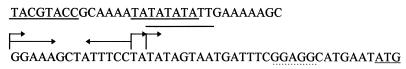Abstract
The glnA gene in the domains Bacteria and Archaea encodes glutamine synthetase, a universally distributed enzyme that functions in ammonia assimilation and glutamine synthesis. We investigated the regulation and function of glnA in the methanogenic archaeon Methanococcus maripaludis. The deduced amino acid sequence of the gene demonstrated its membership in class GSI-α of glutamine synthetases. The gene appeared to be expressed as a monocistronic operon. glnA mRNA levels and specific activities of glutamine synthetase were regulated similarly by nitrogen. Three transcription start sites were identified, corresponding to two overlapping nitrogen-regulated promoters and one weaker constitutive promoter. An inverted repeat immediately upstream of the regulated transcription start sites mediated repression under noninducing conditions. Thus, mutations that altered the sequence of the inverted repeat resulted in derepression. The inverted repeat had sequence similarity with a repeat that we previously identified as the nif operator of M. maripaludis, suggesting a common mechanism of nitrogen regulation. Efforts to produce a glnA null mutant failed, suggesting that glnA is an essential gene in M. maripaludis.
Glutamine synthetase (GS), a universally distributed enzyme that functions in ammonia assimilation and glutamine (Gln) synthesis, is known to exist in three distantly related forms (17). GSI, encoded by the glnA gene, is represented by the well-studied enzyme of enteric bacteria and is distributed throughout the domains Bacteria and Archaea. Phylogenetic analysis (7) has revealed two main subdivisions. GSI-α is found in the low G+C gram-positive bacteria, the genus Thermotoga, and the Euryarchaeota (including methanogens and extreme halophiles). GSI-β is found in all remaining Bacteria. GSI is composed of 12 identical subunits (25). GSII occurs mainly in Eucarya but is also found in rhizobia and certain actinomycetes and is composed of eight identical subunits. GSIII, encoded by glnN, is present in a few bacterial species, including certain cyanobacteria (26) and rhizobia, and is composed of six identical subunits (12). A few bacteria, such as Rhizobium meliloti, have all three forms of GS (10).
All three GS forms can be regulated by the nitrogen status of the cell, in keeping with their function in ammonia assimilation. At the transcriptional level, glnA in enteric bacteria is regulated by the nitrogen-sensing regulatory cascade (25). Expression from a second promoter allows for a low level of expression in nitrogen excess (20). In a species of the cyanobacterium Synechocystis, glnA is regulated moderately and glnN markedly by nitrogen (26). At the enzyme modification level, GSI enzymes in the β subdivision are reversibly inhibited by adenylylation at a conserved Tyr residue, a mechanism also involving the nitrogen-sensing regulatory cascade (7, 25). GSIII in R. meliloti has been shown to be inhibited by ADP-ribosylation (19).
GS appears to function universally in ammonia assimilation, producing Gln from glutamate (Glu) and ammonia. Gln is then used by glutamate synthase to produce two moles of Glu, a central amino donor for biosynthesis. However, other pathways for ammonia assimilation exist. In enteric bacteria under conditions of high ammonia concentration, glutamate dehydrogenase produces Glu from 2-ketoglutarate and ammonia (25). It has also been suggested that alanine dehydrogenase can function in ammonia assimilation in certain Bacteria and Archaea, including several methanogenic species (3, 15). A second function of GS is in Gln production for protein synthesis. However, an alternative pathway occurs in some organisms: conversion of glutamyl-tRNA to glutaminyl tRNA. Indeed, some organisms, including Methanococcus jannaschii, appear to lack glutaminyl tRNA synthetase (8).
In the methanogenic Archaea, ammonia is a universal source of nitrogen. Some species can use certain organic nitrogen compounds as well, and some can fix dinitrogen (11). Methanococcus maripaludis grows well with ammonia or alanine (Ala), and can also grow diazotrophically (5, 16, 33). For ammonia assimilation, GS is used in methanogens, where it is known only in the form GSI-α. GSI from Methanobacterium ivanovii was dodecameric and was not regulated by adenylylation (4), which is now known to be the case with all GSI-α enzymes (7). The glnA gene was cloned from Methanococcus voltae and sequenced (23). A comparison of deduced amino acid sequences confirmed that the protein belonged to the GSI-α group and lacked a 29-amino-acid stretch corresponding to a protease-sensitive loop present in GSI-β enzymes (1). Northern analysis indicated that the M. voltae gene was transcribed as a monocistronic operon, and a putative promoter sequence was identified (23). An inverted repeat lay immediately 5′ to the putative TATA box. Interestingly, this inverted repeat is similar to one in the nifH promoter region of M. maripaludis that we have shown mediates regulation by repressor binding (9). In M. voltae, GS activity was partially repressed at high ammonia concentrations (23).
In this paper, we present an analysis of glnA expression and function in M. maripaludis. We identify multiple transcription start sites, and we document regulated expression mediated by an inverted repeat sequence that is similar to the one observed in the glnA promoter region of M. voltae and to the one shown to regulate nifH expression in M. maripaludis. In addition, we attempt to determine the function of glnA in the cell by directed mutagenesis.
MATERIALS AND METHODS
Growth of cultures.
M. maripaludis LL (16) and its derivatives were maintained on McC medium (34) using anaerobic techniques described previously (2). Puromycin (2.5 μg/ml) was added as needed. Growth of cultures for primer extension, Northern analyses, and GS assays was in nitrogen-free medium (5), and the gas atmosphere was 20% N2, 20% CO2, and 60% H2 at a total pressure of three atmospheres. The medium was supplemented as needed with ammonia (10 mM) and/or Gln (10 mM). Cultures were shaken at 37°C. Gln stock solution was adjusted to neutral pH and filter sterilized before storage.
Cloning and sequencing glnA.
DNA fragments of M. maripaludis glnA were obtained by PCR using the forward primer 5′-TTT/C GAC/T GGT/A TCT/A TCA/T AT-3′ or 5′-GCT/A ACA/T TTC/T ATG CCT/A AAA CC-3′ and the reverse primer 5′-CCT/A GGA/T ACT AAT CTT TTG/A TAT/A GAG/A TT-3′. PCR was performed with 200 ng of M. maripaludis genomic DNA (prepared as described previously [9]) and 200 ng of each primer using cloned Pfu polymerase (Stratagene). Amplification was performed for 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 30 sec. A 0.74-kb PCR product was cloned into pGEM7 (Promega). DNA sequencing confirmed that it was part of glnA. The 0.74-kb fragment was used to screen a λ library of M. maripaludis DNA as described previously (5). Positive plaques were suspended in suspension medium (SM) (27) and were used to infect Escherichia coli LE392 (27). Phage DNA was extracted by using the Wizard Lambda Preps DNA Purification System (Promega). The DNA was digested with HindIII, and a 4.7-kb fragment was isolated from an agarose gel by using the Prep-A-Gene DNA Purification Kit (Bio-Rad) and cloned into pGEM7 to yield pGlnA2. The glnA gene was sequenced on both strands using walking primers. Sequencing was performed with the ABI Prism dRhodamine Terminator Cycle Sequencing Reaction Kit (PE Applied Biosystems) and analyzed at the DNA Sequencing Facility of the Department of Biochemistry at the University of Washington. Sequence comparisons were done using the pileup and gap functions of the Genetics Computer Group software.
Primer extension analysis.
Cells were grown on nitrogen-free medium plus ammonia to an optical density at 660 nm (OD660) of approximately 0.3, centrifuged anaerobically in growth tubes at 750 × g for 10 min, resuspended in nitrogen-free medium with the appropriate nitrogen source, and incubated overnight. Cells grown on N2 were harvested at an OD660 of 0.3 to 0.6, and cells on NH4+ or NH4+ plus Gln at an OD660 of 0.5 to 0.8. Cells were harvested by centrifuging aerobically at 4°C and processed as previously described (16) to obtain RNA. Primer extension analysis was performed as previously described (9) using the primer TCTTCCTCACCAGCAGCACC.
GS activity assay.
Cells were grown to an OD660 between 0.3 and 0.5, pelleted at 4°C, and resuspended in cold buffer containing 10 mM imidazole · HCl (pH 7.2), 0.3 mM MnCl2, and 1 mM 2-mercaptoethanol. Cells were disrupted by sonication, and debris was removed by centrifugation. GS assays were performed by a modified γ-glutamyltransferase assay (29) at pH 7.2 with a Mn2+ concentration of 0.3 mM. Mg2+ was reported to inhibit the activity of the adenylylated E. coli enzyme in the transferase assay (29). However, in preliminary experiments, Mg2+ between 5 mM and 60 mM inhibited the activity equally in M. maripaludis extracts from cells grown with either N2 or NH4+, so Mg2+ was subsequently omitted. Protein in cell extracts was measured by the Coomassie blue method (28) using bovine serum albumin as standard. Assays were performed with triplicate cultures for each condition, and standard errors of the means were calculated.
Construction of M. maripaludis mutants.
The plasmid pGlnA2 (described above) contains the glnA gene near the right end (5′ to 3′ orientation) of a 4.7-kb HindIII fragment of the M. maripaludis genome (Fig. 1). The 4.7-kb HindIII fragment was recloned into a pGEM derivative from which the EcoRI site had been removed (9). Then an internal portion of glnA was deleted by removing a 615-bp EcoRI fragment (nucleotides 576 to 1190 in the GenBank entry) to produce pCM1. Two mutations were made in pCM1, one changing the GGAA in the first half of the inverted repeat (Fig. 2) to CCTT (pCM2), and the other changing in addition the TTCC in the second half of the repeat to AAGG (pCM3). For this purpose, we used the Statagene QuikChange Site-Directed Mutagenesis kit with the primers CCGCAAAATATATATATTGAAAAAGCCCTTAGCTATTTCCTATATAGTAATGATTTCGGA and GCCTCCGAAATCATTACTATATAGGAAATAGCTAAGGGCTTTTTCAATATATATATTTTG for the first mutation and CGTACCGCAAAATATATATATTGAAAAAGCCCTTAGCTATAAGGTATATAGTAATGATTT and CATGCCTCCGAAATCATTACTATATACCTTATAGCTAAGGGCTTTTTCAATATATATATT for the second mutation. PCR was performed with an initial incubation at 95°C for 30 sec followed by 18 cycles of 95°C for 30 sec, 40°C for 1 min, and 68°C for 18 min. The mutations were confirmed by sequencing. The resulting 4.2-kb HindIII fragments (from pCM1, pCM2, and pCM3) were cloned into pJK3 (22), which contains a puromycin resistance marker for selection in methanogenic Archaea, to produce the plasmids pCM11, pCM12, and pCM13. All three constructs contained the insert in the same orientation, with transcription of glnA in the same direction as the transcription of the puromycin resistance gene. The three plasmids were transformed into M. maripaludis as previously described (32) to create mutants 1, 2, and 3 (strains Mm311, Mm312, and Mm313) respectively. Southern analysis confirmed that in all cases a single recombination event had occurred to the 5′ side of the deletion mutation.
FIG. 1.
Restriction map of the glnA region of the M. maripaludis genome. R, EcoRI; H, HindIII. Construction of a mutation by replacement of an internal fragment of glnA with a puromycin resistance cassette is shown.
FIG. 2.
Nucleotide sequence of the glnA promoter region. Shown are three TATA box promoter sequences (underlined), three transcription start sites (bent arrows), an inverted repeat (overlined with arrows), a putative ribosome binding site (underlined with dots), and the translation start (underlined).
To confirm that the three M. maripaludis transformants contained the desired mutations, the 5′ portions of glnA including the promoter regions were amplified from genomic DNA prepared as previously described (5). PCR was performed using the forward primer GATACATTTACGTACCGG (upstream of the promoter region) and the reverse primer GCTGCAAGCATTACTGTG (downstream of the deletion site). PCR was performed with cloned Pfu polymerase (Statagene) by using an initial incubation at 94°C for 45 sec followed by 25 cycles of 94°C for 45 sec, 50°C for 45 sec, and 72°C for 3 min, and a final incubation at 72°C for 10 min. Two hundred nanograms of template DNA and of each primer was used. For each mutant, PCR products of the two expected sizes, corresponding to the full length and shortened (by deletion) glnA fragments, were obtained. Sequencing of the PCR products confirmed that the mutants contained the expected mutations in the inverted repeats preceding the shortened glnA genes.
An insertion mutation in the glnA coding region was constructed as follows. The 4.7-kb HindIII fragment was recloned into a pGEM7 derivative in which the EcoRI site had been eliminated (9). The 0.6-kb EcoRI fragment internal to glnA was replaced with a puromycin resistance cassette (Fig. 1) (13). This construct was linearized with HindIII and introduced into M. maripaludis by transformation, and transformants were plated on McC agar containing puromycin and Gln (10 mM).
Northern analysis.
Freshly grown cultures in McC were used to inoculate N-free medium with and without added NH4+. After overnight growth at an OD600 between 0.1 and 0.35, cells were harvested anaerobically at 4°C by spinning at 750 × g for 10 min in the growth tubes. RNA was obtained using the RNeasy kit (Qiagen) following manufacturer’s directions, except that cells were suspended in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate). Northern analysis was performed essentially as described (16). The probe, a SnaBI-EcoRI fragment corresponding to the 5′ portion of glnA, was labeled using the Random Primed DNA Labeling Kit (Boehringer Mannheim). Band intensities were determined by exposure on a phosphorimager and integration.
Southern analysis and colony hybridization.
Genomic DNA was obtained as described previously (5), and 2.5 μg of the DNA was digested and run on a gel. DNA was transferred to a Zeta-Probe Blotting Membrane (Bio-Rad) and probed following the manufacturer’s instructions. Colony hybridization was done using Colony/Plaque Screen Hybridization Transfer Membrane (Du Pont/NEN) according to the manufacturer’s directions. Probes were labeled with the Random Primed DNA Labeling Kit (Boehringer Mannheim).
Nucleotide sequence accession number.
The nucleotide sequence of the M. maripaludis glnA gene, with promoter and terminator regions, is available at GenBank under accession no. AF062391.
RESULTS
Cloning and sequencing of glnA.
In order to clone glnA from M. maripaludis, we designed degenerate primers (two forward and one reverse) based on conserved coding regions from a variety of Bacteria and M. voltae (23). PCR reactions yielded DNA with the two expected sizes, 0.74 and 0.22 kb. Probing a Southern blot of M. maripaludis genomic DNA with the 0.74-kb fragment indicated that a single copy of glnA was present (not shown). The 0.74-kb fragment was used to identify a 15-kb clone from a genomic library of M. maripaludis that contained glnA. The gene was located on a 4.7-kb HindIII fragment (Fig. 1), which was cloned.
The sequence of the glnA gene revealed an open reading frame (ORF) of 446 amino acids. The amino acid sequence aligned most closely with other glnA genes of the GSI-α group and lacked the 28-amino-acid sequence corresponding to a protease sensitive loop present in GSI-β enzymes (references 1 and 23 and data not shown). Amino acid sequence identities shared with glnA of other organisms were 77% with M. voltae, 54% with Bacillus subtilis, and 40% with E. coli.
The glnA gene was preceded by a putative ribosome binding site and overlapping putative promoters (Fig. 2). The ribosome binding site resembled others in Methanococcus (6). It was located only one nucleotide upstream from the first in-frame ATG codon and seven nucleotides from the second, suggesting that the latter ATG, which aligns with the first codon of M. voltae glnA, is the translational start site. The overlapping promoters resembled closely the consensus for methanogenic Archaea, TTTA(T/A)ATA (24). Between the ribosome binding site and the promoter, an inverted repeat was found (Fig. 2). The glnA ORF was followed by two oligo(dT) sequences that may signal termination (24).
Transcription and regulation of glnA.
We studied the initiation of glnA transcription and its regulation by primer extension analysis. Cells were grown with N2 only, or supplemented with Gln or NH4+ plus Gln. Primer extension analysis indicated three different 5′ ends for glnA mRNA (Fig. 3). The nucleotide positions corresponding to the two more downstream 5′ ends each occurred 28 bp from the centers of the overlapping promoter sequences identified above (Fig. 2). This spacing follows precedent for promoters of methanogens (24). The most upstream end was also positioned 28 bp from a putative promoter sequence, albeit with weaker similarity to the consensus (Fig. 2). The most likely explanation for these results is that transcription initiates at three separate locations, each corresponding to a separate TATA box promoter.
FIG. 3.
Primer extension analysis of glnA transcripts in wild-type M. maripaludis. N2, N2 alone; gln, N2 + Gln; gln + NH3, N2 + Gln + NH4+.
We expected that cells grown on N2 would be nitrogen limited and would therefore express higher levels of glnA in order to maximize the assimilation of ammonia. In contrast, cells grown with NH4+ should express lower levels of glnA. With N2, the transcripts originating from the two overlapping downstream sites were stronger than the transcript from the upstream site (Fig. 3). The same pattern was observed with cells grown with Gln, consistent with the apparent lack of ability of Gln to serve as a nitrogen source (see below). In contrast, when cells were grown with NH4+, transcription from the downstream sites was markedly decreased relative to the upstream site. These results suggest that transcription from the overlapping downstream promoters is regulated by nitrogen, while the upstream promoter may be constitutive. The regulation of glnA mRNA levels by nitrogen was reflected in GS activity levels. Thus, GS activity in N2-grown cells was 203 ± 67 nmol min−1 mg protein−1 while in NH4+-grown cells it was only 44 ± 11 nmol min−1 mg protein−1.
The inverted repeat located immediately 5′ to the regulated start sites resembles an operator site (CGGAAAGAAGCTTCCG [underlining indicates inverted repeat]) in the promoter region of nifH that we have shown to be involved in repression (9). Therefore, we hypothesized that the repeat in the glnA promoter region would function similarly. To test this hypothesis, we made mutations in the inverted repeats. We made three mutants of M. maripaludis by introducing altered glnA gene regions and allowing the introduced DNA to recombine into the genome (see Materials and Methods). This procedure yielded in each strain two copies of glnA separated by vector sequences. In each strain one copy of glnA was unaltered, and the other copy was shortened by an in-frame internal deletion. The shortened gene would produce an mRNA that would be smaller than the wild-type gene and hence would be distinguishable in Northern blots. In the three mutants the glnA gene bearing the internal deletion was preceded either by an unaltered glnA promoter region (mutant 1), by a promoter region in which the first half of the inverted repeat was altered (GGAAAGCTATTTCC→CCTTAGCTATTTCC [mutant 2]), or by a promoter region in which both halves of the repeat were altered (→CCTTAGCTATAAGG [mutant 3]). The sequence of each of these promoter regions was confirmed after PCR amplification from the genome.
Cultures were prepared under conditions of nitrogen excess (10 mM NH4+) and nitrogen limitation (N2 only) and Northern analysis of the glnA transcripts was performed (Fig. 4). As expected, two bands were observed for each mutant. The top band corresponded to wild-type glnA. The bottom band corresponded to glnA with the in-frame internal deletion, transcribed from the unaltered promoter region (mutant 1) or from the promoter region with mutations in the inverted repeat (mutants 2 and 3). Northerns were performed on wild-type M. maripaludis too, and a single full-length band was observed (not shown). A comparison of the full-length transcript under the two conditions indicated that for each strain, induction occurred under nitrogen limiting conditions (Fig. 4). Roughly the same level of induction occurred in the wild-type strain (not shown). This observation is consistent with the limited use of the downstream transcription start sites in the presence of NH4+. In order to evaluate the effects of the mutations in the inverted repeat, we compared the intensities of the two bands in each lane by taking the ratio of the lower band to the upper band (Fig. 4). This calculation eliminated the effects of unwanted differences between lanes. Thus, variations in the amount of RNA loaded onto the gel or differences in the exact conditions of cultures at harvest time were controlled for. The results with mutants 2 and 3 were then compared to mutant 1. Strikingly, the mutation in the first half of the inverted repeat (mutant 2) increased transcription under nitrogen excess, confirming the hypothesis that the repeat is involved in repression under noninducing conditions. The mutation affecting both halves of the inverted repeat (mutant 3) also resulted in derepression, but to a lesser extent than in mutant 2. This result can be explained if the mutation in mutant 3 weakened the downstream promoters, which is not unlikely given the close proximity of the second half of the inverted repeat to the transcription start sites. Consistent with this interpretation, transcription of the altered glnA gene in mutant 3 was lower under inducing conditions than in mutant 1, which has an unaltered promoter region. The experiment was repeated with different cultures, and the same results were observed. These results indicate that the inverted repeat is required for repression of glnA transcription. It is unlikely that the results can be explained by the introduction of artifactual promoter activity, since the increased transcription observed did not occur under inducing conditions.
FIG. 4.
Northern analysis of glnA transcripts in three mutants in the presence and absence of ammonia. The intensity ratios of the lower and upper bands are indicated.
Evidence that glnA is an essential gene in M. maripaludis.
In order to test directly whether GlnA is necessary for ammonia assimilation or Gln synthesis in M. maripaludis, we attempted to make a glnA null mutant. We hypothesized that such a mutant would be unable to grow on NH4+ and would require Gln as a nitrogen source. In the 4.7-kb HindIII fragment, the internal EcoRI fragment of glnA was replaced with a puromycin resistance cassette (Fig. 1). The resulting construct (glnA::Pur) was introduced into M. maripaludis by transformation. In this procedure double recombination (one recombination event on each side of Purr) would yield a glnA null mutant. Ordinarily, we easily obtain double recombinants in M. maripaludis in nonessential genes (5, 9). Transformants were plated on McC agar medium containing puromycin and Gln (10 mM). Five transformants were examined by Southern blot to determine whether double recombinants were obtained (Fig. 5). However, in no case had the mutant glnA region replaced the wild-type gene. All five transformants contained both 4.7- and 5.9-kb HindIII fragments that hybridized to the glnA region, indicating that both wild-type and mutant glnA genes were present. All five transformants also contained 0.6-kb as well as 2.0- and 5.7-kb EcoRI fragments (Fig. 1 and 5), indicating the presence of the wild-type glnA region (the mutant glnA gene would not be detected in an EcoRI digest because the Purr cassette was not probed for). Three of these transformants (Fig. 5, lanes 8, 10, and 11) also contained a 7.1-kb EcoRI fragment that indicated the presence of the 3.0-kb vector between two glnA regions. These transformants apparently arose by a single recombination event involving circularized transforming DNA. However, the two remaining transformants (lanes 7 and 9) lacked the 7.1-kb EcoRI fragment, suggesting that double recombination had taken place but that a wild-type copy of the glnA region was retained, perhaps on a separate copy of the chromosome.
FIG. 5.
Southern analysis of glnA::Pur transformants. Lanes: 1 to 6, HindIII digests of genomic DNA; 7 to 12, EcoRI digests of genomic DNA. Lanes 1 and 7, 2 and 8, 3 and 9, 4 and 10, 5 and 11, five separate mutants after transformation of wild-type M. maripaludis with the glnA::Pur construct. Lanes 6 and 12, wild-type M. maripaludis. The probe was the 4.7-kb wild-type HindIII fragment (Fig. 1).
A second attempt to generate glnA null mutants was made. In this case, Southern analysis indicated that four of four transformants contained both wild-type and mutant glnA regions without evidence of vector sequences. In addition, 100 transformants were screened by colony hybridization and appeared to retain the 0.6-kb EcoRI fragment internal to glnA. These results suggested that Gln supplied in the culture medium could not substitute for GS, perhaps because of an inability of M. maripaludis to transport Gln. Consistent with this interpretation, Gln did not appear to function as sole nitrogen source for M. maripaludis. Thus, we obtained only poor growth of wild-type M. maripaludis on Gln, and this growth could be attributed to NH4+ derived from Gln by chemical degradation.
If Gln could not be transported by M. maripaludis, it seemed possible that a different, transportable nitrogen source might substitute for Gln. M. maripaludis grows on Ala as well as it does on NH4+ (18, 33), so Ala can be transported. We hypothesized that Ala might provide the nitrogen needs of the cell by amino transfer to produce Glu. The necessary enzyme, alanine aminotransferase, is present in the related species M. jannaschii as evidenced by genome sequencing (8), and a low level of alanine aminotransferase activity was detected in Methanococcus aeolicus (35). In addition, glutaminyl tRNA might be produced from glutamyl tRNA as may be the case in M. jannaschii, which apparently lacks glutaminyl tRNA synthetase (8). Therefore, we grew four transformants from the first attempt at mutagenesis for three cycles in McC medium in the presence of Ala (10 mM) to see whether glnA null mutants could then be obtained. These mutants might arise in the single recombinants by a second recombination event to eliminate the wild-type gene, or in the transformants that apparently retained a wild-type chromosome copy by elimination of that copy. In either case, such mutants should then require Ala as a nitrogen source and should be unable to use NH4+. After the third cycle of growth with Ala, each transformant was plated on McC medium plus Ala to obtain isolated colonies. Fifty colonies from each transformant were then replica plated onto medium without Ala. However, no Ala-dependent derivatives were obtained. Our inability to obtain a glnA null mutant suggests that M. maripaludis cannot transport Gln and that glnA in M. maripaludis is an essential gene even in the presence of alternative nitrogen sources.
DISCUSSION
We have cloned the gene for GS from M. maripaludis and have confirmed that it belongs to the GSI-α class of GS enzymes along with the protein from other methanogenic Archaea. As in M. voltae (23), glnA in M. maripaludis appears to constitute a monocistronic operon, as evidenced by the size and 5′ end of the transcript and the apparent presence of terminator sequences immediately downstream of the stop codon.
Our results suggest that GS is an essential enzyme in M. maripaludis. Since the provision of external Gln was insufficient to render GS dispensable, M. maripaludis may be unable to transport Gln. Gln is turn may be essential either as a nitrogen donor for nitrogen metabolism (via Glu) or for protein synthesis or both. Each of these possibilities suggests a different (but mutually compatible) model: if Gln is an essential nitrogen donor for nitrogen metabolism, the results suggest that only GS can serve to assimilate ammonia, and its function cannot be replaced by a glutamate dehydrogenase or alanine dehydrogenase. Gln also cannot be obtained from glutaminyl tRNA. Even with Ala as a nitrogen source, GS is still essential, suggesting that Ala is converted to ammonia by an alanine dehydrogenase rather than to Glu by an aminotransferase. GS then assimilates the ammonia obtained from Ala. If Gln is essential for protein synthesis, this could suggest that a conventional synthesis of glutaminyl tRNA with glutaminyl tRNA synthetase occurs, and that the system implicated in M. jannaschii (direct conversion of glutamyl tRNA to glutaminyl tRNA) (8) does not function in M. maripaludis. Alternatively, the direct conversion could operate but the amido donor for the reaction could be Gln itself.
M. maripaludis glnA mRNA levels were regulated by nitrogen as in E. coli. GS activity levels showed the same trends (similar to those in M. voltae [23]), so no evidence of posttranscriptional regulation was obtained. However, it should be noted that the γ-glutamyltransferase assay for GS activity does not always distinguish between modified (inactive) and unmodified (active) forms of the enzyme (29).
In E. coli, glnA is transcribed from two promoters. The stronger, downstream promoter is regulated by nitrogen while the weaker, upstream promoter is constitutive (20). M. maripaludis appears to have a similar arrangement. The overlapping promoters corresponding to the downstream start sites may be considered together, and constitute the stronger, downstream, nitrogen-regulated promoter set (Fig. 2). A weaker promoter further upstream appears to be constitutive. This arrangement may allow for the dual function of GS, ammonia assimilation for the cell and the synthesis of Gln for protein synthesis.
However, the mechanism of regulation in M. maripaludis contrasts with the mechanism in enteric bacteria. The work presented here indicates that in M. maripaludis nitrogen regulation occurs by repression and depends on an inverted repeat sequence. This mechanism contrasts with nitrogen regulation of glnA in E. coli, which occurs by activation via an upstream enhancer element (20). This situation parallels the regulation of nif gene expression. We reported that in M. maripaludis nif gene transcription is regulated by repression, also involving an inverted repeat adjacent to the transcription start site (9). This mechanism contrasts with nif regulation in Klebsiella pneumoniae, which occurs by an activation mechanism (21). Thus, in each organism the regulation of glnA and nif shows similarities, but contrasts with the other organism. It should be noted however that in B. subtilis, glnA transcription is regulated by repression mediated by two inverted repeat sequences in the promoter region (14).
The similarity between glnA and nif regulation in M. maripaludis is underscored by the similar nucleotide sequences of the inverted repeats. Thus, the inverted repeat in the nif promoter region, CGGAAAGAAGCTTCCG (9) compares with the one in the glnA promoter region, CGGAAAGCTATTTCCT. Both genes are regulated by nitrogen, and the same repressor protein may bind to both sites. Taking glnA and nif together, the positions of the inverted repeats with respect to the promoters may be instructive. The regulation of glnA mediated by the inverted repeat presumably acts on the downstream promoters, since these are regulated in the wild-type strain while the upstream promoter is constitutive. Therefore, in the regulation of both glnA and nif, where the inverted repeat is effective in repression, it is adjacent to the transcription start site, just upstream from it in the case of glnA and just downstream from it in the case of nif. In cases where the inverted repeat is further downstream from the promoter, more distant from the transcription start site, it does not appear to play a significant role in repression. Thus, the inverted repeat in glnA does not appear to mediate repression of transcription from the upstream promoter, and a second, similar repeat in the nif region downstream from the first also does not appear to mediate significant repression (9). These observations may indicate that repression operates at some step in transcription initiation and cannot block transcription once it has initiated.
These studies are only the beginning in our understanding of nitrogen regulation of transcription in M. maripaludis. It remains to identify the repressor proteins themselves and to elucidate the overall mechanism of nitrogen sensing and regulation. Whatever the overall mechanism, it is notable that similarities are indicated in other species and genera of methanogens. Inverted repeats with sequence similarity to the one described here (consensus GGAAN6TTCC) are found in the promoter regions of glnA of other methanococci (M. voltae [23] and M. jannaschii [8]) and even of Methanobacterium thermoautotrophicum ΔH (30). A similar inverted repeat is also found in the nif promoter regions of M. maripaludis (9) and Methanococcus thermolithotrophicus (31). These observations suggest a common mechanism of nitrogen regulation and may provide a way to identify at least one class of nitrogen-regulated promoters in Archaea.
ACKNOWLEDGMENTS
We thank Peter Kessler and an anonymous reviewer for valuable suggestions. We thank William Metcalf for pJK3.
This work was supported by grant 96-35305-3891 from the U.S. Department of Agriculture.
REFERENCES
- 1.Almassy R J, Janson C A, Hamlin R, Xuong N H, Eisenberg D. Novel subunit-subunit interactions in the structure of glutamine synthetase. Nature. 1986;323:304–309. doi: 10.1038/323304a0. [DOI] [PubMed] [Google Scholar]
- 2.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatnagar L, Jain M K, Zeikus J G, Aubert J-P. Isolation of auxotrophic mutants in support of ammonia assimilation via glutamine in Methanobacterium ivanovii. Arch Microbiol. 1986;144:350–354. [Google Scholar]
- 4.Bhatnagar L, Zeikus J G, Aubert J-P. Purification and characterization of glutamine synthetase from the archaebacterium Methanobacterium ivanovi. J Bacteriol. 1986;165:638–643. doi: 10.1128/jb.165.2.638-643.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blank C E, Kessler P S, Leigh J A. Genetics in methanogens: transposon insertion mutagenesis of a Methanococcus maripaludis nifH gene. J Bacteriol. 1995;177:5773–5777. doi: 10.1128/jb.177.20.5773-5777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown J R, Daniels C J, Reeve J N. Gene structure, organization, and expression in Archaebacteria. Crit Rev Microbiol. 1989;16:287–337. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- 7.Brown J R, Masuchi Y, Robb F T, Doolittle W F. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J Mol Evol. 1994;38:566–576. doi: 10.1007/BF00175876. [DOI] [PubMed] [Google Scholar]
- 8.Bult C J, et al. Complete genome sequence of the methanogenic Archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 9.Cohen-Kupiec R, Blank C, Leigh J A. Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc Natl Acad Sci USA. 1997;94:1316–1320. doi: 10.1073/pnas.94.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bruijn F J, Rossbach S, Schneider M, Ratet P, Messmer S, Szeto W W, Ausubel F M, Schell J. Rhizobium meliloti 1021 has three differentially regulated loci involved in glutamine biosynthesis, none of which is essential for symbiotic nitrogen fixation. J Bacteriol. 1989;171:1673–1682. doi: 10.1128/jb.171.3.1673-1682.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMoll E. Nitrogen and phosphorus metabolism of methanogens. In: Ferry J G, editor. Methanogenesis. New York, N.Y: Chapman and Hall, Inc.; 1993. pp. 473–489. [Google Scholar]
- 12.García-Dominguez M, Reyes J C, Florencio F J. Purification and characterization of a new type of glutamine synthetase from cyanobacteria. Eur J Biochem. 1997;244:258–264. doi: 10.1111/j.1432-1033.1997.00258.x. [DOI] [PubMed] [Google Scholar]
- 13.Gernhardt P, Possot O, Foglino M, Sibold L, Klein A. Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol Gen Genet. 1990;221:273–279. doi: 10.1007/BF00261731. [DOI] [PubMed] [Google Scholar]
- 14.Gutowski J C, Schreier H J. Interaction of the Bacillus subtilis glnRA repressor with operator and promoter sequences in vivo. J Bacteriol. 1992;174:671–681. doi: 10.1128/jb.174.3.671-681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenealy W R, Thompson T E, Schubert K R, Zeikus J G. Ammonia assimilation and synthesis of alanine, aspartate, and glutamate in Methanosarcina barkeri and Methanobacterium thermoautotrophicum. J Bacteriol. 1982;150:1357–1365. doi: 10.1128/jb.150.3.1357-1365.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessler P S, Blank C, Leigh J A. The nif gene operon of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol. 1998;180:1504–1511. doi: 10.1128/jb.180.6.1504-1511.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumada Y, Benson D R, Hillemann D, Hosted T J, Rochefort D A, Thompson C J, Wohlleben W, Tateno Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc Natl Acad Sci USA. 1993;90:3009–3013. doi: 10.1073/pnas.90.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leigh, J. A. 1998. Unpublished results.
- 19.Liu Y, Kahn M L. ADP-ribosylation of Rhizobium meliloti glutamine synthetase III in vivo. J Biol Chem. 1995;270:1624–1628. doi: 10.1074/jbc.270.4.1624. [DOI] [PubMed] [Google Scholar]
- 20.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1344–1356. [Google Scholar]
- 21.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metcalf W M, Zhang J K, Apolinario E, Sowers K R, Wolfe R S. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci USA. 1997;94:2626–2631. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Possot O, Sibold L, Aubert J-P. Nucleotide sequence and expression of the glutamine synthetase structural gene, glnA, of the archaebacterium Methanococcus voltae. Res Microbiol. 1989;140:355–371. doi: 10.1016/0923-2508(89)90012-0. [DOI] [PubMed] [Google Scholar]
- 24.Reeve J N. Molecular biology of methanogens. Annu Rev Microbiol. 1992;46:165–191. doi: 10.1146/annurev.mi.46.100192.001121. [DOI] [PubMed] [Google Scholar]
- 25.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 391–407. [Google Scholar]
- 26.Reyes J C, Muro-Pastor M I, Florencio F J. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J Bacteriol. 1997;179:2678–2689. doi: 10.1128/jb.179.8.2678-2689.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 28.Sedmak J J, Grossberg S E. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal Biochem. 1977;79:544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro B M, Stadtman E R. Glutamine synthetase (Escherichia coli) Methods Enzymol. 1970;17A:910–922. doi: 10.1016/s0076-6879(85)13032-6. [DOI] [PubMed] [Google Scholar]
- 30.Smith D R, et al. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Souillard N, Sibold L. Primary structure, functional organization and expression of nitrogenase structural genes of the thermophilic archaebacterium Methanococcus thermolithotrophicus. Mol Microbiol. 1989;3:541–551. doi: 10.1111/j.1365-2958.1989.tb00200.x. [DOI] [PubMed] [Google Scholar]
- 32.Tumbula D L, Makula R A, Whitman W B. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol Lett. 1994;121:309–314. [Google Scholar]
- 33.Whitman W B. Order II. Methanococcales Balch and Wolfe 1981, 216VP. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey’s Manual of Systematic Bacteriology. Vol. 3. Baltimore, Md: Williams and Wilkins; 1989. pp. 2185–2190. [Google Scholar]
- 34.Whitman W B, Shieh J, Sohn S, Caras D S, Premachandran U. Isolation and characterization of 22 mesophilic methanococci. Syst Appl Microbiol. 1986;7:235–240. [Google Scholar]
- 35.Xing R, Whitman W B. Characterization of amino acid aminotransferases of Methanococcus aeolicus. J Bacteriol. 1992;174:541–548. doi: 10.1128/jb.174.2.541-548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]