Abstract
While most nanomaterials are designed to assist tumor therapy, some inorganic nanoparticles have been reported to impede cancer development. We assume that the immune response elicited by these foreign nanoparticles might be associated with the remodeling of immune landscape in the tumor microenvironment (TME). We studied representative inorganic nanoparticles widely used in the biomedical field and first demonstrated that needle-shaped hydroxyapatite (n-nHA), granule-shaped hydroxyapatite, and silicon dioxide can effectively impair tumor progression in vivo. Substantial multinucleated giant cells (MNGCs) were formed around these antitumor nanoparticles, while the ratio of monocytes and macrophages was decreased in the TME. We found that high expression of the STXBP6 protein induced by n-nHA–treated macrophages triggers autophagy, which markedly promotes macrophage fusion into MNGCs. In this way, extensive depletion of tumor-associated macrophages in the TME was achieved, which suppressed tumor growth and metastasis. This intrinsic antitumor immunity of inorganic nanoparticles should not be neglected when designing future nanomedicines to treat cancer.
Inorganic nanoparticles induce multinucleated giant cell formation and deplete tumor-associated macrophages to delay tumor growth.
INTRODUCTION
Owing to advances in cancer detection and treatment, the awareness and survival rate of cancer individuals continue to increase (1). Nevertheless, cancer remains the predominant cause of death and an intractable barrier to extending life expectancy globally (2). In 2020, new cancer cases and deaths worldwide were estimated to be up to 19.3 million and 10.0 million, respectively (3). Clinically, the universal therapy for solid tumors is surgical resection combined with chemotherapy or radiation (4, 5), where nanotechnology, particularly nanomaterials, has emerged as a considerable tool to deliver cytotoxic medicine (6) or enhance radioactive signals (7, 8). However, these nanomedicines, usually packaged in polymeric nanoparticles, have made only moderate contributions to the survivorship issues of patients with cancer. Studies conducted from 1995 to 2019 covering 10,958 individuals with multiple types of cancer confirmed that only 2.76% of patients gained a longer survival period after polymeric nanomedicine administration (9).
In contrast with polymeric nanoparticles as drug vehicles, inorganic nanoparticles are primarily used in tumor imaging and to intensify physiotherapy (e.g., Cornell Dots, AuroLase, and NBTXR3/Hensify) (7). Recently, several studies discovered that these inorganic nanoparticles might have an inherent antitumor ability (10). Various mechanisms have been proposed on the basis of in vitro evidence. Nondegradable metallic nanoparticles with surface charge, e.g., titanium dioxide nanoparticles (n-TiO2) and silver nanoparticles (n-Ag), are reported to disrupt cell morphology and interfere with important proteins for cell migration to prevent metastasis (10, 11). Nonmetallic and partially degradable nanoparticles, e.g., hydroxyapatite nanoparticles (n-HA), calcium carbonate nanoparticles (n-CaCO3), and silica nanoparticles (n-SiO2), are recognized to raise the local pH value to neutralize the acidic tumor microenvironment (TME) and trigger oxidative stress (12–14). Our research group has focused on the antitumor effect of n-HA on melanoma, osteosarcoma, and hepatic carcinoma since 2007 (12, 15–19). In one of our clinical trials, we applied an n-HA–based composite material as a bone void filler in the defects created by benign tumors (20). No tumor reoccurrence was observed among 21 patients, and an alteration in immune cell constitution was observed in their blood test results. Note that several recent animal studies also discovered that a variety of immune cells gathered around n-HA in the TME, including multinucleated giant cells (MNGCs), lymphocytes, and neutrophils (19, 21). However, the relationship between the recruitment of immune cells and the antitumor ability of these inorganic nanoparticles is still unknown.
We know that the host immune response can be provoked by inorganic nanoparticles in certain clinical scenarios. For example, the oral implant alloy Ti6Al4V released n-TiO2 debris years after implantation, which resulted in mucositis in nearly 80% of patients with increased serum levels of interleukin-1β (IL-1β), IL-6, and tumor necrosis factor–α (TNF-α) (22, 23). Similarly, it was reported that long-term exposure to n-SiO2 via intravenous injection during medical imaging activated the Janus kinase 2/signal transducer and activator of transcription 3 pathway and accelerated pulmonary inflammation (24). Furthermore, a moderate inflammatory phase induced by n-HA is necessary for bone regeneration when it is used as a bone graft (25). During this phase, elevated secretion levels of TNF-α, macrophage inflammatory protein 1, and granulocyte colony-stimulating factor were measured in the local tissue, which allows polarization of macrophages and thus expedites osteogenic differentiation (26, 27). Coincidentally, the abovementioned inflammatory cytokines induced by inorganic nanoparticles also play vital roles in tumor inhibition (28). These findings provided the rationale to assume that the inflammatory response caused by inorganic nanoparticles might be correlated with their inherent antitumor ability. Therefore, we investigated the potential inorganic nanoparticle-induced immune landscape change at the cellular and transcriptional levels in the TME and combined both levels to unveil their inherent antitumor mechanism. We enrolled inorganic nanoparticles commonly applied in the biomedical field, including n-HA, n-CaCO3, n-SiO2, n-TiO2, and n-Ag. We aim to build a comprehensive understanding of the biological response caused by inorganic nanoparticles in the TME and present a remarkable opportunity to harness this power in modern tumor therapy.
RESULTS
Discrepancies in antitumor efficacy in vitro and in vivo
In this work, six nanoparticles, including two n-HAs, n-CaCO3, n-SiO2, n-TiO2, and n-Ag, were used. Previous studies indicated that the antitumor performance of n-HA is highly dependent on particle morphology (26, 29). Thus, we synthesized two n-HAs with different shapes, needle-shaped n-HA (n-nHA), and granule-shaped n-HA (n-gHA). The chemical compositions of all six nanoparticles enrolled were confirmed by x-ray diffraction and Fourier transform infrared spectroscopy (figs. S1 and S2). Transmission electron microscopy images showed that all samples had a primary size within the nanoscale (fig. S3). The hydrodynamic size of these nanoparticles evaluated using dynamic light scattering ranged between 250 and 1000 nm (fig. S4, A and B). All studied nanoparticles showed a negative zeta potential of approximately −15 mV (fig. S4C). First, the in vitro 4T1 tumor inhibition efficacy of a wide concentration range (0 to 1000 μg/ml) of these nanoparticles was examined by Cell Counting Kit-8 (CCK-8) assay. In accordance with the literature, we observed a dose-dependent boost of 4T1 cell killing in the coculture of all nanoparticles with distinctly different antitumor abilities among the different nanoparticles (fig. S5). For all groups, tumor cell viability was significantly reduced when the cells were incubated with nanoparticles at 1000 μg/ml. On the basis of this result, we adopted this dose of nanoparticles for in vivo studies to ensure the inhibitory effect of the nanoparticles on the proliferation of 4T1 cells.
Next, we used an in vivo tumor model to assess the therapeutic effect of these nanoparticles. The model was established by inoculating rodent 4T1 breast cancer cells into the right flanks of immunocompetent BALB/c mice. We observed that tumor growth was significantly inhibited only in mice treated with n-nHA, n-gHA, and n-SiO2 (Fig. 1, A to C, and fig. S6, A to C). However, the tumor inhibition effect of n-CaCO3, n-TiO2, and n-Ag was not as prominent as that detected in the cell coculture experiment. This great discrepancy in antitumor effect between in vitro and in vivo studies can be attributed to the involvement of the animal immune system. Furthermore, unlimited nutrients and more adhesion sites in the body favored tumor growth, and lung metastases were even observed in the n-CaCO3–treated mice, which showed the weakest in situ tumor inhibition effect (fig. S7).
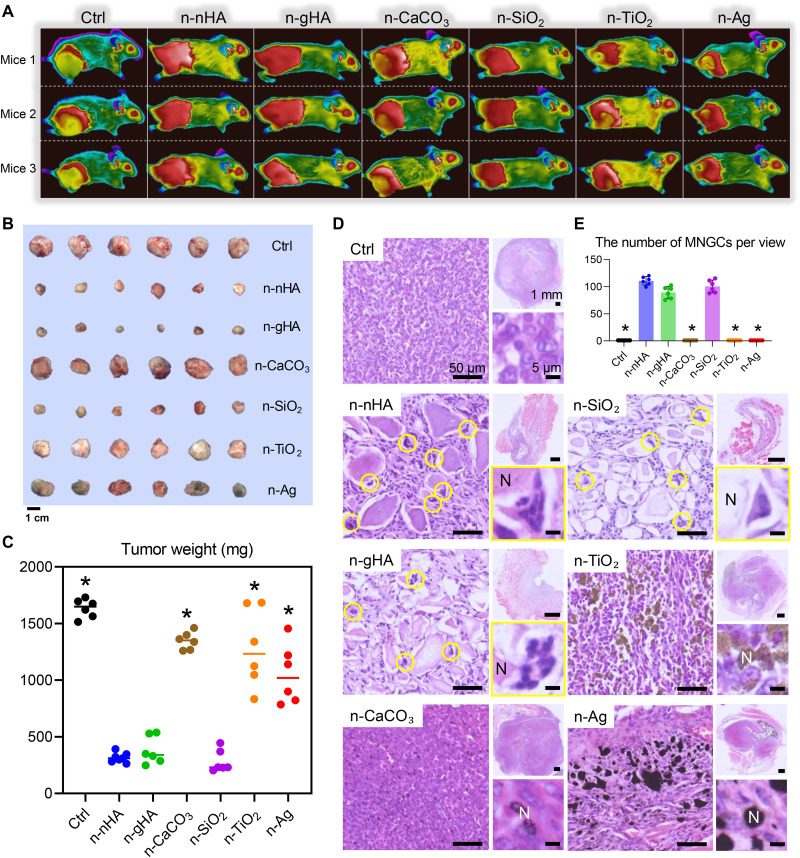
Fig. 1. In vivo antitumor effects of six inorganic nanoparticles.
(A) Thermal images of tumor-bearing mice after a single injection of different inorganic nanoparticles. Three representative mice from each group are shown at day 28. Figure S6A shows mouse thermal images at other time points. Panels B and C of fig. S6 show the measurements of mouse body weight and tumor volumes during the experimental window. (B) Photographs and (C) the measurement of weight of excised tumors at the end of in vivo studies. Scale bar, 1 cm. *P < 0.05, significant difference was assessed by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test compared to n-nHA, n-gHA, and n-SiO2 groups. (D) Representative hematoxylin and eosin (H&E) staining images of tumor tissues excised at day 28. Top-right images show tumor tissues at low magnification. Scale bars, 1 mm. Left images show the appearance of nanoparticle aggregates in tumors. Scale bars, 50 μm. Bottom-right images show the highly magnified cells surrounding the nanoparticle aggregates. Scale bars, 5 μm. N, aggregates of nanoparticles. Circles indicate the MNGCs. (E) Average number of MNGCs counted from 100-fold histology images of tumors representative of three independent mice. *P < 0.05, significant difference was assessed by one-way ANOVA followed by Tukey’s post hoc test compared to n-nHA, n-gHA, and n-SiO2 groups. Ctrl, control.
Nanoparticles encouraged MNGC formation and macrophage depletion
Within tumor tissues treated with the antitumor nanoparticles n-nHA, n-gHA, and n-SiO2, we detected a remarkable population of MNGCs. They were identified by hematoxylin and eosin (H&E) staining and later confirmed by CD86high, CD206low, and TRAPhigh immunohistochemistry (IHC) staining (Fig. 1D and fig. S8) (30). MNGCs are derived from monocyte-macrophage lineage cell fusion for phagocytosis and removal of large foreign particles, which individual macrophages cannot eliminate (31). MNGCs only accumulated around the antitumor nanoparticle aggregates, not in n-CaCO3–, n-TiO2–, and n-Ag–treated tumors (Fig. 1E). Despite ultrasonic dispersion of the material before injection, the nanoparticles still formed “soft” aggregates in the animal body, which is due to the low zeta potential of the particles (fig. S4). The higher the absolute value of zeta potential, the more stable the system. When the zeta potential is low, the attraction usually exceeds the repulsive force and the colloid tends to condense. The zeta potential of the nanoparticles involved in this study in a liquid environment is around −15 mV. Thus, the system is in an unstable state that is prone to aggregation with a bigger hydrodynamic size than primary size. This phenomenon of considerable MNGC generation around the nanoparticle aggregates demonstrated that n-nHA, n-gHA, and n-SiO2 might induce an alteration of immune constitution at least within the TME. To examine the extent and impact of this alteration, we analyzed the hemogram of the peripheral blood of tumor-bearing mice treated with nanoparticles. In the groups with tumor progression—the n-CaCO3, n-TiO2, and n-Ag groups—the portion of leukocytes was abnormally high, which was in line with the results of breast cancer studies (32). However, n-nHA, n-gHA, and n-SiO2 partially restored this cancer indicator back to a normal level (fig. S9A). Similarly, we observed a healthy sized spleen in the groups treated with antitumor nanoparticles (fig. S9B). Immune perturbations occur with tumors (33). An enlarged spleen is always seen in preclinical solid tumor models, which indicates an impaired peripheral immune system due to tumor-driven hematopoiesis (34, 35). However, our findings suggested that the antitumor nanoparticles can revert the tumor subverted systematic immunity back to a normal status.
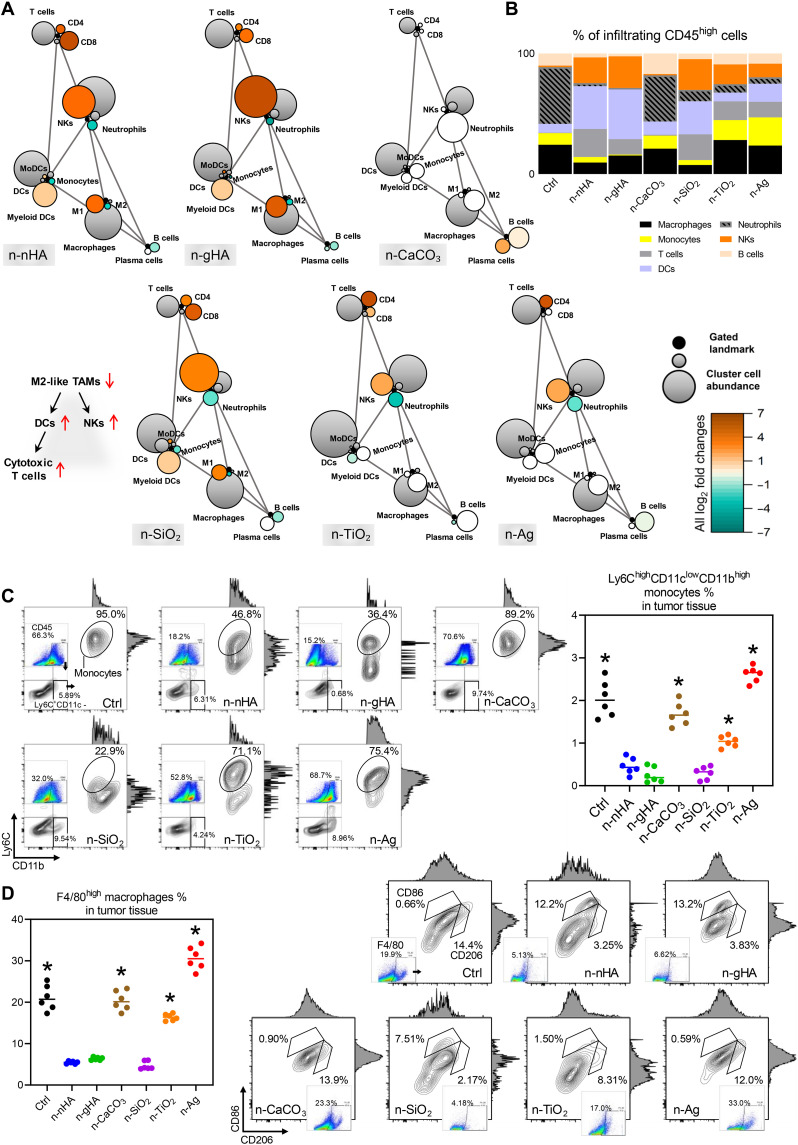
Next, we carried out flow cytometry analyses on different types of immune cells to investigate to what extent these inorganic nanoparticles might alter the TME. We found that the global immune constitution in tumors varied with the types of nanoparticles used (Fig. 2, A to C, and figs. S10 to S15). In the antitumor groups—the n-nHA, n-gHA, and n-SiO2 groups—minimal frequencies of monocytes (Ly6ChighCD11clowCD11bhigh) and macrophages (F4/80high) were generally detected (Fig. 2C). Furthermore, a specific decrease in CD206high tumor-associated macrophages (TAMs) was found preferentially in the TME of tumors treated with these antitumor nanoparticles (Fig. 2D). TAMs are M2-like macrophages that can engineer an immunosuppressive TME, triggering tumoricidal immunocyte dysfunction to promote tumor growth and metastasis. Furthermore, depleting TAMs is a pursuit of modern immune therapy to limit tumor development (36). In the antitumor nanoparticle groups, the evident decrease in monocytes and macrophages observed by flow cytometry coincided with the considerable MNGC formation visualized by histological staining. This finding suggests that antitumor nanoparticles can reduce TAM infiltration by promoting macrophage fusion into MNGCs. In addition, more T cells (CD8high), natural killer cells (NKs; CD45highNK1.1high), myeloid dendritic cells (DCs; Ly6ClowCD11chigh), and monocyte-derived DCs (Ly6ChighCD11chighCD11bhigh) were discovered in the antitumor groups. This finding is consistent with the observation in TAM exhaustion chemotherapies, which result in DC maturation and T cell and NK activation (37, 38). Meanwhile, there was a dense infiltration of neutrophils (CD45highLy6Ghigh) in the TME of the control and n-CaCO3–treated groups, which highly correlated with tumor promotion and metastasis, in line with the observed lung metastasis (fig. S7) (39). Together, the above findings revealed that n-nHA, n-gHA, and n-SiO2 might positively change the immune constitution of the TME, encouraging MNGC formation to exhaust macrophages, the source of TAMs.
Fig. 2. Remodeled tumor immune landscape evoked by nanoparticles.
(A) Statistical scaffold maps of tumor immune frequencies relative to the negative Ctrl group with no nanoparticle treatment. Nodes in black reflect cell populations distinguished manually, and nodes with other colors denote unsupervised clustering of immunocytes. The size of the node indicates the abundance of the cell subset in the parent immunocyte. The grade of coloring indicates the log2 fold change: Orange color represents significantly higher population frequency. Green color represents significantly lower frequency. White color represents cell frequency with no significant difference compared to the Ctrl group. (B) Composition of immunocyte infiltrations in the TME with manual gating. Representative scatter plots and quantification of (C) Ly6ChighCD11clowCD11bhigh monocytes and (D) F4/80high macrophages in the tumor masses at day 28. *P < 0.05, compared to the n-nHA, n-gHA, and n-SiO2 groups.
Antitumor nanoparticles elevated STXBP6 expression in MNGCs
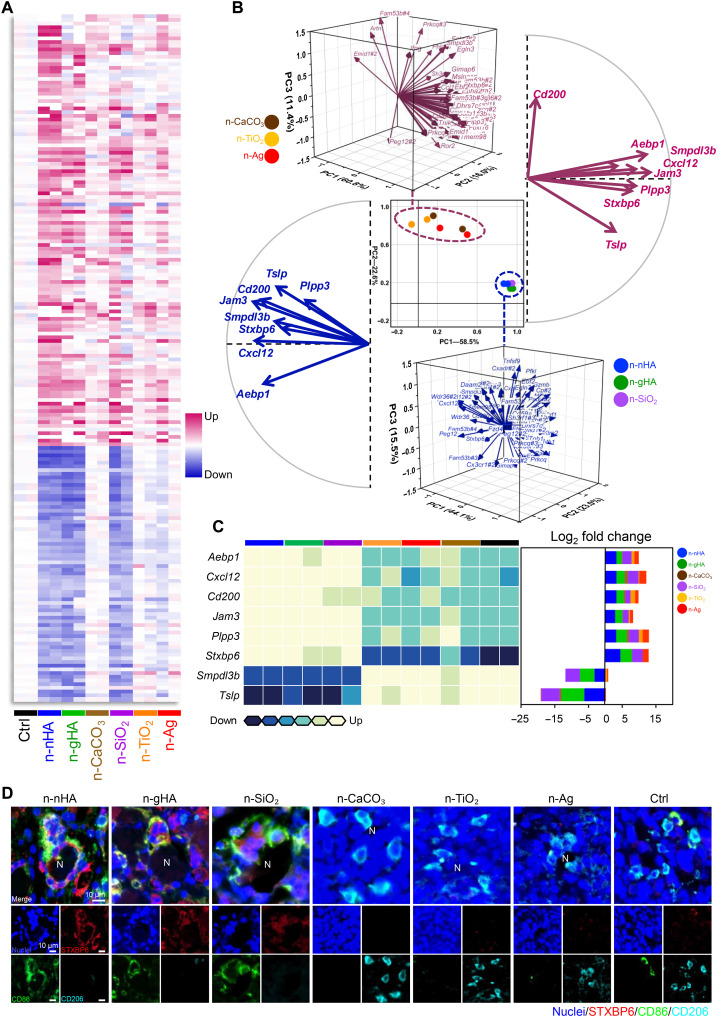
We then tried to decipher the mechanisms that trigger the formation of MNGCs preferentially in the n-nHA–, n-gHA–, and n-SiO2–treated TME. The transcriptional landscape of tumor tissue from all six nanoparticle groups was profiled with a gene microarray (Fig. 3A). Hundreds of differentially expressed genes were discovered between antitumor nanoparticles and their non-antitumor peers (P < 0.05, fold change > 2), indicating profound modulation at the transcriptional level in the TME (fig. S16, A and B). We identified 62 enriched genes overlapping among the n-nHA, n-gHA, and n-SiO2 groups compared with the control tumor-bearing mice (P < 0.05, fold change > 5; fig. S17). These 62 genes are all immune related and reported to be important for extravasation, migration, and adhesion of leukocytes (Jam3, Cx3cr1, and Cx3cl1); functional regulation of the mononuclear phagocyte system (Aebp1, Mgll, Ackr2, Padi4, Smpdl3b, and Ahr); development, differentiation, and activation of T and B cells (Cxcl12, Cd200, Prkcq, Tmem98, Satb1, Cd59a, Lmcd1, Fbxl16, Tslp, Cxadr, Il12rb1, Tnfsf9, Sh3rf1, Phlda1, Wdr36, Batf2, and Ifng); and autophagosome formation (Stxbp6, Dhrs7c, and Synpo2). Principal components analysis (PCA) was performed to group these genes. PCA confirmed that these enriched genes can perfectly discriminate the tumor inhibition ability of these nanoparticles. In Fig. 3B (middle), the antitumor groups—n-nHA, n-gHA, and n-SiO2—spontaneously clustered together, separating from the non-antitumor groups, n-CaCO3, n-TiO2, and n-Ag. The contributing features of these genes are shown in the three-dimensional vector plots of Fig. 3B. We found eight genes with opposite expression regulation between the antitumor and non-antitumor groups, including Aebp1, Cxcl12, Cd200, Jam3, Plpp3, Stxbp6, Tslp, and Smpdl3b (Fig. 3C).
Fig. 3. Transcriptional analysis for screening key genes in the nanoparticle-mediated antitumor immune response.
(A) Heatmap of immune-associated genes differentially expressed in tumors of the n-nHA group compared to the Ctrl group. Tumors were excised at day 28 after nanoparticle treatment. Fold change > 5. P < 0.05. (B) PCA of overlapping genes differentially expressed among the antitumor groups, i.e., the n-nHA, n-gHA, and n-SiO2 groups, compared to the Ctrl group. The scatter plot (center) with principal components (PCs) 1 and 2 shows the variation in gene profiles among mice treated with different nanoparticles. Three-dimensional vector plots in blue and pink show the gene contributions to PCs 1 and 2 of the antitumor groups and other ineffective groups, respectively. Two-dimensional vector plots in blue and pink show the eight genes with reversed contributions between the antitumor and ineffective groups, respectively. (C) Heatmap (left) of the eight genes with reversed contributions between the antitumor and ineffective groups. Row normalized to each gene’s maximum positive expression. Stacked bar plot (right) of the log2 fold change in gene expression relative to the Ctrl, colored by nanoparticles. (D) Representative IF images of tumor tissues treated with different nanoparticles. Tumors were collected at day 28 after nanoparticle treatment. Red, STXBP6; green, CD86; cyan, CD206; blue, nuclei [4′,6-diamidino-2-phenylindole (DAPI)]. Scale bars, 10 μm.
We further performed quantitative real-time polymerase chain reaction (qPCR) to validate the expression of the eight identified genes. With specially designed primers, only five genes presented consistent expression results by gene microarray analysis, Cxcl12, Jam3, Stxbp6, Smpdl3b, and Tslp (fig. S18). Next, we performed IHC staining to confirm and explore the localization of the corresponding proteins of these five genes in the TME. CXCL12, JAM3, SMPDL3B, and TSLP were uniformly expressed in the whole tumor tissues (fig. S19A). Distinctively, syntaxin binding protein 6 (STXBP6) showed MNGC-specific positive staining, and this was further supported by the immunofluorescent (IF) staining finding of colocalization of STXBP6 (red), CD86 (green), and CD206 (cyan), indicating that STXBP6 resided exclusively in the cytoplasm of MNGCs adhered to the antitumor nanoparticles n-nHA, n-gHA, and n-SiO2 (Fig. 3D and fig. S19, A and B). In contrast, only CD206-positive TAMs without STXBP6 staining were observed around the non-antitumor nanoparticles.
To further confirm the presence of STXBP6 in nanoparticle-induced tumor suppression, n-nHA was chosen as the representative nanoparticle to carry out the subsequent experiments. First, RAW 264.7 macrophages, a widely available cell line progenitor of MNGCs, were cocultured with n-nHA for 3 days (fig. S20A). Macrophages in the n-nHA coculture generally showed a larger size with multiple filopodial extensions. These extensions are called fusopods in monocyte-macrophage lineage cell fusion and allow for the contact and fusing of macrophages (40). The increase in cell area and cell number was further quantified (fig. S20B), which indicated a significant difference between n-nHA–treated and nontreated macrophages. As determined by IF staining and qPCR, we found more STXBP6 protein and gene expression in macrophages cocultured with n-nHA. We then used macrophage/n-nHA–conditioned medium (MnCM) for 4T1 tumor cell culture. A marked decrease in proliferation was discovered in 4T1 tumor cells cultured in MnCM compared to those cultured in macrophage-conditioned medium without n-nHA interaction (CM; fig. S20C). These findings revealed that macrophages are one of the target cells responsible for the antitumor ability of the nanoparticles. Upon nanoparticle stimulation, regulation of STXBP6 expression in macrophages is activated. We confirmed that the nanoparticles promoted macrophage fusion, excluding other factors. Biological cues released as a result of nanoparticle-stimulated macrophage fusion are necessary for nanoparticles to exert their tumor-suppressive effect.
Stxbp6 silencing abolished the antitumor effect of n-nHA
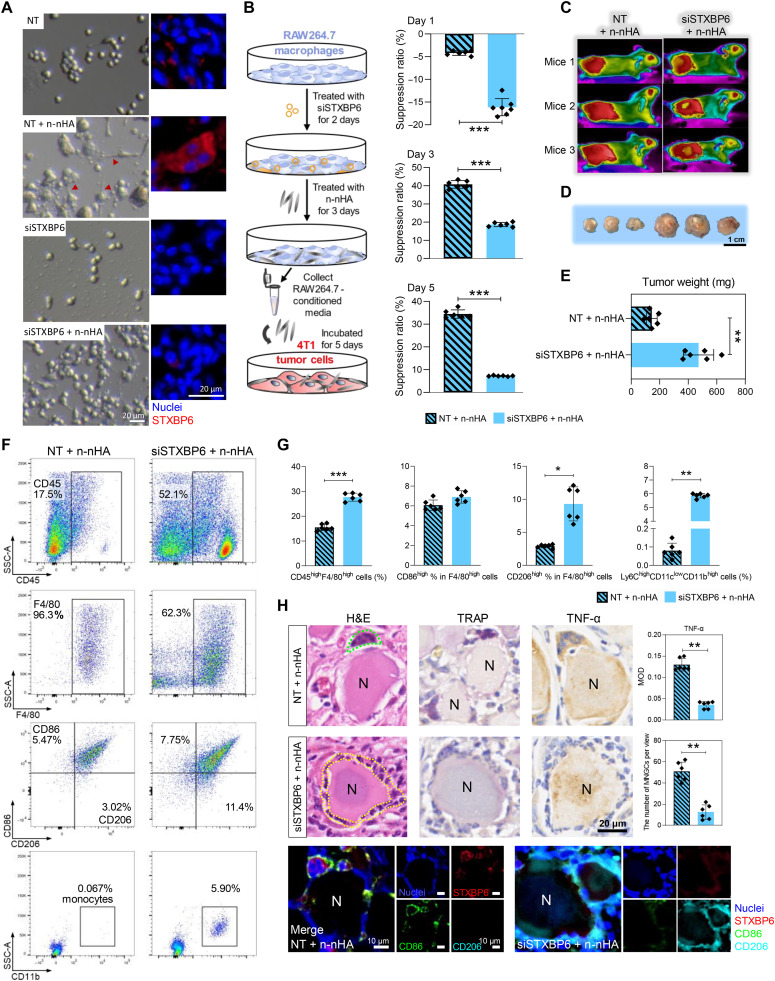
To further validate the role of Stxbp6 in nanoparticle-induced tumor suppression, we used small interfering RNA (siRNA) to silence Stxbp6 expression in vitro and in vivo. Three candidate siRNAs were designed, and the sequence with the maximal knockdown rate (∼70%) was selected (siSTXBP6; fig. S21A). siRNA-mediated gene silencing successfully inhibited Stxbp6 expression in macrophages cocultured with n-nHA (fig. S21B). We observed that siSTXBP6 abolished n-nHA–evoked fusopod formation and cell spreading of macrophages, indicating reduced cell fusion (Fig. 4A and fig. S22). Furthermore, when siSTXBP6 was added to MnCM, the MnCM, its antitumor effect on 4T1 cells diminished (Fig. 4B). These in vitro findings indicate that STXBP6 is indispensable for nanoparticle-guided macrophage fusion and tumor cell suppression effects.
Fig. 4. Stxbp6 knockdown inhibited macrophage fusion to form MNGCs and abolished the antitumor efficacy of n-nHA in vitro and in vivo.
(A) Representative photographs (left) and immunofluorescence (right) of macrophages cultured with no siRNA treatment (NT group), n-nHA but no siRNA treatment (NT + n-nHA group), Stxbp6-silenced treatment (siSTXBP6 group) alone, and n-nHA plus Stxbp6-silenced treatment (siSTXBP6 + n-nHA group). Red, STXBP6; blue, nuclei (DAPI). Scale bars, 20 μm. Figure S22 shows the ratio of cells with fusopods and the quantification of macrophage area and length. (B) Experimental scheme (left) and suppression ratios (right) of tumor cells cultured in MnCM of the NT + n-nHA and siSTXBP6 + n-nHA groups. **P < 0.01 and ***P < 0.001. (C) Thermal images of n-nHA–treated tumor-bearing mice with saline (no siRNA) or siSTXBP6 intratumoral injection twice every 3 days. Three representative mice from each group are shown at day 28. (D) Photographs and (E) the measurement of weight of the excised tumors 28 days after siRNA injection. Scale bar, 1 cm. **P < 0.01. (F) Representative scatter plots and (G) quantification of CD45high lymphocytes, F4/80highCD86high M1-like macrophages, F4/80highCD206high M2-like macrophages, and Ly6ChighCD11clowCD11bhigh monocytes in tumor tissues collected at day 28. *P < 0.05, **P < 0.01, and ***P < 0.001. (H) Representative H&E, IHC, and IF staining images of tumor tissues excised at day 28. The average number of MNGCs counted from 100-fold histology images of tumors representative of three independent mice. IHC staining of TRAP and TNF-α shows the location and cytokine expression of MNGCs. In IF staining, red, STXBP6; green, CD86; cyan, CD206 (cyan); and blue, nuclei (DAPI). Black scale bar, 20 μm. White scale bars, 10 μm. **P < 0.01.
Next, we performed in vivo experiments to examine whether Stxbp6 silencing in the TME hampers nanoparticle-mediated tumor suppression. To test this hypothesis, the siRNA selected previously was methylated and modified with cholesterol to increase its half-life and stability and prevent it from being degraded by nuclease in vivo. Tumor-bearing BALB/c mice received either n-nHA treatment or n-nHA treatment plus siSTXBP6 administration (Fig. 4, C to E). After 28 days, we found that the addition of siSTXBP6 abrogated the in vivo antitumor efficacy of n-nHA. We further examined the potential immune constitution change in the TME with STXBP6 knockdown. A significant increase in the frequencies of monocytes and macrophages was detected in the TME with the addition of siSTXBP6 compared to n-nHA administration alone (Fig. 4, F and G, and fig. S23). This finding reveals that the n-nHA–promoted monocyte-macrophage lineage depletion was eliminated by the knockdown of STXBP6. Consequently, in the absence of the depletion effect, TAMs (CD206high) reoccurred in the siSTXBP6-treated TME. Because MNGCs cannot be detected by flow cytometry, we used H&E, IHC, and IF staining to further observe the changes in MNGCs (Fig. 4H). We observed that siSTXBP6 treatment eliminated 78% of MNGCs that previously attached to the n-nHA aggregates (P < 0.01). Instead, more TAMs (cyan) infiltrated the TME. Collectively, Stxbp6 silencing prevented macrophages from fusing into MNGCs, thereby increasing TAM infiltration in tumors and abolishing the tumor inhibition effect of n-nHA.
Autophagy participated in n-nHA–evoked macrophage fusion
We then examined the function of STXBP6 in modulating cellular events. In the literature, STXBP6 is identified as a regulator of autophagosome formation (41, 42). In our microarray analysis, we also found that the genes involved in various stages of autophagosome formation were preferentially up-regulated by the antitumor nanoparticles (fig. S24). Autophagy can be linked to multinuclear cell formation in various ways (43, 44), with one being the facilitation of macrophage phagocytosis by fusion into a larger cell (45). We presumed that the n-nHA–guided macrophage fusion event observed in this study might be related to autophagy. To examine the presence of autophagy, we performed IHC staining and Western blotting of microtubule-associated protein light chain 3 (LC3), one of the critical hallmarks of autophagy (46). We found that LC3 was positively expressed in the MNGC cytoplasm of the n-nHA–treated TME. However, upon the addition of siSTXBP6, no LC3 expression was observed at the same location (fig. S25). Furthermore, we detected considerable autophagosome formation in the MNGC via transmission electron microscopy (fig. S26). Next, we used a molecular promoter [everolimus (EVR)] and inhibitor [3-methyladenine (3-MA)] to intentionally modulate autophagy. In vitro, under autophagy-inhibited conditions (AUT−/n-nHA), even with the addition of n-nHA, we found that only a few macrophage fusion events occurred, reflected by a decrease in fusopods and cell area, compared to the pure n-nHA–treated group (blank/n-nHA). In contrast, n-nHA–guided macrophage fusion was further accentuated under autophagy-promoting conditions (AUT+/n-nHA; fig. S27, A to C). These two macrophage-conditioned media with autophagy modulation were then collected for culturing of 4T1 tumor cells. We found that AUT−/n-nHA conditioned medium completely lost its antitumor effect (fig. S27D).
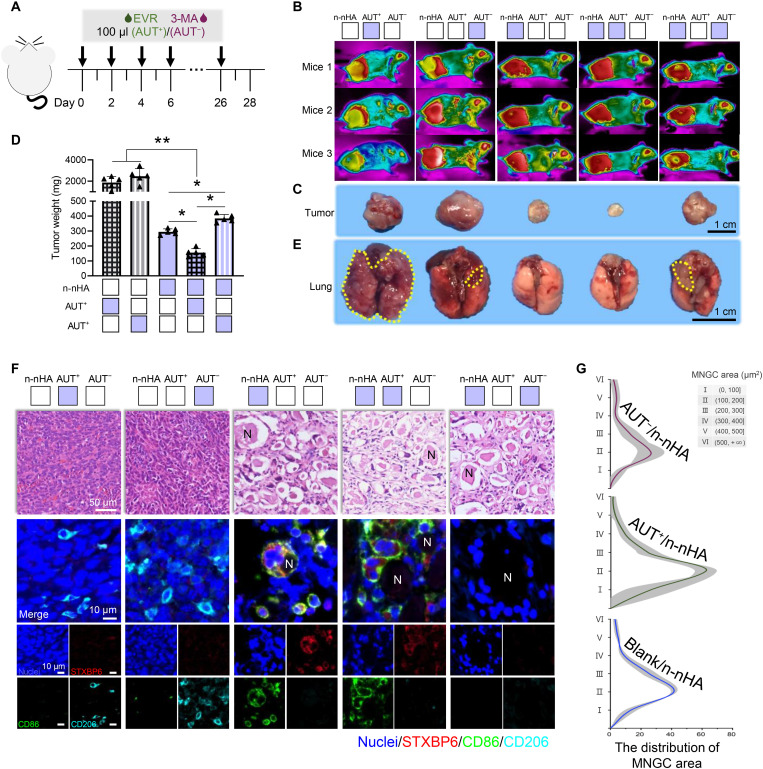
On the basis of the above prominent in vitro findings, we further investigated the role of autophagy in n-nHA–guided MNGC formation in vivo. Every 2 days, autophagy interferents together with n-nHA were intratumorally administered to the 4T1 model (Fig. 5A). Without n-nHA, neither autophagy promotion nor inhibition alone had any effect on solid tumor suppression (Fig. 5, B to D). Within the nanoparticle-treated groups, tumor weight and size were in the order of AUT+/n-nHA < n-nHA < AUT−/n-nHA. The in vivo inhibition of autophagy partially impaired the antitumor ability of n-nHA. Notably, the autophagy promoter and n-nHA together presented a synergistic suppressive effect on tumors. Grayish-white tumor masses showing almost no vascularization were harvested in the AUT+/n-nHA group. Accordingly, lung metastases were found in the mice treated with AUT−/n-nHA and in the mice administered autophagy modulation agents only (Fig. 5E and fig. S29). In H&E and IF colocalization staining (Fig. 5, F and G), we further proved that n-nHA–induced MNGC formation is highly autophagy dependent. Larger MNGCs were present in the AUT+/n-nHA group, while smaller MNGCs were observed in the AUT−/n-nHA group. Together, these results confirmed that autophagy is important for antitumor nanoparticle-induced MNGC formation in the TME.
Fig. 5. STXBP6-regulated autophagy drives MNGC generation.
(A) Experimental timeline and treatments of the in vivo study. BALB/c mice were inoculated with 4T1 cells and n-nHA on day 0. The tumor-bearing mice were then intratumorally injected with EVR or 3-MA every other day until day 28. (B) Thermal images of n-nHA–treated tumor-bearing mice with EVR or 3-MA injection for autophagy promotion (AUT+) and inhibition (AUT−), respectively. Tumor-bearing mice treated with EVR or 3-MA alone were used as Ctrl groups. Three representative mice from each group are shown at day 28. (C) Photographs and (D) the measurement of weight of the excised tumors at day 28. Scale bar, 1 cm. *P < 0.05 and **P < 0.01. (E) Representative lung images of different groups. Lung metastases are indicated within yellow dotted lines. Scale bar, 1 cm. Figure S29 shows the corresponding H&E staining images of lung tissues. (F) Representative H&E (top row) and IF staining images (bottom rows) of tumor tissues excised at day 28. Scale bar in H&E staining, 50 μm. In IF staining, red, STXBPT; green, CD86; cyan, CD206; and blue, nuclei (DAPI). Scale bars in IF staining, 10 μm. (G) The distribution of the MNGC area counted from 100-fold histology images of tumors representative of three independent mice. Blank/n-nHA, n-nHA treatment with vehicle phosphate-buffered saline (PBS) injection; AUT+/n-nHA, n-nHA treatment with EVR injection; AUT−/n-nHA, n-nHA treatment with 3-MA injection.
DISCUSSION
Several lines of clinical and preclinical evidence reveal that emerging nanotechnologies, especially polymeric nanoparticle-based anticancer agents, have encouraging prospects in cancer therapy. However, these nanomedicines have merely benefited a minority of patients and moderately contributed to individual survivorship (6, 9). Recently, inorganic nanoparticles commonly used as auxiliary agents, i.e., drug or gene carriers and photothermal or radiation signal enhancers in cancer therapies, have been confirmed to have an innate antitumor capability for a variety of cell lines, including osteosarcoma, breast cancer, melanoma, and glioma (8). Moreover, in our previous study, a large amount of accumulated immunocytes was observed around inorganic nanoparticles preventing tumor progression in vivo (19, 21). However, the relationship between the inorganic nanoparticle-induced immune response and its inherent antitumor ability remains obscure.
Here, we first identified three inorganic nanoparticles with in vivo antitumor ability—n-nHA, n-gHA, and n-SiO2—out of six commonly used biomedical nanoparticles. We observed that these antitumor particles encouraged the fusion of macrophages to form MNGCs in the TME and significantly reduced the infiltration of M2-like TAMs. Previous studies have confirmed that TAMs can induce tumoricidal immunocyte dysfunction and build an immunosuppressive TME to facilitate cancer progression (36). In this study, the immune alteration, depletion of TAMs, triggered by these particles was found to enable the recruitment and activation of cytotoxic T cells and NKs and subsequently reversed the immunosuppressive TME to impair tumor progression. In summary, MNGC generation induced by these three antitumor inorganic nanoparticles not only significantly impeded TAM accumulation but also reverted the tumor-subverted systematic immunity back to a normal status. Meanwhile, this immune war in the local TME could also profoundly reduce lung metastasis and reverse the enlargement of the spleen seen in tumor-bearing individuals.
Furthermore, through bioinformatic analysis, we identified a key protein, STXBP6, which resides exclusively in the cytoplasm of MNGCs. We suspect that it is probably involved in nanoparticle-mediated macrophage fusion. We found that Stxbp6 knockdown prevented macrophages from fusing into MNGCs, even with antitumor nanoparticle interference. Furthermore, knockdown of Stxbp6 increased TAM infiltration in tumors and abolished the tumor-suppressive effect of n-nHA. However, how STXBP6 is involved in regulating MNGC generation is still unknown. Other research groups have demonstrated that STXBP6 is a regulator of autophagosome formation (41, 42). We then tried to explore the role of autophagy in antitumor nanoparticle-induced MNGC formation. Our results demonstrated that the antitumor nanoparticle-treated TME shows up-regulated expression of genes involved in various stages of autophagosome formation. In addition, MNGCs highly expressing STXBP6 have an elevated level of the autophagosome marker LC3 and an ultramicrostructure with substantial autophagosome formation. Autophagy can be linked to multinuclear cell formation in various ways. One is to facilitate macrophage phagocytosis by fusion into a larger cell (43–45). Thus, we presumed that STXBP6 might have promoted the formation of MNGCs by mediating macrophage autophagy. When autophagy was inhibited, even with the addition of n-nHA, we observed a sharp decrease in the formation of macrophage fusopods and cell fusion in vitro. Meanwhile, the in vivo obstruction of autophagy partially impaired the tumor suppression effect of n-nHA. In contrast to autophagy inhibition, autophagy promotion in vivo in combination with n-nHA treatment significantly reduced the infiltration of TAMs, considerably increased MNGC accumulation, and further decreased the size of solid tumors.
Together, we demonstrated that inorganic nanoparticles with inherent antitumor properties can spontaneously remodel the immune constitution of the TME by promoting MNGC formation and TAM depletion to suppress tumor growth (Fig. 6). This is an intriguing and significant cancer-related finding that lays the foundation to provide a novel avenue for nanotechnology applications in tumor therapy. In future investigation, we are preparing an in situ sprayed anti-recurrence gel for post-surgical cancer treatment clinically. The antitumor inorganic nanoparticles and autophagy promotion agents will be dispersed in the fibrin gel and then sprayed and mixed with the thrombin solution in the tumor resection cavity to form an anti-recurrence fibrin gel in situ. Many open questions remain; for example, what factors from the material aspect of inorganic nanoparticles drive STXBP6 overexpression in macrophages and what causes STXBP6-mediated autophagy to preferentially promote cell fusion rather than programmed cell death? Addressing these questions can offer new opportunities to further explore inorganic nanomaterial-mediated TAM depletion for cancer immunotherapy.
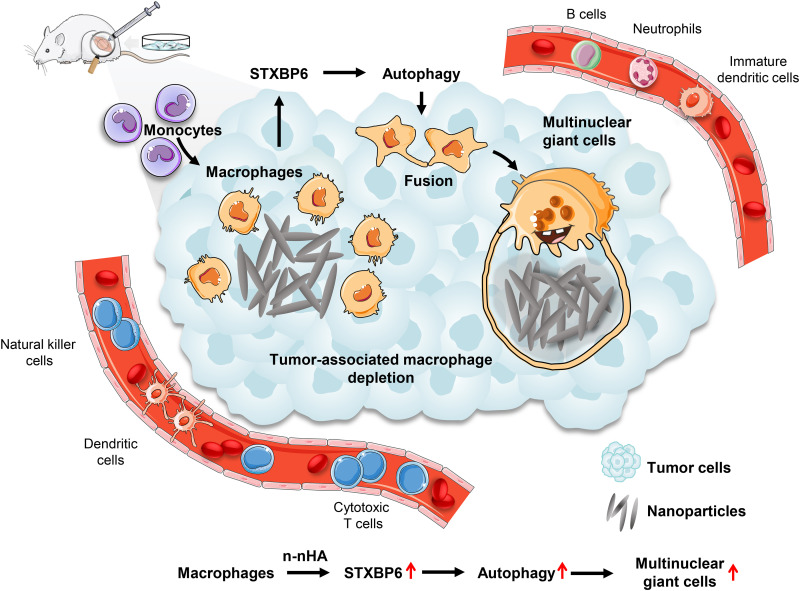
Fig. 6. Schematic illustration of the antitumor immunity evoked by n-nHA.
The intervention of n-nHA countered tumor-mediated inflammation and engineered an antitumor immune microenvironment mitigating tumor growth. n-nHA injected intratumorally into tumor-bearing mice effectively promoted the fusion of macrophages to form substantial MNGCs surrounding the nanoparticle aggregates. This considerable fusion event of macrophages resulted from the elevated expression of STXBP6 induced by n-nHA, followed by the activation of macrophage autophagy. Through the formation of MNGCs, n-nHA induced extensive depletion of tumor-associated macrophages and permitted more tumor-killing immunocytes, including cytotoxic T cells, DCs, and NKs, to infiltrate into the TME, which notably impaired tumor progression.
MATERIALS AND METHODS
Study design
This study focused on the immune responses elicited by inorganic nanoparticles in TME to investigate their inherent antitumor mechanism. We specifically enrolled the nanoparticles already applied in the biomedical field, including n-HA, n-CaCO3, n-SiO2, n-TiO2, and n-Ag. Because of the morphology-dependent antitumor property of n-HA, two n-HAs with different morphologies, n-nHA and granule-shaped n-gHA, were included in the current study. To assess the tumor cell inhibition ability of these nanoparticles excluding host response, rodent 4T1 breast cancer cells were cultured with a wide concentration range (0 to 1000 μg/ml) of nanoparticles in vitro (n = 4 to 8 per group per time point). To investigate the immune response that arose from these inorganic nanoparticles, an in vivo tumor model was established by inoculating 4T1 cells into the right flanks of immunocompetent BALB/c mice, which were then randomly divided into seven groups with intratumoral injection with six different nanoparticles (n = 8). The body weight and tumor volume of each mouse were measured every 2 days. After 4 weeks, the mice were euthanized and tumor weight was quantified. Next, we carried out immunohistochemical and flow cytometry analyses of the excised tumor tissues to explore the inorganic nanoparticle-altered immune constitution of the TME (n = 6). We then tried to decipher the gene level change that triggers the modification of immune composition of the TME. The transcriptional landscape of tumor tissues from all six nanoparticle groups was profiled with a gene microarray. qPCR analysis and IHC staining were used to verify the expression of several identified genes and their proteins (n = 6). On the basis of the results of the above experiments, we identified the role of one critical cell type, MNGC, and an essential protein, STXBP6, involved in the tumor suppression of antitumor nanoparticles. n-nHA was chosen as the representative nanoparticle to carry out the subsequent experiments. RAW 264.7 macrophages, a widely available cell line progenitor of MNGCs, were cocultured with n-nHA (n = 3 to 6). In addition, we used siRNA to silence Stxbp6 expression to validate the role of Stxbp6 in nanoparticle-induced macrophage fusion and antitumor effect (n = 3 to 6). Last, to examine the biological function of STXBP6 in the fusion event of MNGCs, a molecular promoter (EVR) and an inhibitor (3-MA) were used to intentionally modulate autophagy in vivo and in vitro (n = 6). All experiments were repeated at least three times, and experimental findings were reproducible. Cells or mice were randomly assigned to different groups before the nanoparticle treatment. All the investigators were blinded to group assignment in the course of data collection and analysis. No data were excluded.
Preparation and characterization of nanoparticles
n-nHA was prepared via the wet chemical route. Calcium nitrate tetrahydrate (99%; Chengdu Kelong Chemical Reagent Co. Ltd. China) and dibasic ammonium phosphate (99%; Chengdu Kelong Chemical Reagent Co. Ltd. China) were mixed at a Ca/P ratio of 1.67 and agitated at 70°C for 4 hours. Ammonia solution (25 to 28%; Chengdu Kelong Chemical Reagent Co. Ltd. China) was added dropwise to the slurry and dynamically monitored with a pH meter to ensure that the pH of the system was 10. The suspension, aged for 24 hours, was then washed twice with deionized water, followed by filtering to acquire the precipitate. The precipitate was lyophilized for 24 hours and calcinated for 2 hours at 300°C before milling and then passed through a 200-mesh sieve to obtain n-nHA. n-gHA was a gift from Sichuan Baiameng Bioactive Material Co. Ltd. n-CaCO3, n-SiO2, n-TiO2, and n-Ag were obtained from Sigma-Aldrich. These six nanoparticles were sterilized by gamma irradiation before in vitro cell evaluation and in vivo animal experiments. The phase compositions, primary sizes and shapes, characteristic functional groups, hydrodynamic sizes, and zeta potentials of each nanoparticle were determined by x-ray diffraction (D8 Advance, Bruker, Germany), transmission electron microscopy (TEM; Tecnai G2 F30 S-TWIN, FEI, USA), Fourier transform infrared spectroscopy (INVENIO, Bruker, Germany), and dynamic light scattering (Zetasizer Nano ZS, Malvern, UK), respectively. TEM images of more than 50 sharply marginated nanoparticles were measured with Image-Pro Plus 6.0 software to determine the average particle size.
Cell lines
Murine breast cancer 4T1 cells (Chinese Academy of Sciences, Shanghai, China) were cultured in RPMI 1640 medium (Gibco, Thermo Fisher Scientific, China) supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco, Invitrogen, Australia) and 1% (v/v) penicillin-streptomycin cocktail (HyClone, Logan, USA). Mouse RAW 264.7 macrophage cells (Chinese Academy of Sciences, Shanghai, China) were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher Scientific, China) supplemented with 10% (v/v) FBS and 1% (v/v) penicillin-streptomycin cocktail. Cells were maintained in a humidified incubator (Thermo Scientific, Thermo Fisher Scientific, China) with a constant temperature (37°C) and CO2 content (5%).
In vitro evaluation of nanoparticle treatment
4T1 tumor cells cocultured with nanoparticles
To assess the particle cytotoxicity effect, 4T1 cells were seeded in 24-well plates (1 × 104 cells per well) and incubated with different concentrations of nanoparticles (0, 100, 250, 500, and 1000 μg/ml) for 1, 3, and 5 days. A CCK-8 assay (Biosharp, Beijing Labgic Technology Co., Ltd., China) was performed to quantify the survival of these particle-treated tumor cells. Briefly, the medium was gently removed, and the cells were further incubated in RPMI with 10% (v/v) CCK-8 dye for 2 hours at 37°C protected from light. Cell viability was determined by normalizing the optical density (OD) value at 450 nm detected via a microplate reader (Multiskan GO, Thermo Fisher, Thermo Fisher Scientific, USA).
To validate the impact of macrophages on the tumor killing capability of n-nHA, 4T1 cells were cultured in MnCM supplemented with n-nHA (1000 μg/ml). Cell proliferation was examined via CCK-8 assay. The suppression ratio was determined as follows: suppression ratio = (Awo − Aw)/(Awo − Ab) × 100% (Aw, absorbance of the group with n-nHA treatment; Awo, absorbance of the group without n-nHA treatment; and Ab, absorbance of the blank group).
RAW 264.7 macrophages cocultured with n-nHA
RAW 264.7 cells (1 × 105 per well) were seeded in 24-well plates, incubated overnight, and treated with n-nHA (1000 μg/ml) for 3 days. MnCM was collected for the in vitro antitumor assay of n-nHA. qPCR analysis was conducted to assess Stxbp6 expression in n-nHA–treated macrophages. The primer sequences designed to evaluate the gene expression levels were as follows: Gapdhmouse, 5′-CCT CGT CCC GTA GAC AAA ATG-3′ (forward) and 5′-TGA GGT CAA TGA AGG GGT CGT-3′ (reverse) and Stxbp6mouse, 5′-CAG GCA TCT ATC ACG AAG GTC A-3′ (forward) and 5′- GAA TCC CGA TTA GGA TCA ATA CC-3′ (reverse).
For Stxbp6 knockdown, 1 × 105 RAW 264.7 macrophages were transfected with three candidate siRNA sequences (RiboBio, Guangzhou, China) at a concentration of 50 nM in 24-well plates following the operation manual. Two days later, transfected cells were collected, and gene expression was detected using qPCR analysis to identify the optimal siRNA for Stxbp6 knockdown in vitro. Then, these Stxbp6-silenced macrophages were incubated with n-nHA for 3 days to obtain conditioned medium (siSTXBP6-MnCM) for the antitumor assay in vitro (fig. S22). Macrophages cultured with negative siRNA (siNEG) and no siRNA treatment (NT) served as control groups. The three candidate siRNA sequences were CCATTCAGCAGCTGATAGT (siRNA#1), GAACAGTGCCCAGCAGTTT (siRNA#2), and AGCAGTTATTACAAGACGT (siRNA#3).
To evaluate the impact of autophagy on macrophage fusion, EVR (0.1 μg/ml; HY-10218, MedChemExpress, USA) or 3-MA (0.75 mg/ml; HY-19312, MedChemExpress, USA) suspended in phosphate-buffered saline (PBS) was added to the macrophage/n-nHA coculture system to promote or inhibit autophagy, respectively. Likewise, the conditioned medium was collected for the antitumor assay in vitro (blank, vehicle PBS treatment; AUT+, EVR treatment; and AUT−, 3-MA treatment.
To observe these n-nHA–treated macrophages, trypan blue staining and IF staining of STXBP6 were performed. Images were obtained using a stereo microscope (ZEISS, Germany) and an inverted fluorescence microscope (DMi8, Leica, Germany). Moreover, the ratio of cells with fusopods, macrophage area, and length were quantified.
Animals
All animal experiments (ethical approval code KS2020308) were performed according to the procedure of the Institutional Animal Care and Use Committee of Sichuan University (Chengdu, China). Female 4-week-old BALB/c mice were purchased from Chongqing Ensiweier Biotechnology Co. Ltd. (China) and fed in cages with sufficient food and water. All in vivo experiments were conducted after 1 week of acclimatization of the animals.
In vivo evaluation of nanoparticle treatment
The tumor model used in this study was established by injecting 1 × 106 4T1 cells suspended in 30 μl of Matrigel Matrix (5 mg/ml; Corning, USA) into the right flank of the mouse. To investigate the antitumor efficacy of six nanoparticles, 56 BALB/c mice with tumor implantation were randomly divided into seven groups (Ctrl, n-nHA, n-gHA, n-CaCO3, n-SiO2, n-TiO2, and n-Ag groups; eight mice per group) and intratumorally injected with nanoparticles suspended in 20 μl of Matrigel Matrix.
To examine the tumor suppression ability of n-nHA in a Stxbp6-silenced environment, six tumor-bearing mice treated with n-nHA were injected with 5 nmol siSTXBP6 (siSTXBP6 + n-nHA) twice a week for 4 weeks. As a positive control, another six tumor-bearing mice were injected with n-nHA alone with no siRNA treatment (NT + n-nHA).
To evaluate the effect of autophagy on the antitumor capability of n-nHA, 15 tumor-bearing mice treated with n-nHA were injected with 0.1 ml of EVR (1 mg/kg), 3-MA (15 mg/kg), or vehicle PBS (Fig. 6A). Ten tumor-bearing mice injected with EVR or 3-MA alone without n-nHA administration served as the control groups.
The body weight and tumor volume of each mouse were measured every 2 days for 4 weeks, and the tumor volume (in cm3) was acquired on the basis of the formula: V = 0.5 × (length × width2). A thermal imaging camera (FLIR T460, Teledyne FLIR, USA) was used to monitor the expansion of the tumor masses weekly. After 4 weeks, the mice were killed by intraperitoneal injection with an overdose of pentobarbital, and mouse blood, tumor, spleen, and lung samples were harvested.
Histological analyses
For H&E staining, tumor and lung tissue samples resected at week 4 were fixed in 4% formaldehyde for more than 48 hours, dehydrated with graded concentrations of ethanol (50, 70, 80, 90, and 100%), and then embedded in paraffin. Each paraffin-embedded specimen was sectioned into 5-μm-thick sections, dried in an oven at 45°C for 24 hours, and stained with H&E. Panoramic images of these histology samples were obtained and analyzed with a scanner (WS-10, Wisleap, China).
For IHC analysis, 5-μm tumor sections were immersed in xylene, dewaxed, and washed with graded ethanol (50, 70, 80, 90, and 100%). The sections were placed into ethylene diamine tetraacetic acid solution at pH 9.0 for antigen retrieval, followed by endogenous peroxidase blockage using 3.0% hydrogen peroxide solution at room temperature for 25 min. Blocking buffer, 5% bovine serum albumin (A8010, Solarbio, China), was applied to prevent the nonspecific binding of antibodies. The sections were then incubated with primary antibodies (anti-CD86, anti-CD206, anti-TRAP, anti-JAM3, anti-STXBP6, anti-TSLP, anti-CXCL12, and anti-SMPDL3B; 1:200 dilution) at 4°C for 12 hours, washed three times with PBS solution, and stained with secondary antibodies (Dako, Denmark) for 1 hour at room temperature. A diaminobenzidine substrate kit (DA1010-10, Solarbio, China) was used for the chromogenic reaction. The nuclei were counterstained with hematoxylin (517-28-2, Solarbio, China) for 3 min. A slide scanner (WS-10, Wisleap, China) and Image-Pro Plus 6.0 software were used for imaging and quantification, respectively. The antibodies used for IHC were anti-mouse CD86 (NBP2-25208, Novus, USA), anti-rabbit CD206 (AF2535, Novus, USA), anti-TRAP (387A-1KT, Sigma-Aldrich, USA), anti-goat JAM3 (AF1213, Novus, USA), anti-goat STXBP6 (NB100-1227, Novus, USA), anti-rabbit TSLP (ab188766, Abcam, UK), anti-mouse CXCL12 (MAB350-100, Novus, USA), and anti-mouse SMPDL3B (sc-137113, Santa, USA).
For IF staining, tumor sections dried at room temperature were washed with PBS three times and blocked in donkey serum at 37°C for 30 min. These specimens were incubated with primary antibodies diluted in donkey serum in PBS at 4°C for 20 hours. Then, the slides were washed with PBS three times and incubated with secondary antibodies at room temperature for 1 hour. Again, these slides were washed with PBS three times. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. IF micrographs were obtained using a SLIDEVIEW (VS200, Olympus, Japan). The following primary antibodies were used: anti-goat STXBP6, anti-mouse CD86, and anti-rabbit CD206.
Flow cytometry
Tumor tissues freshly resected at week 4 were immersed in a solution mixed with 15% (w/v) hyaluronidase grade I (1141MG100, BioFroxx, Germany) and 75% (w/v) collagenase IV (2091MG100, BioFroxx, Germany) at 37°C for 45 min to hydrolyze the extracellular matrix. The tissue fragments were filtered with 70-μm nylon cell strainers (352350, FALCON, Corning, USA) to obtain a single-cell suspension, followed by centrifugation to collect the lower cell extract. The extraction was incubated with 1 ml of red cell lysis solution (BL503A, Biosharp, Beijing Labgic Technology Co. Ltd., China) for 2 to 3 min. Cells were added to 9 ml of PBS and centrifuged to remove the redundant red cells. The collected cells were incubated with flow cytometry staining buffer (00-4222-26, eBioscience, Invitrogen, USA) containing FBS for 5 min to minimize the nonspecific binding of antibodies. The cells were then incubated with the antibodies for 30 min and protected from exposure to light. Flow cytometry (BD LSRFortessa, USA) was used to identify immune cell subsets, and FlowJo v.10.6.2 was used to analyze the data. Moreover, a statistical scaffold map was introduced to further visualize the classifications and frequencies of immune cell clusters. Frequencies of immune cell subsets were calculated as a proportion of the parent immune subset, and fold changes were depicted with a double-gradient spectrum. All results presented in scaffold maps were relative to the negative control group with no nanoparticle treatment. Nodes in black denote cell populations distinguished manually, and nodes with other colors represent unsupervised clustering of lymphocytes. The grade of coloring indicated the log2 fold change: Orange reflected a higher population frequency relative to the Ctrl, and green reflected a lower population frequency relative to the Ctrl (*P < 0.05, **P < 0.01, and ***P < 0.001). The size of the node indicates the abundance of the cell subset in the parent immunocyte. The antibodies used for flow cytometry were as follows: BrilliantViolet421 DAPI (catalog no. 564907, BD Pharmingen, USA), FITC CD45 (clone 30-F11; catalog no. 553079, BD Pharmingen, USA), PE NK1.1 (clone PK136; catalog no. 108707, BioLegend, USA), APC Ly-6G (clone 1A8; catalog no. 560599, BD Pharmingen, USA), PE CD19 (clone 1D3; catalog no. 557399, BioLegend, USA), APC CD45R/B220 (clone RA3-6B2; catalog no. 553092, BD Pharmingen, USA), FITC CD3 (clone 145-2C11; catalog no. 553061, BD Pharmingen, USA), APC CD4 (clone RM4-5; catalog no. 553051, BD Pharmingen, USA), PE CD8 (clone 53-6.7; catalog no. 553032, BD Pharmingen, USA), PE-Cy7 CD86 (clone GL-1; catalog no. 105014, BioLegend, USA), APC CD206 (clone MR6F3; catalog no. 17-2061-82, eBioscience, USA), PE F4/80 (clone T45-2342; catalog no. 565410, BD Pharmingen, USA), FITC CD11b (clone M1/70; catalog no. 101206, BioLegend, USA), APC CD11c (clone N418; catalog no. 117310, BioLegend, USA), and PE Ly-6C (clone HK1.4; catalog no. 128007, BioLegend, USA).
Whole mouse genome oligo microarrays
Freshly harvested tumor tissues were quickly frozen with liquid nitrogen, cryopulverized (BioPulverizer, BioSpec, USA), and homogenized (MiniBeadbeater-16, BioSpec, USA). To dissociate the nucleoprotein complexes thoroughly, the homogenate was incubated for 5 min at room temperature. Following chloroform addition and centrifugation, the mixture separated, and RNA remained predominantly in the colorless upper aqueous phase. The aqueous phase was mixed with isopropyl alcohol to precipitate the RNA. An RNeasy Mini Kit (QIAGEN 74104, QIAGEN, Germany) was used for RNA extraction, quantification, and quality control. RNA hybridization was conducted by the Gene Expression Hybridization Kit (Agilent 5188-5242, Agilent, USA) at 65°C for 17 hours after purifying the labeled/amplified RNA. The fluorescence intensity of the microarray read by a microarray scanner (GenePix 4000B, Molecular Devices, USA) was converted into digital data for storage and analyzed with the supporting software. For gene expression levels, differences with fold changes greater than 2 and P values less than 0.05 were considered statistically significant. To investigate the variations in the overlapping gene expression induced by different nanoparticles treated in vivo, PCA was applied for in-depth assessment and graphical visualization of the data.
qPCR
To further accurately estimate the effect of the selected genes on tumor suppression, real-time qPCR was applied to assess gene expression at the mRNA level. The resected tumor samples were frozen with liquid nitrogen, and then, the total RNA was extracted with an RNeasy Mini Kit (QIAGEN 74104, QIAGEN, Germany) on the basis of the manufacturer’s recommendations. A spectrophotometer (ND-1000, NanoDrop, USA) was used to qualify and quantify the total RNA. The extracted RNA was reverse-transcribed into cDNA with a Reverse Transcriptase kit (SuperScript III, Invitrogen, USA) and a PCR system (GeneAmp 9700, Applied Biosystems, USA). Then, a real-time PCR system (ViiA 7, Applied Biosystems, USA) was used to conduct qPCR. The primer sequences designed to evaluate the gene expression levels are as follows: Gapdhmouse, 5′-CAC TGA GCA AGA GAG GCC CTA T-3′ (forward) and 5′-GCA GCG AAC TTT ATT GAT GGT ATT-3′ (reverse); Cd200mouse, 5′-CTC TCC ACC TAC AGC CTG ATT-3′ (forward) and 5′-AGA ACA TCG TAA GGA TGC AGT TG-3′ (reverse); Aebp1mouse, 5′-TTG GAA ACG CTG GAT CGG TTA-3′ (forward) and 5′-CTT GAC CTT GCC AGG CAT TT-3′ (reverse); Jam3mouse, 5′-TCT CAA ATC CAG CAA CCG AAA C-3′ (forward) and 5′-GTC CGT AAT GAT GCA AGA CAA T-3′ (reverse); Plpp3mouse, 5′-GCA TCA AGT ATC CCC TGA AAG TC-3′ (forward) and 5′-CAT ACG GGT TCT GAG TGG TGG A-3′ (reverse); Stxbp6mouse, 5′-CTC TTG ATG AAA GAA TGC TGG GA-3′ (forward) and 5′-TGA CCT TCG TGA TAG ATG CCT-3′ (reverse); Smpdl3bmouse, 5′-CAG GGG CTC AAC TAG GGA G-3′ (forward) and 5′-GGG CCA GCA TTT AGC ACA G-3′ (reverse); Cxcl12mouse, 5′-ACC TCG GTG TCC TCT TGC TG-3′ (forward) and 5′-CGT TGG CTC TGG CGA TGT G-3′ (reverse); and Tslpmouse, 5′-CAG GCG ACA GCA TGG TTC TT-3′ (forward) and 5′-GGC AGC CAG GGA TAG GAT TG-3′ (reverse).
Statistical analysis
Each experiment was conducted in at least three replicates. Statistical analysis was performed using Origin 2021b software, and all data are expressed as the means ± SE. One-way analysis of variance (ANOVA) with Tukey’s correction was carried out for comparisons with more than two groups. Student’s t test was used for comparisons between two groups. A P < 0.05 was considered statistically significant.
Acknowledgments
We thank L. Chen from the Analytical & Testing Center of Sichuan University for assistance with imaging technique.
Funding: This work was supported by the National Key Research and Development Program of China (2022YFC2409705), National Natural Science Foundation of China (grant no. 81971755), and Sichuan Science and Technology Program (2020YFS0038).
Author contributions: X.Y., X. Zhu, and X. Zhang conceived and guided the study. X.Y. and S.C. designed the experiments and provided insight into the final interpretation of the findings. S.C. performed the experiments and conducted data analysis. Z.X., M.G., and R.Z. helped in animal studies and sample collection. X.Y. and S.C. co-wrote the manuscript. All authors participated in result discussion, provided feedback, and helped optimize the research.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S29
Table S1
REFERENCES AND NOTES
- 1.Miller K. D., Nogueira L., Mariotto A. B., Rowland J. H., Yabroff K. R., Alfano C. M., Jemal A., Kramer J. L., Siegel R. L., Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 69, 363–385 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Laversanne M., Weiderpass E., Soerjomataram I., The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 127, 3029–3030 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Isakoff M. S., Bielack S. S., Meltzer P., Gorlick R., Osteosarcoma: Current treatment and a collaborative pathway to success. J. Clin. Oncol. 33, 3029–3035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta G. K., Collier A. L., Lee D., Hoefer R. A., Zheleva V., Siewertsz V., Tangtan A. M., Guye M. L., Chang D. Z., Winston J. S., Samli B., Jansen R. J., Petricoin E. F., Goetz M. P., Bear H. D., Tang A. H., Perspectives on triple-negative breast cancer: Current treatment strategies, unmet needs, and potential targets for future therapies. Cancer 12, 2392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meel R. V. D., Sulheim E., Shi Y., Kiessling F., Mulder W. J. M., Lammers T., Smart cancer nanomedicine. Nat. Nanotechnol. 14, 1007–1017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anselmo A. C., Mitragotri S., Nanoparticles in the clinic: An update. Bioeng. Transl. Med. 4, e10143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang H., Feng W., Chen Y., Shi J., Inorganic nanoparticles in clinical trials and translations. Nano Today 35, 100972 (2020). [Google Scholar]
- 9.Martin J. D., Cabral H., Stylianopoulos T., Jain R. K., Improving cancer immunotherapy using nanomedicines: Progress, opportunities and challenges. Nat. Rev. Clin. Oncol. 17, 251–266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Shabib N. A., Husain F. M., Qais F. A., Ahmad N., Khan A., Alyousef A. A., Arshad M., Noor S., Khan J. M., Alam P., Albalawi T. H., Shahzad S. A., Phyto-mediated synthesis of porous titanium dioxide nanoparticles from Withania somnifera root extract: Broad-spectrum attenuation of biofilm and cytotoxic properties against HepG2 cell lines. Front. Microbiol. 11, 1680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovács D., Igaz N., Marton A., Rónavári A., Bélteky P., Bodai L., Spengler G., Tiszlavicz L., Rázga Z., Hegyi P., Vizler C., Boros I. M., Kónya Z., Kiricsi M., Core–shell nanoparticles suppress metastasis and modify the tumour-supportive activity of cancer-associated fibroblasts. J. Nanobiotechnol. 18, 1–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H., Wu C., Gao D., Chen S., Zhu Y., Sun J., Luo H., Yu K., Fan H., Zhang X., Antitumor effect by hydroxyapatite nanospheres: Activation of mitochondria-dependent apoptosis and negative regulation of phosphatidylinositol-3-kinase/protein kinase B pathway. ACS Nano 12, 7838–7854 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Som A., Raliya R., Tian L., Akers W., Ippolito J. E., Singamaneni S., Biswas P., Achilefu S., Monodispersed calcium carbonate nanoparticles modulate local pH and inhibit tumor growth in vivo. Nanoscale 8, 12639–12647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Setyawati M. I., Leong D. T., Mesoporous silica nanoparticles as an antitumoral-angiogenesis strategy. ACS Appl. Mater. Interfaces 9, 6690–6703 (2017). [DOI] [PubMed] [Google Scholar]
- 15.B. Guo, B. Li, X. L. Wang, M. Zhang, N. Yan, X. D. Zhang, in Key Engineering Materials. (Trans Tech Publications, 2007), vol. 330-332, pp. 279–282. [Google Scholar]
- 16.Qing F., Wang Z., Hong Y., Liu M., Guo B., Luo H., Zhang X., Selective effects of hydroxyapatite nanoparticles on osteosarcoma cells and osteoblasts. J. Mater. Sci. Mater. Med. 23, 2245–2251 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Zhang H., Qing F., Zhao H., Fan H., Liu M., Zhang X., Cellular internalization of rod-like nano hydroxyapatite particles and their size and dose-dependent effects on pre-osteoblasts. J. Mater. Chem. B 5, 1205–1217 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Wu H., Hua Y., Wu J., Zeng Q., Yang X., Zhu X., Zhang X., The morphology of hydroxyapatite nanoparticles regulates clathrin-mediated endocytosis in melanoma cells and resultant anti-tumor efficiency. Nano Res. 15, 6256–6265 (2022). [Google Scholar]
- 19.Zhang K., Zhou Y., Xiao C., Zhao W., Wu H., Tang J., Li Z., Yu S., Li X., Min L., Yu Z., Wang G., Wang L., Zhang K., Yang X., Zhu X., Tu C., Zhang X., Application of hydroxyapatite nanoparticles in tumor-associated bone segmental defect. Sci. Adv. 5, eaax6946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Y., Ren C., Zhang B., Yang H., Lang Y., Min L., Zhang W., Pei F., Yan Y., Li H., Mo A., Tu C., Duan H., Analyzing the behavior of a porous nano-hydroxyapatite/polyamide 66 (n-HA/PA66) composite for healing of bone defects. Int. J. Nanomedicine 9, 485–494 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L., Liang Z., Chen C., Yang X., Fu D., Bao H., Li M., Shi S., Yu G., Zhang Y., Zhang C., Zhang W., Xue C., Sun B., Engineered hydroxyapatite nanoadjuvants with controlled shape and aspect ratios reveal their immunomodulatory potentials. ACS Appl. Mater. Interfaces 13, 59662–59672 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Nelson K., Hesse B., Addison O., Morrell A. P., Gross C., Lagrange A., Suárez V. I., Kohal R., Fretwurst T., Distribution and chemical speciation of exogenous Micro- and nanoparticles in inflamed soft tissue adjacent to titanium and ceramic dental implants. Anal. Chem. 92, 14432–14443 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Cao X., Han Y., Gu M., Du H., Song M., Zhu X., Ma G., Pan C., Wang W., Zhao E., Goulette T., Yuan B., Zhang G., Xiao H., Foodborne titanium dioxide nanoparticles induce stronger adverse effects in obese mice than non-obese mice: Gut microbiota dysbiosis, colonic inflammation, and proteome alterations. Small 16, e2001858 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Yu Y., Zhu T., Li Y., Jing L., Yang M., Li Y., Duan J., Sun Z., Repeated intravenous administration of silica nanoparticles induces pulmonary inflammation and collagen accumulation via JAK2/STAT3 and TGF-β/Smad3 pathways in vivo. Int. J. Nanomedicine Volume 14, 7237–7247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu Z., Zhong Y., Hu H., Shao D., Haag R., Schirner M., Lee J., Sullenger B., Leong K. W., Design of therapeutic biomaterials to control inflammation. Nat. Rev. Mater. 7, 557–574 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebre F., Sridharan R., Sawkins M. J., Kelly D. J., O’Brien F. J., Lavelle E. C., The shape and size of hydroxyapatite particles dictate inflammatory responses following implantation. Sci. Rep. 7, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., Wang M., Chen F., Wang J., Li X., Liang J., Fan Y., Xiao Y., Zhang X., Correlations between macrophage polarization and osteoinduction of porous calcium phosphate ceramics. Acta Biomater. 103, 318–332 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Xia Y., Rao L., Yao H., Wang Z., Ning P., Chen X., Engineering macrophages for cancer immunotherapy and drug delivery. Adv. Mater. 32, 2002054 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Yuan Y., Liu C., Qian J., Wang J., Zhang Y., Size-mediated cytotoxicity and apoptosis of hydroxyapatite nanoparticles in human hepatoma HepG2 cells. Biomaterials 31, 730–740 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Miron R. J., Zohdi H., Fujioka-Kobayashi M., Bosshardt D. D., Giant cells around bone biomaterials: Osteoclasts or multi-nucleated giant cells? Acta Biomater. 46, 15–28 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Kloc M., Subuddhi A., Uosef A., Kubiak J. Z., Ghobrial R. M., Monocyte–macrophage lineage cell fusion. Int. J. Mol. Sci. 23, 6553 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen B. M., Hiam K. J., Burnett C. E., Venida A., De Barge R., Tenvooren I., Marquez D. M., Cho N. W., Carmi Y., Spitzer M. H., Systemic dysfunction and plasticity of the immune macroenvironment in cancer models. Nat. Med. 26, 1125–1134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garner H., de Visser K. E., Immune crosstalk in cancer progression and metastatic spread: A complex conversation. Nat. Rev. Immunol. 20, 483–497 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Coffelt S. B., Kersten K., Doornebal C. W., Weiden J., Vrijland K., Hau C.-S., Verstegen N. J. M., Ciampricotti M., Hawinkels L. J. A. C., Jonkers J., de Visser K. E., IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strauss L., Sangaletti S., Consonni F. M., Szebeni G., Morlacchi S., Totaro M. G., Porta C., Anselmo A., Tartari S., Doni A., Zitelli F., Tripodo C., Colombo M. P., Sica A., RORC1 regulates tumor-promoting "emergency" granulo-monocytopoiesis. Cancer Cell 28, 253–269 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Cassetta L., Pollard J. W., Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 17, 887–904 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Li Z., Ding Y., Liu J., Wang J., Mo F., Wang Y., Chen-Mayfield T.-J., Sondel P. M., Hong S., Hu Q., Depletion of tumor associated macrophages enhances local and systemic platelet-mediated anti-PD-1 delivery for post-surgery tumor recurrence treatment. Nat. Commun. 13, 1–15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P., Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faget J., Groeneveld S., Boivin G., Sankar M., Zangger N., Garcia M., Guex N., Zlobec I., Steiner L., Piersigilli A., Xenarios I., Meylan E., Neutrophils and snail orchestrate the establishment of a pro-tumor microenvironment in lung cancer. Cell Rep. 21, 3190–3204 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., Brooks P. J., Jang J. J., Silver A. S., Arora P. D., McCulloch C. A., Glogauer M., Role of actin filaments in fusopod formation and osteoclastogenesis. Biochim. Biophys. Acta Mol. Cell Res. 1853, 1715–1724 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Lenka G., Shan J., Halabi N. M., Abuaqel S. W. J., Goswami N., Schmidt F., Zaghlool S., Romero A. R., Subramanian M., Boujassoum S., Al-Bozom I., Gehani S., Khori N. A., Bedognetti D., Suhre K., Ma X., Dömling A., Rafii A., Chouchane L., STXBP6, reciprocally regulated with autophagy, reduces triple negative breast cancer aggressiveness. Clin. Transl. Med. 10, e147 (2020). [Google Scholar]
- 42.Scales S. J., Hesser B. A., Masuda E. S., Scheller R. H., Amisyn, a novel syntaxin-binding protein that may regulate SNARE complex assembly. J. Biol. Chem. 277, 28271–28279 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Kakanj P., Bhide S., Moussian B., Leptin M., Autophagy–mediated plasma membrane removal promotes the formation of epithelial syncytia. EMBO J. 41, e109992 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takanezawa Y., Nakamura R., Sone Y., Uraguchi S., Kiyono M., An autophagy deficiency promotes methylmercury-induced multinuclear cell formation. Biochem. Biophys. Res. Commun. 511, 460–467 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Sanjuan M. A., Dillon C. P., Tait S. W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J. L., Withoff S., Green D. R., Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Tanida I., Ueno T., Kominami E., LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 36, 2503–2518 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S29
Table S1