Abstract
The polarization of tumor-associated macrophages (TAMs) from M2 to M1 phenotype demonstrates great potential for remodeling the immunosuppressive tumor microenvironment (TME) of hepatocellular carcinoma (HCC). d-lactate (DL; a gut microbiome metabolite) acts as an endogenous immunomodulatory agent that enhances Kupffer cells for clearance of pathogens. In this study, the potential of DL for transformation of M2 TAMs to M1 was confirmed, and the mechanisms underlying such polarization were mainly due to the modulation of phosphatidylinositol 3-kinase/protein kinase B pathway. A poly(lactide-co-glycolide) nanoparticle (NP) was used to load DL, and the DL-loaded NP was modified with HCC membrane and M2 macrophage-binding peptide (M2pep), forming a nanoformulation (DL@NP-M-M2pep). DL@NP-M-M2pep transformed M2 TAMs to M1 and remodeled the immunosuppressive TME in HCC mice, promoting the efficacy of anti-CD47 antibody for long-term animal survival. These findings reveal a potential TAM modulatory function of DL and provide a combinatorial strategy for HCC immunotherapy.
Modulation of M2 tumor-associated macrophages by a D-lactate nanoformulation promotes immunotherapy for liver cancer.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and increases annually with new morbidity and mortality more than 900,000 and 800,000, respectively, worldwide (1). Immunotherapy provides great promise for different cancers. However, the liver is highly exposed to antigens and endotoxins (2), and the mechanisms underlying such homeostasis cause the prevention of immune responses against specific antigens (known as immunological tolerance) in the liver (3). Moreover, hepatocyte carcinogenesis results often from chronic inflammation, fibrosis, and cirrhosis, and these pathological conditions generate a state in which fully activated effector immune cells fail to induce productive immune responses (known as immunological ignorance) against hepatocyte transformation and tumor cell proliferation (4). For these reasons, HCC demonstrates an immunosuppressive tumor microenvironment (TME) that is resistant to immunotherapy.
Macrophages are known as a heterogeneous cell population and can be generally classified to two subpopulations that are termed classically activated macrophages (M1 or M1-like phenotype) and alternatively activated macrophages (M2 or M2-like phenotype) (for readability, M1 and M2 will be used in this study for the nomenclature) (5). Macrophages within the tumor, also known as tumor-associated macrophages (TAMs), are a critical regulator of immunosuppressive TME for immune escape and tumor development (6). Most TAMs present M2 phenotype and produce immunosuppressive factors [e.g., transforming growth factor–β (TGF-β) and interleukin-10 (IL-10)] to support other immunosuppressive cells [e.g., myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs)] (7). In contrast to M2 TAMs, M1 ones generate immunostimulatory factors [e.g., IL-1β, IL-12, and tumor necrosis factor–tumor necrosis factor–α (TNF-α)] to induce other immunostimulatory cells [e.g., cytotoxic T lymphocytes (CTLs) and natural killer (NK) cells] (8). Thus, approaches used to polarize TAMs from M2 to M1 have demonstrated great potential for reversing the immunosuppressive TME (9–12).
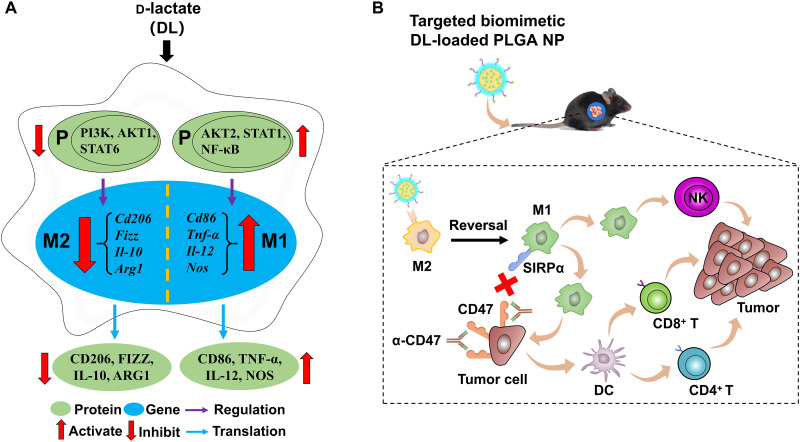
Recently, it has been reported that d-lactate (DL), a gut microbiome small-molecule metabolite, reached the liver through the portal vein and enhanced the capacity of Kupffer cells (the liver-resident macrophages) for promoting the clearance of pathogens from the bloodstream (13). Because DL augments the phagocytotic function of Kupffer cells (a critical feature of M1 macrophages), we hypothesize that DL may polarize M2 TAMs toward M1 ones and remodel the immunosuppressive TME of HCC. In this study, DL switched TAMs from M2 to M1, which was mainly due to the inhibition of phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathway and the activation of nuclear factor κB (NF-κB) pathway (Fig. 1A).
Fig. 1. DL modulates TAMs and remodels the immunosuppressive TME for HCC.
(A) The proposed mechanisms of DL-mediated TAM modulation. (B) Delivery of DL using a targeted biomimetic PLGA NP achieves immunotherapy in combination with anti-CD47 antibody (α-CD47).
In addition, an M2 macrophage-binding peptide (M2pep)–targeted HCC membrane–coated poly(lactide-co-glycolide) (PLGA) nanoparticle (NP) was developed in this study for delivery of DL to M2 TAMs within HCC (Fig. 1B). The resultant nanoformulation (termed DL@NP-M-M2pep) accumulated inside the tumor via HCC membrane–associated homing function and transported DL to M2 TAMs via M2pep-mediated targeting capacity, collectively resulting in transformation of M2 TAMs to M1 ones and remodeling the immunosuppressive TME in allograft and carcinogen-induced orthotopic HCC mouse models, respectively. Furthermore, the combination of anti-CD47 antibody and DL@NP-M-M2pep achieved long-term survival of carcinogen-induced orthotopic HCC mice, providing a promising combinatory approach for HCC.
RESULTS
Polarization of M2 macrophages to M1 by DL
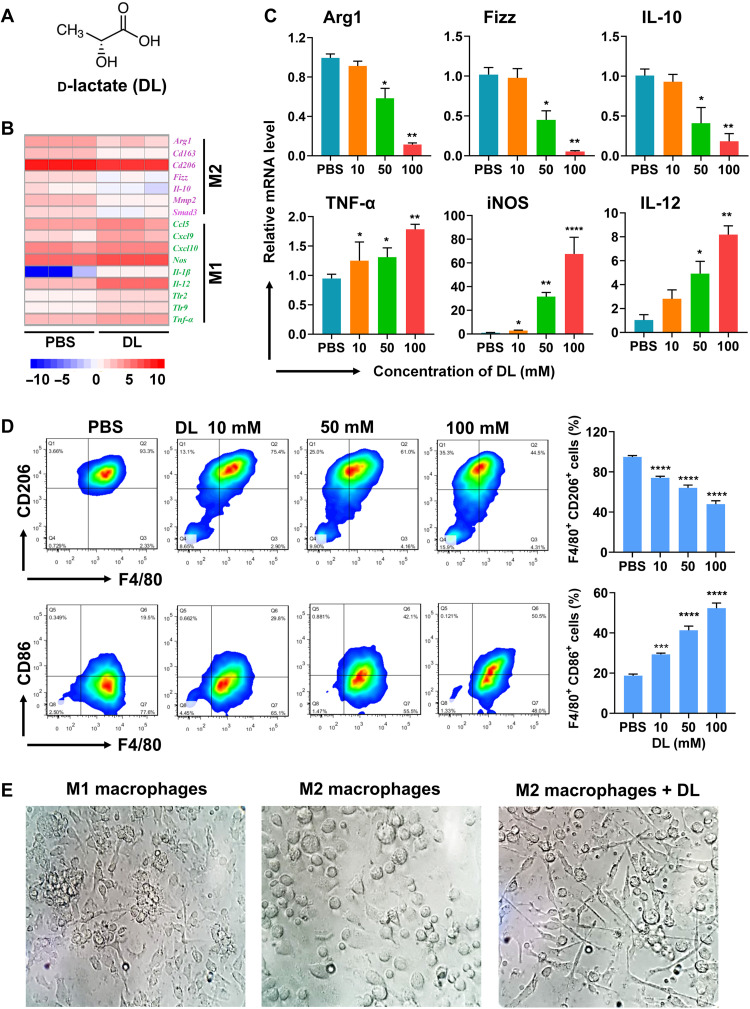
In this study, the capacity of DL (Fig. 2A) for macrophage polarization was evaluated using bone marrow (BM)–derived macrophages (BMDMs) that were stimulated by either IL-4 (M2 macrophages) or interferon-γ (IFN-γ) and lipopolysaccharide (LPS; M1 macrophages). These M2 or M1 macrophages may resemble M2 or M1 TAMs, therefore, have been widely used for TAM-related studies (14–16). RNA sequencing profiling results indicated that, when M2 macrophages were treated with DL, M2-associated genes (e.g., Arg1, Cd163, Cd206, Fizz, Il-10, Mmp2, and Smad3) were significantly down-regulated (Fig. 2B), but M1-associated genes (e.g., Ccl5, Cxcl9, Cxcl10, Nos, Il-1β, IL-12, Tlr2, Tlr9, and Tnf-α) were significantly up-regulated (Fig. 2B). Furthermore, the expression of genes associated with macrophage polarization was confirmed using the quantitative reverse transcription polymerase chain reaction (RT-PCR) (Fig. 2C). When M2 macrophages were treated with DL, the expression of M2 functional markers [arginine-1 (Arg-1), Fizz, and IL-10] was significantly (P < 0.01, P < 0.01, and P < 0.01) down-regulated, but the expression of M1 functional markers [TNF-α, iNOS (inducible nitric oxide synthase), and IL-12] was significantly (P < 0.01, P < 0.01, and P < 0.0001) up-regulated (Fig. 2C).
Fig. 2. DL regulates the polarization of M2 macrophages to M1 ones.
(A) Chemical structure of DL. (B) The regulation of M1- and M2-associated genes in IL-4–stimulated bone marrow (BM)–derived macrophages (BMDMs) (M2 macrophages) with the treatment of phosphate-buffered saline (PBS) or DL (50 mM) was shown in the hierarchical cluster heatmap. (C) Quantitative reverse transcription polymerase chain reaction (RT-PCR) was performed to examine the mRNA expression in DL-treated M2 macrophages (n = 3; *P < 0.05, **P < 0.01, and ****P < 0.0001 to PBS). (D) Phenotypic change in M2 macrophages treated with PBS or DL was analyzed using flow cytometry (F4/80+ CD86+ for M1 and F4/80+ CD206+ for M2) (n = 3; ***P < 0.001 and ****P < 0.0001 to PBS). (E) The morphology of interferon-γ (IFN-γ) and lipopolysaccharide (LPS) costimulated BMDMs (M1 macrophages), M2 macrophages, and DL (50 mM)–treated M2 macrophages.
Flow cytometric results indicated that, when M2 macrophages were treated with DL, the CD206+ F4/80+ population (M2 phenotype) was significantly (P < 0.0001) decreased (Fig. 2D), while the CD86+ F4/80+ population (M1 phenotype) was significantly (P < 0.0001) increased (Fig. 2D). In addition, M1 and M2 macrophages demonstrated distinct morphological structures (Fig. 2E), which were similar to those previously reported for M1 and M2 ones (17, 18). Note that DL changed the morphology of M2 macrophages that became similar to that of M1 ones (Fig. 2E). Thus, results in Fig. 2 indicated that DL transforms M2 macrophages to M1 ones.
Macrophage polarization by DL via inhibition of PI3K/Akt pathway and activation of NF-κB pathway
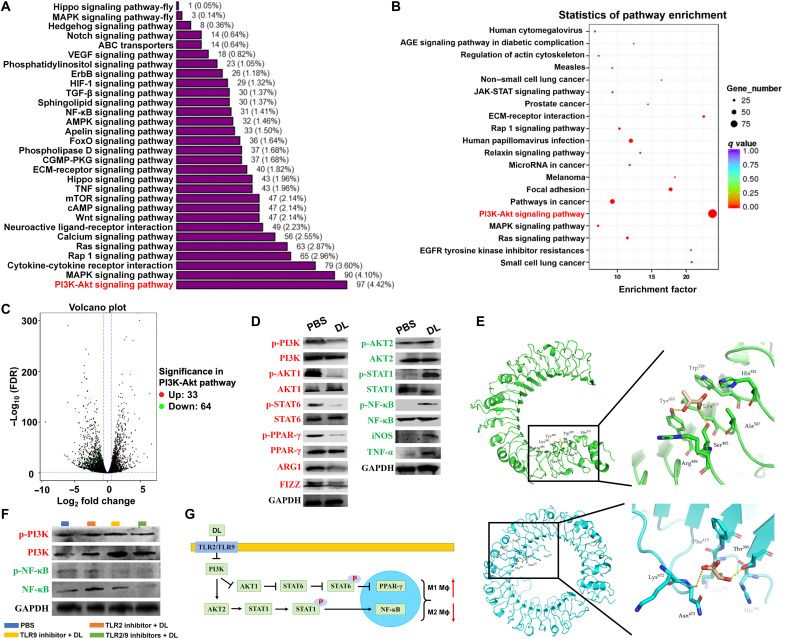
The mechanisms underlying the efficacy of DL on macrophage polarization were subsequently investigated using M2 macrophages. The functional analysis by Gene Ontology (GO; Fig. 3A) and Kyoto Encyclopedia of Genes and Genomes (KEGG; Fig. 3B) demonstrated the enrichment of DL-regulated gene products on the PI3K/Akt pathway. The gene products associated with the PI3K/Akt pathway were significantly varied in DL-treated M2 macrophages (Fig. 3C), showing that 33 genes were up-regulated (red dots) and 64 genes were down-regulated (green dots) in this pathway. These results indicated that the PI3K/Akt pathway is closely associated with DL-mediated macrophage polarization.
Fig. 3. DL induces the polarization of M2 macrophages to M1 ones by inhibiting the PI3K/Akt pathway and activating the NF-κB pathway.
(A) Gene Ontology (GO) classification of expressed genes in DL (50 mM)–treated M2 macrophages. MAPK, mitogen-activated protein kinase. VEGF, vascular endothelial growth factor; ABC, adenosine 5′-triphosphate–binding cassette; HIF-1, hypoxia-inducible factor 1; ECM, extracellular matrix; cAMP, adenosine 3′,5′-monophosphate; AMPK, adenosine 5'-monophosphate-activated protein kinase; CGMP-PKG, cyclic guanosine monophosphate-protein kinase G; mTOR, mammalian target of rapamycin. (B) Signaling pathway enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) in DL (50 mmol)–treated M2 macrophages. (C) The up-regulated (red dots) and down-regulated (green dots) genes associated with the PI3K/Akt signaling pathway in DL (50 mM)–treated M2 macrophages were presented using the volcano plot. JAK-STAT, Janus kinase–signal transducer and activator of transcription; EGFR, epidermal growth factor receptor; AGE, advanced glycation end product. (D) The activity of PI3K/Akt and NF-κB signaling pathways in PBS- or DL (50 mM)–treated M2 macrophages was analyzed using Western blot assay. The quantification was shown in fig. S1. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (E) The binding of DL to macrophage surface receptors (TLR2 indicated in green; TLR9 indicated in blue) was analyzed using molecular docking technique. (F) After blocking TLR2/TLR9, the expression of PI3K and NF-κB in PBS- and DL (50 mM)–treated M2 macrophages was analyzed using Western blot assay. The quantification was shown in fig. S2. (G) The schematic of signaling pathways associated with DL-induced macrophage polarization from M2 to M1.
The activity of PI3K and AKT1 was significantly (P < 0.001 and P < 0.01) suppressed in DL-treated M2 macrophages (Fig. 3D and fig. S1). Two downstream transcription factors of AKT1, signal transducer and activator of transcription 6 (STAT6) and peroxisome proliferator–activated receptor γ (PPAR-γ), were also significantly (P < 0.001 and P < 0.0001) deactivated (Fig. 3D and fig. S1). The down-regulation of PI3K/Akt1 pathway also significantly (P < 0.001 and P < 0.05) suppressed the expression of ARG-1 and FIZZ (two critical M2 functional factors) (Fig. 3D and fig. S1). In contrast, the activity of AKT2 was significantly (P < 0.05) enhanced in M2 macrophages following DL treatment (Fig. 3D and fig. S1). Two downstream transcription factors of AKT2, STAT1 and NF-κB, were also significantly (P < 0.05, P < 0.0001, and P < 0.01) activated (Fig. 3D and fig. S1). The expression of iNOS and TNF-α (two critical M1 functional factors) was also significantly (P < 0.01 and P < 0.05) up-regulated (Fig. 3D and fig. S1).
The binding conformations of DL in Toll-like receptor 2 (TLR2) and TLR9 were predicted using molecular docking (19, 20). The results showed that DL may interact with TLR2 through hydrophobic interactions (Trp529, His531, Ala507, Lys505, and Arg486) and hydrogen bonds (Tyr483 and Ser485) (Fig. 3E, green); DL may interact with TLR9 through hydrophobic interactions (His394, Arg418, Phe419, and Lys472) and hydrogen bonds (Thr395 and Asn473) (Fig. 3E, blue). The interaction of DL with TLR2 and/or TLR9 was further confirmed in M2 macrophages by pretreatment of TLR2 and/or TLR9 inhibitors. The results showed that either TLR2 inhibitor, TLR9 inhibitor, or both significantly abolished the effects of DL on the PI3K and NF-κB activities (Fig. 3F and fig. S2). Thus, the results in Fig. 3 indicated that DL interacts with TLR2 and/or TLR9, which induce the inhibition of PI3K/Akt pathway and the activation of NF-κB pathway, facilitating macrophage polarization from M2 to M1 (Fig. 3G).
Preparation and physicochemical characterization of nanoformulation
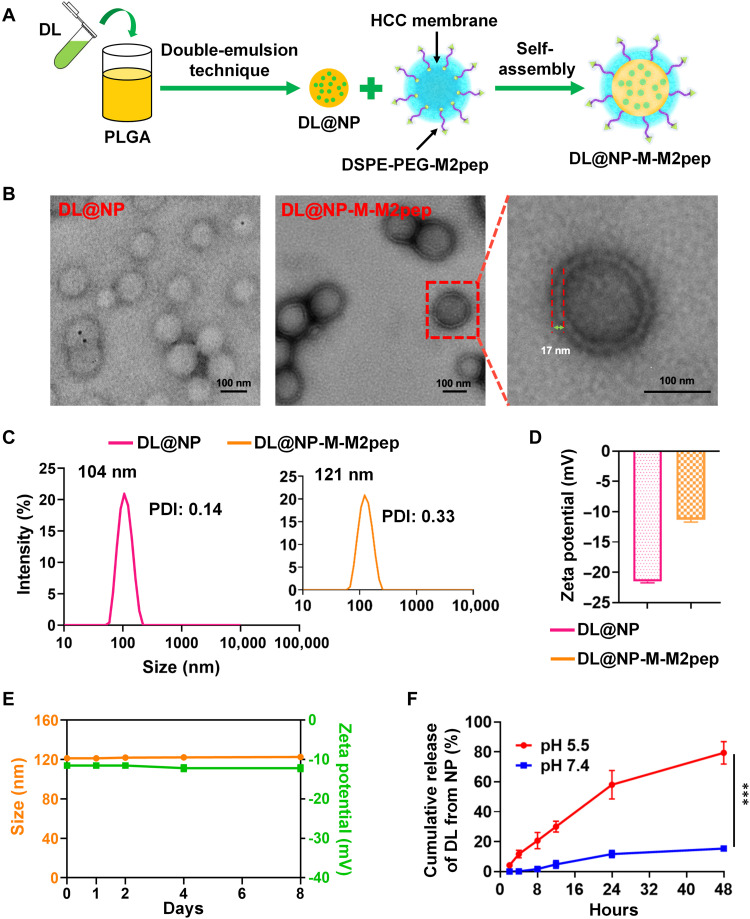
In this study, an M2pep-targeted HCC membrane–coated PLGA NP was developed (Fig. 4A) to harness in vivo delivery of exogenous DL to M2 TAMs within the TME. Following the surface coating of DL-loaded NPs (termed DL@NP) with M2pep-inserted HCC membrane (fig. S3), a targeted biomimetic nanoformulation (termed DL@NP-M-M2pep) demonstrated the “membrane-core” nanostructure (Fig. 4B), which was similar to nontargeted counterpart (DL@NP-M). The membrane thickness of DL@NP-M-M2pep (as well as DL@NP-M) was ~17 nm, which was similar to the thickness of natural cancer cell membrane (21). In addition, DL@NP-M-M2pep achieved the particle size of ⁓120 nm [polymer dispersity index (PDI) ≈ 0.3] and the surface charge of ⁓−11 mV (Fig. 4, C and D), which were similar to those of DL@NP-M. Notably, DL@NP-M-M2pep showed a larger particle size (~120 nm) than DL@NP (~105 nm, PDI ≈ 0.1; Fig. 4C), and the difference (~15 nm) was similar to the membrane thickness (~17 nm) as observed in transmission electron microscopy (TEM) (Fig. 4B). These results indicated the successful coating of DL@NP for forming DL@NP-M and DL@NP-M-M2pep. In addition, DL@NP, DL@NP-M, and DL@NP-M-M2pep achieved similar loading capacity (LC) (~7 wt %), indicating that the integrity of nanoformulations was not affected by the HCC membrane coating. When DL@NP-M-M2pep was incubated within neutral phosphate-buffered saline (PBS) for the stability evaluation, the size and charge did not fluctuate significantly (Fig. 4E).
Fig. 4. Preparation and physicochemical characterization of DL@NP-M-M2pep.
(A) Formulation schematic of DL@NP-M-M2pep. (B) TEM images of DL@NP and DL@NP-M-M2pep. (C) The size and PDI of DL@NP and DL@NP-M-M2pep (n = 3). (D) The charge of DL@NP and DL@NP-M-M2pep (n = 3). (E) The change in size and charge of DL@NP-M-M2pep incubated in PBS over 1 week (n = 3). (F) The in vitro DL release from DL@NP-M-M2pep when incubated in release medium (pH 5.5 and 7.4) (n = 3; ***P < 0.001).
The results in Fig. 4F showed that ~2% of DL were released from DL@NP-M-M2pep after 8 hours of incubation in neutral environment (pH 7.4), whereas the release of DL was significantly (P < 0.001) increased (~20%) in acidic environment (pH 5.5). Following incubation for 24 hours, ~10 and 60% of DL were released from DL@NP-M-M2pep at pH 7.4 and 5.5, respectively (Fig. 4F). After 48 hours, ~15 and 80% of DL were released from DL@NP-M-M2pep at pH 7.4 and 5.5, respectively (Fig. 4F). The pH-sensitive drug release is likely caused by the degradation of PLGA-based NPs in low-pH environments through autocatalysis. Moreover, DL@NP-M-M2pep displayed similar release profiles to nontargeted counterpart (DL@NP-M).
Ex vivo studies of nanoformulation
It was reported that IL-4–stimulated BMDM-derived M2 macrophages have been used to investigate the delivery efficacy of M2pep-targted NPs (22–24). In this study, targeted delivery ability of DL@NP-M-M2pep was assessed in M2 macrophages using flow cytometry (Fig. 5A). The results showed that DL@NP-M-M2pep achieved significantly (P < 0.001) higher cellular uptake in M2 macrophages as compared to PBS and DL@NP-M (Fig. 5A). Moreover, targeted delivery ability of DL@NP-M-M2pep was assessed in M2 and M1 macrophages using confocal laser scanning microscopy (CLSM) (fig. S4). As a consequence, DL@NP-M-M2pep achieved significantly (P < 0.001) higher internalization to M2 macrophages than M1 ones (fig. S4). These results confirmed the M2pep-mediated drug delivery potential.
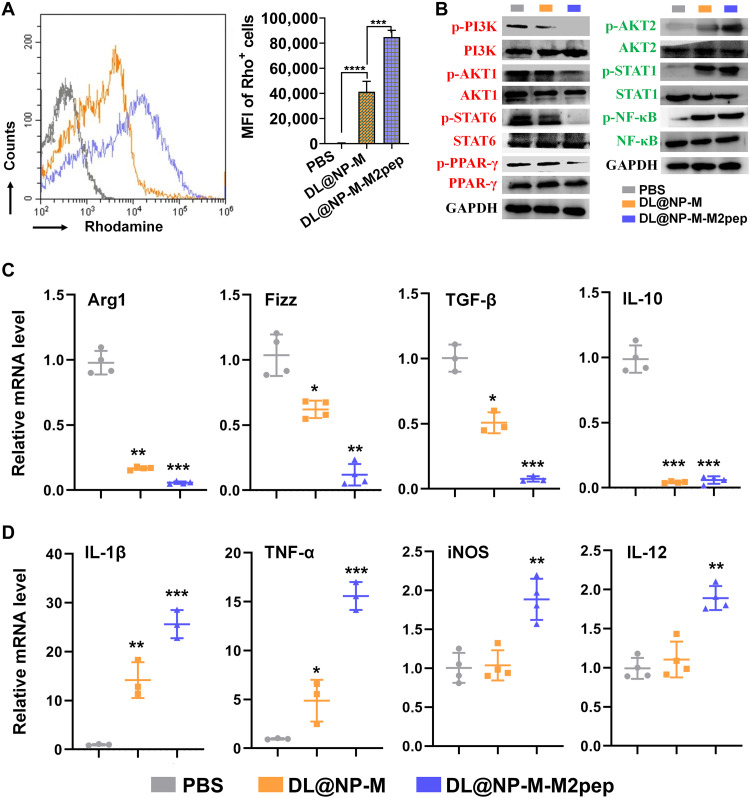
Fig. 5. DL@NP-M-M2pep induces the polarization of M2 macrophages to M1 ones.
(A) Cellular uptake of rhodamine-labeled DL@NP-M and DL@NP-M-M2pep (50 mM DL, same as below) was assessed using flow cytometry (n = 3; ***P < 0.001 and ****P < 0.0001). (B) The activity of PI3K/Akt and NF-κB pathways in M2 macrophages treated with PBS, DL@NP-M, and DL@NP-M-M2pep was analyzed using Western blot assay. The quantification was shown in fig. S5. (C) Quantitative RT-PCR was performed to examine the mRNA expression of M2-associated genes in M2 macrophages treated with PBS, DL@NP-M, and DL@NP-M-M2pep (n = 4; *P < 0.05, **P < 0.01, and ***P < 0.001 to PBS). (D) Quantitative RT-PCR was performed to examine the mRNA expression of M1-associated genes in M2 macrophages treated with PBS, DL@NP-M, and DL@NP-M-M2pep (n = 4; *P < 0.05, **P < 0.01, and ***P < 0.001 to PBS).
Subsequently, the efficacy of DL@NP-M-M2pep for macrophage polarization was confirmed in M2 macrophages. The activity of PI3K/Akt pathway was significantly (P < 0.0001) down-regulated by DL@NP-M-M2pep as compared to PBS and DL@NP-M, in terms of the activities of PI3K, AKT1, STAT6, and PPAR-γ (Fig. 5B and fig. S5). In contrast, the activity of NF-κB pathway was significantly (P < 0.0001) up-regulated by DL@NP-M-M2pep as compared to PBS and DL@NP-M, in terms of the activities of AKT2, STAT1, and NF-κB (Fig. 5B and fig. S5). Following the inhibition of PI3K/Akt pathway, the expression of M2-associated factors (e.g., Arg1, Fizz, TGF-β, and IL-10) was significantly (P < 0.001, P < 0.01, P < 0.001, and P < 0.001) down-regulated by DL@NP-M-M2pep (Fig. 5C). In contrast, after the activation of NF-κB pathway, the expression of M1-associated factors (e.g., Il-1β, TNF-α, iNOS, and IL-12) was significantly (P < 0.001, P < 0.001, P < 0.01, and P < 0.01) up-regulated (Fig. 5D). Therefore, results in Fig. 5 confirmed the potential of DL@NP-M-M2pep for M2 macrophage-specific delivery of DL for M2-to-M1 transition.
Toxicity, pharmacokinetics, and biodistribution of nanoformulation
The in vivo toxicity of DL@NP-M-M2pep was investigated using healthy mice (n = 6). The results showed that no significant body weight loss was caused following multiple intravenous injections of DL@NP-M-M2pep (100 mM DL) as compared to PBS and free DL (a gut microbiome metabolite, generally considered nontoxic) (Fig. 6A). In addition, no obvious damage was observed in major organs (the heart, liver, spleen, lung, and kidneys) of mice treated with DL@NP-M-M2pep as compared to PBS (Fig. 6B). Meanwhile, the liver/kidney functions were analyzed to further assess the systemic toxicity. The level of alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), and creatinine (CRE) in the serum was not significantly increased by DL@NP-M-M2pep as compared to PBS (Fig. 6C). These results confirmed the safety of our nanoformulation for in vivo application.
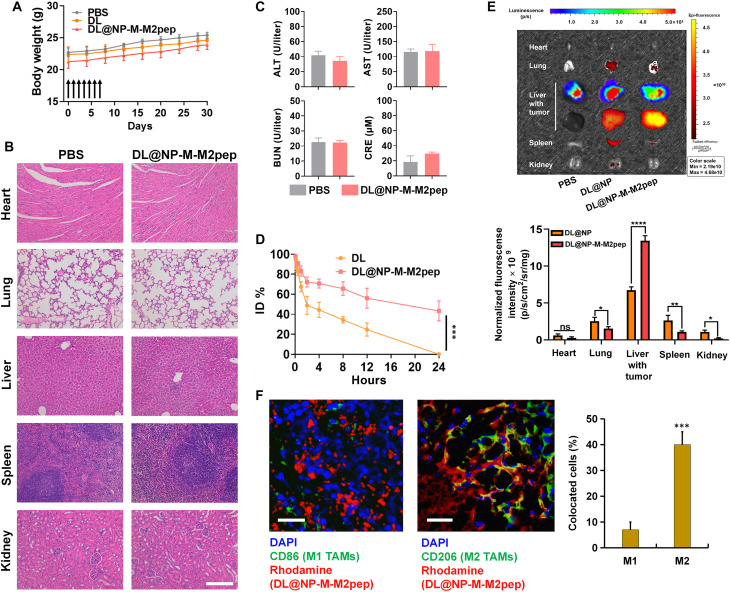
Fig. 6. Toxicity, half-life, and biodistribution of DL@NP-M-M2pep.
(A) Body weight of healthy mice within a 30-day period following intravenous treatments (n = 6). (B) Major organs were assessed using hematoxylin and eosin staining assay on day 30 following intravenous treatments (scale bar, 100 μm). (C) The blood analysis for the liver/kidney functions including alanine aminotransferase (ALT), alanine aminotransferase (AST), blood urea nitrogen (BUN), and creatinine (CRE) was determined on day 30 following the treatments (n = 6). (D) The curve of injected drug concentration (ID %) versus time point was plotted in Hepa1-6-luc–derived orthotopic HCC mice (n = 4; ***P < 0.001). (E) Biodistribution of DiR-labeled nanoformulations in major organs and liver tumors at 24 hours after intravenous injection in Hepa1-6-luc–derived orthotopic HCC mice (n = 4; *P < 0.05, **P < 0.01, and ****P < 0.0001; ns, not significant). (F) Following the biodistribution as described above, the level of rhodamine-labeled DL@NP-M-M2pep inside M1 and M2 TAMs was detected in tumors using immunofluorescent staining assay (blue, cell nucleus; green, CD86+ or CD206+ cells; and red, rhodamine) (scale bars, 100 μm; n = 3; ***P < 0.001). DAPI, 4′,6-diamidino-2-phenylindole.
Subsequently, the half-life of free DL and DL loaded in nanoformulation was evaluated in Hepa1-6-luc–derived orthotopic HCC mice (Fig. 6D). The results showed that the blood circulation time of DL within DL@NP-M-M2pep was significantly prolonged [half-time (t1/2) of DL ≈ 12 hours] as compared to that of free DL (t1/2 of DL ≈ 2 hours) (Fig. 6D). In addition, the biodistribution of DL@NP-M-M2pep was also assessed using Hepa1-6-luc–derived orthotopic HCC mice (Fig. 6E). The major organs and liver tumors were imaged at 24 hours after one intravenous injection of DiR-loaded nanoformulations (Fig. 6E). The results showed that DL@NP-M-M2pep was significantly (P < 0.0001, more than twofolds) observed inside the liver tumor as compared to DL@NP, which was confirmed by the delivery of nanoformulation (labeled by DiR) into the tumor (indicated by the luminescence due to the oxidation of luciferin catalyzed by the luciferase in HCC cells) (Fig. 6E). In contrast, DL@NP-M-M2pep was significantly (P < 0.05, ~2-folds) less observed in the lung, spleen, and kidneys as compared to DL@NP (Fig. 6E). Moreover, DL@NP-M demonstrated similar half-life and biodistribution with those of DL@NP-M-M2pep. Furthermore, TAM-targeted delivery capacity of DL@NP-M-M2pep was investigated using CLSM (Fig. 6F). As shown in Fig. 6F, the level of DL@NP-M-M2pep inside M2 TAMs was significantly (P < 0.05) higher than that inside M1 counterparts. These results confirmed that DL@NP-M-M2pep, due to HCC membrane–based and M2pep-targeted modification, improves the blood circulation, tumor homing, and TAM targeting (21–24).
Polarization of TAMs and remodeling of immunosuppressive TME by nanoformulation in HCC
To confirm the hypothesis that DL may polarize M2 TAMs toward M1 ones and remodel the immunosuppressive TME in HCC, therapeutic studies were first investigated in the Hepa1-6-luc–derived orthotopic HCC mouse model (Fig. 7A). The results showed that free DL (exogenous DL on its own) slightly (P > 0.05) slowed down tumor growth as compared to PBS (Fig. 7, B and C), while DL@NP-M significantly (P < 0.01) retarded tumor development as compared to PBS and free DL (Fig. 7, B and C). Notably, DL@NP-M-M2pep further (P < 0.001) inhibited tumor growth within a 21-day period in comparison with free DL and DL@NP-M (Fig. 7, B and C). Consequently, DL@NP-M-M2pep significantly improved the animal survival (6 of the 10 mice > 90 days) as compared to PBS (median survival ≈ 13 days), free DL (median survival ≈ 16 days), and DL@NP-M (1 of the 10 mice > 90 days) (Fig. 7D). Note that tumor growth was not influenced by blank NPs (DL@NP-M-M2pep without DL) (fig. S6), indicating that therapeutic efficacy of DL@NP-M-M2pep is due to DL but not to blank NPs.
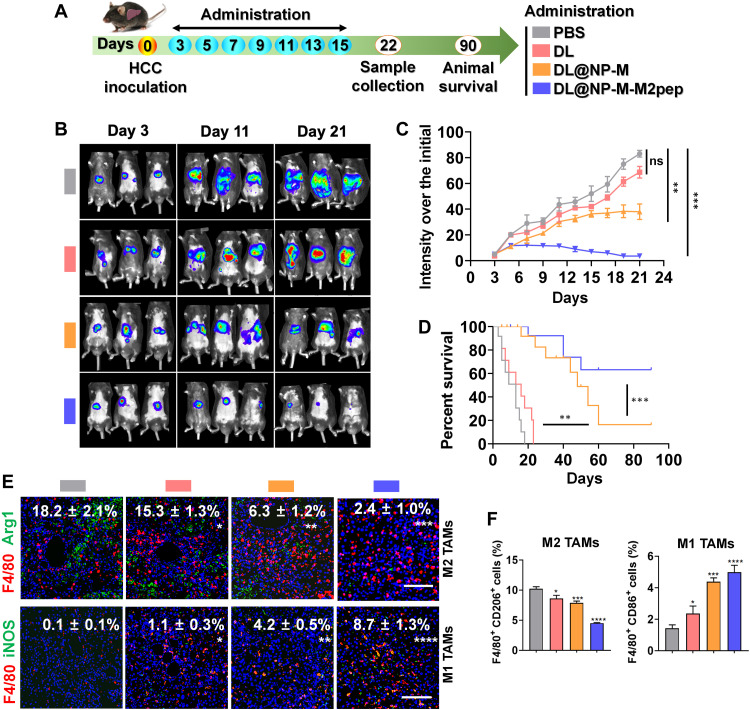
Fig. 7. DL@NP-M-M2pep achieves antitumor effects by polarizing TAMs from M2 to M1 in Hepa1-6-luc–derived orthotopic HCC mice.
(A) Tumor inoculation and treatment scheme. (B) The representative images of animals with different treatments. (C) The HCC development over a 21-day period (n = 6). (D) Animal survival (n = 10). (E) The M2 and M1 TAMs were detected using immunofluorescent staining assay (scale bars, 100 μm; n = 4). (F) Flow cytometric analysis of M2 and M1 TAMs (fig. S8) (n = 4) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns to PBS).
Subsequently, the efficacy of DL@NP-M-M2pep on macrophage modulation for anti-HCC therapy was confirmed. Immunofluorescent staining results demonstrated that DL@NP-M-M2pep significantly (P < 0.001) diminished the population of F4/80+ Arg1+ M2 TAMs (~2%) within the tumor as compared to PBS (~18%), free DL (~15%), and DL@NP-M (~6%) (Fig. 7E); in contrast, DL@NP-M-M2pep significantly (P < 0.0001) elevated the number of F4/80+ iNOS+ M1 ones (~9%) within the tumor as compared to PBS (~0.1%), free DL (~1%), and DL@NP-M (~4%) (Fig. 7E). In addition, different markers for M2 and M1 TAMs were used to further confirm the efficacy of DL@NP-M-M2pep on macrophage modulation. As shown in fig. S7, DL@NP-M-M2pep significantly (P < 0.0001) diminished the population of F4/80+ CD206+ M2 TAMs (~2%) within the tumor as compared to PBS (~28%), free DL (~21%), and DL@NP-M (~8%); in contrast, DL@NP-M-M2pep significantly (P < 0.0001) elevated the number of F4/80+ CD86+ M1 ones (~25%) within the tumor as compared to PBS (~0.2%), free DL (~5%), and DL@NP-M (~12%) (fig. S7). Furthermore, flow cytometric results indicated that DL@NP-M-M2pep significantly (P < 0.0001) reduced the amount of F4/80+ CD206+ M2 TAMs within the tumor as compared to PBS, free DL, and DL@NP-M (Fig. 7F and fig. S8); in contrast, DL@NP-M-M2pep significantly (P < 0.0001) increased the population of F4/80+ CD86+ M1 ones within the tumor as compared to other controls (Fig. 7F and fig. S8). These results indicated that DL@NP-M-M2pep polarizes M2 TAMs toward M1 ones within the TME.
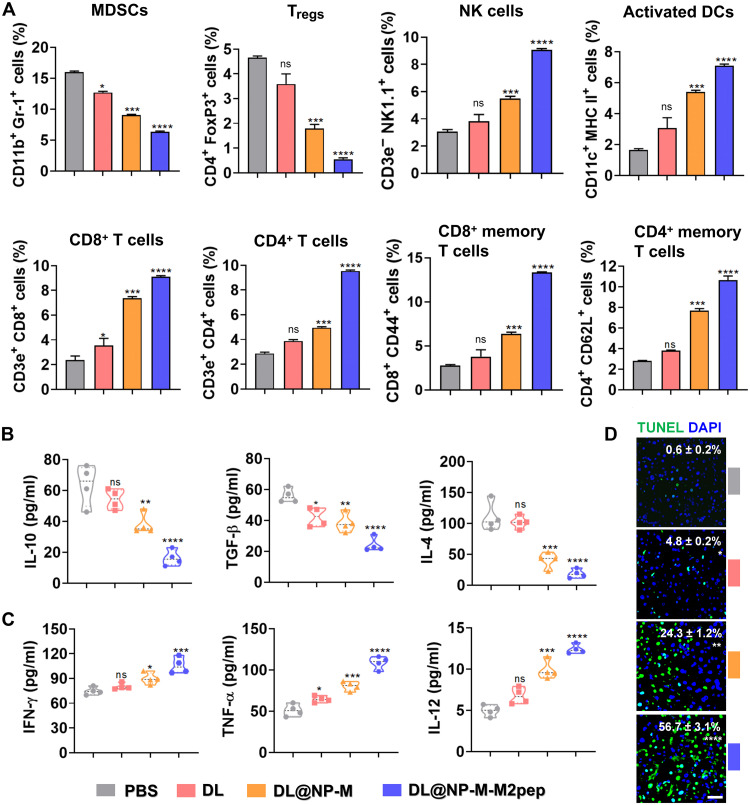
Following TAM polarization, the capacity of DL@NP-M-M2pep for remodeling the immunosuppressive TME was assessed (Fig. 8). Flow cytometric results showed that immunosuppressive cells such as MDSCs and Tregs were significantly (P < 0.0001) down-regulated inside tumors by DL@NP-M-M2pep as compared to PBS, free DL, and DL@NP-M (Fig. 8A and fig. S9); in contrast, immunostimulatory cells, including NK cells, activated dendritic cells (DCs), CD8+ cytotoxic and memory T cells, and CD4+ helper and memory T cells, were significantly (P < 0.0001) up-regulated inside tumors by DL@NP-M-M2pep as compared to other controls (Fig. 8A and figs. S10 to S13). Accordingly, the down-regulation of immunosuppressive cells achieved by DL@NP-M-M2pep significantly (P < 0.0001) reduced the expression of immunoinhibitory factors such as IL-10, TGF-β, and IL-4 (Fig. 8B), and the up-regulation of immunostimulatory cells achieved by DL@NP-M-M2pep significantly (P < 0.0001) increased the expression of immunostimulatory factors such as IFN-γ, TNF-α, and IL-12 (Fig. 8C). In addition, the remodeling of immunosuppressive TME achieved by DL@NP-M-M2pep significantly (P < 0.0001) resulted in tumor apoptosis (~57%) as compared to PBS, free DL (~5%), and DL@NP-M (~24%) (Fig. 8D). These results confirmed the capacity of DL@NP-M-M2pep for remodeling the immunosuppressive TME and achieving anti-HCC immunotherapy, which is mainly due to the efficacy of TAM modulation (Fig. 7).
Fig. 8. DL@NP-M-M2pep remodels the immunosuppressive TME in Hepa1-6-luc–derived orthotopic HCC.
(A) The immunosuppressive cells such as MDSCs and Tregs in tumors (fig. S9). The immunostimulatory cells as NK cells, activated DCs, and effector T cells in tumors (figs. S10 to S13) (n = 4). (B) The expression of immunosuppressive cytokines (IL-10, TGF-β, and IL-4) in tumors was measured using enzyme-linked immunosorbent assay (ELISA) (n = 4). (C) The expression of immunostimulatory cytokines (IFN-γ, TNF-α, and IL-12) in tumors was measured using ELISA (n = 4). (D) The average number of apoptotic cells per high-power field was detected by terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) and analyzed by ImageJ. (scale bar, 50 μm; n = 3) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns to PBS).
Eradication of HCC by anti-CD47 antibody and nanoformulation
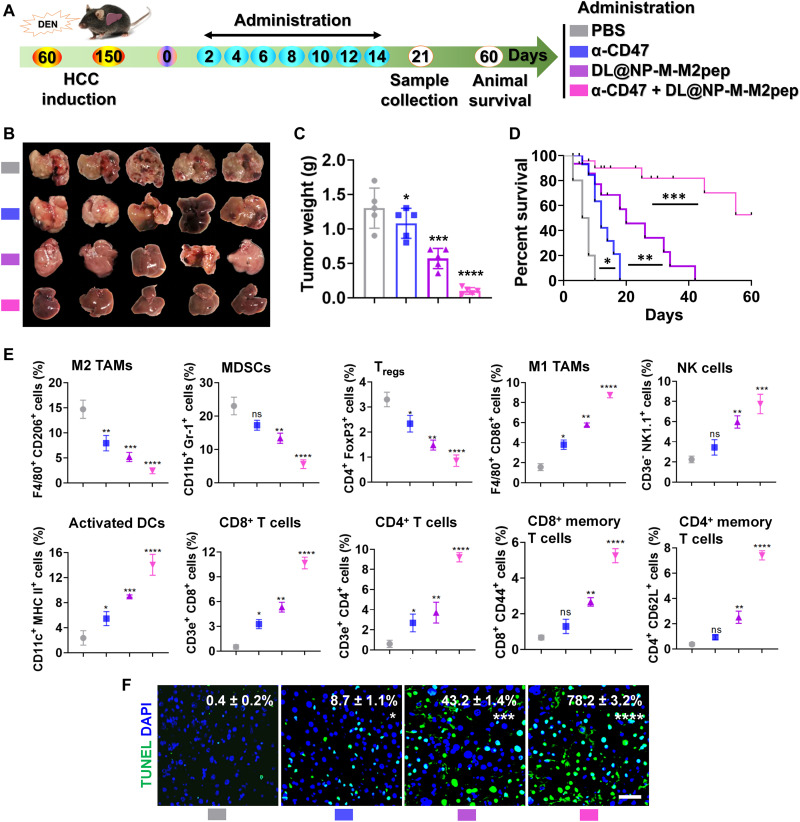
As confirmed above, DL@NP-M-M2pep reprogrammed the TAM polarization from M2 to M1 and remodeled the immunosuppressive TME in HCC. Subsequently, the combinative efficacy of anti-CD47 antibody (α-CD47, it blocks the “do not eat me” signal) and our nanoformulation was assessed using carcinogen-induced orthotopic HCC mice (Fig. 9A). The results showed that tumor development was significantly (P < 0.05) slowed down by α-CD47 as compared to PBS, while DL@NP-M-M2pep achieved significantly (P < 0.001) better anti-HCC efficacy than α-CD47 (Fig. 9, B and C). Notably, the combination of α-CD47 and DL@NP-M-M2pep further (P < 0.0001) suppressed tumor growth relative to monotherapies (Fig. 9, B and C), resulting in longer animal survival (5 of the 10 mice > 60 days) than PBS (median survival ≈ 10 days), α-CD47 (median survival ≈ 18 days), and DL@NP-M-M2pep (median survival ≈ 40 days) (Fig. 9D).
Fig. 9. The combination of anti-CD47 antibody and DL@NP-M-M2pep promotes antitumor efficacy in carcinogen-induced orthotopic HCC mice.
(A) Tumor induction and treatment scheme. (B) The representative images of HCC with different treatments. (C) Tumor weight after subtracting liver weight of healthy mice (n = 5). (D) Animal survival (n = 10). (E) Flow cytometric analysis of immune cells (n = 5) (figs. S14 to S19). MHC, major histocompatibility complex. (F) The average number of apoptotic cells per high-power field was detected by TUNEL staining and analyzed by ImageJ (scale bar, 50 μm; n = 3) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns to PBS).
Flow cytometric results indicated that DL@NP-M-M2pep significantly (P < 0.001) reduced the population of M2 TAMs (F4/80+ CD206+) inside tumors as compared to PBS and α-CD47 (Fig. 9E and fig. S14), while the combination of DL@NP-M-M2pep and α-CD47 further (P < 0.0001) decreased the number of M2 TAMs (Fig. 9E and fig. S14). In addition, DL@NP-M-M2pep significantly (P < 0.01) elevated the amount of M1 ones (F4/80+ CD86+) inside tumors as compared to PBS and α-CD47 (Fig. 9E and fig. S14), while the combination of DL@NP-M-M2pep and α-CD47 further (P < 0.0001) increased the population of M1 ones (Fig. 9E and fig. S14). Following macrophage polarization, our combination strategy significantly remodeled the immunosuppressive TME to immunostimulatory, which was accompanied with the down-regulation of MDSCs and Tregs (P < 0.0001 and P < 0.0001) (Fig. 9E and fig. S15) and with the up-regulation of NK cells, activated DCs, and effector T cells (P < 0.001, P < 0.0001, and P < 0.0001) (Fig. 9E and figs. S16 to S19). Consequently, the conversion of immunosuppressive TME achieved by our combination significantly (P < 0.0001) induced tumor apoptosis (~78%) as compared to PBS, α-CD47 (~9%), and DL@NP-M-M2pep (~43%) (Fig. 9F). These results confirmed that our nanoformulation is able to modulate macrophage polarization for remodeling the immunosuppressive TME, which can significantly improve anti-HCC efficacy in combination with α-CD47, providing a promising combinatorial approach for HCC.
DISCUSSION
The HCC progression is highly associated with the development of immunosuppressive TME, in which a complex cross-talk between different cell types (e.g., tumor cells and immune cells) exists. Among immune cells, TAMs, which are characterized as M2 (or M2-like) phenotype, are commonly perceived protumoral (25). M2 TAMs maintain the immunosuppressive TME, resulting in tumor progression and immunotherapy resistance (25). Therefore, strategies that transform M2 TAMs to M1 (or M1-like, generally considered antitumoral) may reverse the immunosuppressive TME for HCC therapy (for readability, M1 and M2 are used in this study for the nomenclature).
It becomes increasingly clear that the microbes exert vital functions in human health and disease by means of regulating the critical processes in metabolism, inflammation, and immunity (26). Accumulating evidence confirms that gut microbes and their metabolites regulate the development of HCC through the communication between the intestine and the liver via the hepatic portal vein (termed the “gut–gut microbe–liver” axis) (27, 28). The modulation of gut microbes and their metabolites associated with the gut–gut microbe–liver axis has demonstrated therapeutic promise for HCC (29, 30). Recently, it has been reported that DL (a gut microbe small-molecule metabolite; Fig. 2A) entered the liver through the hepatic portal vein and restored Kupffer cell–mediated pathogen clearance in germ-free mice (13). DL changed the morphology of macrophages by increasing their surface area and volume, which enhanced the phagocytotic capacity of Kupffer cells (it is an essential function of M1 macrophages) (13). Thus, we hypothesize that DL facilitates the transition of TAMs from M2 to M1 and reverses the immunosuppressive TME for HCC therapy.
In this study, M2-associated genes (e.g., Arg1, Fizz, and Il-10) in M2 macrophages were down-regulated following the treatment of DL (Fig. 2, B and C), while M1-associated genes (e.g., Tnf-α, Nos, and Il-12) were up-regulated under the same conditions (Fig. 2, B and C). In addition, DL decreased the CD206+ F4/80+ population (M2 phenotype) in M2 macrophages (Fig. 2D) but increased the CD86+ F4/80+ population (M1 phenotype) under the same conditions (Fig. 2D). Moreover, following the treatment of DL, the morphological structure of M2 macrophages became similar to that of M1 ones (Fig. 2E). These results indicated that DL transforms M2 macrophages to M1 ones.
It has been reported that the PI3K/Akt pathway profoundly contributes to macrophage polarization (23, 31–33). The activation of this pathway induces TAMs into M2 phenotype and generates anti-inflammatory factors resistant to M1 polarization (34), which are highly associated with poor clinical outcomes of tumors (35). Because the correlation of PI3K/Akt pathway and DL-mediated polarization (Fig. 3, A to C), we hypothesize that DL inhibit this pathway to reprogram M2 macrophages to M1 ones.
The isoforms of AKT, namely, AKT1 and AKT2, exhibit opposing functions in macrophage polarization; for example, the loss of AKT1 generates M1 phenotype, but the deficiency of AKT2 induces M2 phenotype (36). In this study, DL suppressed the activity of PI3K, AKT1, STAT6, and PPAR-γ in M2 macrophages (Fig. 3D). Two downstream transcription factors of AKT1, STAT6 and PPAR-γ, are known to restrict M1 polarization but drive M2 polarization (37, 38). Consequently, the expression of ARG-1 and FIZZ (two critical M2 functional factors) was down-regulated by the treatment of DL (Fig. 3D). In contrast, DL enhanced the activity of AKT2, STAT1, and NF-κB in M2 macrophages (Fig. 3D). As two downstream transcription factors of AKT2, STAT1 is positively correlated with the M1 polarization (39), but the role of NF-κB in macrophage polarization is controversy. Previous studies indicated a protumoral role of NF-κB pathway in TAMs (40, 41), and the NF-κB inhibition reprogrammed M2 TAMs to M1 ones (42). In contrast, increasing studies demonstrate that M1-associated proinflammatory components and their anticancer activities are achieved on the NF-κB activation (43), and the transition of M2 TAMs to M1 ones is facilitated via activating the NF-κB pathway (44–46). In this study, DL enhanced the activity of NF-κB in M2 macrophages, which were accompanied with the inhibition of M2-associated anti-inflammatory factors (e.g., ARG-1 and FIZZ; Fig. 3D) and the activation of M1-associated proinflammatory factors (e.g., iNOS and TNF-α; Fig. 3D), indicating the connection of NF-κB activation and M2-to-M1 polarization achieved by DL treatment. The inhibition of PI3K/Akt pathway and the activation of NF-κB pathway were achieved following the interaction of DL with TLR2 and/or TLR9 on the macrophages (Fig. 3, E and F). These results confirmed that DL converts M2 macrophages to M1 ones via modulating the PI3K/Akt pathway (Fig. 3G), demonstrating great potential for remodeling the TME of HCC.
However, note that the in vivo application of exogenous DL for TAM polarization can be seriously impeded by delivery barriers such as poor pharmacokinetics, low tumor accumulation, and nonspecific cell targeting. Emerging nano delivery systems are developed to transport drugs into TAMs by circumventing these delivery barriers (47, 48). For example, NPs with surface coating of tumor cell membranes are known to improve blood circulation and tumor homing (also known as homologous targeting) (49). M2pep, a peptide developed by Pun and co-workers, shows specific targeting for both M2 primary macrophages (e.g., IL-4–stimulated BMDMs) and TAMs but demonstrates low affinity to other leukocytes (including M1 counterparts) (50). This peptide has been therefore used to enhance drug delivery to M2 primary macrophages and TAMs (22–24, 50, 51). PLGA (a copolymer of lactic and glycolic acids) is one of the well-defined drug delivery carriers due to biodegradable characteristics, controllable drug release, and surface functionalization and has been investigated for development of nano drug delivery systems (52, 53). In this study, an M2pep-targeted HCC membrane–coated PLGA NP was produced for delivery of exogenous DL, and the resultant nanoformulation (DL@NP-M-M2pep) demonstrated favorable physicochemical features, with respect to particle size, surface charge, stability, and pH-sensitive drug release (Fig. 4), compliant with the requirements for successful in vivo delivery (54).
Notably, DL@NP-M-M2pep achieved more specific delivery to M2 macrophages than nontargeted counterpart (DL@NP-M) (Fig. 5A) and demonstrated preferential targeting to M2 macrophages relative to M1 ones (fig. S4). These are mainly due to the capacity of M2pep for specifically targeting M2 macrophages (50). As compared to DL@NP-M, DL@NP-M-M2pep significantly suppressed the activity of PI3K, AKT1, STAT6, and PPAR-γ but enhanced the activity of AKT2, STAT1, and NF-κB in M2 macrophages (Fig. 5B). Consequently, DL@NP-M-M2pep attenuated the expression of Arg1, Fizz, TGF-β, and IL-10 (Fig. 5C), while it increased the expression of IL-1β, TNF-α, iNOS, and IL-12 (Fig. 5D). These results confirmed the capacity of DL@NP-M-M2pep for targeting M2 macrophages and converting them into M1 ones.
It has been reported that NPs with the cancer cell membrane coating maintain the biological functionalities of cancer cells (49); therefore, biomimetic NPs are perceived as the “self” and avoid the recognition of immune system and the quick clearance from the bloodstream (55). In this study, DL@NP-M-M2pep achieved sixfold longer blood circulation time of DL in the allograft HCC mouse model as compared to that of free DL (Fig. 6D). In addition, because of the tumor homing function achieved by HCC cell membrane coating (56, 57), tumor accumulation was improved by DL@NP-M-M2pep, which was evident with the delivery of nanoformulation (labeled by DiR) into the tumor (indicated by the luminescence due to the oxidation of luciferin catalyzed by the luciferase in HCC cells) (Fig. 6E). Oppositely, DL@NP-M-M2pep was significantly less observed in the healthy organs (e.g., the lung, spleen, and kidneys) (Fig. 6E). Furthermore, DL@NP-M-M2pep achieved preferential delivery to M2 TAMs relative to M1 ones (Fig. 6F), which is mainly due to the capacity of M2pep for specifically targeting M2 TAMs (50). Together, these results confirmed that our nanoformulation is potentially able to overcome the delivery barriers associated with DL as described above.
Following the successful in vivo delivery, DL@NP-M-M2pep efficiently suppressed tumor growth in the allograft HCC mouse model (Fig. 7, A to C), resulting in long-term animal survival (6 of the 10 mice > 90 days) (Fig. 7D). The anti-HCC efficacy was mainly due to the polarization of M2 TAMs to M1 (Fig. 7, E and F). It is well established that M2 TAMs, as an immunoinhibitory regulator within the TME, support immune suppressive cells (e.g., MDSCs and Tregs) (7) but suppress immune stimulatory cells (e.g., NK cells, activated DCs, and effector T cells) (8). In this study, DL@NP-M-M2pep reduced the amount of MDSCs and Tregs (Fig. 8A), along with the reduced level of immune inhibitory cytokines (IL-10, TGF-β, and IL-4) within the TME (Fig. 8B). In contrast, DL@NP-M-M2pep elevated the amount of NK cells, activated DCs, CD8+ cytotoxic/memory T cells, and CD4+ helper/memory T cells (Fig. 8A), along with the increased level of immune stimulatory cytokines (IFN-γ, TNF-α, and IL-12) within the TME (Fig. 8C). Consequently, DL@NP-M-M2pep induced tumor apoptosis for treating HCC (Fig. 8D). These results confirmed that transformation of TAMs from M2 to M1 successfully remodeled the immunosuppressive “cold” tumor into the immunostimulatory “hot” one.
CD47, also termed as integrin-associated protein, is a transmembrane glycoprotein widely expressed on the surface of various cells (58). Accumulating studies indicate that CD47 is overexpressed by several cancers (e.g., HCC), and the interaction of CD47 and its ligand [signal regulatory protein α (SIRPα)] on macrophages and DCs induces anti-phagocytic “do not eat me” function and causes immune escape (59). Now, antibodies against CD47 and SIRPα are being under investigation in a number of clinical trials for hematological cancers and solid tumors (60). In this study, α-CD47 was able to retard tumor growth in the carcinogen-induced orthotopic HCC mouse model (Fig. 9, A to C) that is a preclinical model known to provide a higher level of genomic instability and form a more “physiologically and clinically relevant” TME (61). This was mainly due to the fact that α-CD47 primes antitumor immune responses via reinforcing the phagocytic function of macrophages and DCs for tumor cells and tumor-associated antigens [see discussion in (62–65)], which was confirmed in this study by the up-regulation of M1 TAMs, activated DCs, and CD8+/CD4+ T cells (Fig. 9E). However, note that α-CD47 on its own was not sufficient to provide long-term animal survival (Fig. 9D). Monotherapy that blocks the interaction of CD47 and SIRPα fails to generate substantial efficacy in clinical trials tested to date (60). Accordingly, other cancer therapies have been used to improve the efficacy of CD47 blockade (66).
In this study, the combination of α-CD47 with DL@NP-M-M2pep profoundly inhibited tumor growth (Fig. 9, A to C) and achieved long-term animal survival (5 of the 10 mice > 60 days) (Fig. 9D). This combinative efficacy was mainly due to the mechanisms as follows (Fig. 1B): (i) DL@NP-M-M2pep transformed M2 TAMs to M1 ones and up-regulated the activated DCs (Fig. 9E). The α-CD47 promotes the efficacy of M1 TAMs and activated DCs for phagocytosis of tumor cells and tumor-associated antigens (62–65), facilitating the recruitment and activation of NK cells and effector T cells (mainly CTLs) for eradicating the residual tumor cells (Fig. 9F) (67). (ii) DL@NP-M-M2pep reversed the cold tumor to the hot one, which was confirmed by the down-regulation of immune suppressive cells and the up-regulation of immune stimulatory cells (Fig. 9E). The efficacy of α-CD47 for antitumor immunity can be augmented within the hot tumor, which is similar to the results have been previously reported in other immune checkpoints [e.g., programmed death 1 (PD-1)/programmed death ligand 1 (PD-L1)] (68–70). Together, the mutually positive feedback between α-CD47 and DL@NP-M-M2pep provides the combinative immunotherapeutic effects for eradicating HCC.
In conclusion, the capacity of DL for TAM modulation was confirmed for the first time (Fig. 2). DL interacted with TLR2 and/or TLR9 on macrophages, which induced the inhibition of PI3K/Akt pathway and the activation of NF-κB pathway, facilitating macrophage polarization from M2 to M1 (Fig. 3). A M2pep-targeted HCC membrane–coated PLGA NP was developed to load DL, and the resultant nanoformulation (DL@NP-M-M2pep) demonstrated favorable physicochemical and ex vivo characteristics (Figs. 4 and 5). DL@NP-M-M2pep significantly improved half-life, tumor distribution, and M2 TAM targeting, without eliciting significant toxic signs (Fig. 6). Consequently, DL@NP-M-M2pep reprogrammed M2 TAMs to M1 ones and reversed the immunosuppressive TME, achieving antitumor outcome in allograft and carcinogen-induced orthotopic HCC mouse models, respectively (Figs. 7 to 9). Notably, the efficacy of α-CD47 was significantly augmented by DL@NP-M-M2pep, generating combinative efficacy in carcinogen-induced orthotopic HCC mice (Fig. 9). These results reveal a potential TAM modulatory function of DL and provide a proof of concept for “DL and anti-CD47 antibody” combinatorial strategy to HCC immunotherapy. In the future, other formulation materials and techniques should be investigated to improve the efficiency and practicality of DL-based nanotherapeutics, aiming to fulfill the combinatorial strategy for clinical application.
MATERIALS AND METHODS
Materials
DL (catalog no. 71716) was bought in Sigma-Aldrich. PLGA [20 kDa, 50:50 lactic acid (LA):glycolic acid (GA)] and 1,2-Distearoyl-sn-glycero-3-phosphorylethanolamine (DSPE)-polyethylene glycol 2000 (PEG2000)-M2pep (DSPE-PEG-M2pep) (M2pep sequence: YEQDPWGVKWWY) were obtained in Xi’an Ruixi Biotechnology Company, China. The list of antibodies, enzyme-linked immunosorbent assay (ELISA) kits, and primers is provided in tables S1, S2, and S3, respectively. The rest of materials and reagents were bought in Sigma-Aldrich unless other mentioned.
Animals
Male and female 5- to 6-week-old C57BL/6J mice (RRID: MGI: 3028467) were bought in Beijing Vital River Laboratory Animal Technology Company. Experiments were approved by the Experimental Animal Ethics Committee of Jilin University (ref. no. 20210002) and were accomplished on the basis of the Guide for the Care and Use of Laboratory Animals.
Cell culture
Mouse HCC Hepa1-6 cells (RRID: CVCL_0327) were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Thermo Fisher Scientific). Mouse HCC Hepa1-6-luc cells (stably expressing luciferase; RRID: CVCL_0327) were cultured in DMEM containing 10% FBS, 1% penicillin-streptomycin, and puromycin (1 μg/ml; Thermo Fisher Scientific). Cells were maintained at 37°C, 5% CO2, and 95% relative humidity.
Ex vivo characterization of DL
BMDMs were prepared as previously reported (71). In brief, BM cells were harvested from the femur and tibia of 5- to 6-week-old C57BL/6J mice. Then, BM cells were cultured in the RPMI 1640 medium (Corning) and 10% FBS and 1% penicillin-streptomycin, supplemented with macrophage colony-stimulating factor (20 ng/ml; Thermo Fisher Scientific) for 5 days to achieve BMDMs. On day 5, BMDMs were cultured for 48 hours in fresh growth medium with either IFN-γ (20 ng/ml; Thermo Fisher Scientific) and LPS (100 ng/ml) or IL-4 (20 ng/ml; Thermo Fisher Scientific) to induce BMDMs to differentiate into M1 or M2 macrophages. Subsequently, macrophages (2 × 105 per well) were seeded overnight in six-well plates and incubated with/without DL [concentration(c) = 50 mM] for 24 hours. The morphological structures were imaged under the light microscope.
In addition, macrophages (2 × 105 per well) were with/without DL ([c] = 10, 50, and 100 mM) in six-well plates for 24 hours. The F4/80+ CD206+ (M2) or F4/80+ CD86+ (M1) populations were determined using flow cytometry (BD FACSCalibur) (72), and the results were analyzed using FlowJo software (version 7.6).
Macrophages (2 × 105 per well) were also incubated with/without DL ([c] = 50 mM) in six-well plates for 24 hours. To study the gene expression profiling, RNA was collected with the RNeasy MiniElute Kit (Qiagen), reverse-transcribed to complementary DNA (cDNA), and sequenced using next-generation sequencing (Illumina). The differential expression analysis was carried out by Biomarker Technologies Co. Ltd. The gene set enrichment analysis (GSEA) was performed with GSEA by false discovery rate q value < 1. The GO and KEGG were performed using the bioinformatics database in BMKCloud (www.biocloud.net/).
Macrophages (2 × 105 per well) were also incubated with/without DL ([c] = 10, 50, and 100 mM) in six-well plates for 24 hours. RNA was extracted using TRIzol reagent (Invitrogen) and reverse-transcribed to cDNA using the Reverse Transcription Master Kit (YEASEN Biotech, China). The quantitative RT-PCR was performed using a 7500 real-time PCR system (Applied Biosystems) under conditions as previously described (68). The mRNA levels were normalized by glyceraldehyde-3-phosphate dehydrogenase.
Macrophages (2 × 105 per well) were also incubated with/without DL ([c] = 50 mM) in six-well plates for 24 hours. Proteins were extracted and qualified as previously reported (70). A sample containing 40 μg of proteins was used for Western blot experiments. Following the incubation with appropriate antibodies, the protein bands were visualized and quantified as previously reported (70).
The DL was docked to the TLR2 and TLR9 proteins using AutoDock 4.2 (73). The three-dimensional structure of DL (compound CID: 61503) was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov), and the protein structures of TLR2 and TLR9 (Protein Data Bank ID: 3A7C and 3WPF) were obtained from the RCSB Protein Data Bank (www.rcsb.org). The grid parameter was generated using the AutoDock Tools (73). The box size was 60 Å by 60 Å by 60 Å with a standard spacing of 0.375 Å. During the docking process, all the relevant torsion angles of ligand were treated as rotatable to allow searching for more conformational space. An initial population was set to 100 randomly placed individuals, and the maximum number of energy evaluations was set to 2,500,000. Last, a total of 100 docking poses were generated, and the pose with the lowest binding free energy was selected as the optimal pose. The same procedure was conducted for the docking simulations of DL with TLR9. In addition, macrophages (2 × 105 per well) were pretreated with TLR2 inhibitor (TL2-C29, InvivoGen) and/or TLR9 inhibitor (ODN 2088, InvivoGen) in six-well plates for 2 hours. After PBS wash, cells were incubated with DL ([c] = 50 mM) for 24 hours. The activity of PI3K and NF-κB was determined using Western blot assay as described above.
Preparation of nanoformulation
PLGA NPs were achieved using the double-emulsion technique. Briefly, 200 μl of PBS containing 150 mg of DL was used as the internal aqueous phase. Subsequently, 1 ml of methylene chloride containing 10 mg of PLGA was used as the oil phase. The internal aqueous phase and oil phase were mixed under a sonication (Digital Sonifier 450, Branson Ultrasonics Corp., USA) for 40 s to prepare the primary emulsion. The primary emulsion was added to 3 ml of external aqueous phase [2% (w/v) sodium cholate] to form a double-emulsion suspension under the sonication within an ice bath for 2 min. Subsequently, the double emulsion was added to 6 ml of aqueous phase containing 0.5% (w/v) sodium cholate, which were then solidified into NPs (termed DL@NP) by evaporating methylene chloride for 4 hours at room temperature.
The HCC membrane was prepared as previously reported (74). Briefly, 2 × 108 Hepa1-6 cells were suspended in 2 ml of Hepes B buffer [Hepes, 2.38 g/liter; MgCl2, 0.476 g/liter; EDTA, 0.292 g/liter, dithiothreitol, 0.154 g/liter; and KCl, 0.746 g/liter (pH 7.6)] with 1% Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific). Cells were enucleated in a disperser (IKA T18), and the supernatant was added on a discontinuous sucrose density gradient composed of 55, 40, and 30% (w/v) sucrose in Hepes B buffer. Following ultracentrifugation (28,000 rpm, 30 min, and 4°C), the HCC membrane was collected, quantified using bicinchonininc acid (BCA) assay (74), and stored in PBS at −20°C. Subsequently, the M2pep-modifed HCC membrane was prepared as previously reported (75). Briefly, 100 μg of DSPE-PEG-M2pep were added to 1 mg of HCC membrane for 30 min at 4°C, and the content was passed once through the extruder (Avanti Polar Lipids) equipped by a 0.2-μm polycarbonate porous membrane, forming M2pep-modifed HCC membrane. After this, 5 mg of DL@NP was added by 1 mg of M2pep-modifed HCC membrane ([c] was determined on the basis of the proteins), and the content was passed nine times through the extruder as mentioned above, forming M2pep-targeted HCC membrane–coated DL-loaded PLGA nanoformulation (termed DL@NP-M-M2pep). The nontargeted nanoformulation (termed DL@NP-M) was prepared as mentioned above without the addition of DSPE-PEG-M2pep. In addition, rhodamine (or DiR)–loaded (0.05 wt %) nanoformulations were prepared as mentioned above with “DL + rhodamine (or DiR)”–coloaded NPs.
Physicochemical and ex vivo characterization of nanoformulation
The morphologic structures of nanoformulations were detected using TEM (JEM-1230) (76). The particle size, surface charge, and stability of nanoformulations were assessed using Malvern Nano-ZS (77). The LC of DL was determined using the d-Lactate Colorimetric Assay Kit (BioVision, USA) as previously described (78), and LC (%) = (the weight of encapsulated drug/the weight of nanoformulation) × 100%.
The cellular uptake of nanoformulations was studied using flow cytometry. The M2 macrophages (2 × 105 per well) were seeded in six-well plates overnight. Cells were then incubated with rhodamine-loaded (0.05 wt %) nanoformulations ([c] of DL = 50 mM). Following 6 hours of incubation, the population of rhodamine-positive cells (%) was determined by flow cytometry.
In addition, M2 macrophages (2 × 105 per well) were seeded in six-well plates overnight and subsequently incubated with nanoformulations ([c] of DL = 50 mM). Following 24 hours of incubation, the Western blot assay was used to evaluate the activity of PI3K/Akt and NF-κB pathways, and the quantitative RT-PCR assay was used to determine the expression of key functional markers.
Safety, half-life, and biodistribution of nanoformulation
The toxicity was assessed in healthy mice (n = 6) following intravenous injections of PBS, free DL (100 mM), and nanoformulation (100 mM DL) (Fig. 6A), and the body weight was regularly recorded. The histopathology of major organs (the heart, liver, spleen, lung, and kidneys) was determined on day 30 using hematoxylin and eosin staining assay (70). The liver and kidney functions (day 30), including ALT, AST, BUN, and CRE, were determined by Wuhan Servicebio Technology Co. Ltd.
The allograft orthotopic HCC model was established as previously reported (79). Briefly, the spleen was exposed, tied in the middle, and cut into two parts with intact vascular pedicle at both sides. Subsequently, 1 × 106 Hepa1-6-luc cells were inoculated into the liver via the vascular vessel in the half spleen, and the other half was returned to the abdomen to keep the immune systems competent (day 0). After this, 90 μl of luciferin (10 μg/μl; Pierce) were intraperitoneally given to animals for monitoring tumor development (IVIS System, PerkinElmer). As an intensity of ~5 × 108 to 10 × 108 p/s/cm2/sr was reached, animals were used for pharmacokinetics and tissue distribution. Mice (n = 4) were intravenously administrated with nanoformulations (100 mM DL), and the concentration of DL in the serum at different time points was determined using the d-Lactate Colorimetric Assay Kit. In addition, mice (n = 4) were intravenously injected with DiR-loaded nanoformulations (0.05 wt % DiR; 100 mM DL). One day after intravenous injection, biodistribution of DiR-loaded nanoformulations was analyzed using the IVIS System (748 nm/780 nm).
In vivo efficacy of nanoformulation
The allograft orthotopic HCC model was established as described above. When an intensity of ~5 × 108 to 10 × 108 p/s/cm2/sr was reached, animals (n = 10) were intravenously injected with PBS, free DL (100 mM), DL@NP-M (100 mM DL), and DL@NP-M-M2pep (100 mM DL) (Fig. 7A). The tumor development and animal survival were assessed using the IVIS System. In addition, animals were euthanized 1 week after treatments, and the tumors were collected for the following experiments: (i) Tumor apoptosis. Tumor tissues were fixed in 4% paraformaldehyde (PFA) and prepared in paraffin-embedded slides. The dewaxed sections (8 μm) were permeabilized, before the staining using the DeadEnd Fluorometric TUNEL system (Promega) and 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific) for CLSM analysis (Olympus FV3000) (79). (ii) Immunofluorescent staining assay. Tumor tissues were fixed in 4% PFA and prepared in paraffin-embedded slides. The immunofluorescent staining assay was performed by Wuhan Servicebio Technology Co. Ltd. Briefly, the dewaxed sections (8 μm) were performed with the antigen retrieval, permeabilization, and blocking (5% bovine serum albumin), before the use of appropriate antibodies for identification of M1 and M2 TAMs using CLSM. (iii) Detection of immune cells. The single cells from tumors were prepared as previously described (69). Following the removal of blood red cells with the Ammonium-Chloride-Potassium (ACK) buffer (Gibco), cells were treated using appropriate antibodies for flow cytometry to determine TAMs, activated DCs, effector T cells, NK cells, MDSCs, and Tregs (69). (iv) Measurement of cytokines and chemokines. Tumors were homogenized using a tissue grinder, and the mRNA level of cytokines and chemokines in the supernatant was determined using the quantitative RT-PCR as described above. In addition, the protein level of cytokines and chemokines in the supernatant was measured using ELISA kits according to the manufacturer’s instruction.
In addition, the carcinogen-induced orthotopic HCC mouse model was established as previously described (80). In brief, after male and female C57BL/6J mice were mated, 2-week-old male mice were intraperitoneally injected once with 100 μl of diethylnitrosamine (DEN; [c] = 10 mg/ml). One week later, mice freely received the drinking water with DEN (2 mg/ml) for 5 months. Subsequently, HCC mice (n = 10) were intravenously injected with different treatments as described above (Fig. 9A). In addition, mice were euthanized 1 week after treatments, and the tumors were collected for the experiments as described above.
Statistical analysis
Each experiment has been repeated twice at least, and all data were shown as means ± SEM. The sample size for all experiments was determined to qualify statistical analysis. Significance of two groups was determined by a two-tailed unpaired Student’s t test. Significance between multiple groups was determined by one-way analysis of variance (ANOVA; Tukey’s multiple comparisons). P < 0.05 was considered statistically significant.
Acknowledgments
We thank L. Huang from the Eshelman School of Pharmacy, University of North Carolina, Chapel Hill, for helpful comments on the manuscript.
Funding: This work was supported by the National Natural Science Foundation of China (82222074, 82074154, and 81774240); the Siming Scholar from Shanghai Shuguang Hospital (SGXZ-201904); the Youth Tip-top Talent Program from Shanghai Municipal Commission of Health; the Constant-eminent Program from Shanghai Municipal Commission of Health; the Xinglin Youth Scholar from Shanghai University of Traditional Chinese Medicine; the Science and Technology Department of Jilin Province (20230402044GH and 20230101170JC); the Norman Bethune Program of Jilin University, China (2022B40); and the “Medicine + X” Interdisciplinary Innovation Team of Norman Bethune Health Science Center of Jilin University, China (2022JBGS05).
Author contributions: Conceptualization: Z.Y. and J.G. Supervision: Z.Y. and J.G. Data curation: S.H., X.B., Y.Z., L.W., Y.L., L.Y., A.L., and X.Z. Investigation: S.H., X.B., Y.Z., L.W., Y.L., L.Y., A.L., and X.Z. Validation: S.H., X.B., Y.Z., L.W., Y.L., L.Y., A.L., and X.Z. Formal analysis: S.H., X.B., Y.Z., L.W., Y.L., L.Y., A.L., X.Z., X.J., D.L., Y.D., and Q.-C.Z. Writing (original draft): S.H., Z.Y., and J.G. Writing (review and editing): S.H., Z.Y., and J.G. Resources: X.J., D.L., Y.D., and Q.-C.Z. Funding acquisition: S.H., Z.Y., and J.G.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S19
Tables S1 to S3
REFERENCES AND NOTES
- 1.H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, F. Bray, Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.P. J. Trivedi, D. H. Adams, Gut-liver immunity. J. Hepatol. 64, 1187–1189 (2016). [DOI] [PubMed] [Google Scholar]
- 3.F. Heymann, F. Tacke, Immunology in the liver—From homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 13, 88–110 (2016). [DOI] [PubMed] [Google Scholar]
- 4.C. N. Jenne, P. Kubes, Immune surveillance by the liver. Nat. Immunol. 14, 996–1006 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Y. Yang, J. Guo, L. Huang, Tackling TAMs for cancer immunotherapy: It’s nano time. Trends Pharmacol. Sci. 41, 701–714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Y. Komohara, Y. Fujiwara, K. Ohnishi, M. Takeya, Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv. Drug Deliv. Rev. 99, 180–185 (2016). [DOI] [PubMed] [Google Scholar]
- 7.C. Ngambenjawong, H. H. Gustafson, S. H. Pun, Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv. Drug Deliv. Rev. 114, 206–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Y. Xia, L. Rao, H. Yao, Z. Wang, P. Ning, X. Chen, Engineering macrophages for cancer immunotherapy and drug delivery. Adv. Mater. 32, e2002054 (2020). [DOI] [PubMed] [Google Scholar]
- 9.L. Rao, L. Wu, Z. Liu, R. Tian, G. Yu, Z. Zhou, K. Yang, H. G. Xiong, A. Zhang, G. T. Yu, W. Sun, H. Xu, J. Guo, A. Li, H. Chen, Z. J. Sun, Y. X. Fu, X. Chen, Hybrid cellular membrane nanovesicles amplify macrophage immune responses against cancer recurrence and metastasis. Nat. Commun. 11, 4909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.X. Cao, B. Li, J. Chen, J. Dang, S. Chen, E. G. Gunes, B. Xu, L. Tian, S. Muend, M. Raoof, C. Querfeld, J. Yu, S. T. Rosen, Y. Wang, M. Feng, Effect of cabazitaxel on macrophages improves CD47-targeted immunotherapy for triple-negative breast cancer. J. Immunother. Cancer 9, e002022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.G. R. Gunassekaran, S. M. Poongkavithai Vadevoo, M. C. Baek, B. Lee, M1 macrophage exosomes engineered to foster M1 polarization and target the IL-4 receptor inhibit tumor growth by reprogramming tumor-associated macrophages into M1-like macrophages. Biomaterials 278, 121137 (2021). [DOI] [PubMed] [Google Scholar]
- 12.K. Yang, W. Han, X. Jiang, A. Piffko, J. Bugno, C. Han, S. Li, H. Liang, Z. Xu, W. Zheng, L. Wang, J. Wang, X. Huang, J. P. Y. Ting, Y. X. Fu, W. Lin, R. R. Weichselbaum, Zinc cyclic di-AMP nanoparticles target and suppress tumours via endothelial STING activation and tumour-associated macrophage reinvigoration. Nat. Nanotechnol. 17, 1322–1331 (2022). [DOI] [PubMed] [Google Scholar]
- 13.B. McDonald, A. Z. Zucoloto, I. L. Yu, R. Burkhard, K. Brown, M. B. Geuking, K. D. McCoy, Programing of an intravascular immune firewall by the gut microbiota protects against pathogen dissemination during infection. Cell Host Microbe 28, 660–668.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 14.X. Zhou, Y. Liu, M. Hu, M. Wang, X. Liu, L. Huang, Relaxin gene delivery modulates macrophages to resolve cancer fibrosis and synergizes with immune checkpoint blockade therapy. Sci. Adv. 7, eabb6596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. A. Hoeksema, Z. Shen, I. R. Holtman, A. Zheng, N. J. Spann, I. Cobo, M. Gymrek, C. K. Glass, Mechanisms underlying divergent responses of genetically distinct macrophages to IL-4. Sci. Adv. 7, eabf9808 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.X. Cao, J. Chen, B. Li, J. Dang, W. Zhang, X. Zhong, C. Wang, M. Raoof, Z. Sun, J. Yu, M. G. Fakih, M. Feng, Promoting antibody-dependent cellular phagocytosis for effective macrophage-based cancer immunotherapy. Sci. Adv. 8, eabl9171 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Li, H. Beauchemin, N. Popovic, A. Peterson, E. d'Hennezel, C. A. Piccirillo, C. Sun, C. Polychronakos, The common, autoimmunity-predisposing 620Arg > Trp variant of PTPN22 modulates macrophage function and morphology. J. Autoimmun. 79, 74–83 (2017). [DOI] [PubMed] [Google Scholar]
- 18.W. Zhao, D. Xu, L. Zhang, H. Meng, Q. Zheng, J. Wang, Anti-inflammation of torachrysone-8-O-β-d-glucoside by hurdling over morphological changes of macrophages. Int. Immunopharmacol. 105, 108548 (2022). [DOI] [PubMed] [Google Scholar]
- 19.S. Roy, A. Mukherjee, B. Paul, O. Rahaman, S. Roy, G. Maithri, B. Ramya, S. Pal, D. Ganguly, A. Talukdar, Design and development of benzoxazole derivatives with toll-like receptor 9 antagonism. Eur. J. Med. Chem. 134, 334–347 (2017). [DOI] [PubMed] [Google Scholar]
- 20.S. R. Natala, A. Habas, E. M. Stocking, A. Orry, D. L. Price, M. B. Gill, D. W. Bonhaus, R. Abagyan, W. Wrasidlo, Structure based design and synthesis of novel Toll-like receptor 2 (TLR 2) lipid antagonists. Bioorg. Med. Chem. Lett. 40, 127861 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Y. Zeng, S. Li, S. Zhang, L. Wang, H. Yuan, F. Hu, Cell membrane coated-nanoparticles for cancer immunotherapy. Acta Pharm. Sin. B 12, 3233–3254 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Y. Qian, S. Qiao, Y. Dai, G. Xu, B. Dai, L. Lu, X. Yu, Q. Luo, Z. Zhang, Molecular-targeted immunotherapeutic strategy for melanoma via dual-targeting nanoparticles delivering small interfering RNA to tumor-associated macrophages. ACS Nano 11, 9536–9549 (2017). [DOI] [PubMed] [Google Scholar]
- 23.M. Li, M. Li, Y. Yang, Y. Liu, H. Xie, Q. Yu, L. Tian, X. Tang, K. Ren, J. Li, Z. Zhang, Q. He, Remodeling tumor immune microenvironment via targeted blockade of PI3K-γ and CSF-1/CSF-1R pathways in tumor associated macrophages for pancreatic cancer therapy. J. Control. Release 321, 23–35 (2020). [DOI] [PubMed] [Google Scholar]
- 24.M. Yang, B. Wang, Y. Yin, X. Ma, L. Tang, Y. Zhang, Q. Fan, T. Yin, Y. Wang, PTN-PTPRZ1 signaling axis blocking mediates tumor microenvironment remodeling for enhanced glioblastoma treatment. J. Control. Release 353, 63–76 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Q. Yang, N. Guo, Y. Zhou, J. Chen, Q. Wei, M. Han, The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm. Sin. B 10, 2156–2170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.V. M. Juarez, A. N. Montalbine, A. Singh, Microbiome as an immune regulator in health, disease, and therapeutics. Adv. Drug Deliv. Rev. 188, 114400 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L. X. Yu, R. F. Schwabe, The gut microbiome and liver cancer: Mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 14, 527–539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R. F. Schwabe, T. F. Greten, Gut microbiome in HCC – Mechanisms, diagnosis and therapy. J. Hepatol. 72, 230–238 (2020). [DOI] [PubMed] [Google Scholar]
- 29.G. Ji, L. Ma, H. Yao, S. Ma, X. Si, Y. Wang, X. Bao, L. Ma, F. Chen, C. Ma, L. Huang, X. Fang, W. Song, Precise delivery of obeticholic acid via nanoapproach for triggering natural killer T cell-mediated liver cancer immunotherapy. Acta Pharm. Sin. B 10, 2171–2182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.G. Ji, X. Si, S. Dong, Y. Xu, M. Li, B. Yang, Z. Tang, X. Fang, L. Huang, W. Song, X. Chen, Manipulating liver bile acid signaling by nanodelivery of bile acid receptor modulators for liver cancer immunotherapy. Nano Lett. 21, 6781–6791 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Q. Luo, N. Zheng, L. Jiang, T. Wang, P. Zhang, Y. Liu, P. Zheng, W. Wang, G. Xie, L. Chen, D. Li, P. Dong, X. Yuan, L. Shen, Lipid accumulation in macrophages confers protumorigenic polarization and immunity in gastric cancer. Cancer Sci. 111, 4000–4011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.C. Yang, R. Dou, C. Wei, K. Liu, D. Shi, C. Zhang, Q. Liu, S. Wang, B. Xiong, Tumor-derived exosomal microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM interaction to facilitate CRC metastasis. Mol. Ther. 29, 2088–2107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. D. Troutman, J. F. Bazan, C. Pasare, Toll-like receptors, signaling adapters and regulation of the pro-inflammatory response by PI3K. Cell Cycle 11, 3559–3567 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D. A. Fruman, H. Chiu, B. D. Hopkins, S. Bagrodia, L. C. Cantley, R. T. Abraham, The PI3K pathway in human disease. Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J. K. Sa, N. Chang, H. W. Lee, H. J. Cho, M. Ceccarelli, L. Cerulo, J. Yin, S. S. Kim, F. P. Caruso, M. Lee, D. Kim, Y. T. Oh, Y. Lee, N. G. Her, B. Min, H. J. Kim, D. E. Jeong, H. M. Kim, H. Kim, S. Chung, H. G. Woo, J. Lee, D. S. Kong, H. J. Seol, J. I. Lee, J. Kim, W. Y. Park, Q. Wang, E. P. Sulman, A. B. Heimberger, M. Lim, J. B. Park, A. Iavarone, R. G. W. Verhaak, D. H. Nam, Transcriptional regulatory networks of tumor-associated macrophages that drive malignancy in mesenchymal glioblastoma. Genome Biol. 21, 216 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. Arranz, C. Doxaki, E. Vergadi, Y. Martinez de la Torre, K. Vaporidi, E. D. Lagoudaki, E. Ieronymaki, A. Androulidaki, M. Venihaki, A. N. Margioris, E. N. Stathopoulos, P. N. Tsichlis, C. Tsatsanis, Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc. Natl. Acad. Sci. U.S.A. 109, 9517–9522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.T. Yu, S. Gan, Q. Zhu, D. Dai, N. Li, H. Wang, X. Chen, D. Hou, Y. Wang, Q. Pan, J. Xu, X. Zhang, J. Liu, S. Pei, C. Peng, P. Wu, S. Romano, C. Mao, M. Huang, X. Zhu, K. Shen, J. Qin, Y. Xiao, Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat. Commun. 10, 4353 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. A. Bouhlel, B. Derudas, E. Rigamonti, R. Dievart, J. Brozek, S. Haulon, C. Zawadzki, B. Jude, G. Torpier, N. Marx, B. Staels, G. Chinetti-Gbaguidi, PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 6, 137–143 (2007). [DOI] [PubMed] [Google Scholar]
- 39.T. Lawrence, G. Natoli, Transcriptional regulation of macrophage polarization: Enabling diversity with identity. Nat. Rev. Immunol. 11, 750–761 (2011). [DOI] [PubMed] [Google Scholar]
- 40.E. Pikarsky, R. M. Porat, I. Stein, R. Abramovitch, S. Amit, S. Kasem, E. Gutkovich-Pyest, S. Urieli-Shoval, E. Galun, Y. Ben-Neriah, NF-κB functions as a tumour promoter in inflammation-associated cancer. Nature 431, 461–466 (2004). [DOI] [PubMed] [Google Scholar]
- 41.S. Maeda, H. Kamata, J. L. Luo, H. Leffert, M. Karin, IKKβ couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell 121, 977–990 (2005). [DOI] [PubMed] [Google Scholar]
- 42.J. Khalife, J. Ghose, M. Martella, D. Viola, A. Rocci, E. Troadec, C. Terrazas, A. R. Satoskar, E. G. Gunes, A. Dona, J. F. Sanchez, P. L. Bergsagel, M. Chesi, A. Pozhitkov, S. Rosen, G. Marcucci, J. J. Keats, C. C. Hofmeister, A. Krishnan, E. Caserta, F. Pichiorri, MiR-16 regulates crosstalk in NF-κB tolerogenic inflammatory signaling between myeloma cells and bone marrow macrophages. JCI Insight 4, e129348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Mantovani, F. Marchesi, A. Malesci, L. Laghi, P. Allavena, Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Z. Yu, Y. Li, Y. Li, J. Zhang, M. Li, L. Ji, Y. Tang, Y. Zheng, J. Sheng, Q. Han, F. Li, J. Guo, L. Wang, X. Sun, Y. Gao, H. Feng, Bufalin stimulates antitumor immune response by driving tumor-infiltrating macrophage toward M1 phenotype in hepatocellular carcinoma. J. Immunother. Cancer 10, e004297 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.S. Wang, M. Lu, W. Wang, S. Yu, R. Yu, C. Cai, Y. Li, Z. Shi, J. Zou, M. He, W. Xie, D. Yu, H. Jin, H. Li, W. Xiao, C. Fan, F. Wu, Y. Li, S. Liu, Macrophage polarization modulated by NF-κB in polylactide membranes-treated peritendinous adhesion. Small 18, e2104112 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Y. Xu, Q. Wang, J. Chen, J. Wu, J. Su, W. Gao, W. Chen, S. Shi, S. Wang, H. Wang, An arabinogalactan isolated from Pollen Typhae induces the apoptosis of RKO cells by promoting macrophage polarization. Carbohydr. Polym. 299, 120216 (2023). [DOI] [PubMed] [Google Scholar]
- 47.M. Sylvestre, C. A. Crane, S. H. Pun, Progress on modulating tumor-associated macrophages with biomaterials. Adv. Mater. 32, e1902007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.C. Zhao, X. Pang, Z. Yang, S. Wang, H. Deng, X. Chen, Nanomaterials targeting tumor associated macrophages for cancer immunotherapy. J. Control. Release 341, 272–284 (2022). [DOI] [PubMed] [Google Scholar]
- 49.R. H. Fang, Y. Jiang, J. C. Fang, L. Zhang, Cell membrane-derived nanomaterials for biomedical applications. Biomaterials 128, 69–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.M. Cieslewicz, J. Tang, J. L. Yu, H. Cao, M. Zavaljevski, K. Motoyama, A. Lieber, E. W. Raines, S. H. Pun, Targeted delivery of proapoptotic peptides to tumor-associated macrophages improves survival. Proc. Natl. Acad. Sci. U.S.A. 110, 15919–15924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.C. Ngambenjawong, M. Cieslewicz, J. G. Schellinger, S. H. Pun, Synthesis and evaluation of multivalent M2pep peptides for targeting alternatively activated M2 macrophages. J. Control. Release 224, 103–111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. Guo, L. Huang, Nanodelivery of cGAS-STING activators for tumor immunotherapy. Trends Pharmacol. Sci. 43, 957–972 (2022). [DOI] [PubMed] [Google Scholar]
- 53.J. Guo, Y. Zou, L. Huang, Nano delivery of chemotherapeutic ICD inducers for tumor immunotherapy. Small Methods 7, e2201307 (2023). [DOI] [PubMed] [Google Scholar]
- 54.J. Guo, L. Huang, Membrane-core nanoparticles for cancer nanomedicine. Adv. Drug Deliv. Rev. 156, 23–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. Wang, M. Zhu, G. Nie, Biomembrane-based nanostructures for cancer targeting and therapy: From synthetic liposomes to natural biomembranes and membrane-vesicles. Adv. Drug Deliv. Rev. 178, 113974 (2021). [DOI] [PubMed] [Google Scholar]
- 56.R. Yang, J. Xu, L. Xu, X. Sun, Q. Chen, Y. Zhao, R. Peng, Z. Liu, Cancer cell membrane-coated adjuvant nanoparticles with mannose modification for effective anticancer vaccination. ACS Nano 12, 5121–5129 (2018). [DOI] [PubMed] [Google Scholar]
- 57.J. Gan, G. Du, C. He, M. Jiang, X. Mou, J. Xue, X. Sun, Tumor cell membrane enveloped aluminum phosphate nanoparticles for enhanced cancer vaccination. J. Control. Release 326, 297–309 (2020). [DOI] [PubMed] [Google Scholar]
- 58.A. van Duijn, S. H. Van der Burg, F. A. Scheeren, CD47/SIRPα axis: Bridging innate and adaptive immunity. J. Immunother. Cancer 10, e004589 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A. Veillette, J. Chen, SIRPα–CD47 immune checkpoint blockade in anticancer therapy. Trends Immunol. 39, 173–184 (2018). [DOI] [PubMed] [Google Scholar]
- 60.M. Feng, W. Jiang, B. Y. S. Kim, C. C. Zhang, Y. X. Fu, I. L. Weissman, Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat. Rev. Cancer 19, 568–586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.B. Olson, Y. Li, Y. Lin, E. T. Liu, A. Patnaik, Mouse models for cancer immunotherapy research. Cancer Discov. 8, 1358–1365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.X. Liu, Y. Pu, K. Cron, L. Deng, J. Kline, W. A. Frazier, H. Xu, H. Peng, Y. X. Fu, M. M. Xu, CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat. Med. 21, 1209–1215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.S. Wang, Q. Wu, T. Chen, R. Su, C. Pan, J. Qian, H. Huang, S. Yin, H. Xie, L. Zhou, S. Zheng, Blocking CD47 promotes antitumour immunity through CD103+ dendritic cell-NK cell axis in murine hepatocellular carcinoma model. J. Hepatol. 77, 467–478 (2022). [DOI] [PubMed] [Google Scholar]
- 64.H. L. Matlung, K. Szilagyi, N. A. Barclay, T. K. van den Berg, The CD47-SIRPα signaling axis as an innate immune checkpoint in cancer. Immunol. Rev. 276, 145–164 (2017). [DOI] [PubMed] [Google Scholar]
- 65.W. B. Yu, Z. H. Ye, X. Chen, J. J. Shi, J. J. Lu, The development of small-molecule inhibitors targeting CD47. Drug Discov. Today 26, 561–568 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Q. Chen, C. Wang, X. Zhang, G. Chen, Q. Hu, H. Li, J. Wang, D. Wen, Y. Zhang, Y. Lu, G. Yang, C. Jiang, J. Wang, G. Dotti, Z. Gu, In situ sprayed bioresponsive immunotherapeutic gel for post-surgical cancer treatment. Nat. Nanotechnol. 14, 89–97 (2019). [DOI] [PubMed] [Google Scholar]
- 67.A. Mantovani, P. Allavena, The interaction of anticancer therapies with tumor-associated macrophages. J. Exp. Med. 212, 435–445 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.J. Guo, Z. Yu, M. Das, L. Huang, Nano codelivery of oxaliplatin and folinic acid achieves synergistic chemo-immunotherapy with 5-fluorouracil for colorectal cancer and liver metastasis. ACS Nano 14, 5075–5089 (2020). [DOI] [PubMed] [Google Scholar]
- 69.J. Guo, Z. Yu, D. Sun, Y. Zou, Y. Liu, L. Huang, Two nanoformulations induce reactive oxygen species and immunogenetic cell death for synergistic chemo-immunotherapy eradicating colorectal cancer and hepatocellular carcinoma. Mol. Cancer 20, 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.D. Sun, Y. Zou, L. Song, S. Han, H. Yang, D. Chu, Y. Dai, J. Ma, C. M. O’Driscoll, Z. Yu, J. Guo, A cyclodextrin-based nanoformulation achieves co-delivery of ginsenoside Rg3 and quercetin for chemo-immunotherapy in colorectal cancer. Acta Pharm. Sin. B 12, 378–393 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.C. W. Shields IV, M. A. Evans, L. L. Wang, N. Baugh, S. Iyer, D. Wu, Z. Zhao, A. Pusuluri, A. Ukidve, D. C. Pan, S. Mitragotri, Cellular backpacks for macrophage immunotherapy. Sci. Adv. 6, eaaz6579 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.H. Yang, L. Song, B. Sun, D. Chu, L. Yang, M. Li, H. Li, Y. Dai, Z. Yu, J. Guo, Modulation of macrophages by a paeoniflorin-loaded hyaluronic acid-based hydrogel promotes diabetic wound healing. Mater. Today Bio 12, 100139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell, A. J. Olson, AutoDock4 and autoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Y. Lv, M. Liu, Y. Zhang, X. Wang, F. Zhang, F. Li, W. E. Bao, J. Wang, Y. Zhang, W. Wei, G. Ma, L. Zhao, Z. Tian, Cancer cell membrane-biomimetic nanoprobes with two-photon excitation and near-infrared emission for intravital tumor fluorescence imaging. ACS Nano 12, 1350–1358 (2018). [DOI] [PubMed] [Google Scholar]
- 75.H. Wang, S. Han, L. Wang, T. Yang, G. Zhang, L. Yu, Y. Zhao, Dual-function baicalin and baicalin-loaded poly(lactic-co-glycolic acid) nanoparticles: Immune activation of dendritic cells and arrest of the melanoma cell cycle at the G2/M phase. Particuology 37, 64–71 (2018). [Google Scholar]
- 76.Q. Liu, X. Chen, J. Jia, W. Zhang, T. Yang, L. Wang, G. Ma, pH-Responsive poly(d,L-lactic-co-glycolic acid) nanoparticles with rapid antigen release behavior promote immune response. ACS Nano 9, 4925–4938 (2015). [DOI] [PubMed] [Google Scholar]
- 77.J. Guo, J. R. Ogier, S. Desgranges, R. Darcy, C. O'Driscoll, Anisamide-targeted cyclodextrin nanoparticles for siRNA delivery to prostate tumours in mice. Biomaterials 33, 7775–7784 (2012). [DOI] [PubMed] [Google Scholar]
- 78.V. Gopalakrishnan, C. N. Spencer, L. Nezi, A. Reuben, M. C. Andrews, T. V. Karpinets, P. A. Prieto, D. Vicente, K. Hoffman, S. C. Wei, A. P. Cogdill, L. Zhao, C. W. Hudgens, D. S. Hutchinson, T. Manzo, M. P. de Macedo, T. Cotechini, T. Kumar, W. S. Chen, S. M. Reddy, R. S. Sloane, J. Galloway-Pena, H. Jiang, P. L. Chen, E. J. Shpall, K. Rezvani, A. M. Alousi, R. F. Chemaly, S. Shelburne, L. M. Vence, P. C. Okhuysen, V. B. Jensen, A. G. Swennes, F. McAllister, E. M. R. Sanchez, Y. Zhang, E. Le Chatelier, L. Zitvogel, N. Pons, J. L. Austin-Breneman, L. E. Haydu, E. M. Burton, J. M. Gardner, E. Sirmans, J. Hu, A. J. Lazar, T. Tsujikawa, A. Diab, H. Tawbi, I. C. Glitza, W. J. Hwu, S. P. Patel, S. E. Woodman, R. N. Amaria, M. A. Davies, J. E. Gershenwald, P. Hwu, J. E. Lee, J. Zhang, L. M. Coussens, Z. A. Cooper, P. A. Futreal, C. R. Daniel, N. J. Ajami, J. F. Petrosino, M. T. Tetzlaff, P. Sharma, J. P. Allison, R. R. Jenq, J. A. Wargo, Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Z. Yu, J. Guo, M. Hu, Y. Gao, L. Huang, Icaritin exacerbates mitophagy and synergizes with doxorubicin to induce immunogenic cell death in hepatocellular carcinoma. ACS Nano 14, 4816–4828 (2020). [DOI] [PubMed] [Google Scholar]
- 80.B. Hu, M. Yu, X. Ma, J. Sun, C. Liu, C. Wang, S. Wu, P. Fu, Z. Yang, Y. He, Y. Zhu, C. Huang, X. Yang, Y. Shi, S. Qiu, H. Sun, A. X. Zhu, J. Zhou, Y. Xu, D. Zhu, J. Fan, IFNα potentiates anti-PD-1 efficacy by remodeling glucose metabolism in the hepatocellular carcinoma microenvironment. Cancer Discov. 12, 1718–1741 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S19
Tables S1 to S3