Abstract
Background
Cardiovascular disease risk prediction models underestimate CVD risk in people living with HIV (PLWH). Our goal is to derive a risk score based on protein biomarkers that could be used to predict CVD in PLWH.
Methods and Results
In a matched case–control study, we analyzed normalized protein expression data for participants enrolled in 1 of 4 trials conducted by INSIGHT (International Network for Strategic Initiatives in Global HIV Trials). We used dimension reduction, variable selection and resampling methods, and multivariable conditional logistic regression models to determine candidate protein biomarkers and to generate a protein score for predicting CVD in PLWH. We internally validated our findings using bootstrap. A protein score that was derived from 8 proteins (including HGF [hepatocyte growth factor] and interleukin‐6) was found to be associated with an increased risk of CVD after adjustment for CVD and HIV factors (odds ratio: 2.17 [95% CI: 1.58–2.99]). The protein score improved CVD prediction when compared with predicting CVD risk using the individual proteins that comprised the protein score. Individuals with a protein score above the median score were 3.10 (95% CI, 1.83–5.41) times more likely to develop CVD than those with a protein score below the median score.
Conclusions
A panel of blood biomarkers may help identify PLWH at a high risk for developing CVD. If validated, such a score could be used in conjunction with established factors to identify CVD at‐risk individuals who might benefit from aggressive risk reduction, ultimately shedding light on CVD pathogenesis in PLWH.
Keywords: cardiovascular disease, HIV, Olink, protein biomarkers, proteomics
Subject Categories: Functional Genomics, Risk Factors, Epidemiology
Nonstandard Abbreviations and Acronyms
- ENet
elastic net method
- CCL25
C‐C motif chemokine 25
- HGF
hepatocyte growth factor
- IDI
integrated discriminant improvement
- NRI
net reclassification index
- PLA2G7
platelet‐activating factor acetylhydrolase
- PLSDA
partial least squares discriminant analysis
- PLWH
people living with HIV
- SCGB3A2
secretoglobin family 3A member
- VIP
variable importance in projection
Clinical Perspective.
What Is New?
As a strategy to identify potential protein biomarkers that can improve cardiovascular disease risk prediction in people living with HIV, we applied state‐of‐the‐art statistical methods to proteomics data from participants enrolled in 1 of 4 trials by INSIGHT (International Network for Strategic Initiatives in Global HIV Trials).
We discovered 8 proteins that when combined into a score was strongly associated with cardiovascular disease risk in people living with HIV.
What Are the Clinical Implications?
A panel of blood biomarkers was found to be more predictive of cardiovascular disease risk than the individual proteins.
The protein score could be used in conjunction with established factors to identify individuals at risk of cardiovascular disease who might benefit from aggressive risk reduction.
With effective combination antiretroviral therapy (ART), people living with HIV (PLWH) are living longer, shifting the primary driver of morbidity and mortality for PLWH from opportunistic infections to chronic age‐related diseases, such as cardiovascular diseases (CVD). 1 , 2 , 3 PLWH are estimated to have a 1.5‐ to 2‐fold increased risk for CVD when compared with people without HIV. 4 Even in virally suppressed individuals who are HIV‐positive, CVD risk is higher than in a population that is HIV negative. 5 Therefore, HIV infection itself is considered a significant risk factor for CVD. Yet CVD screening and prevention methods are currently developed in the general population and do not perform well in a population that is HIV‐positive. 6 , 7 With a high proportion of people with HIV living to older ages, it is imperative to develop CVD risk models tailored to a population that is HIV‐positive.
Protein levels could improve prediction of cardiovascular health and as such, the development and use of protein panels could better predict CVD outcomes. 8 In the general population, a proteomic risk score has been shown to perform better than the Framingham risk score for individuals with stable coronary heart disease. 9 , 10 Protein biomarkers in inflammation and metabolic regulation pathways such as CRP (C‐reactive protein), insulin‐like growth factor, and leptin have been found to be related to CVD outcomes. 11 Proteins capture more variability than standard clinical characteristics and thus have the potential to contribute to a more individualized CVD risk assessment. 9 The development of a proteomic risk score for CVD in an ethnically and racially diverse population that is HIV‐positive has not yet been explored. We derive a proteomic risk score in PLWH to aid in HIV‐specific risk assessment. We hypothesize that by using proteomics data we will identify novel proteins and that a derived protein score will be able to more accurately discriminate CVD cases from controls compared with the individual proteins alone.
METHODS
Requests for data can be made through the INSIGHT (International Network for Strategic Initiatives in Global HIV Trials) website at http://insight‐trials.org/. Proposals are revised by the INSIGHT Scientific Steering Committee, all individuals consented to studies of genomics, which covers the institutional review board approval and consent for this project. Codes used in the analysis may be requested from the corresponding author.
Population
Participants were enrolled in 1 of 4 trials conducted by the Community Programs for Clinical Research on AIDS and INSIGHT (International Network for Strategic Initiatives in Global HIV Trials): FIRST (Flexible Initial Retrovirus Suppressive Therapies), 12 , 13 ESPRIT (Evaluation of Subcutaneous Proleukin [Interleukin‐2] in a Randomized International Trial), 14 SMART (Strategies for Management of Antiretroviral Therapy), 15 or START (Strategic Timing of Antiretroviral Therapy). 16 Enrollment in these trials occurred from 1999 to 2013. The results and population under study for each of these trials have been published previously and are reviewed here. 12 , 13 , 14 , 15 , 16 For all studies, race and ethnicity were self‐reported. FIRST enrolled ART‐naive participants at sites across the United States who were randomly assigned in a ratio of 1:1:1 to a protease inhibitor strategy, a nonnucleoside reverse transcriptase inhibitor strategy, or a 3‐class strategy with both a protease inhibitor and a nonnucleoside reverse transcriptase inhibitor. ESPRIT enrolled patients with HIV who had CD4+ cell counts 300 or more per cubic millimeter and were randomly assigned to receive interleukin‐2 plus ART or ART alone from sites across the world including Africa, Asia, Australia, Europe, Israel, North America, South America, and Mexico. SMART randomly assigned people with HIV who had a CD4+ cell count of more than 350 per cubic millimeter to the continuous use of ART or the episodic use of ART from sites across the world including Africa, Asia, Australia, Europe, Israel, North America, South America, and Mexico. START randomly assigned people with HIV who had a CD4+ count of more than 500 cells per cubic millimeter to start ART immediately or to defer it until the CD4+ count decreased to 350 cells per cubic millimeter or until the development of the acquired immunodeficiency syndrome or another condition that dictated the use of antiretroviral therapy. Individuals were enrolled from Africa, Asia, Australia, Europe, Israel, North America, South America, and Mexico. CVD case was defined as stroke, myocardial infarction, coronary revascularization, coronary artery disease (CAD) requiring surgery, or death from CVD, including unwitnessed deaths with unknown cause. 17 , 18 For all studies, an end point review committee reviewed documentation provided by clinical sites using prespecified criteria. 18 Myocardial infarction was defined following the universal definition. 19 Strokes were defined using 5 criteria: (1) acute onset with clinically compatible course; (2) computed tomography or magnetic resonance imaging compatible with diagnosis of stroke and current neurologic signs and symptoms; (3) stroke diagnosed as cause of death at autopsy; (4) positive lumbar puncture compatible with subarachnoid hemorrhage; and (5) death certificate or death note from medical record listing stroke as the cause of death. Coronary revascularization or CAD requiring surgery required a medical record from the hospitalization where the procedure was performed. A participant was considered as having a stroke if the first and second criteria were met, the third was met, the first and fourth criteria were met, or the first and fifth criteria were met. Cause of death was coded using documentation of the death provided by the clinical sites using the Coding of Death in HIV system. 20 In all studies, cause of death was coded using documentation of the death provided by the clinical sites using the Coding of Death in HIV system. 20 Each CVD case was matched with 2 controls based on age (±5 years) at baseline, treatment arm of study, and randomization date (±90 days). The matching within treatment arm accounts for ART differences between treatment regimens during the study alongside ART differences between and within the studies included. When controls were not found during the first round of matching, the age and randomization date criteria were widened. All cases and controls consented to the collection of DNA and have genomics data. Stored plasma samples were used to measure protein biomarkers for a case–control CVD substudy.
Proteomics Data From Olink, Normalization, and Quality Control
We used a baseline plasma specimen from consenting individuals to measure protein biomarkers from 5 Olink multiplex panels (Cardiovascular II, Cardiovascular III, Immune Response, Inflammation, and Cardiometabolic). Each panel has 92 target proteins. Detection and sample analysis of proteins from Olink is performed using proximity extension analysis with the target protein detected through high‐throughput real‐time polymerase chain reaction. 21 The proximity extension analysis assay by Olink has been used in the general population for identifying risk factors for CVD, and inclusion of proteins in the specified panel were chosen based on previously established risk factors and potential risk factors, with collaboration from leading experts in the fields. 22
The data were intensity normalized, meaning that the data are adjusted to make the median value for each assay on each plate equal to the median for that of the other plates. The data are presented as normalized protein expression values and are reported on the log2 scale. To monitor the quality of assay performance and the quality of each individual sample, 4 internal controls for each sample are added. There are 2 steps of quality control: (1) each sample plate with above 0.2 normalized protein expression value when evaluated on the SD of the internal controls pass this first step, and (2) samples that deviate <0.3 normalized protein expression value from the median value of the controls for each individual sample pass this step.
Risk Factors
CVD‐specific risk factors considered were sex; age; history of CVD at baseline, defined as prior CAD requiring treatment, prior myocardial infarction, prior stroke, or prior CAD requiring surgery; lipid‐lowering medication at baseline; blood pressure lowering medication at baseline; self‐reported race or ethnicity, which was categorized for SMART, START, and FIRST as either Black, Hispanic, Asian, White, or Other, with ESPRIT as either Black, Asian, White, or Other; and diabetes. Any individual who did not identify as Black was categorized as non‐Black. A patient was considered to have diabetes if they received a diagnosis of diabetes requiring drug treatment. One risk factor not included was smoking status at baseline, because it was not measured in ESPRIT. We do consider supplementary analysis on a subgroup with smoking status measurements in Data S1. HIV‐specific baseline factors that were considered were ART use, CD4+ values, and HIV RNA values.
Statistical Analysis
Protein Biomarker Identification
We searched for candidate proteins that are able to differentiate CVD cases from controls using bootstrap, partial least squares discriminant analysis (PLSDA), and PLSDA with regularized logistic regression via the elastic 23 net (PLSDA+ ENet). Before bootstrapping, we filtered out proteins with low potential to distinguish between cases and controls via a multivariable conditional logistic regression model predicting CVD case status using each protein while adjusting for age, sex, and Black race and ethnicity. We retained proteins that had P values <0.05. We then created 200 bootstrap data sets by randomly sampling the data (n=390) with replacement, keeping the ratio of CVD cases and controls. For each bootstrapped set, we fit a 2‐component PLSDA model. We used PLSDA variable importance in projection (VIP) scores to identify proteins that contributed most to the differentiation of CVD cases from controls. Following previous studies, we used cutoff values of 1, 1.5, and 2 for the average (over the 200 VIP scores) VIP score. Because there were many proteins with average VIP scores >1, we implemented a logistic regression model with an ENet penalty 24 on each bootstrap data set to reduce the list of potential protein biomarkers. To do this, we ranked proteins based on the frequency of nonzero coefficients out of the 200 ENet models. Proteins ranked in the top 25th percentile were considered as having more potential for differentiating CVD cases from controls. Note that instead of performing all the analyses on the observed data we used bootstrap for stability in the proteins identified. Further, this allowed the data to be used for the protein score and multivariable model developments to be different from the data used for biomarker identification.
Protein Score Development
We developed a protein score to assess the contribution of the identified protein biomarkers in a single combined measure. For each bootstrap data set and each protein, we used a conditional logistic regression model to obtain the log odds ratio of the protein in predicting CVD case status. A weighted mean (over the 200 log‐odds ratios from the conditional logistic regression models) was then used to obtain an overall log‐odds ratio for each protein. We used the full data set and the weighted mean for each protein to derive the protein score. We estimated the protein score for each individual as the sum of the products of the weighted mean for each protein and the corresponding protein data for that individual.
Model Development, Performance, and Internal Validation
We used multivariable conditional logistic regression models to investigate the following models: (1) baseline, (2) baseline+individual proteins, and (3) baseline+protein score. The baseline model included both HIV‐related factors—specifically CD4 and RNA values at baseline—and CVD‐related factors—specifically sex, age, body mass index, diabetes status, race, prior history of CVD, and lipid and blood pressure‐lowering medication variables all measured at baseline. For prediction estimates, we assessed the area under the curve (AUC) from the receiver operating characteristic curve and reported classification accuracy, sensitivity, and specificity estimates. These estimates were obtained from the optimal cutoff point on the receiver operating characteristic curve (ie, Youden's index). We assessed whether adding the protein score to the baseline model (ie, model 3) improved prediction more than adding each individual protein included in the score to the baseline model (ie, model 2). We assessed the improvements in the performance of model 2 over 1 and of model 3 over 1 using the difference in the area under the receiving operating characteristic curve, the continuous or categorical free net reclassification improvement (NRI), 25 , 26 and the integrated discriminant improvement (IDI). 25 , 27 The NRI 25 , 26 is based on the notion that a valuable new biomarker will tend to increase predicted risks for cases and decrease predicted risks for controls. The IDI 25 , 27 gives the difference in discrimination slopes between the models compared. For the computations of NRI and IDI, we used the CVD events and predicted probabilities based on the conditional logistic regression models. The continuous NRI was used because that does not depend on prespecified risk categories. In sensitivity analyses, we adjusted for the inflammatory biomarker, high‐sensitivity CRP (hsCRP), and the coagulation biomarker, D‐dimer, as PLWH with higher levels of these inflammatory biomarkers are more likely to experience fatal CVD events. We carried out separate analyses for individuals who were on ART at baseline, those who did not have any prior history of CVD at baseline, and those with data on smoking status at baseline. In the absence of external validation data, we internally validated our findings using bootstrap. For internal validation, we repeated each step of the model development from candidate protein biomarker selection to protein score development to model development and finally to performance assessment. Please refer to Data S1 for details and Figure S1 for flow chart of our analyses process. We performed all analyses using R software version 3.6.0 (The R Foundation). 28 Statistical significance was ascertained at a 2‐sided significance level of 0.05, and because these are exploratory analyses we did not perform any adjustment for multiplicity.
RESULTS
Study Population
Of the 390 individuals with and without CVD included in our analyses, the median (interquartile range) age at baseline was 47 (41–54) years and median (interquartile range) body mass index at baseline was 24.6 (22.2–27.2) kg/m2. Thirteen percent of participants were female, and 17% were Black. At baseline, 4.9% of participants had a history of a CVD event, 69% were on ART, 18% were taking blood pressure medication, and 19% were taking lipid lowering medication. Baseline demographic and other characteristics of the study participants by case/control status are presented in Table 1. Compared with those who did not develop CVD during follow‐up, participants who developed CVD during follow‐up were more likely at baseline to have a history of CVD at baseline, to be male, and be taking blood pressure‐ and lipid‐lowering medications. Refer to Table S1 for the breakdown of CVD events.
Table 1.
Baseline Characteristics for Cases and Controls
| Overall | Case | Control | P value | |
|---|---|---|---|---|
| N | 390 | 131 | 259 | |
| Study: N (%) | 1.00 | |||
| Evaluation of Subcutaneous Proleukin [Interleukin‐2] in a Randomized International Trial | 228 (58.46) | 77 (58.78) | 151 (58.30) | |
| Flexible Initial Retrovirus Suppressive Therapies | 24 (6.15) | 8 (6.11) | 16 (6.18) | |
| Strategies for Management of Antiretroviral Therapy | 48 (12.31) | 16 (12.21) | 32 (12.36) | |
| Strategic Timing of Antiretroviral Therapy | 90 (23.08) | 30 (22.90) | 60 (23.17) | |
| Region: N (%) | 0.73 | |||
| Africa | 6 (1.54) | 2 (1.53) | 4 (1.54) | |
| Asia | 12 (3.08) | 2 (1.53) | 10 (3.86) | |
| Australia/New Zealand | 33 (8.46) | 13 (9.92) | 20 (7.72) | |
| Europe | 164 (42.05) | 55 (41.98) | 109 (42.08) | |
| North/South America | 175 (44.87) | 59 (45.04) | 116 (44.79) | |
| Male sex: N (%) | 341 (87.44) | 120 (91.60) | 221 (85.33) | 0.11 |
| Non‐Black race*: N (%) | 324 (83.08) | 109 (83.21) | 215 (83.01) | 1.00 |
| History of cardiovascular disease: N (%) | 19 (4.87) | 16 (12.21) | 3 (1.16) | <0.01 |
| Body mass index, kg/m2: mean (SD) | 25.38 (5.03) | 25.81 (5.58) | 25.16 (4.72) | 0.23 |
| Age, y: mean (SD) | 47.69 (9.14) | 48.60 (9.57) | 47.23 (8.89) | 0.16 |
| Diabetes diagnosis: N (%) | 0.07 (0.25) | 0.09 (0.29) | 0.06 (0.23) | 0.22 |
| CD4+ count, cells/mm3: mean (SD) | 547.79 (215.29) | 541.02 (233.76) | 551.22 (205.71) | 0.66 |
| HIV RNA, copies/mL: mean (SD) | 22540.37 (138511.71) | 35856.41 (224730.34) | 15779.12 (57288.42) | 0.18 |
| On antiretroviral therapy: N (%) | 271 (69.49) | 93 (70.99) | 178 (68.73) | 0.73 |
| On lipid‐lowering treatment: N (%) | 72 (19.10) | 32 (25.20) | 40 (16.00) | 0.04 |
| On blood pressure treatment: N (%) | 66 (17.51) | 33 (25.98) | 33 (13.20) | <0.01 |
| Proteomic risk score: mean (SD) | 0.00 (1.00) | 0.50 (0.95) | −0.25 (0.93) | <0.01 |
| High‐sensitivity C‐reactive protein, μg/mL: mean (SD) | 3.70 (6.23) | 4.49 (8.46) | 3.30 (4.68) | 0.08 |
| D‐dimer, μg/mL: mean (SD) | 0.41 (0.56) | 0.51 (0.87) | 0.37 (0.28) | 0.02 |
P values comparing cases and controls for continuous variables are obtained using Kruskal–Wallis tests, and P values for binary/categorical variables are obtained using chi‐square tests. History of cardiovascular disease at baseline defined as prior coronary artery disease (CAD) requiring treatment, prior myocardial infarction, prior stroke, or prior CAD requiring surgery. On antiretroviral therapy is defined as on therapy at baseline or at study initiation. Race was self‐reported race.
Non‐Black race includes people who self‐identified as White, Asian, Hispanic, or Other races or ethnicities.
Proteins Identified by PLSDA and PLSDA+ENet
Out of the 459 proteins passing the proteomics quality control step, 107 proteins passed the filtering step with a P value <0.05. A protein that appears on multiple panels passed if it met this criterion for at least 1 panel. The number of significant proteins from the individual panels (ignoring overlaps) were as follows: CVD3 (28), immune response (20), cardiometabolic (16), CVD2 (17), and inflammation (26). Table S2 gives all statistically significant proteins used in PLSDA.
Twenty‐nine proteins were identified as being more able to distinguish between CVD cases and controls with average VIP scores >1 (Table S2). A protein that appeared on multiple panels is selected if it has VIP>1 for at least 1 panel. After using a logistic regression with an ENet penalty on the 29 proteins identified with average VIP scores >1, 8 proteins (FAM3B, ITGA11 [integrin ɑ11], IL6 [interleukin‐6], HGF [hepatocyte growth factor], CCL25 [C‐C motif chemokine 25], gastrotropin, PLA2G7 [platelet‐activating factor acetylhydrolase], SCGB3A2 [secretoglobin family 3A member]) were identified as having more potential for differentiating CVD cases from controls. Table S3 gives the count and proportions of times these 8 proteins had nonzero coefficients from the ENet model across all 200 bootstrap sets. A protein score consisting of these 8 proteins was then developed. When the average VIP score cutoff criterion was set to 1.5 or 2, only 1 protein (gastrotropin) was selected. Figure S2 gives the correlation of these protein biomarkers. The correlation between the proteins ranged from −0.13 to 0.36, and none of the proteins was highly correlated (ie, >0.5) with the other proteins.
Comparison of Baseline and Protein Models
A multivariable conditional logistic regression model predicting CVD case status using the protein score with adjustment for CVD‐ and HIV‐related risk factors was statistically significant (P<0.05 [95% CI, 1.58–2.99]; Figure and Table S4). A 1‐SD increase in the protein score was associated with an odds ratio (OR) for CVD of 2.17. The proteins FAM3B, IL6, HGF, gastrotropin, PLA2G7, and SCGB3A2 were each statistically significant (P<0.05) when added to the baseline model (Table 2) and their corresponding ORs were lower than the OR for the protein score in the protein score+baseline model (we are able to directly compare the ORs of the individual proteins to that of the protein score because these proteins and the protein score have each been standardized to have variance 1). Further, IL6 and PLA2G7 had AUC values that were somewhat higher than the other proteins (Table 3), and similar to the AUC from the protein score+baseline model. However, the NRIs for IL6 and PLA2G7 were 0.40 (95% CI, 0.19–0.61) and 0.28 (95% CI, 0.06–0.49), respectively, as compared with 0.66 (95% CI, 0.46–0.86) for the baseline+protein score model. The protein score+baseline model had an AUC of 0.73 (95% CI, 0.65–0.81), which was a 5.8% improvement in AUC over the baseline model (AUC=0.69 [95% CI, 0.60–0.78]), or a 17.4% (0.73–0.69)/(0.73–0.50) improvement in prediction ability (Table 3). The prediction accuracy, sensitivity, and specificity estimates (evaluated at the optimal cutoff point) for the protein score+baseline model were 0.75, 0.78, and 0.73, respectively, compared with 0.65, 0.81, and 0.57 for the baseline model. The NRI (Table 4) was 0.66 (95% CI, 0.46–0.86); this was driven more by net proportion of cases assigned a higher risk (NRI of cases is 0.41 [95% CI, 0.25–0.57]) than in net proportion of controls assigned a lower risk (NRI of controls is 0.25 [95% CI, 0.13–0.37]). The IDI was estimated to be statistically significant (IDI = 0.09 [95% CI, 0.06–0.14]). Three of the 8 proteins in the protein score (FAM3B, ITGA11, IL6) were found on the immune response Olink panel; 2 (HGF and CCL25) were from the inflammation panel; gastrotropin, SCGB3A2, and PLA2G7 were from the CVD2, CVD3, and cardiometabolic panels, respectively. A description of each protein is found in Data S1.
Figure . Conditional logistic regression model of CVD on standardized protein score.
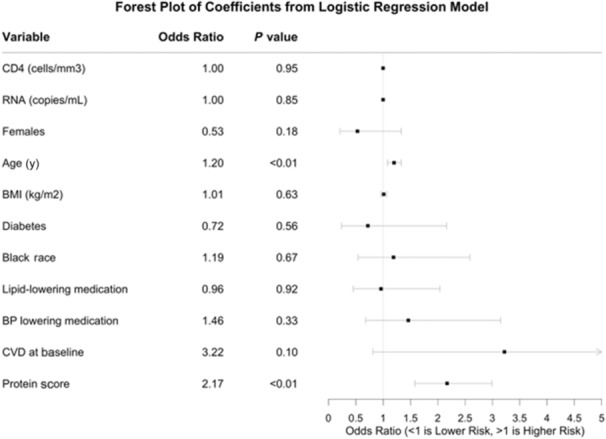
Odds ratios for women compare women to men; Black race is compared with non‐Black race or ethnicity. BMI indicates body mass index; BP, blood pressure; CD4, CD4+ count at baseline; and CVD, cardiovascular disease. CVD at baseline was defined as prior coronary artery disease (CAD) requiring treatment, prior myocardial infarction, prior stroke, or prior CAD requiring surgery. Race was self‐reported race.
Table 2.
Conditional Logistic Regression Model of CVD on Each of the Standardized Protein Used to Develop the Protein Score
| Variable | Odds ratio | SE | P‐value | Lower CI | Upper CI |
|---|---|---|---|---|---|
| Baseline | |||||
| + FAM3B | 1.48 | 0.14 | 0.01 | 1.12 | 1.95 |
| + Integrin ɑ11 | 0.85 | 0.13 | 0.20 | 0.66 | 1.09 |
| + Interleukin‐6 | 1.54 | 0.13 | 0.00 | 1.18 | 2.00 |
| + Hepatocyte growth factor | 1.63 | 0.15 | 0.00 | 1.22 | 2.17 |
| + C‐C motif chemokine 25 | 1.26 | 0.12 | 0.06 | 0.99 | 1.61 |
| + Gastrotropin | 0.72 | 0.14 | 0.02 | 0.55 | 0.95 |
| + Platelet‐activating factor acetylhydrolase | 1.48 | 0.14 | 0.01 | 1.12 | 1.95 |
| + Secretoglobin family 3A member | 1.32 | 0.13 | 0.03 | 1.03 | 1.69 |
Here, n=375 (cases=126 and controls=249) as there were 15 samples with missing values. Baseline is a model with the following variables: CD4, RNA, sex, age, body mass index, diabetes status at baseline, prior history of CVD, lipid‐lowering medication, and blood pressure‐lowering medication. CVD indicates cardiovascular disease.
Table 3.
Incremental Contribution of Individual Proteins and Protein Score to CVD Risk When Added to Baseline Model (n=375)
| Model | AUC | Lower CI | Upper CI | Change in AUC from baseline | NRI among cases | NRI among controls | Overall NRI |
|---|---|---|---|---|---|---|---|
| Baseline* | 0.69 | 0.60 | 0.78 | … | … | … | … |
| + FAM3B | 0.71 | 0.63 | 0.80 | 0.02 | 0.22 (0.05, 0.39) | 0.04 (−0.08, 0.17) | 0.27 (0.06, 0.48) |
| + Integrin ɑ11 | 0.69 | 0.61 | 0.78 | 0.004 | 0.13 (−0.05, 0.30) | −0.00 (−0.13, 0.12) | 0.12 (−0.09, 0.34) |
| + Interleukin‐6 | 0.73 | 0.65 | 0.81 | 0.04 | 0.24 (0.07, 0.41) | 0.17 (0.04, 0.29) | 0.40 (0.19, 0.61) |
| + Hepatocyte growth factor | 0.71 | 0.62 | 0.80 | 0.02 | 0.18 (0.00, 0.35) | 0.12 (−0.01, 0.24) | 0.29 (0.08, 0.50) |
| + C‐C motif chemokine 25 | 0.71 | 0.62 | 0.80 | 0.02 | 0.06 (−0.11, 0.24) | 0.14 (0.02, 0.26) | 0.20 (−0.01, 0.42) |
| + Gastrotropin | 0.70 | 0.62 | 0.79 | 0.01 | 0.22 (0.05, 0.39) | 0.12 (−0.01, 0.24) | 0.34 (0.13, 0.55) |
| + Platelet‐activating factor acetylhydrolase | 0.72 | 0.64 | 0.80 | 0.03 | 0.14 | 0.13 | 0.28 (0.06, 0.49) |
| (−0.03, 0.32) | (0.01, 0.26) | ||||||
| + Secretoglobin family 3A member | 0.70 | 0.62 | 0.79 | 0.01 | 0.18 (0.00, 0.35) | 0.16 (0.03, 0.28) | 0.33 (0.12, 0.54) |
| + Protein score from all 8 proteins | 0.73 | 0.65 | 0.81 | 0.04 | 0.41 (0.25, 0.57) | 0.25 (0.13, 0.37) | 0.66 (0.46, 0.86) |
Baseline* is a model with the following variables: CD4, RNA, sex, age, body mass index, diabetes status at baseline, prior history of cardiovascular disease, lipid‐lowering medication, and blood pressure‐lowering medication. The proteins FAM3B, IL6, HGF, gastrotropin, PLA2G7, and SCGB3A2 were each statistically significant (P<0.05) when added to the baseline model. Protein score was statistically significant (P<0.01) when added to the baseline model. Odds ratio for protein score: 2.17 (CI, 1.58–2.97). AUC indicates area under the curve; CCL25, C‐C motif chemokine 25; FAM3B, protein FAM3B; HGF, hepatocyte growth factor; IL6, interleukin‐6; NRI, net reclassification index; PLA2G7, platelet‐activating factor acetylhydrolase; and SCGB3A2, secretoglobin family 3A member.
Table 4.
Reclassification Table Using Category‐Free NRI. Model With Protein Score in Addition to Baseline Variables
| All | Reclassified upwards n (proportion) | Reclassified downwards N (proportion) | NRI | ||
|---|---|---|---|---|---|
| Number of cases | 126 | 88.96 (0.71) | 37.04 (0.29) | Among cases | 0.41 (0.25–0.57) |
| Number of controls | 249 | 92.88 (0.37) | 153.88 (0.62) | Among controls | 0.25 (0.13–0.37) |
| Overall NRI (CI) | 0.66 (0.46–0.86) | ||||
Net reclassification index (NRI) of cases is greater than NRI of controls. This suggests that addition of the protein score helps increase predicted risk for those with cases more than it decreases predicted risk for controls.
Categorization of Protein Score and Association With CVD Risk
To further assess the impact of the protein score on CVD risk, we categorized the protein score and considered associations of the categorized score with CVD. When the score was categorized above and below the median protein score, individuals with a protein score above the median were 3.1 times more likely to have CVD compared with individuals with a score below the median (Table S5). When the score was dichotomized as those individuals with scores in the top 25% versus those with scores in the bottom 75%, we found that those with scores in the top 25% were 2.9 times more likely to have CVD compared with those with scores in the bottom 75% (Table S5).
DISCUSSION
Our assessment of the contribution of protein biomarkers to CVD risk prediction showed that a protein risk score developed using 8 proteins—FAM3B, ITGA11, IL6, HGF, CCL25, gastrotropin, PLA2G7, SCGB3A2—was associated with CVD risk. A description of current literature for each protein is found in Data S1. Of these 8 proteins, IL6 is the only one to have previously been associated with CVD in both PLWH and the general population. 17 , 29 , 30 HGF and PLA2G7 have both been found to be associated with coronary heart disease in the general population. 31 , 32 , 33 , 34 The results here indicate that both HGF and PLA2G7 are potentially also associated with CVD in PLWH. In the general population, dysregulation of FAM3B is associated with diabetes, a known risk factor for CVD. 35 CCL25 is related to T cells, which are involved in the development and progression of CVDs in the general population. 36 One novel protein is ITGA11, which has not been extensively studied in humans. 37 , 38 Another novel protein is gastrotropin, a member of the fatty acid‐binding protein family, which is thought to serve an integral role in metabolic function. 39 , 40 SCGB3A2 has been found in a case–control study in a Korean population to contribute to susceptibility to asthma. 41 , 42
The protein score predicted CVD better when added to both HIV and CVD risk factors compared with models with the individual proteins and HIV and CVD risk factors. The model with the protein score and HIV and CVD risk factors showed better prediction performance compared with the HIV and CVD factors only model. When the protein risk score was dichotomized, our analyses showed that individuals with a score above the median score were 3 times more likely to develop CVD, which is illustrated in Table S5. The inclusion of D‐dimer and hsCRP in the baseline model did not alter our findings, illustrated in Tables S6 and S7, respectively. The following subgroup analyses were done: the first, restricted to individuals on ART at baseline, is shown in Table S8, the second, restricted to individuals with no prior history of CVD, is shown in Table S9, and the third, restricted to individuals with smoking data at baseline, is shown in Tables S10 and S11, all again demonstrated the ability of the protein score to discriminate between CVD cases and controls. Model performance statistics from the sensitivity analyses were comparable to model performance statistics from the full population. Using resampling techniques, we were able to internally validate our findings; internal validation results are shown in Table S12. Please refer to Data S1 for results on sensitivity, subgroup, and internal validation analyses.
A proteomic risk score has been shown to perform better than the Framingham risk score for individuals with stable coronary heart disease. 9 , 10 Proteins are able to capture variability beyond standard clinical characteristics and thus could contribute to an individualized CVD risk assessment. 9 This article identified candidate proteins and derived a proteomic risk score in PLWH to aid in HIV‐specific risk assessment.
The incremental AUC (5.8%), the NRI (0.66), and the IDI (0.09) of the protein score, based on published risk estimates, indicate a moderate improvement in risk prediction. Similar improvements in the AUC were shown in a 9‐protein risk score developed and validated for coronary heart disease in Ganz et al. 10 with similar improvement in risk prediction with a C statistic (or AUC) that ranges from 0.64 for refit Framingham to 0.71 for refit Framingham plus 9‐protein risk score, which is a 10.9% improvement in risk prediction. This is similar to our improvement from a 0.69 for the baseline model to 0.73 for baseline plus our proteomic risk score, which is a 5.8% improvement in risk prediction.
Our article has several strengths. One is the use of an ethnically and racially diverse population, which provides information on understudied groups. However, there was not sufficient sample size to evaluate if the predictive value of the protein score was similar or different by race and ethnicity. A second strength is the use of a population that is HIV‐positive. PLWH are at an increased risk for CVDs with substantial variability in CVD risk that is left unexplained by established risk factors. This study begins to characterize some of the heterogeneity within PLWH. Third is the use of a protein score because of its potential clinical relevance. Polygenic risk scores are often criticized for lack of clinical relevance due to the need to use complex machinery and statistical methods to genotype every individual, which is not yet readily available in the clinical setting. Comparatively the measurement of proteins and this protein score, which involves only 8 proteins, is potentially more clinically applicable as it is more easily measurable and thus more clinically relevant. A final strength is the use of state‐of‐the‐art statistical methods for biomarker identification.
One limitation is a small (n=390) study sample size, limiting power for detecting modest associations and inhibiting our ability to validate the risk score on an independent sample, instead we rely on resampling techniques for validation. We did not have controls who were HIV negative, so it is unclear whether the increased risk is specific to HIV infection. Further, only 69% of participants were on ART so it is likely that our findings will change if all participants were on ART. We note that because we matched cases and controls within treatment arms, ART differences between treatment regimens during the study are minimized. When we restricted our analysis to participants on ART at baseline (n=269), the protein score was again statistically significant, and improved prediction beyond the baseline model. We observed that unlike in the combined data where IL6 was likely driving the effect of the protein score, in this subpopulation, no individual protein had an AUC that was comparable to the protein score+baseline model. As another limitation, we did not account for social determinants of health conditions such as socioeconomic status and neighborhood environment variables, which have been suggested to affect CVD in the general population. 43 Finally, current smoking status at baseline was not measured in all the studies. As such, we did supplementary subgroup analyses on the individuals with current smoking status measurements. However, these results are limited due to small sample size.
CONCLUSIONS
We find that a proteomic risk score developed in a multiethnic and multiracial cohort that is HIV‐positive is a potentially beneficial approach for capturing heterogeneity above and beyond established risk factors. We developed a protein risk score that has high potential for benefit in CVD risk prediction in PLWH. We also provide a statistical approach to protein score development that can be applied for more individualized risk prediction.
Sources of Funding
This study was funded by National Institutes of Health (NIH) grants UM1‐AI068641, UM1‐AI120197, and U01‐AI136780 (for START); NIH grants UM1‐AI068641 and U01‐AI46957 (for ESPRIT); NIH grants U01‐AI042170 and U01AI046362 (for FIRST); and NIH grants UM1‐AI068641, U01‐AI046362, and U01‐AI042170 (for SMART). Sandra Safo was partially supported by grant numbers 5KL2TR002492‐04 from NIH and 13XS134T11 from Leidos and NIH. Lillian Haine was supported by the NIH under award number T32HL129956 and in part by the NIH's National Center for Advancing Translational Sciences (NCATS) grants TL1R002493 and UL1TR002494. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH's NCATS.
Disclosures
None.
Supporting information
Data S1–S2
Tables S1–S12
Figures S1–S2
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.122.027273
This article was sent to Pamela N. Peterson, MD, Deputy Editor, for review by expert referees, editorial decision, and final disposition.
For Sources of Funding and Disclosures, see page 9.
REFERENCES
- 1. Smith CJ, Ryom L, Weber R, Morlat P, Pradier C, Reiss P, Kowalska JD, de Wit S, Law M, el Sadr W, et al; D:A:D Study Group . Trends in underlying causes of death in people with HIV from 1999 to 2011 (D:a:D): a multicohort collaboration. Lancet. 2014;384:241–248. doi: 10.1016/S0140-673660604-8 [DOI] [PubMed] [Google Scholar]
- 2. Farahani M, Mulinder H, Farahani A, Marlink R. Prevalence and distribution of non‐AIDS causes of death among HIV‐infected individuals receiving antiretroviral therapy: a systematic review and meta‐analysis. Int J STD AIDS. 2017;28:636–650. doi: 10.1177/0956462416632428 [DOI] [PubMed] [Google Scholar]
- 3. Drozd DR, Kitahata MM, Althoff KN, Zhang J, Gange SJ, Napravnik S, Burkholder GA, Mathews WC, Silverberg MJ, Sterling TR, et al. Increased risk of myocardial infarction in HIV‐infected individuals in North America compared with the general population. J Acquir Immune Defic Syndr. 2017;75:568–576. doi: 10.1097/QAI.0000000000001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shah ASV, Stelzle D, Lee KK, Beck EJ, Alam S, Clifford S, Longenecker CT, Strachan F, Bagchi S, Whiteley W, et al. Global burden of atherosclerotic cardiovascular disease in people living with HIV systematic review and meta‐analysis. Circulation. 2018;138:1100–1112. doi: 10.1161/CIRCULATIONAHA.117.033369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Longenecker CT, Sullivan C, Baker JV. Immune activation and cardiovascular disease in chronic HIV infection. Curr Opin HIV AIDS. 2016;11:216–225. doi: 10.1097/COH.0000000000000227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thompson‐Paul AM, Lichtenstein KA, Armon C, Palella FJ Jr, Skarbinski J, Chmiel JS, Hart R, Wei SC, Loustalot F, Brooks JT, et al. Cardiovascular disease risk prediction in the HIV outpatient study. Clin Infect Dis. 2016;63:1508–1516. doi: 10.1093/cid/ciw615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Triant VA, Perez J, Regan S, Massaro JM, Meigs JB, Grinspoon SK, D'Agostino RB Sr. Cardiovascular risk prediction functions underestimate risk in HIV infection. Circulation. 2018;137:2203–2214. doi: 10.1161/CIRCULATIONAHA.117.028975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lindsey ML, Mayr M, Gomes AV, Delles C, Arrell DK, Murphy AM, Lange RA, Costello CE, Jin YF, Laskowitz DT, et al. Transformative impact of proteomics on cardiovascular health and disease: a scientific statement from the American Heart Association. Circulation. 2015;132:852–872. doi: 10.1161/CIR.0000000000000226 [DOI] [PubMed] [Google Scholar]
- 9. Sze SK. Artificially intelligent proteomics improves cardiovascular risk assessment. EBioMedicine. 2019;40:23–24. doi: 10.1016/j.ebiom.2019.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, Sterling DG, Williams SA. Development and validation of a protein‐based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. 2016;315:2532–2541. doi: 10.1001/jama.2016.5951 [DOI] [PubMed] [Google Scholar]
- 11. Ho JE, Lyass A, Courchesne P, Chen G, Liu C, Yin X, Hwang SJ, Massaro JM, Larson MG, Levy D. Protein biomarkers of cardiovascular disease and mortality in the community. J Am Heart Assoc. 2018;7:7. doi: 10.1161/JAHA.117.008108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. MacArthur RD, Chen L, Mayers DL, Besch CL, Novak R, van den Berg‐Wolf M, Yurik T, Peng G, Schmetter B, Brizz B, et al. The rationale and design of the CPCRA (Terry Beirn Community Programs for Clinical Research on AIDS) 058 FIRST (Flexible Initial Retrovirus Suppressive Therapies) trial. Control Clin Trials. 2001;22:176–190. doi: 10.1016/S0197-2456(01)00111-8 [DOI] [PubMed] [Google Scholar]
- 13. MacArthur RD, Novak RM, Peng G, Chen L, Xiang Y, Hullsiek KH, Kozal MJ, van den Berg‐Wolf M, Henely C, Schmetter B, et al. A comparison of three highly active antiretroviral treatment strategies consisting of non‐nucleoside reverse transcriptase inhibitors, protease inhibitors, or both in the presence of nucleoside reverse transcriptase inhibitors as initial therapy (CPCRA 058 FIRST study): a long‐term randomised trial. Lancet. 2006;368:2125–2135. doi: 10.1016/S0140-6736(06)69861-9 [DOI] [PubMed] [Google Scholar]
- 14. Emery S, Abrams DI, Cooper DA, Darbyshire JH, Lane HC, Lundgren JD, Neaton JD, ESPRIT Study Group . The Evaluation of Subcutaneous Proleukin® (interleukin‐2) in a Randomized International Trial: rationale, design, and methods of ESPRIT. Control Clin Trials. 2002;23:198–220. doi: 10.1016/S0197-2456(01)00179-9 [DOI] [PubMed] [Google Scholar]
- 15. Strategies for Management of Antiretroviral Therapy (SMART) Study Group , El‐Sadr WM, Lundgren J, Neaton JD, Gordin F, Abrams D, Arduino RC, Babiker A, Burman W, Clumeck N, et al. CD4+ count–guided interruption of antiretroviral treatment. N Engl J Med. 2006;355:2283–2296. doi: 10.1056/nejmoa062360 [DOI] [PubMed] [Google Scholar]
- 16. INSIGHT START Study Group , Lundgren JD, Babiker AG, Gordin F, Emery S, Grund B, Sharma S, Avihingsanon A, Cooper DA, Fätkenheuer G, et al. Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med. 2015;373:795–807. doi: 10.1056/NEJMoa1506816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nordell AD, McKenna M, Borges AH, Duprez D, Neuhaus J, Neaton JD. Severity of cardiovascular disease outcomes among patients with HIV is related to markers of inflammation and coagulation. J Am Heart Assoc. 2014;3. doi: 10.1161/JAHA.114.000844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lifson AR, INSIGHT Endpoint Review Committee Writing Group , Belloso WH, Davey RT, Duprez D, Gatell JM, Hoy JF, Krum EA, Nelson R, Pedersen C, et al. Development of diagnostic criteria for serious non‐AIDS events in HIV clinical trials. HIV Clin Trials. 2010;11:205–219. doi: 10.1310/HCT1104-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA. White HD; Executive Group on behalf of the Joint European Society of Cardiology (ESC)/American College of Cardiology (ACC)/American Heart Association (AHA)/World Heart Federation (WHF) Task Force for the Universal Definition of Myocardial Infarction. Fourth universal definition of myocardial infarction (2018). Circulation. 2018;138:e618–e651. doi: 10.1161/CIR.0000000000000617 [DOI] [PubMed] [Google Scholar]
- 20. Kowalska JD, Friis‐Møller N, Kirk O, Bannister W, Mocroft A, Sabin C, Reiss P, Gill J, Lewden C, Phillips A, et al. The Coding Causes of Death in HIV (CoDe) Project: initial results and evaluation of methodology. Epidemiology. 2011;22:516–523. doi: 10.1097/EDE.0B013E31821B5332 [DOI] [PubMed] [Google Scholar]
- 21. Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, et al. Homogenous 96‐plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. doi: 10.1371/journal.pone.0095192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olink CVD III. protein biomarker panel for cardiovascular disease studies. Accessed April 26, 2021 https://www.olink.com/products/target/cvd‐iii‐panel/.
- 23. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. doi: 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zou H, Hastie T. Regularization and variable selection via the elastic net. J Roy Stat Soc, Series B (Statistical Methodology). 2005;67:301–320. doi: 10.1111/j.1467-9868.2005.00503.x. http://www.jstor.org/stable/3647580 [DOI] [Google Scholar]
- 25. Pencina MJ, d'Agostino RB, d'Agostino RB, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–172. doi: 10.1002/sim.2929 [DOI] [PubMed] [Google Scholar]
- 26. Pencina MJ, D'Agostino RB, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30:11–21. doi: 10.1002/sim.4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kerr KF, McClelland RL, Brown ER, Lumley T. Evaluating the incremental value of new biomarkers with integrated discrimination improvement. Am J Epidemiol. 2011;174:364–374. doi: 10.1093/aje/kwr086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. R Core Team . R: a language and environment for statistical computing. 2019.
- 29. Duprez DA, Neuhaus J, Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, et al. Inflammation, coagulation and cardiovascular disease in HIV‐infected individuals. PLoS ONE. 2012;7:e44454. doi: 10.1371/journal.pone.0044454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsu DC, Ma YF, Hur S, Li D, Rupert A, Scherzer R, Kalapus SC, Deeks S, Sereti I, Hsue PY. Plasma IL‐6 levels are independently associated with atherosclerosis and mortality in HIV‐infected individuals on suppressive antiretroviral therapy. AIDS. 2016;30:2065–2074. doi: 10.1097/QAD.0000000000001149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bielinski SJ, Berardi C, Decker PA, Larson NB, Bell EJ, Pankow JS, Sale MM, Tang W, Hanson NQ, Wassel CL, et al. Hepatocyte growth factor demonstrates racial heterogeneity as a biomarker for coronary heart disease. Heart. 2017;103:1185–1193. doi: 10.1136/heartjnl-2016-310450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bell EJ, Decker PA, Tsai MY, Pankow JS, Hanson NQ, Wassel CL, Larson NB, Cohoon KP, Budoff MJ, Polak JF, et al. Hepatocyte growth factor is associated with progression of atherosclerosis: the Multi‐Ethnic Study of Atherosclerosis (MESA). Atherosclerosis. 2018;272:162–167. doi: 10.1016/j.atherosclerosis.2018.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hurt‐Camejo E, Camejo G, Peilot H, Öörni K, Kovanen P. Phospholipase A2 in vascular disease. Circ Res. 2001;89:298–304. doi: 10.1161/hh1601.095598 [DOI] [PubMed] [Google Scholar]
- 34. Caslake MJ, Packard CJ. Lipoprotein‐associated phospholipase A2 (platelet‐activating factor acetylhydrolase) and cardiovascular disease. Curr Opin Lipidol. 2003;14:347–352. doi: 10.1097/00041433-200308000-00002 [DOI] [PubMed] [Google Scholar]
- 35. Zhang W, Chen S, Zhang Z, Wang C, Liu C. FAM3B mediates high glucose‐induced vascular smooth muscle cell proliferation and migration via inhibition of miR‐322‐5p. Sci Rep. 2017;7:7. doi: 10.1038/s41598-017-02683-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jung MK, Shin EC. Aged T cells and cardiovascular disease. Cell Mol Immunol. 2017;14:1009–1010. doi: 10.1038/cmi.2017.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Talior‐Volodarsky I, Connelly KA, Arora PD, Gullberg D, McCulloch CA. α11 integrin stimulates myofibroblast differentiation in diabetic cardiomyopathy. Cardiovasc Res. 2012;96:265–275. doi: 10.1093/cvr/cvs259 [DOI] [PubMed] [Google Scholar]
- 38. Romaine A, Sørensen IW, Zeltz C, Lu N, Erusappan PM, Melleby AO, Zhang L, Bendiksen B, Robinson EL, Aronsen JM, et al. Overexpression of integrin α11 induces cardiac fibrosis in mice. Acta Physiologica. 2018;222:222. doi: 10.1111/apha.12932 [DOI] [PubMed] [Google Scholar]
- 39. Thumser AE, Moore JB, Plant NJ. Fatty acid binding proteins: tissue‐specific functions in health and disease. Curr Opin Clin Nutr Metab Care. 2014;17:124–129. doi: 10.1097/MCO.0000000000000031 [DOI] [PubMed] [Google Scholar]
- 40. Fisher E, Grallert H, Klapper M, Pfäfflin A, Schrezenmeir J, Illig T, Boeing H, Döring F. Evidence for the Thr79Met polymorphism of the ileal fatty acid binding protein (FABP6) to be associated with type 2 diabetes in obese individuals. Mol Genet Metab. 2009;98:400–405. doi: 10.1016/j.ymgme.2009.08.001 [DOI] [PubMed] [Google Scholar]
- 41. Inoue K, Wang X, Saito J, Tanino Y, Ishida T, Iwaki D, Fujita T, Kimura S, Munakata M. Plasma UGRP1 levels associate with promoter G‐112A polymorphism and the severity of asthma. Allergol Int. 2008;57:57–64. doi: 10.2332/allergolint.O-07-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou Z, Zuo CL, Li XS, Ye XP, Zhang QY, Wang P, Zhang RX, Chen G, Yang JL, Chen Y, et al. Uterus globulin associated protein 1 (UGRP1) is a potential marker of progression of Graves' disease into hypothyroidism. Mol Cell Endocrinol. 2019;494:110492. doi: 10.1016/j.mce.2019.110492 [DOI] [PubMed] [Google Scholar]
- 43. Powell‐Wiley TM, Baumer Y, Baah FO, Baez AS, Farmer N, Mahlobo CT, Pita MA, Potharaju KA, Tamura K, Wallen GR. Social determinants of cardiovascular disease. Circ Res. 2022;130:782–799. doi: 10.1161/CIRCRESAHA.121.319811 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1–S2
Tables S1–S12
Figures S1–S2


