Abstract
Deinococcus radiodurans is able to resist and survive extreme DNA damage induced by ionizing radiation and many other DNA-damaging agents. It is believed that it possesses highly efficient DNA repair mechanisms. To characterize the repair pathway of oxidized purines in this bacteria, we have purified, from crude extracts, proteins that recognize these oxidized bases. We report here that D. radiodurans possesses two proteins excising the oxidized purines (formamidopyrimidine and 8-oxoguanine) by a DNA glycosylase–a purinic/apyrimidine lyase mechanism. Moreover, one of those proteins is endowed with a thymine glycol DNA glycosylase activity. One of these proteins could be the homolog of the Escherichia coli Fpg enzyme, which confirms the existence of a base excision repair system in this bacteria.
Deinococcus radiodurans is a bacterium which is extremely resistant to the lethal and mutagenic effects of ionizing radiation (1, 3) as well as other physical and chemical DNA-damaging agents (including mitomycin C and UV [33, 35, 38]). For example, an exponential culture of wild-type D. radiodurans R1 can withstand from 5 to 15 kGy of γ radiation (depending on the culture conditions) with neither loss of viability nor increased mutagenesis (10, 32), while an Escherichia coli culture is 100 times less resistant (40). It has been suggested that this elevated resistance of D. radiodurans is due to unusually efficient DNA repair mechanisms (30, 33), although these hypotheses have not yet been investigated in detail.
To protect themselves from the oxidative DNA damage caused by ionizing radiation cells have evolved efficient and accurate repair systems to remove these lesions from DNA (11). In E. coli, three DNA glycosylases are known to recognize oxidized purines and pyrimidines present in DNA. The Fpg protein (formamidopyrimidine-DNA glycosylase) recognizes oxidized purines such as 2,6-diamino-4-hydroxy-5N-methylformamidopyrimidine (Fapy) and 7,8-dihydro-8-oxoguanine (8-oxoG) residues and removes them from DNA. 8-oxoG is the major oxidative lesion produced in DNA by reactive oxygen species (17) and has been shown to be highly mutagenic in vivo and in vitro (37, 45); if not repaired, it may mispair with adenine during DNA replication, causing G-C to T-A transversion mutations (9, 23, 29). Noncoding oxidized pyrimidines (such as thymine glycol, cytosine glycol, and N-substituted urea [15, 24)) are recognized and removed by the Nth (2, 11) and/or Nei (21, 27) proteins, which share a common range of substrates. These three glycosylases have an apurinic/apyrimidine (AP) lyase activity which cleaves the DNA backbone by a β-elimination mechanism for the Nth protein (leaving an α,β-unsaturated aldehyde at the 3′ side of the nick [6, 22, 27]) and a β,δ-elimination mechanism for the Fpg and Nei proteins (leaving a gap bordered by 3′ and 5′ phosphoryl group [21, 41]). In addition to these activities, Fpg and Nei proteins also catalyze the excision of 5′-terminal deoxyribose phosphate from DNA (DNA deoxyribose phosphodiesterase [dRpase] activity [16, 21]).
Thus far, only four D. radiodurans proteins have been associated with ionizing radiation resistance: the recA and pol gene products, homologs of E. coli RecA (19) and DNA polymerase I (18), respectively, and the irrB and irrI gene products, whose biochemical functions have yet to be determined (26, 44). An activity that cleaves DNA containing thymine glycol adducts and a deoxyribophosphodiesterase have been identified in partially purified extracts of D. radiodurans (33). However, the enzymes involved in the repair of the oxidized purines, such as 8-oxoG, caused in DNA by ionizing radiation has not yet been identified.
In attempt to investigate the mechanisms involved in the resistance of D. radiodurans to the mutagenic effects of reactive oxygen species, we have purified from crude extracts of this bacterium enzymatic activities that recognize oxidized purines. The substrate used to identify these activities is a duplex polynucleotide containing [3H]Fapy residues. We have identified two different enzymes excising Fapy residues, both of which have associated 8-oxoG glycosylase and AP lyase activities, and also present the biochemical characterization and determination of the substrate specificities of these two enzymes.
MATERIALS AND METHODS
Growth of organisms and preparation of cells.
D. radiodurans ATCC 13939 (American Type Culture Collection) was grown at 32°C in TGY broth (10 g of tryptone, 5 g of yeast extract, and 1 g of glucose per liter). The cells were harvested in the late exponential phase (optical density at 600 nm of ∼0.9) by centrifugation, washed with 80% ethanol (vol/vol) in Tris-EDTA buffer, then washed with Tris-EDTA buffer. This treatment was optimized to obtain quantitative lysis of the bacteria.
Enzymes assays. (i). Fapy-DNA glycosylase assay and identification of reaction products.
Fapy-DNA glycosylase activity was measured with [3H]Fapy–poly(dG-dC) (6) as the substrate. The standard incubation mixture (total volume, 50 μl) contained 70 mM HEPES-KOH (pH 8.5), 70 mM KCl, 2 mM β-mercaptoethanol, 2 mM Na2EDTA, 10% glycerol, 2,000 cpm of [3]Fapy–poly(dG-dC) (3,106 cpm/pmol), and limiting amounts of enzyme. After incubation at 32°C, the reaction mixture was ethanol precipitated and the ethanol-soluble radioactivity was measured by scintillation counting (5). To identify the reaction products, the ethanol supernatant was supplemented with authentic Fapy and analyzed by high-pressure liquid chromatography using a C18μ Bondapack column as previously described (4).
(ii) 8-oxoG-DNA glycosylase assay.
A 34-mer oligonucleotide containing a single 8-oxoG residue at position 16 (5′-GGCTTCATCGTTATT8-oxoGATGACCTGGTGGATACCG3′) was used as the substrate in 8-oxoG-DNA glycosylase assays. The 5′ end of this oligonucleotide was 32P labeled with T5 polynucleotide kinase in the presence of [γ-32P]ATP (3,000 Ci/mmol; ICN) (8) and purified by using a Nensorb cartridge (Du Pont). Annealing of the 34-mer with an oligonucleotide of complementary sequence, but with a C, T, G, or A opposite the 8-oxoG, was at a 1:2 molar ratio (labeled/unlabeled) at 90°C for 10 min followed by slow cooling to room temperature. Analysis of the resultant duplex oligonucleotide by native polyacrylamide gel electrophoresis (PAGE) on a 10% gel showed that more than 95% of the labeled oligonucleotide was in a duplex. The standard assay mixture (10 μl, final volume) contained 70 mM HEPES-KOH (pH 8.5), 70 mM NaCl, 2 mM Na2EDTA, 2 mM β-mercaptoethanol, 100 fmol of 32P-end-labeled duplex oligonucleotide containing 8-oxoG, and limiting amounts of enzyme. After incubation for 10 min at 32°C, the reaction was stopped by adding 10 μl of formamide dye solution; then the mixture was heated at 90°C for 5 min and loaded onto a denaturing 20% polyacrylamide gel containing 7 M urea. After electrophoresis, the gels were autoradiographed at −20°C, and the corresponding bands were identified and quantified with a PhosphorImager (Storm 840; Molecular Dynamics).
(iii) Thymine glycol glycosylase assay.
DNA containing thymine glycol residues was obtained (27) by exposing 150 μg of pBR322 DNA to 0.1% OsO4 for 30 min at 70°C. The modified DNA was phenol extracted, ethanol precipitated, and then purified by electrophoresis on an agarose gel (1%). Covalently closed circular DNA was phenol extracted from the agarose and ethanol precipitated. The enzymatic reaction was carried out with 500 ng of DNA in 10 μl of 70 mM HEPES-KOH (pH 8.5)–2 mM Na2EDTA–70 mM NaCl–2 mM β-mercaptoethanol–10% glycerol at 32°C for 10 min. The products of the reactions were separated by electrophoresis on a 0.9% agarose gel, the gel was stained with ethidium bromide, and photographed, and plasmid nicking was quantified by microdensitometric analysis of a photographic negative (Chromoscan 3; Jocye-Loebl).
(iv) Assay for AP nicking activity by using an oligonucleotide containing a unique abasic site.
A 50-mer oligonucleotide containing a single hypoxanthine (HX) residue at position 26 (5′GACTACAAATACATCGTCACCTGGGHXCATGTTGCAGATCCTTCCAGTGCG3′) was 32P labeled at the 5′ end, annealed with the complementary sequence, and analyzed as described above. The AP site was produced by excision of HX residues from 0.4 pmol of 32P-labeled duplex, using saturated amounts of AlkA protein in 20 μl of 25 mM sodium phosphate (pH 7.2)–1 mM Na2EDTA–100 mM KCl–100 μg of bovine serum albumin per ml) for 25 min at 37°C (35). The reaction was cooled for 10 min at 4°C. To measure the nicking activity at the AP site, limited amounts of enzyme were added and incubated for 10 min at 32°C. The reaction was stopped by adding 10 μl of formamide dye mixture, and the products were analyzed by PAGE (20% gel) in the presence of 7 M urea. All other steps were as described above.
Purification of the Fapy-DNA glycosylase.
All purification procedures were performed at 4°C. Protease inhibitors were added in all buffers at the following concentrations: antipain, leupeptin, and aprotinin (Boehringer Mannheim), each at 3 μg/ml; and phenylmethylsulfonyl fluoride (Boehringer Mannheim), 1 mM. Cell pellets (490 g) were lysed in 2 volumes of buffer A (20 mM HEPES-KOH [pH 8.5], 1 mM Na2EDTA, 2 mM β-mercaptoethanol, 10% [vol/vol] glycerol) containing 0.3 mg of lysozyme per ml and 0.25 M NaCl. The lysate was centrifuged at 10,000 × g for 30 min at 4°C. The supernatant was the crude extract (fraction I). To remove nucleic acids, fraction I was loaded onto a QMA anion-exchange column (Waters Acell) equilibrated with buffer A containing 0.25 M NaCl. The fractions that were not retained on the QMA column (fraction II) contained the Fapy-DNA glycosylase activity. Fraction II was dialyzed against buffer A containing 0.02 M NaCl and loaded onto a Phospho-Ultrogel A6 (Pharmacia LKB Biotechnology Inc.) column. The column was developed by using a linear gradient of buffer A without glycerol from 0.02 to 1 M NaCl. Fractions containing Fapy-DNA glycosylase activity were pooled (fraction III), ammonium sulfate was added to 1.7 M, and the mixture was loaded onto a fast protein liquid chromatography (FPLC) Phenyl Superose HR 5/5 column (Pharmacia). The column was washed with buffer B (20 mM HEPES-KOH [pH 8.5], 1 mM Na2EDTA, 2 mM β-mercaptoethanol, 0.25 M NaCl, 1.7 M ammonium sulfate) and eluted with a linear gradient (1.7 to 0 M ammonium sulfate in buffer B). The eluate from this column gave two peaks of Fapy-DNA glycosylase activity eluting at 0.75 and 0.35 M ammonium sulfate. These peaks were separately pooled and further purified (fractions IVa and IVb). Those two fractions were dialyzed against buffer A containing 0.02 M NaCl and loaded onto an FPLC MonoS HR 5/5 column (Pharmacia). The active fractions eluted at 0.35 M (fraction V) and 0.43 M (fraction VI) NaCl, respectively, using a 0.02 to 1 M NaCl gradient in buffer A.
Immunoblotting.
The proteins were separated by sodium dodecyl sulfate (SDS)-PAGE (12% gel) and transferred onto a nitrocellulose membrane treated with 5% nonfat milk in TBS-T buffer (20 mM Tris base, 137 mM NaCl, 0.1% Tween 20 [Bio-Rad [pH 7.5]) for 1 h as described by Towbin et al. (43). The membrane was then incubated for 1 h with serum from rabbit immunized with the Fpg protein of E. coli (1/5,000 [vol/vol] in TBS-T buffer). After three washes, the membrane was incubated for 1 h with peroxidase-coupled anti-immunoglobulin G gamma chain (anti-rabbit; Sanofi Pasteur) diluted 1/5,000. Peroxidase activity was detected with the Boehringer Mannheim chemiluminescence blotting substrate PDO. The membrane was then stripped of bound antibodies (using 100 mM β-mercaptoethanol–2% SDS–62.5 mM Tris-HCl [pH 7.5] at 50°C for 30 min), washed three times with TBS-T, and reprobed with the rabbit serum containing antibodies against E. coli Nth (5% nonfat milk in TBS-T for 1 h and rabbit serum for 1 h); then immunoreactive bands were detected as described above.
Protein concentration determination.
Protein concentrations were determined by the method of Bradford (7), using bovine serum albumin as the standard. Since the enzymatic preparations obtained in the last step of the purification were not homogeneous, we used the following approach to evaluate protein concentrations. Proteins were separated by SDS-PAGE (12% gel) and stained with Coomassie blue. A photograph of the gel was taken, and the intensities of protein bands were quantified by microdensitometry (Chromoscan 3; Joyce-Loebl) of the photographic negative. The total amount of protein run on the gel was then used to estimate the relative protein concentrations of each protein band. Identification of the protein bands reacting with antibodies allowed evaluation of the concentrations of the two enzymes.
Formation of enzyme-DNA complexes in the presence of NaBH4.
The enzymes were incubated with 100 fmol of the 32P-end-labeled 8-oxoG-containing duplex (34-mer) at 32°C for 20 min in 20 μl of a solution containing 25 mM sodium phosphate (pH 8.5), 1 mM Na2EDTA, 70 mM NaCl, 100 μg of bovine serum albumin per ml, and 100 mM NaBH4 (a 2 M NaBH4 stock solution in water was prepared immediately prior to use). The reaction was stopped by addition of SDS (0.5%, final concentration) and heating for 10 min at 37°C. The reaction products were separated by SDS-PAGE (15% gel), and the gel was dried and autoradiographed. The bands corresponding to DNA-protein complex or to free DNA were quantified with a PhosphorImager as described above. The formation of a covalent complex between DNA containing an AP site and proteins was achieved by using the 50-mer oligonucleotide containing an abasic site as described above.
RESULTS
Purification of the Fapy-DNA glycosylase activities in D. radiodurans.
To identify the enzymes repairing oxidized purines in D. radiodurans, we could assay either the 8-oxoG or the Fapy-DNA glycosylase activity. However, preliminary experiments showed that 8-oxoG/C glycosylase activity was not detectable in crude extracts; consequently, we used [3H]Fapy–poly(dG-dC) as the substrate, although the Fapy-DNA glycosylase activity is barely detectable in the crude lysate of D. radiodurans (Table 1). Given the low level of activity, we purified the enzyme from 200 liters (490 g) of the wild-type strain, using a series of column chromatography steps. Two peaks of Fapy-DNA glycosylase activity were separated by FPLC on the hydrophobic column (fractions IVa and IVb [Table 1]). Those peaks were then separately pooled and further purified by FPLC using an ion-exchange column (fractions V and VI). The two preparations obtained in the final steps were purified more than 300- and 2,000-fold, respectively. However, these values are undoubtedly underestimates due to the difficulty in obtaining reliable enzymatic activity determinations when the crude extract is used. The radioactive materials released from [3H]Fapy–poly(dG-dC) by the two D. radiodurans Fapy-DNA glycosylases coelute with authentic Fapy marker molecules in two peaks, corresponding to the two rotameric forms of the free Fapy base (data not shown). Therefore, the two purified enzymatic activities are DNA glycosylases.
TABLE 1.
Purification of D. radiodurans Fapy-DNA glycosylase activities
| Step | Fraction | Protein (mg) | Fapy-DNA glycosylase sp act (U/mg)a | Yield (%) | Purification factor (fold) |
|---|---|---|---|---|---|
| Crude extract | I | 6,256 | 0.6 | 100 | 1 |
| QMA-Acell | II | 4,955 | 0.6 | 94 | 1 |
| Phospho-Ultrogel | III | 857 | 8.2 | 64 | 13 |
| Phenyl Superose-FPLC | IVa | 39.5 | 6.8 | 7 | 11.3 |
| IVb | 35 | 24 | 22 | 40 | |
| MonoS FPLC | |||||
| Fraction IVa | V | 0.5 | 182 | 2.4 | 300 |
| Fraction IVb | VI | 0.31 | 1,210 | 10 | 2,000 |
Measured with [3H]Fapy–poly(dG-dC) as the substrate. One unit excises I pmol of Fapy residues in 5 min at 32°C.
Presence of a 8-oxoG-DNA glycosylase activity.
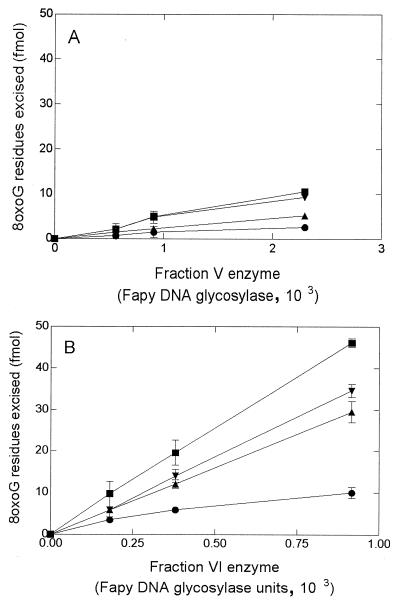
Since the E. coli Fpg protein excises both Fapy and 8-oxoG residues in DNA, we tested whether an 8-oxoG-DNA glycosylase activity is also present in fractions V and VI, using as substrate a duplex oligonucleotide containing a single 8-oxoG residue. The two most purified fractions nicked this duplex oligonucleotide in a concentration-dependent manner (Fig. 1). We have also found that in each case, the activities excising Fapy residues and cleaving the 8-oxoG-containing duplex coelute during MonoS chromatography (data not shown), strongly suggesting that both enzymes carry these two activities, albeit with different ratios. Note that fraction VI exhibits a much higher 8-oxoG glycosylase activity relative to Fapy excision.
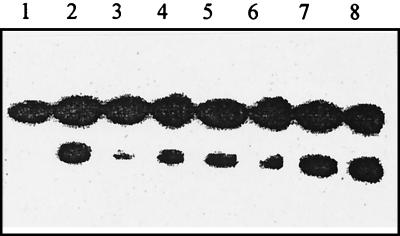
FIG. 1.
Cleavage of the 8-oxoG/C duplex by the following: lane 1, no enzyme, lane 2, 5 ng of E. coli Fpg protein; lanes 3, 4, and 5, 0.2, 0.4, and 1 U of Fapy DNA glycosylase (fraction V); lanes 6, 7, and 8, 0.2, 0.4, and 1 U of Fapy DNA glycosylase (fraction IV). The duplex (100 fmol/reaction) was incubated with the enzyme at 32°C for 10 min.
Use of antibodies against E. coli Fpg or Nth to characterize the D. radiodurans enzymes.
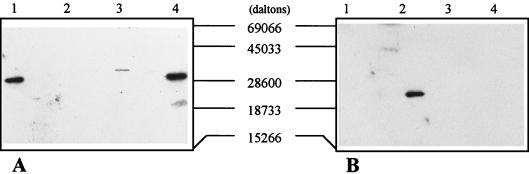
The results presented above show that D. radiodurans has two enzymes with common activities (Fapy- and 8-oxoG-DNA glycosylases). We have further investigated whether these enzymes could be structural homologs of the E. coli Fpg protein by immunoblotting the two proteins with anti-E. coli Fpg protein antibodies. The results (Fig. 2A) show that both fractions possess a protein reacting with these antibodies. Since we have used the same amounts of enzyme (measured with Fapy-DNA glycosylase units), Fig. 2A shows that the fraction V protein reacts weakly and fractions VI protein exhibits a stronger reaction. Moreover, these proteins have different apparent molecular masses (∼38 and ∼34 kDa for the fraction V and VI proteins, respectively).
FIG. 2.
Immunoblotting of fractions V and VI protein with anti-Fpg (A) and anti-Nth (B) antibodies. Lane 1, E. coli Fpg protein (100 ng); lane 2, E. coli Nth protein (100 ng); lane 3, fraction V (0.5 U of Fapy-DNA glycosylase); lane 4, fraction VI (0.5 U of Fapy-DNA glycosylase).
Recently, a Fapy-DNA glycosylase activity in yeast has been found in a non-Fpg protein-like enzyme, Ntg1 (14). Ntg1 shows a sequence similarity with E. coli Nth (2, 14), and excises oxidized pyrimidines (thymine glycols) from yeast DNA. It also removes Fapy residues but not 8-oxoG, whereas in E. coli, Nth removes oxidized pyrimidines but not Fapy residues. Therefore, we investigated whether the D. radiodurans enzymes could be recognized by antibodies against the E. coli Nth protein by reprobing the immunoblot described above with anti-E. coli Nth protein antibodies. The results show that there is no detectable reaction with either fraction V or fraction VI (Fig. 2B).
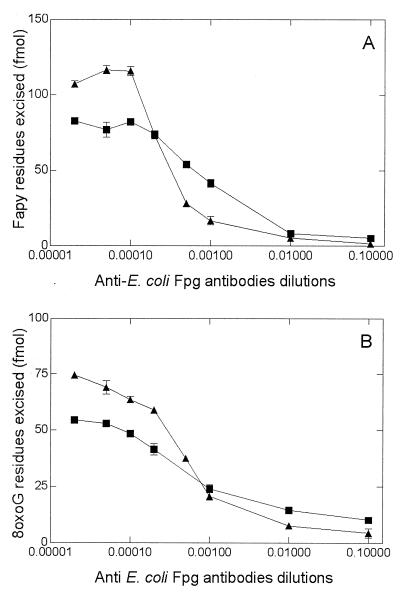
We further investigated the inhibitory effects of the antibodies toward the enzymatic activities of the two proteins. The results (Fig. 3) show that the various antibodies inhibit the two enzymatic activities but with different patterns. The anti-Fpg antibody inhibited the enzymatic activities of the fraction VI protein more efficiently than those of the fraction V protein. The Western blot analysis (Fig. 2) also showed that anti-Fpg antibodies react better with the enzyme presents in fraction VI than with the fraction V enzyme. Taken together with the fact that the two enzymatic activities have different apparent molecular weights, these results suggest that two different enzymes having structural homologies with the E. coli Fpg protein are responsible for the excision of the Fapy residues and incision of 8-oxoG-containing DNA in D. radiodurans.
FIG. 3.
D. radiodurans Fapy and 8-oxoG DNA glycosylase activities in the presence of various concentrations of anti-Fpg antibodies. Fraction V (0.04 U of Fapy DNA glycosylase; ■) and fraction VI (0.04 U of Fapy DNA glycosylase; ▴) were incubated for 10 min at 32°C with 700 fmol of [3H]Fapy–poly(dG-dC) or 100 fmol of 8-oxoG/C-containing oligonucleotide (34-mer) (B).
Repair of Fapy lesions by the D. radiodurans enzymes.
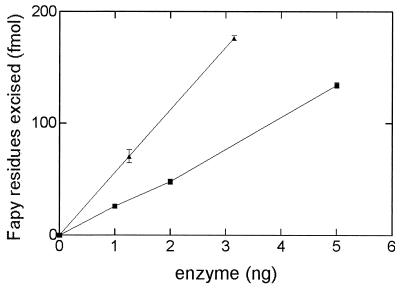
The optimum pH for both D. radiodurans enzymes in Fapy excision was pH 8.4, using a HEPES-KOH buffer containing 70 mM NaCl (data not shown). The abilities of the two enzymes to excise Fapy residues was analyzed. As noted above, both enzymes release Fapy residues, with the fraction VI enzyme being the most efficient (Fig. 4). Analysis of the kinetic parameters for the release of [3H]Fapy residues gave an apparent Km value for the fraction VI enzyme which is comparable to that of the E. coli Fpg protein, while the fraction V enzyme had a higher Km value (Table 2). Comparison of the two enzymes Kcat/Km values for excision of Fapy residues (Table 2) shows that these values are similar to those of the E. coli Fpg protein.
FIG. 4.
Fapy-DNA glycosylase activities of fraction V (■) and fraction VI (▴). Increasing amounts of each enzyme were incubated in the presence of [3H]Fapy–poly(dG-dC) at 32°C for 10 min, and the [3H]Fapy residues released were measured (for details, see Materials and Methods).
TABLE 2.
Kinetic parameters for excision of Fapy and 8-oxoG (from oligonucleotides) residues for D. radiodurans fraction V and fraction VI enzymes
| Protein | Fapy-DNA glycosylase activity
|
8-oxoG-DNA glycosylase activity
|
||||
|---|---|---|---|---|---|---|
| Km (nM) | kcat (1/min) | kcat/Km (1/nM · min) | Km (nM) | kcat (1/min) | kcat/Km (1/nM · min) | |
| Fraction V | 22.1 | 0.33 | 0.16 | 83.3 | 0.2 | 0.0024 |
| Fraction VI | 5.3 | 0.49 | 0.1 | 10 | 0.6 | 0.06 |
| E. coli Fpg protein | 10 | 0.5 | 0.05 | 4 | 0.43 | 0.11 |
Repair of 8-oxoG residues by the D. radiodurans enzymes.
The efficiency of the two enzymes to excise 8-oxoG residues as a function of the base opposite to the lesion was analyzed by using a 34-mer duplex DNA containing an 8-oxoG residue opposite one of the four bases. To compare the two enzymatic activities on 8-oxoG residues, the reactions were standardized by Fapy units as a measure of the two enzymes concentrations. The results show that both enzymes incise the various substrates with different efficiencies (Fig. 5). Fraction V enzyme incises the four duplexes with a low efficiency and has a slight preference for 8-oxoG/C and 8-oxoG/T mismatches. Fraction VI enzyme incises 8-oxoG residues from all the mismatches with a higher efficiency, and the 8-oxo-G residue is better eliminated when opposite a pyrimidine; however, the 8-oxoG/A mismatch is recognized at a detectable rate. Fraction VI enzyme exhibits Km and Kcat/Km values for the 8-oxoG-DNA glycosylase activity comparable to those of the E. coli Fpg protein, while fraction V enzyme has a much (10-fold) lower apparent Km and a 20-fold-reduced Kcat/Km (Table 2).
FIG. 5.
Cleavage of DNA duplexes containing a single 8-oxoG mismatched with one of the four bases C (■), T (▾), G (▴), and A (•). The various duplexes (100 fmol) were incubated with increasing amounts of proteins standardized as Fapy-DNA glycosylase activity of fraction V (A) and fraction VI (B) for 10 min at 32°C.
Mechanisms of action of the enzymes in repair of 8-oxoG residues.
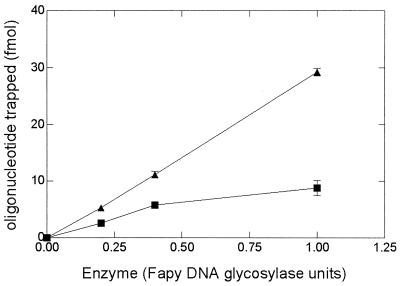
Following the enzymatic treatment of the 8-oxo-G-containing duplex, further incubation with 10% piperidine (which nicks DNA at AP sites) leads to no increase in the amount of incised substrate (data not shown), thus showing that our assay did not underestimate the excision of 8-oxoG. This result implies either that an AP nicking activity contaminates both enzyme preparations or that for both enzymes, an associated nicking activity, presumably AP lyase activity, is involved in repair of the 8-oxoG lesions. If the latter is the case, it should be possible to isolate the covalent Schiff base intermediate as an enzyme-substrate complex as shown for the E. coli Fpg protein (42), as this complex can be reduced in the presence of sodium borohydride to yield a stable covalent complex (37, 42). We therefore tested the ability of the two D. radiodurans enzymes to trap 8-oxoG/C DNA in the presence of NaBH4. As shown in Fig. 6, both enzymes generate covalent complexes with DNA, suggesting that they have an intrinsic β-AP lyase activity associated with their DNA glycosylase activity (see below).
FIG. 6.
NaBH4 trapping of 8-oxoG-containing DNA (200 fmol) treated with different amounts of proteins standardized as Fapy-DNA glycosylase activity from fraction V (■) and fraction VI (▴). The reactions were performed at 32°C for 30 min (for details, see Materials and Methods).
Nicking activities at a abasic sites of the D. radiodurans enzymes.
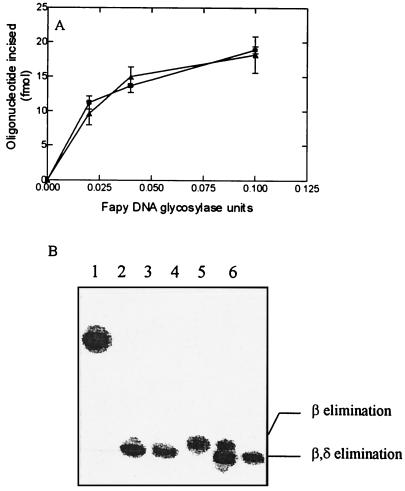
The nicking activities at AP sites by the two enzymes was further characterized by using as substrate a duplex oligonucleotide containing a unique abasic site at a defined position. The results (Fig. 7A) show that for both enzymes, the AP lyase activities can operate independently of the glycosylase activity. The mechanism of the AP lyase activity was determined by comparing the products of the reaction obtained with fraction V and VI enzymes with the products of the reaction of the E. coli Fpg (β,δ-elimination) and Nth (β-elimination) proteins. Figure 7B shows that the products of the reaction obtained with fractions V and VI migrate at the same position as those obtained with the E. coli Fpg protein. These results suggest that the two D. radiodurans enzymes use the same mechanism for nicking DNA at abasic sites: a β,δ elimination.
FIG. 7.
Cleavage of an oligonucleotide containing a unique abasic site. Oligonucleotide (100 fmol) was incubated for 10 min at 32°C with enzyme. The products of the reaction were separated by SDS-PAGE, and the results were quantified with PhosphorImager. (A) Increasing amounts of fraction V (■) and fraction VI (▴). (B) Lane 1, duplex oligonucleotide containing an AP site. lane 2, like lane 1 but incubated with 0.2 M NaOH before analysis; lane 3, like lane 1 but incubated with 10 ng of E. coli Fpg protein; lane 4, like lane 1 but incubated with 10 ng of E. coli Nth protein; lane 5, like lane I but incubated with 10 ng of fraction V enzyme; lane 6, like lane 1 but incubated with 10 ng of fraction VI enzyme.
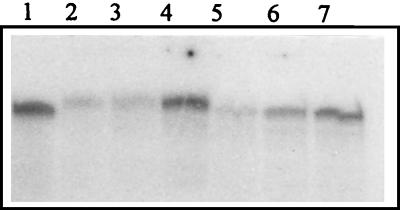
We also tested whether these two enzymes could trap DNA containing an AP site in the presence of NaBH4. The results (Fig. 8) show that both enzymes generate covalent complexes with DNA containing ATP sites. The migration positions of the two covalent complexes are also in good agreement with the enzymes’ apparent molecular masses as measured above, compared to the migration of the complex formed between the E. coli Fpg protein (30.2 kDa) and the AP site-containing DNA duplex.
FIG. 8.
NaBH4 trapping of AP site-containing DNA (100 fmol) with 50 ng of E. coli Fpg protein (lane 1), 10, 20 50 ng of fraction V enzyme (lanes 2, 3, and 4), and 10, 20, and 50 ng of fraction VI enzyme (lanes 5, 6, and 7) for 10 min at 32°C.
Incision of osmium tetroxide-damaged DNA by the two enzymes.
To further characterize the substrate specificities of the two deionococcal Fapy- and 8-oxoG-DNA glycosylases, we tested their ability to recognize oxidized pyrimidines, using as substrate plasmid DNA treated with OsO4, which predominantly generates thymine glycols in DNA (14, 24). The fraction V enzyme efficiently cleaves the OsO4-treated DNA, whereas fraction VI enzyme does not (Fig. 9A). The results are compatible with a two-step mechanism: the N-glycosylase activity excised the oxidized base then the AP lyase activity leads to the strand cleavage at the ATP site. The unoxidized DNA was resistant to enzymatic cleavage when treated with the fraction V enzyme, thus ruling out possible contamination of the preparation by a nonspecific endonuclease.
FIG. 9.
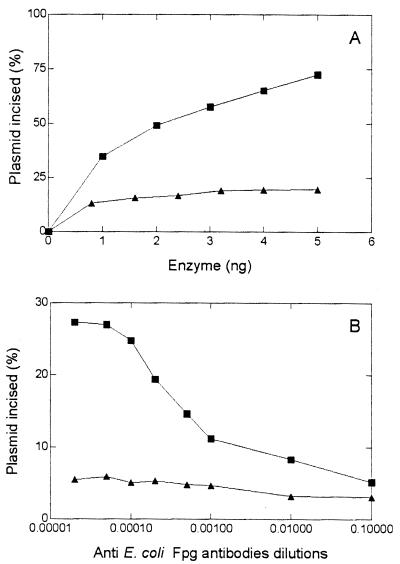
Incision of OsO4-treated DNA by the D. radiodurans enzymes. (A) 500 ng of OsO4-treated DNA was incubated at 32°C for 10 min with increasing amounts of fraction V (■) and fraction VI (▴) enzymes. The products of the reaction were separated on an agarose gel and quantified by microdensitometric analysis (for details, see Materials and Methods). (B) Thymine glycol DNA glycosylase activities of fraction V (■) and fraction VI (▴) enzymes in the presence of different dilutions of anti E. coli Fpg antibodies.
To confirm that this thymine glycol DNA glycosylase activity is carried by the Fapy- or 8-oxoG-DNA glycosylase–AP lyase protein present in fraction V, OsO4-treated-DNA cleavage reactions were performed in the presence of various amounts of anti-Fpg protein antibodies. The results (Fig. 9B) show that only anti-Fpg antibodies are able to inhibit the enzymatic reactions, suggesting that the thymine glycol glycosylase activity is carried by the same enzyme that cleaves at Fapy, 8-oxoG, and AP lesions.
DISCUSSION
D. radiodurans has an extraordinary ability to withstand both the lethal and the mutagenic effects of ionizing radiation. This bacterium is multigenomic (20), with stationary-phase cells carrying an estimated four genome equivalents. It has been proposed (10) that preexisting alignment between homologous sequences of the chromosomes facilitates the association of these regions after X irradiation, leading to rapid recombinational repair of the complete genome. However, this model cannot explain the resistance of the bacterium to the mutagenic effects of ionizing radiation, believed to be due to the generation of oxidized bases in DNA (17). The effects of these modified bases are eliminated in E. coli by a set of DNA repair enzymes including the Fpg, MutY, and MutT proteins, coordinated in the GO repair system (12, 13, 29).
Due to the potential importance of the mechanisms that counteract the mutagenic effects of oxidized purines, our goal was to identify the enzymes involved in the repair of these oxidized bases in D. radiodurans, in particular to purify from bacterial extracts the proteins that excise Fapy residues since it was not possible to detect 8-oxoG repair activity in crude extracts of D. radiodurans (data not shown). Our results show that two protein fractions from D. radiodurans exhibit Fapy-DNA glycosylase activity. The two proteins differ in both hydrophobicity and charge, since they are eluted on the FPLC MonoS or hydrophobic columns at different ionic strengths. The proteins also have different catalytic parameters. Fraction VI enzyme exhibits robust 8-oxoG-DNA glycosylase and AP lyase activities and has very efficient kinetic parameters (Km and Kcat) for action on Fapy and 8-oxoG residues similar to those of the E. coli Fpg protein (6). The mechanisms of action of this fraction VI enzyme appears to involve a transient Schiff base intermediate with 8-oxoG or AP site containing DNA that can be trapped upon reduction with NaBH4, with the AP lyase activity proceeding by a β,δ elimination. Anti-E. coli Fpg antibodies recognize the fraction VI enzyme, and these antibodies strongly inhibit its glycosylase and AP lyase activities, suggesting significant structural similarity between the E. coli and D. radiodurans proteins. The E. coli Fpg protein is unable to incise 8-oxoG/A complex, which is a substrate for the MutY protein in the GO repair system (8, 28). In contrast, the fraction VI enzyme is able to recognize and repair the 8-oxoG/A mismatch, although only at a slow rate. This activity is also inhibited by anti-Fpg antibodies (data not shown), suggesting that it is carried by the same protein. This result suggest that D. radiodurans may have a GO repair system different from that in E. coli. Indeed, we have been unable to detect a MutY like activity in the crude extract or in the Phosphor-Ultrogel fractions from D. radiodurans (data not shown).
The second protein (fraction V enzyme) shows a weaker affinity for Fapy residues and a low 8-oxoG/C-DNA glycosylase activity. As described for the fraction VI enzyme, fraction V enzyme also has an AP lyase activity, which proceeds by a β,δ-elimination mechanism; it also forms a transient Schiff base intermediate with 8-oxoG or AP site containing DNA. A thymine glycol DNA glycosylase associated with fraction V enzyme has also been identified. Anti-E. coli Fpg antibodies inhibit all of those activities, but less efficiently than for the fraction VI enzyme. This suggests that the fraction V protein has some structural similarities with the Fpg protein, although less marked than for the fraction VI enzyme. A thymidine glycol DNA glycosylase has previously been partially purified from D. radiodurans extracts (34), but the relationship between this ∼30-kDa protein and the ∼38-kDa fraction V protein is not clear.
The fact that D. radiodurans possesses two glycosylases, active both on Fapy and 8-oxoG residues, raises the possibility that the smaller one (fraction V) is derived from the larger one by limited proteolysis. Indeed, they have the same optimum pH, similar sizes, and intrinsic AP lyase activities, both functioning by β,δ elimination, and both cross-react with anti-E. coli Fpg antibodies but not with anti-E. coli Nth antibodies. However, only fraction V protein has a thymine glycol DNA glycosylase activity. It is formally possible that the smaller protein (fraction VI) is a proteolytic fragment of the larger (fragment V) one and that their different thymine glycol glycosylase activities are due to the fact that one activity is a fragment of the other. However, the fact that only the smaller protein excised oxidized pyrimidines favors the presence of two distinct unrelated proteins. Furthermore, in testing for activity on 8-oxoG containing-DNA, we found that the smaller protein (fraction VI) was more active than the larger one (fraction V), and immunoblotting results show that the E. coli anti-FPG antibodies binds much more weakly to fraction V than to fraction VI (consistent with the fact that fraction VI is more inhibited by these antibodies than fraction V in both Fapy and 8-oxoG activities). Furthermore, regardless of which base is opposite the 8-oxoG lesion in a DNA duplex, the smaller protein (fraction VI) is in all cases more active than the larger one (fraction V). These results together are most consistent with the two proteins being independent gene products.
Part of the sequence of the genome of D. radiodurans is currently available (TIGR database). A computer search for homologies to the E. coli Fpg protein reveals one sequence (fragment gdr 31) coding for a putative protein having a molecular mass of 34.8 kDa (39% identity and 52.6% similarity with the E. coli Fpg protein amino acids sequence) and containing a four-cysteine zinc finger motif. These finding suggest that this sequence could code for the fraction VI enzyme. However, we have been unable to detect any other D. radiodurans sequence encoding a Fpg homolog that could code for the fraction V enzyme, although the genomic sequence is not yet complete. Concerning potential homologs of the E. coli Nth protein, genomic fragment gdr 4 encodes a putative protein of 226 amino acids (molecular weight of 28,288, with 40% identity and 54% similarity with the E. coli Nth protein amino acids sequence), significantly smaller than the fraction V enzyme that we have purified and characterized. This putative protein could be the ∼30-kDa thymine glycol DNA glycosylase that has been partially purified from D. radiodurans extracts (34).
In conclusion, D. radiodurans is the first bacterium that appears to contain two different proteins excising 8-oxoG residues. One of them shares significant homology with the E. coli Fpg protein and a common range of enzymatic activities. This protein could be a homolog of the E. coli Fpg protein, which confirms the existence of a base excision repair system in D. radiodurans (25). Further characterization of these two proteins and of their corresponding genes will be necessary to determine their exact roles in the unusual resistance of D. radiodurans to ionizing radiation and other DNA-damaging agents.
ACKNOWLEDGMENTS
This work was supported by grants from Electricité de France (Radioprotection), the Association pour la Recherche contre le Cancer, and fellowships (to C.B.) from la Ligue contre le Cancer and the Association pour la Recherche contre le Cancer.
REFERENCES
- 1.Anderson A W, Norden H C, Cain R F, Parrish G, Duggan D. Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 1956;10:575–578. [Google Scholar]
- 2.Asahara H, Wistort P M, Bank J F, Bakerian R H, Cunningham R P. Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry. 1989;28:4444–4449. doi: 10.1021/bi00436a048. [DOI] [PubMed] [Google Scholar]
- 3.Boiteux S, Belleney J, Roques B P, Laval J. Two rotameric forms of imidazole ring-opened 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res. 1984;6:3673–3683. doi: 10.1093/nar/12.13.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boiteux S, Belleney J, Roques B P, Laval J. Two rotameric forms of imidazole ring-opened 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res. 1984;6:3673–3683. doi: 10.1093/nar/12.13.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boiteux S, O’Connor T R, Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987;6:3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boiteux S, O’Connor T R, Lederer F, Gouyette A, Laval J. Homogeneous Escherichia coli FPG. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidine sites. J Biol Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Castaing B, Geiger A, Seliger H, Nehls P, Laval J, Zelwer C, Boiteux S. Cleavage and binding of a DNA fragment containing a single 8-oxoguanine by wild type and mutant FPG proteins. Nucleic Acids Res. 1993;21:2899–2905. doi: 10.1093/nar/21.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng K C, Cahill D S, Kasai H, Nishimura S, Loeb L A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G-T and A-C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 10.Daly M J, Ouyang L, Fuchs P, Minton K W. In vivo damage and RecA-dependent repair of plasmid and chromosomal DNA in the radiation-resistant bacterium Deinococcus radiodurans. J Bacteriol. 1994;176:3508–3517. doi: 10.1128/jb.176.12.3508-3517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 12.Dianov G, Lindahl T. Reconstitution of the DNA base excision-repair pathway. Curr Biol. 1994;4:1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 13.Dianov G, Sedgwick B, Daly G, Olsson M, Lovett S, Lindahl T. Release of 5′-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res. 1994;22:993–998. doi: 10.1093/nar/22.6.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eide L, Bjors M, Pirovano M, Alseth I, Berdal K G, Seeberg E. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. Proc Natl Acad Sci USA. 1997;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans J M, Maccabee M, Hatahet Z, Courcelle J, Bockrath R, Ide H, Wallace S. Thymine ring saturation products: lesion bypass, misinsertion and implications for mutagenesis. Mutat Res. 1993;299:147–156. doi: 10.1016/0165-1218(93)90092-r. [DOI] [PubMed] [Google Scholar]
- 16.Graves R J, Felzenswalb I, Laval J, O’Connor T R. Excision of the 5′-terminal deoxyribose phosphate from damaged DNA is catalyzed by the FPG protein of Escherichia coli. J Biol Chem. 1992;267:14429–14435. [PubMed] [Google Scholar]
- 17.Grollman A P, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 18.Gutman P D, Fuchs P, Ouyane L, Minton K W. Identification, sequencing, and targeted mutagenesis of a DNA polymerase gene required for the extreme radioresistance of Deinococcus radiodurans. J Bacteriol. 1993;175:3581–3590. doi: 10.1128/jb.175.11.3581-3590.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutman P D, Carroll J D, Masters C I, Minton K W. Sequencing, targeted mutagenesis and expression of a recA gene required for the extreme radioresistance of Deinococcus radiodurans. Gene. 1994;141:31–37. doi: 10.1016/0378-1119(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 20.Hansen M T. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J Bacteriol. 1978;134:71–75. doi: 10.1128/jb.134.1.71-75.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang D, Hatahet Z, Blaisdell J O, Melamede R J, Wallace S S. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Linn S. The mechanisms of action of E. coli endonuclease III and T4 UV endonuclease (endonuclease V) at AP sites. Nucleic Acids Res. 1988;16:1135–1141. doi: 10.1093/nar/16.3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Mura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 24.Maccabee M, Evans J S, Glackin M P, Hatahet Z, Wallace S. Pyrimidine ring fragmentation products. Effects of lesion structure and sequence context on mutagenesis. J Mol Biol. 1994;256:514–530. doi: 10.1006/jmbi.1994.1162. [DOI] [PubMed] [Google Scholar]
- 25.Masters I C, Moseley B E B, Minton K W. AP endonuclease and uracil DNA glycosylase activities in Deinococcus radiodurans. Mutat Res. 1991;254:263–272. doi: 10.1016/0921-8777(91)90065-w. [DOI] [PubMed] [Google Scholar]
- 26.Mattimore V, Udupa K S, Berne G A, Battista J R. Genetic characterization of forty ionizing radiation-sensitive strains of Deinococcus radiodurans: linkage information from transformation. J Bacteriol. 1995;177:5232–5237. doi: 10.1128/jb.177.18.5232-5237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melamede R J, Hatahet Z, Kow Y W, Ide H, Wallace S S. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 28.Michaels M L, Tchou J, Grollman A P, Miller J H. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 29.Michaels M L, Miller J H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minton K W. DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol Microbiol. 1994;13:9–15. doi: 10.1111/j.1365-2958.1994.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 31.Minton K W. Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mutat Res. 1996;363:1–7. doi: 10.1016/0921-8777(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 32.Moseley B E B, Mattingly A .Proc. Natl. Acad. Sci. USA. Repair of irradiated transforming deoxyribonucleic acid in wild-type and radiation-sensitive mutant of Micrococcus radiodurans. J Bacteriol. 1971;105:976–983. doi: 10.1128/jb.105.3.976-983.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moseley B E B. Photobiology and radiobiology of Micrococcus (Deinococcus) radiodurans. Photochem Photobiol Rev. 1983;7:223–274. [Google Scholar]
- 34.Mun C, Del Rowe J, Sandigursky M, Minton K, Franklin W A. DNA deoxyribophosphodiesterase and an activity that cleaves DNA containing rhymine glycol adducts in Deinococcus radiodurans. Radiat Res. 1994;138:282–285. [PubMed] [Google Scholar]
- 35.Murray R G E, Brooks B W. Deinococcus. In: Sneath P, Mair N, Sharpe M, Holt J, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins; 1986. pp. 1035–1043. [Google Scholar]
- 36.Saparbaev M, Laval J. Excision of the hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkypurine DNA glycosylases. Proc Natl Acad Sci USA. 1994;91:5873–5878. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shibutani S, Takeshita M, Grollman A P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 38.Smith M D, Masters C I, Moseley B E B. Molecular biology of radiation resistant bacteria. In: Herbert R A, Sharp R J, editors. Molecular biology and biotechnology of extremophiles. New York, N.Y: Chapman & Hall; 1992. pp. 258–280. [Google Scholar]
- 39.Sun B, Latham K A, Dodson M L, Lloyd R S. Studies of the catalytic mechanism of five DNA glycosylases intermediates. Probing for enzyme-DNA imino J. Biol Chem. 1995;270:19501–19508. doi: 10.1074/jbc.270.33.19501. [DOI] [PubMed] [Google Scholar]
- 40.Sweet D M, Moseley B E B. The resistance of Micrococcus radiodurans to killing and mutation by agents which damage DNA. Mutat Res. 1976;34:175–186. doi: 10.1016/0027-5107(76)90122-6. [DOI] [PubMed] [Google Scholar]
- 41.Tchou J, Kasai H, Shibutani S, Chung M H, Laval J, Grollman A P, Nishimura S. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tchou J, Grollman A P. The catalytic mechanism of Fpg protein. Evidence for a Schiff base intermediate and amino terminus localization of the catalytic site. J Biol Chem. 1995;270:11671–11677. doi: 10.1074/jbc.270.19.11671. [DOI] [PubMed] [Google Scholar]
- 43.Towbin H T, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Udupa K S, O’Cain P A, Mattimore V, Battista J R. Novel ionizing radiation-sensitive mutants of Deinococcus radiodurans. J Bacteriol. 1994;176:7439–7446. doi: 10.1128/jb.176.24.7439-7446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood M L, Dizgaroglu M, Gajewski E, Essigmann J M. Mechanistic studies of ionizing radiation and oxydative mutagenesis: effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique abasic site in a viral genome. Biochemistry. 1990;26:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]