Abstract
The Methanococcus jannaschii gene MJ1392 was cloned, and its protein product was hyperexpressed in Escherichia coli. The resulting protein was purified and shown to catalyze the condensation of pyruvate and acetyl coenzyme A, with the formation of (R)-citramalate. Thus, this gene (cimA) encodes an (R)-citramalate synthase (CimA). This is the first identification of this enzyme, which is likely involved in the biosynthesis of isoleucine.
Citramalate (2-methylmalate) is a biochemical intermediate known to be involved in several aspects of bacterial metabolism, including, among others, the anaerobic metabolism of glutamate via the methylaspartate pathway of Clostridium tetanomorphum (2). In this pathway, glutamate is converted via l-threo-β-methylaspartate [(2S,3R)-3-methylaspartate] to mesaconate [(E)-2-methyl-2-butenedionic acid], which is then hydrated by citramalate hydrolyase (15, 16) to l-(+)-citramalate (S-citramalate). The resulting citramalate is then cleaved by citramalate lyase to pyruvate and acetate (1). Hydration of citraconic acid [(Z)-2-methyl-2-butenedionic acid] to d-(−)-citramalate (R-citramalate) has also been described (13, 18). A similar pathway for the metabolism of itaconate, via itaconyl coenzyme A (CoA) and citramalyl-CoA, to acetyl-CoA and pyruvate has also been reported (5, 6). The formation of either d-(−)- or l-(+)-citramalate by the direct condensation of acetyl-CoA and pyruvate appears never to have been directly observed, but both of these reactions have been proposed as a key step in a number of biosynthetic pathways. Among these is the formation of isoleucine via the pyruvate pathway, which, based on 13C nuclear magnetic resonance labeling studies (7–9, 15), is the major pathway for isoleucine formation in many species of methanogenic archaea. Labeling studies in other organisms have produced similar results (4, 23).
As part of studies aimed at establishing the identity of genes in the methanogens that were possibly involved in the biosynthesis of coenzymes, we cloned and overexpressed the Methanococcus jannaschii gene MJ1392 and identified the reaction catalyzed by the encoded enzyme. We report here that this gene product is (R)-citramalate synthase. This is the first report of the identification of such an enzyme, which we now designate CimA, which is the product of the cimA gene.
Identification, cloning, and high-level expression of the gene product.
The approach that was used to identify this gene was the outcome of other work aimed at identifying the reactions catalyzed by archaeal enzymes with sequences homologous to homocitrate synthase (NifV) (24). The searches for these genes in the genomes of the methanogens M. jannaschii (3) and Methanobacterium thermoautotrophicum ΔH (16) were performed with the blast_to_rpraze method (The Institute for Genomic Research, Rockville, Md.). The genome of M. jannaschii was found to contain three genes homologous to the homocitrate synthase gene (nifV) (MJ0503, MJ1392, and MJ1543), and the M. thermoautotrophicum ΔH genome had three genes homologous to the homocitrate synthase gene (nifV) (MTH1630, MTH723, and MTH1481). The protein products from one or more of these genes were targets for the enzymes involved in the biosynthesis of α-ketosuberate (10) and thus coenzyme B (7-mercaptoheptanoylthreonine phosphate). Since we did not know the reactions catalyzed by the protein products of each of these genes, we cloned each gene to determine the reactions involved.
Plasmid construct AMJGS60, containing the M. jannaschii gene MJ1392, and the PUC18 vector were obtained from The Institute of Genomic Research/American Type Culture Collection microbial genome special collection. The oligonucleotide primers used to direct the PCR formation of a specific gene cartridge had the following sequences: 5′ CATGCATATGATGGTAAGGATATTTGAT 3′ and 5′ GATCGGATCCTTAATTCAATAACATATTGAT 3′.
The high-level expression of the MJ1392 gene product in the Escherichia coli host strain BL21(DE3) was accomplished by constructing a gene cartridge in vitro and cloning this cartridge into the pT7-7 plasmid (20) such that the gene expression is controlled by the T7 phage transcriptional and translational regulatory elements, which in turn are regulated by the lac control elements. The recombinant plasmid was transformed into the host strain BL21(DE3) E. coli cells, and the BL21(DE3) E. coli cells containing the pT7-7 plasmid, with insert, were grown in LB medium supplemented with 100 mg of ampicillin per ml at 30°C to an absorbance at 600 nm of 1.0. Protein production was then induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After the addition of IPTG, the cells were cultured for another 2 h, harvested by centrifugation, and frozen at −20°C until used.
Preparation of cell extract and purification of citramalate synthase.
The activity of the hyperexpressed M. jannaschii enzyme was measured in E. coli cell extracts prepared by sonication of the E. coli cell pellets (100 to 200 mg [wet weight]) in 2 ml of 0.1 M TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid], pH 7.5, buffer. High expression of the desired protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% polyacrylamide) of the SDS-soluble cellular proteins, which showed a band at the expected molecular mass of 50 kDa, in agreement with the known gene sequence. The amino-terminal sequence of MJ1392 was calculated to be MMVRIFDTTL on the basis of the DNA sequence. The sequence of the overexpressed protein band was found to be MLVRIFDTTL. The difference observed in the second amino acid in these sequences would result from a single base pair substitution in the third base of the methionine codon and could be the result of an error in the gene sequence reported. The similarity between these sequences further supports the likelihood that the desired protein was obtained.
SDS-PAGE analysis of the proteins in both the soluble extract and the resulting insoluble pellet showed that most of the desired protein was in the pellet. Attempts to recover active enzyme from the insoluble protein in the pellet by the methods of Sambrook et al. (14) were not successful. The enzyme was thus purified from the soluble fraction. Induction of protein synthesis in the presence of 1 mM 2-mercaptoethanol or by growth and induction protein synthesis at 30°C did not allow greater solubilization of the desired protein. The protein band or enzymatic activity was not present in the E. coli cells that did not contain the gene insert.
Protein purification was initiated by the addition of solid ammonium sulfate (to 45% saturation) to the soluble protein fraction. The sample was then centrifuged at 16,000 × g for 2 min; the resulting supernatant was then brought to 55% ammonium sulfate saturation and centrifuged for 5 min at 16,000 × g. At this point, CimA was located in the pellet, which was then dissolved in buffer containing 20 mM TES buffer (pH 7.0), 5 mM MgCl2, and 150 mM KCl.
The resuspended ammonium sulfate sample was then heated at 60°C for 10 min and centrifuged at 16,000 × g for 4 min, and the supernatant was placed on a Pharmacia Superose 6 column equilibrated in the same buffer at a flow rate of 0.5 ml/min. The elution time of the enzyme corresponded to a molecular mass of 93 to 128 kDa, indicating that its functional conformation is that of a dimer. This has been observed for other enzymes analogous to the nifV gene product (24). Activity assays, discussed below, were used to monitor the purification steps (Table 1).
TABLE 1.
Purification of citramalate synthase
| Purification step | Vol (ml) | Total amt of protein (mg) | Total activity (nmol/min) | Sp act (nmol/min/mg) | Yield (%) | Fold purification |
|---|---|---|---|---|---|---|
| Crude extract | 1 | 7.6 | 818 | 108 | 100 | 1.0 |
| 45–55% ASa precipitate | 1 | 2.3 | 204 | 89 | 25 | 0.8 |
| Superose 6 fractionb | 0.5 | 0.008 | 23 | 2,870 | 2.8 | 26.5 |
Ammonium sulfate saturation.
200 μl heated at 60°C for 10 min before separation.
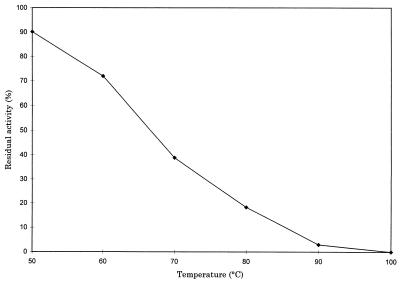
The thermostability of the enzyme was measured by incubating samples (100 to 200 μl) at various temperatures for 10 min. The denatured protein was removed by centrifugation, and the supernatant was assayed for enzymatic activity as discussed below. The enzyme showed moderate stability to elevated temperatures (Fig. 1) but did not exhibit the stability expected when compared with other thermophilic proteins expressed by gene cloning (19). Increasing the ionic strength with potassium chloride did not alter the thermostability. Other factors must be present for this protein to maintain its activity at the growth temperatures of the thermophilic M. jannaschii.
FIG. 1.
Residual enzymatic activity after heat treatment at different temperatures for 10 min.
Enzymatic activity and substrate specificity.
The enzyme activity was assayed by monitoring the production of CoA over time by a procedure similar to that used by Srere (17). Samples to be assayed were brought to a final volume of 100 μl by mixing with TES buffer (0.1 M, pH 7.5), and made 1 mM acetyl-CoA and 1 mM pyruvate by the addition of 0.1 M solutions of these substrates. The resulting solutions were then incubated at 50°C for 1 h. To the resulting incubation mixture were added 50 μl of 10 mM 5,5′-dithio-bis(2-nitrobenzoic acid) in 0.1 Tris-HCl (pH 8.0), 70 μl of 1 M Tris-HCl (pH 8.0), and distilled water to a total volume of 0.9 ml. The absorbance at 412 nm was recorded and blanked against an identical incubation sample without the pyruvate. The micromoles of HS-CoA produced were calculated from a standard curve generated with known concentrations (0 to 110 μM) of 2-mercaptoethanol. The production of CoA was found to be linear over the 1-h time period of the assay, and product formation was a linear function of the amount of enzyme added. The protein concentration of each of the samples was determined by the bicinchoninic acid method (Pierce Scientific Co.), as described by the manufacturer, with bovine serum albumin serving as the standard.
The specific activity of CimA was 2.9 μmol/min/mg of protein, which agrees with that observed for other, similar, enzymes (24). The Kms for pyruvate and for acetyl-CoA have been determined to be 0.85 and 0.14 mM, respectively. The enzyme was specific for acetyl-CoA and pyruvate. Incubation of the enzyme with α-ketoglutarate, α-ketoadipate, α-ketopimelate, α-ketoisovalerate, and acetyl-CoA (each at 2.8 mM) produced no detectable amount of possible condensation products, as assayed by gas chromatography-mass spectrometry (GC-MS) of the methyl ester derivatives of the possible products (10). Likewise, incubation of the enzyme with acetyl-CoA (1 mM) and propionyl-CoA (0.9 mM) failed to produce 2,3-dimethylmalate. The finding that α-ketoisovalerate was not a substrate allows this enzyme to be distinguished from 2-isopropylmalate synthase, which is also able to catalyze the condensation of acetyl-CoA and pyruvate (11) to form an isomer of citramalate, but with a Km 2 orders of magnitude larger than that of α-ketoisovalerate. Thus, on the basis of the substrates used by this enzyme, the earlier putative identification of the MJ1392 gene product as isopropylmalate synthase was incorrect.
Analysis of products.
The citramalate product was confirmed by GC-MS of its dimethyl ester derivative, as previously described (10). The product was found to be (R)-citramalate by GC-MS with a type G-TA Chiraldex column (0.25 mm by 40 m; Advanced Separation Technologies Inc., Whippany, N.J.) programmed from 95°C to 180°C at 3°C per min. On the Chiraldex GC column, dimethyl (S)-citramalate was found to elute before dimethyl (R)-citramalate.
The determination that this enzyme is a citramalate synthase further extends the number of natural products that are known to be derived from acetyl-CoA and α-keto acids.
Acknowledgments
This work was supported by National Science Foundation grant MCB 963086.
REFERENCES
- 1.Barker H A. Citramalate lyase of Clostridium tetanomorphum. Arch Mikrobiol. 1967;59:4–12. doi: 10.1007/BF00406311. [DOI] [PubMed] [Google Scholar]
- 2.Buckel W, Barker H A. Two pathways of glutamate fermentation by anaerobic bacteria. J Bacteriol. 1974;117:1248–1260. doi: 10.1128/jb.117.3.1248-1260.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1997;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 4.Charon N W, Johnson R C, Peterson D. Amino acid biosynthesis in the spirochete Leptospira: evidence for a novel pathway of isoleucine biosynthesis. J Bacteriol. 1974;117:203–211. doi: 10.1128/jb.117.1.203-211.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper R A, Kornberg H L. The utilization of itaconate by Pseudomonas sp. Biochem J. 1964;91:82–91. doi: 10.1042/bj0910082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper R A, Itiaba K, Kornberg H L. The utilization of aconate and itaconate by Micrococcus sp. Biochem J. 1965;94:25–31. doi: 10.1042/bj0940025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eikmanns B, Jaenchen R, Thauer R K. Unusual pathway of isoleucine biosynthesis in Methanobacterium thermoautotrophicum. Arch Microbiol. 1983;136:106–110. [Google Scholar]
- 8.Ekiel I, Sprot G D, Patel G B. Acetate and CO2 assimilation by Methanothrix concilii. J Bacteriol. 1985;162:905–908. doi: 10.1128/jb.162.3.905-908.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekiel I, Smith I C P, Sprott G D. Biosynthesis of isoleucine in methanogenic bacteria: a 13C NMR study. Biochemistry. 1984;23:1683–1687. [Google Scholar]
- 10.Howell D M, Harich K, Xu H, White R H. The α-keto acid chain elongation reactions involved in the biosynthesis of coenzyme B (7-mercaptoheptanoylthreonine phosphate) in methanogenic Archaea. Biochemistry. 1998;37:10108–10117. doi: 10.1021/bi980662p. [DOI] [PubMed] [Google Scholar]
- 11.Kohlhaw G B, Leary T R. α-Isopropylmalate synthase (Salmonella typhimurium) Methods Enzymol. 1970;17a:771–777. [Google Scholar]
- 12.Oda Y, Suzuki S, Katsuki H. Physiological roles of two enzymes with fumarase activity in two pseudomonads. Biochem Int. 1987;14:871–878. [PubMed] [Google Scholar]
- 13.Raghavendra Rao M R, Subramanian S S, Rahatekar H I, Paranjape S V. Enzymatic hydration of citraconate to (−)citramalate. Biochem Biophys Res Commun. 1963;12:78–82. doi: 10.1016/0006-291x(63)90417-0. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Schafer S, Paame T, Vilu R, Fuchs G. 13C-NMR study of acetate assimilation in Thermoproteus neutrophilus. Eur J Biochem. 1989;186:695–700. doi: 10.1111/j.1432-1033.1989.tb15262.x. [DOI] [PubMed] [Google Scholar]
- 16.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J, Rice P, Nölling J, Reeve J N. The complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srere P A. Citrate synthase. Methods Enzymol. 1969;13:3–11. [Google Scholar]
- 18.Subramanian S S, Raghavendra Rao M R. Purification and properties of citraconase. J Biol Chem. 1968;243:2367–2372. [PubMed] [Google Scholar]
- 19.Suzuki T, Inoki Y, Yamagishi A, Iwasaki T, Wakagi T, Oshima T. Molecular and phylogenetic characterization of isopropylmalate dehydrogenase of a thermoacidophilic archaeon, Sulfolobus sp. strain 7. J Bacteriol. 1997;179:1174–1179. doi: 10.1128/jb.179.4.1174-1179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka K, Nakamura K, Mikami E. Fermentation of S-citramalate, citrate, mesaconate, and pyruvate by a gram-negative strictly anaerobic non-spore-former, Formivibrio citricus gen. nov., sp. nov. Arch Microbiol. 1991;155:491–495. [Google Scholar]
- 22.Wang C C, Barker H A. Activation of l-citramalate hydrolyase from Clostridium tetanomorphum. J Biol Chem. 1969;244:2527–2538. [PubMed] [Google Scholar]
- 23.Westfall H N, Charon J W, Peterson D E. Multiple pathways for isoleucine biosynthesis in the spirochete Leptospira. J Bacteriol. 1983;154:846–853. doi: 10.1128/jb.154.2.846-853.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng L, White R H, Dean D R. Purification of Azotobacter vinelandii nifV-encoded homocitrate synthase. J Bacteriol. 1997;197:5963–5966. doi: 10.1128/jb.179.18.5963-5966.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]