Visual Abstract
Abstract
Deleterious germ line DDX41 variants confer risk for myeloid neoplasms (MNs) and less frequently for lymphoid malignancies, with autosomal dominant inheritance and an estimated prevalence of 3% among MNs. Germ line DDX41 variants include truncating alleles that comprise about two-thirds of all alleles, missense variants located preferentially within the DEAD-box domain, and deletion variants. The identification of a truncating allele on tumor-based molecular profiling should prompt germ line genetic testing because >95% of such alleles are germ line. Somatic mutation of the wild-type DDX41 allele occurs in about half of MNs with germ line DDX41 alleles, typically in exons encoding the helicase domain and most frequently as R525H. Several aspects of deleterious germ line DDX41 alleles are noteworthy: (1) certain variants are common in particular populations, (2) MNs develop at older ages typical of de novo disease, challenging the paradigm that inherited cancer risk always causes disease in young people, (3) despite equal frequencies of these variants in men and women, men progress to MNs more frequently, suggesting a gender-specific effect on myeloid leukemogenesis, and (4) individuals with deleterious germ line DDX41 variants develop acute severe graft-versus-host disease after allogeneic hematopoietic cell transplantation with wild-type donors more than others unless they receive posttransplant cyclophosphamide, suggesting a proinflammatory milieu that stimulates donor-derived T cells. Biochemical studies and animal models have identified DDX41’s ability to interact with double-stranded DNA and RNA:DNA hybrids with roles in messenger RNA splicing, ribosomal RNAs or small nucleolar RNAs processing, and modulation of innate immunity, disruption of which could promote inflammation and drive tumorigenesis.
Routine use of next-generation sequencing of hematologic malignancies has greatly expanded the representation of hereditary predisposition syndromes among patients with leukemia. Introduced by Associate Editor Mario Cazzola, this Review Series highlights 4 such genetic predisposition syndromes and provides strong support for the need to include germ line genetic testing for patients with myelodysplasia and myeloid leukemia.
Clinical observations
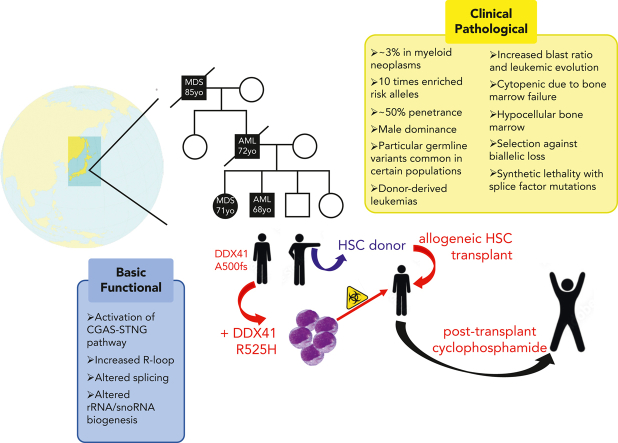
Myeloid neoplasms (MNs) with deleterious germ line DDX41 variants (Table 1) constitute an autosomal dominant familial myelodysplastic syndrome (MDS)/acute myeloid leukemia (AML) disorder as described by the World Health Organization,21 occurring at an estimated prevalence of 3% among all MNs regardless of the family history.1 As such, DDX41 is the most common germ line predisposition gene leading to MNs in adults. Importantly, most cases with this disorder report no family history of hematopoietic malignancy (HM) at diagnosis.1, 2, 3, 4, 5, 6 After the initial descriptions,7,8 germ line and somatic DDX41 mutations have garnered the attention of hematologists throughout the world, identifying both deleterious and benign variants and unique clinical features of the MNs that develop in these individuals.1, 2, 3, 4, 5, 6,9,13,14 Although a complete understanding of the disease risk and prognostication is currently lacking, accumulated genetic data afford informed clinical decisions for these patients and their families.
Table 1.
Description of HMs arising from deleterious germ line DDX41 variants in WHO classification vs recent updates
| Topics | WHO classification | Updates by recent studies |
|---|---|---|
| Prevalence | 1.5% of MNs | High in high-risk MDS/sAML (5%-8%), low in MPN (<1%)1, 2, 3, 4, 5, 6, 7, 8, 9/10 times more enrichment in MNs1 |
| Germ line mutations | Half of all the mutations | 70%-80%, two-thirds are truncating4,10, 11, 12 |
| Age at onset | Long latency (mean 62 yo) | Long latency (mean 66 yo)1, 2, 3, 4, 5, 6, 7, 8, 9,11,13, 14, 15, 16, 17 |
| Male predominance | No description | Yes (1.7-3.1 times), compared with WT cases in any disease phenotype1,2,14 |
| Hematologic findings | Leukopenia/erythroid dysplasia | Hypocellular bone marrow/higher blast count in MDS1 |
| Penetrance | Not fully established | 0% of penetrance at 40 yo/50% lifelong penetrance1 |
| Karyotype | Frequent normal karyotype | Normal karyotype is 2-4 times more frequent, compared with WT cases1,7,13,14 |
| Concomitant mutations | No description | Mutations in CUX1 and GNAS are more frequent compared with WT cases1 Mutations in ASXL1, TET2, DNMT3A, and TP53 are frequent but the frequencies are not significantly different compared with those in WT cases1,2,7,18 |
| Leukemic evolution | No description | Faster leukemic evolution1,19 |
| Prognosis | Poor in cases with somatic deletion | Better in cases with pathogenic variants1,2,4,14,17,20 |
| Therapeutics | Lenalidomide may be effective | Hypomethylating agents may be effective1 |
MPN, myeloproliferative neoplasm; WT, wild-type.
DDX41 variant types
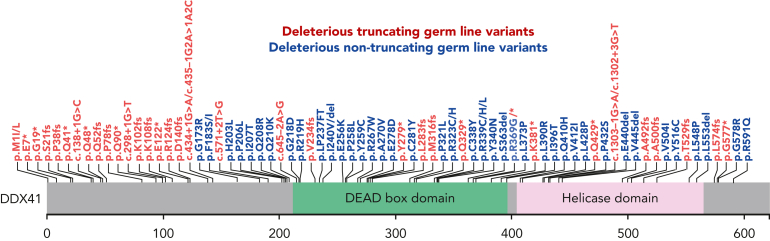
DDX41-mutated MNs define a unique clinical subtype that is distinct from wild-type ones.1 Among the DDX41 mutations identified in thousands of MNs, more than 80% are germ line in nature and about 15% are somatic.1 Deleterious germ line DDX41 variants include truncating alleles, which comprise about two-thirds of such variants, missense variants encoding changes within the DEAD-box domain, and deletions (Figure 1).1,4,10, 11, 12 Interestingly, very rare congenital cases caused by biallelic deleterious germ line variants have been reported.22 When molecular profiling panels for MNs include DDX41, the identification of a DDX41 variant should prompt further assessment. Because >95% of truncating alleles are germ line, individuals with such alleles should be prioritized for germ line genetic testing. Increasingly, germ line missense DDX41 variants are recognized as deleterious,1 so these should also prompt further testing. Importantly, the c.3G>A, p.M1? start-loss allele, one that to date has only been seen as a germ line allele, often gets detected at a variant allele frequency (VAF) of about 30% due to a common polymorphism that results in decreased binding of polymerase chain reaction primers made according to the standard DNA sequence. Therefore, the identification of this allele at VAFs in this range should also prompt proper germ line testing.
Figure 1.
Deleterious DDX41 germ line variants that confer risk to MNs. The DDX41 protein schematic is shown, including the DEAD-box (green) and helicase (pink) domains. Truncating variants are shown in red, and missense variants in blue.
As with other cancer predisposition genes, DDX41 variants within MNs are often biallelic, with somatic mutations being detected in about half of the cases with germ line DDX41 predisposition variants.1 When they occur, missense somatic mutations are mainly found within the helicase domain.1, 2, 3, 4, 5, 6, 7,9,13,14 Among somatic DDX41 mutations, R525H is identified in 80% of MNs, G530D/S in 9%, P321L in 7%, and T227A/M in 3%, with less than 5% being truncating alleles.1 For individuals in whom only somatic DDX41 mutations are identified within MNs, testing for germ line risk alleles should include the capacity to detect large deletions of multiple exons to avoid missing a deleterious germ line DDX41 allele.1
Population distribution of DDX41 variants
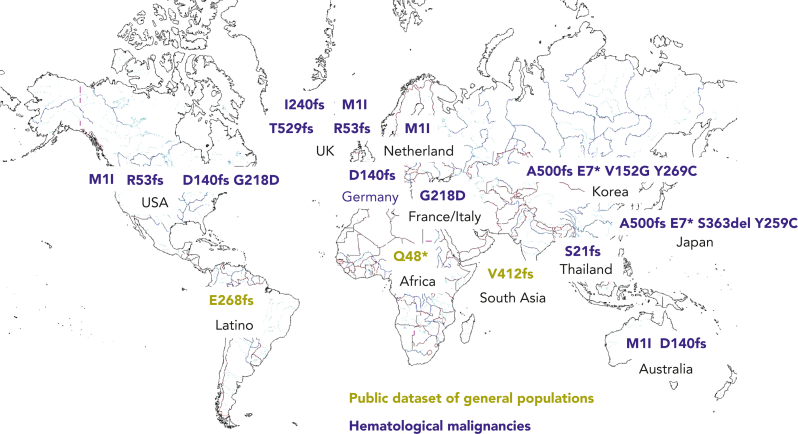
Deleterious germ line DDX41 variants have been reported throughout the world, including Europe, the United States, Australia, the United Kingdom, France, Japan, Korea, and Thailand (Figure 2).2, 3, 4, 5, 6, 7, 8, 9,13,15, 16, 17,20,23 Certain variants are more frequent within particular populations. For example, the c.3G>A, p.M1? and c.415_418dup, p.D140Gfs∗2 alleles are more common in people of Northern European descent, whereas the c.1496dup, p.Ala500Cysfs∗9 allele is more frequent in Japanese and Korean individuals, suggesting that these are founder variants. With the availability of large-scale public sequencing databases of healthy people, several novel rare truncating DDX41 variants can be seen, such as in South America, Africa, and India, which likely represent additional germ line variants more common in those populations (Figure 2, olive font). We look forward to germ line testing within those populations for confirmation of those alleles as germ line variants. Most of the germ line variants are not found in both Asian and European populations, but both Asian and European variants are identified in people living in the United Kingdom and the United States, probably because of immigration from Asian areas. For this reason, enrichment of germ line variants should be calculated from patients with MNs vs healthy controls while controlling for the genetic background. Recently, the enrichment of Asian pathogenic variants was assessed by comparing more than 4000 MNs to more than 20 000 healthy controls within Japanese individuals, yielding an enrichment of deleterious germ line DDX41 variants with an odds ratio (OR) of 10.6 (95% confidence interval: 4.5-22.8).1
Figure 2.
Ethnic diversity of deleterious germ line DDX41 alleles. A world map is displayed with common deleterious germ line DDX41 alleles present within particular geographic areas. Standard variant nomenclature is used. Deleterious germ line variants identified within HMs are shown in blue type, and those identified from public databases are given in olive type.
Age distribution MNs derived from deleterious germ line DDX41 variants
Deleterious germ line DDX41 variants challenge the notion that inheritance of a cancer predisposition allele incites tumorigenesis at a younger age. Typically, germ line DDX41-mutated MNs develop with long latency at an average age of about 68 years old (yo), which is similar to de novo AML.2, 3, 4, 5, 6, 7, 8, 9,11,13, 14, 15, 16, 17 For this reason, the age of onset of MNs should not be a criterion for prioritizing germ line genetic testing.
MN penetrance
Importantly however, the penetrance of MNs derived from deleterious germ line DDX41 variants has not been established.24 Because such information is necessary for appropriate genetic counseling, researchers have used the kin (first-degree relatives) cohort method recently to estimate the penetrance of DDX41-mutated MNs, as was performed for BRCAmut-associated breast cancer. This calculation of disease incidence considers competing risks associated with other causes of death using the Fine-Gray model, which theoretically attributes more precise risks typically in older populations. For deleterious DDX41 germ line variants, the risk of any MN is almost negligible before 40 yo, but rapidly increases to 49% by the age of 90.1
Gender distribution of MNs derived from deleterious germ line DDX41 variants
Several groups have demonstrated that men with deleterious DDX41 variants are more likely to develop MNs than women with such alleles,2,14 despite equal frequencies of pathogenic germ line DDX41 variants in men vs women, suggesting a gender-specific effect on myeloid leukemogenesis. The male gender is more frequent in DDX41-mutated cases than in wild-type ones, with an OR ranging from 1.7 to 3.1. Accordingly, the enrichment of DDX41 pathogenic variants is more remarkable in men (OR, 20.7) than in women (OR, 5.0), although both OR are significant compared with healthy individuals. The penetrance is also (∼4 times) higher in men than in women.1
Disease phenotypes and co-occurring secondary mutations
Within specific types of MNs, deleterious germ line DDX41 variants are found in 3% to 5% in MDS, 5% to 9% in secondary AML, 1% to 2% in MDS/myeloproliferative neoplasms, and <0.5% in myeloproliferative neoplasms. Compared with individuals without deleterious germ line DDX41 variants, patients with MDS and AML are 10 times more enriched for such alleles.1 The most common preceding diagnosis in those who developed secondary AML was MDS.1 Patients with such DDX41 alleles who develop MNs usually present with leukopenia and hypocellular bone marrow with prominent erythroid dysplasia. An initial sequencing study of erythroleukemia revealed that the category of DDX41 mutant cases is a subset with distinct genetic profile,25 but such variants were less frequent in another study of erythroleukemia.26 Among MDS subtypes, refractory anemia with excess blasts is the most frequent. In accordance with the phenotypic features, deleterious germ line DDX41 variants are significant risk factors of leukemic evolution,19 which is confined to truncating alleles, but very rarely observed in the cases with missense variants.1 Despite higher risk phenotypes, the most frequent cytogenetic pattern in the MNs that develop is a normal karyotype, confirmed in phenotype-matched subgroup analysis.1,7,13,14 Most frequent co-occurring somatic mutations within the MNs occur in ASXL1, CUX1, TP53, DNMT3A, and TET2,2,7,9,18 rather than mutations in genes typically associated with secondary AML, such as NRAS, FLT3, and NPM1. Among these, only CUX1 mutations are preferentially observed in DDX41-mutated cases compared to wild-type ones.1 Of note, the clone sizes of co-occurring somatic mutations are smaller than somatic DDX41 mutations, suggesting that their acquisition occurs after the somatic DDX41 mutation.1 Although MNs are the overwhelming majority of HMs diagnosed in these individuals, lymphoid malignancies have been observed, also in the adult age range.2,3,8,16,27 Solid tumors have also been observed in individuals with deleterious germ line DDX41 variants, although the contributions of these variants to tumorigenesis is unclear.
Cellular functions of DDX41
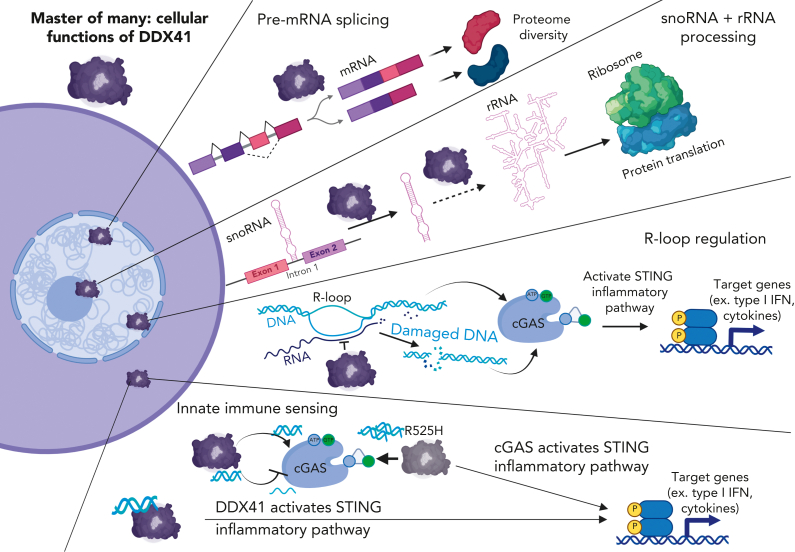
DDX41 is a member of the DEAD-box RNA helicase family, although unlike the name implies, it interacts with double-stranded DNA (dsDNA) and RNA:DNA hybrids.28 Expressed widely across many tissues,29 DDX41 is found in both the cytoplasm and nucleus with distinct and pleiotropic functions in each cellular compartment (Figure 3).30 Our understanding of how DDX41 insufficiency contributes to the development of HMs is still in its infancy.
Figure 3.
Master of many: cellular functions of DDX41. DDX41 localizes to both cytoplasmic and nuclear compartments and plays distinct roles in each location. In the nucleus, DDX41 functions in at least 3 processes. Via its association with the catalytically active spliceosome, DDX41 regulates pre-mRNA splicing. Here, it also modulates snoRNA processing, potentially via promoting excision of snoRNA-containing introns found in ribosomal protein genes. DDX41 can also modify pre-rRNA processing, either indirectly via its control of snoRNA processing or perhaps directly via an unknown mechanism. In the nucleus, DDX41 also interacts with R-loops, which comprise RNA:DNA hybrids and ssDNAs. DDX41 insufficiency leads to R-loop accumulation and subsequent increased dsDNA breaks that trigger a cGAS-STING–mediated type I IFN response. Cytoplasmically localized DDX41 can bind to infection-derived or damaged endogenous dsDNA and then promote STING activation and type I IFN response. In addition, DDX41 can modulate cGAS activity via its dsDNA unwinding capabilities by altering the relative amounts of dsDNA-to-ssDNA. For example, the R525H DDX41 mutant, which has intact dsDNA binding activity but diminished unwinding activity, can overstimulate cGAS leading to a heightened type I IFN response. Created with BioRender.com. DDX41, DEAD-box helicase 41; ssDNA, single-stranded DNA.
DDX41 as a splicing regulator
DDX41 is a metazoan-specific spliceosomal component that is part of the catalytically active complex C spliceosome (Figure 3).7 In line with this connection, spliceosomal components are among the top DDX41-interacting proteins identified in both Caenorhabditis elegans and human cells (Table 2).7,34 Several groups have identified significant alternative splicing changes in DDX41 mutant or knockdown hematopoietic (hematopoietic stem and progenitor cells [HSPCs], erythroid progenitors, and leukemic cells) and nonhematopoietic cell types across multiple organisms (C elegans, zebrafish, mice, and humans) indicating high conservation for DDX41 in splicing regulation.7,31,32,34,37 The functional significance of the specific splicing alterations identified in these studies remains untested. However, 1 study showed patients with deleterious DDX41 variants rarely have somatic mutations in spliceosomal components.7 This negative selection for mutational co-occurrence supports the model that the function of DDX41 in splicing is critical for hematopoiesis. Consistent with the mutual exclusivity observed in patients, genetic interaction studies in C elegans sacy-1 (DDX41 homolog) mutants revealed synthetic lethality with splicing factor mutants.34 Combined, these findings indicate that although the precise function of DDX41 in premessenger RNA (mRNA) processing is unknown, it is essential for cellular survival.
Table 2.
Contrast of phenotypes in animal vs human studies
| Animal studies | Human observations | |
|---|---|---|
| Effect on HSPCs | Hypomorphic zebrafish ddx41 mutants display aberrant HSPC expansion, with deregulation of type I IFN pathway components and targets31 | Patients present with leukopenia, hypocellular bone marrow, and erythroid dysplasia1 |
| Mouse Ddx41 KO HSPC show diminished survival and transplantation capacity32,33 | ||
| Mouse HSPCs with Ddx41 heterozygous KO show competitive advantage33 | ||
| Mice with Ddx41 heterozygous KO show age-dependent hematopoietic defects including anemia, thrombocytopenia, and BM hypocellularity33 | ||
| Animals with heterozygous KO of Ddx41 function do not develop hematopoietic malignancies31, 32, 33 | Patients with deleterious germ line DDX41 variants develop hematopoietic malignancies, mostly of myeloid lineage, with late onset1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,20,22, 23, 24 | |
| DDX41-interacting proteins | Spliceosomal components (C elegans)34 | Spliceosomal components7 |
| R-loop–interacting proteins35,36 | ||
| Alternative splicing changes in DDX41-mutant or knockdown cells | Increased alternative exon usage, intron retention, alternative 5' and 3' splice site usage (C elegans, zebrafish, mice)31,34,37 | Increased exon inclusion and intron retention7 |
| Mutual exclusivity with splicing mutations? | Yes. C elegans sacy-1 mutants show synthetic lethality with germ line splicing factor mutants34 | Yes. Co-occurrence of somatic mutations in genes encoding splicing factors is rare, suggesting that DDX41's splicing function is critical7 |
| Role in ribosome biogenesis (snoRNA and pre-rRNA processing) | Ddx41-deficient HSPCs display increased snoRNA levels33 | Cells overexpressing DDX41 R525H display defects in pre-rRNA processing and had diminished cellular proliferation30 |
| DDX41-interacting RNAs are enriched for snoRNAs, suggesting that DDX41 could remove snoRNA-containing introns via splicing regulation33 | ||
| R-loop interactions | Increased RNA:DNA hybrids result from insufficient Ddx41 levels, which induces a cGAS-STING-mediated inflammatory response that directs HSPC expansion (zebrafish)31 | Increased RNA:DNA hybrids and consequently elevated dsDNA breaks result from insufficient DDX41 levels35 |
| cGAS-STING interactions | Murine fetal liver Ddx41-null HSPCs display diminished inflammatory gene expression32 | Loss of DDX41 in monocytic cells lowered dsDNA-triggered activation of the cGAS-STING pathway. However, expression of DDX41 R525H, with intact dsDNA binding but dampened helicase activity, activates the STING pathway38 |
| Human hematopoietic cells show paradoxical increased type I IFN signaling upon DDX41 knockdown35 |
BM, bone marrow; DDX41, DEAD-box helicase 41; KO, knockout.
DDX41 in ribosomal RNA (rRNA) and small nucleolar RNA (snoRNA) processing
In addition to a role in pre-mRNA splicing, DDX41 affects the processing of RNAs critical for ribosome assembly and function (Figure 3).30,33 Human cells overexpressing a common somatic hypomorphic mutant form of DDX41 (R525H) displayed defects in pre-rRNA processing, which correlated with diminished cellular proliferation.30 Pre-rRNAs are transcribed and processed in the nucleolus, where they are then assembled into ribosomes. Part of their processing and maturation is mediated by snoRNA. Defects in snoRNAs will alter pre-rRNA cleavage and pseudouridylation, which are required for proper ribosome functioning.39 As an evolutionarily conserved coupling mechanism, most snoRNAs are located within the introns of ribosomal protein genes.40 Thus, the first step in snoRNA processing requires proper splicing. A recent study demonstrated that DDX41 could be a previously unrecognized player involved in snoRNA processing.33 Ddx41-deficient murine HSPCs displayed a marked increase in snoRNA levels. Moreover, the identification of DDX41-interacting RNAs revealed a strong enrichment for snoRNAs, suggesting that the effect could be direct, potentially via the removal of snoRNA-containing introns via splicing regulation. Consistent with this function being important for hematopoiesis, knockdown of several snoRNAs in murine HSPCs diminished cell viability and differentiation.33 Currently, it remains unclear if the effect of DDX41 on ribosome biogenesis is via direct regulation to both snoRNAs and pre-rRNAs or due to snoRNA defects that then alter pre-rRNA processing. In addition, the relative contribution of splicing defects in protein coding vs noncoding genes on DDX41-mutant hematopoietic pathology is an unanswered question in the field.
DDX41 in innate immunity
DEAD-box helicases also play critical roles in immunity.41 This is best characterized in myeloid effector cells such as macrophages and dendritic cells that activate type I interferon (IFN) signaling in response to either infection or exposure to DNA-type pathogen-associated molecular patterns.41 DDX41 is one of the pattern recognition receptors that is stimulated by these pathogen-associated molecular patterns, specifically bacterial cyclic dinucleotides as well as viral dsDNA and RNA:DNA hybrids.28,42,43 It is thought that cytoplasmically localized DDX41 initiates the type I IFN signaling cascade, starting with the stimulator of IFN genes (STING, also known as TMEM173, MITA, MPYS, or ERIS), which then activates TANK-binding kinase 1, IFN response factor 3, and nuclear factor κB (Figure 3).42,43
HMs, especially MNs, have a strong connection to inflammatory dysfunction.44 Because of the connection between DDX41 and type I IFN, this pathway has been examined in DDX41-mutant hematopoiesis.31,32,35 Functionally hypomorphic zebrafish ddx41 mutants display aberrant HSPC expansion.31 Gene expression analysis of the ddx41 mutant compared with normal HSPCs revealed deregulation of type I IFN pathway components and targets. Similar results were observed in human hematopoietic cells upon DDX41 knockdown.35 Surprisingly, type I IFN signaling appeared upregulated, not downregulated in these studies. This result is seemingly contradictory with a function of DDX41 as an activator of this pathway, thus another role of DDX41 might be driving this inflammation.
Within the nucleus, DDX41 interacts with R-loops, which are structural DNA variants comprising single-stranded DNAs and RNA:DNA hybrids (Figure 3).35,36 DDX41 is a helicase that is stimulated by RNA:DNA hybrid binding, suggesting it could unwind and destroy these molecules.28 In line with this model, both zebrafish and human cells with insufficient DDX41 levels accumulate RNA:DNA hybrids, which result in elevated dsDNA breaks.31,35 Since the discovery of DDX41 as a DNA pattern recognition receptor, new data indicate the main DNA sensor in most cells is cyclic guanosine monophosphate- adenosine 5′-monophosphate (GMP-AMP) synthase (cGAS).45 cGAS is activated by many infectious and damage-associated stimuli, including dsDNA, RNA:DNA hybrids, micronuclei, and damaged mitochondria, among others.45 Ligand binding to cGAS stimulates its enzymatic activity, leading to the production of the second messenger cyclic GMP-AMP, which can bind to several proteins, including STING, leading to an inflammatory signaling cascade that results ultimately in activation of the type I IFN pathway.45 In the zebrafish ddx41-mutant model, R-loops and/or the ensuing damaged DNA orchestrated a cGAS-STING–mediated inflammatory response that promoted HSPC expansion.31 These data imply that DDX41 regulation of R-loop levels is important in hematopoietic homeostasis.
Other connections between DDX41 and cGAS are emerging. DDX41 binds the bacterial second messengers cyclic-di-guanosine monophosphate and cyclic-di-adenosine monophosphate, which are structurally similar to cGAS-produced cyclic GMP-AMP, suggesting cGAS could modulate DDX41 activity.42 In addition, a recent study uncovered a mechanism for DDX41 modulation of cGAS activity.38 Loss of DDX41 in monocytic cells lowered dsDNA-triggered activation of the cGAS-STING pathway. In contrast, expression of the R525H hypomorphic mutant DDX41, which displays dampened helicase activity with intact dsDNA binding, increased pathway activation (Figure 3). The findings indicate a complex interplay among the nucleic-acid sensing pathways that could have a significant impact on hematopoiesis and immunity.
Interplay of DDX41 functions in hematopoiesis
The examination of how the pleiotropic cellular functions of DDX41 affect hematopoiesis and how they cross talk is in its infancy. The differences among the studies in various hematopoietic and nonhematopoietic cells could be explained by cell-type selective functions and dependencies. In addition, DDX41 isoforms may localize to distinct cellular compartments.8,30 Full-length DDX41 appears largely nuclear, whereas a shorter isoform with a truncated N-terminus is found predominantly in the cytoplasm.30 The relative levels of these 2 isoforms could alter how a particular cell deploys DDX41. Expression of the corresponding transcripts across hematopoietic cell types and during HSPC development is unknown.
Management implications
MN pathogenesis
Deleterious germ line DDX41 variants challenge the accepted paradigm that germ line predisposition variants cause cancer at ages younger than expected because MNs rarely develop in people younger than 40 years and typically in the late 60s.2, 3, 4, 5, 6, 7,9,13,14 This inverted age distribution of MNs suggests a unique pathogenesis. Cascade testing in families has identified many relatives who share the deleterious familial DDX41 allele. These individuals are being closely followed decades before MNs develop, which will allow the natural history and clonal progression of these malignancies to be described. Because we postulate that MNs arise as the sequential acquisition of somatic mutations, beginning with the phenomenon of clonal hematopoiesis (CH), an open question is whether CH precedes the development of MNs in those with deleterious germ line DDX41 alleles. If so, it is possible that the presence of CH represents a molecular clock that marks the initiation of MN development. Future studies will demonstrate whether CH-associated somatic mutations occur within expected genes (eg, DNMT3A, TET2, ASXL1, and others) or within a gene set unique to germ line DDX41-associated MNs. Because MNs occur within the expected age range, the role of the pathogenic germ line DDX41 variant throughout most of an individual’s lifetime and its impact on any CH that might develop remain a mystery. If these questions could be answered by future studies, we might be able to design specific interventions that could be instituted decades before MN development to delay or prevent these cancers.
Multiple recent studies have shown that deleterious germ line DDX41 variants are associated with a better prognosis than de novo MNs.2,4,14,17,20 Initially, however, MNs arising in those with deleterious germ line DDX41 variants were described as having a poor prognosis, likely because cases with somatic deletion of the DDX41 locus, typically caused by the deletion of the telomeric region of chromosome 5q with complex karyotypes and biallelic TP53 mutations, were included in the mutated cohort. Table 1 contrasts initial descriptions of MNs arising in those with deleterious germ line DDX41 variants46 with a more updated understanding of the features of these malignancies.2, 3, 4, 5, 6,9,13,14 A recent detailed study showed that the bone marrow blast percent is higher in DDX41-mutated MDS compared with DDX41–wild-type MDS. In contrast, the bone marrow blast percent is lower in DDX41-mutated secondary AML compared with such cases with wild-type DDX41, suggesting that with deleterious germ line DDX41 variants, bone marrow blasts increase until leukemic evolution (at ∼20%) but tend not to progress rapidly after that point.1
Therefore, the discrepancy between frequent leukemic evolution and better prognosis should be addressed. Another possible explanation of this discrepancy between frequent leukemic evolution and better overall survival is that DDX41-mutated cases respond to specific therapy. Early data suggest that patients may respond to lenalidomide,7 and the most recent study showed that hypomethylating agents might be effective and improve overall survival in DDX41-mutant MN cases (hazard ratios: 0.25-0.60) compared with wild-type DDX41 ones.1 This retrospective study also showed that treatment with hypomethylating agents before hematopoietic stem cell transplantation (HSCT) dramatically improved overall survival in DDX41-mutated cases compared with wild-type cases, which should be validated by prospective clinical trials.
Implications for allogeneic HSCT
For most individuals with germ line predisposition variants, the risk of developing a HM is relieved by allogeneic HSCT using a donor who lacks the deleterious familial allele. However, given the common nature of some germ line DDX41 variants within particular populations, several instances have been described in which allogeneic donors, either related or unrelated, had a deleterious germ line DDX41 variant that was transferred to the recipient during allogeneic HSCT.47, 48, 49, 50 For this reason, we advocate the following: (1) the addition of DDX41 to all molecular profiling panels being used to prognosticate for HMs, because the identification of a truncating DDX41 allele at VAF >30% is likely to be germ line; however, the use of such panels should not replace proper germ line testing, because these panels are generally unable to detect all types of germ line alleles;51,52 and (2) testing of all allogeneic stem cell donors for germ line predisposition variants, including DDX41.
We advocate transplantation for individuals diagnosed with MNs in clinical remission when they have deleterious germ line DDX41 variants (ie, likely pathogenic or pathogenic variants). Like in other germ line cancer predisposition disorders, only deleterious germ line variants are clinically actionable. Germ line variants deemed to be of uncertain significance, likely benign, or benign do not prompt further studies or cascade testing in families unless the variant gets upgraded into likely pathogenic or pathogenic classification with additional functional or segregation data.
Observational studies have shown that after allogeneic HSCT using wild-type donors, individuals with deleterious germ line DDX41 variants suffer more graft-versus-host disease (GVHD) than others.53,54 One study showed that patients with deleterious germ line DDX41 variants had more stage 3 to 4 acute GVHD (38%) than those with deleterious CHEK2 variants (0%), other hereditary HM variants (12%), or patients without such germ line variants (9%) unless they had received posttransplant cyclophosphamide as GVHD prophylaxis.53 We postulate that individuals with deleterious germ line DDX41 variants have a proinflammatory milieu generated from the germ line allele that remains in all of their nontransplanted organs, which stimulates donor-derived T cells and causes severe GVHD. Based on these observations, we advocate consideration of lower intensity transplantation regimens, optimizing donor selection, and use of posttransplant cyclophosphamide as GVHD prophylaxis when individuals with deleterious germ line DDX41 variants undergo allogeneic HSCT, even when using wild-type donors.
Population screening and monitoring
Because truncating and deleterious germ line DDX41 variants can be identified in screening sequencing databases from populations across the world (Figure 2), we recognize that it is possible to do population-based screening for deleterious germ line DDX41 variants. Currently, we advocate that once an individual is identified as having a deleterious germ line DDX41 variant, baseline blood work, including a complete blood cell count and differential, and a bone marrow biopsy be performed to test for the presence of cytopenias and/or bone marrow dysplasia, respectively. It is important to establish the degree of bone marrow dysplasia before MN progression so that an accurate diagnosis of MDS can be made over time for patients. We advise including molecular profiling that includes DDX41 and cytogenetic analysis at the time of bone marrow biopsy because the presence of the DDX41 R525H somatic mutation and/or del(5q) could signal impending or overt MN. The age at which baseline studies are performed and when surveillance monitoring begins should be decided ideally in collaboration with genetic counselors experienced in hereditary hematopoietic malignancies and discussions that consider the family history and the personal preferences of the affected individuals. Once the baseline studies have been performed, we advise repeating a complete blood cell count and differential 3 to 4 times per year with consideration of an annual bone marrow examination. Evidence for progression toward MN includes the identification of somatic mutations and/or karyotypic abnormalities, and some advocate for preemptive allogeneic HSCT if such changes are demonstrated above baseline studies even in the absence of increasing blast burden in the bone marrow. The hope is that future studies will define the molecular progression to MNs for individuals with germ line DDX41 predisposition alleles, allowing us to intercede and slow or prevent MN development without the need for allogeneic HSCT.
Conclusion
Identified in about 3% of MNs, deleterious germ line DDX41 variants constitute the most common germ line predisposition disorder causing myeloid, and less often lymphoid, malignancies in adults. Several unique features of these alleles are important for optimal patient management, as follows: (1) the vast majority of truncating DDX41 alleles are germ line, and others, such as the c.3G>A, p.M1? start-loss allele, have only been seen as germ line alleles; therefore, when such alleles are detected on tumor-based molecular profiling platforms, they should prioritize that individual for proper germ line testing; (2) MNs that develop in people with deleterious germ line DDX41 alleles occur at ages typical of de novo disease, challenging the paradigm that inherited cancer risk always causes disease at young ages; (3) men with these alleles progress to MNs more frequently, suggesting a gender-specific effect on myeloid leukemogenesis; (4) because certain variants are quite common in particular populations, unrelated allogeneic HSCT donors with these alleles have inadvertently been used, resulting in the transfer of a deleterious germ line DDX41 variant into recipients; The presence of these common DDX41 variants may allow donor-based and/or population-based screening in the future; (5) when wild-type donors are used in allogeneic HSCT for those with deleterious germ line DDX41 variants, posttransplant cyclophosphamide appears to be an effective means of posttransplant GVHD prophylaxis that prevents the otherwise severe GVHD that occurs in these individuals. The exact mechanism(s) by which these variants confer risk to MNs is unclear, but biochemical studies and animal models demonstrate that DDX41 interacts with dsDNA and RNA:DNA hybrids with roles in mRNA splicing, rRNAs/snoRNAs processing, and modulation of innate immunity, which could lead to an inflammatory milieu that drives tumorigenesis.
Conflict-of-interest disclosure: L.A.G. receives royalties from UptoDate, Inc for a coauthored article on germ line predisposition to hematopoietic malignancies. The remaining authors declare no competing financial interests.
Acknowledgments
The authors thank their patients and their patients’ families as well as the members of their respective laboratories for their continued dedication to studies of germ line predisposition to hematopoietic malignancies.
Authorship
Contribution: H.M., T.V.B., and L.A.G. wrote and edited the manuscript and its tables and figures.
References
- 1.Makishima H, Saiki R, Nannya Y, et al. Germ line DDX41 mutations define a unique subtype of myeloid neoplasm. Blood. 2023;141(5):534–549. doi: 10.1182/blood.2022018221. [DOI] [PubMed] [Google Scholar]
- 2.Quesada AE, Routbort MJ, DiNardo CD, et al. DDX41 mutations in myeloid neoplasms are associated with male gender, TP53 mutations and high-risk disease. Am J Hematol. 2019;94(7):757–766. doi: 10.1002/ajh.25486. [DOI] [PubMed] [Google Scholar]
- 3.Goyal T, Tu ZJ, Wang Z, Cook JR. Clinical and pathologic spectrum of DDX41-mutated hematolymphoid neoplasms. Am J Clin Pathol. 2021;156(5):829–838. doi: 10.1093/ajcp/aqab027. [DOI] [PubMed] [Google Scholar]
- 4.Wan Z, Han B. Clinical features of DDX41 mutation-related diseases: a systematic review with individual patient data. Ther Adv Hematol. 2021;12 doi: 10.1177/20406207211032433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi EJ, Cho YU, Hur EH, et al. Unique ethnic features of DDX41 mutations in patients with idiopathic cytopenia of undetermined significance, myelodysplastic syndrome, or acute myeloid leukemia. Haematologica. 2022;107(2):510–518. doi: 10.3324/haematol.2020.270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang F, Long N, Anekpuritanang T, et al. Identification and prioritization of myeloid malignancy germline variants in a large cohort of adult patients with AML. Blood. 2022;139(8):1208–1221. doi: 10.1182/blood.2021011354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polprasert C, Schulze I, Sekeres MA, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27(5):658–670. doi: 10.1016/j.ccell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewinsohn M, Brown AL, Weinel LM, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127(8):1017–1023. doi: 10.1182/blood-2015-10-676098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebert M, Passet M, Raimbault A, et al. Germline DDX41 mutations define a significant entity within adult MDS/AML patients. Blood. 2019;134(17):1441–1444. doi: 10.1182/blood.2019000909. [DOI] [PubMed] [Google Scholar]
- 10.Churpek JE, Smith-Simmer K. In: GeneReviews. Adam MP, Ardinger HH, Pagon RA, et al., editors. University of Washington; 2021. DDX41-associated familial myelodysplastic syndrome and acute myeloid leukemia. [Google Scholar]
- 11.Badar T, Chlon T. Germline and somatic defects in DDX41 and its impact on myeloid neoplasms. Curr Hematol Malig Rep. 2022;17(5):113–120. doi: 10.1007/s11899-022-00667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinriki S, Matsui H. Unique role of DDX41, a DEAD-box type RNA helicase, in hematopoiesis and leukemogenesis. Front Oncol. 2022;12:992340. doi: 10.3389/fonc.2022.992340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P, Brown S, Williams M, et al. The genetic landscape of germline DDX41 variants predisposing to myeloid neoplasms. Blood. 2022;140(7):716–755. doi: 10.1182/blood.2021015135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li P, White T, Xie W, et al. AML with germline DDX41 variants is a clinicopathologically distinct entity with an indolent clinical course and favorable outcome. Leukemia. 2022;36(3):664–674. doi: 10.1038/s41375-021-01404-0. [DOI] [PubMed] [Google Scholar]
- 15.Polprasert C, Takeda J, Niparuck P, et al. Novel DDX41 variants in Thai patients with myeloid neoplasms. Int J Hematol. 2020;111(2):241–246. doi: 10.1007/s12185-019-02770-3. [DOI] [PubMed] [Google Scholar]
- 16.Singhal D, Hahn CN, Feurstein S, et al. Targeted gene panels identify a high frequency of pathogenic germline variants in patients diagnosed with a hematological malignancy and at least one other independent cancer. Leukemia. 2021;35(11):3245–3256. doi: 10.1038/s41375-021-01246-w. [DOI] [PubMed] [Google Scholar]
- 17.Alkhateeb HB, Nanaa A, Viswanatha D, et al. Genetic features and clinical outcomes of patients with isolated and comutated DDX41-mutated myeloid neoplasms. Blood Adv. 2022;6(2):528–532. doi: 10.1182/bloodadvances.2021005738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu S, Li B, Qin T, et al. Molecular and clinical features of myeloid neoplasms with somatic DDX41 mutations. Br J Haematol. 2021;192(6):1006–1010. doi: 10.1111/bjh.16668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernard E, Tuechler H, Greenberg PL, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evidence. 2022;1(7) doi: 10.1056/EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
- 20.Duployez N, Largeaud L, Duchmann M, et al. Prognostic impact of DDX41 germline mutations in intensively treated acute myeloid leukemia patients: an ALFA-FILO study. Blood. 2022;140(7):756–768. doi: 10.1182/blood.2021015328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoury JD, Solary E, Abla O, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36(7):1703–1719. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diness BR, Risom L, Frandsen TL, et al. Putative new childhood leukemia cancer predisposition syndrome caused by germline bi-allelic missense mutations in DDX41. Genes Chromosomes Cancer. 2018;57(12):670–674. doi: 10.1002/gcc.22680. [DOI] [PubMed] [Google Scholar]
- 23.Bannon SA, Routbort MJ, Montalban-Bravo G, et al. Next-generation sequencing of DDX41 in myeloid neoplasms leads to increased detection of germline alterations. Front Oncol. 2021;10:582213. doi: 10.3389/fonc.2020.582213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arber DA, Orazi A, Hasserjian RP, et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140(11):1200–1228. doi: 10.1182/blood.2022015850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iacobucci I, Wen J, Meggendorfer M, et al. Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat Genet. 2019;51(4):694–704. doi: 10.1038/s41588-019-0375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeda J, Yoshida K, Nakagawa MM, et al. Amplified EPOR/JAK2 genes define a unique subtype of acute erythroid leukemia. Blood Cancer Discov. 2022;3(5):410–427. doi: 10.1158/2643-3230.BCD-21-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinberg OK, Kuo F, Calvo KR. Germline predisposition to hematolymphoid neoplasia. Am J Clin Pathol. 2019;152(3):258–276. doi: 10.1093/ajcp/aqz067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stavrou S, Aguilera AN, Blouch K, Ross SR. DDX41 recognizes RNA/DNA retroviral reverse transcripts and is critical for in vivo control of murine leukemia virus infection. mBio. 2018;9(3) doi: 10.1128/mBio.00923-18. e00923-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crown Human Genome Center WIoS. GeneCards. https://www.genecards.org/cgi-bin/carddisp.pl?gene=DDX41&keywords=DDX41 Accessed 18 October 2022.
- 30.Kadono M, Kanai A, Nagamachi A, et al. Biological implications of somatic DDX41 p.R525H mutation in acute myeloid leukemia. Exp Hematol. 2016;44(8):745–754. doi: 10.1016/j.exphem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 31.Weinreb JT, Ghazale N, Pradhan K, et al. Excessive R-loops trigger an inflammatory cascade leading to increased HSPC production. Dev Cell. 2021;56(5):627–640. doi: 10.1016/j.devcel.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J, Mahmud N, Bosland MC, Ross SR. DDX41 is needed for pre- and postnatal hematopoietic stem cell differentiation in mice. Stem Cell Rep. 2022;17(4):879–893. doi: 10.1016/j.stemcr.2022.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chlon TM, Stepanchick E, Hershberger CE, et al. Germline DDX41 mutations cause ineffective hematopoiesis and myelodysplasia. Cell Stem Cell. 2021;28(11):1966–1981. doi: 10.1016/j.stem.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukamoto T, Gearhart MD, Kim S, Mekonnen G, Spike CA, Greenstein D. Insights into the involvement of spliceosomal mutations in myelodysplastic disorders from analysis of SACY-1/DDX41 in Caenorhabditis elegans. Genetics. 2020;214(4):869–893. doi: 10.1534/genetics.119.302973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosler T, Conte F, Longo GMC, et al. R-loop proximity proteomics identifies a role of DDX41 in transcription-associated genomic instability. Nat Commun. 2021;12(1):7314. doi: 10.1038/s41467-021-27530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang IX, Grunseich C, Fox J, et al. Human proteins that interact with RNA/DNA hybrids. Genome Res. 2018;28(9):1405–1414. doi: 10.1101/gr.237362.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinreb JT, Gupta V, Sharvit E, Weil R, Bowman TV. Ddx41 inhibition of DNA damage signaling permits erythroid progenitor expansion in zebrafish. Haematologica. 2022;107(3):644–654. doi: 10.3324/haematol.2020.257246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh RS, Vidhyasagar V, Yang S, et al. DDX41 is required for cGAS-STING activation against DNA virus infection. Cell Rep. 2022;39(8):110856. doi: 10.1016/j.celrep.2022.110856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6(2):225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMahon M, Contreras A, Ruggero D. Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip Rev RNA. 2015;6(2):173–189. doi: 10.1002/wrna.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ullah R, Li J, Fang P, Xiao S, Fang L. DEAD/H-box helicases:anti-viral and pro-viral roles during infections. Virus Res. 2022;309:198658. doi: 10.1016/j.virusres.2021.198658. [DOI] [PubMed] [Google Scholar]
- 42.Parvatiyar K, Zhang Z, Teles RM, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13(12):1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12(10):959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barreyro L, Chlon TM, Starczynowski DT. Chronic immune response dysregulation in MDS pathogenesis. Blood. 2018;132(15):1553–1560. doi: 10.1182/blood-2018-03-784116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowman TV, Trompouki E. Sensing stemness. Curr Stem Cell Rep. 2021;7(4):219–228. doi: 10.1007/s40778-021-00201-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi S, Kobayashi A, Osawa Y, et al. Donor cell leukemia arising from preleukemic clones with a novel germline DDX41 mutation after allogenic hematopoietic stem cell transplantation. Leukemia. 2017;31(4):1020–1022. doi: 10.1038/leu.2017.44. [DOI] [PubMed] [Google Scholar]
- 48.Gibson CJ, Kim HT, Zhao L, et al. Donor clonal hematopoiesis and recipient outcomes after transplantation. J Clin Oncol. 2022;40(2):189–201. doi: 10.1200/JCO.21.02286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feurstein SK, Trottier AM, Estrada-Merly N, et al. Germline predisposition variants occur in myelodysplastic syndrome patients of all ages. Blood. 2022;140(24):2533–2548. doi: 10.1182/blood.2022015790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lahtinen AK, Koski J, Ritari J, et al. Clinically relevant germline variants in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2023;58(1):39–45. doi: 10.1038/s41409-022-01828-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kraft IL, Godley LA. Identifying potential germline variants from sequencing hematopoietic malignancies. Blood. 2020;136(22):2498–2506. doi: 10.1182/blood.2020006910. [DOI] [PubMed] [Google Scholar]
- 52.Trottier AM, Godley LA. Inherited predisposition to haematopoietic malignancies: overcoming barriers and exploring opportunities. Br J Haematol. 2021;194(4):663–676. doi: 10.1111/bjh.17247. [DOI] [PubMed] [Google Scholar]
- 53.Saygin C, Roloff GW, Hahn CN, et al. Allogeneic hematopoietic stem cell transplant outcomes in adults with inherited myeloid malignancies. Blood Adv. 2023;7(4):549–554. doi: 10.1182/bloodadvances.2022008172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baranwal A, Nanaa A, Viswanatha D, et al. Outcomes of allogeneic transplant in patients with DDX41 mutated myelodysplastic syndrome and acute myeloid leukemia. Bone Marrow Transplant. 2022;57(11):1716–1718. doi: 10.1038/s41409-022-01776-6. [DOI] [PMC free article] [PubMed] [Google Scholar]