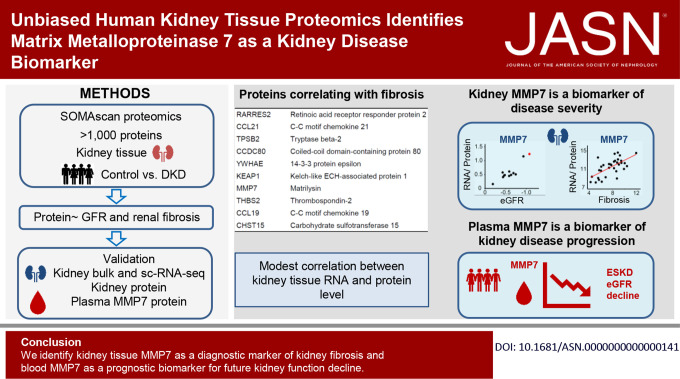
Keywords: CKD, diabetic kidney disease
Abstract
Significance Statement
Although gene expression changes have been characterized in human diabetic kidney disease (DKD), unbiased tissue proteomics information for this condition is lacking. The authors conducted an unbiased aptamer-based proteomic analysis of samples from patients with DKD and healthy controls, identifying proteins with levels that associate with kidney function (eGFR) or fibrosis, after adjusting for key covariates. Overall, tissue gene expression only modestly correlated with tissue protein levels. Kidney protein and RNA levels of matrix metalloproteinase 7 (MMP7) strongly correlated with fibrosis and with eGFR. Single-cell RNA sequencing indicated that kidney tubule cells are an important source of MMP7. Furthermore, plasma MMP7 levels predicted future kidney function decline. These findings identify kidney tissue MMP7 as a biomarker of fibrosis and blood MMP7 as a biomarker for future kidney function decline.
Background
Diabetic kidney disease (DKD) is responsible for close to half of all ESKD cases. Although unbiased gene expression changes have been extensively characterized in human kidney tissue samples, unbiased protein-level information is not available.
Methods
We collected human kidney samples from 23 individuals with DKD and ten healthy controls, gathered associated clinical and demographics information, and implemented histologic analysis. We performed unbiased proteomics using the SomaScan platform and quantified the level of 1305 proteins and analyzed gene expression levels by bulk RNA and single-cell RNA sequencing (scRNA-seq). We validated protein levels in a separate cohort of kidney tissue samples as well as in 11,030 blood samples.
Results
Globally, human kidney transcript and protein levels showed only modest correlation. Our analysis identified 14 proteins with kidney tissue levels that correlated with eGFR and found that the levels of 152 proteins correlated with interstitial fibrosis. Of the identified proteins, matrix metalloprotease 7 (MMP7) showed the strongest association with both fibrosis and eGFR. The correlation between tissue MMP7 protein expression and kidney function was validated in external datasets. The levels of MMP7 RNA correlated with fibrosis in the primary and validation datasets. Findings from scRNA-seq pointed to proximal tubules, connecting tubules, and principal cells as likely cellular sources of increased tissue MMP7 expression. Furthermore, plasma MMP7 levels correlated not only with kidney function but also associated with prospective kidney function decline.
Conclusions
Our findings, which underscore the value of human kidney tissue proteomics analysis, identify kidney tissue MMP7 as a diagnostic marker of kidney fibrosis and blood MMP7 as a biomarker for future kidney function decline.
Introduction
Diabetic kidney disease (DKD) is a leading cause of ESKD. It is estimated that close to 40% of patients with diabetes develop DKD defined as albuminuria or an impairment of kidney function (<60 ml/min per 1.73 m2). Although improved glycemic and blood pressure control have lowered the risk of kidney disease development, we still cannot predict who with diabetes mellitus (DM) will develop DKD or progress to ESKD. The kidney failure risk equation (KFRE) has emerged as an important tool to predict the risk of renal failure in subject with eGFR <60 ml/min per 1.73 m2.1,2 Indeed, just including four variables, the KFRE can predict patients ESKD risk with high precision (c-statistics >0.92). Unfortunately, KFRE does not perform well for subjects with stage 1–2 CKD. The degree of albuminuria shows a strong correlation with kidney disease progression, and it is most frequently used by Phase 2 and 3 clinical studies to enrich subjects who likely develop kidney failure. Recently, we have analyzed histological changes in 859 human kidney tissue samples using an unbiased scoring system.3 We showed that while at advanced CKD stages (CKD 3–5) histological changes strongly correlate with kidney function, this is not the case for early (stage 1–2) CKD. We identified several samples with relatively severe structural damage despite preserved kidney function. Importantly, the degree of fibrosis improved the future kidney function decline estimation, indicating the critical role of fibrosis and tissue analysis for better prognostication.
At present, there is an important need to identify biology-driven biomarkers for DKD subtypes or patients who will progress to ESKD. Blood proteins are considered probably the most ideal biomarkers, as they represent an easily accessible body fluid. Blood protein levels are also tightly regulated resulting in relatively low interindividual variation. Targeted biomarker studies indicated TNF receptor superfamily members 1A and 1B,4 soluble urokinase receptor5 as potential biomarkers for kidney outcomes. Recent studies reported the results from unbiased blood proteomic analyses,6–11 including the results from the Atherosclerosis Risk In Communities (ARIC) Study in which the team reported 12 proteins of inflammation and other processes as prognostic biomarkers.7
Tissue gene expression analysis has been extensively used to understand disease mechanisms and for biomarker discovery. Unbiased RNA sequencing has identified a large number of genes whose levels correlate with kidney function, fibrosis, or glomerulosclerosis.12–16 The recent development in droplet-based sequencing enabled single-cell level gene expression and epigenome analysis and illuminated new genes, cell types, and disease mechanisms. Although gene expression is an important estimator of protein expression, protein levels are also determined by multiple other mechanisms. Indeed, recent studies have indicated relatively poor correlation between gene expression and protein levels in blood samples.17–19 The liver is likely an important source of blood proteins; therefore, the poor correlation of protein and RNA levels in blood samples might be less surprising. Correlations between tissue gene expression and protein levels remain to be established. Protein detection and quantification has been historically challenging. Although improvements in mass spectrometry platforms aided new discoveries for kidney diseases,20,21 much work remains to be conducted to better understand protein expression in the diseased kidney. The recent development of high-throughput aptamer-based proteomics technologies, such as SomaScan, has enabled quantification of over 1000 proteins in a biological sample.22,23 SomaScan was successfully used in large clinical and epidemiologic studies to analyze plasma protein levels of CKD and DKD subjects,6–11 but to the best of our knowledge, unbiased proteomics studies have not been performed in human kidney tissue samples.
Here, we report our first in class study to apply unbiased affinity proteomics to a cohort of control and DKD kidneys. Here, we identify matrix metalloproteinase 7 (MMP7), a member of the matrix metalloproteinase family, as a biomarker of fibrosis in patients with DKD. Single-cell gene expression indicates that tubule cells are the main source of MMP7; furthermore, we show that plasma MMP7 is a biomarker for the risk of kidney failure in a large external cohort.24
Methods
Human Kidney
Kidney tissue samples were obtained from surgical nephrectomies in both DKD and control group. Only the normal, unaffected part of the tissue (taken at least 2 cm away from the cancer) was used for our analysis. Nephrectomies were deidentified, and the corresponding clinical information including age, race, sex, diabetes and hypertension (HTN) status, as well as creatinine values were collected through an honest broker. This study was approved by the Institutional Review Board of the University of Pennsylvania and by the Committee of Human Studies at the Joslin Diabetes Center. No informed consent was obtained because this study was considered “exempt.”
Diagnosis of DKD
Adjacent tissue sample from each kidney was formalin-fixed paraffin-embedded and stained with hematoxylin eosin and periodic acid–Schiff. Kidney sections were evaluated by an expert renal pathologist who was not aware of the clinical information. We used an unbiased scoring system as we published earlier.3 DKD was defined as persistent reduction in eGFR to <60 ml/min per 1.73 m2 in a patient with type 2 diabetes, according to the guidelines of the National Kidney Foundation–Kidney Disease Outcome Quality Initiative25,26 in addition to having mesangial expansion and diabetic glomerulosclerosis on light microscopy, consistent with at least class II diabetic nephropathy of the Renal Pathology Society criteria.27
Tissue Handling
Kidneys were removed, immediately snap-frozen in liquid nitrogen, and stored at −80°C until homogenization. Tissue was cryopulverized using a TissueLyser instrument (QIAGEN, Hilden, Germany), and protein was extracted using a lysis buffer of tissue protein extraction reagent (Thermo Fisher Scientific, Waltham, MA) containing protease inhibitors (Halt; Thermo Fisher Scientific). Protein concentrations were measured using bicinchoninic acid protein assay (Pierce; Thermo Fisher Scientific).
Kidney Tissue Proteomics
We used the SomaScan assay platform (SomaLogic, Boulder, CO) at the Genomics Proteomics Core at Beth Israel Deaconess Medical Center, Harvard affiliate, Boston, MA, for proteomics. Thousand three hundred and five targeted proteins or protein complexes in the primary dataset (n=33, ten controls and 23 DKD tissue samples) and 7596 targeted proteins or protein complexes in the kidney tissue validation dataset (n=186, in preparation) were measured. The SomaScan platform technology and its performance characteristics have been previously described.6,22,23 In brief, the assay uses slow off-rate modified DNA aptamers (SOMAmers) capable of binding to specific protein targets with high sensitivity and specificity. Protein levels are captured in relative fluorescence units. Aptamers used for the primary dataset detecting 1305 proteins are described in Supplemental Spreadsheet 1. To account for variation across kidney extracts, calibrator and buffer samples were added in a 96-well plate. Quality control was performed at the sample and SOMAmer level by the manufacturer's recommendations. The former involved the use of the hybridization controls, whereas the latter involved control SOMAmers for data normalization and calibration samples. The sample data were first normalized to remove within-run hybridization variation followed by adaptive normalization by maximum likelihood with point and variance estimates from a normal US population. The intensity of protein signals was transformed base 2 logarithmic (log2) values for further analysis.
Blood Proteomics Dataset
The validation dataset for blood proteomics was drawn from the ARIC Study, an ongoing cohort of individuals recruited from four US communities: suburbs of Minneapolis, Minnesota; Jackson, Mississippi; Forsyth County, North Carolina; and Washington County, Maryland.24 Enrollment occurred between 1987 and 1989, with subsequent visits in 1990–1992 (visit 2), 1993–1995 (visit 3), 1996–1998 (visit 4), 2011–2013 (visit 5), 2016–2017 (visit 6), and 2018–2019 (visit 7). Participants with (n=1623) or without (n=9407) DM who attended visits 2, were free of ESKD, had eGFR measures, and agreed to participate in research were included. Urine was not collected at the study visits. MMP7 levels were measured using the SomaScan v4 platform. Outcomes evaluated including 50% decline in eGFR occurring at a subsequent study visit and ESKD, defined through linkage with the US Renal Data System.28 Associations between log2-transformed MMP7 and outcomes were assessed using Cox regression. Two models were used: model 1, adjusting for age, sex, and a composite race-center variables, and model 2, additionally adjusting for systolic blood pressure, use of antihypertensive medications, prevalent cardiovascular disease, eGFR on the basis of both creatinine and cystatin, HDL cholesterol, prevalent smoking, and total cholesterol. The last day of follow-up for events was December 31, 2019.
Weighted Gene Coexpression Network Analysis for Proteomics
To extract sets of proteins coexpressed within the dataset, the weighted gene coexpression network analysis (WGCNA) package29 in the R environment (version 1.71) was applied. Clustering protein trees were established based on the similarity of protein expression profiles across samples using the adjacency function with a signed network. Thereafter, the Dynamic Hybrid Tree Cut algorithm was used to cut the hierarchal clustering tree, and modules were defined as branches from the tree cutting. A soft thresholding power of ten was selected based on the scale independence chart, as described in the WGCNA tutorials. The randomly color-labeled each module was summarized by the first principal component of the standardized module expression profiles (referred to as module eigenprotein). We set the minimum module size to 30 proteins, and these modules were merged when the difference between their module eigenprotein profiles was <0.25. The module eigenproteins were then analyzed for correlations with clinical phenotypes, such as eGFR and interstitial fibrosis.
Pathway Analysis
Genes encoding proteins with expression levels showing significant linear correlation with kidney interstitial fibrosis were separately imported into the Database for Annotation, Visualization, and Integrated Discovery (DAVID, version v2022q2) bioinformatics resource (https://david.ncifcrf.gov/),30 where they were analyzed using Gene Ontology. The WGCNA modules were analyzed similarly.
Cluster Analysis
Cluster analysis of proteomics in the WGCNA brown module was performed using Molecular Complex Detection (MCODE) plug-in, which provides a novel clustering algorithm to screen the modules of the protein–protein interaction network through Cytoscape.31,32 MCODE scores of >3 and the number of nodes >3 were set as cut-off criteria with the default parameters (degree cutoff ≥2, node score cutoff ≥2, K-core ≥2, and max depth=100).
Kidney Tissue RNA-seq
RNA isolation, sequencing, and analysis were performed as previously published.33–35 Reads were aligned to the human genome (hg19/GRCh37) using STAR (version 2.7.3a). Gene and isoform expression levels of transcript per million (TPM) were estimated using RNA-Seq by Expectation Maximization (version 1.3.0). Log2-transformed (TPM+1) values were used for further analysis.
Single-Cell RNA-seq Data Analysis
Reanalysis of the single-cell dataset was performed using Seurat (version 4.0.3). The original Seurat object used in this study was obtained from the NIDDK Kidney Precision Medicine Project (KPMP) repository (https://atlas.kpmp.org/repository/).36 This dataset was downloaded as an h5 Seurat object (File name: “521c5b34-3dd0-4871-8064-61d3e3f1775a_PREMIERE_Alldatasets_08132021.h5Seurat”), with quality control and clustering already performed by the KPMP team. The Seurat object contains two types of clustering, one of which named “subclass.l1” having 13 clusters was applied in this study. Living donor (LD) and DKD subjects were subset from this Seurat object and used for further analysis. The DimPlot function was used to generate the Uniform Manifold Approximation and Projection (UMAP) plots. The DotPlot function was used to visualize gene expression, and features were visualized on UMAP plots with function FeaturePlot. Differential expressions in each cluster between LD and DKD subjects were calculated using FindMarkers (min.pct=0.2, logfc.threshold=0.25).
Statistical Analyses
Analyses were performed using RStudio (v4.1.3) (R Development Core Team, Vienna, Austria). For linear regression model, eGFR and interstitial fibrosis score were log2-transformed to approximate the normal distribution. False discovery rate (FDR) was calculated by Benjamin–Hochberg procedure. Proteins or genes with FDR <0.05 were considered significant. In correlation analysis, Pearson correlation coefficient was used. To identify unbiased subgroups on the basis of proteomics, unsupervised hierarchical clustering was performed on the scaled data using the Wards method with Manhattan distances.37 The optimal number of clusters was determined by average silhouette method38,39 in the factoextra function and found to be 2.
Results
Clinical and Histological Characteristics of the Study Population
Our primary dataset contained 33 human kidney cortical samples; 23 samples from subjects with DKD and ten healthy controls. The DKD diagnosis was established based on eGFR and histological lesions. In our dataset, the mean eGFR, calculated by Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) formula,40 was 40±15 ml/min per 1.73 m2 for DKD samples (ranging from 7 to 59 ml/min per 1.73 m2) and 110±18 ml/min per 1.73 m2 for the controls (ranging from 91 to 143 ml/min per 1.73 m2). The mean age was 67±12 years for the DKD group and 44±12 years for the control group. Subjects with DKD were more frequently male (65%), mostly White (96%), and had higher body mass index (BMI). The full clinical information and pathological characterization are presented in Table 1. Medication data and proteinuria were collected from charts; however, they were incomplete and therefore not used in this analysis. We performed histological scoring of the kidney samples by characterizing 19 different histological lesions by a pathologist who was not aware of the clinical information, as we published earlier.3 We found that the degree of interstitial fibrosis exhibited a negative correlation with eGFR (R=−0.59, P < 0.001) (Figure 1A).12 Similarly, glomerulosclerosis negatively correlated with kidney function (R=−0.49, P < 0.01) (Supplemental Figure 1A). Interstitial fibrosis positively correlated with glomerular sclerosis (R=0.87, P = 3.7e-11) (Supplemental Figure 1B).
Table 1.
Patient characteristics of primary database
| Characteristic | Control (n=10) |
DKD (n=23) |
|---|---|---|
| Age, yr (SD) | 44.3 (12.1) | 67.0 (11.9) |
| Male, % | 50.0 | 65.2 |
| Race, n (%) | ||
| White | 5 (50) | 22 (95.6) |
| Black | 5 (50) | 1 (4.4) |
| DM | 0 (0) | 23 (100) |
| HTN | 0 (0) | 22 (95.6) |
| BMI, kg/m2 (SD) | 28.5 (8.2) | 34.2 (9.5) |
| SBP, mm Hg (SD) | 124 (9) | 144 (21) |
| DBP, mm Hg (SD) | 76 (8) | 73 (11) |
| eGFR, ml/min per 1.73 m2 (SD) | 110 (18) | 40 (16) |
| Serum glucose, mg/dl (SD) | 105 (22) | 142 (28) |
| Histology | ||
| Glomerulosclerosis, % (SD) | 1.9 (2.7) | 24.7 (30.8) |
| Interstitial fibrosis, % (SD) | 3.0 (5.0) | 30.4 (30.9) |
Data are presented as mean and standard deviation with the median values or percentage (%). DKD, diabetic kidney disease; DM, diabetes mellitus; HTN, hypertension; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Figure 1.
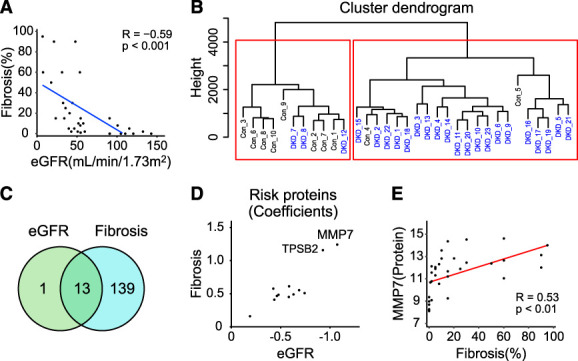
Unbiased proteomics analysis of human DKD kidney samples. (A) Scatterplot of eGFR (ml/min per 1.73 m2) and degree of interstitial fibrosis (Pearson R of −0.59). (B) Hierarchical clustering on the basis of proteomics data. (C) Venn diagram of proteins correlated with eGFR and interstitial fibrosis. (D) Scatterplot of correlation of coefficients between 12 proteins and eGFR and fibrosis. (E) Scatterplots of Matrix Metalloproteinase 7 (MMP7) expression measured by SOMAmer array (x axis) and degree of interstitial fibrosis (y axis) (Pearson R of 0.53). Figure 1 can be viewed in color online at www.jasn.org.
Unbiased Proteomics of Human DKD Samples
To understand changes in protein levels in human kidney samples, we first performed unsupervised hierarchical clustering of the proteomics data using 1305 proteins from the primary dataset (Figure 1B). Our analysis identified two well-separated clusters, one mostly containing control samples while the second mostly containing DKD samples, suggesting global protein expression changes in DKD kidneys.
Then, we set to define protein expression profiles that correlate with eGFR or tubulointerstitial fibrosis in our study group. Using a linear regression model and eGFR as a dependent variable, our analysis identified 14 proteins whose expression showed significant linear association with eGFR (FDR <0.05) after adjusting for age, race, sex, and BMI (Supplemental Spreadsheet 2). Next, we aimed to identify proteins whose expression correlates with tubulointerstitial fibrosis. Using a linear regression model, we found that the level of 152 proteins was associated with tubulointerstitial fibrosis after adjusting for covariates (Table 2 and Supplemental Spreadsheet 3). Proteins whose levels correlated with fibrosis had diverse functions, including cell surface receptor signaling, cellular response–related pathways, developmental pathways, and cell adhesion (Supplemental Figure 1C and Supplemental Spreadsheet 4).
Table 2.
Top ten proteins associated with renal interstitial fibrosis in linear regression model, adjusted for age, sex, race, and body mass index
| Symbol | Protein Name | Coefficient | P Value | FDR |
|---|---|---|---|---|
| RARRES2 | Retinoic acid receptor responder protein 2 | 0.577 | 1.17E-07 | 1.53E-04 |
| CCL21 | C-C motif chemokine 21 | 1.306 | 3.31E-06 | 8.68E-04 |
| TPSB2 | Tryptase β-2 | 1.157 | 2.53E-06 | 8.68E-04 |
| CCDC80 | Coiled-coil domain-containing protein 80 | 1.076 | 3.32E-06 | 8.68E-04 |
| YWHAE | 14-3-3 protein epsilon | −0.418 | 1.95E-06 | 8.68E-04 |
| KEAP1 | Kelch-like ECH-associated protein 1 | −0.109 | 7.74E-06 | 1.68E-03 |
| MMP7 | Matrilysin | 1.242 | 1.77E-05 | 2.30E-03 |
| THBS2 | Thrombospondin-2 | 1.094 | 1.87E-05 | 2.30E-03 |
| CCL19 | C-C motif chemokine 19 | 0.482 | 1.94E-05 | 2.30E-03 |
| CHST15 | Carbohydrate sulfotransferase 15 | 0.462 | 1.90E-05 | 2.30E-03 |
Coefficients were derived using the linear regression model adjusted for age, sex, race, and body mass index, in which fibrosis scores were transformed into log2 (fibrosis+1). P values were adjusted for FDR. FDR, false discovery rate; MMP7, matrix metalloprotease 7.
To understand similarities and differences between eGFR-associated and fibrosis-associated proteins, we found 13 proteins whose levels were associated with both eGFR and interstitial fibrosis (Figure 1C), confirming the relatedness of eGFR and fibrosis12 of those 12 proteins had negative correlation with eGFR and positive correlation with fibrosis (Supplemental Spreadsheet 2 and Supplemental Spreadsheet 3).
Our analysis identified MMP7 as the risk protein with the largest effect estimate for associations with both eGFR and interstitial fibrosis (Figure 1D). MMP7 levels showed a negative correlation with eGFR (Supplemental Figure 1D) and a positive correlation with fibrosis (Figure 1E). The MMP7 level was higher in DKD (Supplemental Figure 1E), indicating that MMP7 is a biomarker of kidney fibrosis. Given the biological relationship between MMP7 and fibrosis, we looked at all matrix metalloproteases (MMPs) and their inhibitor tissue inhibitor of metalloproteinases (TIMPs) in our dataset. MMP7 had the most apparent correlation with eGFR and tubulointerstitial fibrosis (Figure 2). In addition, several MMP7-correlated proteins were included in the list of fibrosis-associated proteins (Supplemental Spreadsheet 3 and Supplemental Spreadsheet 5), highlighting that MMP7 is one of the key proteins in DKD-associated fibrosis.
Figure 2.
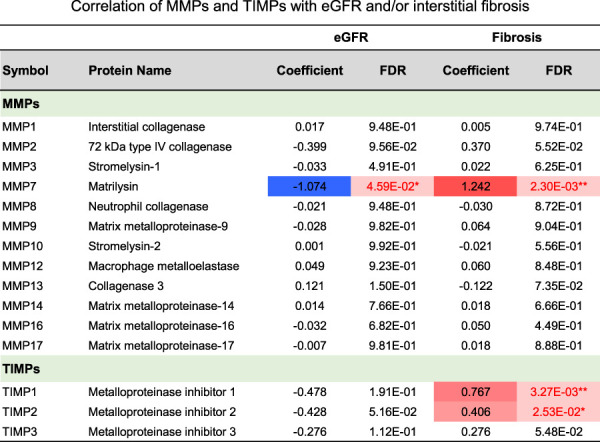
Correlation of matrix metalloproteases (MMPs) and inhibitor tissue inhibitor of metalloproteinases (TIMPs) with eGFR and/or interstitial fibrosis. Coefficients were derived using the linear regression model adjusted for age, sex, race, and BMI; P values were adjusted for FDR. *P < 0.05; **P < 0.01. Figure 2 can be viewed in color online at www.jasn.org.
Unbiased Weighted Gene Correlation Network Analysis Identified the Important Module Correlating with Renal Phenotypes
To identify coexpressed proteins and modules in our dataset in an unbiased manner, we performed WGCNA. WGCNA lead to the identification of 8 coexpressed protein modules (Figure 3, A–D). We next examined the association between these eight modules and clinical, demographics, and histological changes. eGFR and interstitial fibrosis clustered with different sets of protein expression modules, indicating important differences between these disease manifestations (Figure 3C and Supplemental Figure 1F). We further studied the association between each of the modules and clinical and histological traits. Focusing on eGFR and fibrosis, we found that the brown module, in which MMP7 was included, had the best correlation with both eGFR and fibrosis (Figure 3E). This module was enriched for proteins encoding for immune cell–related pathways (Figure 3F and Supplemental Spreadsheet 4) in the functional annotation and pathway enrichment analysis. To better understand the brown module, the protein–protein interaction network was constructed by STRING database, together with the top significant submodule (Supplemental Figure 1G), in which other MMPs and TIMPs, such as MMP2 and TIMP2, were also included.
Figure 3.
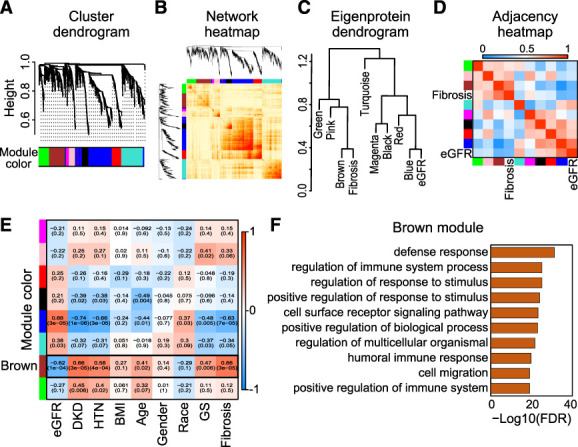
WGCNA of human DKD kidney samples. (A) Hierarchical clustering dendrogram of the proteins. (B) Heatmap representing the topological overlap matrix among all proteins in the analysis. (C) Dendrogram of eight module eigenproteins and two clinical traits (eGFR and interstitial fibrosis). (D) Eigenprotein adjacency heatmap with eGFR and interstitial fibrosis (see Methods section). (E) Correlation of module eigenproteins with clinical characteristics. Each row corresponds to a module eigenprotein, and the columns are clinical traits. The values in the cells are presented as “Pearson R (P value)” and color-coded by direction and degree of the correlation (red=positive correlation; blue=negative correlation). (F) Gene ontology pathway analysis of the top pathways enriched in the protein sets of the WGCNA brown module. Con, control; GS, glomerular sclerosis; HTN, hypertension. Figure 3 can be viewed in color online at www.jasn.org.
Tissue Transcript Levels Show Modest Correlation with Protein Levels
We next investigated the correlation of gene expression levels from RNA sequencing and protein levels analyzed in the same 33 samples. To integrate mRNA expression with protein measurements, only the mRNA corresponding to 1305 SOMAmers (1225 genes) were examined (Supplemental Spreadsheet 6). The mRNA–protein correlation was modest when all sample data were pooled (R=0.46, R2=0.21, P < 1.0e-300) (Figure 4A) and when samples were analyzed individually (R=0.46, R2=0.21, P = 4.5e-64) (Figure 4B). This potentially indicates that a large fraction of the variance in protein expression is not reflected by gene expression, underscoring the importance of protein profiling. Future studies should examine the role of consistency in measurement and normalizations of the aptamer dataset. Regarding the MMP7 expression, the mRNA–protein correlation was higher (R=0.64, P = 6.4e-05) (Figure 4C). We observed a significant correlation between MMP7 transcript and eGFR (R=−0.53, P < 0.01) (Figure 4D) as well as interstitial fibrosis (R=0.52, P < 0.01) (Figure 4E) which was consistent with the patterns in the proteomics data. Furthermore, MMP7 transcript expressions were higher in DKD compared with the control group (Figure 4F).
Figure 4.
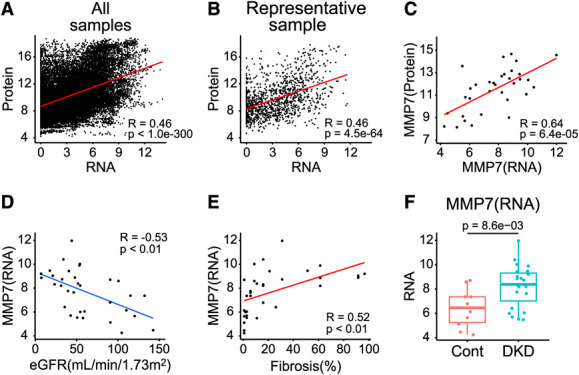
Bulk RNA-seq displayed modest correlation with proteomics. (A) Correlation between kidney bulk RNA-seq and proteomics using all samples. (B) Correlation between bulk RNA-seq and proteomics using a representative sample. (C) Correlation of MMP7 expression between RNA and protein, with Pearson R of 0.64. (D) Correlation between MMP7 expression examined by bulk RNA-seq and eGFR, with Pearson R of −0.53. (E) Correlation between MMP7 expression examined by bulk RNA-seq and interstitial fibrosis, with Pearson R of 0.52. (F) Box plots showing MMP7 expression examined by bulk RNA-seq in DKD and control subjects. P values were calculated with the Wilcoxon rank-sum test (for two group comparison). Figure 4 can be viewed in color online at www.jasn.org.
The Cell Types Expressing MMP7 in Human DKD Kidneys
To understand which cell type expresses MMP7 in human DKD kidneys, we analyzed the transcriptome of 64,333 individual cells from of DKD or LD human kidneys. We obtained information for 14 DKD kidneys (42,870 cells) and 20 LD kidneys (21,463 cells) (Figure 5A) from the NIDDK KPMP repository (https://atlas.kpmp.org/repository/),36 as described in Methods section. The cells were classified into 13 clusters by the KPMP team (Supplemental Spreadsheet 7), and we found that MMP7 was expressed by multiple kidney tubule epithelial cells (Figure 5, B–D). To identify cell types with increased MMP7 levels in DKD, we evaluated the differentially expressed genes in all clusters between DKD and LD. MMP7 was significantly (FDR <0.05) higher in proximal tubule (PT), connecting tubule (CNT), and principal cell (PC) clusters (Supplemental Spreadsheet 8). PT and CNT cells showed the largest change in gene expression (Figure 5, E and F, Supplemental Figure 2A). Taken together, these results indicate that PT, CNT, and PC are the likely sources of increased MMP7 levels in DKD kidneys.
Figure 5.
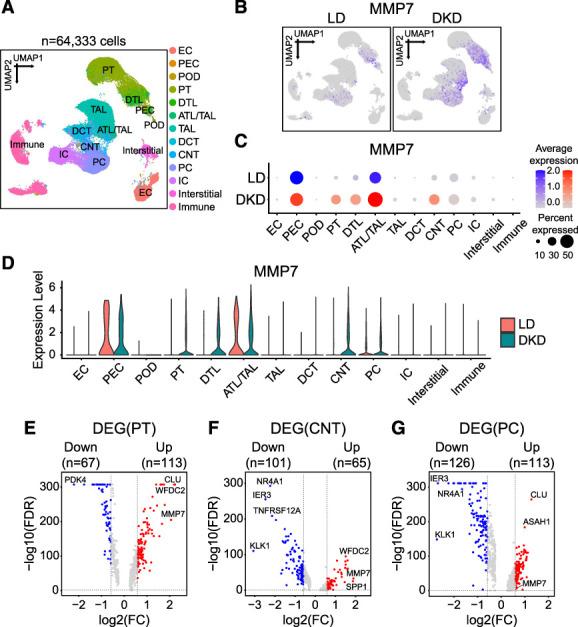
Cellular expression of MMP7 in human kidney single-cell RNA-seq. (A) UMAP showing 13 cell clusters (see Methods section). Assigned cell types are summarized in the Supplemental Spreadsheet 6. (B) Feature plots showing the expression of MMP7 in DKD subjects and LD. (C) Bubble plots showing MMP7 expression across 13 clusters between DKD subjects and LD. The size of the circle indicates the percent of positive cells, and the color indicates the level of expression. (D) Violin plots showing the MMP7 expression across 13 clusters between DKD subjects and LD. (E–G) Volcano plots of DEGs between DKD and LD in PT (E), CNT (F), and PC (G) clusters identified in the single-cell data. The x axis is log2-FC, and y axis is the statistical significance FDR=−log10. FDR <0.05 and |log2FC|>0.58 (=FC of 1.5) were considered criteria for DEG selection. Genes above the horizontal dotted gray line had FDR <0.05. DEG, differentially expressed gene; FC, fold change. Figure 5 can be viewed in color online at www.jasn.org.
Kidney Tissue MMP7 Gene and Protein Levels Correlate with eGFR and Fibrosis in External Datasets
Correlations between MMP7 gene and protein expression and kidney function and fibrosis were investigated in external datasets. Gene expression data (RNA-seq) from 433 microdissected human kidney tubule samples were analyzed, including healthy controls, diabetes, HTN, CKD, and DKD with varying degrees of fibrosis and kidney function41 (validation dataset 1, Supplemental Table 1). MMP7 expression strongly and significantly correlated with the severity of interstitial fibrosis (R=0.55, P = 2.3e-34) (Figure 6A) and negatively correlated with the level of eGFR (R=−0.35, P = 1.7e-13) (Supplemental Figure 2B) in this dataset.
Figure 6.
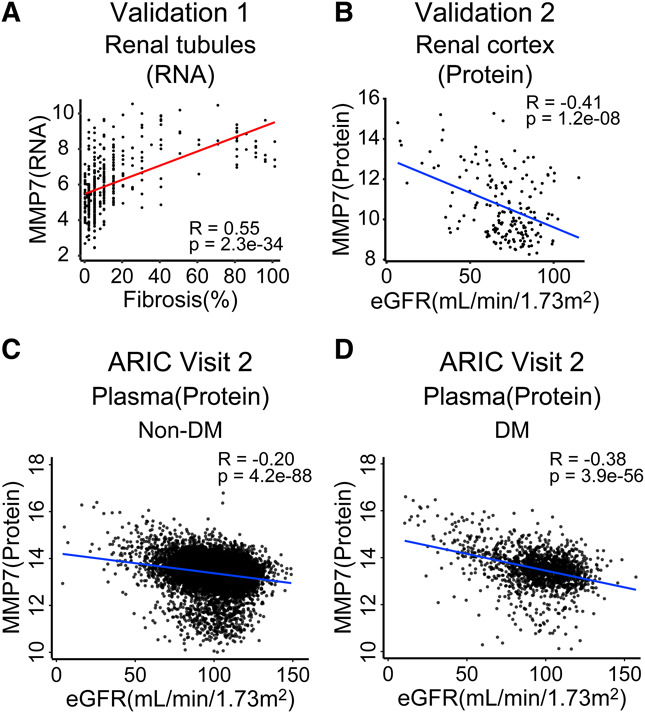
Validation of the association of MMP7 with eGFR and interstitial fibrosis in other human kidney datasets. (A) Scatterplots of the MMP7 transcript level and the degree of fibrosis in 433 microdissected human kidney tubules (validation dataset 1). (B) Scatterplots of MMP7 protein expression and eGFR (ml/min per 1.73 m2) in 186 human DKD and control kidneys (validation dataset 2). (C) Scatterplot of plasma MMP7 protein levels and eGFR (ml/min per 1.73 m2) in nondiabetic participants of ARIC (validation dataset 3). (D) Scatterplot of plasma MMP7 protein levels and eGFR (ml/min per 1.73 m2) in diabetic participants of ARIC (validation dataset 3). Figure 6 can be viewed in color online at www.jasn.org.
In addition, the protein level of MMP7 was examined in an external dataset of 186 human kidney samples (34 from DKD, 152 from control) (validation dataset 2, Supplemental Table 1). The MMP7 protein level showed a negative correlation with eGFR (R=−0.41, P = 1.2e-08) (Figure 6B) in this dataset as well. Together, these findings confirm the consistent correlations between kidney MMP7 protein and RNA expression, interstitial fibrosis, and eGFR across multiple validation datasets.
Circulating MMP7 Predicts Kidney Disease Progression in Patients with or without Diabetes
Given the strong association between tissue MMP7 protein and fibrosis, a strong predictor of kidney failure, we investigated whether circulating MMP7 could serve as a biomarker for kidney disease progression. We analyzed the association between MMP7 levels and eGFR decline in 1623 participants with diabetes and 9407 participants without diabetes enrolled in the ARIC Study (validation dataset 3, Table 3). The mean eGFR, estimated by the 2021 CKD-EPI equation on the basis of creatinine and cystatin C,42 was 95±20 and 99±16 ml/min per 1.73 m2, respectively. Over an average follow-up of 17 years, 354 diabetic and 1190 nondiabetic participants experienced eGFR decline by 50%, whereas 153 diabetic and 131 nondiabetic participants developed ESKD.
Table 3.
Summary of Atherosclerosis Risk In Communities visit 2 participants characteristics
| Characteristic | Non-DM (n=9407) |
DM (n=1623) |
|---|---|---|
| Age, yr (SD) | 56.8 (5.7) | 58.1 (5.7) |
| Male (%) | 4108 (43.7) | 977 (45.5) |
| Black, n (%) | 1849 (19.7) | 646 (39.8) |
| DM, n (%) | 0 (0) | 1623 (100) |
| HTN, n (%) | 2948 (31.3) | 950 (58.5) |
| Antihypertensive medication, n (%) | 2139 (22.7) | 808 (49.8) |
| Prevalent CVD, n (%) | 595 (6.3) | 241 (14.8) |
| Current smoker, n (%) | 2128 (22.6) | 302 (18.6) |
| Former smoker, n (%) | 3543 (37.7) | 625 (38.5) |
| Total cholesterol, mmol/L (SD) | 5.4 (1.0) | 5.5 (1.2) |
| HDL cholesterol mmol/L (SD) | 1.3 (0.4) | 1.1 (0.4) |
| BMI, kg/m2 (SD) | 27.3 (5.0) | 31.2 (5.9) |
| SBP, mm Hg (SD) | 120 (18) | 128 (20) |
| eGFRcrcys, ml/min per 1.73 m2 (SD) | 99 (16) | 95 (20) |
| eGFR 50% decline, n (%) | 1190 (12.7) | 354 (21.8) |
| Follow-up to eGFR 50% decline years (range) | 24.6 (17.8–27.7) | 18.3 (10.4–24.2) |
| Incident ESKD, n (%) | 131 (1.4) | 153 (9.4) |
| Follow-up to ESKD years (range) | 25.7 (18.0–27.9) | 18.5 (11.0–25.5) |
Data are presented as mean (SD), median (range), or n (%). DM, diabetes mellitus; HTN, hypertension; CVD, cardiovascular disease; BMI, body mass index; SBP, systolic blood pressure; eGFRcrcys, eGFR on the basis of creatinine and cystatin C.
Plasma MMP levels in participants with or without diabetes at baseline negatively correlated with eGFR (R=−0.38 for diabetic and R=−0.20 for nondiabetic participants) (Figure 6, C and D). In Cox regression analyses among diabetic individuals, plasma MMP7 concentration was significantly associated with a 2.3-fold greater risk of experiencing eGFR decline by 50% or more (hazard ratio [HR], 2.28; 95% confidence interval [CI], 1.92 to 2.70) and a 3.8-fold greater risk of developing kidney failure (HR, 3.78; 95% CI, 3.00 to 4.77), respectively (Table 4). After adjusting for additional covariates, including eGFR and smoking status, plasma MMP7 concentration remained significantly associated with greater risk of 50% decline of eGFR (HR, 1.77; 95% CI, 1.48 to 2.11) and a kidney failure (HR, 2.01; 95% CI, 1.54 to 2.62) (Table 4). In nondiabetic individuals, higher plasma MMP7 concentration was also significantly associated with greater risk of a kidney failure (HR, 2.15; 95% CI, 1.60 to 2.89) (Table 4), after adjusting for additional covariates. In conclusion, blood MMP7 concentration can identify individuals with or without diabetes who are at increased risk of progressing to kidney failure in the general population.
Table 4.
Circulating matrix metalloprotease 7 was associated with greater risk of eGFR decline by 50% or ESKD in Atherosclerosis Risk In Communities participants at visit 2
| Outcome | Model | Non-DM (n=9407) | DM (n=1623) | ||||
|---|---|---|---|---|---|---|---|
| Events | HR (95% CI) | P Value | Events | HR (95% CI) | P Value | ||
| eGFR 50% decline | Model 1 | 1190 | 1.35 (1.23 to 1.48) | 3.20E-10 | 354 | 2.28 (1.92 to 2.70) | 4.44E-21 |
| eGFR 50% decline | Model 2 | 1190 | 1.24 (1.13 to 1.35) | 3.66E-06 | 354 | 1.77 (1.48 to 2.11) | 2.33E-10 |
| eGFR 50% decline | Model 3 | 1190 | 1.17 (1.05 to 1.29) | 2.94E-03 | 354 | 1.68 (1.40 to 2.01) | 1.79E-08 |
| ESKD | Model 1 | 131 | 3.94 (3.09 to 5.03) | 3.93E-28 | 153 | 3.78 (3.00 to 4.77) | 1.95E-29 |
| ESKD | Model 2 | 131 | 2.01 (1.51 to 2.69) | 1.96E-06 | 153 | 2.01 (1.54 to 2.62) | 3.27E-07 |
| ESKD | Model 3 | 131 | 2.15 (1.60 to 2.89) | 3.23E-07 | 153 | 1.77 (1.35 to 2.32) | 3.26E-05 |
Model 1 was adjusted for age, sex, and race/center; model 2 was additionally adjusted for systolic blood pressure, antihypertensive medications, prevalent cardiovascular disease, smoking status, eGFRcrcys, HDL cholesterol levels, and total cholesterol levels; model 3 was adjusted for all previously mentioned covariates along with the first five principal components of the proteomics data. All hazard ratios are expressed per doubling of the protein level. DM, diabetes mellitus; HR, hazard ratio; CI, confidence interval.
Discussion
This study presents the first unbiased human kidney tissue proteomics dataset for healthy and DKD samples. The analysis identified 14 proteins that correlated with eGFR and 152 proteins that correlated with fibrosis, with MMP7 showing the largest effect size. The correlation between tissue MMP7 protein expression and kidney function was confirmed in an external human kidney tissue dataset. Kidney tissue MMP7 transcript levels also correlated with fibrosis and eGFR in both primary and in an independent external dataset. Proximal tubules CNTs, and collecting duct cells being the most likely sources of MMP7 in DKD. In addition, plasma MMP7 levels were found not only correlated with kidney function but also predicted future kidney function decline indicating that MMP7 is a prognostic biomarker.
The lack of unbiased human kidney proteomics has been a major gap in our understanding of human DKD development. Mass spectrometry-based tissue proteomics has provided important initial insight into protein levels, but it has been challenging to accurately quantify protein levels in large cohorts. The new aptamer-based methods can quantify thousands of proteins in multiple samples. Protein quantification is critical as previous datasets analyzing blood samples indicated that blood protein levels show a poor correlation with RNA levels. This might not be surprising as most blood proteins are synthesized in the liver. Paired tissue protein and RNA datasets are rare, so the RNA–protein correlation in tissue samples is not well understood. Our dataset shows a modest association between transcript and protein expression in paired kidney tissue samples, but further studies are needed to better understand the relationship between transcript and protein levels in the kidney. Orthogonal validation will also be essential, as it has also been reported that some aptamer probes may not be specific or unable to accurately capture protein-level changes.
The development of kidney fibrosis is characterized by the proliferation and transformation of fibroblasts as well as accumulation of extracellular matrix (ECM), which is the pathological hallmark of progressive kidney disease.43 However, the role of tissue ECM in CKD is poorly understood. Recent studies indicate that stromal cells are responsible for the expression of ECM components and that the ECM undergoes important remodeling with MMPs being the main proteinases involved in ECM degradation.44 MMP7 cleaves collagen III/IV/V/IX/X/XI and proteoglycans45 and has been extensively studied in relation to kidney disease development. Studies have shown that MMP7 is regulated by the Wnt/β catenin pathway and that its expression is increased in renal fibrosis46 and AKI.47,48 Mice with genetic deletion of MMP7 were found to be protected from AKI. Therefore, the unbiased discovery of MMP7 in human samples is strongly supported by previous mechanistic studies.
It is important to note that previous studies have investigated MMP7 as a DKD biomarker in patients through targeted biomarker studies. These studies have shown that the serum MMP7 level was increased in patients with proteinuric DKD subjects,49 and kidney biopsies from patients with IgA nephropathy, lupus nephritis, and DKD showed higher MMP7 mRNA or protein levels.46,50–52 Urine MMP7 concentration was associated with mortality in patients with proteinuria and DKD.53 In addition, circulating MMP7 was associated with DKD progression in type 2 diabetic individuals with eGFR >60 ml/min per 1.73 m2 in the Joslin Kidney Study.54 In this study, we identified MMP7 in unbiased tissue proteomics studies and validated it in external tissue datasets. Our findings provide new insights into the role and source of kidney tissue MMP7 protein, and point to MMP7 as a potential biomarker of DKD, as demonstrated by its correlation with kidney function and prediction of future kidney function decline in a community-based diabetic population with varying degrees of kidney function.
In this study, TNF superfamily member 12 (TNFSF12), a type II transmembrane glycoprotein of the TNF superfamily also known as TNF-like weak inducer of apoptosis (TWEAK),55 was the only protein that correlated with eGFR but not with interstitial fibrosis in the kidney. Previous studies have shown that circulating TNFSF12 concentrations were lower in subjects with type 2 diabetes56 and DKD57 and were strongly associated with progressive renal decline58 and the risk of kidney failure.6 However, there are conflicting reports on the proinflammatory effects of TNFSF12 on tubular epithelial cells and its role in nondiabetic AKI and CKD. TNFSF12 might have proinflammatory effects on tubular epithelial cells in vitro and in vivo, but TNFSF12 blockade reduced tubulointerstitial inflammation in AKI mice,59 and TNFSF12 might promote nondiabetic AKI and CKD.59,60 Further animal and clinical investigations are needed to better understand the role of TNFSF12 in the development and progression of DKD.
In this study, several limitations should be noted. First, there is a lack of proteinuria data in the primary kidney tissue proteomics study and in ARIC participants, which makes it unclear whether MMP7 would improve outcome precision after accounting for proteinuria. In addition, the small sample size of the primary dataset limits the assessment of additional covariates beyond age, sex, race, and BMI in the linear regression model. Although these are important limitations, the small sample size only allows for adjustment of a limited number of covariates without overfitting. Furthermore, there is no follow-up information for the kidney tissue cohort, and samples (both control and DKD) were obtained from unaffected portions of nephrectomies. Finally, there is no information available regarding whether these associations extend to subjects with type 1 diabetes and kidney disease.
In summary, here, we present the first in class unbiased affinity proteomics of DKD kidneys with evaluation of gene expression by RNA-seq, multiple validations of tissue MMP7 levels in different DKD population, and the recognition of blood MMP7 as a biomarker of future eGFR decline in subjects with diabetes and with varying degree of kidney function.
Supplementary Material
Acknowledgments
The authors thank the staff and participants of the ARIC study for their important contributions. Some of the data reported here have been supplied by the US Renal Data System.
Disclosures
J. Coresh reports Consultancy: SomaLogic and Research Funding: National Institutes of Health. M.E. Grams reports Advisory or Leadership Role: American Journal of Kidney Diseases, CJASN, JASN Editorial Board, ASN Publication Committee, KDIGO Executive Committee (cochair elect), NKF Scientific Advisory Board, and USRDS Scientific Advisory Board and Other Interests or Relationships: Grant funding from NKF, which receives funding from multiple pharmaceutical companies, grant funding from NIH, payment from academic institutions for grand rounds, and payment from NephSAP. A. Karihaloo is an employee of Novo Nordisk. M.A. Niewczas has provided consulting for Otsuka. M.A. Niewczas also reports Patents or Royalties: Coinventor of the TNF-R1 and TNF-R2 issued patent for predicting, risk of ESKD. This patent was licensed by the Joslin Diabetes Center to EKF Diagnostics. M.B. Palmer reports Advisory or Leadership Role: Editorial Board, American Journal of Kidney Disease. K. Susztak reports Consultancy: Astra Zeneca, GSK, Novo Nordisk, and Pfizer; Ownership Interest: Jnana; Research Funding: Astra Zeneca, Bayer, Boehringer Ingelheim, Calico, Gilead, GSK, Jnana, Kyowa Kirin Genentech, Maze, Novartis, Novo Nordisk, ONO Pharma, Regeneron, and Variant Bio; Honoraria: Astra Zeneca, Bayer, Jnana, Maze, and Pfizer; and Advisory or Leadership Role: Editorial board, Cell Metabolism, EBioMedicine, JASN, Journal of Clinical Investigation, Kidney International, Med, Jnana, Pfizer. The laboratory of K. Susztak is supported by BIPI, Calico, Genentech, Gilead, GSK, Maze, Novartis, Novo Nordisk, and Regeneron. K. Susztak in on the SAB of Pfizer and Jnana. All remaining authors have nothing to disclose.
Funding
The authors' work described in this article was supported by the National Institutes of Health (NIH Grant Nos. R01 DK105821, R01 DK087635, and R01 DK076077 to K.S.; R01 DK123459 to M.A.N.; R01 DK124399 to M.E.G.; and P30 DK036836 to Joslin Diabetes Center). This work was also supported by research grant from Manpei Suzuki Diabetes Foundation to D. Hirohama and by the mentored fellowship from the William Randolph Hearst Foundation to Z.M. Dom in M.A. Niewczas laboratory.
The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (under Contract Nos HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700005I, and HHSN268201700004I). The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by NHLBI contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). The interpretation and reporting of these data are the responsibility of the authors and in no way should be seen as an official policy or interpretation of the US Government.
Author Contributions
Conceptualization: Monika A. Niewczas, Katalin Susztak.
Data curation: Amin Abedini, Simon T. Dillon, Zaipul Md Dom, Daigoro Hirohama, Salina Moon, Matthew B. Palmer, Allison Vassalotti.
Formal analysis: Amin Abedini, Josef Coresh, Tomohito Doke, Morgan E. Grams, Daigoro Hirohama, Anil Karihaloo, Hongbo Liu, Victor Martinez, Aditya Surapaneni.
Funding acquisition: Monika A. Niewczas, Katalin Susztak.
Investigation: Matthew B. Palmer.
Methodology: Monika A. Niewczas, Katalin Susztak.
Project administration: Daigoro Hirohama, Katalin Susztak.
Resources: Katalin Susztak.
Supervision: Monika A. Niewczas, Katalin Susztak.
Validation: Morgan E. Grams.
Visualization: Daigoro Hirohama.
Writing – original draft: Daigoro Hirohama, Katalin Susztak.
Writing – review & editing: Salina Moon, Monika A. Niewczas.
Data Sharing Statement
SomaScan kidney tissue proteomics data of primary dataset have been deposited in Mendeley https://data.mendeley.com/datasets/83k89shdx5/ Regarding Blood Proteomics data, preexisting data access policies for each of the parent cohort studies specify that research data requests can be submitted to each steering committee; these will be promptly reviewed for confidentiality or intellectual property restrictions and will not unreasonably be refused. Please refer to the data sharing policies of these studies. Individual-level patient or protein data may further be restricted by consent, confidentiality, or privacy laws/considerations. These policies apply to both clinical and proteomic data.
All codes used for the analysis are provided using a “GitHub repository (https://github.com/dhirohama/Human_Kidney_Proteomics).”
Supplemental Material
This article contains the following supplemental material online at http://links.lww.com/JSN/E407, http://links.lww.com/JSN/E408, http://links.lww.com/JSN/E409, http://links.lww.com/JSN/E410, http://links.lww.com/JSN/E411, http://links.lww.com/JSN/E412, http://links.lww.com/JSN/E413, http://links.lww.com/JSN/E414, and http://links.lww.com/JSN/E415.
Supplemental Spreadsheet 1. List of 1305 proteins measured using SomaScan.
Supplemental Spreadsheet 2. Proteins associated with eGFR in linear regression model, with or without adjustment for age, sex, race, and BMI.
Supplemental Spreadsheet 3. Proteins associated with renal interstitial fibrosis in linear regression model, with or without adjustment for age, sex, race, and BMI.
Supplemental Spreadsheet 4. DAVID gene ontology analysis for renal interstitial fibrosis-associated proteins and proteins in the brown module of WGCNA.
Supplemental Spreadsheet 5. Proteins associated with MMP7 in linear regression model.
Supplemental Spreadsheet 6. Paired expression of 1225 mRNA and protein levels in human kidney samples.
Supplemental Spreadsheet 7. Cell numbers of 13 clusters between LD and DKD in the KPMP dataset.
Supplemental Spreadsheet 8. DEGs of all 13 clusters between LD and DKD in the KPMP dataset.
Supplemental Table 1. Patient characteristics of validation database.
Supplemental Figure 1. Correlation of clinical parameters with MMP7.
Supplemental Figure 2. MMP7 expression by single-cell RNA-seq and validation of MMP7 expression by bulk RNA-seq in kidneys.
References
- 1.Tangri N Stevens LA Griffith J, et al. A predictive model for progression of chronic kidney disease to kidney failure. JAMA. 2011;305(15):1553–1559. doi: 10.1001/jama.2011.451 [DOI] [PubMed] [Google Scholar]
- 2.Tangri N Grams ME Levey AS, et al. Multinational assessment of accuracy of equations for predicting risk of kidney failure: a meta-analysis. JAMA. 2016;315(2):164–174. doi: 10.1001/jama.2015.18202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinn GZ Abedini A Liu H, et al. Renal histologic analysis provides complementary information to kidney function measurement for patients with early diabetic or hypertensive disease. J Am Soc Nephrol. 2021;32(11):2863–2876. doi: 10.1681/ASN.2021010044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niewczas MA Gohda T Skupien J, et al. Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J Am Soc Nephrol. 2012;23(3):507–515. doi: 10.1681/ASN.2011060627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayek SS Sever S Ko YA, et al. Soluble urokinase receptor and chronic kidney disease. N Engl J Med. 2015;373(20):1916–1925. doi: 10.1056/nejmoa1506362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niewczas MA Pavkov ME Skupien J, et al. A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med. 2019;25(5):805–813. doi: 10.1038/s41591-019-0415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grams ME Surapaneni A Chen J, et al. Proteins associated with risk of kidney function decline in the general population. J Am Soc Nephrol. 2021;32(9):2291–2302. doi: 10.1681/ASN.2020111607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen TK Surapaneni AL Arking DE, et al. APOL1 kidney risk variants and proteomics. Clin J Am Soc Nephrol. 2022;17(5):684–692. doi: 10.2215/CJN.14701121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Z Jin J Tin A, et al. Polygenic risk scores for kidney function and their associations with circulating proteome, and incident kidney diseases. J Am Soc Nephrol. 2021;32(12):3161–3173. doi: 10.1681/ASN.2020111599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi H Looker HC Satake E, et al. Neuroblastoma suppressor of tumorigenicity 1 is a circulating protein associated with progression to end-stage kidney disease in diabetes. Sci Transl Med. 2022;14(657):eabj2109. doi: 10.1126/scitranslmed.abj2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi H Looker HC Satake E, et al. Results of untargeted analysis using the SOMAscan proteomics platform indicates novel associations of circulating proteins with risk of progression to kidney failure in diabetes. Kidney Int. 2022;102(2):370–381. doi: 10.1016/j.kint.2022.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckerman P Qiu C Park J, et al. Human kidney tubule-specific gene expression based dissection of chronic kidney disease traits. EBioMedicine. 2017;24:267–276. doi: 10.1016/j.ebiom.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju W Greene CS Eichinger F, et al. Defining cell-type specificity at the transcriptional level in human disease. Genome Res. 2013;23(11):1862–1873. doi: 10.1101/gr.155697.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Connell PJ Zhang W Menon MC, et al. Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: a multicentre, prospective study. Lancet. 2016;388(10048):983–993. doi: 10.1016/s0140-6736(16)30826-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J Shrestha R Qiu C, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360(6390):758–763. doi: 10.1126/science.aar2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson PC Wu H Kirita Y, et al. The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc Natl Acad Sci U S A. 2019;116(39):19619–19625. doi: 10.1073/pnas.1908706116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwanhäusser B Busse D Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- 18.Cuadrado E van den Biggelaar M de Kivit S, et al. Proteomic analyses of human regulatory T cells reveal adaptations in signaling pathways that protect cellular identity. Immunity. 2018;48(5):1046–1059.e6. doi: 10.1016/j.immuni.2018.04.008 [DOI] [PubMed] [Google Scholar]
- 19.Hukelmann JL Anderson KE Sinclair LV, et al. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat Immunol. 2016;17(1):104–112. doi: 10.1038/ni.3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummins TD Korte EA Bhayana S, et al. Advances in proteomic profiling of pediatric kidney diseases. Pediatr Nephrol. 2022;37(10):2255–2265. doi: 10.1007/s00467-022-05497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Randles MJ Lausecker F Kong Q, et al. Identification of an altered matrix signature in kidney aging and disease. J Am Soc Nephrol. 2021;32(7):1713–1732. doi: 10.1681/ASN.2020101442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gold L Ayers D Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams SA Kivimaki M Langenberg C, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med. 2019;25(12):1851–1857. doi: 10.1038/s41591-019-0665-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright JD Folsom AR Coresh J, et al. The ARIC (atherosclerosis risk in communities) study: JACC focus seminar 3/8. J Am Coll Cardiol. 2021;77(23):2939–2959. doi: 10.1016/j.jacc.2021.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Kidney Foundation. KDOQI clinical practice guideline for diabetes and CKD: 2012 update. Am J Kidney Dis. 2012;60(5):850–886. doi: 10.1053/j.ajkd.2012.07.005 [DOI] [PubMed] [Google Scholar]
- 26.Tervaert TWC Mooyaart AL Amann K, et al. Pathologic classification of diabetic nephropathy. J Am Soc Nephrol. 2010;21(4):556–563. doi: 10.1681/ASN.2010010010 [DOI] [PubMed] [Google Scholar]
- 27.Johansen KL Chertow GM Gilbertson DT, et al. US renal data system 2021 annual data report: Epidemiology of kidney disease in the United States. Am J Kidney Dis. 2022;79(4):A8–A12. doi: 10.1053/j.ajkd.2022.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1):559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman BT Hao M Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–W221. doi: 10.1093/nar/gkac194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4(1):2. doi: 10.1186/1471-2105-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandettini WP Kellman P Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson. 2012;14(1):83. doi: 10.1186/1532-429x-14-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu C Huang S Park J, et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med. 2018;24(11):1721–1731. doi: 10.1038/s41591-018-0194-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H Doke T Guo D, et al. Epigenomic and transcriptomic analyses define core cell types, genes and targetable mechanisms for kidney disease. Nat Genet. 2022;54(7):950–962. doi: 10.1038/s41588-022-01097-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng X Guan Y Ma Z, et al. Mapping the genetic architecture of human traits to cell types in the kidney identifies mechanisms of disease and potential treatments. Nat Genet. 2021;53(9):1322–1333. doi: 10.1038/s41588-021-00909-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen J Sealfon R Menon R, et al. ; Kidney Precision Medicine Project. A reference tissue atlas for the human kidney. Sci Adv. 2022;8(23):eabn4965. doi: 10.1126/sciadv.abn4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimes PK, Liu Y, Neil Hayes D, Marron JS. Statistical significance for hierarchical clustering. Biometrics. 2017;73(3):811–821. doi: 10.1111/biom.12647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lengyel A, Botta-Dukát Z. Silhouette width using generalized mean-A flexible method for assessing clustering efficiency. Ecol Evol. 2019;9(23):13231–13243. doi: 10.1002/ece3.5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer MB Abedini A Jackson C, et al. The role of glomerular epithelial injury in kidney function decline in patients with diabetic kidney disease in the TRIDENT cohort. Kidney Int Rep. 2021;6(4):1066–1080. doi: 10.1016/j.ekir.2021.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levey AS Stevens LA Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604–612. doi: 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung KW Dhillon P Huang S, et al. Mitochondrial damage and activation of the STING pathway lead to renal inflammation and fibrosis. Cel Metab. 2019;30(4):784–799.e5. doi: 10.1016/j.cmet.2019.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inker LA Eneanya ND Coresh J, et al. New creatinine- and cystatin C-based equations to estimate GFR without race. N Engl J Med. 2021;385(19):1737–1749. doi: 10.1056/nejmoa2102953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y. Cellular and molecular mechanisms of renal fibrosis. Nat Rev Nephrol. 2011;7(12):684–696. doi: 10.1038/nrneph.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161–174. doi: 10.1038/nrc745 [DOI] [PubMed] [Google Scholar]
- 44.Ke B, Fan C, Yang L, Fang X. Corrigendum: matrix metalloproteinase-7 and kidney fibrosis. Front Physiol. 2017;8:192. doi: 10.3389/fphys.2017.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He W Tan RJ Li Y, et al. Matrix metalloproteinase-7 as a surrogate marker predicts renal Wnt/β-catenin activity in CKD. J Am Soc Nephrol. 2012;23(2):294–304. doi: 10.1681/ASN.2011050490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wozniak J, Floege J, Ostendorf T, Ludwig A. Key metalloproteinase-mediated pathways in the kidney. Nat Rev Nephrol. 2021;17(8):513–527. doi: 10.1038/s41581-021-00415-5 [DOI] [PubMed] [Google Scholar]
- 47.Liu Z, Tan RJ, Liu Y. The many faces of matrix metalloproteinase-7 in kidney diseases. Biomolecules. 2020;10(6):960. doi: 10.3390/biom10060960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ban CR Twigg SM Franjic B, et al. Serum MMP-7 is increased in diabetic renal disease and diabetic diastolic dysfunction. Diabetes Res Clin Pract. 2010;87(3):335–341. doi: 10.1016/j.diabres.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 49.Zhou D Tian Y Sun L, et al. Matrix metalloproteinase-7 is a urinary biomarker and pathogenic mediator of kidney fibrosis. J Am Soc Nephrol. 2017;28(2):598–611. doi: 10.1681/ASN.2016030354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reich HN Landolt-Marticorena C Boutros PC, et al. Molecular markers of injury in kidney biopsy specimens of patients with lupus nephritis. J Mol Diagn. 2011;13(2):143–151. doi: 10.1016/j.jmoldx.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen CD Lindenmeyer MT Eichinger F, et al. Improved elucidation of biological processes linked to diabetic nephropathy by single probe-based microarray data analysis. PLoS One. 2008;3(8):e2937. doi: 10.1371/journal.pone.0002937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afkarian M Zelnick LR Ruzinski J, et al. Urine matrix metalloproteinase-7 and risk of kidney disease progression and mortality in type 2 diabetes. J Diabetes Complications. 2015;29(8):1024–1031. doi: 10.1016/j.jdiacomp.2015.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ihara K Skupien J Kobayashi H, et al. Profibrotic circulating proteins and risk of early progressive renal decline in patients with type 2 diabetes with and without albuminuria. Diabetes Care. 2020;43(11):2760–2767. doi: 10.2337/dc20-0630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chicheportiche Y Bourdon PR Xu H, et al. TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J Biol Chem. 1997;272(51):32401–32410. doi: 10.1074/jbc.272.51.32401 [DOI] [PubMed] [Google Scholar]
- 55.Kralisch S Ziegelmeier M Bachmann A, et al. Serum levels of the atherosclerosis biomarker sTWEAK are decreased in type 2 diabetes and end-stage renal disease. Atherosclerosis. 2008;199(2):440–444. doi: 10.1016/j.atherosclerosis.2007.10.022 [DOI] [PubMed] [Google Scholar]
- 56.Liu SY, Chen J, Li YF. Clinical significance of serum interleukin-8 and soluble tumor necrosis factor-like weak inducer of apoptosis levels in patients with diabetic nephropathy. J Diabetes Investig. 2018;9(5):1182–1188. doi: 10.1111/jdi.12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Md Dom ZI Satake E Skupien J, et al. Circulating proteins protect against renal decline and progression to end-stage renal disease in patients with diabetes. Sci Transl Med. 2021;13(600):eabd2699. doi: 10.1126/scitranslmed.abd2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanz AB Justo P Sanchez-Niño MD Blanco-Colio LM Winkles JA Kreztler M, et al. The cytokine TWEAK modulates renal tubulointerstitial inflammation. J Am Soc Nephrol. 2008;19(4):695–703. doi: 10.1681/ASN.2007050577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanz AB Izquierdo MC Sanchez-Niño MD, et al. TWEAK and the progression of renal disease: clinical translation. Nephrol Dial Transplant. 2014;29(suppl 1):i54–i62. doi: 10.1093/ndt/gft342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernardi S Voltan R Rimondi E, et al. TRAIL, OPG, and TWEAK in kidney disease: biomarkers or therapeutic targets? Clin Sci. 2019;133(10):1145–1166. doi: 10.1042/cs20181116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
SomaScan kidney tissue proteomics data of primary dataset have been deposited in Mendeley https://data.mendeley.com/datasets/83k89shdx5/ Regarding Blood Proteomics data, preexisting data access policies for each of the parent cohort studies specify that research data requests can be submitted to each steering committee; these will be promptly reviewed for confidentiality or intellectual property restrictions and will not unreasonably be refused. Please refer to the data sharing policies of these studies. Individual-level patient or protein data may further be restricted by consent, confidentiality, or privacy laws/considerations. These policies apply to both clinical and proteomic data.
All codes used for the analysis are provided using a “GitHub repository (https://github.com/dhirohama/Human_Kidney_Proteomics).”