Abstract
Peptidoglycan from Deinococcus radiodurans was analyzed by high-performance liquid chromatography and mass spectrometry. The monomeric subunit was: N-acetylglucosamine–N-acetylmuramic acid–l-Ala–d-Glu-(γ)–l-Orn-[(δ)Gly-Gly]–d-Ala–d-Ala. Cross-linkage was mediated by (Gly)2 bridges, and glycan strands were terminated in (1→6)anhydro-muramic acid residues. Structural relations with the phylogenetically close Thermus thermophilus are discussed.
The gram-positive bacterium Deinococcus radiodurans is remarkable because of its extreme resistance to ionizing radiation (14). Phylogenetically the closest relatives of Deinococcus are the extreme thermophiles of the genus Thermus (4, 11). In 16S rRNA phylogenetic trees, the genera Thermus and Deinococcus group together as one of the older branches in bacterial evolution (11). Both microorganisms have complex cell envelopes with outer membranes, S-layers, and ornithine-Gly-containing mureins (7, 12, 19, 20, 22, 23). However, Deinococcus and Thermus differ in their response to the Gram reaction, having positive and negative reactions, respectively (4, 14). The murein structure for Thermus thermophilus HB8 has been recently elucidated (19). Here we report the murein structure of Deinococcus radiodurans with similar detail.
D. radiodurans Sark (23) was used in the present study. Cultures were grown in Luria-Bertani medium (13) at 30°C with aeration. Murein was purified and subjected to amino acid and high-performance liquid chromatography (HPLC) analyses as previously described (6, 9, 10, 19). For further analysis muropeptides were purified, lyophilized, and desalted as reported elsewhere (6, 19). Purified muropeptides were subjected to plasma desorption linear time-of-flight mass spectrometry (PDMS) as described previously (1, 5, 16, 19). Positive and negative ion mass spectra were obtained on a short linear 252californium time-of-flight instrument (BioIon AB, Uppsala, Sweden). The acceleration voltage was between 17 and 19 kV, and spectra were accumulated for 1 to 10 million fission events. Calibration of the mass spectra was done in the positive ion mode with H+ and Na+ ions and in the negative ion mode with H− and CN− ions. Calculated m/z values are based on average masses.
Amino acid analysis of muramidase (Cellosyl; Hoechst, Frankfurt am Main, Germany)-digested sacculi (50 μg) revealed Glu, Orn, Ala, and Gly as the only amino acids in the muramidase-solubilized material. Less than 3% of the total Orn remained in the muramidase-insoluble fraction, indicating an essentially complete solubilization of murein.
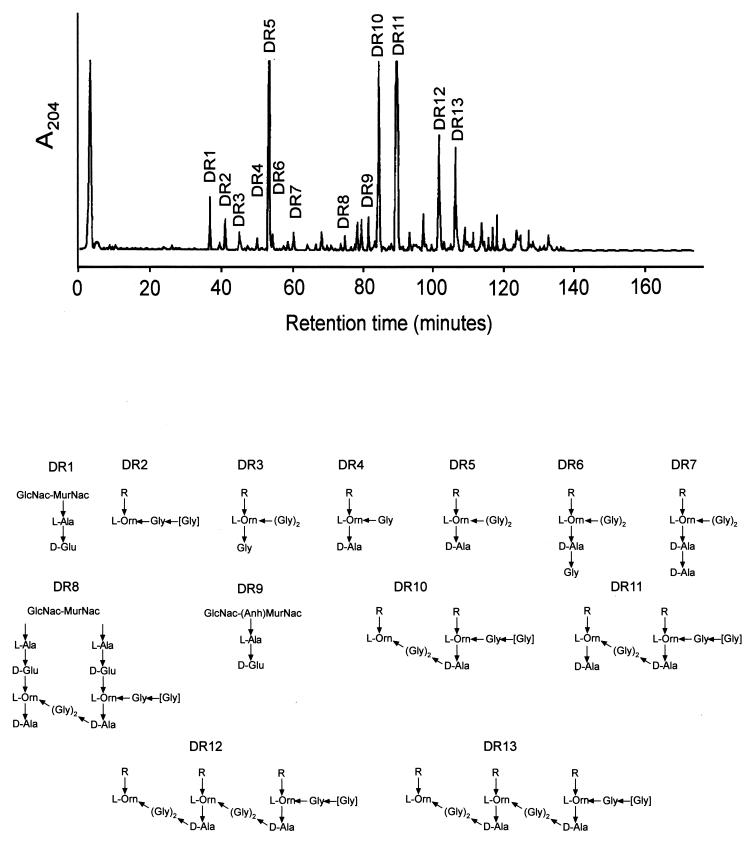
Muramidase-digested murein samples (200 μg) were analyzed by HPLC as described in reference 19. The muropeptide pattern (Fig. 1) was relatively simple, with five dominating components (DR5 and DR10 to DR13 [Fig. 1]). The muropeptides resolved by HPLC were collected, desalted, and subjected to PDMS. The results are presented in Table 1 compared with the m/z values calculated for best-matching muropeptides made up of N-acetylglucosamine (GlucNAc), N-acetylmuramic acid (MurNAc), and the amino acids detected in the murein. The more likely structures are shown in Fig. 1. According to the m/z values, muropeptides DR1 to DR7 and DR9 were monomers; DR8, DR10, and DR11 were dimers; and DR12 and DR13 were trimers. The best-fitting structures for DR3 to DR8, DR11, and DR13 coincided with muropeptides previously characterized in T. thermophilus HB8 (19) and had identical retention times in comparative HPLC runs. The minor muropeptide DR7 (Fig. 1) was the only one detected with a d-Ala–d-Ala dipeptide and most likely represents the basic monomeric subunit. The composition of the major cross-linked species DR11 and DR13 confirmed that cross-linking is mediated by (Gly)2 bridges, as proposed previously (20).
FIG. 1.
HPLC muropeptide elution patterns of murein purified from D. radiodurans. Muramidase-digested murein samples were subjected to HPLC analysis, and the A204 of the eluate was recorded. The most likely structures for each muroeptide as deduced by PDMS are shown. The position of residues in brackets is the most likely one as deduced from the structures of other muropeptides but could not be formally demonstrated. R = GlucNac–MurNac–l-Ala–d-Glu-(γ)→.
TABLE 1.
Calculated and measured m/z values for the molecular ions of the major muropeptides from D. radiodurans
| Muropeptidea | Ion |
m/z
|
Δmb | Error (%)c | Muropeptide composition
|
Muropeptide abundance (mol%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calculated | Measured | NAGd | NAMe | Glu | Orn | Ala | Gly | |||||
| DR1 | [M+H]+ | 699.69 | 700.1 | 0.41 | 0.06 | 1 | 1 | 1 | 0 | 1 | 0 | 12.0 |
| DR2 | [M+H]+ | 927.94 | 928.3 | 0.36 | 0.04 | 1 | 1 | 1 | 1 | 1 | 2 | 5.7 |
| DR3 | [M+Na]+ | 1,006.97 | 1,007.5 | 0.53 | 0.05 | 1 | 1 | 1 | 1 | 1 | 3 | 3.0 |
| DR4 | [M+Na]+ | 963.95 | 964.6 | 0.65 | 0.07 | 1 | 1 | 1 | 1 | 2 | 1 | 2.5 |
| DR5 | [M+H]+ | 999.02 | 999.8 | 0.78 | 0.08 | 1 | 1 | 1 | 1 | 2 | 2 | 27.7 |
| [M−H]− | 997.00 | 997.3 | 0.30 | 0.03 | ||||||||
| DR6 | [M+Na]+ | 1,078.5 | 1,078.8 | 0.75 | 0.07 | 1 | 1 | 1 | 1 | 2 | 3 | 2.4 |
| DR7 | [M+H]+ | 1,070.09 | 1,071.0 | 0.90 | 0.08 | 1 | 1 | 1 | 1 | 3 | 2 | 2.2 |
| DR8 | [M+Na]+ | 1,520.53 | 1,521.6 | 1.08 | 0.07 | 1 | 1 | 2 | 2 | 4 | 4 | 2.2 |
| DR9 | [M+Na]+ | 701.64 | 702.1 | 0.46 | 0.03 | 1 | 1f | 1 | 0 | 1 | 0 | 5.0 |
| DR10 | [M+H]+ | 1,907.94 | 1,907.8 | 0.14 | 0.01 | 2 | 2 | 2 | 2 | 3 | 4 | 10.1 |
| [M−H]− | 1,905.92 | 1,906.6 | 0.68 | 0.04 | ||||||||
| DR11 | [M+H]+ | 1,979.01 | 1,979.1 | 0.09 | 0.01 | 2 | 2 | 2 | 2 | 4 | 4 | 19.1 |
| [M−H]− | 1,977.00 | 1,977.3 | 0.30 | 0.02 | ||||||||
| DR12 | [M+H]+ | 2,887.93 | 2,886.5 | −1.43 | −0.05 | 3 | 3 | 3 | 3 | 5 | 6 | 4.4 |
| [M−H]− | 2,885.91 | 2,885.8 | −0.11 | −0.01 | ||||||||
| DR13 | [M+H]+ | 2,959.00 | 2,957.8 | −1.20 | −0.04 | 3 | 3 | 3 | 3 | 6 | 6 | 3.6 |
| [M−H]− | 2,956.99 | 2,955.9 | −1.09 | −0.04 | ||||||||
DR5 and DR10 to DR13 were analyzed in both the positive and negative ion modes. Muropeptides DR1 to DR4 and DR6 to DR9 were analyzed in the positive mode only due to the small amounts of sample available.
Mass difference between measured and calculated quasimolecular ion values.
[(Measured mass−calculated mass)/calculated mass] × 100.
N-Acetylglucosamine.
N-Acetylmuramitol.
(1→6)Anhydro-N-acetylmuramic acid.
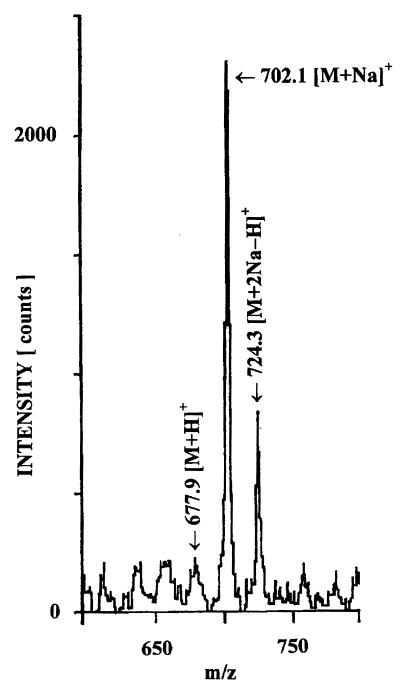
Structural assignments of muropeptides DR1, DR2, DR8 to DR10, and DR12 deserve special comments. The low m/z value measured for DR1 (700.1) fitted very well with the value calculated for GlucNAc–MurNAc–l-Ala–d-Glu (699.69). Even smaller was the mass deduced for DR9 from the m/z value of the molecular ion of the sodium adduct (702.1) (Fig. 2). The mass difference between DR1 and DR9 (19.9 mass units) was very close indeed to the calculated difference between N-acetylmuramitol and the (1→6)anhydro form of MurNAc (20.04 mass units). Therefore, DR9 was identified as GlucNAc–(1→6)anhydro-MurNAc–l-Ala–d-Glu (Fig. 1). Muropeptides with (1→6)anhydro muramic acid have been identified in mureins from diverse origins (10, 15, 17, 19), indicating that it might be a common feature among peptidoglycan-containing microorganisms.
FIG. 2.
Positive-ion linear PDMS of muropeptide DR9. Muropeptide DR9 was purified, desalted by HPLC, and subjected to PDMS to determine the molecular mass. The masses for the dominant molecular ions are indicated.
The measured m/z value for the [M+Na]+ ion of DR8 was 1,521.6, very close to the mass calculated for a cross-linked dimer without one disaccharide moiety (1,520.53) (Fig. 1; Table 1). Such muropeptides, also identified in T. thermophilus HB8 and other bacteria (18, 19), are most likely generated by the enzymatic clevage of MurNAc–l-Ala amide bonds in murein by an N-acetylmuramyl–l-alanine amidase (21). In particular, DR8 could derive from DR11. The difference between measured m/z values for DR8 and DR11 was 478.7, which fits with the mass contribution of a disaccharide moiety (480.5) within the mass accuracy of the instrument.
The m/z values for muropeptides DR2, DR10, and DR12 supported the argument for structures in which the two d-Ala residues from the d-Ala–d-Ala C-terminal dipeptide were lost, leaving Orn as the C-terminal amino acid.
The position of one Gly residue in muropeptides DR2, DR8, and DR10 to DR13 could not be formally demonstrated. One of the Gly residues could be at either the N- or the C-terminal positions. However, the N-terminal position seems more likely. The structure of the basic muropeptide (DR7), with a (Gly)2 acylating the δ-NH2 group of Orn, suggests that major muropeptides should present a (Gly)2 dipeptide. The scarcity of DR3 and DR6, which unambiguously have Gly as the C-terminal amino acid (Fig. 1), supports our assumption.
Molar proportions for each muropeptide were calculated as proposed by Glauner et al. (10) and are shown in Table 1. For calculations the structures of DR10 to DR13 were assumed to be those shown in Fig. 1. The degree of cross-linkage calculated was 47.2%. Trimeric muropeptides were rather abundant (8 mol%) and made a substantial contribution to total cross-linkage. However, higher-order oligomers were not detected, in contrast with other gram-positive bacteria, such as Staphylococcus aureus, which is rich in such oligomers (8). The proportion of muropeptides with (1→6)anhydro-muramic acid (5 mol%) corresponded to a mean glycan strand length of 20 disaccharide units, which is in the range of values published for other bacteria (10, 17).
The results of our study indicate that mureins from D. radiodurans and T. thermophilus HB8 (19) are certainly related in their basic structures but have distinct muropeptide compositions. In accordance with the phylogenetic proximity of Thermus and Deinococcus (11), both mureins are built up from the same basic monomeric subunit (DR7 in Fig. 1), are cross-linked by (Gly)2 bridges, and have (1→6)anhydro-muramic acid at the termini of glycan strands. Most interestingly, Deinococcus and Thermus are the only microorganisms identified at present with the murein chemotype A3β as defined by Schleifer and Kandler (20). Nevertheless, the differences in muropeptide composition were substantial. Murein from D. radiodurans was poor in d-Ala–d-Ala- and d-Ala–Gly-terminated muropeptides (2.2 and 2.4 mol%, respectively) but abundant in Orn-terminated muropeptides (23.8 mol%) and in muropeptides with a peptide chain reduced to the dipeptide l-Ala–d-Glu (18 mol%). In contrast, neither Orn- nor Glu-terminated muropeptides have been detected in T. thermophilus HB8 murein, which is highly enriched in muropeptides with d-Ala–d-Ala and d-Ala–Gly (19). Furthermore, no traces of phenyl acetate-containing muropeptides, a landmark for T. thermophilus HB8 murein (19), were found in D. radiodurans. Cross-linkage was definitely higher in D. radiodurans than in T. thermophilus HB8 (47.4 and 27%, respectively), largely due to the higher proportion of trimers in the former.
The similarity in murein basic structure suggests that the difference between D. radiodurans and T. thermophilus HB8 with respect to the Gram reaction may simply be a consequence of the difference in the thickness of cell walls (2, 3, 23). Interestingly, D. radiodurans murein turned out to be relatively simple for a gram-positive organism, possibly reflecting the primitive nature of this genus as deduced from phylogenetic trees (11). Our results illustrate the phylogenetic proximity between Deinococcus and Thermus at the cell wall level but also point out the structural divergences originated by the evolutionary history of each genus.
Acknowledgments
The technical assistance of J. de la Rosa is greatly appreciated.
This work was supported by grant BIO94-0789 from the CICYT to M.A.P. and an institutional grant from the Fundación Ramón Areces. The mass spectrometry work was supported by grant 11183 from the Austrian Science Foundation (to G.A.). Travel money was provided by the Acción Integrada Austria-España 22B and the Mixed Committee for Scientific and Technical Cooperation Austria—Spain. J.C.Q. was supported by a fellowship from the Fundación Rich, Madrid, Spain.
REFERENCES
- 1.Allmaier G, Rodríguez M C, Pittenauer E. Optimization of sample deposition for plasma desorption mass spectrometry. Rapid Commun Mass Spectrom. 1992;6:284–288. [Google Scholar]
- 2.Baumeister W, Kübler O. Topographic study of the cell surface of Micrococcus radiodurans. Proc Natl Acad Sci USA. 1978;75:5525–5528. doi: 10.1073/pnas.75.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brock T D, editor. Thermophilic microorganisms and life at high temperatures. New York, N.Y: Springer-Verlag; 1978. pp. 72–91. [Google Scholar]
- 4.Brock T D. Genus Thermus Brock and Freeze 1969, 295AL. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 333–337. [Google Scholar]
- 5.Caparrós M, Pittenauer E, Schmid E R, de Pedro M A, Allmaier G. Molecular weight-determination of biosynthetically modified monomeric and oligomeric muropeptides from Escherichia coli by plasma desorption mass spectrometry. FEBS Lett. 1993;316:181–185. doi: 10.1016/0014-5793(93)81211-h. [DOI] [PubMed] [Google Scholar]
- 6.Caparrós M, Pisabarro A G, de Pedro M A. Effect of d-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J Bacteriol. 1992;174:5549–5559. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castón J R, Berenguer J, de Pedro M A, Carrascosa J L. S-layer protein from Thermus thermophilus HB8 assembles into porin-like structure. Mol Microbiol. 1993;9:65–75. doi: 10.1111/j.1365-2958.1993.tb01669.x. [DOI] [PubMed] [Google Scholar]
- 8.De Jonge B L M, Tomasz A. The muropeptide composition of the peptidoglycan of Staphylococcus aureus determined with reversed-phase high performance liquid chromatography. In: de Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis: metabolism and structure of the bacterial sacculus. New York, N.Y: Plenum Press; 1993. pp. 77–82. [Google Scholar]
- 9.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;182:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 10.Glauner B, Höltje J-V, Schwarz U. The composition of the murein of Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 11.Hensel R, Demharter O, Kandler O, Kroppenstedt R M, Stackebrandt E. Chemotaxonomic and molecular-genetic studies of the genus Thermus: evidence for a phylogenetic relationship of Thermus aquaticus and Thermus ruber to the genus Deinococcus. Intl J System Bacteriol. 1986;36:444–453. [Google Scholar]
- 12.Lancy P, Jr, Murray R G. The envelope of Micrococcus radiodurans: isolation, purification, and preliminary analysis of the wall layers. Can J Microbiol. 1978;24:162–186. doi: 10.1139/m78-029. [DOI] [PubMed] [Google Scholar]
- 13.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 14.Murray R G E. Genus Deinococcus Brock and Murray 1981, 354vp. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1035–1043. [Google Scholar]
- 15.Pfanzagl B, Zenker A, Pittenauer E, Allmaier G, Martínez-Torrecuadrada J, Schmid E R, de Pedro M A, Löffelhardt W. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J Bacteriol. 1996;188:332–339. doi: 10.1128/jb.178.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pittenauer E, Schmid E R, Allmaier G, Pfanzagl B, Löffelhardt W, Quintela-Fernández C, de Pedro M A, Stanek W. Structural characterization of the cyanelle peptidoglycan of Cyanophora paradoxa by 252Cf plasma desorption mass spectrometry and fast atom bombardment tandem mass spectrometry. Biol Mass Spectrom. 1993;22:524–536. doi: 10.1002/bms.1200220906. [DOI] [PubMed] [Google Scholar]
- 17.Quintela J C, Caparrós M, de Pedro M A. Variability of peptidoglycan structural parameters in Gram-negative bacteria. FEMS Microbiol Lett. 1995;125:95–100. doi: 10.1111/j.1574-6968.1995.tb07341.x. [DOI] [PubMed] [Google Scholar]
- 18.Quintela J C, de Pedro M A, Zöllner P, Allmaier G, García-del Portillo F. Peptidoglycan structure of Salmonella typhimurium growing within cultured mammalian cells. Mol Microbiol. 1997;23:693–704. doi: 10.1046/j.1365-2958.1997.2561621.x. [DOI] [PubMed] [Google Scholar]
- 19.Quintela J C, Pittenauer E, Allmaier G, Arán V, de Pedro M A. Structure of peptidoglycan from Thermus thermophilus HB8. J Bacteriol. 1995;177:4947–4962. doi: 10.1128/jb.177.17.4947-4962.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shockman G D, Höltje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Elsevier Science Publisher B.V., Amsterdam, The Netherlands. 1994. pp. 131–166. [Google Scholar]
- 22.Thompson B G, Murray R G. Isolation and characterization of the plasma membrane and the outer membrane of Deinococcus radiodurans strain Sark. Can J Microbiol. 1981;27:729–734. doi: 10.1139/m81-111. [DOI] [PubMed] [Google Scholar]
- 23.Work E, Griffits H. Morphology and chemistry of cell walls of Micrococcus radiodurans. J Bacteriol. 1968;95:641–657. doi: 10.1128/jb.95.2.641-657.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]