Abstract
Short prokaryotic argonaute (pAgo) and toll/interleukin-1 receptor/resistance protein (TIR)–analog of PAZ (APAZ) form a heterodimeric SPARTA complex that provides immunity to its prokaryotic host through an abortive infection mechanism. Monomeric SPARTA senses foreign RNA/DNA duplexes to assemble an active tetramer resulting in cell death by nicotinamide adenine dinucleotide (oxidized form) (NAD) depletion via an unknown mechanism. We report nine structures of SPARTA in different functional states at a resolution range of 4.2 to 2.9 angstroms, revealing its activation mechanism. Inactive SPARTA monomers bind to RNA/DNA duplexes to form symmetric dimers mediated by the association of Ago subunits. The initiation of tetramer assembly induces flexibility of the TIR domains enabling a symmetry-breaking rotational movement of a TIR domain in the dimer units which facilitates the TIR oligomerization, resulting in the formation of the substrate binding pocket and the activation of the SPARTA complex’s NADase activity. Our findings provide detailed structural and mechanistic insights into activating a short argonaute defense system.
Nine cryo-EM structures of the SPARTA complex in different functional states reveal its activation mechanism for abortive immunity.
INTRODUCTION
The Argonaute (Ago) protein family is highly conserved in all domains of life (1). Eukaryotic Ago proteins (eAgos) are part of the RISC (RNA-induced silencing complex), catalyzing RNA-guided RNA cleavage in a process known as RNA interference (1–3). Prokaryotic Ago proteins (pAgos) form complexes with single-stranded RNA (ssRNA) or DNA guides to repress the expression and replication of complementary DNA derived from plasmid and phage infection via nuclease activity or other mechanisms (4–8). Long pAgos are structurally similar to eAgos having P-element Induced WImpy (PIWI), middle (MID), N and PAZ domains (9, 10). However, short pAgos lack the PAZ and N domains found in canonical or long Agos but include the MID and PIWI domains required for guide recognition. The PIWI domain, albeit present in short pAgos, lacks the DEDX catalytic tetrad required for nuclease activity (11). Short pAgos associate with other protein effectors to compensate for catalytic inactivity, particularly proteins containing an APAZ domain fused to different enzymatic domains based on which these complexes form distinct phylogenetic clades (8, 12, 13).
Recent studies on short pAgos complexes from different clades have revealed that these systems provide host immune defense via abortive infection or suicide mechanisms (14–16). In the S2A clade, short Agos from Maribacter polysiphoniae and Crenotalea thermophila associate with the toll/interleukin-1 receptor/resistance protein (TIR)–analog of PAZ (APAZ) protein to assemble into a heterodimeric complex of approximately 100-kDa molecular weight, forming a host defense system known as SPARTA (short pAgo and the associated TIR-APAZ). Upon host infection by phages or plasmids, mRNAs expressed from the foreign DNA are processed into small [15 to 50 nucleotides (nt)] 5′-phosphorylated RNA guides that bind to SPARTA and complementary target single-stranded DNA. The recognition of the RNA/DNA duplexes triggers SPARTA tetramerization from inactive monomers and the activation of NADase activity of the TIR domains. This results in nicotinamide adenine dinucleotide (oxidized form) (NAD) depletion and host cell death, thereby limiting infection propagation (14). The SPARSA (short pAgo and the associated Sir2) complex from Geobacter sulfurreducens, which functions via a similar mechanism, has a Sir2 domain fused to APAZ instead of TIR and does not require oligomerization for activity (15). Hence, the oligomerization-dependent SPARTA activation driven by sequence-specific nucleic acid recognition reveals an unprecedented mechanism for Ago-mediated host defense. Detailed structural information on the different steps of assembly and activation is required to understand the SPARTA mechanism of action (12).
RESULTS
The overall structure of an active MapSPARTA tetramer
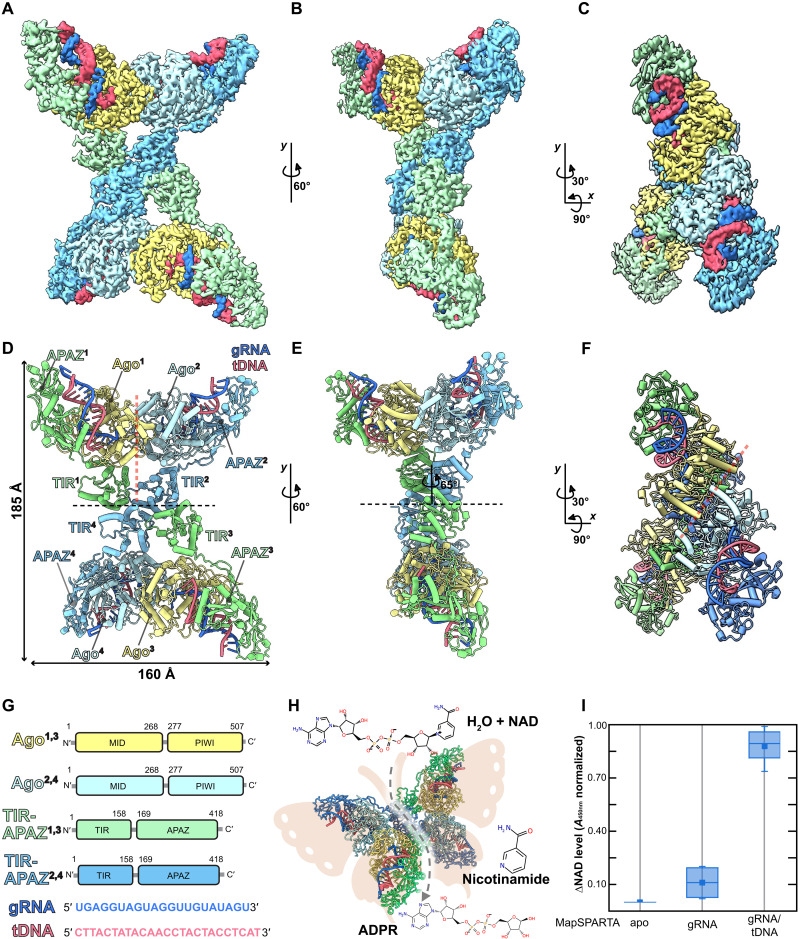
To gain insights into the mechanism of assembly and activation of a SPARTA tetramer by an RNA/DNA heteroduplex, we reconstituted full-length M. polysiphoniae SPARTA (MapSPARTA) bound to RNA/DNA heteroduplexes (fig. S1, A and B). Consistent with previous data, MapSPARTA becomes catalytically active as a NADase in the presence of the guide RNA/target DNA heteroduplex (Fig. 1I) (14). We used cryo–electron microscopy (cryo-EM) to determine the structure of the MapSPARTA complex at an overall resolution of 3.1 Å (Fig. 1, A to F, figs. S2 and S13, table S1, and movie S1). The maps obtained reveal a tetramer with apparent densities for four copies each of the 18-nt RNA/16-nt DNA heteroduplex, Ago (residues 1 to 507), APAZ (residues 159 to 418), and TIR (1 to 158) domains (4:4:4:4) (Fig. 1, A to F, and fig. S2).
Fig. 1. The overall structure of the M. polysiphoniae SPARTA (MapSPARTA) tetramer.
(A to C) Cryo–electron microscopy (cryo-EM) densities of the MapSPARTA tetramer in three views. (D to F) Cartoon representation of the MapSPARTA tetramer in three views corresponding to the cryo-EM densities in (A), (B), and (C). The black dashed line indicates the tetramer interface, while the red dashed line indicates the pseudo-symmetric dimer interface. (G) Domain organization of the SPARTA complex. (H) Cartoon representation of an active SPARTA tetramer showing its butterfly shape. (I) Quantification of the change in nicotinamide adenine dinucleotide (oxidized form) (NAD) levels in the presence of apo, guide RNA (gRNA) bound, and heteroduplex-bound MapSPARTA. tDNA, target DNA; ADPR, adenosine 5´-diphosphoribose.
These are arranged as pseudo-symmetric dimers. The association of TIR domains from each pseudo-symmetric dimer unit mediates the tetramerization of the complex (Fig. 1D). Along the axis crossing the tetramer interface, the pseudo-symmetric dimers are arranged such that there is an approximately 65° relative rotational difference between monomers (Fig. 1E). This arrangement results in a structure resembling a “butterfly” with the thorax composed of four TIR domains. Both wings consist of two copies of the heteroduplex-bound Ago subunits and APAZ domains. MapSPARTA tetramerization mediated by the TIR domains is required to position and stabilize the TIR domains in a structural arrangement proficient for NADase activity (Fig. 1H).
Mechanism of MapSPARTA activation
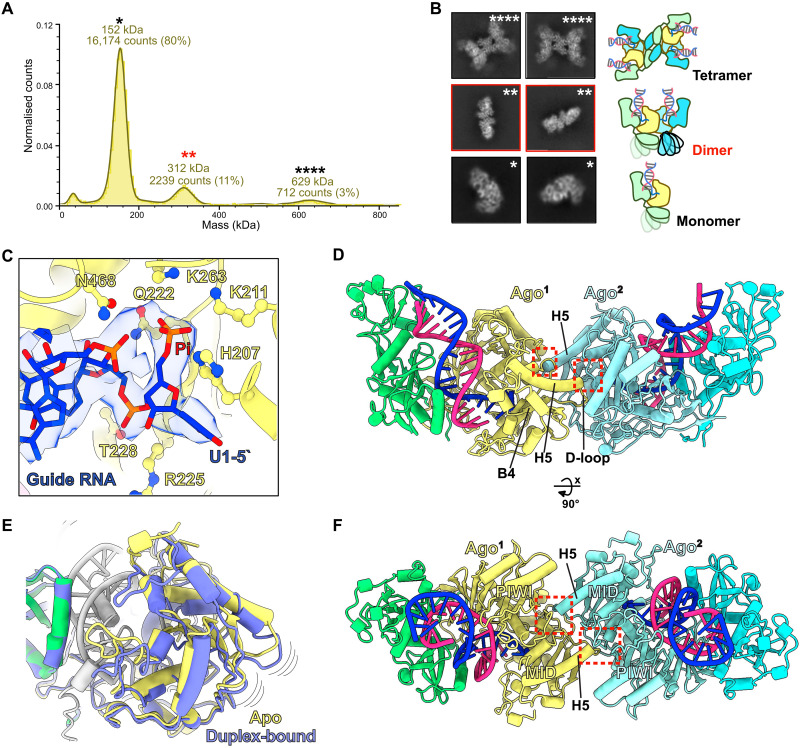
To gain insights into the assembly of MapSPARTA tetramers from a monomeric state, we performed single-molecule molecular mass measurements by mass photometry on MapSPARTA incubated with RNA/DNA heteroduplex (17). Precise signals for single molecules corresponding to the molecular mass of MapSPARTA monomers, dimers, and tetramers were detected (Fig. 2A). Notably, further two-dimensional (2D) and 3D classification of cryo-EM datasets captured additional structural states of the MapSPARTA complex. These resulted in cryo-EM maps for heteroduplex-bound monomeric MapSPARTA and three dimer states resolved at overall resolutions ranging from 2.9 to 4.8 Å (Fig. 2B, figs. S2 and S13, table S1, and movie S2). For comparison, we also collected cryo-EM datasets for the inactive apo form and obtained a cryo-EM map at an overall resolution of 3.7 Å (fig. S4). We resolved a cryo-EM structure of the guide RNA–only bound MapSPARTA, which was likely to be highly dynamic, resulting in a low-resolution 3D reconstruction (fig. S5). These results suggest that MapSPARTA first transitions from a monomer to a dimer before tetramer assembly upon activation (movie S3).
Fig. 2. Assembly of SPARTA dimers upon heteroduplex recognition.
(A) Mass photometry data of the M. polysiphoniae SPARTA (MapSPARTA) incubated with guide RNA (gRNA) and target DNA (tDNA) heteroduplex. The asterisks show molecular weights corresponding to a monomer (*), dimer (**), and tetramer (****). The red asterisks highlight the dimer. (B) Representative two-dimensional class averages of monomer, dimer, and tetramer MapSPARTA. (C) Cartoon and density representation of the 5′ phosphorylated gRNA interacting with residues of the middle (MID) pocket of prokaryotic argonaute (pAgo). (D and F) The structure of dimer state-1 with red dashed boxes indicates the dimerization interface where the C terminus of helix 5 interacts with a pocket formed between the Argonaute (Ago) MID and P-element Induced WImpy (PIWI) domains. (E) Overlay of the Ago MID domain structures in the apo and heteroduplex-bound complexes shows Ago’s structural change upon duplex recognition.
The structure of the MapSPARTA monomer reveals a modular architecture in which Ago and APAZ associate to form the Ago module having the canonical bilobate structure of Ago proteins (figs. S6, A and B, and S7A) (9). The Ago MID and PIWI domains include one lobe and the APAZ domain includes another lobe with similarity to the N domain and L1 and L2 regions of long Agos. Therefore, the complex of Ago and APAZ forms a structure that is highly identical to other reported Agos but together lacks the PAZ domain (fig. S6, A and C). The TIR domain is a separate module tethered to the Ago MID domain. The RNA/DNA heteroduplex-bound structure revealed a similar mode of heteroduplex recognition as previously described in the RsAgo-RNA/DNA ternary complex structure (figs. S6A and S7A) (18). The heteroduplex binds in a positively charged channel formed between the lobes of the Ago module via a network of hydrogen bonds. The 5′-end nucleotide of the guide RNA is unpaired, and conserved residues in the MID domain are essential for its recognition, as already demonstrated for other Agos (4, 18–21). The 5′-end phosphate group makes hydrogen bond interactions with side chains of residues H207, K211, Q222, and K263, which form a pocket in the MID domain that is essential for 5′-phosphate recognition (Fig. 2C and fig. S7, A and B). The MID domain residues R225 and T228 also interact with the 2′-hydroxy group of nucleotide 1 and the phosphate group of nucleotide 2 of the guide RNA and are essential for RNA-specific guide recognition (Fig. 2C). Side-chain interactions between R152 and uracil 1 of the guide RNA mediate 5′-end nucleotide specificity (fig. S7B). We observed that the 16-nt target DNA was bound to the complex by Watson-Crick base-pairing with the guide RNA to form an A-form duplex (fig. S7A). In the region of the APAZ domain analogous to the N domain in long Agos, residue side chains (K211, N212, R263, Q267, and K271) make hydrogen bonds with the phosphate backbones to stabilize the heteroduplex binding (fig. S7C). Still, this region does not cap the end of the duplex, thus providing a rationale for activating MapSPARTA by heteroduplexes up to 60 base pairs (bp) in length, as previously reported (14). A comparison of the MapSPARTA monomer’s apo and heteroduplex-bound form revealed that a helix from the C terminus of the APAZ domain (inhibitory helix) in the apo structure occupied the positively charged channel between the two lobes of the Ago module (figs. S4 and S8). This suggests that the apo form represents an auto-inhibited state of the complex. The inhibitory helix would have to be displaced out of the channel to facilitate nucleic acid recognition in the heteroduplex-bound form. In all heteroduplex-bound SPARTA structures, the apparent density for the inhibitory helix is absent, suggesting that it is flexible when displaced out of the nucleic acid binding channel by the heteroduplex. Heteroduplex binding further results in conformational changes in secondary structure elements in the Ago MID and PIWI domains, which have implications for dimer assembly as observed in the structure of the MapSPARTA dimer (Fig. 2E).
The structures of the MapSPARTA dimer revealed that each dimer contained two copies each of RNA/DNA duplexes, Ago, and APAZ domains (2:2:2:2) with a similar arrangement in all three states but with main differences in the conformation of the TIR domains (Fig. 2, D and F; figs. S2, S9, and S13; and table S1). All three dimer states use the same dimerization interface in which charged amino acids from the C terminus of alpha helix five (H5) and a loop (D-loop) before beta sheet four (B4) of the two Ago MID domains interact with amino acids formed by a charged pocket between the MID and PIWI domains of the adjacent of the Ago monomer. This arrangement forms an interlocking dimer with a surface area of 1294 Å2 for the dimer interface (Fig. 2, D and F, fig. S10, and movie S2). In one dimer state (dimer-1), MapSPARTA assembles as a symmetric dimer with no apparent densities for the two TIR domains suggesting that the TIR domains are highly flexible in this state (fig. S9A). In the second dimer state (dimer-2), low-resolution density (7 to 8 Å) for one of the TIR domains becomes apparent, suggesting the initiation of dimerization of TIR domains but with conformational flexibility (fig. S9B).
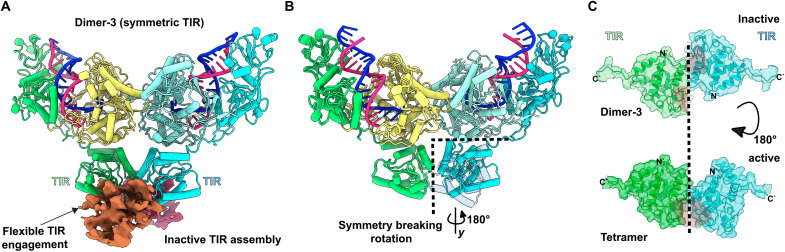
The third dimer state (dimer-3) reveals a symmetric arrangement of MapSPARTA, including the TIR domains resolved in the cryo-EM map (Fig. 3A and fig. S9C). The conformation of TIR domains in dimer-3 is antagonistic to MapSPARTA tetramer assembly. In the active tetramer, the TIR domains are arranged in a conformation in which one of the TIR domains is approximately 180° rotated relative to the other when compared to the symmetric TIR arrangement in dimer-3 (Fig. 3, B and C). This rotational movement results in symmetry breaking facilitated by the oligomerization of the TIR domains for tetramer assembly, as shown in the pseudo-symmetric dimer units of the SPARTA tetramer (Figs. 1D and 4A). Noticeably, density for a third TIR domain plausibly from another partially engaged MapSPARTA dimer interacts and stabilizes the dim-3 arrangement (Fig. 3A). However, this arrangement is averse to the formation of an active tetramer. These structural transitions suggested that these states were intermediates toward MapSPARTA tetramer assembly. Therefore, we propose that RNA/DNA duplex binding induces conformational changes in the Ago MID and PIWI domains, generating the dimerization interface between Ago monomers arranged in an antiparallel orientation. The initiation of MapSPARTA tetramer assembly induces flexibility of the two TIR domains, which then associate in a parallel orientation resulting from a symmetry-breaking rotational movement of one TIR domain relative to the other. These structural movements drive the association with the TIR domains from another MapSPARTA dimer resulting in the assembly of a MapSPARTA tetramer capable of substrate binding (movie S3). These structures reveal a unique mechanism by which sequence-specific nucleic acid recognition by a guide-bound Ago complex induces Ago dimerization to modulate the oligomerization and activity of a coupled effector enzyme.
Fig. 3. Toll/interleukin-1 receptor/resistance protein (TIR) adaptation is required for M. polysiphoniae SPARTA (MapSPARTA) tetramer assembly.
(A) Structure of dimer state-3 showing symmetric TIR arrangement in the cartoon with additional TIR density forming an inactive TIR assembly. (B) Representation of the 180° rotation of a TIR domain in the MapSPARTA dimer upon tetramerization. (C) Cartoon and surface representation show complete catalytic site formation (in red) upon TIR rotation.
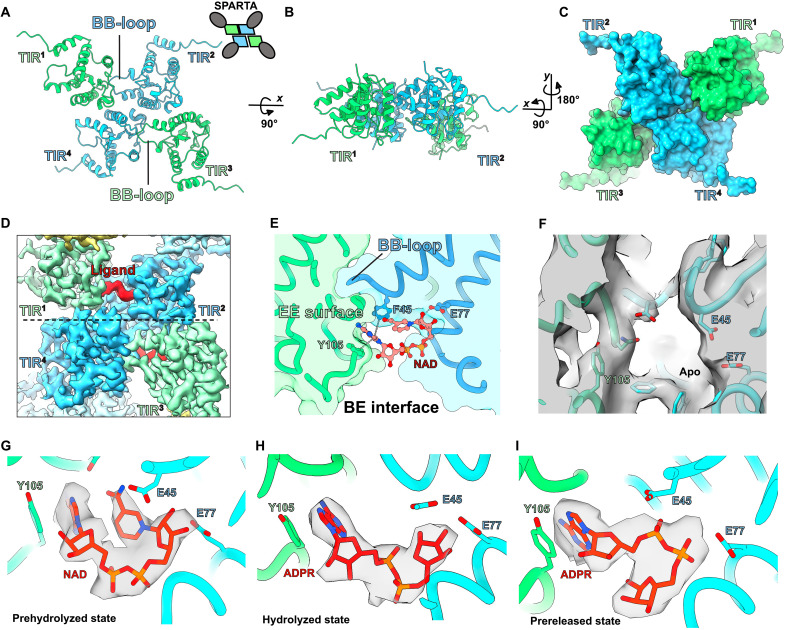
Fig. 4. Snapshots of the M. polysiphoniae SPARTA (MapSPARTA) NADase activity.
(A to C) The structural arrangement of toll/interleukin-1 receptor/resistance protein (TIR) domains from the MapSPARTA tetramer. (D) Cryo–electron microscopy (cryo-EM) density of a ligand/substrate bound (in red) in the TIR domains (in green and blue). (E) Details of the interaction between the bound nicotinamide adenine dinucleotide (oxidized form) (NAD) and residues of the TIR domain. (F) Cryo-EM density showing empty ligand/substrate binding pocket in apo MapSPARTA tetramer (G to I) Cryo-EM density for NAD and adenosine 5´-diphosphoribose (ADPR) in the prehydrolyzed, hydrolyzed, and prereleased states.
Mechanism of MapSPARTA NADase activity
MapSPARTA heteroduplex-induced tetramerization leads to the activation of its NADase activity. To gain insights into the catalytic activity of MapSPARTA, we determined cryo-EM structures of tetrameric MapSPARTA in the presence of NAD. Three-dimensional classification and local refinement resulted in three structures of ligand-bound MapSPARTA at resolution ranges of 3.2 to 3.7 Å (fig. S2). In all structures, the associated TIR domains from a MapSPARTA dimer (“intrastrand” in filamentous TIRs) adopt a head-to-tail arrangement mediated by interactions between the BB-loop and EE surface of adjacent TIR domains (BE interface) similar to that described for AbTir-TIR, MAL-TIR, and MyD88-TIRs but differing from another tetrameric TIR arrangement from plant NbTIR-NLR ROQ1 (Fig. 4, A to C, and fig. S11) (22–24). In the cryo-EM maps, we observed an apparent density for NAD or the reaction product adenosine 5´-diphosphoribose (ADPR) at the BE interface that is absent in the map of the apo MapSPARTA (Fig. 4, D to F). The BE interface forms a catalytic site with the bound ligand conformation similar to SARM1-TIR and AbTir-TIR (22, 25) (Fig. 4, D and E, and fig. S11). Therefore, two NAD molecules bind per tetramer. The three structures revealed that the ligand bound in a prehydrolyzed state with NAD in the catalytic site, a hydrolyzed state resulting from removing the nicotinamide moiety, and a prereleased state as a result of a flip of the ribose moiety of ADPR, possibly required before it exits the catalytic site (Fig. 4, G to I, fig. S12, and movie S4). Interactions with residues in the BE interface stabilize NAD binding in the catalytic site. Notably, the conserved aromatic residues F45 and Y105 make pi-pi interactions with the adenine base both from NAD and ADPR and the highly conserved residue E77, shown to be essential for catalysis (14), makes hydrogen bond interactions with the C2 and C3 hydroxyls of NAD before cleavage (Fig. 4, E and G). This interaction is lost upon the ribose flipping of ADPR, preparing the site for the release of ADPR and the binding of another molecule of NAD (Fig. 4I). These structures give insight into the catalytic mechanism of the MapSPARTA NADase activity.
DISCUSSION
Ago and TIR domain-containing proteins play essential functions in innate immunity against foreign genetic elements in eukaryotic and prokaryotic hosts. Studies over the past two decades have resulted in detailed insights into the independent functional mechanisms of these proteins (4, 26). However, the prokaryotic SPARTA complex, which includes the cooperative action of short pAgos and TIR-APAZ proteins in a single functional complex, presents a recently discovered immune defense complex. The interaction between the Ago and APAZ domains results in a structure very similar to long Agos to act as the “sensor” module, while the TIR is a separate “effector” module of the SPARTA complex.
Here, we describe the structural mechanism by which RNA guide–mediated DNA recognition by the SPARTA sensor Ago module regulates the activity of the effector TIR module. The activation mechanism involves choreography of structural transitions involving conformational movements in the Ago and TIR subunits which are essential to activate its catalytic activity to facilitate defense against invaders such as foreign plasmids (Fig. 5A). A critical structural intermediate was the unpredicted formation of SPARTA dimers mediated by the Ago MID and PIWI domains upon RNA/DNA heteroduplex recognition. Although dimerization of AfAgo, a long B clade pseudo-short pAgo, was previously reported in another study, the functional relevance of the dimerization, in that case, remains unknown (27). Here, we show a functionally important Ago dimerization mechanism mediated primarily by the Ago sensor module to regulate the assembly and activation of the TIR effector module. Our findings suggest that Ago dimerization could be used in other short Ago systems to regulate the effector modules. However, it seems unlikely that all short Agos use this mechanism because the Ago of the SPARSA did not require oligomerization to activate the effector enzyme (Sir2) (15). Therefore, high-resolution structural studies on other short Ago systems could reveal mechanisms of Ago-mediated effector regulation, opening options for further engineering these systems for biotechnological applications (28). For instance, the modular arrangement of the SPARTA complex suggests that nucleic acid detection by the Ago module could regulate other enzymatic or functional modules that require dimerization or oligomerization for activity. This being the case, interchanging the effector module with other dimerization-dependent effectors, e.g., dimerization-dependent fluorescent proteins, proteases, kinases, transcriptional regulators, and scaffold proteins, could result in designer molecular machines with broad-range applications (Fig. 5B). In conclusion, our study reports an Ago functional mechanism that paves the way for engineering the SPARTA system for biotechnological and therapeutic applications.
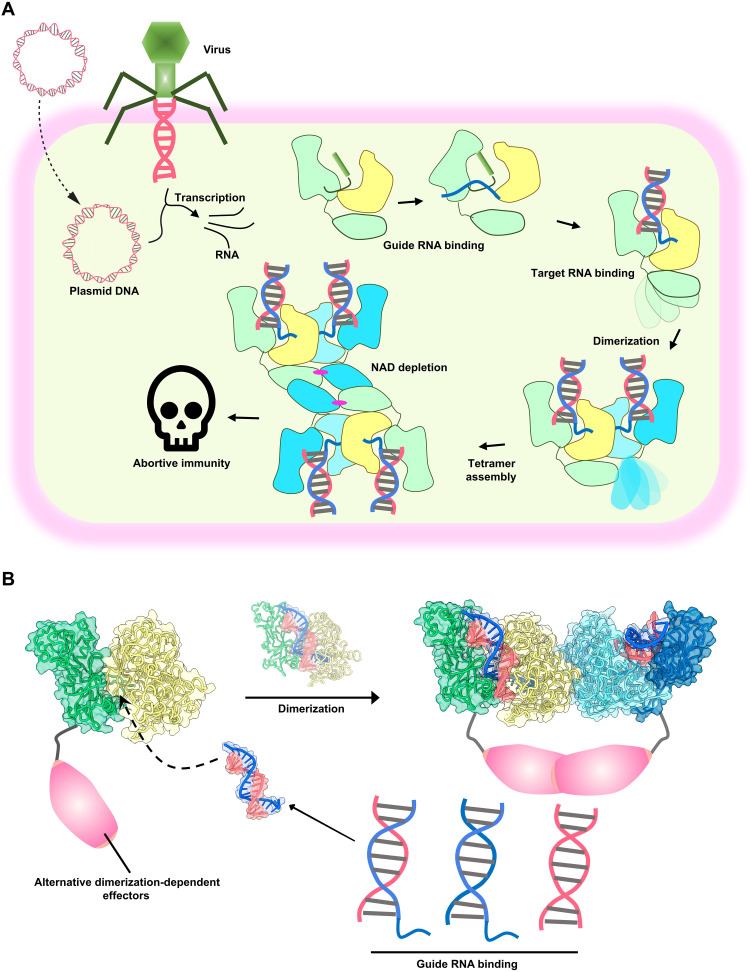
Fig. 5. Model of SPARTA activity in prokaryotic immunity and potential biotechnological applications.
(A) Assembly pathway of M. polysiphoniae SPARTA (MapSPARTA) upon binding guide RNA (gRNA) generated from transcription of invading plasmid or viral DNA which leads to nicotinamide adenine dinucleotide (oxidized form) (NAD) depletion and abortive immunity. (B) Cartoon of the formation of SPARTA dimers upon nucleic acid recognition with the TIR domain replaced with other dimerization-dependent effectors for biotechnological applications.
MATERIALS AND METHODS
Plasmid constructs design and cloning
To reconstitute the MapSPARTA complex, M. polysiphoniae pAgo was coexpressed with M. polysiphoniae TIR-APAZ using two plasmids. Full-length M. polysiphoniae pAgo codon-optimized for Escherichia coli was synthesized with a sequence for an N-terminal tobacco etch virus (TEV) cleavage site and cloned (GenScript Biotech) into pMAL-c4x plasmid to be in-frame with an N-terminal maltose binding protein (MBP) tag. Full-length M. polysiphoniae TIR-APAZ codon-optimized for E. coli was synthesized with a sequence for an N-terminal 6xHis tag and TEV cleavage site and cloned (GenScript Biotech) into pET24a plasmid. In this way, both genes were inserted downstream of a LacI-repressed T7 promoter to facilitate inducible expression using isopropyl-β-d-thiogalactopyranoside (IPTG).
Protein expression and purification
The MapSPARTA complex composed of MapAgo and MapTIR-APAZ was coexpressed and purified from E. coli BL21(DE3) Star. For expression, chemically competent E. coli BL21(DE3) Star was transformed with both, the pMAL-c4x plasmid containing the ampicillin resistance gene for expression of MapAgo and the pET24a plasmid containing kanamycin resistance gene for the expression of MapTIR-APAZ, and grown overnight at 37°C on LB agar plates containing both selection antibiotics [kanamycin (50 μg/ml) and carbenicillin (100 μg/ml)]. The colonies obtained were transferred to 100 ml of 2xYT media containing selection antibiotics and grown overnight at 37°C with shaking. Sixty milliliters of the overnight culture was used to inoculate 6 liters of 2xYT media containing selection antibiotics. The cultures were grown until they reached an absorbance at 600 nm of 0.5 to 0.7, and then incubated at 4°C for 1 hour, before inducing protein expression with 0.5 mM IPTG for 18 hours at 20°C. The cultures were harvested by centrifugation at 3000g for 30 min at 4°C. The resulting supernatant was discarded, and the pellet was resuspended in 100 ml of precooled lysis buffer [25 mM Hepes-NaOH (pH 7.5), 500 mM NaCl, 10% glycerol, 25 mM imidazole, and 1 mM β-mercaptoethanol] supplemented with two tablets of cOmplete EDTA-free Protease Inhibitor Cocktail (Roche) before lysis by sonication. The lysate was clarified by centrifugation for 30 min at 70,560g at 4°C in an Optima XPN Ultracentrifuge (Beckman Coulter) using a Type 45 Ti rotor. The supernatant, which contained soluble 6xHis-tagged MapTIR-APAZ in complex with MBP-tagged MapAgo, was loaded onto 5 ml of HisPur Ni-NTA Resin (Thermo Fisher Scientific) pre-equilibrated with wash buffer [25 mM Hepes-NaOH (pH 7.5), 500 mM NaCl, 10% glycerol, 25 mM imidazole, and 1 mM β-mercaptoethanol] in an XK16/20 column (Cytiva Life Sciences). After loading, the column was washed with 50 ml of wash buffer and eluted with 10 ml of elution buffer [25 mM Hepes-NaOH (pH 7.5), 500 mM NaCl, 10% glycerol, 400 mM imidazole, and 1 mM β-mercaptoethanol]. Pooled elution fractions were loaded onto 3 ml of amylose resin (New England Biolabs) pre-equilibrated with amylose wash buffer [25 mM Hepes (pH 7.5), 200 mM NaCl, 10% glycerol, and 1 mM dithiothreitol (DTT)]. Upon complete loading, the column was washed with amylose wash buffer and eluted with 10 ml of amylose elution buffer [25 mM Hepes (pH 7.5), 200 mM NaCl, 10% glycerol, 1 mM DTT, and 20 mM maltose]. The pooled elution fraction was concentrated to ∼500 μl in 50K Amicon Ultra-15 concentrators (Millipore, Billerica, MA) and further purified by gel filtration chromatography on a Superdex 200 10/300 GL gel filtration column (Cytiva Life Sciences) in gel filtration buffer [25 mM Hepes-NaOH (pH 7.5), 150 mM NaCl, 10% glycerol, and 1 mM DTT]. Peak fractions as determined by the chromatograms at UV280 generated from the Unicorn software (version 7.1) containing complete SPARTA complexes of subunits MapAgo and MapTIR-APAZ as determined by SDS–polyacrylamide gel electrophoresis (PAGE) analysis were pooled, concentrated, flash-frozen, and stored at −80°C.
For the reconstitution of guide RNA–bound SPARTA complex, 2 μM purified SPARTA was mixed with 2 μM guide RNA in reconstitution buffer [25 mM Hepes-NaOH (pH 7.5), 125 mM NaCl, 3 mM MgCl2, and 1 mM DTT] and incubated at 37°C for 15 min. After incubation, the sample was loaded onto Superdex 200 10/300 GL gel filtration column (Cytiva Life Sciences) in gel filtration buffer [25 mM Hepes-NaOH (pH 7.5), 125 mM NaCl, 3 mM MgCl2, and 1 mM DTT]. The sample was concentrated to absorption at 280 nm (A280nm) of 0.8 using a 50K Amicon Ultra-15 concentrator (Millipore, Billerica, MA) and was ready for cryo-EM experiments. For the reconstitution of heteroduplex-bound SPARTA complex after incubation with guide RNA as previously described, the sample was further incubated with target DNA for 30mins at 37°C. Following incubation, the sample was loaded onto 10/300 GL Superose 6 gel filtration column (Cytiva Life Sciences) in gel filtration buffer [25 mM Hepes-NaOH (pH 7.5), 125 mM NaCl, 2 mM MgCl2, and 1 mM DTT], gel filtration peaks corresponding to heteroduplex-bound SPARTA tetramers and monomers were separately concentrated to A280nm of 0.8 in 50K Amicon Ultra-15 concentrators (Millipore, Billerica, MA) and were ready to be used for cryo-EM experiments. Samples not used for cryo-EM were mixed with an equal volume of buffer containing 25 mM Hepes-NaOH (pH 7.5), 200 mM NaCl, 20% glycerol, and 1 mM DTT and were flash-frozen for storage at −80°C.
SDS-PAGE analysis
To evaluate the purity of the SPARTA complex and the presence of the individual pAgo and TIR-APAZ subunits in the complex, an SDS-PAGE analysis was performed. Twenty microliters of protein sample was supplemented with 5 μl of 4X NuPAGE LDS sample buffer (Thermo Fisher Scientific). Samples were incubated at 95°C for 10 min before loading on 4 to 12% SurePAGE bis-tris precast gels (Witec AG). Spectra Multicolor Broad Range Protein Ladder (10 to 180 kDa) was also loaded on the gel to run as a size marker. Gels were run in 1× tris-MOPS SDS running buffer (Witec AG) at 200 V for 30 min, washed briefly in Milli-Q water, and stained for 2 hours with QuickBlue Protein Stain (LuBioScience GmbH) with shaking. Gels were washed in Milli-Q water before imaging on an iBright FL1500 Imaging System (Thermo Fisher Scientific).
Urea-PAGE analysis
To evaluate the presence of bound nucleic acids in the ssRNA or heteroduplex-bound SPARTA complex, urea-PAGE analysis was performed on peak fractions from the gel filtration of the complexes. Following gel filtration, samples from the peak fraction were incubated with urea and proteinase K (Thermo Fisher Scientific) at final concentrations of 0.1 M and 1 μg/μl, respectively, and incubated at 50°C for 15 min. Loading dye (2×) [1× tris-borate-EDTA (TBE), 12% Ficoll, 7 M urea) was added to the reaction to a final 1× concentration, followed by heating at 95°C for 5 min before loading on a 15% Novex TBE-Urea gel (Thermo Fisher Scientific). For nuclease digestion, 20 μl of heteroduplex-bound MapSPARTA with absorption at A280nm of 0.8 was digested with excess amounts of ribonuclease A (RNAse A) (Thermo Fisher Scientific), micrococcal nuclease (New England Biolabs), RNase III (New England Biolabs), and deoxyribonuclease I (Sigma-Aldrich) for 1 hour at 37°C before proteinase K treatment and gel loading. The gel was run at 200 V for 45 min and stained with 1× GelRed nucleic acid gel stain for 30 min before imaging on an iBright FL1500 Imaging System (Thermo Fisher Scientific).
NAD/NADH quantification
SPARTA samples at a concentration of 0.5 μM were incubated with 10 μM reduced form of NAD+ (NADH) for 2 hours at 37°C. Fifty microliters of each sample was used as input for the NAD/NADH Quantitation Kit (Sigma-Aldrich, MAK037-1KT) according to the instructions provided by the manufacturer. The measurements were repeated in at least triplicates.
Mass photometry
Mass photometry experiments were carried out using a Refeyn TwoMP (Refeyn Ltd., Oxford, UK) MP system. AcquireMP and DiscoverMP software packages were used to record movies and analyze data, respectively, using standard settings. Microscope coverslips (high precision glass coverslips, Marienfeld) were cleaned following the Refeyn Ltd. individual rinsing procedure. Reusable self-adhesive silicone culture wells (Grace Bio-Labs reusable CultureWell gaskets) were used to keep the sample droplet shape. Contrast-to-mass calibration was carried out using BSA (Albumin, Bovine Serum Fraction V, Low Heavy Metals; Millipore), giving molecular weights of 66, 132, 198, and 264 kDa. Immediately before the measurements, protein stocks were diluted directly in a buffer containing 25 mM Hepes-NaOH (pH 7.5), 125 mM NaCl, and 2 mM MgCl2. To this end, 1 to 2 μl of protein solution was added into 18 to 19 μl of analysis buffer to reach a final drop volume of 20 μl.
Cryo-EM sample preparation and data collection
Cryo-EM grids for the apo SPARTA monomer, guide RNA–bound SPARTA monomer, heteroduplex-bound SPARTA monomer, and heteroduplex-bound SPARTA tetramer were prepared by applying 3 μl of concentrated sample onto 400-mesh R1.2/1.3 UltrAuFoil grids (Quantifoil Micro Tools GmbH), which were rendered hydrophilic by glow discharging at 15 mA for 90 s with a PELCO easiGlow device (Ted Pella Inc.). The sample was adsorbed for 30 s on the grids, followed by blotting and plunge-freezing into liquid ethane using a Vitrobot Mark IV plunge freezer (Thermo Fisher Scientific). For the heteroduplex-bound SPARTA tetramer supplemented with NAD, after application of 3 μl of the concentrated sample on the grid, 0.5 μl of 50 mM beta-NAD sodium salt (Sigma-Aldrich GmbH) was added directly to the sample followed by immediate plunge-freezing. Cryo-EM data were collected using the automated data acquisition software EPU (Thermo Fisher Scientific) on a Titan Krios G4 transmission electron microscope (Thermo Fisher Scientific), operating at 300 kV and equipped with a cold field-emission gun electron source and a Falcon4 direct detection camera. Images were recorded in counting mode at a nominal magnification of ×96,000, corresponding to a physical pixel size of 0.726 and 0.5082 Å at the sample level. Datasets were collected at a defocus range of 0.8 to 2.5 μm with a total electron dose of 60 e−/Å2. Image data were saved as Electron Event Recordings.
Cryo-EM image processing, model building, and refinement
The cryo-EM image processing was performed using cryoSPARC v3.4 (29). The EM movie stacks were aligned and dose-weighted using patch-based motion correction (cryoSPARC implementation). Contrast transfer function (CTF) estimation was performed using the patch-based option as well. For the data of the apo SPARTA monomer, a blob picker was used for initial particle picking, which resulted in 200,000 particles from the initial 1000 images. These particles were used for two rounds of 2D classification to generate templates for template-based particles picking on the full dataset, resulting in 2,821,100 particles. Multiple rounds of 2D classifications were performed and junk particles were deselected. Ab initio reconstruction and hetero-refinement yielded multiple 3D classes. The best 3D class consisting of 85,650 particles used for nonuniform refinement yielded a cryo-EM map at 3.74-Å overall resolution in C1 symmetry (fig. S4).
The 2D classes generated from the apo SPARTA monomer were used for template picking for the guide RNA–bound SPARTA monomer datasets. This resulted in 1,941,707 particles. Several rounds of 2D classification, ab initio reconstruction, and hetero-refinement resulted in a map at 7.11-Å overall resolution in C1 symmetry from 59,152 particles (fig. S5). The SPARTA tetramer samples were processed using the blob picker for particle picking, 2D classification, template generation, and template picking. Multiple rounds of 3D classification and hetero-refinement resulted in three distinct states of the SPARTA dimer at an overall resolution of 3.3- to 3.9-Å overall resolution in C2 symmetry. Dimer states 1, 2, and 3 consisted of 59,663, 36,541, and 25,123 particles, respectively (fig. S2). The duplex-bound monomer was also resolved from 44,404 particles at 4.8-Å overall resolution (fig. S2). The apo SPARTA tetramer could also be resolved to 3.9 Å from 14,276 particles using a similar processing strategy (fig. S4). The NAD-supplemented tetramer was also processed with a similar approach. Three-dimensional classification resulted in three structures that contained three distinct ligand conformations. These three states were referred to as the prehydrolyzed, hydrolyzed, and prereleased states. Each state contained 65,473, 50,287, and 52,117 particles respectively at an overall resolution of 3.2 to 3.7 Å in C1 symmetry (fig. S2).
Atomic models for all nine were mostly manually built de novo in Coot 0.9.4 (30). For low-resolution or low-quality regions, an AlphaFold2 (ColabFold implementation) prediction was used as a guide for amino acid assignment and backbone tracing (31). Real-space refinement for all built models was performed using Phenix, version 1.19.2-4158 by applying a general restraints setup (32).
Data visualization
Structural alignments and superpositions were performed with PyMOL (The PyMOL Molecular Graphics System, version 1.8.2.0, Schrödinger, LLC) and UCSF ChimeraX v1.4. Gel images were processed and prepared on ImageJ (version 1.53k). Figures were rendered using PyMOL, UCSF Chimera, ChimeraX (33), and Adobe Illustrator (https://adobe.com/products/illustrator).
Acknowledgments
We thank A. Myasnikov, B. Beckert, S. Nazarov, and E. Uchikawa from the Dubochet Center for Imaging of the EPFL, the University of Lausanne, and the University of Geneva for support with cryo-EM data collection; and Y. Duhoo and K. Lau from the EPFL, Protein Production and Structure Core Facility, for help with mass photometry experiments.
Funding: This work was supported by the Swiss National Science Foundation (grants CRSII5_177195, IZLCZ0_206089 and 310030_188548).
Author contributions: Conceptualization: D.N. and B.E. Methodology: B.E., D.N., X.L., and H.S. Investigation: D.N., B.E., and X.L. Funding acquisition: H.S. Project administration and supervision: D.N. and B.E. Writing—original draft: D.N. and B.E. Writing—review and editing: D.N., B.E., and H.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The reconstructed maps are available from the EMDB database under access codes EMDB-17299, -17300, -17310, -17308, -17307, -17306, -17301, -17302, -17305, -17303, and -17304. The atomic models are available in the PDB database under access codes PDB-ID 8OZ6, 8OZI, 8OZG, 8OZF, 8OZE, 8OZD, and 8OZC.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S13
Table S1
Legends for movies S1 to S4
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S4
REFERENCES AND NOTES
- 1.D. C. Swarts, K. Makarova, Y. Wang, K. Nakanishi, R. F. Ketting, E. V. Koonin, D. J. Patel, J. van der Oost, The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 21, 743–753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.G. Hutvagner, M. J. Simard, Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32 (2008). [DOI] [PubMed] [Google Scholar]
- 3.M. Ghildiyal, P. D. Zamore, Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 10, 94–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L. Lisitskaya, A. A. Aravin, A. Kulbachinskiy, DNA interference and beyond: Structure and functions of prokaryotic Argonaute proteins. Nat. Commun. 9, 5165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D. C. Swarts, M. M. Jore, E. R. Westra, Y. Zhu, J. H. Janssen, A. P. Snijders, Y. Wang, D. J. Patel, J. Berenguer, S. J. J. Brouns, J. van der Oost, DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D. C. Swarts, M. Szczepaniak, G. Sheng, S. D. Chandradoss, Y. Zhu, E. M. Timmers, Y. Zhang, H. Zhao, J. Lou, Y. Wang, C. Joo, J. van der Oost, Autonomous generation and loading of DNA guides by bacterial Argonaute. Mol. Cell 65, 985–998.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Kuzmenko, A. Oguienko, D. Esyunina, D. Yudin, M. Petrova, A. Kudinova, O. Maslova, M. Ninova, S. Ryazansky, D. Leach, A. A. Aravin, A. Kulbachinskiy, DNA targeting and interference by a bacterial Argonaute nuclease. Nature 587, 632–637 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Y. Chen, Z. Zeng, Q. She, W. Han, The abortive infection functions of CRISPR-Cas and Argonaute. Trends Microbiol. 31, 405–418 (2023). [DOI] [PubMed] [Google Scholar]
- 9.J.-J. Song, S. K. Smith, G. J. Hannon, L. Joshua-Tor, Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Y. Wang, S. Juranek, H. Li, G. Sheng, T. Tuschl, D. J. Patel, Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456, 921–926 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J.-B. Ma, Y.-R. Yuan, G. Meister, Y. Pei, T. Tuschl, D. J. Patel, Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.B. Koopal, S. K. Mutte, D. C. Swarts, A long look at short prokaryotic Argonautes. Trends Cell Biol. 33, 605–618 (2023). [DOI] [PubMed] [Google Scholar]
- 13.K. S. Makarova, Y. I. Wolf, J. van der Oost, E. V. Koonin, Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 4, 29 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.B. Koopal, A. Potocnik, S. K. Mutte, C. Aparicio-Maldonado, S. Lindhoud, J. J. M. Vervoort, S. J. J. Brouns, D. C. Swarts, Short prokaryotic Argonaute systems trigger cell death upon detection of invading DNA. Cell 185, 1471–1486.e19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Zaremba, D. Dakineviciene, E. Golovinas, E. Zagorskaitė, E. Stankunas, A. Lopatina, R. Sorek, E. Manakova, A. Ruksenaite, A. Silanskas, S. Asmontas, A. Grybauskas, U. Tylenyte, E. Jurgelaitis, R. Grigaitis, K. Timinskas, Č. Venclovas, V. Siksnys, Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD+ depletion. Nat. Microbiol. 7, 1857–1869 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Z. Zeng, Y. Chen, R. Pinilla-Redondo, S. A. Shah, F. Zhao, C. Wang, Z. Hu, C. Wu, C. Zhang, R. J. Whitaker, Q. She, W. Han, A short prokaryotic Argonaute activates membrane effector to confer antiviral defense. Cell Host Microbe 30, 930–943.e6 (2022). [DOI] [PubMed] [Google Scholar]
- 17.A. Sonn-Segev, K. Belacic, T. Bodrug, G. Young, R. T. VanderLinden, B. A. Schulman, J. Schimpf, T. Friedrich, P. V. Dip, T. U. Schwartz, B. Bauer, J.-M. Peters, W. B. Struwe, J. L. P. Benesch, N. G. Brown, D. Haselbach, P. Kukura, Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat. Commun. 11, 1772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.T. Miyoshi, K. Ito, R. Murakami, T. Uchiumi, Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 7, 11846 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.E. Kaya, K. W. Doxzen, K. R. Knoll, R. C. Wilson, S. C. Strutt, P. J. Kranzusch, J. A. Doudna, A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. U.S.A. 113, 4057–4062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Y. Liu, D. Esyunina, I. Olovnikov, M. Teplova, A. Kulbachinskiy, A. A. Aravin, D. J. Patel, Accommodation of helical imperfections in rhodobacter sphaeroides Argonaute ternary complexes with guide RNA and target DNA. Cell Rep. 24, 453–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.J. W. Hegge, D. C. Swarts, S. D. Chandradoss, T. J. Cui, J. Kneppers, M. Jinek, C. Joo, J. van der Oost, DNA-guided DNA cleavage at moderate temperatures by Clostridium butyricum Argonaute. Nucleic Acids Res. 47, 5809–5821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. K. Manik, Y. Shi, S. Li, M. A. Zaydman, N. Damaraju, S. Eastman, T. G. Smith, W. Gu, V. Masic, T. Mosaiab, J. S. Weagley, S. J. Hancock, E. Vasquez, L. Hartley-Tassell, N. Kargios, N. Maruta, B. Y. J. Lim, H. Burdett, M. J. Landsberg, M. A. Schembri, I. Prokes, L. Song, M. Grant, A. DiAntonio, J. D. Nanson, M. Guo, J. Milbrandt, T. Ve, B. Kobe, Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling. Science 377, eadc8969 (2022). [DOI] [PubMed] [Google Scholar]
- 23.T. Ve, P. R. Vajjhala, A. Hedger, T. Croll, F. DiMaio, S. Horsefield, X. Yu, P. Lavrencic, Z. Hassan, G. P. Morgan, A. Mansell, M. Mobli, A. O’Carroll, B. Chauvin, Y. Gambin, E. Sierecki, M. J. Landsberg, K. J. Stacey, E. H. Egelman, B. Kobe, Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling. Nat. Struct. Mol. Biol. 24, 743–751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S. Ma, D. Lapin, L. Liu, Y. Sun, W. Song, X. Zhang, E. Logemann, D. Yu, J. Wang, J. Jirschitzka, Z. Han, P. Schulze-Lefert, J. E. Parker, J. Chai, Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370, eabe3069 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Y. Shi, P. S. Kerry, J. D. Nanson, T. Bosanac, Y. Sasaki, R. Krauss, F. K. Saikot, S. E. Adams, T. Mosaiab, V. Masic, X. Mao, F. Rose, E. Vasquez, M. Furrer, K. Cunnea, A. Brearley, W. Gu, Z. Luo, L. Brillault, M. J. Landsberg, A. DiAntonio, B. Kobe, J. Milbrandt, R. O. Hughes, T. Ve, Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules. Mol. Cell 82, 1643–1659.e10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.K. Essuman, J. Milbrandt, J. L. Dangl, M. T. Nishimura, Shared TIR enzymatic functions regulate cell death and immunity across the tree of life. Science 377, eabo0001 (2022). [DOI] [PubMed] [Google Scholar]
- 27.E. Golovinas, D. Rutkauskas, E. Manakova, M. Jankunec, A. Silanskas, G. Sasnauskas, M. Zaremba, Prokaryotic Argonaute from Archaeoglobus fulgidus interacts with DNA as a homodimer. Sci. Rep. 11, 4518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.E. V. Kropocheva, L. A. Lisitskaya, A. A. Agapov, A. A. Musabirov, A. V. Kulbachinskiy, D. M. Esyunina, Prokaryotic Argonaute proteins as a tool for biotechnology. Mol. Biol. 56, 854–873 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. Punjani, J. L. Rubinstein, D. J. Fleet, M. A. Brubaker, cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 30.P. Emsley, B. Lohkamp, W. G. Scott, K. Cowtan, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli, D. Hassabis, Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D. Liebschner, P. V. Afonine, M. L. Baker, G. Bunkóczi, V. B. Chen, T. I. Croll, B. Hintze, L. W. Hung, S. Jain, A. J. McCoy, N. W. Moriarty, R. D. Oeffner, B. K. Poon, M. G. Prisant, R. J. Read, J. S. Richardson, D. C. Richardson, M. D. Sammito, O. V. Sobolev, D. H. Stockwell, T. C. Terwilliger, A. G. Urzhumtsev, L. L. Videau, C. J. Williams, P. D. Adams, Macromolecular structure determination using x-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. D. Goddard, C. C. Huang, E. C. Meng, E. F. Pettersen, G. S. Couch, J. H. Morris, T. E. Ferrin, UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S13
Table S1
Legends for movies S1 to S4
Movies S1 to S4