisolated from pMT3000 and inserted into pIJ5950, replacing the sigE-containing XhoI-Asp718 fragment. The resulting plasmid, pIJ5951, contains ΔsigE flanked by 0.85 kb of DNA upstream and 0.7 kb downstream. ΔsigE was isolated as a BamHI-BglII fragment and inserted into BamHI-digested pDH5 to give pIJ5952. pIJ5952 was passaged through the nonmethylating E. coli strain ET12567, then used to transform S. coelicolor M600, with selection for thiostrepton resistance (Thior). A representative transformant was subcultured on nonselective media for one round of sporulation, and the resulting spores were plated out to allow identification of Thios colonies. Thios colonies were screened for the presence or absence of the sigE gene by Southern hybridization of digested chromosomal DNA. A representative Thios isolate in which sigE had been deleted was designated J2130.
FIG. 1.
Restriction map of the S. coelicolor 2.05-kb PvuII insert containing sigE in pIJ5950. The sigE open reading frame is represented by an open arrow; the position of the hyg insertion mutation (A) and the extent and sequence of the ΔsigE in-frame deletion mutation (B) are shown.
A disruption mutation was made in sigE by inserting a hygromycin resistance cassette (hyg) into the internal NcoI site. The 1.7-kb hyg cassette (63) was blunt ended and cloned into pIJ5950 which had been digested with NcoI and filled in, to give pIJ5954. The sigE::hyg mutant allele was isolated as a BglII fragment and cloned into pSET151. The resulting plasmid, pIJ5955, was used to transform S. coelicolor M600, with selection for hygromycin resistance (Hygr). All transformants were also Thior, indicating a single crossover event. sigE::hyg mutants were identified by screening for Hygr Thios colonies after a round of nonselective growth, and their structures were confirmed by Southern hybridization. A representative sigE::hyg mutant of M600 was designated J2141.
Conjugation.
pSET152 and its derivatives were introduced by transformation into ET12567 containing the RK2 derivative pUZ8002 (62). pUZ8002 can supply transfer functions to oriT-carrying plasmids, such as pSET152, but is not efficiently transferred itself because of a mutation in its own oriT. However, a low level of self-transfer allowed pUZ8002 to be introduced into ET12567. Conjugations between E. coli (pUZ8002) and S. coelicolor were carried out essentially as described previously (34).
RNA isolation.
RNA was isolated from liquid-grown mycelium as described elsewhere (28). However, rather than exhaustive phenol-chloroform extraction and DNase treatment, RNA was purified from contaminating DNA and protein by CsCl gradient centrifugation following an initial phenol-chloroform extraction, as described previously (26). RNA was isolated from solid media by scraping mycelium and spores from cellophane-covered plates and extracting as described above except that, after the addition of phenol-chloroform, the mixture was heated at 65°C for 10 min prior to vortex mixing.
S1 nuclease transcription mapping.
The hrdD promoter region was mapped by using a probe generated by PCR from pIJ2036 with a 5′-end-labelled oligonucleotide primer internal to hrdD (HD1; 5′-TTCAGCGGGTGGTCCGGTGGAC-3′) and the reverse sequencing primer. HD1 (30 pmol) was labelled with [γ-32P]ATP (3,000 Ci mmol−1; Dupont-NEN) (57). The end-labelled PCR product was purified from an agarose gel by using a gel extraction kit (Qiagen). The dagA promoter region was mapped by using a 560-bp SmaI-AvaII fragment isolated from pIJ2027, labelled uniquely at the 5′ end of the AvaII site, as described elsewhere (8). The protected DNA fragments were quantified with a phosphorimager (Fujix BAS1000).
Lysozyme sensitivity test.
Sensitivity to lysozyme was tested by spotting 5 μl of lysozyme in 10 mM Tris-HCl (pH 8) at various concentrations on confluent lawns of spores on agar (2 × 106 spores per plate). Other cell wall-lytic enzymes tested were Cellosyl (a gift from R. Marquardt, Hoechst AG, Frankfurt, Germany), mutanolysin (Sigma), or CwlA amidase (20).
Isolation and analysis of peptidoglycan.
Peptidoglycan was prepared by a modification of the method of Atrih et al. (3). Fifty milliliters of mid- to late-exponential cultures in NMMP plus glucose were centrifuged for 2 min at 5,000 × g. The mycelium was resuspended in 10 ml of extraction buffer (50 mM Tris-HCl, 2 mM EDTA, 10 mM dithiothreitol [pH 7]), heated in a boiling-water bath for 20 min to inactivate autolysins, and then transferred to ice. The mycelium was disrupted by using a French press, and then 10% sodium dodecyl sulfate was added to a final concentration of 4%, followed by incubation at 37°C for 40 min. Crude walls were collected at room temperature by centrifugation at 11,000 × g for 20 min, then washed four times with warm water. The pellet was resuspended in 6 ml of Tris-HCl (pH 7) and treated with pronase (0.5 mg/ml; Sigma) at 60°C for 90 min. Cell walls were collected as before, resuspended in 5 ml of extraction buffer containing 4% sodium dodecyl sulfate, and boiled for 16 min. Purified walls were collected and washed four times with water as above, then resuspended in MilliQ water and stored at −20°C.
Cell walls were treated with aqueous hydrofluoric acid to remove the accessory polymers, then digested with Cellosyl and reduced with sodium borohydride (at a final concentration of 2 mg/ml) as previously described (3). The muropeptide profile was investigated by applying Cellosyl-digested peptidoglycan to a TSK SWXL2000 gel filtration column (7.8 mm by 30 cm; Anachem, Luton, United Kingdom). The muropeptides were eluted with 40 mM sodium phosphate (pH 6.5) at a flow rate of 0.3 ml min−1, and the eluted compounds were detected by the absorbance at 202 nm (A202). Muropeptide separations carried out at a higher temperature (45°C versus the standard 40°C) or over a longer gradient (180 min versus the standard 160 min) resulted in similar profiles. Amino acid analysis was performed by the Pico Tag method (3). The cross-linking index was determined as described elsewhere (42).
RESULTS
sigE mutants are sensitive to cell wall-hydrolytic enzymes.
To investigate the biological role of sigE in S. coelicolor, two mutant alleles were constructed in vitro: one with an in-frame internal deletion mutation and one with a hygromycin resistance gene (hyg) insertion mutation (Fig. 1). These mutant alleles were used to replace the wild-type allele in M600, a plasmid-free derivative of the wild-type strain, creating J2130 (ΔsigE) and J2141 (sigE::hyg). The former mutation should be nonpolar on potential downstream genes, whereas the hyg insertion mutation might affect the expression of any genes cotranscribed with sigE.
Compared to M600, the sigE mutants grew well and sporulated with equal vigor on MM, R2, R2YE, and SMMS media. They produced normal levels of the pigmented antibiotics actinorhodin and undecylprodigiosin and of the calcium-dependent lipopeptide antibiotic CDA. The sigE mutants were also unaffected in their resistance to heat, their ability to produce siderophores, and their resistance to oxidizing agents—phenotypes controlled by ECF sigma factors in other bacteria (44).
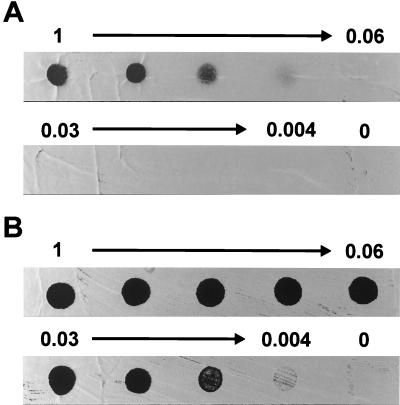
However, the sigE mutants were up to 50 times more sensitive to egg white lysozyme (Fig. 2), an enzyme which hydrolyzes the β1-4 linkage between adjacent N-acetylmuramic acid and N-acetylglucosamine units in the glycan backbone of peptidoglycan. In our standard assay, lysozyme was spotted onto freshly plated confluent lawns of spores and zones of clearing were noted after 48 h. Under these conditions, lysozyme sensitivity probably arises soon after germination because Streptomyces spores are lysozyme resistant, whereas germlings are especially sensitive (55). Indeed, identical results were obtained by spotting lysozyme on newly germinated spores. Vegetative mycelia from sigE mutants were also more sensitive to lysozyme than that from the wild type, as judged by spotting lysozyme on 12-h-old confluent plates. To rule out the possibility that lysozyme sensitivity was caused by polar effects on possible downstream genes, sigE was reintroduced into the mutants by using the vector pSET152, which integrates site-specifically at the phage φC31 attB site (5). A derivative of pSET152 carrying a 2.05-kb PvuII fragment containing sigE restored lysozyme resistance to J2130 and J2141, whereas pSET152 carrying the ΔsigE mutation in the same fragment did not (data not shown).
FIG. 2.
Lysozyme sensitivity of M600 (sigE+) (A) compared to that of J2130 (ΔsigE) (B). Spores of each strain were plated on Difco nutrient agar to give confluent lawns and then a twofold dilution series of egg white lysozyme (from 1 mg ml−1, as indicated) was spotted onto the plates immediately after plating. Zones of clearing, seen here as dark circles, were photographed after 2 days of incubation at 30°C.
In other organisms, resistance to lysozyme has been attributed to O acetylation of peptidoglycan at the C-6 hydroxyl moiety of muramyl residues (18). However, the sigE mutants were also much more sensitive than their parent to Cellosyl and mutanolysin, muramidases which are insensitive to this type of O acetylation, suggesting that the C-6 position is unaltered in acetylation.
To see if the sigE mutants were also susceptible to cell wall-lytic enzymes that cut other linkages in the peptidoglycan, we tested their sensitivity to the CwlA amidase from B. subtilis (20), which cleaves the peptide side chain from the glycan backbone. By the plate assay, the sigE mutants were found to be at least 50-fold more sensitive than the wild type to the CwlA amidase (data not shown). Sensitivity to more than one type of cell wall-hydrolytic enzyme suggested that the mutants have an altered cell wall structure, which allowed hydrolytic enzymes increased access to the peptidoglycan.
Muropeptide analysis of sigE mutant cell walls indicates an altered composition.
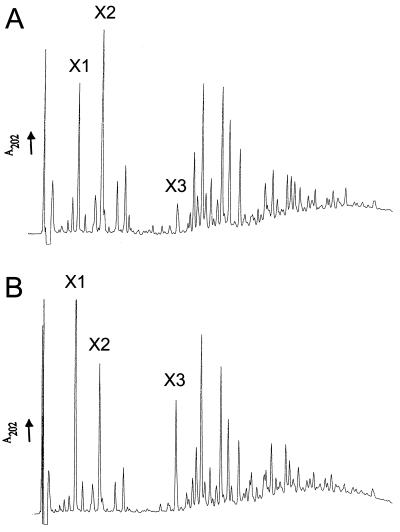
In an attempt to detect possible changes in cell wall structure, the muropeptide profiles of a sigE mutant and its congenic parent were determined. Cell walls were isolated from exponentially growing cultures and subjected to enzymatic hydrolysis followed by reverse-phase high-performance liquid chromatography (RP-HPLC). The muropeptide profiles obtained after RP-HPLC for M600 (sigE+) and J2130 (ΔsigE) are shown in Fig. 3A and B, respectively. For each strain, the profiles were reproducible both in peak retention time and in the relative amounts of different muropeptides for three independent cultures. The two strains presented an identical complement of muropeptides but showed differences in the abundances of certain muropeptides. For example, muropeptides X1 and X3, most likely a monomer and a dimer, respectively, are present in larger amounts in J2130, while muropeptide X2 is less abundant in J2130. Amino acid analysis revealed, in addition to glucosamine and muramic acid, comparable ratios of glutamic acid, glycine, alanine, and diaminopimelic acid in both strains. These amino acids have been identified previously in the peptidoglycan of streptomycetes (58). One possible explanation for the altered ratio of muropeptides would be a difference in the cross-linking of the peptidoglycan. To determine whether the sigE mutant was affected in peptidoglycan cross-linking and in the distribution of oligomers, muropeptides digested with Cellosyl were separated by gel permeation HPLC. The cross-linking index, as determined by the method of Martin and Gmeiner (42), was 47 for M600 and 46.4 for J2130, indicating no significant difference between the two strains.
FIG. 3.
Analysis of peptidoglycan composition by RP-HPLC. Samples of peptidoglycan isolated from M600 (sigE+) (A) or J2130 (ΔsigE) (B) were digested with Cellosyl and subjected to HPLC analysis, and the A202 values of the eluates were monitored. Muropeptides that show particularly different abundances in the two strains are marked (X1 through X3).
sigE mutants conditionally overproduce actinorhodin on media deficient in Mg2+.
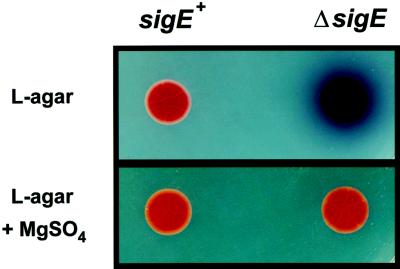
Although the sigE mutants appeared identical to the parent strain on most solid media, they overproduced the blue-pigmented antibiotic actinorhodin on certain complex media such as L agar (Fig. 4) and MS agar (data not shown). On these media the colonies sporulated very poorly and also had a crenellated appearance, which was not caused by actinorhodin overproduction, because it remained in a constructed sigE act double mutant (data not shown). Again, the overproduction of actinorhodin and the crenellation could be complemented in trans by integrating a functional copy of sigE into the chromosome at the φC31 attB site by using pSET152.
FIG. 4.
The ΔsigE mutant, J2130, overproduces the blue-pigmented antibiotic actinorhodin on medium deficient in Mg2+. Ten microliters of a spore suspension (108 spores ml−1) was spotted on L agar alone or on L agar plus 1.6 mM MgSO4. Plates were photographed after 3 days of incubation at 30°C.
The media on which the SigE phenotype was apparent (L agar and MS agar) lack added Mg2+, in contrast to the media on which the phenotype was not manifested (SMMS, R2YE, R2, and MM). When MgSO4 (1.6 mM) was included in the L-agar plates, the ΔsigE mutant J2130 no longer overproduced actinorhodin (Fig. 4) and the colony surface was no longer crenellated (data not shown). Also, when the concentration of MgSO4 in SMMS agar was reduced from the usual 5 mM to less than 1 mM, the sigE mutants produced actinorhodin earlier and in greater amounts than the parental wild type (data not shown). The suppression effect was caused by Mg2+ and not by the counterion, since it was observed with either MgSO4 or MgCl2. The addition of Ca2+ could also suppress the overproduction of actinorhodin and colony crenellation in the sigE mutants, although to a slightly lesser extent.
However, Mg2+ could not suppress the lysozyme sensitivity phenotype of sigE mutants. The presence of 5 mM Mg2+ in SMMS agar increased the resistance to lysozyme of both the sigE mutants and the parent, M600, but the sigE mutants remained substantially more sensitive to lysozyme than M600 in the presence of high (5 mM) or low (50 μM) concentrations of Mg2+.
The hrdDp1 promoter is ςE dependent.
To facilitate in vitro analysis of ςE, the protein was overproduced in E. coli and purified to homogeneity. A sigE overexpression plasmid, pIJ2078, based on the T7 expression vector pET11c, was constructed as described in Materials and Methods. Active ςE was recovered from inclusion bodies by using a minor modification of the method of Nguyen et al. (48). This involved the purification of the inclusion bodies, their solubilization with 0.25% (wt/vol) Sarkosyl, extensive dialysis to remove the detergent, and further purification by Mono-Q anion-exchange column chromatography.
Previous work showed that the ςE holoenzyme (EςE) purified from S. coelicolor can direct transcription from the dagAp2 promoter in vitro (9). An alignment of streptomycete promoters compiled by Bourn and Babb (6) revealed that the hrdDp1 promoter is identical to dagAp2 at 5 of 6 and 4 of 6 bases in the putative −35 and −10 regions, respectively (Fig. 5). hrdDp1 is one of two promoters that drive expression of the gene encoding ςHrdD, one of three S. coelicolor sigma factors that are very closely related in amino acid sequence and promoter specificity to ςHrdB, the principal, essential sigma factor of this species. However, the function of ςHrdD is unknown; hrdD null mutants are apparently unaffected in growth, morphological development, and antibiotic production (10). To see if ςE could direct transcription from hrdDp1 in vitro, recombinant ςE was added to core RNA polymerase and used to transcribe hrdDp1-containing templates. The two templates used, HindIII-NarI and HindIII-SstI, would be expected to lead to the formation of runoff products of 316 and 456 nucleotides (nt), respectively. Products of the expected sizes were obtained in a ςE-dependent manner (Fig. 6). In contrast, no transcription from hrdDp2 was detected. In vitro transcription using total RNA polymerase isolated from YEME-grown cultures of S. coelicolor M145 produced a low level of hrdDp1 transcripts but abundant transcription from hrdDp2 (Fig. 6).
FIG. 5.
The three classes of promoter described in this paper. dagAp2 and phsAp are recognized by EςE in vitro, but their activities are unaffected in sigE null mutants; hrdDp1 is recognized by EςE in vitro and is highly sigE dependent in vivo; hrdDp2 is not recognized by EςE in vitro but is partially sigE dependent in vivo. The putative −35 and −10 regions are underlined, and the conserved sequences are shown in boldface. The transcription start points are italicized and boldfaced.
FIG. 6.
In vitro transcription of hrdD. Core RNA polymerase (core), core RNA polymerase plus recombinant ςE (core + ςE), or total RNA polymerase isolated from S. coelicolor M145 (holo) was used in in vitro transcription reactions. Transcription from hrdDp1 would be expected to lead to the formation of a runoff product of 316 or 456 nt with either a HindIII-NarI or a HindIII-SstI fragment, respectively, used as a template. Transcription from hrdDp2 would be expected to lead to the formation of a runoff product of 204 nt with the HindIII-SstI template. ETE, end-to-end transcription. The mobilities of the size markers (M) are indicated on the left.
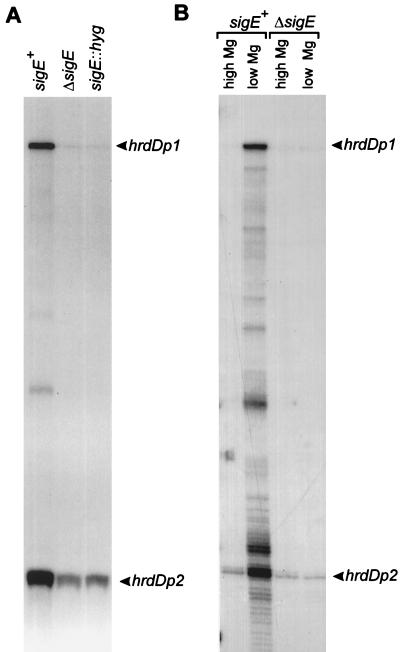
To see if EςE also transcribed hrdDp1 in vivo, S1 nuclease mapping of hrdD was performed with RNA isolated from M600 and the sigE mutants J2130 and J2141. For this experiment, RNA was isolated from surface-grown MS agar cultures because growth on this medium gave a clear mutant phenotype, suggesting that ςE was active in the wild-type strain under these conditions. RNA was isolated from 36-h cultures; by this time aerial mycelium was present and a few spores could be seen in both M600 and the sigE mutants. Representative results (Fig. 7A) showed that the level of transcription from hrdDp1 was severely reduced (∼20-fold) in J2130 and J2141 compared to that in M600. However, weak promoter activity could still be detected in RNA samples from both sigE mutants. Taken together with the in vitro data, these data show that hrdDp1 is an in vivo target for EςE but that another form of RNA polymerase holoenzyme also contributes to transcription from hrdDp1. Interestingly, the level of hrdDp2 activity was also significantly lower (approximately threefold) in RNA isolated from the sigE mutants, suggesting that hrdDp2 also depends partially on sigE. There is considerable similarity between hrdDp1 and hrdDp2, with 4 of 6 nt in both the proposed −35 and −10 promoter recognition sequences being identical (Fig. 5), but since EςE cannot direct transcription from hrdDp2 in vitro, it is not clear if EςE recognizes hrdDp2 in vivo.
FIG. 7.
(A) S1 mapping of the hrdD promoter region using RNA isolated from S. coelicolor M600 (sigE+), J2130 (ΔsigE), and J2141 (sigE::hyg) grown on MS agar. (B) S1 mapping of the hrdD promoter region using RNA isolated from S. coelicolor M600 (sigE+) and J2130 (ΔsigE) grown in NMMP liquid medium with 2 mM (high Mg) or 50 μM (low Mg) MgCl2. The uniquely 5′-end-labelled probe was prepared as described in Materials and Methods. Protected fragments corresponding to initiation at hrdDp1 and hrdDp2 are indicated.
Expression of hrdD is induced in cultures deficient in Mg2+ in a ςE-dependent manner.
The actinorhodin overproduction and altered colony morphology of the sigE mutants could be suppressed by the addition of Mg2+ to the medium. To see if ςE-directed expression of hrdD varied in response to changing Mg2+ concentrations, RNA was isolated from NMMP liquid cultures containing high (2 mM) or low (50 μM) concentrations of Mg2+, and transcription was assessed by S1 nuclease mapping. In the sigE+ strain, M600, the levels of hrdDp1 and hrdDp2 transcripts increased approximately 12- and 4-fold, respectively, in Mg2+-deficient medium (Fig. 7B), whereas in J2130 (ΔsigE) both promoters were transcribed at low basal levels and were insensitive to Mg2+ concentrations.
ςE is not essential for dagAp2 transcription in vivo.
ςE was originally identified by its ability to direct transcription from the dagAp2 promoter in vitro (9). To see if dagAp2 required ςE for activity in vivo, transcript levels were investigated by S1 nuclease mapping. There was no difference between J2130 (ΔsigE) and its parent, M600, in the level of the dagAp2 transcript, indicating that ςE does not contribute significantly to dagAp2 transcription in vivo, at least under the growth conditions used here (data not shown). To see if ςE was required when the copy number of dagAp2 was increased, S1 nuclease mapping was performed on RNA isolated from J2130 or M600 carrying the dagA gene on a multicopy plasmid (pIJ2020). Again, there was no difference in the level of the dagAp2 transcript between the two strains (data not shown).
DISCUSSION
The evidence presented here suggests that sigE is required for normal cell wall structure in S. coelicolor. sigE null mutants are particularly sensitive to cell wall-lytic enzymes, including muramidases and amidases, and their peptidoglycan has an altered muropeptide composition. Although the ΔsigE mutant showed alterations in the ratio of certain muropeptides, a wild-type complement of muropeptides appeared to be maintained. One possible explanation for this is a change in the activity of autolysins, enzymes that hydrolyze peptidoglycan and play important roles in cell wall growth, cell separation, and differentiation (60). However, because the activities of autolysins can be affected by changes in the cell wall itself (see below), it would be difficult to identify the root cause of the altered muropeptide composition.
Sensitivity to cell wall-lytic enzymes often correlates with increased sensitivity to autolysins and can be attributed to a range of different changes in the cell wall, including O acetylation of the peptidoglycan (18) and modification of the accessory polymers (see, e.g., references 43 and 61). A change in the degree of O acetylation of the C-6 hydroxyl moiety of muramyl residues is unlikely to be the cause of increased sensitivity to lysozyme because sigE mutants are also more sensitive to the muramidases Cellosyl and mutanolysin, enzymes which are unaffected by this type of acetylation. In addition, sigE mutants are also more sensitive to the CwlA amidase, which cuts peptidoglycan at a different position (20). Preliminary investigations into possible changes in the accessory polymers have not revealed any differences in the teichoic acid content and composition of sigE mutants (53). The future identification of genes under the control of ςE should help define its precise role in cell wall structure. Helmann and colleagues have used consensus-based computer searches of the complete B. subtilis genome sequence to identify promoters, and hence genes, under the control of the ECF sigma factors ςX and ςW (29, 32). The ongoing S. coelicolor genome sequencing project (www.sanger.ac.uk/Projects/S_coelicolor/) should permit the same approach to be used in identifying candidate members of the ςE regulon.
Interestingly, B. subtilis ςX is involved in transcribing a number of genes concerned with cell wall structure, including lytR, a regulator of autolysin expression, and csbB, which encodes a putative membrane-bound glycosyl transferase, possibly involved in peptidoglycan biosynthesis (29). It is therefore conceivable that ςE and ςX play related roles in S. coelicolor and B. subtilis, respectively.
sigE mutants required Mg2+ or Ca2+ for normal growth and sporulation. Divalent cations can stabilize cell walls (17, 36, 41, 47) and can protect cells from autolysis as well as from exogenous lytic enzymes (36, 61). Thus, it seems likely that Mg2+ stabilizes the defect in the cell walls of sigE mutants, thereby suppressing the phenotype. However, even in the presence of Mg2+, the defect in the cell walls of the sigE mutants remains because they are still more sensitive to cell wall-lytic enzymes than the parental wild-type strain. It is not clear why the sigE mutants overproduce the antibiotic actinorhodin on medium with low levels of Mg2+. Antibiotic production can be induced by a number of different stresses (4), and it is possible that the overproduction of actinorhodin seen in sigE mutants is an indirect consequence of stresses induced by the cell wall defect when it is not stabilized by Mg2+.
ςHrdD is one of three S. coelicolor sigma factors that are very closely related in amino acid sequence and promoter specificity to ςHrdB, the principal, essential sigma factor of this species. However, the function of ςHrdD is unknown; hrdD null mutants are apparently unaffected in growth, morphological development, and antibiotic production (10). Expression of hrdD was found to be partially dependent on sigE, but since hrdD mutants do not resemble sigE mutants, the sigE mutant phenotype is not manifested through hrdD. Expression of hrdD increased in cultures grown under conditions of Mg2+ deficiency in a sigE-dependent manner. However, it is not clear whether the cells were responding directly to Mg2+ deficiency or to possible changes in the cell wall resulting from this deficiency. Magnesium has been shown to affect the expression of cell wall proteins in Bacillus brevis 47 (1) and to affect lipopolysaccharide composition in Salmonella typhimurium via the PhoP-PhoQ virulence regulatory system (23). However, only in the case of the PhoP-PhoQ system has Mg2+ been shown to act directly as an extracellular signal (21).
In sigE mutants, transcription from hrdDp1 is severely reduced but not abolished, showing that another RNA polymerase holoenzyme also contributes to hrdDp1 transcription in vivo. Given that hrdDp1 is clearly similar to cognate promoters of other ECF sigma factors, it is likely that this holoenzyme contains another ECF sigma factor. In addition to ςE, there are at least another five ECF sigma factors encoded by the S. coelicolor genome (46, 49, 51). Considering that there are 10 ECF sigma factors in another actinomycete, Mycobacterium tuberculosis (not including ςF, which was classified as an ECF sigma factor by Cole et al. [12] but is more similar to the sporulation subfamily [15]), we suspect that many more ECF sigma factors will be discovered in S. coelicolor by genome sequencing. It will be interesting to establish the extent to which different ECF sigma factors have overlapping promoter specificity and thus overlapping function. Indeed, there is ample evidence, from this work and from work with B. subtilis (29, 31), that more than one ECF sigma factor can recognize certain promoters.
Although transcription from both hrdDp1 and hrdDp2 was reduced in the sigE mutants, only hrdDp1 was recognized by EςE in vitro. This could mean that the dependence of hrdDp2 on ςE is indirect or that transcription from hrdDp2 in vivo requires an accessory factor absent from the in vitro reactions, or that the in vitro reaction conditions do not reflect the in vivo conditions in some other way. It is feasible that ςE can direct transcription from hrdDp2 in vivo, because the promoter is very similar to hrdDp1 in both the −35 and −10 recognition sequences (Fig. 5). The residual activity of hrdDp2 in a sigE null mutant is due, at least in part, to a newly identified ECF sigma factor, ςR (51). ςR directs transcription from hrdDp2 in vitro (35), and hrdDp2 depends partially on ςR in vivo (52). The construction of a sigE sigR double mutant will be needed to establish if yet other holoenzymes are involved in the transcription of hrdDp2 in vivo.
Although ςE was originally identified by its ability to direct transcription from dagAp2 in vitro, ςE did not contribute to transcription from dagAp2 in vivo, at least under the conditions used here. Similarly, in a related paper we showed that while EςE could direct transcription of the phsA gene of S. antibioticus in vitro, sigE was not required for normal levels of phsA transcription in vivo (34). Thus, three classes of promoter have been described in this paper: dagAp2 and phsAp are recognized by EςE in vitro, but their activities are unaffected in sigE null mutants; hrdDp1 is recognized by EςE in vitro and is highly sigE dependent in vivo; hrdDp2 is not recognized by EςE in vitro but is partially sigE dependent in vivo. Although all the promoters are quite similar in both the −35 and −10 recognition regions, there are some clear differences (Fig. 5). Major challenges for the future will be to establish which nucleotides in these promoter regions determine sigma specificity and to establish the extent to which promoter dependence is affected by the growth conditions used.
ACKNOWLEDGMENTS
We thank Keith Chater, David Hopwood, and Tobias Kieser for critical reading of the manuscript and Andrew Philipson, Ian Hancock, and Maureen McCann for unpublished cell wall analyses.
This work was funded by BBSRC grant GR/J67994 (to M.J.B.), by a Lister Institute research fellowship (to M.J.B.), by a Royal Society University Research fellowship (to S.J.F.), by BBSRC grant 50/F08202 (to S.J.F.) and by a grant-in-aid to the John Innes Centre from the BBSRC.
REFERENCES
- 1.Adachi T, Yamagata H, Tsukagoshi N, Udaka S. Repression of the cell wall protein gene operon in Bacillus brevis 47 by magnesium and calcium ions. J Bacteriol. 1991;173:4243–4245. doi: 10.1128/jb.173.13.4243-4245.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angerer A, Enz S, Ochs M, Braun V. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. FecI belongs to a new subfamily of ς70-type factors that respond to extracytoplasmic stimuli. Mol Microbiol. 1995;18:163–174. doi: 10.1111/j.1365-2958.1995.mmi_18010163.x. [DOI] [PubMed] [Google Scholar]
- 3.Atrih A, Zöllner P, Allmaier G, Foster S J. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J Bacteriol. 1996;178:6173–6183. doi: 10.1128/jb.178.21.6173-6183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibb M J. The regulation of antibiotic production in Streptomyces coelicolor A3(2) Microbiology. 1996;142:1335–1344. doi: 10.1099/13500872-142-6-1335. [DOI] [PubMed] [Google Scholar]
- 5.Bierman M, Logan R, O’Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 6.Bourn W R, Babb B. Computer-assisted identification and classification of streptomycete promoters. Nucleic Acids Res. 1995;23:3696–3703. doi: 10.1093/nar/23.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buttner, M. J. Unpublished data.
- 8.Buttner M J, Fearnley I M, Bibb M J. The agarase gene (dagA) of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis. Mol Gen Genet. 1987;209:101–109. doi: 10.1007/BF00329843. [DOI] [PubMed] [Google Scholar]
- 9.Buttner M J, Smith A M, Bibb M J. At least three different RNA polymerase holoenzymes direct transcription of the agarase gene (dagA) of Streptomyces coelicolor A3(2) Cell. 1988;52:599–607. doi: 10.1016/0092-8674(88)90472-2. [DOI] [PubMed] [Google Scholar]
- 10.Buttner M J, Chater K F, Bibb M J. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A(3)2. J Bacteriol. 1990;172:3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakraburtty R, Bibb M J. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole S T, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Danese P N, Silhavy T J. The ςE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 14.De Las Peñas A L, Connolly, Gross C A. The ςE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of ςE. Mol Microbiol. 1997;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- 15.DeMaio J, Zhang Y, Ko C, Young D, Bishai W. A stationary phase stress-response sigma factor from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1996;93:2790–2794. doi: 10.1073/pnas.93.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deretic V, Schurr M J, Boucher J C, Martin D W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyle R J. How cell walls of gram-positive bacteria interact with metal ions. In: Beveridge T J, Doyle R J, editors. Metal ions and bacteria. New York, N.Y: John Wiley & Sons; 1989. pp. 275–293. [Google Scholar]
- 18.Dupont C, Clarke A J. Dependence of lysozyme-catalysed solubilization of Proteus mirabilis peptidoglycan on the extent of O-acetylation. Eur J Biochem. 1991;195:763–769. doi: 10.1111/j.1432-1033.1991.tb15764.x. [DOI] [PubMed] [Google Scholar]
- 19.Floriano B, Bibb M J. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:385–396. doi: 10.1046/j.1365-2958.1996.6491364.x. [DOI] [PubMed] [Google Scholar]
- 20.Foster S J. Cloning, expression, sequence analysis and biochemical characterization of an autolytic amidase of Bacillus subtilis 168 trpC2. J Gen Microbiol. 1991;137:1987–1998. doi: 10.1099/00221287-137-8-1987. [DOI] [PubMed] [Google Scholar]
- 21.García Véscovi E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 22.Gorham H C, McGowan S J, Robson P R H, Hodgson D A. Light-induced carotogenesis in Myxococcus xanthus: light-dependent membrane sequestration of ECF sigma factor CarQ by anti-sigma factor CarR. Mol Microbiol. 1996;19:171–186. doi: 10.1046/j.1365-2958.1996.360888.x. [DOI] [PubMed] [Google Scholar]
- 23.Guo L, Lim K B, Gunn J S, Bainbridge B, Darveau R P, Hackett M, Miller S I. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 24.Hershberger C D, Ye R W, Parsek M R, Xie Z D, Chakrabarty A M. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative ς factor (ςE) Proc Natl Acad Sci USA. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hillemann D, Pühler A, Wohlleben W. Gene disruption and gene replacement in Streptomyces via single stranded DNA transformation of integration vectors. Nucleic Acids Res. 1991;19:727–731. doi: 10.1093/nar/19.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindle Z, Smith C P. Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol Microbiol. 1994;12:737–745. doi: 10.1111/j.1365-2958.1994.tb01061.x. [DOI] [PubMed] [Google Scholar]
- 27.Hobbs G, Frazer C, Gardner D C J, Cullum J, Oliver S G. Dispersed growth of Streptomyces in liquid culture. Appl Microbiol Biotechnol. 1989;31:272–277. [Google Scholar]
- 28.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 29.Huang X, Helmann J D. Identification of target promoters for the Bacillus subtilis ςX factor using a consensus-directed search. J Mol Biol. 1998;279:165–173. doi: 10.1006/jmbi.1998.1765. [DOI] [PubMed] [Google Scholar]
- 30.Huang X, Decatur A, Sorokin A, Helmann J D. The Bacillus subtilis ςX protein is an extracytoplasmic function ς factor contributing to survival at high temperature. J Bacteriol. 1997;179:2915–2921. doi: 10.1128/jb.179.9.2915-2921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X, Fredrick K L, Helmann J D. Promoter recognition by Bacillus subtilis ςW: autoregulation and partial overlap with the ςX regulon. J Bacteriol. 1998;180:3765–3770. doi: 10.1128/jb.180.15.3765-3770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. Identification of target promoters for the Bacillus subtilis extracytoplasmic function ς factor, ςW. Mol. Microbiol., in press. [DOI] [PubMed]
- 33.Janssen G R, Bibb M J. Derivatives of pUC18 that have BglII sites flanking a multiple cloning site and that retain ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 34.Jones G H, Paget M S B, Chamberlin L, Buttner M J. Sigma-E is required for the production of the antibiotic actinomycin in Streptomyces antibioticus. Mol Microbiol. 1997;23:169–178. doi: 10.1046/j.1365-2958.1997.2001566.x. [DOI] [PubMed] [Google Scholar]
- 35.Kang J-G, Hahn M-Y, Ishihama A, Roe J-H. Identification of sigma factors for growth phase-related promoter selectivity of RNA polymerases from Streptomyces coelicolor A3(2) Nucleic Acids Res. 1997;25:2566–2573. doi: 10.1093/nar/25.13.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kihm D J, Leyer G J, An G-H, Johnson E A. Sensitization of heat-treated Listeria monocytogenes to added lysozyme in milk. Appl Environ Microbiol. 1994;60:3854–3861. doi: 10.1128/aem.60.10.3854-3861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunst F, et al. The complete genome sequence of Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 38.Lonetto M A, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacNeil D J, Occi J L, Gewain K M, MacNeil T, Gibbons P H, Ruby C L, Danis S J. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene. 1992;115:119–125. doi: 10.1016/0378-1119(92)90549-5. [DOI] [PubMed] [Google Scholar]
- 41.Marquis R E. Salt-induced contraction of bacterial cell walls of gram-positive cocci. J Bacteriol. 1968;95:775–781. doi: 10.1128/jb.95.3.775-781.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin H H, Gmeiner J. Modification of peptidoglycan structure by penicillin action in the cell walls of Proteus mirabilis. Eur J Biochem. 1979;95:487–495. doi: 10.1111/j.1432-1033.1979.tb12988.x. [DOI] [PubMed] [Google Scholar]
- 43.Massidda O, Kariyama R, Daneo-Moore L, Shockman G D. Evidence that the PBP 5 synthesis repressor (psr) of Enterococcus hirae is also involved in the regulation of cell wall composition and other cell wall-related properties. J Bacteriol. 1996;178:5272–5278. doi: 10.1128/jb.178.17.5272-5278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 45.Missiakas D, Mayer M P, Lemaire M, Georgopoulos C, Raina S. Modulation of the Escherichia coli ςE (RpoE) heat-shock transcription factor activity by the RseA, RseB and RseC proteins. Mol Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- 46.Molle, V., and M. J. Bibb. Personal communication.
- 47.Murray T, Popham D L, Setlow P. Bacillus subtilis cells lacking penicillin-binding protein 1 require increased levels of divalent cations for growth. J Bacteriol. 1998;180:4555–4563. doi: 10.1128/jb.180.17.4555-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen L H, Jensen D B, Burgess R R. Overproduction and purification of ς32, the Escherichia coli heat shock transcription factor. Protein Expr Purif. 1993;4:425–433. doi: 10.1006/prep.1993.1056. [DOI] [PubMed] [Google Scholar]
- 49.Paget, M. S. B., and M. J. Buttner. Unpublished data.
- 50.Paget M S B, Hintermann G, Smith C P. Construction and application of streptomycete promoter probe vectors which employ the Streptomyces glaucescens tyrosinase-encoding gene as reporter. Gene. 1994;146:105–110. doi: 10.1016/0378-1119(94)90842-7. [DOI] [PubMed] [Google Scholar]
- 51.Paget M S B, Kang J-G, Roe J-H, Buttner M J. ςR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2) EMBO J. 1998;17:5776–5782. doi: 10.1093/emboj/17.19.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paget, M. S. B., J.-G. Kang, J.-H. Roe, and M. J. Buttner. Unpublished data.
- 53.Philipson, A., and I. Hancock. Personal communication.
- 54.Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the ςE (ς24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14:1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodicio M-R, Manzanal M-B, Hardisson C. Protoplast formation during spore germination in Streptomyces. Curr Microbiol. 1978;1:89–92. [Google Scholar]
- 56.Rouvière P E, De Las Peñas A, Mecsas J, Lu C Z, Rudd K E, Gross C A. rpoE, the gene encoding the second heat-shock sigma factor, ςE, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 58.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 60.Ward J B, Williamson R. Bacterial autolysins: specificity and function. In: Nombela C, editor. Microbial wall synthesis and autolysis. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1984. pp. 159–166. [Google Scholar]
- 61.Wecke J, Madela K, Fischer W. The absence of d-alanine from lipoteichoic acid and wall teichoic acid alters surface charge, enhances autolysis and increases susceptibility to methicillin in Bacillus subtilis. Microbiology. 1997;143:2953–2960. doi: 10.1099/00221287-143-9-2953. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, J., and D. H. Figurski. Personal communication.
- 63.Zalacaín M, Gonzalez A, Guerrero M C, Mattaliano R J, Malpartida F, Jimenez A. Nucleotide sequence of the hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Nucleic Acids Res. 1986;14:1565–1581. doi: 10.1093/nar/14.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]