Abstract
Microbial communities that form surface-attached biofilms must release and disperse their constituent cells into the environment to colonize fresh sites for continued survival of their species. For pathogens, biofilm dispersal is crucial for microbial transmission from environmental reservoirs to hosts, cross-host transmission, and dissemination of infections across tissues within the host. However, research on biofilm dispersal and its consequences in colonization of fresh sites remain poorly understood. Bacterial cells can depart from biofilms via stimuli-induced dispersal or disassembly due to direct degradation of the biofilm matrix, but the complex heterogeneity of bacterial populations released from biofilms rendered their study difficult. Using a novel 3D-bacterial “biofilm-dispersal-then-recolonization” (BDR) microfluidic model, we demonstrated that Pseudomonas aeruginosa biofilms undergo distinct spatiotemporal dynamics during chemical-induced dispersal (CID) and enzymatic disassembly (EDA), with contrasting consequences in recolonization and disease dissemination. Active CID required bacteria to employ bdlA dispersal gene and flagella to depart from biofilms as single cells at consistent velocities but could not recolonize fresh surfaces. This prevented the disseminated bacteria cells from infecting lung spheroids and Caenorhabditis elegans in on-chip coculture experiments. In contrast, EDA by degradation of a major biofilm exopolysaccharide (Psl) released immotile aggregates at high initial velocities, enabling the bacteria to recolonize fresh surfaces and cause infections in the hosts efficiently. Hence, biofilm dispersal is more complex than previously thought, where bacterial populations adopting distinct behavior after biofilm departure may be the key to survival of bacterial species and dissemination of diseases.
Subject terms: Biofilms, Population dynamics, Microbial ecology, Bacterial physiology, Antibiotics
Introduction
Unicellular prokaryotes and lower eukaryotes spend most of their time in the biofilm life cycle, which cycles between planktonic cell adhesion, biofilm formation, maturation, and dispersal. The biofilm lifecycle is highly dependent on c-di-GMP signalling found in most bacterial species, where c-di-GMP is synthesized by diguanylate cyclase (DGC) to form biofilms, and c-di-GMP is degraded by phosphodiesterases (PDE) to achieve biofilm dispersion [1].
As the final stage of biofilm life cycle, biofilm dispersal is the detachment of bacteria from the biofilm and dissemination into the environment. The biological dispersal is essential for continued survival of microbial species in the environment. In microbial ecology, dispersal is important for nutrient cycling and bacteriovorous predator-bacterial prey interactions [2]. For pathogenic microbes, biofilm dispersal plays important roles in disease transmission, where microbes can be disseminated from environmental reservoirs to hosts, cross-host transmission, and across tissues and organs within the host. In mammalian hosts, bacteria dispersed from biofilms can spread into the bloodstream, leading to fatal infections in other tissues and sepsis [3–5]. Although biofilm formation and maturation are well studied [6, 7], the population dynamics of biofilm dispersal, its impact on recolonization of fresh sites and even dissemination of infection remain poorly understood, necessitating in-depth investigation of the last step in biofilm life cycle.
Similar to other stages of biofilm life cycle, dispersal is a complex process that involves different environmental stimuli, bacterial pathways and effectors. There are two strategies adopted by microbes to achieve biofilm dispersal: (1) chemically-induced dispersal (CID) of biofilms using dispersal cues, such as nitric oxide [8, 9], and (2) enzymatic disassembly (EDA) of the biofilm matrix to break down biofilms and release bacterial cells passively [10, 11]. Environmental cues, such as nitric oxide, can induce the expression of PDEs, such as DipA and NdeA [8], leading to down-regulation of c-di-GMP signalling, expression of motility apparatus, and degradation of biofilm matrix components, thereby enabling the CID of biofilms [12]. During this transient phase, bacterial cells have the unique physiology of biofilms and planktonic cells, which have high virulence potential but are susceptible to iron limitation [13]. Depleting nutrients such as pyruvate can also induce biofilm diffusion [14]. In contrast, biofilm disintegration caused by enzymatic degradation of the biofilm substrate can expel bacteria from the biofilm. Enzymes include glycosidases, proteases, and DNases [15–17], which degrade their respective biofilm matrix components. Both CID and EDA are also promising anti-biofilm approaches for eliminating biofilm diseases in humans or biofouling in the environment, since bacteria released from biofilms are susceptible to antimicrobial removal and immune attacks [18]. However, little is known about the population dynamics of bacteria released during biofilm dispersal or disassembly, which hinders the practical application of antibiofilm approaches in environmental and clinical settings.
Here we show that Pseudomonas aeruginosa populations generated from biofilm dispersal and disassembly adopt different fates in dissemination and recolonization of new areas, with contrasting implications in host infection. P. aeruginosa is a ubiquitous biofilm-forming bacterial species found in most environmental ecosystems [19] and in opportunistic infections of the organ systems (lung and gastrointestinal system) [20, 21] and medical devices [22, 23]. Moreover, it could disseminate from the origin of infection to other organs, leading to bacteremia and sepsis [24, 25]. Our findings were achieved by developing a dual-chamber “biofilm-dispersal-then-recolonization” (BDR) microfluidic lab-on-a-chip platform that rapidly cultures and disperses 3D-biofilms in the primary chamber for subsequent colonization of biofilm-released bacterial populations in a secondary chamber. Moreover, human lung 3D-spheroid culture or Caenorhabditis elegans models were incorporated in the secondary chamber to evaluate disease dissemination.
In CID, the bacteria activate the biofilm dispersal gene bdlA and leave the biofilm as single cells in a flagella-dependent manner but are surprisingly unable to colonize fresh surfaces. This has a significant implication, as these dispersed cells could not effectlvely establish an infection on the lung spheroids and Caenorhabditis elegans in on-chip coculture experiments. In contrast, EDA caused by enzymatic degradation of Psl, the major exopolysaccharide in P. aeruginosa biofilm infections [26, 27], could cause the release of small immotile bacterial aggregates at high initial speeds. The passive-released bacterial aggregates then colonize new surfaces and establish effective infections on the hosts.
Our work demonstrated that biofilm dispersal is more complex than previously thought, where adopting distinct behavior and population dynamics after biofilm departure may increase the chances of bacterial survival. Our work also provides insights into bacterial dissemination and colonization of fresh sites in microbial ecology and diseases.
Results
Establishment of the 3D-bacterial “biofilm dispersion-and-recolonization” (BDR) microfluidic model
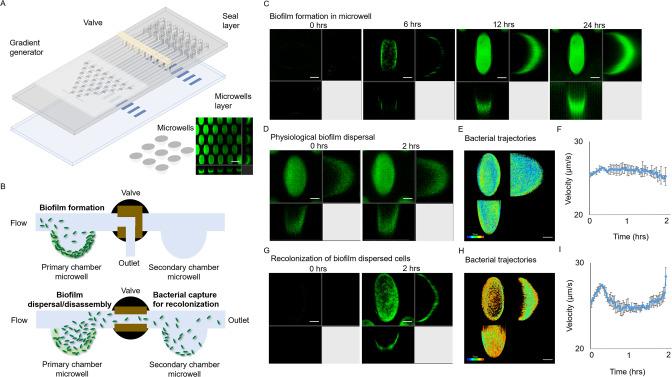
Existing bacterial models, such as the flow chamber and Calgary biofilm assay, do not fully recapitulate the 3D structure of biofilms found in vivo [28], and cannot examine dispersion and recolonization in a single device. To overcome these limitations, we developed the BDR platform (Fig. 1A) to evaluate both biofilm dispersal and recolonization. The BDR platform comprised of a primary chamber for initial biofilm formation dispersion and a secondary chamber for capturing biofilm-released cells for recolonization under continuous flow (Fig. 1B). Each channel contains up to 300 tapered microwells, compared to flat surfaces commonly used in conventional biofilm cultures [29], which enabled P. aeruginosa to rapidly colonize and form biofilms within 12 h under continuous flow as 3D cultures (Fig. 1C, Supplementary Fig. S1).
Fig. 1. Tracking of physiological biofilm dispersal using a bacterial “biofilm-dispersal-and-recolonization” (BDR) microfluidic chip.
A Biochip design, containing gradient generator, primary channel and downstream secondary channel. Each channel includes 300 microwells, where bacteria could colonize and grow biofilms in the microwells. B Experimental design of biofilm formation in the primary chamber, followed by biofilm dispersal and subsequent recolonization in the secondary chamber. C gfp-tagged WT biofilm formation over 24 h. Representative images are shown, where the scale bar is 50 µm. D Physiological biofilm dispersal over 2 h. E Time-resolved cell trajectories of physiologically biofilm dispersed cells. Legend: color scheme from purple to red indicated chronological order from 0 to 2 h. F Average velocity of physiologically biofilm dispersed cells. G Colonization of physiological biofilm dispersed cells in recolonization chamber over 2 h. H Time-resolved cell trajectories of physiologically biofilm dispersed cells upon recolonization. Legend: color scheme from purple to red indicated chronological order from 0 to 2 h. I Average velocity of physiological biofilm dispersed cells upon recolonization. The means and s.d. from triplicate experiments were shown.
It is important to note that the tapered microwells with dimensions 150 × 250 × 150 μm (length × width × depth), are different from the larger microwells from microplates. These tapered microwells in microfluidics devices are typically used for establishment of 3D cultures, which recapitulates the physiology and behavior of cells in humans [30–32]. A large number of microwells in each channel also provided simultaneous observation of multiple technical replicate wells, enabling us to confirm the reproducibility of our observations (Fig. 1A). We showed that the bacterial communities cultivated in each microwell are true biofilms, as they possessed increased expression of biofilm markers (pcdrA-gfp biosensor and c-di-GMP) [13, 33] in bacterial cells (Supplementary Fig. S2). Hence, both biofilm markers indicated that the microwells can be used to cultivate 3D biofilms in BDR device which have similar physiology as a typical biofilm cultivated in vitro or in vivo.
After biofilm cultivation in the primary chamber for 12 h, we evaluated the dispersion process and the performance of the secondary chamber in capturing biofilm-released cells by turning on the valve that enables media flow from the primary chamber into the secondary chamber. Under normal conditions under media flow without antibiofilm treatment, the biofilms remained intact after 2 h, while basal levels of biofilm dispersal occurred continuously (Fig. 1D, Supplementary Video 1). Freshly dispersed cells would enter the media space of the microwell for microscopic observation and eventually exit the microwell (Fig. 1E). The tapered microwells provided sufficient confined boundaries to retain newly dispersed cells for observation, allowing long-term monitoring of cell velocity immediately after dispersion (Fig. 1F). We demonstrated bacterial velocities immediately after dispersion (Fig. 1F), where the average velocity of bacteria released from biofilm in general remained consistent over time with a value of ~30 µm s−1. Although this corroborated with the flagellar-dependent velocities of planktonic bacteria [34], our goal was not to track individual bacteria due to limitations of the confocal microscopy in capturing 3D images, but to demonstate the general trend of how bacteria departed from the biofilm over time.
To recolonize bacterial cells released from biofilms in the primary chamber, we then flowed the bacterial suspension from the primary chamber into the secondary chamber, where microwells in the secondary recolonization chamber could capture biofilm-dispersed cells for recolonization (Fig. 1G, Supplementary Video 2). Time-resolved cell trajectories revealed the consistent recolonization of dispersed cells (Fig. 1H), which maintained consistent velocities when they were freshly dispersed from the biofilm (Fig. 1I).
CID induces rapid release of bacterial cells
Bacterial cells released from biofilms under physiological conditions can comprise mixed populations of actively dispersed cells departing on their own and passively dispersed cells released from biofilms. Passive dispersal can occur due to biofilm matrix degradation or shear stress [10]. Hence, we next aim to generate homogenous populations of biofilm-dispersed cells and biofilm-disassembled cells for subsequent study.
To study the dynamics of these two modes and their implications in disease dissemination, we employed two antibiofilm strategies, namely the [1] induced biofilm dispersal and [2] biofilm disassembly. To evaluate strategies for inducing dispersal in biofilms, we generated dispersed cells from biofilms by reducing c-di-GMP based on chemical stimuli with nitric oxide (NO) in the form of sodium nitroprusside (SNP) or by inducing the expression of plasmid-encoded YhjH PDE to degrade c-di-GMP [35] (Supplementary Fig. S3A). This would eliminate the possibility of pleiotropic effects caused purely by SNP treatment or YhjH PDE expression. For biofilm disassembly, we generated biofilm disassembled cells by enzymatic treatment with 2 different enzymes (cellulase and pectinase) [21] (Supplementary Fig. S3B). Pectinase could degrade Psl exopolysaccharide in P. aeruginosa biofilms (Supplementary Fig. S4). The Psl-containing ΔpelA mutant biofilm would then be dispersed by pectinase, while Psl-deficient ΔpslBCD mutant biofilm remained intact after pectinase treatment (Supplementary Fig. S5). Exogenous addition of Psl to ΔpelAΔpslBCD mutants, which were biofilm-deficient, enabled biofilm formation. Biofilm formation could then be abrogated by pectinase treatment (Supplementary Fig. S6), indicating that biofilm disassembly required enzymes that specifically degrade biofilm matrix components.
During the induced biofilm dispersal, bacterial cells left the biofilms within 2 h (Fig. 2A, Supplementary Videos 3 and 4). Mathematical models for inducing biofilm dispersal revealed an exponential decay in biofilm biomass, indicating that many bacteria dispersed in the initial stage and slowed down in the later stage (Fig. 2C). The majority of bacteria dispersed from the biofilm and entered the media space of the microwells within the 1st hr, but completely departed from the microwell within the 2nd hr (Fig. 2D). We observed a similar trend of CID bacterial motility as compared to that of physiologically dispersed cells during the initial phase, but CID bacterial motility decreased significantly over time, which was attributed to the majority of bacteria leaving the microwells by the end of 2 h (Fig. 2E).
Fig. 2. Dynamics of CID mediated by c-di-GMP signalling reduction.
A Representative images of biofilm dispersal over time, where the scale bar is 50 µm. B Reduction of biofilm biomass (GFP intensity) over time. C Mathematical modeling revealed biofilm dispersal follows an exponential decay. D Time-resolved cell trajectories of induced biofilm dispersal. Legend: color scheme from purple to red indicates chronological order from 0 to 2 h. E Reduced speed of biofilm dispersed cells over time. The means and s.d. from triplicate experiments were shown.
EDA leads to the explosive release of bacterial aggregates
We found that EDA resulted in a continuous reduction of biofilm from top to bottom of the microwells (Fig. 3A, B, Supplementary Videos 5 and 6), indicating the action by enzymes from the exterior to the interior of biofilms. Mathematical models for inducing biofilm disassembly revealed a polynomial decay in biofilm biomass, indicating that bacteria remained in the microwell longer and left the microwell slower than induced biofilm dispersal (Fig. 3C). Bacteria were retained in the microwells after biofilm EDA (Fig. 3D). The rate of biofilm disassembly accelerated, which was then followed by a rapid decrease in rate (Fig. 3E), resembling an initial explosive release of cells from the biofilms.
Fig. 3. Dynamics of biofilm EDA.
A Representative images of biofilm disassembly over time. Representative images are shown, where the scale bar is 50 µm. B Reduction of biofilm biomass (GFP intensity) over time. C Mathematical modeling revealed biofilm disassembly follows a polynomial decay. D Time-resolved cell trajectories of biofilm disassembly. Legend: The color scheme from purple to red indicated chronological order from 0 to 2 h. E The initial explosive increase in biofilm disassembled cells, followed by a rapid decline in velocity. The means and s.d. from triplicate experiments were shown.
Bacteria undergo distinct spatiotemporal dynamics between CID and EDA
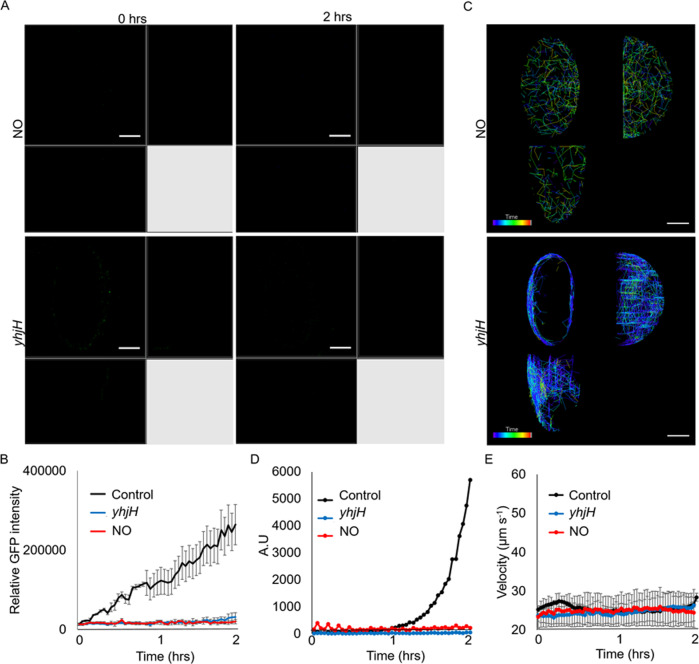
Our next aim was to determine the mechanism underlying the difference between induced biofilm dispersal and biofilm disassembly, where CID appeared to be actively undertaken by bacteria. In contrast, EDA is a passive expulsion of bacteria from the matrix. There are several reasons to support this hypothesis. We observed a decrease in intracellular c-di-GMP levels in bacteria during the induction of biofilm diffusion, while, biofilm-disassembling bacteria retained c-di-GMP levels similar to biofilm cells. This was supported by observing and measuring the GFP fluorescence intensity of the PAO1/pcdrA-gfp biosensor (Fig. 4A, B), and quantifying c-di-GMP levels with the ELISA assay (Fig. 4C).
Fig. 4. Bacterial cells released by CID and EDA possessed differing physiologies.
A Biofilm dispersed WT/pcdrA-gfp cells downregulated GFP expression, while biofilm-disassembled cells retained GFP expression. Representative images are shown, where the scale bar is 10 µm. B Relative GFP intensity of WT/pcdrA-gfp cells after CID or EDA. C ELISA revealed reduced c-di-GMP levels in biofilm dispersed cells and high c-di-GMP levels in biofilm-disassembled cells. Means and s.d. from triplicate experiments are shown. **p < 0.01. D No biofilm dispersal in ΔbdlA mutant. E Effective biofilm disassembly in ΔbdlA mutant. F No biofilm dispersal in ΔfliM mutant. G Effective biofilm disassembly in ΔfliM mutant. H Biofilm-dispersed cells primarily exist as single cells, while biofilm-disassembled cells exist as small multicellular aggregates. Representative images are shown, where the scale bar is 10 µm.
This was also reflected in the role of the c-di-GMP signalling-controlled gene bdlA in biofilm dispersal [36, 37]. Therefore, we found that the ΔbdlA mutant could not undergo induced biofilm dispersal but could still be released by pectinase during biofilm disassembly (Fig. 4D, E, Supplementary Fig. S7A, Supplementary Videos 7 and 8). On the other hand, we observed the restoration of biofilm dispersal phenotype in the ΔbdlA/plac-bdlA complementation strain, as it could undergo induced biofilm dispersal by SNP (Supplementary Fig. S7B).
We also demonstrated another difference related to flagellar motility, which we previously observed in biofilm-dispersed cells, as they expressed flagella to actively depart from the biofilm. The flagella-deficient ΔfliM mutants lost the ability to leave the microwells during induced biofilm dispersal, but ΔfliM could still be released during biofilm disassembly (Fig. 4F, G, Supplementary Fig. S8A, Supplementary Videos 9 and 10). The ΔfliM/plac-fliM complementation strain could undergo induced biofilm dispersal by SNP, indicating that flagella is important in active biofilm dispersal (Supplementary Fig. S8B).
Furthermore, we observed that biofilm disassembled bacteria present as small aggregates instead of biofilm dispersed bacteria, which were predominantly single cells (Fig. 4H). This observation further suggested that bacteria released biofilm disintegration retains some aspects of their previous life as a biofilm.
Bacteria generated from CID cannot recolonize fresh surfaces
The biofilm life cycle usually involves bacterial recolonization of new areas. Since biofilm dispersal differs from biofilm disassembly, we asked if bacterial cells generated from both approaches would have differing recolonization dynamics. It is important to note that bacteria released from biofilm dispersal and disassembly remained viable, where PI dead stain did not label the bacterial cells (Supplementary Fig. S9), indicating that their recolonization was not affected by cell death. While bacteria derived from physiological biofilm dispersal could rapidly colonize secondary chambers, bacteria released due to c-di-GMP-reduced CID could not colonize the microwells (Fig. 5A–D, Supplementary Videos 11 and 12), despite the ability of biofilm dispersed bacteria to maintain consistent speeds in general (Fig. 5E). This could be attributed to the fact that induced biofilm dispersed cells maintained their physiology for at least 6 h [13], so they could not recolonize the microwells.
Fig. 5. Recolonization dynamics of CID-released bacteria.
A Representative images of recolonization of CID-released cells over time, where the scale bar is 50 µm. B No increase in biofilm biomass (GFP intensity) over time. C Time-resolved cell trajectories of recolonization by induced biofilm dispersed cells. Legend: color scheme from purple to red indicated chronological order from 0 to 2 h. D Mathematical modeling revealed biofilm dispersed cells recolonization follows no pattern. E Consistent velocities of biofilm dispersed cells over time. The means and s.d. from triplicate experiments were shown.
Bacterial aggregates from biofilm disassembly slowly recolonized new areas
EDA-released bacteria could colonize the secondary chamber, albeit at a lower magnitude than physiological dispersal over time (Fig. 6A, B, Supplementary Videos 13 and 14). Our mathematical model showed exponential recolonization of biofilm disassembled bacteria, but still at a lower magnitude than physiological dispersal (Fig. 6C). Recolonization of these bacteria occurred mainly in the 2nd hr (Fig. 6D), as they required some time to migrate from the primary chamber to the secondary chamber. This corresponded to faster recolonization of bacteria expelled from the biofilm (Fig. 6E), when they were initially released from the biofilm at high speeds.
Fig. 6. Recolonization dynamics of EDA-released bacteria.
A Representative images of recolonization of biofilm-disassembled cells over time, where the scale bar is 50 µm. B Partial increase in biofilm biomass (GFP intensity) over time. C Mathematical modeling revealed biofilm disassembled cell recolonization follows an exponential pattern. D Time-resolved cell trajectories of recolonization by biofilm-disassembled cells. Legend: color scheme from purple to red indicated chronological order from 0 to 2 h. E High velocities of biofilm disassembled cells over time. The means and s.d. from triplicate experiments were shown.
Similar dispersal and recolonization behavior adopted by P. aeruginosa clinical isolates
To evaluate if our findings in PAO1 could be applicable to other P. aeruginosa strains, we tested the effects of biofilm CID and EDA on PA14 and 2 other clinical isolates (CF173 and CF273) [38]. For PA14, CID could induce biofilm dispersal, with minimum recolonization of dispersed bacteria. However, as PA14 possessed the ability to produce Pel but not Psl [39], pectinase-mediated EDA was not very effective in breaking down the biofilm matrix, resulting in minimal bacterial aggregates which recolonized the surface of secondary chamber (Supplementary Fig. S10a). Till date, no Pel-degrading enzymes was commercially available for testing. As for CF173 and CF273, they were pro-biofilm-forming clinical isolates collected from cystic fibrosis patients, so we employed higher concentrations of antibiofilm agents to elicit CID and EDA. Nonetheless, we observed similar trends between both clinical isolates and PAO1, where CID-generated bacteria could not recolonize fresh areas and EDA-generated bacterial aggregates could recolonize the secondary chamber (Supplementary Fig. S10b, c). This indicated that our findings are common across different P. aeruginosa strains, albeit different concentrations of antibiofilm agents may be required.
Unique characteristics of biofilm-released bacteria determine the fate of recolonization and dissemination of diseases in vivo
Since CID and EDA could lead to different fates in bacterial migration and subsequent recolonization of fresh surfaces, we then evaluate if disseminated bacteria could cause infections in both in vitro human lung 3D-spheroids and in vivo C. elegans models. The lung spheroid culture model is a clinically relevant model for personalized medicine and it can recapitulate similar findings in animals and humans [40, 41]. C. elegans is a common animal model used to study P. aeruginosa pathogenesis [42, 43], and its small size and transparency were advantageous in establishing animal models in microfluidics models [44, 45].
Under normal conditions, P. aeruginosa bacteria freshly released from biofilms could colonize the lung spheroid cells (Fig. 7A–C). The EDA cells could colonize on the lung spheroids at higher numbers and cause infections similarly to control cells (Fig. 7A–C). However, CID cells could not colonize on the eukaryotic models at lower numbers, thereby causing minimal infections and killing cells (Fig. 7A–C). These in vitro data were similar to the C. elegans infection model, where we observed lesser killing of animals by CID cells than in control (Fig. 7D). This was attributed to minimal colonization of bacteria within the C. elegans intestine (Fig. 7E, F). Hence, this provides insights into both antibiofilm strategies, which could be adopted in clinical settings.
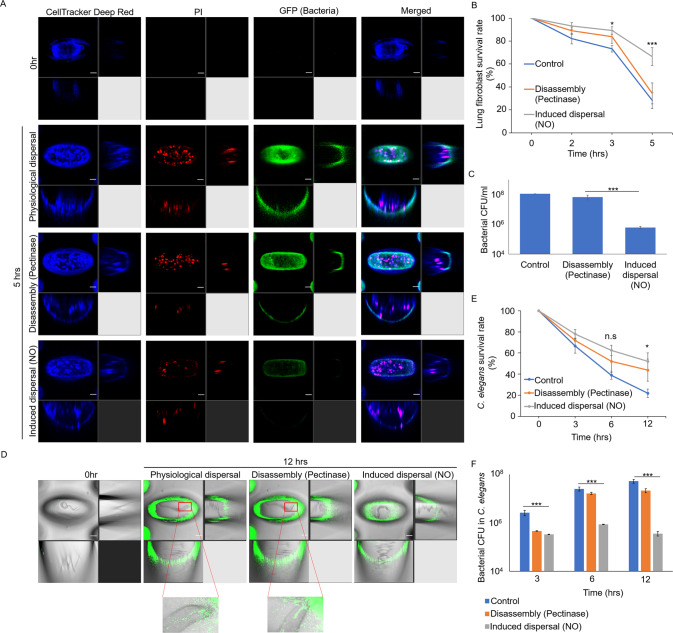
Fig. 7. Virulence of bacterial cells released from biofilm CID and EDA in eukaryotic models.
A Representative images of human lung spheroids in association with bacteria released from CID or EDA, where the scale bar is 50 µm. B Survival rate of lung spheroids after exposure to bacteria released from biofilm dispersal or disassembly.; ***p < 0.001; n.s not significant. C Bacterial numbers on lung spheroids over time. D Representative images of C. elegans in association with bacteria released from biofilm dispersal or disassembly, where the scale bar is 50 µm. Inset images showed the localization of gfp-tagged bacteria in the animal intestine. E Survival rate of C. elegans after exposure to bacteria released from biofilm dispersal or disassembly. The means and s.d. from triplicate experiments were shown. *p < 0.05; n.s not significant. F Bacterial numbers in the C. elegans intestine over time. The means and s.d. from triplicate experiments were shown. ***p < 0.001.
Discussion
In this study, we tracked the dynamics of the spread and recolonization of biofilms in space and time, to provide insights into how multicellular structures return to unicellular form for migration. This was achieved by using the BDR microfluidic-based platform to monitor biofilm dispersal and recolonization. Advances in microfluidics enable better control and precision of miniaturized biofilm growth at a lower cost than conventional biofilm cultivation approaches [33, 46, 47], so our BDR platform has several advantages over conventional biofilm cultivation approaches (Supplementary Table S1). Moreover, we could incorporate in vitro human cell spheroid cultures or in vivo animal models within the device, indicating the added flexibility of evaluating bacterial virulence. The BDR platform can also be applied for high-throughput cultivation and antimicrobial testing of biofilms. In combination with a gradient generator with the BDR platform, different concentrations of antibiotics can be generated against biofilms to evaluate their antimicrobial efficacy (Supplementary Figs. S11 and S12).
Biofilm dispersal has been touted as a strategy to combat biofilm-mediated infections in clinical settings. Still, questions remained regarding the differences between CID and EDA [10, 11]. Here, we unveiled the new mechanisms underlying biofilm CID and EDA by employing spatiotemporal imaging, mutagenesis, biosensor quantification and mathematical modeling (Fig. 8, Supplementary Table S2). Bacteria resulting from CID are usually single-celled, where they require bdlA gene for dispersal and employ flagella to leave biofilms. They cannot immediately recolonize on fresh surfaces, which corroborated our previous study that the dispersed cells maintained their physiology for 6 h [13]. However, EDA of biofilm releases “biofilm-like” bacteria aggregates, bypassing the need for activating biofilm dispersal genes and motility apparatus. They can recolonize fresh surfaces, albeit at lower efficiencies than physiological biofilm dispersal, possibly due to the low metabolism of biofilm cells.
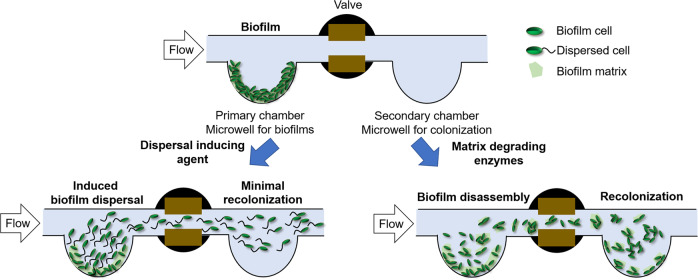
Fig. 8. Schematic diagram displaying the differences between biofilm chemical-induced dispersal (CID) and biofilm enzymatic disassembly (EDA).
Biofilms are first cultivated in the primary chamber, followed by CID (left) and EDA (right) treatments. CID caused by dispersal inducing agent leads to the release of active bacteria from the biofilm, resulting in minimal recolonization in the secondary chamber. EDA caused by matrix degrading enzymes results in the passive release of small bacterial aggregates which can recolonize the secondary chamber.
We also showed that antibiofilm strategies adopting either biofilm dispersal or disassembly have varying implications for pathogenesis and disease dissemination. As biofilm-dispersed cells could not colonize on host surfaces, there were lower levels of the killing of eukaryotic hosts. Although this appeared to contradict our previous work showing that CID cells possessed heightened virulence factors against the host macrophages upon direct exposure in a static infection model [13], our current work showed that CID-released bacteria could not colonize on the host and inflict virulence on the host, while EDA-relased bacteria could colonize and kill the hosts more efficiently in continuous flow. It could be possible that the larger EDA-released aggregates could settle downward onto the lung spheroids more efficiently than CID-released bacteria (Supplementary Fig. S13), and the presence of flow could cause the unattached CID-released bacteria to be washed off from the lung spheroids, resulting in higher contact-based killing of lung spheroids by the EDA-released aggregates.
Our work supported the findings from prior studies using mouse models of biofilm infection, where the animals could tolerate CID [48], whereas lethal septicaemia could occur in the animals with EDA [3]. This indicated that biofilm dispersal might be a better antibiofilm strategy than biofilm disassembly, where there was lower disease dissemination and damage to other organs by biofilm-dispersed cells than biofilm biofilm-disassembled cells.
Our work has several limitations. Firstly, the biofilms were exposed to continuous flow of liquid, enabling the introduction of fresh nutrients and oxygen, and removal of waste and bacterial cells. While continuous flow is relevant to natural aquatic environments with shear stress [49], or human infections such as catheter-related infections and urinary tract infections [50, 51], we did not evaluate our results in static conditions with limited nutrient supply and retainment of waste. Bacteria released by CID or EDA may behave differently in static conditions, especially in recolonization of fresh areas and infections. Next, we observed the effects of CID and EDA on biofilms for short periods of time, which may not be sufficient to evaluate long-term effects on biofilms. Lastly, we did not test other types of antibiofilm agents which may impose different effects on biofilms, such as nanoparticles, bacteriophages, photosensitizer and N-acetyl-cysteine [52–55]. This indicated that it is crucial to evaluate each antibiofilm agent carefully in different applications.
In summary, our work demonstrated that multicellular communities that undergo induced dispersal or disassembly, experience different fates in migration and colonization of new areas. The principles defining these collective cell movement patterns may be relevant to the multicellular organization and development of other prokaryotic and eukaryotic systems, and the establishment of suitable antibiofilm strategies adapted for different situations.
Methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Supplementary Table S3. E. coli DH5a strain was used for standard DNA manipulations. LB medium (Difco, Becton Dickinson and Company, USA) was used to cultivate E. coli, and P. aeruginosa strains for bacterial growth. For experiments, P. aeruginosa strains were grown in ABTGC (ABT minimal medium supplemented with 2 g L−1 glucose and 2 g L−1 casamino acids at 37 °C [35]. For plasmid maintenance in E. coli, the medium was supplemented with 100 μg ml−1 ampicillin and 15 μg ml−1 gentamicin. For marker selection in P. aeruginosa, 30 μg ml−1 gentamicin and 100 μg ml−1 streptomycin were used.
Fabrication of device
The CAD software designed the microfluidic device as four components using standard fabrication methods [33]. The master mold was fabricated by diffuser back-side lithography procedures [46]. The base layer is composed of an array of 300 microwells with dimensions 150 × 250 × 150 μm (length × width × depth) [30]. The second 8-channel layer comprises the primary and serpentine secondary chamber, each with a 6 mm depth separated by a valve. The third layer comprised a tree-shaped gradient generator composed of two inlet channels and eight output channels [30]. The material for the fabrication of the microfluidic device was PDMS (Polydimethylsiloxane), prepared using a Sylgard 184 silicone elastomer kit (Dow Corning, USA) by thoroughly mixing the base resin and curing agent in a ratio of 10:1 by weight [56]. The cured PDMS molds were plasma treated and exposed to trichloro(1H,1H,2H,2H-perfluorooctyl) silane (Sigma-Aldrich, #448931, Germany) overnight in a vacuum desiccator [30, 57]. The different layers were bonded together using plasma technology and placed in a 70 °C oven for 2 h.
Calibration of the gradient generator
The device was primed with ethanol and checked to ensure no trapped air bubbles. The ddH2O and 10 μM SYTO-9 fluorescent dye (Invitrogen, USA) were pumped into the gradient generator at 30 μl min−1 through both inlets separately with a syringe pump (New Era Pump Systems, Farmingdale, USA). Liquid samples were collected from each outlet for fluorescence density measurement with a microplate reader (Tecan, Infinite M1000 Pro, Switzerland). To visualize the gradient distribution generated in the device, blue and red food dyes were introduced separately into both inlets with the syringe pump. The simulated flow profile was generated using the multiphysics modeling software (COMSOL).
Cultivation of biofilms in device
P. aeruginosa biofilms were cultivated in an ABTGC medium at 37 °C. The device was supplied with a medium flow using the syringe pump, while the waste medium was removed into a waste beaker. Each channel was inoculated with 100 μl of a 100X diluted overnight culture using a syringe and needle, followed by incubation without flow for 20 min to allow bacteria to colonize the microwells. The medium flow was started for continuous flow and maintained at a velocity of 0.2 mm/s by the syringe pump (New Era Pump Systems, Farmingdale, USA) for 12 h at 37 °C.
Biofilm CID and EDA
The device comprises primary and secondary chambers. The primary chamber was used for biofilm cultivation and subsequent dispersal, and the secondary chamber filled with only media was left isolated via a closed valve. For biofilm dispersal and disassembly, the valve was opened to connect both chambers, enabling the movement of bacteria from the primary to the secondary chamber.
During CID, the 150 µM sodium nitroprusside dihydrate (SNP, Sigma-Aldrich, Germany) was used for P. aeruginosa wild-type PAO1 strain, while 5% (w/v) L-arabinose (Sigma-Aldrich, Germany) was used for PAO1/pBAD-yhjH strain. For biofilm EDA, the 100 units/ml cellulase (from Trichoderma reesei, Sigma-Aldrich, Germany) and 12.5 units/ml pectinase (from Aspergillus aculeatus, Sigma-Aldrich, Germany) were employed as biofilm disrupting agents. The syringe pump flowed the chemicals into the device at a constant speed for 2 h at 37 °C. Biofilm-released bacterial cells were collected from the waste outlet for further analysis.
Microscopy and video acquisition of biofilms
All microscopic images of biofilms and dispersed cells were captured by a Leica TCS SP8 MP Multiphoton/Confocal Microscope system using a 10× objective and secondary lens zoom of 40× to monitor brightfield and GFP fluorescence in Z-stacks. For closer observation of biofilm-released PAO1/pcdrA-gfp cells, these cells were collected from the device and directly observed using the Leica TCS SP8 MP Multiphoton/Confocal Microscope system with a 63× objective. ImageJ, LAS X, and Imaris software (Bitplane AG, Zurich, Switzerland) were used to process the images. The formula used to quantify GFP fluorescence levels was corrected total cell fluorescence (CTCF) = integrated density − (area of selected cells × mean fluorescence of background readings).
Bacterial biofilm video capturing
Time-lapse photography was performed using the biofilm tracking function in Imaris software and ImageJ according to the manufacturer’s instructions. Cell movement trajectories and associated data such as average fluorescent intensities and average velocities were exported for analysis.
Mathematical modeling
Mathematical modeling of the bacterial fluorescence signals comprises video processing using a Python program and data processing using MATLAB. The modeling program can be divided into two parts: 1. video processing and 2. data processing. During video processing, the program grayscales the photo, and then counts all the pixels with brightness. Most of the background noise is at 0–30, while Fig. 2A showed that most of the fluorescence signal was within range of 60–90.
For data processing, the program will count the number of white pixels in each image and save the value into a txt file. The program would normalize the starting value to the same intensity during data processing and use cftool in MATLAB to fit data. The regression formulas used by MATLAB are polynomial and exponential regression, as follows.
First-order polynomial regression:
| 1 |
Second-order polynomial regression:
| 2 |
Fourth-order polynomial regression:
| 3 |
Exponential regression:
| 4 |
a, b, c, d and e are the parameter that needs to be fitted. x is the number of each frame in video, y is the intensity after normalization.
The R2 is a value to measure how well a model can predict the data. The higher the value of R2, the better the model is at predicting the data. The value of R2 can be calculate as the equation below:
| 5 |
the calculated values of regression equation, is the mean value of y.
Quantification of bacterial numbers by colony-forming units (CFU)
Biofilm-released bacteria were collected directly from the waste outlet of the secondary chamber. 1 ml of 0.9% NaCl (w/v) saline was flushed repeatedly into each chamber to dislodge biofilms and retrieve biofilm bacteria from primary and secondary chambers. Biofilm cells were then homogenized by sound sonication (Elmasonic P120H, power = 50%, frequency = 37 KHz) in an ice-cold water bath for 15 min, followed by vigorous vortexing for 5 min. As previously described [28], cell suspensions were serially diluted in saline and transferred to LB agar plates (5 replicates) for incubation for 16 h at 37 °C. Colonies were enumerated, where the CFU ml−1 is tabulated by (colony number × dilution factor)/volume.
Quantification of c-di-GMP levels in bacterial biofilms by ELISA
As described previously [29], bacterial samples were collected and then sonicated at 40% amplitude for 5 min through a sonicate machine (SFX 550, SSE-1, Branson, Emerson, USA) with 45 s on/60 s off output to lyse bacteria. The c-di-GMP concentration was tested by a c-di-GMP ELISA kit (LMAI, Shanghai, China) according to the manufacturer’s protocol and measured at OD490 by a microplate reader (Tecan, Infinite M1000 Pro, Switzerland).
Isolation and quantification of exopolysaccharide concentration
As previously described [58], Psl was extracted by growing ΔpelA/plac-YedQ static biofilms on standard Petri dishes containing 15 ml LB supplemented with appropriate antibiotics at 37 °C for 16 h. The biofilms were centrifuged for separation from the supernatant at 10,000 × g for 5 min. The cell pellet was resuspended in 5 ml of 0.9% NaCl and subjected to mild water-bath sonication (Elmasonic P120H, power = 50%, frequency = 37 kHz, 5 min) to separate the cells from the surface-associated matrix. Centrifugation separated the cells from the matrix, leaving the crude matrix extract behind in the supernatant. The crude extract was then further treated by removing eDNA by precipitation with 25% ethanol and 0.1 M CaCl2, followed by degradation of extracellular proteins by 0.5 mg ml−1 Proteinase K at 60 °C for 1 h and inactivation at 80 °C for 30 min. The crude extract was then filtered with a centrifugal filter (<3 kDa) to remove the metabolites. The extract was then lyophilized and resuspended in sterile ddH2O. The polysaccharide concentration was determined by the phenol-sulfuric acid colorimetric method [59].
Biofilm disruption assay in 24 wells plate
Bacteria were cultivated in 1 ml ABTGC medium in each well of a 24-well cell culture plate (SPL Life Science Co., Ltd) at 37 °C for 12 h for biofilm formation. The biofilms were washed trice with 0.9% NaCl to remove planktonic bacteria, followed by pectinase treatment at various concentrations in an ABTGC medium. After further incubation at 37 °C for 12 h, the biofilms were washed three times with 0.9% NaCl, scraped from the well surface with a cell scraper, and resuspended in 1 ml 0.9% NaCl for CFU assay.
Exogenous addition and coating of exopolysaccharides on microwells
The Pel and Psl exopolysaccharides were extracted as described previously [42]. The extracted 10 ug ml−1 Pel or Psl were evenly coated on the surface of the microwells at 4 °C for 16 h and dried. The ΔpelAΔpslBCD/plac-gfp biofilm was then cultivated on the microwells coated with Pel or Psl for further analysis.
Cultivation of human lung fibroblasts and 3D spheroids
Human lung fibroblasts (ATCC PCS-201-013, HLF) were cultivated in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) at a density of 1.0 × 106 cells ml−1 in a T25 flask (SPL, Korea) at 37 °C, 5% CO2, 99% humidity for 72 h. For the generation of 3D spheroids as previously described [60], the cell droplets (20 μl) were placed on the lid of a hydrated 60 × 15 mm cell culture dish and incubated at 37 °C, 5% CO2, 99% humidity for 24 h. The spheroids were transferred into the secondary chamber of the BDR platform using a sterile syringe and needle, followed by incubation at 37 °C, 5% CO2, and 99% humidity for 20 min to enable attachment to the bottom of microwells.
Infection of human lung spheroids by biofilm-released bacteria
After the biofilm growth and bacteria dispersion in the primary chamber, the gfp-tagged bacterial cells were introduced into the secondary chamber containing the lung spheroids at a flow rate of 4 ml hr−1. The infection was established at 37 °C, 5% CO2, 99% humidity for 0 h, 2 h and 5 h. Lung spheroids were stained with 3 um CellTracker Deep Red Fluorescent Stain (Invitrogen, USA) and Propidium Iodide (PI, Invitrogen, USA) to observe the infection directly using the confocal microscope (Leica TCS SP8 MP) with a 40× objective. ImageJ, LAS X, and Imaris software (Bitplane AG, Zurich, Switzerland) were used to process the images. The fluorescence intensities of Deep Red and PI were measured using ImageJ. The PI fluorescence/CellTracker deep-red fluorescence ratio × 100% was quantified to tabulate the percentage of dead cells in the lung spheroids.
Cultivation of C. elegans
As previously described [42], the E. coli OP50 strain was prepared as nematode feed by growing the bacterial lawn on Nematode growth medium (NGM) agar plates at 37 °C for 16 h. The C. elegans N2 strain was cultivated on an OP50 lawn at room temperature for 72 h to expand the population.
Cultivation and infection of C. elegans in the BDR platform
Prior to the experiment, the L3 stage animals were introduced into the secondary chamber of the BDR platform with syringe injection, where at least one worm was introduced into each microwell of the channels. After biofilm growth and bacteria dispersion in the primary chamber, the gfp-tagged bacterial cells were flown into the secondary chamber containing the lung spheroids at a flow rate of 4 ml hr−1. The infection was established at 37 °C, 5% CO2, 99% humidity for 0 h, 3 h, 6 h and 12 h. The number of live (moving) and dead (non-motile) nematodes from the microwells were observed and tabulated under a stereomicroscope (Zeiss). For imaging, the C. elegans-biofilm model was observed using the confocal microscope (Leica TCS SP8 MP) with a 40× objective.
The live nematodes were collected from the secondary chamber and washed 3 times with 0.9% NaCl to quantify the PAO1 numbers in the animals. The animals were then ground by the microtube pellet pestle (Sigma-Aldrich, Germany) for release of bacteria and resuspension in 1 ml 0.9% NaCl. The bacterial suspension was serially diluted in 0.9% NaCl and plated on Pseudomonas isolation agar (PIA) for CFU assay.
Statistical analysis
The results were expressed as means ± standard deviation. Data groups were analyzed using the one-way ANOVA and Student’s t test to evaluate associations between independent variables, and the p values were calculated. Three independent trials were conducted in triplicates for each experiment, where the results were shown as the mean ± standard deviation.
Supplementary information
Supplementary Tables figures and video legends
Acknowledgements
This research is supported by The Hong Kong Polytechnic University, Department of Applied Biology and Chemical Technology Startup Grant (BE2B), Departmental General Research Fund (UALB), One-line account (ZVVV), Environmental and Conservation Fund (ECF-48/2019 and ECF-84/2021), Health and Medical Research Fund (HMRF-20190302), and State Key Laboratory of Chemical Biology and Drug Discovery Fund (1-BBX8). This work was also supported by the City University of Hong Kong [7005208,7005464,7020002,9610430,9667220]; Hong Kong Center for Cerebro- Cardiovascular Health Engineering (COCHE); Research Grants Council of the Hong Kong Special Administrative Region [21200921]; Pneumoconiosis Compensation Fund Board [9211276]; and the Hetao Shenzhen-Hong Kong Science and Technology Innovation Cooperation Zone Shenzhen Park Project (HZQB-KCZYZ-2021017).
Author contributions
SLC and BLK designed methods and experiments. YM and YD performed laboratory experiments and analyzed the data. YM, YD, BLK and SLC interpreted the results and wrote the paper. All authors have contributed to, seen, and approved the manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare the following financial interests/personal relationships, which may be considered as potential competing interests: One or more authors have a pending patent related to this work.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yanlin Deng, Haojun Hua.
Contributor Information
Bee Luan Khoo, Email: blkhoo@cityu.edu.hk.
Song Lin Chua, Email: song-lin.chua@polyu.edu.hk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01446-5.
References
- 1.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–73. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 2.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol. 2012;10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 3.Fleming D, Rumbaugh K. The consequences of biofilm dispersal on the host. Sci Rep. 2018;8:10738. doi: 10.1038/s41598-018-29121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolcott R. Biofilm and catheter-related bloodstream infections. Br J Nurs. 2021;30:S4–s9. doi: 10.12968/bjon.2021.30.8.S4. [DOI] [PubMed] [Google Scholar]
- 5.Minasyan H. Sepsis: mechanisms of bacterial injury to the patient. Scand J Trauma Resusc Emerg Med. 2019;27:19. doi: 10.1186/s13049-019-0596-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin B, Fei C, Bridges AA, Mashruwala AA, Stone HA, Wingreen NS, et al. Cell position fates and collective fountain flow in bacterial biofilms revealed by light-sheet microscopy. Science. 2020;369:71–7. doi: 10.1126/science.abb8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paula AJ, Hwang G, Koo H. Dynamics of bacterial population growth in biofilms resemble spatial and structural aspects of urbanization. Nat Commun. 2020;11:1354. doi: 10.1038/s41467-020-15165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Petrova OE, Su S, Lau GW, Panmanee W, Na R, et al. BdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of Pseudomonas aeruginosa. PLOS Pathog. 2014;10:e1004168. doi: 10.1371/journal.ppat.1004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–53. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rumbaugh KP, Sauer K. Biofilm dispersion. Nat Rev Microbiol. 2020;18:571–86. doi: 10.1038/s41579-020-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu M, Chua SL. Demolishing the great wall of biofilms in Gram-negative bacteria: To disrupt or disperse? Med Res Rev. 2020;40:1103–16. doi: 10.1002/med.21647. [DOI] [PubMed] [Google Scholar]
- 12.Guilhen C, Forestier C, Balestrino D. Biofilm dispersal: multiple elaborate strategies for dissemination of bacteria with unique properties. Mol Microbiol. 2017;105:188–210. doi: 10.1111/mmi.13698. [DOI] [PubMed] [Google Scholar]
- 13.Chua SL, Liu Y, Yam JKH, Chen Y, Vejborg RM, Tan BGC, et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun. 2014;5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 14.Goodwine J, Gil J, Doiron A, Valdes J, Solis M, Higa A, et al. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Sci Rep. 2019;9:3763. doi: 10.1038/s41598-019-40378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu S, Su T, Wu H, Liu S, Wang D, Zhao T, et al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 2015;25:1352–67. doi: 10.1038/cr.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martí M, Trotonda MP, Tormo-Más MÁ, Vergara-Irigaray M, Cheung AL, Lasa I, et al. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 2010;12:55–64. doi: 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Hall-Stoodley L, Nistico L, Sambanthamoorthy K, Dice B, Nguyen D, Mershon WJ, et al. Characterization of biofilm matrix, degradation by DNase treatment and evidence of capsule downregulation in Streptococcus pneumoniae clinical isolates. BMC Microbiol. 2008;8:173. doi: 10.1186/1471-2180-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reffuveille F, de la Fuente-Núñez C, Mansour S, Hancock RE. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob Agents Chemother. 2014;58:5363–71. doi: 10.1128/AAC.03163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crone S, Vives-Flórez M, Kvich L, Saunders AM, Malone M, Nicolaisen MH, et al. The environmental occurrence of Pseudomonas aeruginosa. APMIS. 2020;128:220–31. doi: 10.1111/apm.13010. [DOI] [PubMed] [Google Scholar]
- 20.Wheatley RM, Caballero JD, van der Schalk TE, De Winter FH, Kapel N, Recanatini C, et al. Assessing the contribution of gut-to-lung translocation to bacterial colonization and antibiotic resistance in an ICU patient. medRxiv. 2022, https://www.medrxiv.org/content/10.1101/2022.01.17.22269403v1. [DOI] [PMC free article] [PubMed]
- 21.Chuang C-H, Wang Y-H, Chang H-J, Chen H-L, Huang Y-C, Lin T-Y, et al. Shanghai fever: a distinct Pseudomonas aeruginosa enteric disease. Gut. 2014;63:736–43. doi: 10.1136/gutjnl-2013-304786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerioli M, Batailler C, Conrad A, Roux S, Perpoint T, Becker A, et al. Pseudomonas aeruginosa implant-associated bone and joint infections: experience in a regional reference center in France. Front Med. 2020;7:513242. doi: 10.3389/fmed.2020.513242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah NB, Osmon DR, Steckelberg JM, Sierra RJ, Walker RC, Tande AJ, et al. Pseudomonas prosthetic joint infections: a review of 102 episodes. J Bone Jt Infect. 2016;1:25–30. doi: 10.7150/jbji.15722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Delden C. Pseudomonas aeruginosa bloodstream infections: how should we treat them? Int J Antimicrobial Agents. 2007;30:71–5. doi: 10.1016/j.ijantimicag.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Jugulete G, Luminos M, Visan A, Draganescu A, Merisescu M, Vasile M, et al. Severe sepsis with multiple organ dysfunctions caused by Pseudomonas aeruginosa in an immunocompetent child. Crit Care. 2012;16:P69. [Google Scholar]
- 26.Stoner SN, Baty JJ, Scoffield JA. Pseudomonas aeruginosa polysaccharide Psl supports airway microbial community development. ISME J. 2022;16:1730–9. doi: 10.1038/s41396-022-01221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jennings LK, Dreifus JE, Reichhardt C, Storek KM, Secor PR, Wozniak DJ, et al. Pseudomonas aeruginosa aggregates in cystic fibrosis sputum produce exopolysaccharides that likely impede current therapies. Cell Rep. 2021;34:108782. doi: 10.1016/j.celrep.2021.108782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts AEL, Kragh KN, Bjarnsholt T, Diggle SP. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol. 2015;427:3646–61. doi: 10.1016/j.jmb.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Sternberg C, Tolker-Nielsen T. Growing and analyzing biofilms in flow cells. Curr Protoc Microbiol. 2006;Chapter 1:Unit 1B.2. 10.1002/9780471729259.mc01b02s00. [DOI] [PubMed]
- 30.Khoo BL, Shang M, Ng CH, Lim CT, Chng WJ, Han J. Liquid biopsy for minimal residual disease detection in leukemia using a portable blast cell biochip. NPJ Precis Oncol. 2019;3:30. doi: 10.1038/s41698-019-0102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khoo BL, Grenci G, Jing T, Lim YB, Lee SC, Thiery JP, et al. Liquid biopsy and therapeutic response: Circulating tumor cell cultures for evaluation of anticancer treatment. Sci Adv. 2016;2:e1600274. doi: 10.1126/sciadv.1600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng Y, Fu Y, Chua SL, Khoo BL. Biofilm Potentiates Cancer‐Promoting Effects of Tumor‐Associated Macrophages in a 3D Multi‐Faceted Tumor Model. Small 2023;19:2205904. [DOI] [PubMed]
- 33.Liu YS, Deng Y, Chen CK, Khoo BL, Chua SL. Rapid detection of microorganisms in a fish infection microfluidics platform. J Hazard Mater. 2022;431:128572. doi: 10.1016/j.jhazmat.2022.128572. [DOI] [PubMed] [Google Scholar]
- 34.Shigematsu M, Meno Y, Misumi H, Amako K. The measurement of swimming velocity of Vibrio cholerae and Pseudomonas aeruginosa using the video tracking methods. Microbiol Immunol. 1995;39:741–4. doi: 10.1111/j.1348-0421.1995.tb03260.x. [DOI] [PubMed] [Google Scholar]
- 35.Chua SL, Tan SY, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, et al. Bis-(3’-5’)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:2066–75. doi: 10.1128/AAC.02499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–43. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrova OE, Sauer K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci. 2012;109:16690–5. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chua SL, Ding Y, Liu Y, Cai Z, Zhou J, Swarup S, et al. Reactive oxygen species drive evolution of pro-biofilm variants in pathogens by modulating cyclic-di-GMP levels. Open Biol. 2016;6:160162. doi: 10.1098/rsob.160162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colvin KM, Irie Y, Tart CS, Urbano R, Whitney JC, Ryder C, et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 2012;14:1913–28. doi: 10.1111/j.1462-2920.2011.02657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chimenti I, Pagano F, Angelini F, Siciliano C, Mangino G, Picchio V, et al. Human lung spheroids as in vitro niches of lung progenitor cells with distinctive paracrine and plasticity properties. Stem Cells Transl Med. 2017;6:767–77. doi: 10.5966/sctm.2015-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fong ELS, Toh TB, Yu H, Chow EK-H. 3D culture as a clinically relevant model for personalized medicine. SLAS Technol. 2017;22:245–53. doi: 10.1177/2472630317697251. [DOI] [PubMed] [Google Scholar]
- 42.Chan SY, Liu SY, Seng Z, Chua SL. Biofilm matrix disrupts nematode motility and predatory behavior. ISME J. 2021;15:260–9. doi: 10.1038/s41396-020-00779-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Liu SY, Chan SY, Chua SL. Biofilm matrix cloaks bacterial quorum sensing chemoattractants from predator detection. ISME J. 2022;16:1388–96. doi: 10.1038/s41396-022-01190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark AS, Huayta J, Arulalan KS, San-Miguel A. Chapter 13 - Microfluidic devices for imaging and manipulation of C. elegans. In: Liu X, Sun Y, editors. Micro and nano systems for biophysical studies of cells and small organisms. Academic Press; 2021. p. 295–321.
- 45.Letizia MC, Cornaglia M, Trouillon R, Sorrentino V, Mouchiroud L, Bou Sleiman MS, et al. Microfluidics-enabled phenotyping of a whole population of C. elegans worms over their embryonic and post-embryonic development at single-organism resolution. Microsyst Nanoengineering. 2018;4:6. doi: 10.1038/s41378-018-0003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng Y, Liu SY, Chua SL, Khoo BL. The effects of biofilms on tumor progression in a 3D cancer-biofilm microfluidic model. Biosens Bioelectron. 2021;180:113113. doi: 10.1016/j.bios.2021.113113. [DOI] [PubMed] [Google Scholar]
- 47.Straub H, Eberl L, Zinn M, Rossi RM, Maniura-Weber K, Ren Q. A microfluidic platform for in situ investigation of biofilm formation and its treatment under controlled conditions. J Nanobiotechnology. 2020;18:166. doi: 10.1186/s12951-020-00724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christensen LD, Gennip MV, Rybtke MT, Wu H, Chiang W-C, Alhede M, et al. Clearance of Pseudomonas aeruginosa Foreign-Body Biofilm Infections through Reduction of the Cyclic Di-GMP Level in the Bacteria. Infect Immun. 2013;81:2705–13. doi: 10.1128/IAI.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dang H, Lovell CR. Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev. 2016;80:91–138. doi: 10.1128/MMBR.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin Z, Yang X, Yang L, Jiang J, Ou Y, Molin S, et al. Formation and properties of in vitro biofilms of ica-negative Staphylococcus epidermidis clinical isolates. J Med. Microbiol. 2007;56:83–93. doi: 10.1099/jmm.0.46799-0. [DOI] [PubMed] [Google Scholar]
- 51.Weaver WM, Milisavljevic V, Miller JF, Di Carlo D. Fluid flow induces biofilm formation in Staphylococcus epidermidis polysaccharide intracellular adhesin-positive clinical isolates. Appl Environ Microbiol. 2012;78:5890–6. doi: 10.1128/AEM.01139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao T, Liu Y. N-acetylcysteine inhibit biofilms produced by Pseudomonas aeruginosa. BMC Microbiol. 2010;10:140. doi: 10.1186/1471-2180-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci. 2007;104:11197–202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao Y, Wang W, Tan F, Cai Y, Lu J, Qiao X. Influence of different ions doping on the antibacterial properties of MgO nanopowders. Appl Surf Sci. 2013;284:726–31. [Google Scholar]
- 55.Zhang T, Deng Y, Liu YS, Chua SL, Tang BZ, Khoo BL. Bacterial targeted AIE photosensitizers synergistically promote chemotherapy for the treatment of inflammatory cancer. Chem Eng J. 2022;447:137579. [Google Scholar]
- 56.Chen CK, Zhang J, Bhingarde A, Matotek T, Barrett J, Hardesty BD, et al. A portable purification system for the rapid removal of microplastics from environmental samples. Biochem Eng J. 2022;428:132614.
- 57.Liao JC, Zou SJ, Deng YL, Jiang Y, Chua SL, Khoo BL. Multivariate analysis of liquid biopsies for real-time detection of patients with biofilm-associated infections (BAI). Biochem Eng J. 2023:453;139595.
- 58.Liu SY, Leung MM-L, Fang JK-H, Chua SL. Engineering a microbial ‘trap and release’ mechanism for microplastics removal. Chem Eng J. 2021;404:127079. [Google Scholar]
- 59.Masuko T, Minami A, Iwasaki N, Majima T, Nishimura S, Lee YC. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem. 2005;339:69–72. doi: 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 60.Wang S, Chan SY, Deng Y, Khoo BL, Chua SL. Oxidative stress induced by Etoposide anti-cancer chemotherapy drives the emergence of tumor-associated bacteria resistance to fluoroquinolones. J Adv Res. 2023;S2090-1232(23)00062-0. 10.1016/j.jare.2023.02.011. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables figures and video legends
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.