Abstract
The gene coding for the immunity protein (mceB) and the structural gene of microcin E492 (mceA), a low-molecular-weight channel-forming bacteriocin produced by a strain of Klebsiella pneumoniae, have been characterized. The microcin gene codes for a precursor protein of either 99 or 103 amino acids. Protein sequencing of the N-terminal region of microcin E492 unequivocally identified this gene as the microcin structural gene and indicated that this microcin is synthesized as a precursor protein that is cleaved at either amino acid 15 or 19, at a site resembling the double-glycine motif. The gene encoding the 95-amino-acid immunity protein (mceB) was identified by cloning the DNA segment that encodes only this polypeptide into an expression vector and demonstrating the acquisition of immunity to microcin E492. As expected, the immunity protein was found to be associated with the inner membrane. Analysis of the DNA sequence indicates that these genes belong to the same family as microcin 24, and they do not share structural motifs with any other known channel-forming bacteriocin. The organization of the microcin- and immunity protein-encoding genes suggests that they are coordinately expressed.
Microcin E492 is a low-molecular-weight channel-forming bacteriocin (21) produced by Klebsiella pneumoniae that is active on strains of the family Enterobacteriaceae (7, 8). All of the other low-molecular-weight channel-forming bacteriocins that have been described are from gram-positive bacteria, among them the lantibiotic nisin and lactococcins A and B, which are nonlantibiotic heat-stable bacteriocins (reviewed in references 1, 19, and 33). Recently, there has been a renewed interest in the study of the mechanism of action of different bacteriocins and the effect of chemical modifications because understanding the strategies used by bacteriocins to kill specific bacteria may be useful in the design of antibiotics for pharmaceutical use (3). In order to better understand the mechanism of action, export, processing, and possible posttranslational modification of microcin E492, the genes needed for immunity protein and microcin production, which encompass a 13-kb DNA fragment, were cloned and expressed in Escherichia coli (35). Studies of the genetic determinants of all microcin systems, with the exception of microcins H47 and E492, have shown that these determinants are encoded on plasmids encompassing DNA segments of less than 6 kb. Microcins H47 (15) and E492 (35), however, are both chromosomally encoded on DNA fragments of more than 10 kb. Preliminary studies of random transposition mutagenesis of the cloned microcin E492 system suggest that most of the 13-kb fragment is needed for the production of active microcin, indicating that this system may involve more components for the production of active microcin than other systems already described. In previous work, we demonstrated that the microcin E492 immunity protein is located inside a 3-kb ClaI DNA fragment (35). Here we report that, in addition to the immunity protein gene, the microcin structural gene is also encoded in this region. The deduced amino acids sequence indicates that no similarity exits between this sequence and that of any known channel-forming bacteriocin, and thus, it appears that this microcin defines a novel class of low-molecular-weight pore-forming bacteriocin which is not related to colicin-like channel-forming bacteriocins from gram-negative microorganisms or to channel-forming bacteriocins from gram-positive bacteria. On the other hand, it was found that microcin E492 shares some characteristic cleavage site features with lactococcins and related bacteriocins found in gram-positive bacteria, as in the case of colicin V (12, 17). Insight into how the immunity protein- and microcin-encoding genes are organized will enrich our understanding of the coordinated regulation of these genes during stationary phase (35).
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains, vectors, and recombinant plasmids used in this work are described in Table 1. E. coli K38pGP1-2 and pT7-7 were kindly provided by S. Tabor (Harvard Medical School).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | F−ompT rB mB | Novagen |
| XL1-Blue | recA endA1 gyrA96 thi-1 hsdR-17 supE44 relA1 lac (F′ proAB lacIqZΔM15 Tn10) | Stratagene |
| K38pGP1-2 | HfrC (λ) bearing plasmid encoding cI857 and T7 RNA polymerase gene under control of PL; Kmr | 32 |
| Plasmids | ||
| pBluescript SK(+/−) | Cloning vector; Ampr | Stratagene |
| pT7-7 | T7 expression vector; Ampr | S. Tabor |
| pJAM434 | Encoding immunity and microcin E492 determinants; Ampr | 35 |
| pBSC-47 | 3-kb ClaI fragment from pJAM434 in pBluescript SK | This study |
| p157 | 0.3-kb NdeI-EcoRI fragment (mceB) from pBSC-47 in pT7-7 | This study |
Microcin E492 assay on plates.
To determine the capacity of the strains used to produce microcin E492, 2 × 107 sensitive E. coli BL21(DE3)/p11α2 bacteria were overlaid onto Luria broth-(LB)-ampicillin (100 μg/ml) plates with 3 ml of soft agar. Single colonies to be checked were transferred with a toothpick onto the seeded plates. After overnight incubation, the colonies producing microcin E492 were surrounded by clear growth inhibition zones.
Microcin immunity test.
Aliquots (50 to 300 μl) of cultures to be tested for immunity were grown in LB-ampicillin, mixed with 3 ml of top agar, and overlaid onto LB plates. Five-microliter volumes of serial dilutions of purified microcin were tested (23). Immune cells did not show any inhibition halo. The purified microcin used for this test was prepared as described by de Lorenzo (7).
DNA isolation and manipulation.
Standard techniques were used for the cloning, transformation, preparation, and restriction analysis of plasmid DNA from E. coli (2, 28).
Nucleotide sequencing and analysis.
Both strands were entirely sequenced from plasmid pBSC-47 (Bluescript vector) by using automated sequencing technology from ABI, in accordance with the manufacturer’s directions, at Chiron Corporation (Emeryville, Calif.). Sequence analysis was performed with the University of Wisconsin Genetics Computer Group package, version 9.1 (10), at CEM, Facultad de Ciencias, Universidad de Chile. Sequence comparisons were performed with the TBLASTN, BLASTN, FASTA, and GAP programs.
Expression and localization of immunity protein MceB.
The mceB gene was amplified by PCR from pBSC-47. The forward primer (5′CGGATAAAAC*ATATGACATTACTTTCATTTGG3′) generated a NdeI restriction site at the mceB initiation codon (underlined). The reverse primer (5′AAAGCAAGAA*TTC*AGTCCTTTTGACTAATTTC3′) was designated to be 15 bp downstream of the mceB stop codon and contained an EcoRI site (underlined). Nucleotides with asterisks indicate the changes made to create the restriction sites. The resulting 0.3-kb product was inserted into the corresponding sites of expression vector pT7-7 to give p157 with the correct in-frame reading phase to produce the 95-amino-acid product. The recombinant vector was introduced into E. coli K38pGP1-2, grown to mid-exponential phase in LB plus kanamycin-ampicillin, transferred to M9 minimal medium (24) minus methionine, and incubated for 30 min. The immunity protein was induced by shifting the temperature to 42°C for 20 min. Rifampin was added to a final concentration of 200 μg/ml, and the culture was incubated further for 10 min at 42°C. [35S]methionine (50 μCi; ICN Translabel) was then added, and the incubation was continued for 15 min. The bacterial cells were collected by centrifugation, washed once with TEN buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl), and immediately processed.
To assess the location of the immunity protein in either the soluble or particulate fraction, cells were resuspended in 1/20 volume of TEN buffer plus 2-mg/ml lysozyme and incubated for 30 min to allow cell lysis. The incubation mixture was adjusted to give final concentrations of 2 mM phenylmethylsulfonyl fluoride, 10 mM MgCl2, 100-μg/ml DNase, and 10-μg/ml RNase, and incubation was continued for 20 min on ice. Four cycles of freezing (liquid nitrogen) and thawing (room temperature) were performed. The lysate was centrifuged at 15,000 × g for 30 min; the supernatant corresponded to the soluble fraction, and the pellet corresponded to the particulate fraction. Each fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Schägger and von Jagow (29).
Separation of the inner and outer membrane fractions was performed by selective solubilization with Triton X-100. Cells were grown in LB plus kanamycin-ampicillin to mid-exponential phase at 30°C, induced for 5 h at 42°C, collected by centrifugation, washed twice with TEN buffer, and resuspended in 50 mM Tris-HCl (pH 8.0)–2 mM phenylmethylsulfonyl fluoride–10-mM MgCl2–100-μg/ml DNase–10-μg/ml RNase. Cellular disruption was achieved by sonic oscillation (100 W, 4 × 1 min), and disrupted cells were kept on ice for 30 min. After centrifugation at 2,000 × g for 10 min at 4°C, the supernatant was further centrifuged at 105,000 × g for 120 min at 4°C to obtain the membrane fractions. The supernatant corresponded to the soluble fraction, and the pellet (inner and outer membrane fractions) was resuspended in 50 mM Tris-HCl (pH 8.0)–10 mM MgCl2–2% Triton X-100 and incubated for 45 min at 37°C to allow solubilization of the inner membrane proteins. This suspension was centrifuged at 105,000 × g for 90 min at 4°C. The supernatant contained the inner membrane proteins, while the pellet (Triton X-100-insoluble fraction) corresponded to the outer membrane fraction. Each fraction was analyzed by SDS-PAGE (29).
N-terminal microsequencing.
Microcin E492 was transferred from polyacrylamide gels onto a polyvinylidene difluoride sequencing membrane (Bio-Rad) for N-terminal sequencing by following the manufacturer’s instructions. The portion of the polyvinylidene difluoride membrane containing the microcin band (without staining) was cut out and microsequenced. N-terminal Edman microsequencing was done with an Applied Biosystems 477A sequencer equipped for on-line high-performance liquid chromatography analysis of the phenylthiohydantoin derivatives at the Protein Chemistry Service, Centro de Investigaciones Biológicas, Consejo Superior de Investigaciones Científicas, Madrid, Spain.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been submitted to the GenBank database under accession no. AF063590.
RESULTS AND DISCUSSION
Cloning and sequence analysis of mceA and mceB genes.
In a previous study, we used deletion analysis to demonstrate that the genetic determinant(s) required for immunity to microcin E492 are encoded in a 3-kb ClaI fragment (35). This fragment, obtained from pJAM434 (35), was cloned into the ClaI site of pBluescript (pBSC-47). This DNA segment could be cloned only in one orientation and conferred immunity to microcin E492. The DNA sequence between the EcoNI and BamHI sites of the cloned ClaI fragment is shown in Fig. 1. Two overlapping open reading frames (ORFs) were identified. The first, designated mceB, encoded a 95-amino-acid polypeptide with a theoretical molecular weight of 10,943 and corresponds to the microcin E492 immunity protein. The second, mceA, encoded a polypeptide of either 103 amino acids with a theoretical molecular weight of 10,091 if the first methionine is used as the start codon or a 99-amino-acid polypeptide with a theoretical molecular weight of 9,562 if the methionine at position 5 is used as the start codon. One of these polypeptide (or both) would correspond to nonprocessed microcin E492. The assignment of these genes (see below) was done experimentally. A putative promoter region with characteristic features, such as AT-rich stretches and two likely −10 (AATAAT) and −35 (TTGACG) regions with 17 bp between them, was identified in the 5′ region. The first ORF, corresponding to mceB, has a potential ribosome binding site with a Shine-Dalgarno sequence (AACGGAT) 7 bp upstream of the ATG initiation codon. The two genes (mceA and -B) overlap by either 23 or 11 nucleotides (depending on the start codon), with a +1 shift for the mceA gene. No independent promoter or ribosome binding site for mceA could be identified if the translation starts in the first methionine, but if translation initiation occurs at the methionine in position 5, a good consensus for a Shine-Dalgarno sequence (TGGAGC) 5 bp upstream of this methionine can be found. Thus, it is more likely that translation initiation occurs at the methionine in position 5. It is possible that these genes are transcriptionally and translationally coupled, and this is consistent with the in vivo studies reported by Wilkens et al. (35) showing that microcin and the immunity protein are expressed only during the exponential phase of growth. An exhaustive analysis of the DNA sequence upstream of the putative promoter by using the computer programs FASTA and BLASTN was performed to search for sequence similarities. No obvious known regulation sites were found, although some similarities to E. coli and Bacillus subtilis genomic sequences with unknown functions were observed. However, it is interesting that there are two direct repeats that overlap the putative −35 region indicated between arrowheads in Fig. 1.
FIG. 1.
Nucleotide sequence of the mceAB-containing region. The deduced amino acid sequences for the mceA (microcin E492) and mceB (immunity protein) genes from K. pneumoniae RYC492 are given below the DNA sequences. The putative transcriptional start is indicated by the curved arrow. Deduced −10 and −35 regions in DNA are in boldface, and the proposed consensus Shine-Dalgarno sequences are underlined. Two direct repeats are indicated between the arrowheads. The inferred cleavage site in MceA is represented by a vertical arrow between amino acids 19 and 20. The amino acids in boldface are those confirmed by N-terminal sequencing of the protein.
Regulation of the expression of the microcin- and immunity protein-encoding genes seems to be complex, since this regulation requires the presence of at least a 5-kb DNA segment contiguous to these genes, which is present in plasmid pJI (35). pJI contains a 6-kb DNA fragment of the 13 kb needed for the production of microcin and its immunity protein (35). By Tn5 mutagenesis (4), it was determined that this plasmid codes for two or three cistrons, besides the structural genes for microcin and its immunity protein, that may be involved in the regulation of gene expression. Cells harboring pBSC-47, the plasmid used for sequencing of the immunity protein and microcin genes, lack two of these cistrons and express the immunity protein in the exponential and stationary phases of growth. In contrast, cells harboring pJI do not express the immunity protein in the stationary phase. Further characterization of the regulation of the production of microcin and its immunity protein awaits the study of the other components of this system involved in the expression. However, a possible target for regulatory proteins are the direct repeats found overlapping the putative promoter region.
The genes encoding microcin E492 and its immunity protein are homologous to those of the microcin 24 system.
Analysis of the sequences of mceA and mceB by using the computer programs FASTA and TBLASTN revealed that the two corresponding ORFs belong to a novel family of bacteriocins and that each presents significant similarities to only one protein in the GenBank database. mceA turned out to be homologous to mtfS, the structural gene of microcin 24, isolated from an E. coli uropathogenic strain (25), while mceB is a homolog of mtfI, a gene encoding the microcin 24 immunity protein (25). Information concerning microcin 24 is only available in GenBank. The deduced amino acid sequences were compared by using the GAP program (Fig. 2). The MtfS and MceA proteins are 90 and 103 amino acids (for comparison, we will use the sequence that starts at the first methionine), respectively, with an identity of 52% and a similarity of 59%. On the other hand, MtfI and MceB are very similar in size (93 and 95 amino acids, respectively), with an identity of 39% and a similarity of 56%. These results strongly suggest that both microcins belong to the same family and probably have an ancestor in common. No information is available about the mechanism of action of microcin 24, but due to its similarity to microcin E492, it is possible to speculate that the target of microcin 24 is probably also the cytoplasmic membrane. The five determinants needed for the production of microcin 24 and its immunity protein are encoded in a 5.3-kb DNA segment (25), while the microcin E492 system is encoded in a 13-kb segment. The difference in size may reflect the complexity of the microcin E492 system, an idea supported by preliminary results with random Tn5 mutagenesis that suggest that between six and eight cistrons (besides the structural gene) encoded in the 13-kb DNA fragment are needed for the production of active microcin (4). In this respect, microcin E492 is also different from colicin V, which mechanism of action at the inner membrane level could be related to microcin E492. Only four plasmid genes, spanning 4.5 kb of DNA, are required for colicin V synthesis, export, and immunity (14, 19). The similarity between the sequences of the microcin protein and the immunity protein of microcins 24 and E492 that suggests that these proteins have an ancestor in common is also present at the level of gene organization. In both cases, the immunity protein- and microcin-encoding genes seem to be under the control of the same promoter, and the C-terminal coding region of the immunity protein overlaps the N terminus of microcin, with a frameshift in the ORF of the microcin protein. As with the transcription of the two converging operons in colicin V, transcription of the mtfI/mtfS operon of microcin 24 also could be under the control of the Fur protein, which binds to DNA at a specific sequence called the Fur box (9), because a sequence similar to a Fur box was found in the mtfI/mtfS promoter region (25). Although microcin E492 expression is regulated by the presence of iron in the growth medium (26), no site for the Fur repressor was found in an extensive region upstream (240 bp) of the mceB gene.
FIG. 2.
Comparison of the deduced amino acid sequences of nonprocessed microcin MceA with MtfS (A) and immunity protein MceB with MtfI (B). Identical matches (vertical lines) and strongly (colons) and weakly (dots) conserved amino acid residues are indicated. The comparison was performed with the GAP program. MceA and MtfS displayed an identity of 52% and a similarity of 59%, while MceB and MtfI presented an identity of 39% and a similarity of 56%.
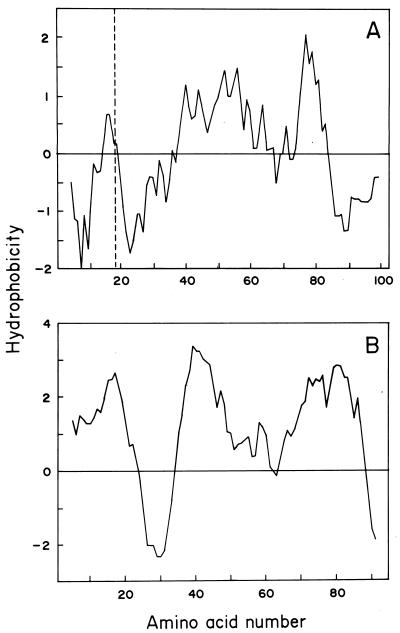
Hydropathy profiles of MceA and MceB are shown in Fig. 3. As expected, the hydropathy profile of immunity protein MceB indicates that it is likely to be an integral membrane protein, with three strong transmembrane regions, comprising amino acids 1 to 21, 35 to 55, and 66 to 90. In contrast, a single proposed transmembrane region is present in the MceA protein, between amino acids 40 and 60 in the nonprocessed protein. These conclusions were based not only on the Kyte-Doolittle (20) profiles shown in Fig. 3 but also on other, independent analyses, such as TMpred (18) and the hydrophobicity profiles obtained by the method of Eisenberg et al. (11). Both sequences were further analyzed, and a comparison of these proteins with several channel-forming bacteriocins and their immunity proteins did not reveal regions with structural homology.
FIG. 3.
Hydrophobicity profiles of MceA (A) and MceB (B). Hydropathy was calculated by the method of Kyte and Doolittle (20) with a span length of 13 amino acid residues and by using a linear weight variation model. The dashed line in panel A shows the inferred processing site for microcin E492.
Microcin E492 resembles colicins from group B in its requirement for tonB and exbB gene products for its uptake into cells (27). TonB-dependent colicins (Ia, Ib, and B) that use the TonB uptake pathway have a sequence motif Asp(Glu)-Thr(Ile)-X-Val(X)-Val, called the TonB box, located near the N-terminal region (6). The possibility that microcin E492 has a TonB box was examined, but it was not possible to draw a definitive conclusion. The only possible TonB box in microcin E492 is the region between amino acids 69 and 73 of the nonprocessed form (Asp-His-Gly-Pro-Val), but this sequence only poorly resembles the consensus.
N-terminal amino acid sequencing of microcin E492.
The amino acid sequence determined by microsequencing of the N-terminal region of microcin E492 cloned in E. coli was X-X-Thr-Asp-Pro-Asn-Thr-Glu-Leu-Leu-Asn-Asp-Leu. The two first amino acids could not be identified due to interference, so it was not until cycle 3 that the amino acids were clearly identified. The same interference was observed with microcin E492 from K. pneumoniae purified by high-performance liquid chromatography (34). This result indicates that the primary translation product of mceA is a precursor that is processed to remove the first 15 or 19 amino acids. Thus, the processed microcin E492 molecule would be an 84-amino-acid polypeptide with a predicted molecular weight of 7,887.
Similarity between cleavage sites of microcin E492 and those of colicin V and lactococcins.
Microcin E492 was found to share many cleavage site features with colicin V and lactococcins and related bacteriocins found in gram-positive bacteria. These features (12, 17) include the presence of Gly residues in position −2 or −1 relative to the processing site. Most of the sequences contain the double-glycine motif, with the exception of lactococcin DR, which contains a glycine-alanine motif (17). Analysis of several leader peptides of the double-glycine motif showed that seven amino acids are found to be conserved in more than half of the aligned leader peptides (17): glycine in position −1, glycine in position −2, isoleucine in position −4, leucine in position −7, glutamic acid in position −8, serine in position −11, and leucine in position −12. From these amino acids, microcin E492 presents glycine in position −2, leucine in position −7, and serine in position −11. The aspartic acid in position −8 is a conservative amino acid substitution for glutamic acid, and isoleucine in position −12 is a conservative amino acid substitution for leucine. Other similarities were found among the cleavage sites of microcin E492, colicin V, and lactococcins and related bacteriocins. The first is a cleaved leader sequence of 15 to 20 amino acids (microcin E492 has an either 15- or 19-amino-acid leader peptide). The second is a predicted α-helical structure over most of the leader sequence. The prediction was made by using the Chou-and-Fasman (5) and Levitt (22) methods (data not shown). This analysis suggested that leader sequence residues 1 to 16 are mostly α-helical and are followed by a β-turn that begins at amino acid 17 (data not shown). It is necessary to point out that these studies were done only for comparison, as these methods are frequently used when comparing these properties of bacteriocins. The third is a predicted β-turn that begins one to three amino acids before the cleavage site (amino acid −3, in this case), which would expose the cleavage site to allow cutting by the specific peptidase. The fourth is the fact that the processed products, in all of these cases, are small, heat-stable protein bacteriocins (43 to 88 amino acids in length) which function by increasing membrane permeability in target cells.
The fact that the secretion of microcin E492 seems to be signal sequence independent, as is that of the colicin V and lactococcin type, leads us to expect that microcin E492 should be secreted by dedicated ABC exporters (13).
Genetic identification and localization of immunity protein MceB.
To demonstrate unequivocally that mceB is the gene that encodes a protein that confers immunity to microcin E492, the DNA fragment containing only the ORF coding for the 95-amino-acid immunity protein was cloned into an expression vector. For this purpose, the pT7-7 expression vector under the control of T7 RNA polymerase was employed. The DNA segment coding only for the immunity protein was PCR amplified and cloned into pT7-7 (see Materials and Methods). Cells transformed with this recombinant vector (p157) acquired immunity to microcin E492, in contrast with the control transformed with pT7-7, which was not immune to microcin, demonstrating that this gene is enough to confer immunity to a high-titer microcin suspension. This construct was first introduced in E. coli XL1-Blue, in order to screen for the cloned immunity gene in a background where no expression was expected, but surprisingly, it was found that these cells acquired immunity to microcin E492. Although expression under the control of a T7 promoter is thought to be completely restricted to cells that express T7 RNA polymerase, there are reports (30) demonstrating that it is possible to find basal T7 RNA polymerase-independent expression of genes situated downstream from a T7 promoter. This very low level of expression would be enough to confer immunity to microcin E492. The pT7 system allows specific radioactive labeling of the protein under the control of the T7 RNA polymerase promoter (32; see Materials and Methods), and we took advantage of this characteristic to determine the localization of the immunity protein. For these experiments, the host strain E. coli BL21(DE3) was not suitable, because these cells segregated the immunity character. This strain does not tightly regulate the expression of T7 RNA polymerase (which is under the control of the lac promoter), resulting in a basal expression of 20 to 30% with respect to that of the fully induced protein. Overexpression of this protein seems to be harmful for the host cells, which, in turn, develop a mechanism to bypass its expression. This interpretation is consistent with the fact that the 3-kb ClaI DNA fragment could be cloned in pBluescript in one only orientation, in which the immunity and microcin genes are opposite to the lac promoter, probably because the cells could not tolerate high-level expression of these proteins. For this reason, we used the tightly regulated system of E. coli K38pGP1-2, in which the T7 RNA polymerase gene is under the control of promoter PL, and thus permitted induction by a temperature shift just before protein labeling (32). E. coli K38pGP1-2 harboring p157 was immune to a microcin suspension with the highest titer available (106 U/ml). Figure 4A shows SDS-PAGE of the soluble and particulate fractions of a bacterial extract in which the immunity protein has been induced and the corresponding autoradiograph (Fig. 4B). This protein was clearly identified in the autoradiograph as the only labeled protein, and as expected, it was located in the particulate fraction, which contains the inner and outer membrane fractions.
FIG. 4.
Expression of the immunity protein from p157. (A) Expression from the T7 promoter of pT7-7 and p157 was assessed by using E. coli K-38pGP1-2 as the bacterial host. Induction of T7 RNA polymerase was performed by incubation at 42°C. Specific transcription from the T7 promoter was performed by addition of rifampin, and proteins were labeled with [35S]methionine. SDS–16% PAGE samples corresponding to E. coli K-38pGP1-2/pT7-7 are in lanes 1, 2, and 3, and those corresponding to E. coli K-38pGP1-2/p157 are in lanes 4, 5, and 6. Total (lanes 1 and 4), soluble (2 and 5), and particulate (3 and 6) fractions of the bacterial extracts were analyzed separately. The arrow indicates the band corresponding to the immunity protein. Molecular weight markers in lane 7 were myoglobin fragments with Mrs of 16,950, 14,440, 8,160, 6,210, and 3,460. (B) Autoradiography of the SDS-PAGE shown in panel A.
The localization of the immunity protein in the inner membrane was achieved by selective solubilization of the cytoplasmic membrane proteins with Triton X-100. Figure 5 shows SDS-PAGE of the inner and outer membrane fractions. The band corresponding to the immunity protein was identified as shown in Fig. 4, and it is only present in the fraction corresponding to the inner membrane. This protein band is absent in the induced control transformed with pT7-7. The estimated molecular weight of the immunity protein from this gel is 11,500, which, within the range of experimental error, is in close agreement with that predicted from the amino acid sequence (10,943).
FIG. 5.
Localization of the immunity protein in the inner membrane. Induction of the T7 RNA polymerase of E. coli K-38pGP1-2 harboring pT7-7 or p157 was performed by incubation at 42°C for 5 h, and the separation of inner and outer membrane fractions was carried out as described in Materials and Methods. SDS–14% PAGE samples corresponding to E. coli K-38pGP1-2/pT7-7 are in lanes 1, 2, and 3, and those corresponding to E. coli K-38pGP1-2/p157 are in lanes 5, 6, and 7. Total (lanes 1 and 5), inner membrane (2 and 6), and outer membrane (3 and 7) fractions of the bacterial extracts were analyzed separately. The arrowhead indicates the band corresponding to the immunity protein. Molecular weight markers in lane 4 were myoglobin fragments with Mrs of 16,950, 14,440, 8,160, 6,210, and 3,460.
The action of immunity proteins of channel-forming colicins, which are always located in the cytoplasmic membrane, is highly specific. Two basic mechanisms have been described. In one, there is a direct interaction between the colicin and the immunity protein that inactivates the function of the pore-forming domain of colicins. Such is the case with colicins A and B (16). Alternatively, the immunity protein exerts its specific effect in the cytoplasmic membrane not only through recognition with the pore-forming domain of colicins but through an interaction with the translocation apparatus, in which the immunity protein would be associated with supermolecular complexes comprised of the receptor and translocation proteins (6). The latter model could explain why low-level expression of an immunity protein, such as the colicin E1 immunity protein (31), would prevent ion channel formation, since the probability of an encounter and association of colicin with the colicin translocation apparatus would be higher. In this respect, this model would fit better the possible mechanism of action of immunity to microcin E492, because extremely low levels of expression of the immunity protein still confer protection to microcin E492.
ACKNOWLEDGMENTS
We thank Catherine Connelly and Stanley Maloy for critical reading of the manuscript and José Manuel Andreu and Juan Evangelio for the facilities provided for microsequencing of microcin E492. We are also indebted to Carlos Medina for helpful discussions and to Claudio Hetz and María José Chiucholo for their help in different stages of this work. The excellent technical assistance of Marcela Vargas is acknowledged.
This work was supported by grant 1961009 from the Fondo Nacional de Desarrollo Científico y Tecnológico.
REFERENCES
- 1.Abee T. Pore-forming bacteriocins of Gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol Lett. 1995;129:1–10. doi: 10.1016/0378-1097(95)00137-T. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 2nd ed. New York, N.Y: Greene Publishing Associates and John Wiley & Sons, Inc.; 1992. [Google Scholar]
- 3.Baba T, Schneewind O. Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol. 1998;6:66–71. doi: 10.1016/S0966-842X(97)01196-7. [DOI] [PubMed] [Google Scholar]
- 4.Baeza, M., C. Hetz, C. Villota, J. E. Villanueva, and R. Lagos. Unpublished data.
- 5.Chou P Y, Fasman G D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 6.Cramer W A, Heymann J B, Schendel S L, Deriy B N, Cohen F S, Elkins P A, Stauffacher C V. Structure-function of the channel-forming colicins. Annu Rev Biophys Biomol Struct. 1995;24:611–641. doi: 10.1146/annurev.bb.24.060195.003143. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo V. Isolation and characterization of microcin E492 from Klebsiella pneumoniae. Arch Microbiol. 1984;139:72–75. doi: 10.1007/BF00692715. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo V, Pugsley A. Microcin E492, a low-molecular-weight peptide antibiotic which causes depolarization of the Escherichia coli cytoplasmic membrane. Antimicrob Agents Chemother. 1985;27:666–669. doi: 10.1128/aac.27.4.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisenberg D, Scharwz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 12.Fath M, Zhang L H, Rush J, Kolter R. Purification and characterization of colicin V from Escherichia coli culture supernatants. Biochemistry. 1994;33:6911–6917. doi: 10.1021/bi00188a021. [DOI] [PubMed] [Google Scholar]
- 13.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fath M J, Skvirky R, Gilson L, Mahanty H K, Kolter R. The secretion of colicin V. In: James R, Lazdunski C, Pattus F, editors. Bacteriocins, microcins, and lantibiotics. Heidelberg, Germany: Springer Verlag; 1992. pp. 331–348. [Google Scholar]
- 15.Gaggero C, Moreno F, Laviña M. Genetic analysis of microcin H47 antibiotic system. J Bacteriol. 1993;175:5420–5427. doi: 10.1128/jb.175.17.5420-5427.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geli V, Lazdunski C. An α-helical hydrophobic hairpin as a specific determinant in protein-protein interaction occurring in Escherichia coli colicin A and B immunity systems. J Bacteriol. 1992;174:6432–6437. doi: 10.1128/jb.174.20.6432-6437.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havarstein L S, Holo H, Nes I F. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by Gram-positive bacteria. Microbiology. 1994;140:2383–2389. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann K, Stoffel W. TMbase—a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 19.Kolter R, Moreno F. Genetics of ribosomally synthesized peptide antibiotics. Annu Rev Microbiol. 1992;46:141–163. doi: 10.1146/annurev.mi.46.100192.001041. [DOI] [PubMed] [Google Scholar]
- 20.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Lagos R, Wilkens M, Vergara C, Cecchi X, Monasterio O. Microcin E492 forms ion channels in phospholipid bilayer membranes. FEBS Lett. 1993;321:145–148. doi: 10.1016/0014-5793(93)80096-d. [DOI] [PubMed] [Google Scholar]
- 22.Levitt M. Conformational preferences of amino acids in globular proteins. Biochemistry. 1978;17:4277–4285. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
- 23.Mayr-Harting A, Hedges A J, Berkeley C W. Methods for studying bacteriocins. Methods Microbiol. 1972;7A:315–422. [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 431–433. [Google Scholar]
- 25.O’Brien, G. J., and H. K. Mahanty. 1996. GenBank accession no. U47048.
- 26.Orellana C, Lagos R. The activity of microcin E492 from Klebsiella pneumoniae is regulated by a microcin-antagonist. FEMS Microbiol Lett. 1996;136:297–303. doi: 10.1111/j.1574-6968.1996.tb08064.x. [DOI] [PubMed] [Google Scholar]
- 27.Pugsley A P, Moreno F, de Lorenzo V. Microcin-E492-insensitive mutants of Escherichia coli K12. J Gen Microbiol. 1986;132:3253–3259. doi: 10.1099/00221287-132-12-3253. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 30.Somerville R L, Ni Shieh T-L, Hagewood B, Cui J. Gene expression from multicopy T7 promoter vectors proceeds at single copy rates in the absence of T7 RNA polymerase. Biochem Biophys Res Commun. 1991;181:1056–1062. doi: 10.1016/0006-291x(91)92044-k. [DOI] [PubMed] [Google Scholar]
- 31.Song H Y, Cramer W A. Membrane topography of ColE1 gene products: the immunity protein. J Bacteriol. 1991;173:2935–2943. doi: 10.1128/jb.173.9.2935-2943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venema K, Venema G, Kok J. Lactococcal bacteriocins: mode of action and immunity. Trends Microbiol. 1995;3:299–304. doi: 10.1016/s0966-842x(00)88958-1. [DOI] [PubMed] [Google Scholar]
- 34.Wilkens, M., and R. Lagos. Unpublished data.
- 35.Wilkens M, Villanueva J E, Cofré J, Chnaiderman J, Lagos R. Cloning and expression in Escherichia coli of genetic determinants for production of and immunity to microcin E492 from Klebsiella pneumoniae. J Bacteriol. 1997;179:4789–4794. doi: 10.1128/jb.179.15.4789-4794.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]