Abstract
The TecA broad-spectrum chlorobenzene dioxygenase of Burkholderia sp. strain PS12 catalyzes the first step in the mineralization of 1,2,4,5-tetrachlorobenzene. The catabolic genes were localized on a small plasmid that belongs to the IncPβ incompatibility group. PCR analysis of the genetic environment of the tec genes indicated high similarity to the transposon-organized catabolic tcb chlorobenzene degradation genes of Pseudomonas sp. strain P51. Sequence analysis of the regions flanking the tecA genes revealed an upstream open reading frame (ORF) with high similarity to the todF 2-hydroxy-6-oxo-2,4-heptadienoate hydrolase gene of Pseudomonas putida F1 and a discontinuous downstream ORF showing high similarity to the todE catechol 2,3-dioxygenase gene of strain F1. Both homologues in strain P51 exist only as deletion remnants. We suggest that different genetic events thus led to inactivation of the perturbing meta-cleavage enzymes in strains P51 and PS12 during the evolution of efficient chlorobenzene degradation pathways. Biochemical characterization of TodF-like protein TlpF and a genetically refunctionalized TodE-like protein, TlpE, produced in Escherichia coli provided data consistent with the proposed relationships.
Considerable quantities of xenobiotics are released each year into the environment. Bacteria that are able to use many of these compounds as sole sources of carbon and energy have been isolated from natural habitats (33). The aerobic metabolism of aromatic compounds is frequently initiated by dioxygenases, followed by a dehydrogenation reaction catalyzed by a cis-dihydrodiol dehydrogenase to give catechols or substituted catechols (17), which serve as substrates for oxygenolytic cleavage of the aromatic ring (21). Chlorocatechols are usually channelled into the Krebs cycle by modified ortho-cleavage pathways, whereas methylcatechols are most commonly metabolized by meta-cleavage routes (27, 32, 34).
Experimental combination of genes encoding the first two enzymes of a toluene degradation pathway with those coding for chlorocatechol degradation produced a functional metabolic sequence for the mineralization of chlorobenzenes via a modified ortho pathway (31). It has been proposed that the 1,2,4-trichlorobenzene degrader Pseudomonas sp. strain P51 evolved by recruitment of the tod pathway genes of the toluene-degrading bacterium Pseudomonas putida F1, followed by mutational drift of the todCBA toluene dioxygenase and todD dehydrogenase genes to yield the tcb genes encoding the first two enzymes in the transformation of chlorobenzenes (40–44). Sequences flanking the tcb genes were suggested to be evolutionary remnants of the todE extradiol dioxygenase and todF hydrolase genes, which had become inactivated by major DNA deletions such that misrouting of catechol into the unproductive meta pathway would no longer be possible (44).
Although a number of chlorobenzene-degrading bacteria have been isolated over the past few years, very few, such as Burkholderia sp. strain PS12, are able to degrade tetrachlorobenzene (5). The evolution of such bacteria able to degrade higher chlorinated xenobiotics is of interest not only from the environmental and biotechnological points of view but also from the genetic point of view. We investigated the localization, organization, and sequences of genes that flank the tecAB chlorobenzene degradation genes of strain PS12, which led us to propose an evolutionary route from the tod toluene degradation gene to the tec and tcb chlorobenzene degradation genes.
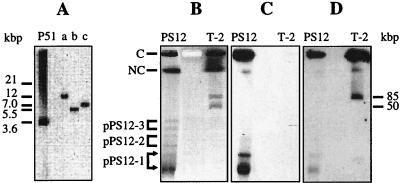
Southern analysis of BamHI-, BglII-, and double-digested PS12 total DNA revealed a single band, indicating that the tecA gene is present at only one locus (Fig. 1A). To analyze whether the tec genes reside on a plasmid, PS12 total DNA prepared as described previously (38) was separated in a 0.9% agarose gel by pulsed-field gel electrophoresis (PFGE) (36). The presence of three plasmids, designated pPS12-1, pPS12-2, and pPS12-3, all of which were present in two forms and all of which are smaller than the 50-kbp reference plasmid of Comamonas testosteroni T-2 used as a size marker (Fig. 1B), was observed. The tecA genes were localized by Southern blotting (Hybond N; Amersham) of the pulsed-field gel and hybridization under stringent conditions with the tecA1 gene probe derived by PCR amplification from plasmid pSTE7 with primers prSTB1 and prSTB2 (forward, atgaatcacaccgacacctcccct; reverse, tcagcgtgtggcgttcagcgcggc). Positive signals were obtained with the two forms of plasmid pPS12-1 (Fig. 1C).
FIG. 1.
Southern analysis of PS12 DNA. Total DNA of 1,2,4,5-tetrachlorobenzene-grown Burkholderia sp. strain PS12 (lane PS12) was digested with BamHI (lane a), BamHI-BglII (lane b), and BglII (lane c) and electrophoretically separated on an agarose gel. The tcbAaAbAcAd dioxygenase genes (lane P51) from Pseudomonas sp. strain P51 were amplified from plasmid pTBCB60 (42) with primers prSTB1 and prSTB4 (forward, atgaatcacaccgacacctcccct [the tecA1 start codon is in boldface]; reverse, tcatgctgagtctccttgttgtgc [the tcbAd stop codon is in boldface]) and were used as a positive control. The gel was analyzed by Southern hybridization with the tecA1 gene probe of PS12 (A). Total DNAs from strain PS12 and toluenesulfonate-grown C. testosteroni T-2 (lane T-2) were subjected to PFGE, stained with ethidium bromide (B), and analyzed by Southern hybridization with the tecA1 gene probe of PS12 (C) and the IncPβ-specific trfA2 gene probe derived from plasmid R751 (D). The bracket arrow indicates the positions of the two PS12-1 plasmid species which gave signals with both probes. The positions of other plasmids are indicated by horizontal bars. The reference plasmids of strain T-2 (pTSA [85 kb, IncPβ] and pT2T [50 kb, unknown incompatibility group]) served as DNA molecular size markers and as controls for probe specificity in Southern experiments. In addition, the positions of the slots containing chromosomal (C) and nicked chromosomal (NC) DNAs are indicated. Excess blank lanes between the PS12 and T-2 samples were digitally removed with Photoshop software (Adobe).
Plasmids of the IncP incompatibility group often carry catabolic genes. In order to determine if strain PS12 contains an IncP plasmid and to classify it, PCR was carried out on PS12 total DNA and reference plasmids RP4 (IncPα) (10) and R751 (IncPβ) (26) by using as templates the previously described conserved primers prSTB59 and prSTB60 (forward, cgaaattcrtrtgggagaagta; reverse, cgyttgcaatgcaccaggtc); prSTB61 and prSTB62 (forward, atgaagaaacggctnaccga; reverse, ttcctgtttyytcttggcgtc), and prSTB63 and prSTB64 (forward, cagcctcgcagagcaggat; reverse, cagccgggcaggataggtgaagt) (18), which are based on replicon-specific DNA regions (trfA2, korA, and oriT). Pairwise sequence alignments (data not shown) of all three PS12-derived PCR products showed higher similarities to the corresponding sequences of IncPβ plasmid R751 (90, 90, and 91% nucleotide sequence identity, respectively) than to those of IncPα plasmid RP4 (85, 77, and 70% nucleotide sequence identity, respectively).
The PFGE Southern blot membrane, after removal of tecA1, was reprobed under stringent conditions with the IncPβ-specific trfA2 probe. The probe hybridized strongly with the positive control (85-kbp plasmid pTSA) and with both forms of plasmid pPS12-1 (Fig. 1D), indicating that the plasmid on which the tec genes reside belongs to the IncPβ subgroup.
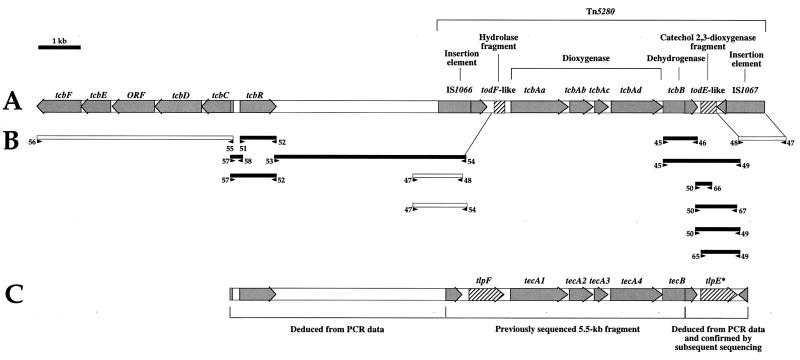
A PCR strategy was used to analyze the genetic organization flanking the previously described 5.5-kb gene cluster of Burkholderia sp. strain PS12 containing the tecA genes, an open reading frame (ORF) encoding a putative protein with similarity to the TodF hydrolase and another ORF for a truncated TecB dehydrogenase (5). Oligonucleotide primers were designed on the basis of known sequences from strain PS12 (5) and Pseudomonas sp. strain P51 (40, 41, 43, 44). The sizes of fragments obtained by PCR with genomic DNA from PS12 as the template (Fig. 2B) suggest an organization in the vicinity of the tec genes similar to that of transposon Tn5280 (43) (Fig. 2C). This transposon (Fig. 2A) harbors the genes encoding TcbA chlorobenzene dioxygenase and TcbB dehydrogenase of strain P51 (42, 44) in the vicinity of the chlorocatechol degradation genes (40) and a LysR-type regulator gene (41). Our experiments indicated that an entire tecB gene is located downstream of the tecA chlorobenzene dioxygenase genes. The distance between tecB and the end of a putative insertion element is approximately 0.4 kb more than was found in strain P51. This suggested the presence of a full-length version of a catechol 2,3-dioxygenase gene, in contrast to the structure in strain P51, which has only a gene remnant. Also, a complete tlpF hydrolase homologue was found upstream of the tecA genes, in contrast to the remnant found in strain P51.
FIG. 2.
PCR analysis of the genetic environment of the tecA genes of Burkholderia sp. strain PS12. The sequence context of a previously characterized 5.5-kb genomic DNA fragment from strain PS12 (5), which is similar to the corresponding sequence of Pseudomonas sp. strain P51 (A), was analyzed by PCR (B). The PS12 sequence comprises the tecA1A2A3A4 genes encoding the tetrachlorobenzene dioxygenase, the truncated tecB gene encoding the cis-chlorobenzene dihydrodiol dehydrogenase, the truncated IS1066 homologue, the tlpF 2-hydroxy-6-oxo-2,4-heptadienoate hydrolase gene, and the inactivated tlpE* catechol 2,3-dioxygenase pseudogene (C). The primers were designed on the basis of known sequences of Pseudomonas sp. strain P51 and Burkholderia sp. strain PS12. PCR products are drawn as black boxes, whereas white boxes indicate failure to obtain a product of the expected size. The numbers and arrows below the boxes refer to prSTB primer numbers and priming direction, respectively, as follows: prSTB45, atgaaactcaaaggtgaagtg, (containing the tecB start codon); prSTB46, tcaagcgaaatgcttgtcgag (containing the tecB stop codon [boldface]); prSTB47, ctagtgtttaattcgtcatg (containing the IS1066 inverted repeat [underlined]); prSTB48, ctagtgtttaattccgtaaattg (containing the IS1066 inverted repeat [underlined] and stop codon [boldface]); prSTB49, caatttacggaattaaacaatag (containing the IS1067 inverted repeat [underlined] and stop codon [boldface]); prSTB50, gctcgacaagcatttcgcttga (containing the tecB stop codon [boldface]); prSTB51, atggaattccggcagctcaag (containing the tcbR start codon [boldface]); prSTB52, tcagtccttcgcggatcgccgc (containing the tcbR stop codon [boldface]); prSTB53, gcggcgatccgcgaaggactga (containing the tcbR stop codon [boldface]); prSTB54, gatcgccgcaatgtggttgcat (containing nucleotides −60 to −82 of the tlpF gene); prSTB55, atgaacgaacgagtgaagcagg (containing the tcbC start codon [boldface]); prSTB56, gtcagggttgcggtggctcc (containing the tcbF stop codon [boldface]); prSTB57, cctgcttcactcgttcgttcat (containing the tcbC start codon [boldface]); prSTB58, cttgagagctgccggaattccat (containing the tcbR start codon [boldface]); prSTB65, catgagcattcaaagattgggctac (containing the todE start codon [boldface]); prSTB66, gtgacctaagccctggtctccag (containing the middle region of todE); prSTB67, gtcaggcgggcgcctggaac (containing the todE stop codon [boldface]). The deduced genetic environment of the 5.5-kb fragment of strain PS12 is indicated 5′ and 3′ with respect to the previously obtained sequence. Gene names are shown above the respective ORFs, which are shown as grey arrows. Hatched boxes indicate sequences exhibiting high similarity to the meta-cleavage pathway todE and todF genes of P. putida F1.
Biochemical analysis of the products of the tcb flanking regions in strain P51 was not possible due to the deleted sequences. However, the presence of complete flanking sequences in strain PS12 (Fig. 2C) permitted such studies to assess evolutionary relationships among strains PS12, P51, and F1.
The TecB dehydrogenase located downstream of the tecA gene and encoded on plasmid pCR12 (Table 1) has been shown to be active against several cis-chlorobenzene dihydrodiols (unpublished data). Downstream of the tecB gene is a 0.9-kb sequence which exhibits high similarity (82%) to the todE catechol 2,3-dioxygenase gene of P. putida F1 (45). This tlpE* (TodE-like protein E) pseudogene is, however, inactive, and the putative functional gene has apparently been inactivated by mutations introducing one frameshift and two premature translational stops. In order to determine whether corrections of these three defects would result in a functional dioxygenase, we repaired them with equivalent sequences from the todE gene by three successive rounds of splicing by overlap extension-PCR (25) using pCR12 as the template and appropriate overlapping oligonucleotide primers prSTB116 and prSTB118 (forward, gaaggagagacaacatgagcattcaaagg [the tlpE* start codon is in boldface]; reverse, ctgaccgatagccgccaggtgcgcg [the codon for Trp that replaces the tlpE* stop codon is in boldface]), prSTB117 and prSTB120 (forward, ccggatcgactcgcgcacctggcggctatc [with the tlpE* stop codon replaced with a codon for Trp [boldface]); reverse, gccgaacgggtccgtacaggaaataag [with the tlpE* frameshift cgct replaced with cg-t [boldface]); prSTB119 and prSTB122 (forward, gggcttatttcctgtacggacccgttc [with the tlpE* frameshift agcg replaced with a-cg [boldface]; reverse, cccagcccttggtatagaaggcgag with the tlpE* stop codon replaced with a codon for Tyr [boldface]); and prSTB121 and prSTB123 (forward, cagcgctcgccttctataccaaggggc [with the tlpE* stop codon replaced with a codon for Tyr [boldface]; reverse, tcatgcgggcggctggaacttgtgc [with the tlpE* stop codon [boldface]), containing the corresponding todE wild-type sequences of strain F1. Gel-purified PCR products served as megaprimers in the subsequent amplification reactions. The resulting refunctionalized catechol 2,3-dioxygenase gene, designated tlpE (TodE-like protein E), was subsequently cloned into pCR2.1 to produce plasmid pCR18 and subcloned into pBluescript II KS(+) to produce plasmid pSTE56.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Sources, reference(s), or structure |
|---|---|---|
| Strains | ||
| E. coli DH5α | deoR endA1 gyrA96 hsdR17 (rK− mK+) | Clontech |
| DH5α | recA1 relA1 supE44 thi-1 Δ(lacZYA-argFV169) φ80δlacZΔM15 F− λ− | |
| E. coli J53 | Met Pro; host for IncP reference plasmids | 9 |
| Burkholderia sp. strain PS12 | Growth on 1,2,4,5-tetrachlorobenzene | 5, 37 |
| C. testosteroni T-2 | pTSA (85 kb, IncPβ), pT2T (50 kb) | 39 |
| Host vectors | ||
| pCR2.1 | Apr Kmr | Invitrogen |
| pBluescript II KS(+) | Apr | Stratagene |
| Plasmids used for cloning and genetic refunctionalization of tlpE* catechol 2,3-dioxygenase pseudogene | ||
| pCR12 | tecB dehydrogenase gene and tlpE* pseudogene | PCR-amplified 1.8-kb fragment from PS12 total DNA with primers prSTB45 and prSTB49 in pCR2.1 |
| pCR18 | tlpE catechol 2,3-dioxygenase gene | 0.9-kb fragment obtained from third round of splicing by overlap extension-pCR in pCR2.1 |
| pSTE3 | tlpF hydrolase gene, tecA1A2A3A4 chlorobenzene dioxygenase gene, 3′-truncated tecB dehydrogenase gene | 5 |
| pSTE56 | Expression of tlpE catechol 2,3-dioxygenase gene | 1.0-kb KpnI-XbaI fragment of pCR18 inserted into KpnI-XbaI site of pBluescript II KS(+) |
| Reference plasmids used for incompatibility group determination | ||
| RP4 | 60 kb; IncPα; Apr Kmr Tcr | 10 |
| R751 | 53 kb; IncPβ; Tpr | 26 |
Antibiotic resistances (solutions in milligrams per liter): Apr, ampicillin (100); Kmr, kanamycin (50); Tcr, tetracycline (20); Tpr, trimethoprim (20). Met, methionine dependent; Pro, proline dependent.
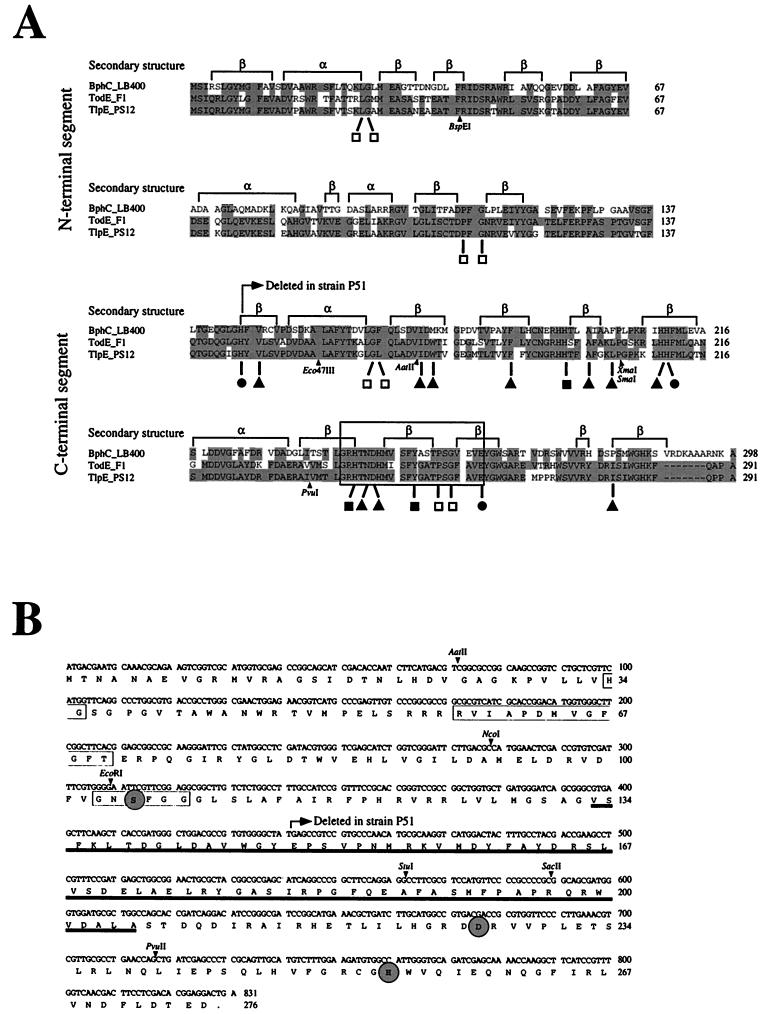
The polypeptide sequences of the refunctionalized TlpE catechol 2,3-dioxygenase and the TodE enzyme exhibited 83% identity and clustered phylogenetically with meta-cleavage enzymes preferring polycyclic aromatic substrates (14). Comparison of the polypeptide sequence with the sequence (24) of the corresponding structure-solved BphC enzyme (14, 19) from Burkholderia sp. strain LB400 (6) revealed that all residues that are iron ligands or play a structural and direct catalytic role (Fig. 3A) are conserved.
FIG. 3.
Sequence alignment and structural features of catechol 2,3-dioxygenases and nucleotide and deduced amino acid sequences of the TlpF hydrolase. (A) Alignment of extradiol dioxygenases of Burkholderia sp. strain LB400 (BphC_LB400) and P. putida F1 (TodE_F1) and the refunctionalized TlpE protein of Burkholderia sp. strain PS12 (TlpE_PS12). Based on the solved structure of the BphC enzyme, two domains (N and C terminal) with similar secondary structures can be distinguished (19), as indicated above the alignment. Identical residues are shaded. The amino acid ligands of the catalytic Fe(II) are marked (•), and those playing a direct catalytic role in the LB400 enzyme are indicated (■). Additional residues that form the substrate binding site in the LB400 enzyme are marked (▴), and conserved residues that play a structural role are indicated (□). The boxed fingerprint region contains the consensus sequence (G or N or T or I or V)-X1-H-X5 or 7-(L or I or V or M or F)-Y-X2-(D or E or N or T or A)-P-X1-(G or P)-X3 or 4-E (14), where Xn indicates n residues of any type, parentheses enclose residues found at one position, and boldface letters indicate the residues found in the TlpE protein. The positions of relevant restriction sites of the corresponding tlpE gene are indicated. (B) The dipeptide His-Gly of the putative oxanion hole, the RVIAPDXXGXGXS motif, and the so-called hydrolase or lipase box with the nucleophile motif Gly103-Xaa-Ser105-Xaa-Xaa-Gly108 (3, 11, 12) of the TlpF hydrolase are boxed. The catalytic residues Ser105, Asp226, and His254, representing the catalytic triad of hydrolases (2, 12, 29) and lipases (8, 11), are circled and in boldface. The region proposed to be involved in determination of substrate specificity (12) is underlined. Relevant restriction sites are indicated.
Cell extracts of Escherichia coli DH5α(pSTE56) cultures grown overnight in Luria-Bertani medium (36) containing 0.1-mg/ml ampicillin and 1.0 mM isopropyl-β-d-thiogalactopyranoside were prepared as previously described, and the protein concentration was determined (7). Enzyme activities were assayed spectrophotometrically by monitoring the product formation from catechol (2-hydroxymuconic semialdehyde [2HMSA]; ɛ375 = 36 mM−1 cm−1), 3-methylcatechol (2-hydroxy-6-oxohepta-2,4-dienoate [HOHDA]; ɛ388 = 16.8 mM−1 cm−1), or 2,3-dihydroxybiphenyl (2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate [HOPDA]; ɛ434 = 13.2 mM−1 cm−1) (15, 35), respectively, in phosphate buffer (50 mM, pH 7.5).
Refunctionalized TlpE* catechol 2,3-dioxygenase was able to oxidize catechol, 3-methylcatechol, and 2,3-dihydroxybiphenyl to products with the expected absorption maxima at 376, 388, and 436 nm, respectively. The highest Vmax was obtained with 3-methylcatechol (1,200 ± 60 μM min−1 g−1), followed by catechol (715 ± 16 μM min−1 g−1) and 2,3-dihydroxybiphenyl (365 ± 38 μM min−1 g−1), which is the same order found with the TodE of strain F1 (23). However, TlpE dioxygenase shows the highest substrate preference, expressed by the relative specificity constant Vmax/Km (16), for 2,3-dihydroxybiphenyl (given as 100), followed by 3-methylcatechol (65) and catechol (0.6), which reflects the low Km values for 3-methylcatechol (1 ± 0.2 μM) and especially 2,3-dihydroxybiphenyl (0.2 ± 0.1 μM) compared to catechol (57 ± 4 μM). As high concentrations of 3-methylcatechol inhibited the TlpE enzyme, a substrate inhibition model (1, 22) was used to calculate the kinetic parameters of 3-methylcatechol oxidation. The inhibition constant Kss was determined to be 130 ± 25 μM. Transformation of 3-chlorocatechol by the TlpE dioxygenase was not observed, although 67 nM 3-chlorocatechol reduced the initial oxidation rate of 50 μM catechol by 50% and 1 μM 3-chlorocatechol completely abolished catechol transformation (4, 28). This inhibition indicates that its original role was in the metabolism of methylbenzenes rather than chlorobenzenes.
An ORF identified upstream of the tecA gene had high similarity, with 87 and 89% identity on the nucleotide and polypeptide levels, respectively (5), to the todF gene from P. putida F1 (30). This gene, designated tlpF, is very similar to the corresponding meta-cleavage pathway gene fragment found in Pseudomonas sp. strain P51 (99.5% similarity in 201 nucleotides) (44). Pairwise sequence comparison with other bacterial hydrolases involved in the degradation of aromatic compounds and a human serine hydrolase indicates that the TlpF hydrolase belongs to the group of serine hydrolases involved in the meta-cleavage pathway for mononuclear aromatics (Fig. 3B).
Functional expression of the recombinant TlpF HOHDA hydrolase in E. coli DH5α was shown by the conversion of 2HMSA, HOHDA, and HOPDA in reaction mixtures containing an extract of E. coli DH5α(pSTE3) cells prepared as described above for E. coli DH5α(pSTE56) cells. The meta-cleavage products which were used as TlpF substrates were produced from catechol, 3-methylcatechol, and 2,3-dihydroxybiphenyl by transformation of 1 mM solutions in phosphate buffer with E. coli DH5α(pSTE56) cells grown and washed as described above and resuspended to an A600 of 10.
The highest Vmax was obtained with HOHDA (3,350 ± 250 μM min−1 g−1), followed by 2HMSA (800 ± 40 μM min−1 g−1), which is the same order found with the 2-hydroxymuconic semialdehyde hydrolase from TOL plasmid pWW0 of P. putida mt-2 (13). The Km values were 4.4 ± 1.1 and 70 ± 5 μM, respectively. Hydrolysis of HOPDA (<5 μM min−1 g−1) was too low for kinetic analysis. In contrast to the substrate preference observed with TlpE dioxygenase, Tlp hydrolase prefers the monocyclic aromatic-derived substrates. Therefore, both sequence analysis and biochemical properties show that the TodF and TlpF proteins cluster with hydrolases involved in the degradation of monocyclic aromatics, whereas the TodE and TlpE enzymes cluster with dioxygenases involved in the degradation of bicyclic aromatics. If polycyclic aromatics that transform enzymes have evolved from those that oxidize monocyclic aromatics, as suggested by Harayama and Rekik (20), the TodF and TlpF hydrolases may be evolutionarily older than the TodE and TlpE dioxygenases. The tod operon itself may thus be a mosaic of genes derived from different pathways for mono- and bicyclic substrates.
In summary, both primary structure analysis and biochemical data strongly suggest that the tec and tcb chlorobenzene degradation genes are closely related and that they diverged rather recently. They seem to have their common origin in the tod genes (or a common precursor) of the toluene degradation pathway and have adapted for efficient chlorobenzene transformation. The flanking evolutionary gene relics are most likely descendants of the corresponding todE and todF genes. Characterization of these evolutionary relics revealed different mutational events—deletions in the case of P51 and point mutations leading to a frameshift and the introduction of stop codons in the case of PS12—having caused inactivation of the meta-cleavage genes disadvantageous for chlorobenzene degradation during subsequent divergence of the two strains.
Nucleotide sequence accession numbers.
The new nucleotide sequences presented here have been deposited in the GenBank database under accession no. AF073901 (trfA2), AF073902 (korA), and AF073903 (oriT) for the IncPβ-specific sequences of plasmid pPS12-1.
The sequence of the refunctionalized tlpE dioxygenase gene is available under GenBank accession no. AF073900. The previously deposited 5.5-kb sequence containing the tlpF HOHDA hydrolase, the tecA chlorobenzene dioxygenase, and the partial tecB cis-chlorobenzene dihydrodiol genes (U78099) has been updated and now includes the entire tecB gene and the tlpE* catechol 2,3-dioxygenase pseudogene.
Acknowledgments
This work was supported by contract BIO4-CT972040 of the BIOTECH program of the EC. K.N.T. expresses gratitude to the Fonds der Chemischen Industrie for generous support.
We thank E. R. B. Moore and his group for generous sequencing support and F. Junker for valuable suggestions concerning PFGE and for providing E. coli(RP4), E. coli(R751), and C. testosteroni T-2 cells. The assistance of I. Plumeier in this work is greatly appreciated. 3,4,6-Trichlorocatechol was kindly provided by H.-A. Arfmann. We are indebted to J. Armengaud and M. W. Klemba for helpful discussions and for critical reading of the manuscript.
REFERENCES
- 1.Adams R H, Huang C M, Higson F K, Brenner V, Focht D D. Construction of a 3-chlorobiphenyl-utilizing recombinant from an intergeneric mating. Appl Environ Microbiol. 1992;58:647–654. doi: 10.1128/aem.58.2.647-654.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad D, Fraser J, Sylvestre M, Larose A, Khan A, Bergeron J, Juteau J M, Sondossi M. Sequence of the bphD gene encoding 2-hydroxy-6-oxo-(phenyl/chlorophenyl)hexa-2,4-dienoic acid (HOP/cPDA) hydrolase involved in the biphenyl/polychlorinated biphenyl degradation pathway in Comamonas testosteroni: evidence suggesting involvement of Ser112 in catalytic activity. Gene. 1995;156:69–74. doi: 10.1016/0378-1119(95)00073-f. [DOI] [PubMed] [Google Scholar]
- 3.Arand M, Grant D F, Beetham J K, Friedberg T, Oesch F, Hammock B D. Sequence similarity of mammalian epoxide hydrolases to the bacterial haloalkane dehalogenase and other related proteins. Implication for the potential catalytic mechanism of enzymatic epoxide hydrolysis. FEBS Lett. 1994;338:251–256. doi: 10.1016/0014-5793(94)80278-5. [DOI] [PubMed] [Google Scholar]
- 4.Bartels A, Knackmuss H J, Reineke W. Suicide inactivation of catechol-2,3-dioxygenase from Pseudomonas putida mt-2 by 3-chlorocatechols. Appl Environ Microbiol. 1984;47:500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beil S, Happe B, Timmis K N, Pieper D H. Genetic and biochemical characterization of the broad spectrum chlorobenzene dioxygenase from Burkholderia sp. strain PS12: dechlorination of 1,2,4,5-tetrachlorobenzene. Eur J Biochem. 1997;247:190–199. doi: 10.1111/j.1432-1033.1997.00190.x. [DOI] [PubMed] [Google Scholar]
- 6.Bopp L H. Degradation of highly chlorinated PCBs by Pseudomonas sp. strain LB400. J Ind Microbiol. 1986;1:23–29. [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Brady L, Brzozowski A M, Derewenda Z S, Dodson E, Dodson G, Tolley S, Turkenburg J P, Christiansen L, Huge-Jensen B, Norskov L, Thim L, Menge U. A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature. 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 9.Clowes R C, Hayes W. Experiments in microbial genetics. Oxford, England: Blackwell Scientific Publications, Ltd.; 1968. [Google Scholar]
- 10.Datta N, Hedges R W, Shaw E J, Sykes R B, Richmond M H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971;108:1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derewenda Z S, Sharp A M. News from the interface: the molecular structures of triacylglyceride lipases. Trends Biochem Sci. 1993;18:20–25. doi: 10.1016/0968-0004(93)90082-x. [DOI] [PubMed] [Google Scholar]
- 12.Diaz E, Timmis K N. Identification of functional residues in a 2-hydroxymuconic semialdehyde hydrolase. A new member of the alpha/beta hydrolase-fold family of enzymes which cleaves carbon-carbon bonds. J Biol Chem. 1995;270:6403–6411. doi: 10.1074/jbc.270.11.6403. [DOI] [PubMed] [Google Scholar]
- 13.Duggleby C J, Williams P A. Purification and some properties of the 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase (2-hydroxymuconic semialdehyde hydrolase) encoded by the TOL plasmid pWW0 from Pseudomonas putida mt-2. J Gen Microbiol. 1986;132:717–726. [Google Scholar]
- 14.Eltis L D, Bolin J T. Evolutionary relationships among extradiol dioxygenases. J Bacteriol. 1996;178:5930–5937. doi: 10.1128/jb.178.20.5930-5937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eltis L D, Hofmann H-J, Hecht H, Lünsdorf H, Timmis K N. Purification and crystallization of 2,3-dihydroxybiphenyl 1,2-dioxygenase. J Biol Chem. 1993;268:2727–2732. [PubMed] [Google Scholar]
- 16.Fersht A. The basic equations of enzyme kinetics. In: Fersht A, editor. Enzyme structure and mechanism. W. C. San Francisco, Calif: Freeman & Co.; 1985. pp. 99–116. [Google Scholar]
- 17.Gibson D T, Subramanian V. Microbial degradation of aromatic hydrocarbons. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 181–252. [Google Scholar]
- 18.Götz A, Pukall R, Smit E, Tietze E, Prager R, Tschäpe H, van Elsas J D, Small K. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl Environ Microbiol. 1996;62:2621–2628. doi: 10.1128/aem.62.7.2621-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han S, Eltis L D, Timmis K N, Muchmore S W, Bolin J T. Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 20.Harayama S, Rekik M. Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J Biol Chem. 1989;264:15328–15333. [PubMed] [Google Scholar]
- 21.Harayama S, Timmis K N. Catabolism of aromatic hydrocarbons by Pseudomonas. In: Hopwood D A, Charter K, editors. Genetics of bacterial diversity. New York, N.Y: Academic Press, Inc.; 1989. pp. 151–174. [Google Scholar]
- 22.Higson F K, Focht D D. Bacterial metabolism of hydroxylated biphenyls. Appl Environ Microbiol. 1989;55:946–952. doi: 10.1128/aem.55.4.946-952.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirose J, Kimura N, Suyama A, Kobayashi A, Hayashida S, Furukawa K. Functional and structural relationship of various extradiol aromatic ring-cleavage dioxygenases of Pseudomonas origin. FEMS Microbiol Lett. 1994;118:273–277. doi: 10.1111/j.1574-6968.1994.tb06840.x. [DOI] [PubMed] [Google Scholar]
- 24.Hofer B, Eltis L D, Dowling D N, Timmis K N. Genetic analysis of a Pseudomonas locus encoding a pathway for biphenyl/polychlorinated biphenyl degradation. Gene. 1993;130:47–55. doi: 10.1016/0378-1119(93)90345-4. [DOI] [PubMed] [Google Scholar]
- 25.Horton R M. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- 26.Jobanputra R S, Datta N. Trimethoprim R factors in enterobacteria from clinical specimens. J Med Microbiol. 1974;7:169–177. doi: 10.1099/00222615-7-2-169. [DOI] [PubMed] [Google Scholar]
- 27.Kaschabek S R, Kasberg T, Müller D, Mars A E, Janssen D B, Reineke W. Degradation of chloroaromatics: purification and characterization of a novel type of chlorocatechol 2,3-dioxygenase of Pseudomonas putida GJ31. J Bacteriol. 1998;180:296–302. doi: 10.1128/jb.180.2.296-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klecka G M, Gibson D T. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida by 3-chlorocatechol. Appl Environ Microbiol. 1981;41:1159–1165. doi: 10.1128/aem.41.5.1159-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau P C, Garnon J, Labbe D, Wang Y. Location and sequence analysis of a 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase-encoding gene (bpdF) of the biphenyl/polychlorinated biphenyl degradation pathway in Rhodococcus sp. M5. Gene. 1996;171:53–57. doi: 10.1016/0378-1119(96)00025-x. [DOI] [PubMed] [Google Scholar]
- 30.Menn F M, Zylstra G J, Gibson D T. Location and sequence of the todF gene encoding 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase in Pseudomonas putida F1. Gene. 1991;104:91–94. doi: 10.1016/0378-1119(91)90470-v. [DOI] [PubMed] [Google Scholar]
- 31.Oltmanns R H, Rast H G, Reineke W. Degradation of 1,4-dichlorobenzene by enriched and constructed bacteria. Appl Microbiol Biotechnol. 1988;28:609–616. [Google Scholar]
- 32.Reineke W, Knackmuss H-J. Microbial degradation of haloaromatics. Annu Rev Microbiol. 1988;42:263–287. doi: 10.1146/annurev.mi.42.100188.001403. [DOI] [PubMed] [Google Scholar]
- 33.Ribbons D W, Eaton R W. Chemical transformation of aromatic hydrocarbons that support the growth of microorganisms. In: Chakrabarty A M, editor. Biodegradation and detoxification of environmental pollutants. Boca Raton, Fla: CRC Press, Inc.; 1982. pp. 60–79. [Google Scholar]
- 34.Rojo F, Pieper D H, Engesser K H, Knackmuss H J, Timmis K N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987;238:1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- 35.Sala-Trepat J M, Evans W C. The meta cleavage of catechol by Azotobacter species. 4-Oxalcrotonate pathway. Eur J Biochem. 1971;20:400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sander P, Wittich R-M, Fortnagel P, Wilkes H, Francke W. Degradation of 1,2,4-trichloro- and 1,2,4,5-tetrachlorobenzene by Pseudomonas strains. Appl Environ Microbiol. 1991;57:1430–1440. doi: 10.1128/aem.57.5.1430-1440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith C L, Warburton P E, Gaal A, Cantor C R. Analysis of genome organization and rearrangements by pulsed field gradient gel electrophoresis. In: Setlow J K, Hollaender A, editors. Genetic engineering. Vol. 8. New York, N.Y: Plenum Publishing Corp.; 1986. pp. 45–70. [Google Scholar]
- 39.Thurnheer T, Köhler T, Cook A M, Leisinger T. Orthanilic acid and analogues as carbon sources for bacteria: growth physiology and enzymatic desulphonation. J Gen Microbiol. 1986;132:1215–1220. [Google Scholar]
- 40.van der Meer J R, Eggen R I, Zehnder A J, de Vos W M. Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J Bacteriol. 1991;173:2425–2434. doi: 10.1128/jb.173.8.2425-2434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Meer J R, Frijters A C, Leveau J H, Eggen R I, Zehnder A J, de Vos W M. Characterization of the Pseudomonas sp. strain P51 gene tcbR, a LysR-type transcriptional activator of the tcbCDEF chlorocatechol oxidative operon, and analysis of the regulatory region. J Bacteriol. 1991;173:3700–3708. doi: 10.1128/jb.173.12.3700-3708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Meer J R, van Neerven A R, de Vries E J, de Vos W M, Zehnder A J. Cloning and characterization of plasmid-encoded genes for the degradation of 1,2-dichloro-, 1,4-dichloro-, and 1,2,4-trichlorobenzene of Pseudomonas sp. strain P51. J Bacteriol. 1991;173:6–15. doi: 10.1128/jb.173.1.6-15.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Meer J R, Zehnder A J, de Vos W M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J Bacteriol. 1991;173:7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werlen C, Kohler H P, van der Meer J R. The broad substrate chlorobenzene dioxygenase and cis-chlorobenzene dihydrodiol dehydrogenase of Pseudomonas sp. strain P51 are linked evolutionarily to the enzymes for benzene and toluene degradation. J Biol Chem. 1996;271:4009–4016. doi: 10.1074/jbc.271.8.4009. [DOI] [PubMed] [Google Scholar]
- 45.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]