Abstract
DoxA is a cytochrome P-450 monooxygenase involved in the late stages of daunorubicin and doxorubicin biosynthesis that has a broad substrate specificity for anthracycline glycone substrates. Recombinant DoxA was purified to homogeneity from Streptomyces lividans transformed with a plasmid containing the Streptomyces sp. strain C5 doxA gene under the control of the strong SnpR-activated snpA promoter. The purified enzyme was a monomeric, soluble protein with an apparent Mr of 47,000. Purified DoxA catalyzed the 13-hydroxylation of 13-deoxydaunorubicin, the 13-oxidation of 13-dihydrocarminomycin and 13-dihydrodaunorubicin, and the 14-hydroxylation of daunorubicin. The pH optimum for heme activation was pH 7.5, and the temperature optimum was 30°C. The kcat/Km values for the oxidation of anthracycline substrates by purified DoxA, incubated with appropriate electron-donating components, were as follows: for 13-deoxydaunorubicin, 22,000 M−1 · s−1; for 13-dihydrodaunorubicin, 14,000 M−1 · s−1; for 13-dihydrocarminomycin, 280 M−1 · s−1; and for daunorubicin, 130 M−1 · s−1. Our results indicate that the conversion of daunorubicin to doxorubicin by this enzyme is not a favored reaction and that the main anthracycline flux through the late steps of the daunorubicin biosynthetic pathway catalyzed by DoxA is likely directed through the 4-O-methyl series of anthracyclines.
Streptomyces sp. strain C5 produces the anthracycline antibiotics ɛ-rhodomycinone, daunorubicin (daunomycin), and baumycins (24). Most daunorubicin-producing strains, including wild-type Streptomyces peucetius, accumulate these three compounds as well as other intermediates of baumycin biosynthesis (24). Arcamone et al. (2), however, isolated a mutant of S. peucetius, which they named S. peucetius subsp. caesius, that produced doxorubicin (14-hydroxydaunorubicin), which added a functionality. There has since been great interest in elucidating the mechanism of doxorubicin biosynthesis and its relationship to daunorubicin biosynthesis (9, 10, 14, 18, 24).
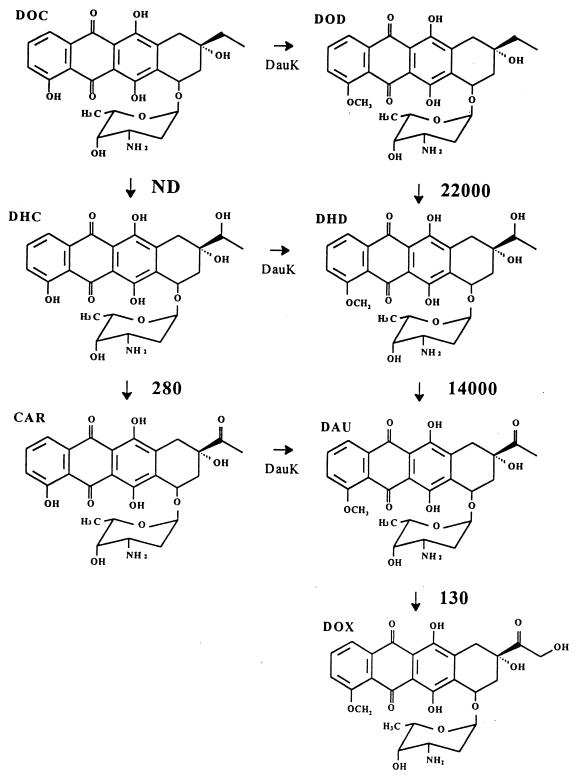
Using both in vivo and in vitro reactions, we have shown previously that DoxA, a cytochrome P-450, catalyzes three oxidation steps in the biosynthesis of doxorubicin: (i) the oxidation of 13-deoxycarminomycin and 13-deoxydaunorubicin to 13-hydroxycarminomycin and 13-dihydrodaunorubicin, respectively; (ii) the oxidation of 13-dihydrocarminomycin and 13-dihydrodaunorubicin to carminomycin and daunorubicin, respectively; and (iii) the 14-hydroxylation of daunorubicin to doxorubicin (9, 24) (Fig. 1). The 13-oxidation of the 13-dihydro anthracyclines was shown to be NADPH and oxygen dependent, indicating that the oxidation step had to proceed through an additional hydroxylation event (9, 24). For that step, we hypothesized that a 13-dihydroxy intermediate was formed which resolved to the 13-keto form (i.e., daunorubicin or carminomycin) via spontaneous dehydration (9, 24).
FIG. 1.
Summary of reactions catalyzed by purified DoxA and their respective kcat/Km values expressed in M−1 · s−1. DOC, 13-deoxycarminomycin; DHC, 13-dihydrocarminomycin; CAR, carminomycin; DOD, 13-deoxydaunorubicin; DHD, 13-dihydrodaunorubicin; DAU, daunorubicin; DOX, doxorubicin; ND, not done (but previously shown to occur [9]). DauK (anthracycline 4-O-methyltransferase) catalyzes the methylation of 4-hydroxyanthracyclines to the 4-methoxy analogues.
Our previous experiments did not delineate the preferred substrates for DoxA, and because they were run with either whole cells or crude extracts, other enzymes present in those cells or extracts (particularly nonspecific 13-keto dehydrogenases) had profound effects on the results obtained, making interpretations about flux and substrate specificities difficult and tentative (9, 10).
Here we report the purification from Streptomyces lividans TK24(pANT195) of recombinant Streptomyces sp. strain C5 DoxA, and we describe its physical properties and kinetic characteristics. The kinetic data obtained in this work indicate that DoxA appears to have a strong preference for 4-methoxy anthracycline intermediates over their 4-hydroxy analogues as well as a preference for hydroxylation of the C-13 position over the C-14 position.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. lividans TK24(pANT195), which contains the Streptomyces sp. strain C5 doxA gene expressed from the strong SnpR-activated snpA promoter (8) in a high-copy-number plasmid, has been described previously (9, 10). Primary seed cultures of S. lividans TK24(pANT195) were incubated for 48 h by rotary shaking (250 rpm; 30°C) in 50 ml of yeast extract-malt extract (YEME) medium, supplemented with 20% (wt/vol) sucrose and 40 μg of thiostrepton · ml−1, in 250-ml shake flasks containing a coiled spring (9). These cultures were used to inoculate 450 ml of the same medium (in 2-liter flasks), which were incubated for 48 h before being used to inoculate a 10-liter (working volume) stirred tank fermentor (MicroFerm; New Brunswick Scientific, New Brunswick, N.J.) containing the same medium. Fermentation conditions were as follows: temperature, 30°C; aeration, constant at 10 liters/min; agitation, 250 rpm; and no pH control. After 100 to 120 h, the mycelium in the fermentation broth was harvested by continuous centrifugation at 16,000 × g at a flow rate of approximately 100 ml · min−1 with a Contifuge 17 RS continuous centrifuge (Heraeus Instruments, Hanau, Germany). Mycelial pellets were stored frozen at −70°C and thawed as required.
Substrates and inhibitors.
Authentic 13-dihydrocarminomycin and rhodomycin D were obtained from the National Cancer Institute, Bethesda, Md. Authentic daunorubicin and doxorubicin were obtained from Calbiochem (La Jolla, Calif.) and Sigma Chemical Co. (St. Louis, Mo.), respectively. Authentic 13-dihydrocarminomycin and carminomycin were donated by Rhône-Poulenc, and 13-dihydrodaunorubicin was a gift from Adria Labs (Dublin, Ohio; now Pharmacia-Upjohn). 13-Deoxydaunorubicin was biosynthesized from rhodomycin D in vitro with recombinant S. lividans(pANT144), containing recombinant Streptomyces sp. strain C5 dauK (encoding anthracycline 4-O-methyltransferase [9, 11]) and dauP (encoding rhodomycin D methylesterase [9, 11]), as described by Dickens et al. (9). 13-Deoxydaunorubicin was then purified by methods previously described and quantified against standards previously generated in our laboratory (9).
The mammalian cytochrome P-450 inhibitors 4-methylpyrazole, quinidine, troleandomycin, and sulfaphenazole were obtained from Sigma. All other chemicals were reagent grade or better.
DoxA monooxygenase assay.
DoxA activity was assayed by the method described previously by Dickens et al. (9). Each assay mixture contained, in a final volume of 500 μl, the following components: cell extract from recombinant S. lividans TK24(pANT195) containing Streptomyces sp. strain C5 DoxA (ca. 0.165 to 3 mg of protein); NADP+ (Sigma Chemical Co.), 0.5 μmol; NADPH (Sigma), 0.5 μmol; glucose-6-phosphate (Sigma), 5 μmol; glucose-6-phosphate dehydrogenase (Sigma), 0.42 U; spinach ferredoxin (Sigma), 22 μg; spinach ferredoxin: NADP+ reductase (Sigma), 0.05 U; 10 mM sodium phosphate buffer (pH 7.5); and anthracycline substrate, 0.25 to 5 μl of stock solutions of various concentrations, ranging from 70 μM to 1.9 mM, dissolved in nanopure water. The NADP+, glucose-6-phosphate, and glucose-6-phosphate dehydrogenase constituted an NADPH-regenerating system. The assay mixtures were incubated at 30°C in 16-mm wells of a cell culture plate (Corning Costar, Cambridge, Mass.), with rotary shaking at 250 rpm for periods ranging from 15 s to 1 h. The enzymatic reactions were stopped by the addition of 125 μl of 100 mM Tris-HCl buffer (pH 10.0), and the reaction products were extracted three times each with 500 μl of chloroform-methanol (9:1). The organic extracts were combined, filtered through 0.2-μm-pore-size nylon Acrodisc 13 filters (Gelman Sciences, Ann Arbor, Mich.), and air dried. The reaction products were resuspended in 10 μl of methanol and analyzed qualitatively and quantitatively by thin-layer chromatography and high-pressure liquid chromatography (HPLC) analysis, respectively, as described in detail previously (9).
Spectral analysis, protein concentration determination, and cytochrome P-450 assay.
Visible and UV absorption spectra of protein samples were obtained with a UV 160-U spectrophotometer (Shimadzu Instruments, Kyoto, Japan). All samples were analyzed in 10 mM phosphate buffer (pH 7.5) containing 20% (vol/vol) glycerol, except that samples for the determination of protein concentration were analyzed in nanopure water. Protein content was quantified by the dye-binding assay developed by Bradford (3). The cytochrome P-450 content of protein samples was assayed spectrophotometrically as described previously by Dickens and Strohl (10) and quantified by the method described by Omura and Sato (21). Reference and experimental samples to be quantified for P-450 content were reduced with a few grains of sodium dithionite, followed by bubbling with CO for 1 min prior to analysis. Spectra were obtained with a Shimadzu UV 160-U spectrophotometer scanning between 400 and 600 nm, and reduced-plus-CO minus reduced difference spectra were obtained by digital subtraction. The difference spectra revealed a strong peak at 450 nm, which was absent in control extracts of S. lividans TK24(pANT849) (recombinant strain containing vector alone [8–10]). An extinction coefficient of 91 cm−1 M−1 was employed for the quantitation of the cytochrome P-450 heme in protein solutions, using the absorbance difference between 450 and 490 nm (21).
Protein purification.
All enzyme purification procedures were carried out at 4°C or performed on ice when applicable. In preliminary experiments, we determined that DoxA represented the only protein in S. lividans(pANT195) possessing a reduced-plus-CO minus reduced spectral peak at 450 nm (9, 10), so purification of DoxA was routinely followed by spectral analysis rather than by HPLC analysis of activity. DoxA activity (as measured by oxidation of 13-dihydrodaunorubicin to daunorubicin) was confirmed at each purification step in fractions containing dithionite-reduced-plus-CO minus reduced maximum absorbance at 450 nm.
Frozen mycelium (150 to 200 g [wet weight]) of S. lividans TK24(pANT195) was thawed and resuspended in 10 mM sodium phosphate buffer (pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride, washed through a centrifugation step at 16,000 × g for 45 min, and resuspended in 10 mM sodium phosphate buffer (pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride and 10% (vol/vol) glycerol (buffer A). The mycelial suspension (approximately 400 ml) was disrupted by using a French pressure cell (American Instrument Co., Urbana, Ill.) at 16,000 lb/in2. The crude mycelial extracts were clarified of unbroken mycelia and insoluble material by centrifugation at 16,000 × g for 45 min, and the proteins in the clarified supernatant were fractionated by precipitation with ammonium sulfate. The 30 to 60% (wt/vol) saturation fraction, found to contain most of the DoxA, was pelleted by centrifugation at 16,000 × g (45 min), resuspended in a minimum amount of 10 mM sodium phosphate buffer (pH 7.5) containing 20% glycerol (vol/vol) (buffer B), and dialyzed overnight against 4 liters of the same buffer. The dialyzed protein solution was loaded onto a Q-Sepharose Fast Flow (Pharmacia, Piscataway, N.J.) column (33 by 2.5 cm) at a flow rate of 2 ml · min−1 by using an EP-1 Econo pump (Bio-Rad Laboratories, Hercules, Calif.) with buffer B and eluted with a linear gradient of 1.0 to 0 M NaCl. Fractions containing the peak P-450 content were collected, pooled, concentrated by precipitation with 80% (wt/vol) ammonium sulfate, and dialyzed as described above against buffer B. The dialyzed protein solution was then loaded onto three 5-ml Econo-Pac Hi-S (Bio-Rad) columns assembled in series and eluted with buffer B at a flow rate of 1 ml · min−1. Fractions containing DoxA were collected, pooled, and concentrated by centrifugation for 1 h at 3,000 × g through Centricon-10 concentrators (Amicon, Beverly, Mass.). The concentrated protein solution was dialyzed overnight against 2 liters of 100 mM sodium phosphate buffer (pH 7.5) containing 20% (vol/vol) glycerol (buffer C), and the dialyzed protein was loaded onto a phenyl Sepharose CL-4B (Pharmacia) column (9 by 2.5 cm) at room temperature to prevent precipitation of the salts. The column was washed with 2 column volumes of 100 mM sodium phosphate buffer (pH 7.5) containing 1.0 M NaCl and 20% glycerol (vol/vol) (buffer D), followed by a 60-ml linear, negative gradient of 1.0 to 0 M NaCl (all run at 1 ml · min−1). DoxA was then eluted from the column by a step gradient down from buffer C to 10 mM sodium phosphate buffer (pH 7.5) containing 20% glycerol (buffer B), concentrated to 2 ml with Centricon-10 concentrators as described above, and dialyzed overnight against buffer B. Approximately 400 μl of the dialyzed protein solution was added to a Superose 6 HR 10/30 fast protein liquid chromatography (FPLC) column (Pharmacia) and fractionated with buffer B at a flow rate of 0.1 ml · min−1; 200-μl fractions were collected. The peak 450-nm fraction contained pure DoxA, as revealed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and activity assays. SDS-PAGE was performed as described by Laemmli (16), and proteins in the gels were detected by silver staining as described by Morrissey (19).
Protein characterization.
The approximate molecular weight of denatured DoxA was determined by SDS-PAGE by comparison to protein standards of known molecular weight (Gibco BRL, Gaithersburg, Md.), using a Mini-Protean II gel apparatus (Bio-Rad) running a constant current of 15 mA per gel. The apparent molecular weight of recombinant, native DoxA was determined by gel filtration chromatography in buffer B with a Superose 6 HR 10/30 column connected to a GP-250 Programmer Plus FPLC system (Pharmacia, Uppsala, Sweden) calibrated with protein molecular weight standards (100 μl each; 2 mg · ml−1) obtained from Boehringer Mannheim (Indianapolis, Ind.). The ratio of the elution volume of peak recombinant DoxA to the void volume of the Superose 6 column was used to interpolate the Mr of recombinant DoxA from the linear plot of the logarithm of the native Mr as a function of the ratio of elution volume to void volume.
Enzyme kinetics.
For enzyme kinetics determinations, DoxA monooxygenase activity was assayed as described above, except that the reactions were reduced in volume to 25 or 50 μl, depending on the substrate used, and carried out in capped 1.5-ml microcentrifuge tubes to avoid evaporation. All components in the kinetic assay mixture were scaled down directly from the 500-μl-volume assay mixture. The reactions were run for periods of 15 s to 1 h and stopped by the addition of 125 μl of 100 mM Tris-HCl buffer (pH 10.0), the reaction products were extracted three times each with 125 μl of chloroform-methanol (9:1), and the organic extracts were combined and filtered as described above. The organic phase was collected and filtered, dried in a Speed-Vac ISS-100 vacuum evaporator (Savant, Farmingdale, N.Y.), and then resuspended in 10 μl of methanol for quantitative analysis via HPLC. DoxA activity was quantified by peak integration of substrate and product as described previously (9).
The assays were optimized with respect to pH and temperature, and the activity of DoxA for each anthracycline substrate was tested for linearity, both with respect to time and protein concentration, by the HPLC quantitation assay. After optimal pH and temperature values were determined and linear ranges were found, a range-finding experiment was run to determine approximate Km values for each substrate. For final determinations, four or five substrate concentrations were used that spanned the concentration range of 0.5 to 10 times the Km. Kinetic data were analyzed, and the kinetic constants Km and Vmax, as well as all statistical analyses, were obtained and calculated by using nonlinear least-squares regression algorithms from Microsoft Excel 7.0. The kinetic constants kcat and kcat/Km were derived. For each concentration of anthracycline substrate tested, four identical assays were run, with each reaction stopped at the respective time points noted above. The errors of the kinetic rates at each substrate concentration were calculated from these data.
Inhibition of DoxA activity.
Known mammalian cytochrome P-450 inhibitors 4-methylpyrazole, quinidine, troleandomycin, and sulfaphenazole (all from Sigma) were prepared by procedures described by Shafiee et al. (23). Additionally, the effects on DoxA activity of 1 and 5 mM rhodomycin D (non-DoxA-metabolized precursor) and doxorubicin (product) were tested. Both troleandomycin and sulfaphenazole were first dissolved in acetone before being added to an aqueous solution. The acetone was removed by bubbling nitrogen gas through the solution until the necessary volume of each stock solution was reached. The inhibition assays were run at 50-μl volumes by using the DoxA kinetic monooxygenase assay for 13-dihydrodaunorubicin oxidation to daunorubicin. Inhibition reactions contained 10 mM 13-dihydrodaunorubicin and 1 or 5 mM inhibitor and were terminated as described above after 15 min. All inhibition reactions were performed in duplicate.
For determination of the kinetics of inhibition of DoxA activity by doxorubicin, the kinetic reactions were run as described above with 13-dihydrodaunorubicin as the substrate. Four concentrations of doxorubicin and four concentrations of substrate were tested in duplicate, and the Ki for doxorubicin inhibition of 13-dihydrodaunorubicin oxidation activity by DoxA was calculated.
RESULTS AND DISCUSSION
Purification of DoxA.
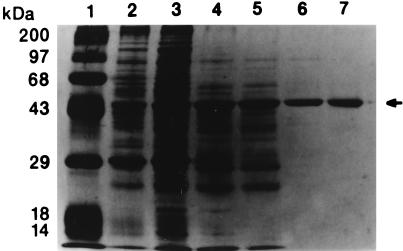
The Streptomyces sp. strain C5 doxA gene was overexpressed in S. lividans TK24 under the control of the SnpR-activated snpA promoter in pANT195 (9, 10), resulting in an observable overproduction of the protein in crude extracts of the recombinant strain (Fig. 2).
FIG. 2.
Denaturing SDS-PAGE gel showing purified DoxA. Lane 1, protein molecular size standards (in kilodaltons) as follows: lysozyme, 14; β-lactoglobulin, 18; carbonic anhydrase, 29; ovalbumin, 43; bovine serum albumin, 68; phosphorylase B, 97; and myosin (heavy chain), 200. Lane 2, S. lividans TK24(pANT195) crude extract. Lane 3, the 30 to 60% ammonium sulfate precipitation concentrate. Lane 4, pooled Q-Sepharose Fast Flow (anion exchange) fractions. Lane 5, pooled Hi-S (cation exchange) fractions. Lane 6, pooled phenyl Sepharose CL-4B (hydrophobic interaction) fractions. Lane 7, the fraction containing purified DoxA from Superose 6 (HPLC gel filtration). Purified DoxA was obtained after Superose 6 gel filtration (denoted by the arrow). The approximate Mr as determined from the SDS-PAGE gel is 47,000.
DoxA was produced as a soluble protein in S. lividans TK24; i.e., all of the activity was present in the supernatant after centrifugation at 100,000 × g for 90 min (data not shown). For simplification, however, this centrifugation step was omitted for subsequent DoxA purifications.
Table 1 shows a representative scheme for purification of DoxA (out of five purifications completed). Nearly all of the DoxA activity was recovered in the 30 to 60% ammonium sulfate fraction, resulting in a twofold purification based on 13-dihydrodaunorubicin 13-oxidation activity. DoxA was eluted from a Q-Sepharose Fast Flow column at 0.25 to 0.3 M NaCl, resulting in an additional 2.2-fold purification. DoxA activity was subsequently eluted in the void volume from the Hi-S cation-exchange column (a negative purification step), resulting in an additional 1.5-fold purification. The enzyme then was bound to the hydrophobic-interaction matrix phenyl Sepharose CL-4B in the presence of high ionic strength buffer (100 mM sodium phosphate [pH 7.5]). After a linear negative gradient of 1.0 to 0 M NaCl, DoxA was still bound to the column. Only after a stepdown from 100 to 10 mM phosphate buffer (pH 7.5) was the enzyme eluted, resulting in a ca.-19-fold purification step. The tight binding of DoxA to this hydrophobic matrix indicates that DoxA is a highly hydrophobic enzyme, even though it was shown not to be membrane bound after high-speed centrifugation. After a final pass through Superose 6, recombinant Streptomyces sp. strain C5 DoxA was purified to electrophoretic homogeneity, as determined by the presence of a single protein band on the silver-stained SDS-PAGE gel (Fig. 2). The overall increase in specific activity was 140-fold over that in crude extracts, with an overall yield of active DoxA of 2.8% (based on heme content).
TABLE 1.
Purification of recombinant Streptomyces sp. strain C5 from S. lividans TK24(pANT195)
| Fraction | Total protein (mg) | Specific content of P-450 (nmol/mg) | C-13 hydroxylation activity (pmol/min/mg) | Ratio of 13-hydroxylation activity per nmol of heme | Recovery (%)a | Purification (fold)b |
|---|---|---|---|---|---|---|
| Crude extract | 4,670 | 0.07 | 4.4 | 61 | 100 | 1 |
| Ammonium sulfate | 2,850 | 0.09 | 8.6 | 94 | 77 | 2.0 |
| Q-Sepharose Fast Flow | 906 | 0.23 | 19.3 | 84 | 62 | 4.5 |
| Econo-Pac High S | 720 | 0.29 | 26.1 | 90 | 63 | 6.1 |
| Phenyl Sepharose CL-4B | 60 | 1.33 | 490 | 368 | 24 | 114 |
| Superose 6 (FPLC) | 7.2 | 1.28 | 603 | 471 | 2.8 | 140 |
Based on P-450 heme content (i.e., active enzyme).
Based on the specific activity of 13-oxidation of 13-dihydrodaunorubicin to daunorubicin.
In the purification scheme shown in Table 1, the ratio of specific 13-dihydrodaunorubicin 13-oxidation activity per specific P-450 heme content is relatively constant at 61/1 to 94/1 throughout the first four steps. In the penultimate and final steps, however, this ratio increases to 368/1 and 471/1, respectively, indicating that a significant portion (approximately 75 to 80%) of the enzyme is not in the 450 nm-absorbing form; i.e., it is inactive. This loss of activity occurred after exposure of the enzyme to 100 mM phosphate buffer (pH 7.5), which we observed to be deleterious to the enzyme. In this purification scheme, approximately 18% of the purified DoxA was calculated to be in the active form.
Stability of purified DoxA.
Oxidation of 13-dihydrodaunorubicin by DoxA was found to occur optimally at pH 7.5 and at a temperature of 30°C (data not shown). This optimal pH, slightly higher than normal physiological conditions, may be necessary to ensure that the proper ionic nature of the zwitterionic anthracycline substrates is maintained for optimal recognition. A similar hypothesis was offered by Connors (4) for explaining this same pH requirement for optimal anthracycline-4-O-methyltransferase (DauK) activity. The presence of 20% (vol/vol) glycerol was shown to be absolutely required for DoxA activity and stability. Purified DoxA showed no appreciable loss of activity even after 7 days at 4°C in the presence of 20% (vol/vol) glycerol (data not shown). Moreover, at room temperature, no discernible loss of activity was observed after 1 h in the presence of 20% glycerol, whereas all activity was lost after 30 min at room temperature in the absence of glycerol (data not shown).
Phenylmethylsulfonyl fluoride (serine protease inhibitor) had no effect on DoxA activity. Dithiothreitol, on the other hand, was strongly inhibitory. Dithiothreitol inhibition was reversible after desalting. High ionic strength buffers, such as buffer C, which contains 100 mM sodium phosphate, also strongly inhibited DoxA activity, but this effect was partially reversible after exchange into low ionic strength buffers, such as 20 mM N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES) or buffer B (10 mM sodium phosphate [pH 7.5]).
Determination of the apparent Mr of recombinant DoxA.
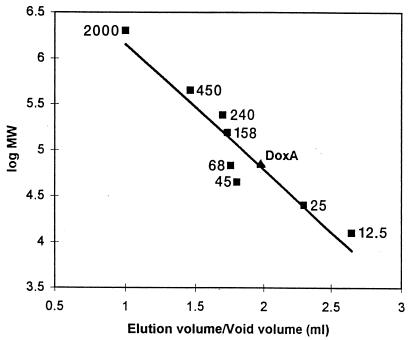
DoxA eluted from a calibrated Superose 6 HR 10/30 FPLC gel filtration at a voided volume/elution volume ratio of 1.79, corresponding to a native protein with an Mr of approximately 49,000 (Fig. 3). Denatured DoxA analyzed by SDS-PAGE was determined to have an apparent Mr of approximately 47,000, indicating that the protein exists in a monomeric form. These results compare favorably with the predicted native Mr of 46,096, obtained by gene sequence analysis (10). Almost all other known streptomycete cytochrome P-450s are also monomers of relatively similar size (20), including P-450Soy from Streptomyces griseus (Mr, ca. 42,000) (25) and P-450ChoP from Streptomyces sp. strain SA-COO (Mr, ca. 47,500) (13).
FIG. 3.
Mr determination of purified, native recombinant DoxA by Superose 6 gel filtration chromatography. The apparent Mr of pure DoxA is 49,000, represented by the triangle. Protein molecular size standards, represented by squares, are (in kilodaltons) as follows: cytochrome c, 12.5; chymotrypsinogen A, 25; albumin (hen egg), 45; bovine serum albumin, 68; aldolase (rabbit muscle), 158; catalase (beef liver), 240; ferritin, 450; and blue dextran, 2,000.
Kinetics of recombinant Streptomyces sp. strain C5 DoxA.
Cytochrome P-450 enzymes require electron transport from NADPH via ferredoxin and ferredoxin:NADP+ oxidoreductase (or a similar system). The genes encoding native Streptomyces strain sp. C5 ferredoxin and ferredoxin:NADP+ reductase have not yet been found (9, 10, 18, 24) and therefore could not be used for the assay of DoxA. Instead, an electron-donating system of spinach ferredoxin and spinach ferredoxin:NADP+ reductase, which has been shown to function adequately for other bacterial cytochrome P-450 activities (1, 23), was used. To ensure that electron shuttling efficiency was not limiting DoxA activity, a relatively high concentration (1 mM) of NADPH was present in all assays and an efficient NADPH regenerating system was used.
Streptomyces sp. strain C5 DoxA oxidizes 13-deoxycarminomycin and 13-deoxydaunorubicin to their respective 13-dihydro forms, with subsequent oxidation of the 13-dihydro moieties to carminomycin and daunorubicin, respectively (9, 24) (Fig. 1). Thus, DoxA exhibits a relatively broad substrate specificity for anthracyclines lacking the 10-carbomethoxyl moiety. This generalized broad substrate specificity exhibited by DoxA is not uncommon for secondary metabolic enzymes (4–6, 9, 24). Anthracyclines possessing functionality at C-10 (e.g., rhodomycin D, 10-carboxy-13-deoxycarminomycin, aklavin, and aclacinomycin A) are not substrates for DoxA, probably due to steric inhibition at the functional end of the molecule (9, 24). Additionally, anthracyclinones (aglycones) are not substrates for DoxA (data not shown), likely indicating a requirement for the sugar moiety (9, 24).
The kinetic constants obtained for DoxA on four anthracycline substrates are shown in Table 2. The Km values for all four substrates are relatively similar, ranging from 0.9 μM for daunorubicin to 4.6 μM for 13-dihydrodaunorubicin. These data suggest that DoxA binds its substrates with relatively similar affinities, irrespective of whether the 4-hydroxyl group is methylated or of the oxidation state at C-13 (methylene, hydroxyl, or keto). 14-Hydroxylation of carminomycin by purified DoxA was not observed, however, even after a 24-h incubation with a high concentration of enzyme. This suggests a stringent specificity for the methoxyl moiety at C-4, perhaps required for proper positioning of the anthracycline side chain, for 14-hydroxylase activity, as we previously suggested (9).
TABLE 2.
Kinetic constants for DoxA substratesa
| DoxA substrateb | Vmax (pmol · min−1 · mg−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 · s−1) |
|---|---|---|---|---|
| DOD | 1,600 ± 100 | 1.1 ± 0.1 | 2.4 × 10−2 ± 2 × 10−3 | 22,000 ± 1,000 |
| DHD | 3,900 ± 100 | 4.6 ± 1.0 | 6.3 × 10−1 ± 1 × 10−2 | 14,000 ± 2,000 |
| DAU | 7.5 ± 0.1 | 0.9 ± 0.1 | 1.2 × 10−4 ± 1 × 10−5 | 130 ± 10 |
| DHC | 19 ± 0.7 | 2.5 ± 0.9 | 2.6 × 10−4 ± 1 × 10−4 | 280 ± 10 |
Data represent kinetics based on total DoxA present in the assays. Our estimate, based on P-450 heme quantitation, is that approximately 20% of the enzyme is in the active form. Data presented represent the means of four data points ±1 standard deviation.
Abbreviations: DOD, 13-deoxydaunorubicin; DHD, 13-dihydrodaunorubicin; DAU, daunorubicin, DHC, 13-dihydrocarminomycin.
The Vmax values for DoxA activity ranged from 7.5 pmol · min−1 · mg−1 for daunorubicin to 3.9 nmol · min−1 · mg−1 for 13-dihydrodaunorubicin, a 520-fold difference. The kcat values were highly substrate dependent. The most rapid turnover was observed when 13-dihydrodaunorubicin was the substrate (6.3 × 10−1 s−1), whereas turnover with daunorubicin as substrate was extremely slow (1.2 × 10−4 s−1). Moreover, the kcat/Km values indicate that 13-deoxydaunorubicin (22,000 M−1 · s−1) and 13-dihydrodaunorubicin (14,000 M−1 · s−1) are by far the preferred substrates, whereas daunorubicin (130 M−1 · s−1) and 13-dihydrocarminomycin (280 M−1 · s−1) are far less preferred. All of these kinetic data reflect activity relative to the total purified DoxA in the assays; nevertheless, the data are affected by the percentage of DoxA in the active (i.e., P-450) form. Based on the ratio of nanomoles of P-450 heme/nanomoles of purified DoxA, we estimate that the DoxA used for the kinetic data was approximately 20% in the P-450 (i.e., active) form.
In the case of mycinamycin biosynthesis, in which two adjacent carbons are oxidized by the same P-450-like enzyme, both reactions appear to be important steps in the biosynthetic pathway in vivo (15). The data presented herein, on the other hand, strongly suggest that 14-hydroxylation of daunorubicin by DoxA is, under wild-type conditions, physiologically irrelevant to the producing organisms. The primary anthracycline biosynthesis products in most daunorubicin-producing strains are actually baumycins, higher glycoside derivatives of daunorubicin (24). In fact, cultures of Streptomyces sp. strain C5 blocked mutants to which radiolabeled daunorubicin was exogenously added converted over 90% of the daunorubicin to baumycins, with no measurable doxorubicin formed (9, 24). Only when doxA was strongly overexpressed in Streptomyces sp. strain C5 did we observe even 10% of exogenously added daunorubicin being converted to doxorubicin (9, 24). From those studies, we concluded that under normal conditions, 14-hydroxylation of daunorubicin could not favorably compete with baumycin biosynthesis from daunorubicin (9, 24), which is supported by the current pure enzyme data. A caveat to this conclusion, however, is the finding by Lomovskaya et al. (18) that DnrV (originally identified as OrfA in Streptomyces sp. strain C5 [10]; the Streptomyces sp. C5 version has recently been renamed DauV [24]), encoded by the gene directly upstream of DoxA, has a dramatic effect on DoxA activity. This may, in the producing organisms, alter significantly the physiological conditions of these reactions. On the other hand, we did not observe that the presence of dauV had any stimulatory effect on DoxA activity in S. lividans TK24 (10). This suggests that additional factors present in daunorubicin-producing strains, but absent from S. lividans TK24, may be required to exacerbate the effect of DnrV (or DauV of Streptomyces sp. C5) on DoxA.
Dickens and Strohl (10) reported that daunorubicin can be reduced back to 13-dihydrodaunorubicin by a putative ketoreductase of low substrate specificity in S. lividans TK24, the heterologous host used in that study. Similarly, Lomovskaya et al. (18) have shown that S. peucetius DnrU (the corresponding gene product in Streptomyces sp. strain C5, originally labeled Orf1 [10], has recently been renamed DauU [24]) possesses 13-ketoreductase activity. Thus, the high Vmax and kcat values for 13-dihydrodaunorubicin likely reveal that DoxA in Streptomyces sp. strain C5 is necessary to compensate for the 13-keto-reductase activity in situ.
Where tested, the catalytic efficiency was dramatically affected by the presence of the methoxy moiety at C-4. The maximal rate of 13-dihydrodaunorubicin (4-methoxy) catalysis is approximately 100-fold higher than that obtained with its 4-hydroxy derivative, 13-dihydrocarminomycin; turnover of the 4-methoxy substrate was 2,423-fold faster than that obtained with the 4-hydroxy analog. Thus, while DoxA binds these two molecules with relatively similar affinities (see above), 13-oxidation of the molecule possessing the 4-methoxy substitution is greatly favored. Based on the kinetic trends obtained in this study, it seems reasonable to suggest that 13-deoxycarminomycin catalysis would proceed at a rate similar to that of 13-dihydrocarminomycin and much slower than that of its 4-methoxy analogue, 13-deoxydaunorubicin. Unfortunately, we cannot confirm this due to the inability to synthesize and stabilize large-enough quantities of 13-deoxycarminomycin for this study (data not shown).
The late steps in daunorubicin biosynthesis, including the (known) calculated kcat/Km values for DoxA, are shown in Fig. 1. Since kcat/Km defines substrate specificity, the data indicate that oxidation of the 4-methoxy anthracyclines may occur preferentially within the producing organism. This would suggest that flux through the daunorubicin (i.e., 4-methoxy) series is preferred over flux through the carminomycin (i.e., 4-hydroxy) series. We recently showed that DauK, anthracycline 4-O-methyltransferase (known as DnrK in S. peucetius [24]), can methylate rhodomycin D, 10-carboxy-13-deoxycarminomycin, 13-deoxycarminomycin, 13-dihydrocarminomycin, and carminomycin (4–6, 9). Although comparative kinetics for these methylation reactions are not known, our current data suggest that the most efficient biosynthesis of daunorubicin would require methylation prior to the formation of carminomycin. We previously had hypothesized that 4-O-methylation of carminomycin might be the penultimate step in daunorubicin biosynthesis (4–6), but the current data suggest that this is probably not the case.
Based on these observations, it appears that the primary function of DoxA is to catalyze the enzymatic conversion of 13-deoxydaunorubicin to daunorubicin via 13-dihydrodaunorubicin.
Inhibition of DoxA.
Known inhibitors of mammalian cytochrome P-450 monooxygenases (1, 7), quinidine, sulfaphenazole, and troleandomycin, inhibited DoxA oxidation of 13-dihydrodaunorubicin by 11, 20, and 35%, respectively, at 1 mM; at 5 mM, all inhibited DoxA activity at >95%. 4-Methylpyrazole did not inhibit DoxA activity at either 1 or 5 mM. Mechanistically, quinidine and sulfaphenazole exhibit reversible heme complexation, with sulfaphenazole acting competitively. Troleandomycin, on the other hand, forms metabolic intermediate heme complexes and thus is considered quasi-irreversible (7). Although these inhibitors function by heme complexation, it is unclear why 4-methylpyrazole had no inhibitory effect on DoxA, especially since the molecule is much smaller than either sulfaphenazole or troleandomycin and thus has less steric hindrance to the heme group. According to Ortiz de Montellano (22), inhibitors that bind not only to the heme group but also to lipophilic regions within the protein are inherently more effective. It is likely that both troleandomycin and sulfaphenazole bind to hydrophobic portions of DoxA, assisting in their inhibition capability, which is noticeably lacking in 4-methylpyrazole.
Rhodomycin D, a precursor anthracycline possessing a 10-carbomethoxyl moiety (9, 24), did not inhibit DoxA activity at either 1 or 5 mM. Although we have no supporting substrate-binding data, it is likely that the large 10-carbomethoxy group present at the reactive end of the anthracycline molecule sterically prevents it from accessing the heme group of the P-450, making it neither a substrate nor an inhibitor.
Doxorubicin completely inhibited DoxA activity at both 1 and 5 mM in preliminary trials. Subsequently, doxorubicin inhibition of DoxA activity was determined to be competitive with respect to 13-dihydrodaunorubicin oxidation, with a Ki of 21 μM. This inhibition constant (equivalent to 11.4 mg of doxorubicin/liter) may be physiologically significant, since high concentrations of doxorubicin may accumulate in the proximity of the enzyme. S. peucetius has been shown to possess an active export mechanism for doxorubicin (12), one of at least three separate mechanisms involved in self-resistance (12, 14, 17, 24). Similarly, Streptomyces sp. strain C5 possesses the same resistance genes, although their functionality has not been proven (24). According to our data, active export of doxorubicin would be important not only as a resistance mechanism but also from a metabolic flux standpoint. If doxorubicin were allowed to accumulate, it might interfere with the biosynthesis of daunorubicin, its immediate precursor, from 10-deoxydaunorubicin. We speculate, therefore, that it is important that doxorubicin be exported from the cell in order for the organism to continue to generate more daunorubicin and doxorubicin.
ACKNOWLEDGMENTS
We thank C. R. Hutchinson for allowing us to preview his manuscript prior to publication. We also thank Jill Johnson, Synthetic Chemistry Laboratory, National Cancer Institute, for authentic samples of carminomycin and rhodomycin D and Rhône-Poulenc and Adria (now Pharmacia-Upjohn) for other authentic anthracycline substrates and standards.
This work was supported by the National Science Foundation under grant no. MCB-94-05730. M.L.D. was supported in part by a presidential fellowship from Ohio State University.
REFERENCES
- 1.Anderson J F, Hutchinson C R. Characterization of Saccharopolyspora erythraea cytochrome P-450 genes and enzymes, including 7-deoxyerythronolide B hydroxylase. J Bacteriol. 1992;174:725–735. doi: 10.1128/jb.174.3.725-735.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcamone F, Cassinelli G, Fantini G, Grein A, Orezzi P, Pol C, Spalla C. Adriamycin, 14-hydroxydaunomycin, a new antitumor antibiotic from S. peucetius var. caesius. Biotechnol Bioeng. 1969;11:1101–1110. doi: 10.1002/bit.260110607. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Connors N C. Studies on the biochemistry and physiology of anthracycline biosynthesis by streptomycetes. Ph.D. dissertation. Columbus: Ohio State University; 1991. [Google Scholar]
- 5.Connors N C, Bartel P L, Strohl W R. Biosynthesis of anthracyclines: carminomycin 4-O-methyltransferase, the terminal enzymic step in the formation of daunomycin. J Gen Microbiol. 1990;136:1895–1898. [Google Scholar]
- 6.Connors N C, Strohl W R. Partial purification and properties of carminomycin 4-O-methyltransferase from Streptomyces sp. strain C5. J Gen Microbiol. 1993;139:1353–1362. doi: 10.1099/00221287-139-6-1353. [DOI] [PubMed] [Google Scholar]
- 7.Correia M A. Rat and human liver cytochrome P450s: substrate and inhibitor specificities and functional markers. In: Ortiz de Montellano P R, editor. Cytochrome P450: structure, mechanism, and biochemistry. 2nd ed. New York, N.Y: Plenum Press; 1995. pp. 607–630. [Google Scholar]
- 8.DeSanti, C. L., and W. R. Strohl. Unpublished results.
- 9.Dickens M L, Priestley N D, Strohl W R. In vivo and in vitro bioconversion of ɛ-rhodomycinone glycoside to doxorubicin: functions of DauP, DauK, and DoxA. J Bacteriol. 1997;179:2641–2650. doi: 10.1128/jb.179.8.2641-2650.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickens M L, Strohl W R. Isolation and characterization of a gene from Streptomyces sp. strain C5 that confers the ability to convert daunomycin to doxorubicin on Streptomyces lividans TK24. J Bacteriol. 1996;178:3389–3395. doi: 10.1128/jb.178.11.3389-3395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickens M L, Ye J, Strohl W R. Analysis of clustered genes encoding both early and late steps in daunomycin biosynthesis by Streptomyces sp. strain C5. J Bacteriol. 1995;177:536–543. doi: 10.1128/jb.177.3.536-543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilfoile P G, Hutchinson C R. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci USA. 1991;88:8553–8557. doi: 10.1073/pnas.88.19.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horii M, Ishizaki T, Paik S Y, Manome T, Murooka Y. An operon containing the genes for cholesterol oxidase and a cytochrome P-450-like protein from a Streptomyces sp. J Bacteriol. 1990;172:3644–3653. doi: 10.1128/jb.172.7.3644-3653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchinson C R. Biosynthetic studies on daunorubicin and tetracenomycin C. Chem Rev. 1997;97:2525–2535. doi: 10.1021/cr960022x. [DOI] [PubMed] [Google Scholar]
- 15.Inouye M, Takada T, Muto N, Beppu T, Horinouchi S. Characterization and expression of a P-450-like mycinamycin biosynthesis gene using a novel Micromonospora-Escherichia coli shuttle cosmid vector. Mol Gen Genet. 1994;245:456–464. doi: 10.1007/BF00302258. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lomovskaya N, Hong S-K, Kim S-U, Fonstein L, Furuya K, Hutchinson C R. The Streptomyces peucetius drrC gene encodes a UvrA-like protein involved in daunorubicin resistance and production. J Bacteriol. 1996;178:3238–3245. doi: 10.1128/jb.178.11.3238-3245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomovskaya N, Otten S L, Doi-Katayama Y, Fonstein L, Liu X-C, Takatsu T, Inventi-Solari A, Filippini S, Torti F, Colombo A L, Hutchinson C R. Doxorubicin overproduction in Streptomyces peucetius: cloning and characterization of the dnrU ketoreductase and dnrV genes and the doxA cytochrome P-450 hydroxylase gene. J Bacteriol. 1999;181:312–325. doi: 10.1128/jb.181.1.305-318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrissey J H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 20.Nelson D R. Directory of P450-containing systems: bacterial P450s. 1996. http://www.icgeb.trieste.it/p450/P450Nom_bacteria.html Website http://www.icgeb.trieste.it/p450/P450Nom_bacteria.html. . [Google Scholar]
- 21.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 22.Ortiz de Montellano P R. Inhibition of cytochrome P450 enzymes. In: Ortiz de Montellano P R, editor. Cytochrome P450: structure, mechanism, and biochemistry. 2nd ed. New York, N.Y: Plenum Press; 1995. pp. 305–366. [Google Scholar]
- 23.Shafiee A, Chen T, Cameron P. Microbial demethylation of immunosuppressant FK-506-specific demethylase showing cytochrome P-450 characteristics from Streptomyces rimosus MA187. Appl Environ Microbiol. 1995;61:3544–3548. doi: 10.1128/aem.61.10.3544-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strohl W R, Dickens M L, Rajgarhia V B, Woo A, Priestley N D. Anthracyclines. In: Strohl W R, editor. Biotechnology of industrial antibiotics. 2nd ed. New York, N.Y: Marcel-Dekker, Inc.; 1997. pp. 577–657. [Google Scholar]
- 25.Trower M K, Lenstra R, Omer C, Buchholz S E, Sariaslani F S. Cloning, nucleotide sequence determination and expression of the genes encoding cytochrome P-450soy (soyC) and ferredoxin soy (soyB) from Streptomyces griseus. Mol Microbiol. 1992;6:2125–2134. doi: 10.1111/j.1365-2958.1992.tb01386.x. [DOI] [PubMed] [Google Scholar]