Abstract
Terminase, an enzyme encoded by the Nu1 and A genes of bacteriophage lambda, is crucial for packaging concatemeric DNA into virions. cosN, a 22-bp segment, is the site on the virus chromosome where terminase introduces staggered nicks to cut the concatemer to generate unit-length virion chromosomes. Although cosN is rotationally symmetric, mutations in cosN have asymmetric effects. The cosN G2C mutation (a G-to-C change at position 2) in the left half of cosN reduces the phage yield 10-fold, whereas the symmetric mutation cosN C11G, in the right half of cosN, does not affect the burst size. The reduction in phage yield caused by cosN G2C is correlated with a defect in cos cleavage. Three suppressors of the cosN G2C mutation, A-E515G, A-N509K, and A-R504C, have been isolated that restore the yield of λ cosN G2C to the wild-type level. The suppressors are missense mutations that alter amino acids located near an ATPase domain of gpA. λ A-E515G, A-N509K, and A-R504C phages, which are cosN+, also had wild-type burst sizes. In vitro cos cleavage experiments on cosN G2C C11G DNA showed that the rate of cleavage for A-E515G terminase is three- to fourfold higher than for wild-type terminase. The A-E515G mutation changes residue 515 of gpA from glutamic acid to glycine. Uncharged polar and hydrophobic residues at position 515 suppressed the growth defect of λ cosN G2C C11G. In contrast, basic (K, R) and acidic (E, D) residues at position 515 failed to suppress the growth defect of λ cosN G2C C11G. In a λ cosN+ background, all amino acids tested at position 515 were functional. These results suggest that A-E515G plays an indirect role in extending the specificity of the endonuclease activity of λ terminase.
λ is a double-stranded DNA phage with a 48.5-kb genome. The packaging of λ DNA into a preformed empty shell, the prohead, requires the phage-encoded enzyme terminase. Terminase is a heteromultimer consisting of gpNu1 (181 residues) and gpA (641 residues). Both subunits contain primary sequences which are characteristic of ATPases (Fig. 1) (22, 25, 45), and kinetic studies demonstrate that the holoenzyme hydrolyzes ATP and dATP (23, 31, 44). It has been found that each of the terminase subunits possesses an ATPase activity (4, 37). gpNu1 contains a DNA-stimulated, low-affinity site with a Km of 469 μM and a kcat of 84 min−1. gpA contains a high-affinity site with a Km of 4.6 μM and a kcat of 38 min−1 (4, 30, 43). DNA lowers the Km and increases the kcat of the gpNu1 ATPase three- and twofold, respectively (43).
FIG. 1.
Linear map of domains in gpNu1 and gpA. Relative positions of mutations which affect the endonucleolytic activity of terminase are shown for gpA. ∗, locations of the A-E515G, A-N509K, and A-R504C mutations in the carboxy half of gpA; x, site of an altered amino acid that renders gpA deficient in nicking activity (17); n, residue 497, the site of the A-K497D mutation (31); ATP, a proposed ATPase center; bZip, the putative basic leucine zipper region; Pro, the specificity domain for binding to proheads. Residues 484 to 505 comprise the Walker A segment of a putative ATPase domain in gpA. The proposed P-loop region of this domain encompasses residues 485 to 497 (25, 45). In gpNu1, HTH indicates the position of a proposed DNA binding HTH motif. gpNu1 and gpA interact via their carboxy and amino termini, respectively, to form the terminase heteromultimer.
At its amino terminus, gpNu1 contains a putative helix-turn-helix (HTH) DNA binding motif which is thought to interact with cosB, the terminase binding site (Fig. 1) (18, 35). Adjacent to the HTH is the putative ATPase center. The carboxy half of gpNu1 has been shown to contain a domain for interaction with the amino-terminal 48 residues of gpA (21). In the C-terminal third of gpA are the putative ATPase center and a putative basic leucine zipper motif (17). The C-terminal 32 amino acids of gpA contain a specificity domain for interaction with the prohead (Fig. 1) (20, 47, 52).
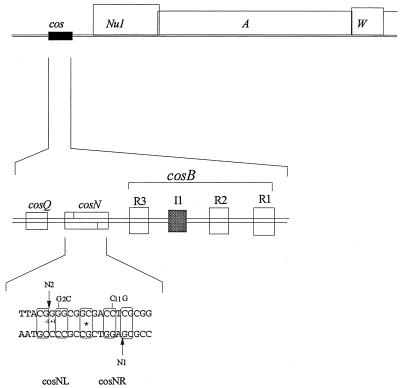
The packaging substrate is a concatemer, or end-to-end multimer, of λ chromosomes which is generated at late times after infection. The site containing the DNA packaging signals is called cos. cos consists of three segments, cosQ, cosN, and cosB (Fig. 2). The first subsite, cosQ, is required for termination of packaging (13). The second segment, cosN, is the site where terminase introduces staggered nicks to generate the 12-base cohesive ends of virion DNA. cosN contains a 16-bp segment showing partial twofold rotational symmetry; the right half-site is called cosNR, and the left half-site is called cosNL (Fig. 2). The twofold rotational symmetry of cosN suggests that symmetrically disposed gpA subunits are responsible for nicking cosNL and cosNR. The third segment, cosB, contains three binding sites for gpNu1, called R3, R2 and R1 (40), and a site, I1, for integration host factor (IHF), a DNA binding-bending protein of the host, Escherichia coli (2, 33, 48, 49). IHF introduces a sharp bend in cosB between R3 and R2 (Fig. 2) (33), and packaging of λ DNA is enhanced threefold by IHF (24, 49). It is believed that IHF facilitates cooperative interactions between terminase bound to R3 and terminases bound to R2 and R1. A second DNA binding-bending protein of the host, HU, has also been proposed to play a role in gpNu1-cos interactions (36). Adjacent to cos are the terminase genes Nu1 and A (Fig. 2).
FIG. 2.
The left end of the λ chromosome and expanded views of cosB and cosN. Nu1 and A, the terminase genes, lie downstream from cos. cos consists of three subsites, cosQ, cosN, and cosB. cosB contains the terminase recognition sequences R3, R2, and R1 and the IHF binding site I1. The 22-bp sequence of cosN has partial rotational symmetry (boxed) around a central point (∗), which divides cosN into cosNL and cosNR. The arrows show the positions of the nicks, N1 and N2, placed by terminase. Base pair 48,502 of the λ sequence is labeled −1.
An initial binding step of terminase to λ DNA is followed by nicking at cosNR and cosNL, in that order. Nicking is stimulated by the presence of ATP (12, 29), but ATP hydrolysis is not required (29). Cue and Feiss (12) found that the presence of ATP does not affect the affinity of terminase for DNA and concluded that ATP increases the velocity of the nicking reaction. The first nick is on the bottom strand, at position N1 in cosNR, and the second nick is on the top strand, at N2 in cosNL (Fig. 2) (29). The nicked strands are then separated, a step which requires ATP hydrolysis (29, 39). After separation of the cohesive ends, a stable complex forms, called Complex I, which consists of terminase bound to the left end of the chromosome (3, 34, 41), presumably through gpNu1-cosB interactions (12, 40). Terminase of Complex I then binds a prohead to form Complex II, a DNA-terminase-prohead ternary complex, and packaging proceeds. Complex II formation is facilitated by gpFI of λ (5, 16).
Mutations in cosN, specifically those in cosNL, affect the nicking activity of terminase; one example is the cosN G2C mutation (Fig. 2). cosN G2C reduces the burst size of λ 10-fold due to a defect in the cos cleavage step of packaging (51). The G2C and C11G mutations are rotationally symmetric (Fig. 2). The burst size of λ cosN G2C C11G is threefold less than that of λ cosN G2C (51). The cosN C11G mutation alone has no significant effect on the λ burst size or on cos cleavage in vitro. In vitro cos cleavage experiments of these and other rotationally symmetric mutants show that defects in the cosNL sequence are more detrimental to the cleavage activity of terminase than are defects in the cosNR sequence. λ cosN G2C and λ cosN G2C C11G mutants form minute plaques on an IHF+ host strain and do not form plaques on an IHF− strain. We describe here suppressors of the cosN G2C C11G defect that have been found in pseudorevertants of λ cosN G2C C11G.
MATERIALS AND METHODS
Strains and plasmids.
A list of the phages, E. coli strains, and plasmids used is given in Table 1, along with relevant markers and references.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Phages | ||
| λ cosN G2C C11G att+ gal+ cI857 | cosN G2C C11G | 51 |
| λ Aam515 cI857 | Used for residue 515 substitution studies | This work |
| λ Aam11am32 cI857 | cos+ Aam515, A-R504C, A-N509K, and A-E515G phage constructions | 20 |
| λ-P1:5R KnrcI857 | Used for rescues and burst size studies | 37 |
| E. coli C strains | ||
| MF1427 | C1a galK sup0 | 42 |
| MF1966 | C-4514 supE | 32 |
| MF1968 | C-4518 supF | 32 |
| MF1972 | C1a sup0himA::Tn10 hip::Cmr IHF− strain | 10 |
| MF2449 | C1a mutD | 19 |
| MF2548 | C-1055 | 46 |
| E. coli K strains | ||
| XL-1 Blue | Host for cloning vectors supE | Stratagene Cloning Systems |
| OR1265 | Host for terminase expression plasmids | 9 |
| Plasmids | ||
| pIBI30, pIBI31 | Cloning vectors | International Biotechnologies, Inc. |
| pBluescript II SK− | Cloning vector | Stratagene Cloning Systems |
| pTSNA-A+, -A-R504C, -A-N509K, or -A-E515G | Used for cosB and cosN rescues; λ cos+A-R504C, A-N509K, and A-E515G constructions; λ DNA 732-2556 in pBluescript | This work |
| pRV-Nu1+, -Nu1ms1, -ms2, or -ms3 | Used for cosB and cosN rescues; λ DNA 194-2819 in pIBI31 | 11 |
| pSX1 | cos+ cleavage substrate; probe for Southern analyses; λ DNA 47942-194 in pUC19 | 51 |
| pUC19 cosNi | cos cleavage substrate; i = G2C C11G, G3C C10G, or A−1T T13A; λ DNA 47942-194 in pUC19 | 51 |
| pCM101 | Wild-type terminase expression plasmid | 9 |
| pCM101 A-E515G | A-E515G terminase expression plasmid | 29a |
| pE515X | Used for residue 515 substitutions: X = D, E, K, R, Q, S, A, F, G, I, V, or Y; λ DNA 2218 to 2815 in pIBI30 | 29a |
| pHW473 | λ cos+Aam515 construction; am mutation at residue 515; λ DNA 48473–5505 in pIBI30 | 29a |
Sequence and amino acid designations.
The numbering convention of Daniels et al. (15) is used; numbering of the λ genome sequence begins with the first base of the left cohesive end (+1) and continues along the l strand (top strand as shown in Fig. 2) in a 5′ to 3′ direction. Base pair 48,502 is referred to as −1. The N1 nick position on the top strand is between bp −1 and 1. The position of the restriction enzyme cleavage sites is given as the first 5′ nucleotide of the recognition sequence. Amino acids are identified by the single-letter designation and numbered according to their position in the open reading frame of the protein.
Media.
Tryptone broth (TB), TB plates, and TB soft agar were prepared as described in Arber et al. (1), except that each was supplemented with 0.01 M MgSO4. Luria-Bertani (LB) broth and LB plates, both without glucose, were also as described in Arber et al. (1). Maltose was added to media at a final concentration of 0.4%, and ampicillin (AMP) and kanamycin (KAN) were added at concentrations of 100 and 50 μg/ml, respectively.
Enzymes, reagents, radiolabel, and DNA manipulations.
Restriction enzymes, T4 DNA ligase, and Vent DNA polymerase were purchased from New England Biolabs. Boehringer Mannheim supplied ATP and the Random-Primed DNA Labeling Kit used to prepare radiolabeled probe for the Southern blot analyses. AmpliTaq DNA polymerase was purchased from Perkin-Elmer, and [α-32P]dCTP was obtained from Amersham Life Science, Inc. Proteinase K, spermidine, putrescine, and β-mercaptoethanol were purchased from Sigma Chemical Co. All enzymes were used according to the manufacturer’s directions. DNA was transformed into cells made competent with 0.1 M CaCl2 (26). DNA sequencing was performed by the University of Iowa DNA Core Facility by using dye terminator cycle sequencing chemistry with AmpliTaq DNA polymerase and FS enzyme. Sequence analysis was done using a 373A Stretch Fluorescent Automated Sequencer (Applied Biosystems).
DNA preparations, PCR, and primers.
λ cosN G2C C11G gal+ att+ cI857 A-E515G DNA was prepared by phenol extraction of CsCl-purified phage lysate as described by Arber et al. (1). Plasmid DNA was prepared with commercial kits (Qiagen, Inc.) or by the method of Birnboim and Doly (6). Diagnostic PCRs were performed on phage DNA supplied in 10 μl of crude lysate under the following conditions: 4.5 mM MgCl2, 0.1% Tween 20, 200 μM (each) dNTP, 0.5 μM (each) primer, 2.5 U of AmpliTaq DNA polymerase, and amplification buffer supplied by the manufacturer. The reactions were run for 30 cycles of 95°C for 1 min, 55°C for 45 s, and 72°C for 1 min, followed by one 7-min extension cycle at 72°C. DNA for cloning was prepared by PCR with the following reaction mixture: 10 μl of crude lysate, 6.0 mM MgSO4, 400 μM (each) dNTP, 0.5 μM (each) primer, 2.0 U of Vent DNA polymerase, and amplification buffer supplied by the manufacturer. The reactions were run as above, except that the annealing temperature was 60 rather than 55°C. Sequencing primers and PCR primers were prepared on a four-column Applied Biosystems DNA synthesizer.
Crosses.
Phage versus plasmid crosses were done with lysogens transformed with plasmid DNA and selected on L-agar plates containing AMP or AMP and KAN (for λ-P1 lysogens) at 31°C. Overnight cultures of the transformed lysogens were diluted 1:100 in LB broth and grown to 5 × 107 cells/ml at 31°C. Induction of prophages at 42°C for 20 min was followed by incubation at 37°C for 60 min. The lysates were treated with chloroform, clarified by centrifugation, and plated on appropriate cells for viable recombinants. Standard phage crosses were performed as described by Arber et al. (1).
mutD mutagenesis.
Phage lysates were grown on MF2449 as described by Fowler et al. (19). Mutant phages were isolated on MF1972.
Terminase preparation and in vitro cos cleavage reactions.
Crude extracts of wild-type and A-E515G terminases were prepared by sonication of cells from induced cultures of MF1427[pCM101] and OR1265[pCM101 A-E515G], respectively, according to the method of Chow et al. (9). Wild-type and A-E515G terminases were purified by a modification (30) of the method of Tomka and Catalano (44).
cos+ and cosN G2C C11G DNA substrates were purified from cosmid pSX1, which carries the wild-type cos segment and its derivative plasmid pUC19 cosN G2C C11G. The plasmids were linearized with BsaI and yield 1.51- and 1.91-kb fragments when cut at cos by terminase. The cos cleavage assay was performed by using the protocol of Chow et al. (9); the 20-μl reactions mixtures contained 30 mM Tris-HCl (pH 9.0), 10 mM MgCl2, 3 mM spermidine, 6 mM putrescine, 7 mM β-mercaptoethanol, 1.5 mM EDTA, 1.5 mM ATP, 10 nM IHF, and 150 nM purified wild-type or A-E515G terminase. After incubation at room temperature, 2 μl of agarose gel loading buffer (50% glycerol, 0.1 M EDTA, 1% sodium dodecyl sulfate, and 0.1% bromophenol blue) was added to stop the reaction.
The samples were heated at 65°C for 10 min and subjected to 1% agarose gel electrophoresis. Following electrophoresis, the DNA was transferred onto a GeneScreen Plus (New England Nuclear) membrane. DNA hybridization was performed with radiolabeled pSX1 as probe. The extent of cleavage was determined by scanning with the Packard InstantImager (Packard Instrument Co., Downers Grove, Ill.) scanning apparatus.
RESULTS
Isolation and mapping of cosN G2C C11G suppressors.
λ cosN G2C C11G gal+ att+ cI857 was mutagenized by infecting MF2449 (mutD), and the resulting lysate was plated on MF1972 (IHF−). Plaque-forming variants were found at a frequency of 2 × 10−5/PFU; nine revertants were chosen for study. The cosN G2C mutation creates an HhaI site. Phage DNA was amplified by PCR, and the presence of the cosN G2C mutation was verified by cutting with HhaI. Chromosomal DNA was prepared from one of the isolates and cut with MluI, and the fragments were individually cloned into pIBI30. To map the suppressor, plasmids containing MluI fragments were used in marker rescue experiments as follows. The plasmids were used to transform MF1427 (λ cosN G2C C11G gal+ att+ cI857) to Apr. The resulting plasmid-bearing lysogens were induced, and the lysates were plated to identify recombinants able to plate on the IHF− strain MF1972. The control cross yielded <10−6 PFU/viable cell, whereas the cross with the fragment containing bp 458 to 5548 produced 10−1 PFU/viable cell (Table 2). This fragment carrying the suppressor was also used in a similar cross with the single cosN mutant, λ cosN G2C; cosN G2C was also suppressed by this fragment (results not shown). The fragment containing the suppressor was then subcloned for further marker rescue crosses, and the suppressor was mapped to the segment extending from bp 2212 to 2815. Sequencing revealed a transition mutation, A2254G, that resulted in changing codon 515 of the A gene from the glutamic acid codon GAA to the glycine codon GGA; A2254G also creates a new HinfI site. The A2254G suppressor mutation was designated A-E515G. For each of the eight remaining isolates, the segment of phage DNA from bp 2212 to 2815 was amplified by PCR and screened by restriction analysis for the HinfI site. Of these eight isolates, seven were HinfI+ and were presumed to be identical to A-E515G. To map the suppressor of the eighth isolate, the DNA segment extending from bp 1818 to 2556 was amplified by PCR and cloned. Marker rescue crosses showed the cloned segment contained a suppressor of cosN G2C C11G (Table 2). Sequencing of the segment revealed two mutations, one at bp 1958 (CCG to CCT, P416S) and one at bp 2220 (CGT to TGT, R504C). These two mutations were separated by subcloning by using the AccI site at bp 2190, and the resulting plasmids were used in a phage versus plasmid cross with λ cosN G2C C11G gal+ att+ cI857. The mutation at bp 1958 (P416S) did not suppress cosN G2C C11G, whereas the mutation at bp 2220 (R504C) did (Table 2). The mutation at bp 2220 (R504C) was named A-R504C.
TABLE 2.
Suppression of the cosN G2C mutation: crosses between λ cosN G2C C11G and cloned DNA segments of λ A+, λ A-E515G, λ A-N509K, or λ A-R504Ca
| Sequence of cloned segment (bp) | Allele | Amino acid change(s) in gpA | PFU/viable cell |
|---|---|---|---|
| 1818–2556 | A+ | None | <5.0 × 10−6 |
| 458–5548 | A-E515G | E515G | 1.1 × 10−1 |
| 2212–2847 | A-N509K | N509K | 4.1 × 10−3 |
| 2190–2556 | A-R504C | R504C | 7.7 × 10−4b |
| 1818–2556 | A-R504C | P416S, R504C | 1.9 × 10−3b |
| 1818–2190 | P416S | <5.3 × 10−5b |
Crosses were performed with plasmids in an MF1427 (λ cosN G2C C11G gal+ att+ cI857) lysogen and were plated on MF1972 for plaques. The higher frequency of recombinants for A-E515G is a reflection of the larger λ DNA insert in the plasmid used (5 kb).
These lysates were plated on MF1427 for recombinants forming normal-sized plaques.
A second lysate of λ cosN G2C C11G, unmutagenized, was plated on MF1972, and plaque-forming variants were found at a frequency of 2 × 10−7/PFU. Six variants were picked for further study. Five of the phages contained a true reversion of the cosN G2C mutation, based on restriction analysis of phage DNA amplified by PCR. DNA from the sixth revertant was amplified by PCR (bp 2212 to 2847), cloned into pBluescript, and shown to contain a suppressor of cosN G2C C11G (Table 2). A transversion was found at bp 2237 (AAC to AAG, N509K); this suppressor was named A-N509K.
The A-E515G, A-N509K, and A-R504C mutations allowed λ cosN G2C C11G to form normal-sized plaques on MF1427 and small plaques on MF1972.
Can the A-E515G, A-N509K, and A-R504C mutations suppress cosB mutations?
We next asked if the A-E515G, A-N509K, and A-R504C suppressors could rescue a cosB defect by using phage versus plasmid crosses. The prophage used was λ-P1:5R cI857 Knr cosB R3−R2−R1−. cosB R3−R2−R1− contains a transition mutation in each of the R sequences of cosB (Fig. 2); phages with these mutations are unable to form plaques (10). Plasmids used were the pTSNA series, which contain λ DNA inserts from bp 732 to 2556 and lack the Nu1 gene but contain the A-E515G, A-N509K, A-R504C, or A+ alleles. The control plasmids were the pRV series, containing λ DNA from bp 194 to 2819, which includes the Nu1 and A genes and the Nu1ms1, Nu1ms2, Nu1ms3, or Nu1+ alleles. The Nu1ms1, Nu1ms2, and Nu1ms3 mutations are suppressors of cosB defects (11). Lysates from the crosses were plated on MF1427; the results are shown in Table 3. The crosses with the positive controls pRV5, pRV39, and pRV7 produced approximately 104 to 106 PFU/ml, whereas crosses with pTSNA A-E515G, pTSNA A-N509K, and pTSNA A-R504C produced 10 to 25 PFU/ml. The crosses with the negative controls, pTSNA and pRV12, produced 10 and 15 PFU/ml, respectively. The A-E515G, A-N509K, and A-R504C mutations were unable to suppress the cosB R3−R2−R1− defect.
TABLE 3.
Suppression of cosB or cosN mutations: crosses between λ cosB or λ cosN and plasmids bearing various Nu1 or A allelesa
| Phage | Plasmid | Allele | PFU/ml |
|---|---|---|---|
| λ-P1 cosB R3− R2− R1− | |||
| pTSNA A-E515G | A-E515G | 10 | |
| pTSNA A-N509K | A-N509K | 15 | |
| pTSNA A-R504C | A-R504C | 25 | |
| pTSNA | A+ | 10 | |
| pRV5 | Nu1ms1 | 7.6 × 105 | |
| pRV39 | Nu1ms2 | 2.0 × 104 | |
| pRV7 | Nu1ms3 | 1.5 × 105 | |
| pRV12 | Nu1+ | 15 | |
| λ-P1 cosN G2C C11G | |||
| pRV5 | Nu1ms1 | 3.9 × 102 | |
| pRV39 | Nu1ms2 | 5.5 × 102 | |
| pRV7 | Nu1ms3 | 7.7 × 102 | |
| pRV12 | Nu1+ | 4.5 × 102 | |
| pTSNA A-E515G | A-E515G | 9.1 × 105 | |
| pTSNA A-N509K | A-N509K | 4.7 × 105 | |
| pTSNA A-R504C | A-R504C | 1.6 × 105 | |
| pTSNA | A+ | 1.5 × 103 |
Phages are in a λ-P1:5R cI857 Knr background. Crosses were performed by inducing plasmid-bearing MF1427 cells lysogenized with cosB R3−R2−R1− or cosN G2C C11G phages. The cross lysates were plated on MF1427. The pRV5, pRV39, and pRV7 plasmids are called rev1, rev9, and rev6, respectively, in reference 11.
Can the cosN G2C C11G defect be suppressed by the cosB suppressors Nu1ms1, Nu1ms2, and Nu1ms3?
We also asked the reciprocal question, whether suppressors of a cosB defect could suppress a cosN defect. Again, phage versus plasmid crosses were done; the prophage used was λ-P1:5R cI857 Knr with the cosN G2C C11G alleles. The plasmids were the pTSNA and pRV series described above, and cross lysates were plated on MF1427 for healthy plaques. Results are shown in Table 3. The yield of PFU in the crosses with pRV5, pRV39, and pRV7, which contain cosB suppressors, was as low as in the crosses with the negative controls pRV12 and pTSNA. The Nu1ms1, Nu1ms2, and Nu1ms3 mutations did not suppress the cosN G2C C11G defect.
Does λ cosN+ A-E515G, -A-N509K, or -A-R504C form a plaque?
Since the A-E515G, A-N509K, and A-R504C mutations were specific to cosN, we asked if they were specific to cosN G2C C11G by trying to construct λ cosN+ A-E515G, -A-N509K, or -A-R504C. We crossed the A-E515G, A-N509K, and A-R504C mutations into λ cosN+. Cells containing pTSNA A-E515G, -A-N509K, or -A-R504C were infected with λ Aam11 am32 cI857. am+ recombinants were found when the resulting lysate was plated on MF1427 (sup0). DNA from the am+ recombinants was amplified by PCR, and sequence analysis of the PCR product confirmed the presence of the A-E515G, A-N509K, and A-R504C mutations. Thus λ cosN+ is viable with A-E515G, A-N509K, and A-R504C.
Burst size studies.
A set of phages in the λ-P1:5R cI857 Knr background was constructed to enable us to examine burst sizes in the presence and absence of IHF. Burst sizes were determined by induction of prophages in either MF1427 (IHF+) or MF1972 (IHF−) (Table 4). The phages were made by phage versus plasmid crosses or by standard phage crosses. The cosN G2C C11G A+ phage had a smaller burst size and was more responsive to IHF than the wild-type phage. The burst size of the wild-type phage increased about 3-fold in MF1427 (IHF+) cells, whereas the burst of the cosN G2C C11G phage increased about 10-fold (Table 4). With A-E515G present, the cosN G2C C11G phage’s burst size was restored to wild-type levels in both MF1427 and MF1972 (Table 4). The burst size of λ cosN G2C C11G in IHF+ cells (MF1427) was also restored to near wild-type levels with A-N509K and A-R504C. The burst sizes of λ cosN G2C C11G A-N509K and λ cosN G2C C11G A-R504C were 10- and 4-fold less than that of λ cosN+ A+ in IHF− cells, respectively (Table 4).
TABLE 4.
Burst sizes of phages with various cosN and A alleles normalized to λ cosN+ A+a
| Genotype | Burst size (PFU/cell) in:
|
|
|---|---|---|
| MF1427 | MF1972 | |
| cosN+ A+ | 1.0 | 0.29 |
| cosN G2C C11G A+ | 0.01b | 8.2 × 10−4 |
| cosN G2C C11G A-E515G | 0.83 | 0.18 |
| cosN G2C C11G A-N509K | 0.69 | 0.03 |
| cosN G2C C11G A-R504C | 0.43 | 0.07 |
| cosN+ A-E515G | 1.01 | 0.23 |
| cosN+ A-N509K | 0.60 | 0.29 |
| cosN+ A-R504C | 0.38 | 0.25 |
λ-P1:5R Knr cI857 lysogens in MF1427 (IHF+) and MF1972 (IHF−) were induced and plated on MF1427. Values are PFU/induced cell, normalized to λ cosN+ A+ induced from MF1427, and are the averages of three experiments. The average burst of λ cosN+ A+ on MF1427 was 243 and that on MF1972 was 71.6.
These lysates were plated on MF2548 to reduce the background of revertants which arise spontaneously on MF1427.
The burst sizes of the λ cosN+ A-E515G, -A-N509K, and -A-R504C phages were near wild-type levels in both MF1427 and MF1972 (Table 4). Thus, the A-E515G, A-N509K, and A-R504C suppressors do not diminish the burst size of λ cosN+.
Comparison of the cos cleavage activities of wild-type and A-E515G terminases.
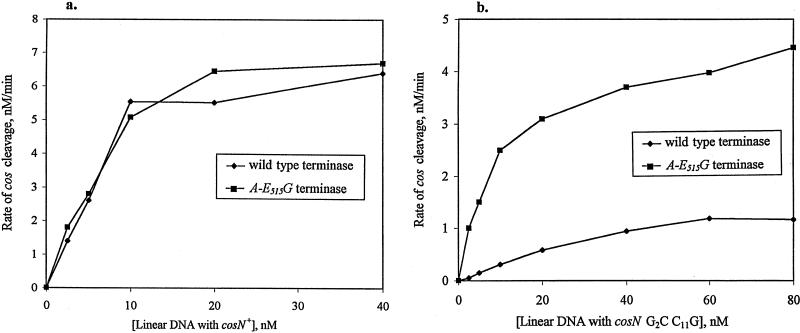
Since it had previously been shown that the cosN G2C C11G mutations affect cos cleavage (51), we asked if the A-E515G mutation improved the ability of terminase to cut DNA containing the cosN G2C C11G mutations. A-E515G was cloned into the terminase expression vector pCM101 (9). Preliminary cos cleavage reactions with crude terminase extracts showed that A-E515G terminase was able to efficiently cut several cosN substrates, including cosN+, cosN G3C C10G, cosN A−1T T13A, and cosN G2C C11G (results not shown). To examine the kinetics of A-E515G terminase, purified wild-type and A-E515G terminases were used in cos cleavage reactions with cosN+ and cosN G2C C11G substrate DNAs. With cosN+ DNA, the initial rates of cleavage are similar for both terminases over a range of DNA substrate concentrations (Fig. 3a). In reactions with the cosN G2C C11G substrate, the A-E515G terminase shows a faster rate of cleavage than the wild-type terminase (Fig. 3b). Thus, the A-E515G mutation significantly improved the ability of terminase to cleave cosN G2C C11G DNA.
FIG. 3.
cos cleavage rates of wild-type terminase and A-E515G terminase. (a) cos cleavage on cosN+ substrate. The reactions used BsaI-linearized pSX1, a cosmid carrying cosN+, as the DNA substrate. Wild-type terminase and A-E515G terminase were used to cleave the DNA substrate in a range of concentrations. (b) cos cleavage on cosN G2C C11G substrate. The reactions used a pSX1 derivative cosmid which carries cosN G2C C11G. This cosmid is linearized by BsaI. Wild-type terminase and A-E515G terminase were used to cleave the DNA substrate in a range of substrate concentrations. Other reaction conditions were as described in Materials and Methods.
Changing gpA residue 515 in λ cosN+ and λ cosN G2C C11G.
To learn about the effects of amino acid substitutions at the residue affected by A-E515G, the pE515X series of plasmids were used in phage versus plasmid crosses. The plasmid inserts are λ DNA from bp 2218 to 2815 and contain various replacements at residue 515 of gpA (29a). Phages containing either cosN+ or cosN G2C C11G were crossed with each of the plasmids. To test the effects of the substitutions in λ cosN+, crosses were done in MF1966 (supE) (λ Aam515 cI857) cells, and the lysates were plated for am+ recombinants on MF1972 (sup0). λ Aam515 cI857 was constructed by a phage versus plasmid cross in MF1968 (supF) cells by using pHW473, which contains the Aam515 marker, and λ Aam11 am32 cI857, which will not grow on a supE host; the cross lysate was plated on MF1966 (supE). The Aam515 change results in a new BfaI site, which was verified by restriction analysis of DNA generated by PCR. To test the effects of the substitutions in λ cosN G2C C11G, crosses were done in MF1427 (λ cosN G2C C11G gal+ att+ cI857) cells, and the lysates were plated for viable phage on MF1972 (IHF−). Residues at position 515 which are acidic, basic, polar uncharged, and hydrophobic allowed plaque formation of λ cosN+ (Table 5). In contrast, only polar uncharged and hydrophobic residues allowed growth of λ cosN G2C C11G; acidic and basic residues at position 515 of gpA did not allow plaque formation. Further, all of the plaque-forming recombinants of λ cosN G2C C11G formed plaques at 31, 37, and 42°C on MF1427.
TABLE 5.
Effect of changing gpA residue 515 on viability of λ cosN+ and λ cosN G2C C11Ga
| Group | Amino acid | Viable recombinants on MF1972 for:
|
|
|---|---|---|---|
| cosN+ | cosN G2C C11G | ||
| Acidic | D | + | − |
| E | + | − | |
| Basic | K | + | − |
| R | + | − | |
| Polar uncharged | Q | + | + |
| S | + | + | |
| Hydrophobic | A | + | + |
| F | + | + | |
| G | + | + | |
| I | + | + | |
| V | + | + | |
| Y | + | + | |
Crosses were performed in an MF1966 (λ Aam515 cI857) lysogen for the cosN+ allele and in an MF1427 (λ cosN G2C C11G gal+ att+ cI857) lysogen for the cosN G2C C11G allele; both were plated on MF1972 for plaques.
DISCUSSION
The A-E515G, A-N509K and A-R504C mutations were isolated as suppressors of the cosN G2C C11G mutations. A number of cos mutations, including cosN G2C C11G, cause plaque formation to be dependent on IHF (reviewed in reference 8). Based on these previous results, we isolated suppressors of the cosN G2C C11G mutations by isolating variants of λ cosN G2C C11G able to form plaques on a host lacking IHF. The suppressors were found to be missense mutations in the A gene. The suppressors result in normal and small-sized plaques by λ cosN G2C C11G on cells with and without IHF, respectively.
cosN mutations affect cos cleavage in vitro, and the in vitro effects of the mutations parallel effects on virus yield (27, 51). Since the in vitro cos cleavage reaction is an analog of the in vivo cos cleavage reaction that initiates packaging of a virus chromosome, the reduced virus yield caused by a cosN mutation is due at least in part to a defect in initial cos cleavage.
The nature of the cosN G2C C11G defect is revealed by our kinetic studies with purified terminase; wild-type terminase has a reduced efficiency in cleaving cosN G2C C11G (Fig. 3b). Considering that the cosN G2C and C11G mutations are located in rotationally symmetric base pairs of cosN, it is likely that symmetrically disposed gpA subunits interact with cosN half-sites, and that the cosN G2C and C11G mutations affect an interaction between gpA and the cosN half-site (39). The A-E515G, A-N509K, and A-R504C suppressors must directly or indirectly affect the gpA-cosN interaction. We next discuss the nature of the suppressor mutations in the light of these possible mechanisms.
The suppressors map in a cluster, affecting gpA residues 504, 509, and 515. Other mutations which specifically alter the endonuclease activity of terminase also affect the C-terminal third of terminase as follows. Mutations affecting residues 401, 403, and 404 abolish the endonuclease activity of terminase; these mutations reside in an amino acid segment which shares homology with DNA polymerases and helicases. Similar loss-of-endonuclease mutations affect residues 586 and 600, located in the putative basic leucine zipper motif (Fig. 1) (17). Finally, a mutation which changes residue 497, which is located in the putative ATPase center of gpA, has been shown to abolish the endonuclease activity of terminase (Fig. 1) (31).
A detailed study of residue 515 and the A-E515G suppressor was carried out. All of the 12 amino acids tested at position 515 were functional in a wild-type background, including acidic (D, E), basic (K, R), polar uncharged (Q, S), and hydrophobic (A, F, G, I, V, Y) sets. This lack of specificity suggests that residue 515 does not form specific interactions with cosN. A broad spectrum of residues at position 515 (polar uncharged and hydrophobic) suppress cosN G2C C11G (Table 5), and the growth of λ cosN G2C C11G with these substitutions is not temperature sensitive. These results suggest that residue 515 indirectly affects interactions of gpA with cosN and is not involved in interactions crucial to the integrity of gpA. Finally, A-E515G terminase has a broadened substrate specificity for the cleavage reaction: cosN+, cosN G3C C10G, cosN A−1T T13A, and cosN G2C C11G are all nicked by A-E515G terminase.
To discuss the effects of A-E515G on cos cleavage we must consider the complex interactions of terminase with cos and with ATP. Terminase is a bivalent DNA binding enzyme; in addition to gpA-cosN interaction, gpNu1 interacts with cosB (7; reviewed in reference 8). This terminase-cosB interaction anchors terminase when cos cleavage occurs at cosN (Fig. 2), and likely decreases the dissociation rate of the overall terminase-cos interaction. The efficiency of cosN nicking by gpA is thereby increased. ATP binding by terminase apparently alters the conformation of terminase in a way that stimulates endonuclease activity and increases the fidelity of cosN nicking (12, 29). ATP does not increase the affinity of terminase for DNA (12). There is a putative ATPase center in gpA; the residues from 485 to 497 are proposed to comprise a P-loop, the glycine-rich, flexible, phosphate binding loop that is terminated by a lysine, which is proposed to be K497 in gpA (Fig. 1) (25). A lethal mutation causing residue 497 to be changed from K to D (A-K497D) decreases the rate of cos cleavage about 2,000-fold from that of the wild type (31) and reduces the affinity of ATP by 14-fold (30). The interpretation of the A-K497D mutation is that communication between the ATP-binding and endonuclease centers has been disrupted. Terminase with the A-K497D change is defective in cos cleavage even at a saturating level of ATP (31).
A-E515G terminase has an endonucleolytic activity that is different from that of wild-type terminase; it cuts a variety of cosN substrates and cuts cosN G2C C11G more efficiently. The ATPase activity of the A-E515G terminase also differs from that of the wild type, with Km being increased twofold and kcat being decreased threefold (29a). Further, A-E515G is also a suppressor of the A-K497D mutation. Studies show that the cos cleavage activity of the A-K497D A-E515G enzyme is ATP stimulated, indicating that the change caused by the A-E515G mutation restores communication between the endonuclease and ATP-binding domains of A-K497D terminase (29a). The ability to alter communication between the ATP and endonuclease centers enables A-E515G terminase to cut cosN G2C C11G more efficiently. Structural studies of A-E515G terminase would increase our understanding of the communication.
It is interesting that the Nu1ms1, Nu1ms2, and Nu1ms3 mutations, which suppress cosB defects (11), do not suppress cosN G2C C11G. The Nu1ms suppressors were found to act at a postcleavage step of packaging, likely involving the formation or stability of DNA-terminase Complex I (7). The suppressors described in this paper affect gpA-cosN interactions. Recently, suppressors of the third cos subsite, cosQ, were described (14). The cosQ suppressors were also distinct; a mild cosQ mutation could be suppressed by slowing the rate of DNA packaging, a suppression mechanism quite distinct from the cosB and cosN suppression mechanisms.
We have shown that the poor growth of λ cosN G2C C11G phage is suppressed by each of three mutations in the carboxy terminus of gpA and that one of them, A-E515G, suppresses at the level of cos cleavage. A variety of mutations have been found to affect the endonuclease function of terminase, and the A-E515G, A-N509K, and A-R504C mutations give further insights into the complexity of the endonucleolytic activity. The study of additional suppressors which are specific to cosN G2C will allow us to determine specific cosN-terminase interactions.
ACKNOWLEDGMENTS
We thank our coworkers, David Cue, Carol Duffy, Hillary Johnson, Jenny Meyer, John Randell, Fyodor Tereshchenko, and Doug Wieczorek, for advice and helpful discussions during the course of this work.
This research was supported by NIH research grant GM 51611.
REFERENCES
- 1.Arber W, Enquist L, Hohn B, Murray N E, Murray K. Experimental methods for use with lambda. In: Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 433–466. [Google Scholar]
- 2.Bear S, Court D, Friedman D. An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J Virol. 1984;52:966–972. doi: 10.1128/jvi.52.3.966-972.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker A, Marko M, Gold M. Early events in the in vitro packaging of bacteriophage λ DNA. Virology. 1977;78:291–305. doi: 10.1016/0042-6822(77)90100-3. [DOI] [PubMed] [Google Scholar]
- 4.Becker A, Gold M. Prediction of an ATP reactive center in the small subunit, Nu1, of the phage lambda terminase enzyme. J Mol Biol. 1988;199:219–222. doi: 10.1016/0022-2836(88)90391-9. [DOI] [PubMed] [Google Scholar]
- 5.Becker A, Murialdo H, Lucko H, Morell J. Bacteriophage lambda DNA packaging. The product of the FI gene promotes the incorporation of the prohead to the DNA-terminase complex. J Mol Biol. 1988;199:597–607. doi: 10.1016/0022-2836(88)90304-x. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Z-H, Hwang Y, Cue D, Catalano C, Feiss M. Mutations in Nu1, the gene encoding the small subunit of bacteriophage λ terminase, suppress the postcleavage DNA packaging defect of cosB mutations. J Bacteriol. 1997;179:2479–2485. doi: 10.1128/jb.179.8.2479-2485.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catalano C E, Cue D, Feiss M. Virus DNA packaging: the strategy used by phage lambda. Mol Microbiol. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 9.Chow S, Daub E, Murialdo H. The overproduction of DNA terminase of coliphage lambda. Gene. 1987;60:177–229. doi: 10.1016/0378-1119(87)90236-8. [DOI] [PubMed] [Google Scholar]
- 10.Cue D, Feiss M. Genetic analysis of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage λ. J Mol Biol. 1992;228:58–71. doi: 10.1016/0022-2836(92)90491-2. [DOI] [PubMed] [Google Scholar]
- 11.Cue D, Feiss M. Genetic analysis of mutations affecting terminase, the bacteriophage λ DNA packaging enzyme, that suppress mutations in cosB, the terminase binding site. J Mol Biol. 1992;228:72–87. doi: 10.1016/0022-2836(92)90492-3. [DOI] [PubMed] [Google Scholar]
- 12.Cue D, Feiss M. The role of cosB, the binding site for terminase, the DNA packaging enzyme of bacteriophage λ, in the nicking reaction. J Mol Biol. 1993;234:594–609. doi: 10.1006/jmbi.1993.1614. [DOI] [PubMed] [Google Scholar]
- 13.Cue D, Feiss M. A site required for termination of packaging of the phage λ chromosome. Proc Natl Acad Sci USA. 1993;90:9290–9294. doi: 10.1073/pnas.90.20.9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cue D, Feiss M. Genetic evidence that recognition of cosQ, the signal for termination of phage λ DNA packaging, depends on the extent of head filling. Genetics. 1997;147:7–17. doi: 10.1093/genetics/147.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels D, Schroeder J, Szybalski W, Sanger F, Coulson A, Hong G, Hill D, Petersen G, Blattner F. Complete annotated lambda sequence. In: Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 519–576. [Google Scholar]
- 16.Davidson A, Gold M. A novel in vitro DNA packaging system demonstrating a direct role for the bacteriophage λ FI gene product. Virology. 1987;161:305–315. doi: 10.1016/0042-6822(87)90122-x. [DOI] [PubMed] [Google Scholar]
- 17.Davidson A R, Gold M. Mutations abolishing the endonuclease activity of bacteriophage λ terminase lie in two distinct regions of the A gene, one of which may encode a “leucine zipper” DNA-binding domain. Virology. 1992;189:21–30. doi: 10.1016/0042-6822(92)90677-h. [DOI] [PubMed] [Google Scholar]
- 18.Feiss M. Terminase and the recognition, cutting and packaging of λ chromosomes. Trends Genet. 1986;2:100–104. [Google Scholar]
- 19.Fowler R G, Degner G E, Cox E C. Mutational specificity of a conditional Escherichia coli mutator, mutD5. Mol Gen Genet. 1974;133:179–191. doi: 10.1007/BF00267667. [DOI] [PubMed] [Google Scholar]
- 20.Frackman S, Siegele D A, Feiss M. A functional domain of bacteriophage λ terminase for prohead binding. J Mol Biol. 1984;180:283–300. doi: 10.1016/s0022-2836(84)80005-4. [DOI] [PubMed] [Google Scholar]
- 21.Frackman S, Siegele D A, Feiss M. The terminase of bacteriophage λ: functional domains for cosB binding and multimer formation. J Mol Biol. 1985;183:225–238. doi: 10.1016/0022-2836(85)90215-3. [DOI] [PubMed] [Google Scholar]
- 22.Fry D, Kuby S, Mildvan A. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci USA. 1986;83:907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold M, Becker A. The bacteriophage λ terminase: partial purification and preliminary characterization of properties. J Biol Chem. 1983;258:14619–14625. [PubMed] [Google Scholar]
- 24.Granston A E, Alessi D M, Eades L, Friedman D. A point mutation in the Nu1 gene of bacteriophage λ facilitates phage growth in Escherichia coli with himA and gyrB mutations. Mol Gen Genet. 1988;212:149–156. doi: 10.1007/BF00322458. [DOI] [PubMed] [Google Scholar]
- 25.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J Mol Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 27.Higgins R R, Becker A. Chromosome end formation in phage lambda, catalyzed by terminase, is controlled by two DNA elements of cos, cosN, and R3 and by ATP. EMBO J. 1994;13:6152–6161. doi: 10.1002/j.1460-2075.1994.tb06962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins R R, Becker A. The lambda terminase enzyme measures the point of its endonucleolytic attack 47 ± 2 bp away from its site of specific DNA binding, the R site. EMBO J. 1994;13:6162–6171. doi: 10.1002/j.1460-2075.1994.tb06963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins R, Lucko H, Becker A. Mechanism of cos DNA cleavage by bacteriophage λ terminase: multiple roles of ATP. Cell. 1988;54:765–775. doi: 10.1016/s0092-8674(88)91021-5. [DOI] [PubMed] [Google Scholar]
- 29a.Hwang, Y., and M. Feiss. Unpublished data.
- 30.Hwang Y, Catalano C E, Feiss M. Kinetic and mutational dissection of the two ATPase activities of terminase, the DNA packaging enzyme of bacteriophage λ. Biochemistry. 1996;35:2796–2803. doi: 10.1021/bi952322z. [DOI] [PubMed] [Google Scholar]
- 31.Hwang Y, Feiss M. Mutations affecting the high affinity ATPase center of gpA, the large subunit of bacteriophage λ terminase, inactivate the endonuclease activity of terminase. J Mol Biol. 1996;261:524–535. doi: 10.1006/jmbi.1996.0480. [DOI] [PubMed] [Google Scholar]
- 32.King R A, Anders D L, Christie G E. Site-directed mutagenesis of an amino acid residue in the bacteriophage P2 ogr protein implicated in interaction with Escherichia coli RNA polymerase. Mol Microbiol. 1992;6:3313–3320. doi: 10.1111/j.1365-2958.1992.tb02199.x. [DOI] [PubMed] [Google Scholar]
- 33.Kosturko L D, Daub E, Murialdo H. The interaction of E. coli integration host factor and lambda cos DNA: multiple complex formation and protein-induced bending. Nucleic Acids Res. 1989;17:317–334. doi: 10.1093/nar/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuzminov A, Schabtach E, Stahl F W. Chi sites in combination with RecA protein increase the survival of linear DNA in Escherichia coli by inactivating the exoV activity of RecBCD nuclease. EMBO J. 1994;13:2764–2776. doi: 10.1002/j.1460-2075.1994.tb06570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kypr J, Mrazek J. Lambda phage protein Nu1 contains the conserved DNA binding fold of repressors. J Mol Biol. 1986;91:139–140. doi: 10.1016/0022-2836(86)90430-4. [DOI] [PubMed] [Google Scholar]
- 36.Mendelson I, Gottesman M, Oppenheim A B. HU and integration host factor function as auxiliary proteins in cleavage of phage lambda cohesive ends by terminase. J Bacteriol. 1991;173:1670–1676. doi: 10.1128/jb.173.5.1670-1676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pal S K, Chattoraj D K. P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J Bacteriol. 1988;170:3554–3560. doi: 10.1128/jb.170.8.3554-3560.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubinchik S, Parris W, Gold M. The in vitro ATPases of bacteriophage λ terminase and its large subunit, gene product A. J Biol Chem. 1994;269:13586–13593. [PubMed] [Google Scholar]
- 39.Rubinchik S, Parris W, Gold M. The in vitro endonuclease activity of gene product A, the large subunit of the bacteriophage lambda terminase, and its relationship to the endonuclease activity of the holoenzyme. J Biol Chem. 1994;269:13575–13585. [PubMed] [Google Scholar]
- 40.Shinder G, Gold M. The Nu1 subunit of bacteriophage lambda terminase binds to specific sites in cos DNA. J Virol. 1988;62:387–392. doi: 10.1128/jvi.62.2.387-392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sippy J, Feiss M. Analysis of a mutation affecting the specificity domain for prohead binding of the bacteriophage λ terminase. J Bacteriol. 1992;174:850–856. doi: 10.1128/jb.174.3.850-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Six E, Klug C A C. Bacteriophage P4: a satellite virus depending on a helper such as prophage P2. Virology. 1973;51:327–344. doi: 10.1016/0042-6822(73)90432-7. [DOI] [PubMed] [Google Scholar]
- 43.Tomka M A, Catalano C E. Kinetic characterization of the ATPase activity of the DNA packaging enzyme from bacteriophage λ. Biochemistry. 1993;32:11992–11997. doi: 10.1021/bi00096a008. [DOI] [PubMed] [Google Scholar]
- 44.Tomka M A, Catalano C E. Physical and kinetic characterization of the DNA packaging enzyme from bacteriophage λ. J Biol Chem. 1993;268:3056–3065. [PubMed] [Google Scholar]
- 45.Walker J, Saraste M, Runswick M, Gay N. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiman M, Bertani G, Kelly B, Sasaki I. Genetic map of E. coli C. Mol Gen Genet. 1970;107:1–31. doi: 10.1007/BF00433220. [DOI] [PubMed] [Google Scholar]
- 47.Wu W-F, Christiansen S, Feiss M. Domains for protein-protein interactions at the N and C termini of the large subunit of bacteriophage λ terminase. Genetics. 1988;119:477–484. doi: 10.1093/genetics/119.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xin W, Feiss M. The interaction of Escherichia coli integration host factor with the cohesive end sites of phages λ and 21. Nucleic Acids Res. 1988;16:2015–2030. doi: 10.1093/nar/16.5.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xin W, Feiss M. Function of IHF in λ DNA packaging. I. Identification of the strong binding site for integration host factor and the locus for intrinsic bending in cosB. J Mol Biol. 1993;230:492–504. doi: 10.1006/jmbi.1993.1166. [DOI] [PubMed] [Google Scholar]
- 50.Xin W, Cai Z-H, Feiss M. Function of IHF in λ DNA packaging. II. Effects of mutations altering the IHF binding site and the intrinsic bend in cosB on λ development. J Mol Biol. 1993;230:505–515. doi: 10.1006/jmbi.1993.1167. [DOI] [PubMed] [Google Scholar]
- 51.Xu S-Y, Feiss M. Structure of the bacteriophage λ cohesive end site: genetic analysis of the site (cosN) at which nicks are introduced by terminase. J Mol Biol. 1991;220:281–292. doi: 10.1016/0022-2836(91)90013-v. [DOI] [PubMed] [Google Scholar]
- 52.Yeo A, Feiss M. Mutational analysis of the prohead binding domain of the large subunit of terminase, the bacteriophage λ DNA packaging enzyme. J Mol Biol. 1995;245:126–140. [PubMed] [Google Scholar]