Abstract
The yeast Saccharomyces cerevisiae has two separate genes (APT1 and APT2) that encode two potentially different forms of adenine phosphoribosyltransferase (APRT). However, genetic analysis indicated that only APT1 could code for a complementing activity. Cloning and expression of both the APT1 and APT2 genes in Escherichia coli showed that although discrete proteins (APRT1 and APRT2) were made by these genes, only APRT1 had detectable APRT activity. Northern and Western blot analyses demonstrated that only APT1 was transcribed and translated under normal physiological conditions in yeast. Phylogenetic analysis revealed that APRT1 and APRT2 are evolutionary closely related and that they arise from a gene duplication event. We conclude that APT1 is the functional gene in S. cerevisiae and that APT2 is a pseudogene.
In most organisms, purine nucleotides can be synthesized de novo by a series of 10 enzymatic reactions that lead to the formation of IMP (4, 5, 20). IMP can be further converted to AMP or GMP to satisfy the purine nucleotide requirements of a growing cell. Alternatively, free-purine bases can be directly recycled from the growth medium and incorporated into purine nucleotide pools. Both sets of reactions involve the action of phosphoribosyltransferases (PRTases), a group of enzymes that have in common the chemistry of transferring the ribose 5-phosphate from α-d-5-phosphoribosyl 1-pyrophosphate (PRPP) onto a nitrogenous base to form an N-riboside monophosphate and pyrophosphate (20). PRTases are essential not only for purine biosynthesis but also for the biosynthesis of pyrimidines, pyridines, histidine, and tryptophan (20).
Adenine PRTase (APRT) is the key enzyme in the direct recycling of free adenine into purine nucleotide pools. APRT catalyzes the Mg2+-dependent transfer of the phosphoribosyl group from PRPP to adenine to form the nucleotide AMP (2, 4, 8, 11, 14, 16, 19, 21, 28, 30). In humans, APRT deficiency causes the accumulation of a poorly soluble adenine derivative, 2,8-dihydroxy adenine, which leads to urolithiasis and renal failure (27).
Typically, APRTs are encoded by constitutively expressed single-copy genes (22). However, two genes encoding two forms of APRT in Arabidopsis thaliana that differ in their nucleotide specificities have recently been reported (18, 25). We have previously identified a gene coding for APRT in Saccharomyces cerevisiae (APT1) (2). In addition, Yuryev and Corden reported the sequence of another potential APT gene in yeast (APT2), based on sequence comparison analysis (32). The finding of two different genes that may encode APRT in yeast has led us to propose that more than one form of this enzyme may also occur in yeast.
To help understand the relationship between APT1 and APT2 and address the possibility that two forms of the enzyme also occur in S. cerevisiae, both genes were used in complementation studies with yeast mutants deficient in APRT activity. Mutations disrupting APT1 and APT2 were made. Whereas disruption of APT1 eliminated APRT activity, disruption of APT2 had no effect on APRT activity or repression of de novo purine biosynthesis by adenine. Both genes were also individually expressed in Escherichia coli cells. Expression of a recombinant APT2 showed that this gene is defective in that it does not encode a functional APRT enzyme. From the complementation and expression studies, evidence is presented to support the hypothesis that APT1 is required and by itself sufficient to code for APRT in S. cerevisiae. In addition, the data presented here lead us to propose that APT2 is in fact a pseudogene, derived from a gene duplication event.
Strains, plasmids, and culture conditions.
E. coli NM522 (GIBCO-BRL) was grown on 2× YT medium supplemented with ampicillin (50 μg/ml of culture) for routine growth of plasmids. E. coli B25 and B26, used in the high-level expression of the APT1 and APT2 genes, were grown on similar medium but also supplemented with kanamycin to select for the lacZ-repressor plasmid, as described in the Qiagen manual.
The following yeast strains were used in this study: Y642 (MATα ura3-52 leu2-3,112 lys2Δ201 his3Δ200 trp1::hisG) (this study), Y643 (MATa ura3-52 leu2-3,112 lys2Δ201 his3Δ200 trp1::hisG) (this study), L3852 (MATα ade2 his3Δ200 leu2-3,112 lys2Δ201 ura3-52) (G. Fink), L4364 (MATa ade2 his3Δ200 leu2-3,112 lys2Δ201 ura3-52) (G. Fink), Y810 (MATα ura3-52 leu2-3,112 lys2Δ201 his3Δ200 trp1::hisG apt2::HIS3) (this study), Y812 (MATa ura3-52 leu2-3,112 lys2Δ201 his3Δ200 trp1::hisG apt2::HIS3) (this study), Y814 (MATα ade2 ihs3Δ200 leu2-3,112 lys2Δ201 ura3-52 apt2::HIS3) (this study), Y808 (MATa ade2 his3Δ200 leu2-3,112 lys2Δ201 ura3-52 apt2::HIS3) (this study), W109-9C (MATa ade2 trp1 ura3 his3 hpt1-27) (R. Woods), DS1.2B/1 (MATα ade2 apt1 aah1 ura3) (R. Woods), Y839 (MATa ura3-52 leu2-3,112 lys2Δ201 his3Δ200 apt2::HIS3 hpt1::URA3) (this study), Y511 (MATa ura3 lys2 leu2 apt1::URA3) (B. Daignan-Fornier), Y350 (MATa ura3 lys3 leu2) (B. Daignan-Fornier), Y842 (MATa ura3-52 leu2-3,112 lys2Δ201 his3Δ200 trp1::hisG apt2::HIS3 aah1::URA3) (this study), X79 (MATa leu2-1 trp1-1) (R. Woods), Y382 (MATa ade2 ade3 ura3 leu2 trp1) (A. Bender), Y388 (MATa ade2 ade3 ura3 leu3 lys1) (A. Bender), Y839 (MATa ura3 trp1 leu2 his3 pep::His3 prb1 can1 GAL) (A. Bender), Y847 (MATa ura3-52 leu2-3,112 lys2Δ201 his3Δ200 apt2::HIS3 apt1::URA3) (this study), and MWT32 (MATa leu2 ade2 ade3 apt1) (M. Taylor).
Yeast cells were transformed by the lithium chloride-triacetin method (17). Bacterial transformations were carried out by the MnCl2 method (24).
Complementation and genetic analysis of APRT.
Transformants were selected by complementation of the URA mutation to the URA+ phenotype (by the plasmid-derived URA marker) in strain DS1.2B and scored for their ability to use adenine as the sole source of purine. Both the APT1 (pRSAPT1, single copy) and APT2 (pRSAPT2, single copy) genes were individually transformed into the DS1.2B mutant and tested for complementation. The APT1 gene complemented the APT mutant phenotype, allowing the APRT-deficient mutant DS1.2B to grow on defined media containing adenine as the sole purine source. The APT2 gene (12) failed to complement the DS1.2B mutant (Table 1). Also, as shown in Table 1, only APT1-transformed cells had wild-type levels of APRT activity.
TABLE 1.
Complementation analysis of an APT− mutant transformed with the APT1 and APT2 genesa
| Strain | Growth on adenine as purine source | APRT sp act (μmol/min/mg) | % of wild-type activity |
|---|---|---|---|
| X79 (MATa leu2-1 trp1 ade3) | + | 6.62 | 100.0 |
| DS1.2B (MATα ura3 apt1 aah1 ade2) | − | 0.005 | 0.07 |
| DS1.2B (pRS416 transformed) | − | 0.004 | 0.04 |
| DS1.2B (pYEPAPT1 transformed) | + | 7.5 | 113.0 |
| DS1.2B (pRSAPT1 transformed) | + | 5.8 | 89.0 |
| DS1.2B (pRSAPT2 transformed) | − | 0.004 | 0 |
The APRT-deficient DS1.2B strain which carries an adenine mutation that blocks the de novo pathway was transformed with various plasmids carrying the APT1 or APT2 gene. This strain is unable to grow on adenine as the sole purine source. The APRT activities in cell extracts from the DS1.2B mutants as well as their parent strain (X79) are given in micromoles of [3H]AMP produced per minute per milligram of protein. Different constructs used to transform DS1.2B are as described in “Complementation and genetic analysis of APRT.” The specific APRT activity of the X79 (wild-type) strain was considered 100% and used for comparison with the specific APRT activities of extracts from the various transformants.
The APT2 gene was disrupted by replacing its entire coding region with the HIS3 gene (6). The resulting plasmid, named P878, carrying the apt2::HIS3 construct was amplified with the following synthetic oligonucleotides: APT23, 5′-GCTACTGTGCATACCGC-3′, and APT24, 5′-GAGGCACTTTGAACGGC-3′. The resulting PCR product was used to transform the yeast strains Y642, Y643, L3852, and L4364. Transformants were selected for histidine prototrophy. Correct integration was verified by PCR. Disruption of the APT2 gene in a wild-type strain does not lead to any obvious growth phenotype. Since apt1 mutants were previously shown to be resistant to 8-azaadenine (23), we tested the resistance of the apt2-disrupted strains to this base analog. We found that the apt2 mutant is as sensitive to 8-azaadenine as the isogenic wild-type strain. Furthermore, the ade2 apt2 double mutant (where the ade2 mutation blocks de novo purine biosynthesis) can use adenine or hypoxanthine as a purine source.
Mutations affecting purine salvage also inhibit adenine repression of the genes encoding enzymes of the purine de novo pathway (7). We have therefore tested whether the apt2-disrupted strain was affected in this regulation process. Expression of an ADE1 (N-succinyl-5-aminoimidazole-4-carboxamide ribotide synthetase, a purine de novo enzyme)-lacZ fusion was assayed in the apt2 mutant and isogenic wild-type strains. The apt2 mutation has no effect on adenine repression of the ADE1-lacZ fusion.
apt2 aah1, apt2 hpt1, and apt1 apt2 double mutants were constructed. All the double mutants grew normally and were phenotypically indistinguishable from the isogenic single mutants in a wild-type APT2 background. apt1 apt2 double mutants salvaged adenine through adenine aminohydrolase. All together, these results suggest that apt2 disruption does not severely affect purine utilization during vegetative growth.
To test whether APT2 encodes a minor isoform of APRT, we introduced the APT2 gene on a multicopy vector (P552) (12, 29) into an apt1 aah1 ade2 triple mutant. This strain is unable to use adenine as a purine source but grows normally through conversion of hypoxanthine into IMP by hypoxanthine-guanine phosphoribosyl transferase. The multicopy vector carrying the APT2 gene is unable to restore adenine utilization to the triple mutant strain, showing that even when overexpressed, APT2 is unable to compensate for the lack of APT1. Similarly, overexpression of APT2 does not restore utilization of hypoxanthine in an hpt1 ade2 double mutant, thus indicating that APRT2 has no significant hypoxanthine PRTase activity.
Our finding that APT1, but not APT2, could complement an APRT-deficient mutant suggests that either APT2 does not encode a functional APRT or that APT2 is indeed required for APRT activity but that it is not by itself sufficient to encode a functional enzyme. However, the fact that disruption of the APT2 gene had no effect on adenine utilization, adenine analog resistance, or regulation of de novo purine biosynthesis further supports the view that APRT2 serves no function in purine recycling in S. cerevisiae.
Expression of recombinant APRT1 and APRT2 proteins.
The APT1 and APT2 genes were individually ligated into His-tag expression vectors (Qiagen, Hilden, Germany), and the pQEAPT1 and pQEAPT2 expression constructs, respectively, were generated. These constructs allow for the expression of the recombinant APRT1 and APRT2 proteins in E. coli with an N-terminal hexahistidine tag. To express the APRT1 and APRT2 proteins, E. coli B25 (Qiagen) was individually transformed with each construct (pQEAPT1 or pQEAPT2) and cells were grown as described in the Qiagen manual. Cells (1L) were routinely grown to mid-log phase (A600, ∼0.5) at 37°C, the point at which IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM. The cells were grown for five additional hours after IPTG addition.
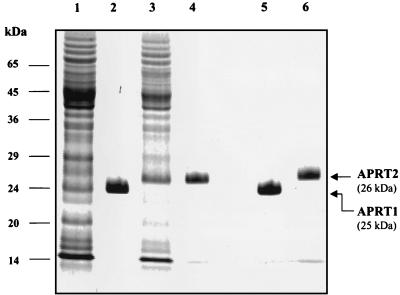
E. coli cells expressing APT1 and APT2 were harvested by centrifugation at 5,000 × g in a Sorvall centrifuge at 4°C for 15 min. The cells were suspended in 30 ml of sonication buffer (50 mM Tris-Cl [pH 7.4], 5 mM MgCl2, 20 mM KCl) and broken by sonication at maximum power with a microtip for cycles of 1 min with a 1-min rest for a total of 10 min. The resulting extract was spun at 20,000 × g for 30 min, and the resulting cell extract was gravity loaded onto a 2-ml Ni2+-nitrilotriacetic acid column. The column was washed with 10 column volumes of sonication buffer (20 ml) containing 50 mM imidazole and eluted with 4 column volumes of the same buffer containing 150 mM imidazole, as described in the Qiagen protein expression manual. The APT1 and APT2 genes were inserted in plasmid vectors under the control of an IPTG-inducible promoter and expressed in E. coli cells (Qiagen). The APT1 gene product (APRT1) migrated as a 25-kDa band, and the APT2 gene product (APRT2) migrated as 26-kDa band in a sodium dodecyl sulfate–12.5% polyacrylamide gel (Fig. 1). These sizes are in agreement with the sizes expected from the deduced coding sequences of the two genes with the His tag at their N termini.
FIG. 1.
Expression of hexahistidine-tagged APRT1 and APRT2 proteins in E. coli cells. Lane 1, total cell extract from E. coli cells transformed with the His-tagged APT1 gene; lane 2, Ni2+-purified recombinant APRT1 protein; lane 3, total cell extract from E. coli cells transformed with the His-tagged APT2 gene; lane 4, Ni2+-purified recombinant APRT2 protein; lanes 5 and 6, purified recombinant APRT1 and APRT2 proteins (respectively) run next to each other to emphasize their relative size difference.
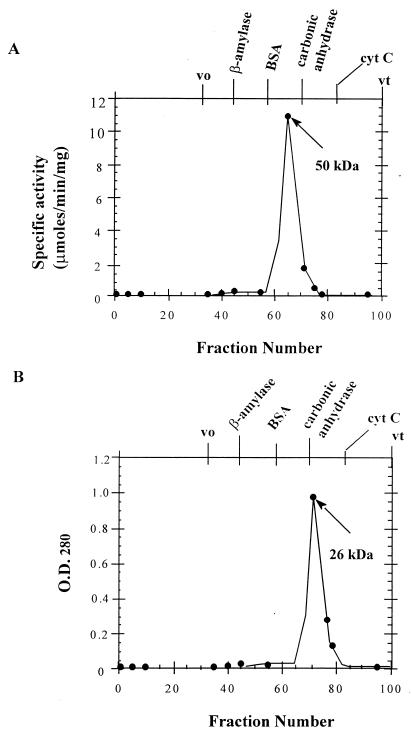
In order to assess the sizes of APRT1 and APRT2 in solution, both recombinant proteins were separately loaded onto a Sephadex G-200 column, calibrated with protein standards, and developed as previously described (1a) (Fig. 2). The purified APRT1 and APRT2 proteins were individually loaded, and the column was developed with the equilibration buffer. Five-milliliter fractions were collected, and their relative protein contents were estimated by measurements of optical density at 280 nm with a model 160 UV spectrophotometer (Shimadzu). Protein-containing fractions were assayed for APRT activity by measuring the incorporation of 3H-labeled adenine into AMP, after the enzyme fraction to be tested was incubated with PRPP and [3H]adenine in the appropriate buffer (15). The fraction with the highest APRT activity (100% relative activity) was used as the peak fraction for determining the native size of the enzyme in solution.
FIG. 2.
Sizing of the APRT1 and APRT2 recombinant proteins. (A) His-tagged APRT1 was chromatographed on a Sephadex G-200 column as described in Materials and Methods. The peak of activity corresponds to that of a 50-kDa protein. (B) The His-tagged APRT2 protein was chromatographed through the same Sephadex column. As no APRT activity was detected with this protein, the peak of absorbance (optical density at 280 nm [O.D.280]) is given and corresponds to that of a 26-kDa protein. β-Amylase (200 kDa), bovine serum albumin (BSA; 66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (cyt C; 13 kDa) were used as standards to calibrate the column. vo, void volume; vt, total volume.
APRT1 eluted between the bovine serum albumin (66 kDa) and carbonic anhydrase (29 kDa) standards. The peak of activity observed coincided with the molecular mass of a 50-kDa protein (Fig. 2A). This molecular mass agrees with that of a homodimer of 25-kDa subunits. When APRT2 was chromatographed through the same column, it eluted in a fraction lacking any detectable APRT activity. The peak of absorbance (A280) corresponded to a 25-kDa protein (Fig. 2B); thus, APRT2 failed to dimerize. The finding that the APRT1 protein had enzymatic activity comparable to that of the native enzyme previously purified in our laboratory suggests that indeed the APT1 gene is sufficient to code for a functional enzyme. Our failure to detect any activity associated with the recombinant APRT2 protein led us to conclude that either the APT2 gene does not encode a functional enzyme or the recombinant APRT2 expressed in E. coli does not fold into a catalytically active conformation. The failure of APRT2 to dimerize when it was separated through a sizing column suggested that the lack of APRT2 activity is due, in part, to its failure to dimerize. In fact, all of the previously characterized APRTs (from mouse, hamster, human, E. coli, and Arabidopsis cells), with the exception of Leishmania donovani APRT (30), are functional as homodimers in solution (20). This failure to dimerize may also be due to improper folding of E. coli-expressed APRT2. However, given the degree of sequence similarity between the two genes (1a), the latter explanation seems unlikely. In addition, high-level expression of APT2 in the APRT-deficient Y847 strain showed no APRT activity (6b).
Analysis of APRT expression.
Total RNA was isolated from various yeast strains by the acid-phenol method (13). For Northern blot analysis, poly(A)+ RNA (5 μg) from strain Y382 or X79 was separated by electrophoresis in a 18% formaldehyde–1.2% agarose gel (Seakem). The gel was run at 100 V for 4 h. The RNA was transferred by means of capillarity to a Zeta-probe membrane as described in the Bio-Rad manual. The membrane was hybridized to the radiolabeled APT1 and/or APT2 probe as described in the Zeta-probe manual. The membrane was exposed to X-ray film (Kodak) for 16 h and developed in an X-Omat machine (Kodak).
Western blot analyses were carried out with cell extracts from various yeast strains, prepared by disruption with glass beads. Samples were electrophoretically separated in a sodium dodecyl sulfate–12% polyacrylamide gel. The proteins were transferred to Trans-Blot transfer membranes by electrophoresis as described in the Bio-Rad manual. Membranes were blocked with 5% nonfat milk (to prevent nonspecific binding of the antibody), and the blots were separately probed with rabbit anti-APRT1 or rabbit anti-APRT2 polyclonal antibodies. For detection, a horseradish peroxidase-linked secondary antibody (Bio-Rad) was hybridized to the same membrane. Bands were visualized by chemiluminescense with an Immuno-Star kit (Bio-Rad). The membrane was exposed to X-ray film, and the film was developed in an X-Omat machine (Kodak).
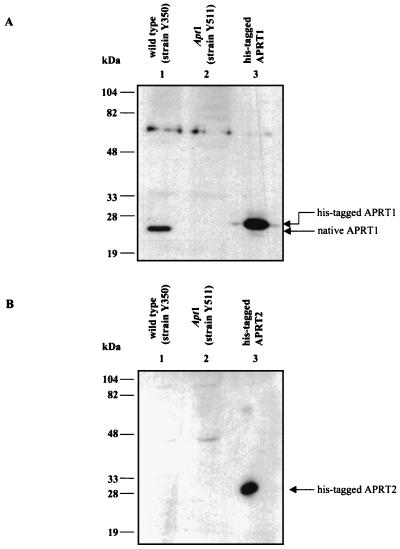
We observed that both the APT1 and APT2 genes were actively transcribed in wild-type yeast cells. However, we failed to detect any APRT2 protein in Western blot experiments using extracts from wild-type yeast and polyclonal antibodies against the recombinant APRT2 protein (Fig. 3B). When similar experiments were carried out with anti-APRT1 antibodies, we could easily detect an APRT1 signal (Fig. 3A). As previously mentioned, the APRT2 protein could still be made but quickly degraded. Regardless, the failure to detect activity and/or the presence of a protein corresponding to APRT2 argues that the bulk of the APRT activity in vivo is provided by APT1 gene expression. These data also suggest that the APT2 gene product either lacks elements important for efficient translation or lacks elements important for proper protein folding and/or stability in vivo. However, overproduction of APRT2 in E. coli and yeast results in a stable nonactive protein (6a).
FIG. 3.
Western blot analysis of native APRT proteins. (A) Western blot probed with polyclonal anti-APRT1 antibody. Lane 1 contains a cell extract from strain Y350 (wild-type strain), lane 2 contains a cell extract from strain Y511 (apt1 mutant strain), and lane 3 contains pure recombinant APRT1. (B) Western blot similar to that shown in panel A but probed with polyclonal anti-APRT2 antibody. Lanes 1 and 2 contain cell extracts from strains Y350 and Y511 (respectively), and lane 3 contains pure recombinant APRT2.
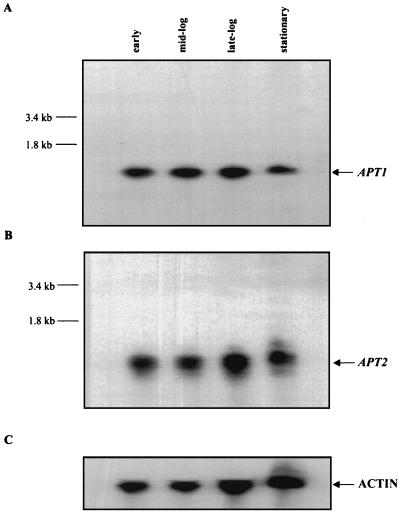
We have hypothesized (1) that the role of the APRT2 protein is to interact with APRT1, thus down-regulating its activity and/or specificity. High levels of purine nucleotides can lead to repression of the de novo purine biosynthetic pathway. We hypothesized that the APRT1-APRT2 interaction would be particularly important during conditions of starvation (i.e., depletion of free adenine), where APRT activity becomes less important as the cell is forced to synthesize purine bases de novo. This hypothesis predicts that the levels of APRT2 transcripts increase as cells deplete free adenine from the medium (i.e., as cells enter stationary phase). However, when Northern blot analysis was performed with radiolabeled DNA probes specific for APT1 or APT2, we detected comparable levels of APT1 and APT2 transcription in wild-type cells regardless of the growth phase of the culture (Fig. 4). APT2 mRNA migrates slightly more slowly than APT1 mRNA. These experiments demonstrate that even if the APT2 gene is actively transcribed, at least under the culture conditions described, it is not actively translated (Fig. 3).
FIG. 4.
Northern blot analysis of the APT1 and APT2 gene products from strain X79 (wild type). (A) Total RNA was extracted at various times during growth and probed with a radiolabeled APT1 probe. (B) Same membrane as that shown in panel A, probed with a radiolabeled APT2 probe. (C) Same membrane as that shown in panel A, probed with a radiolabeled yeast actin used as a loading control.
Phylogenetic comparison analysis of the APRT1 and APRT2 proteins.
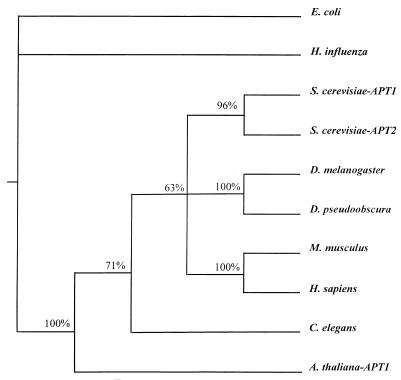
Amino acid sequence alignments obtained from the SeqApp program were used in the PAUP program (version 3.11; Smithsonian) to determine the phylogenetic relationship between various APRTs. The analysis was performed by maximum parsimony analysis with the exhaustive-search method. Bacterial sequences were used as outgroups. The S. cerevisiae APRT1 (GenBank accession no. S49755) and APRT2 (GenBank accession no. L14434) sequences were used in the alignment, together with the homologous APRTs (GenBank accession numbers in parentheses) from E. coli (M14040), Haemophilus influenza (U32748), Drosophila melanogaster (S34831), D. pseudoobscura AF025800), Mus musculus (M86440), Homo sapiens (p07741), Caenorhabditis elegans (U80438), and Arabidopsis thaliana APRT1 (p31166). The phylogenetic analysis was performed on the APRT sequence alignment previously described by us (1a) (Fig. 5). We wished to determine if APT2 was the result of a duplication of APT1 (or vice versa) or if APT2 was more closely related to some other APRT. The phylogenetic analysis showed that APT1 is most closely related to APT2.
FIG. 5.
Comparative phylogenetic reconstruction of APRT1 and APRT2. A phylogenetic tree of various deduced APRT sequences constructed with PAUP version 3.11 is shown. Numbers indicate the probability of two sequences branching together.
Upon completion of the Yeast Genome Project, it was discovered that much of the genome was in fact duplicated. Fifty-five duplicate regions which cover approximately 50% of the S. cerevisiae genome have been found (10). It has been proposed that the entire yeast genome was duplicated and that subsequently many of the duplicated regions were lost (31). APT1 and APT2 are both found in duplicated regions of each other: APT1 on chromosome XIII and APT2 on chromosome IV (31). Southern blot analysis was used to determine if this APT duplication event was found only in S. cerevisiae. The closely related yeast Picchia pastoris was examined for the presence of two APTs. With APT1 and APT2 probes, it appears that P. pastoris also contains two APTs (data not shown).
Taken together, the complementation and the expression data lead us to conclude that APT1 is by itself sufficient to code for APRT in S. cerevisiae. Furthermore, from the phylogenetic comparative analysis, we propose that the APT2 gene is a pseudogene, perhaps derived from an aberrant gene duplication of the APT1 gene. The occurrence of pseudogenes in yeast, although rare, is not unprecendented. It is possible that APT2 codes for an APRT homolog that has diverged to the point of possessing a different function. However, our failure to detect APRT2 protein in various yeast strains makes this possibility unlikely and makes our original proposal of two forms of APRT in yeast untenable.
Acknowledgments
We are grateful to Alan Bender, J. L. Corden, G. Fink, and R. Woods for sending plasmids and strains.
This work was supported by grants from the Fondation pour la Recherche Medicale, Conseil Regional d’Aquitaine, and the CNRS (UPR9026). M.L.G. was supported by a Ministère des Affaire Etrangères fellowship.
REFERENCES
- 1.Alfonzo J D. Ph.D. thesis. Bloomington, Ind: Indiana University; 1995. [Google Scholar]
- 1a.Alfonzo J D, Sahota A, Taylor M W. Purification and characterization of adenine phosphoribosyltransferase from Saccharomyces cerevisiae. Biochim Biophys Acta. 1997;1341:173–182. doi: 10.1016/s0167-4838(97)00068-x. [DOI] [PubMed] [Google Scholar]
- 2.Alfonzo J D, Sahota A, Deeley M C, Ranjekar P, Taylor M W. Cloning and characterization of the adenine phosphoribosyltransferase-encoding gene (APT1) from Saccharomyces cerevisiae. Gene. 1995;161:81–85. doi: 10.1016/0378-1119(95)00239-3. [DOI] [PubMed] [Google Scholar]
- 3.Allen J B, Elledge S J. A family of vectors that facilitate transposon and insertional mutagenesis of cloned genes in yeast. Yeast. 1994;10:1267–1272. doi: 10.1002/yea.320101003. [DOI] [PubMed] [Google Scholar]
- 4.Arnold W J. Adenine phosphoribosyltransferase. Methods Enzymol. 1978;51:568–574. doi: 10.1016/s0076-6879(78)51079-3. [DOI] [PubMed] [Google Scholar]
- 5.Arnold W J, Kelley W N. Human hypoxanthine guanine phosphoribosyltransferase: purification and subunit structure. J Biol Chem. 1971;246:7398–7406. [PubMed] [Google Scholar]
- 6.Benes V, Hostomsky Z, Arnold L, Paces V. M13 and pUC vectors with new unique restriction sites for cloning. Gene. 1993;74:527–534. doi: 10.1016/0378-1119(93)90360-f. [DOI] [PubMed] [Google Scholar]
- 6a.Crother, T. R. Unpublished data.
- 6b.Crother, T. R., and M. W. Taylor. Unpublished results.
- 7.Daignan-Fornier B, Fink G R. Corregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci USA. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deeley M C. Adenine deaminase and adenine utilization in Saccharomyces cerevisiae. J Bacteriol. 1992;174:3102–3110. doi: 10.1128/jb.174.10.3102-3110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert G D. Enzyme kinetics. Bloomington, Ind: dogStar Software; 1989. [Google Scholar]
- 10.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 11.Groth D P, Young L G, Kenimer J G. Adenine phosphoribosyltransferase from rat liver. Methods Enzymol. 1978;51:574–580. doi: 10.1016/s0076-6879(78)51080-x. [DOI] [PubMed] [Google Scholar]
- 12.Guetsova M L, Lecoq K, Daignan-Fornier B. The isolation and characterization of Saccharomyces cerevisiae mutants that constitutively express purine biosynthetic genes. Genetics. 1997;147:383–397. doi: 10.1093/genetics/147.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press; 1991. [Google Scholar]
- 14.Hershey H V, Gutstein R, Taylor M W. Cloning and restriction map of the E. coli apt gene. Gene. 1982;19:89–92. doi: 10.1016/0378-1119(82)90192-5. [DOI] [PubMed] [Google Scholar]
- 15.Hershey H V, Taylor M V. Purification of adenine phosphoribosyltransferase by affinity chromatography. Prep Biochem. 1978;8:453–462. doi: 10.1080/00327487808061662. [DOI] [PubMed] [Google Scholar]
- 16.Hochstadt J. Adenine phosphoribosyltransferase from Escherichia coli. Methods Enzymol. 1978;51:558–567. doi: 10.1016/s0076-6879(78)51078-1. [DOI] [PubMed] [Google Scholar]
- 17.Keszenman-Pereyra D, Hieda K. A colony procedure for transformation of Saccharomyces cerevisiae. Curr Genet. 1988;13:21–23. doi: 10.1007/BF00365751. [DOI] [PubMed] [Google Scholar]
- 18.Moffat B A, McWhinnie E, Burkhart E, Pasternak J J, Rothstein S J. A complete cDNA for adenine phosphoribosyltransferase from Arabidopsis thaliana. Plant Mol Biol. 1992;18:653–662. doi: 10.1007/BF00020008. [DOI] [PubMed] [Google Scholar]
- 19.Moffat B A, McWhinnie E A, Agarwal S K, Schaff D A. The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene. 1994;143:211–216. doi: 10.1016/0378-1119(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 20.Musick D L. Structural features of the phosphoribosyltransferase and their relationship to the human deficiency disorders of purine and pyrimidine metabolism. Crit Rev Biochem. 1981;11:1–34. doi: 10.3109/10409238109108698. [DOI] [PubMed] [Google Scholar]
- 21.Nagy M, Ribet A. Purification and comparative study of adenine and guanine phosphoribosyltransferases from Schizosaccharomyces pombe. J Biochem. 1977;77:77–85. doi: 10.1111/j.1432-1033.1977.tb11643.x. [DOI] [PubMed] [Google Scholar]
- 22.Park J, Taylor M W. Analysis of signals controlling expression of the Chinese hamster ovary APRT gene. Mol Cell Biol. 1988;8:2536–2544. doi: 10.1128/mcb.8.6.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahota A, Ranjekar P K, Alfonzo J, Lewin A S, Taylor M W. Mutants of Saccharomyces cerevisiae deficient in adenine phosphoribosyltransferase. Mutat Res. 1987;180:81–87. doi: 10.1016/0027-5107(87)90069-8. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Schnorr K M, Gaillard C, Biget E, Nygaard P, Laloue M. A second form of adenine phosphoribosyltransferase in Arabidopsis thaliana with relative specificity towards cytokinins. Plant J. 1996;9:891–898. doi: 10.1046/j.1365-313x.1996.9060891.x. [DOI] [PubMed] [Google Scholar]
- 26.Segel I H. Enzyme kinetics. New York, N.Y: John Wiley & Sons; 1975. [Google Scholar]
- 27.Simmonds H A, Van Acker K J, Cameron J S, Snedden W. The identification of 2,8 dihydroxyadenine, a new compound of urinary stones. J Biochem. 1976;157:485. doi: 10.1042/bj1570485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stambrook P J, Tischfield J A, Khan S A, Sikela J M, Dush M K. Nucleotide sequence and organization of the mouse adenine phosphoribosyltransferase gene: presence of a coding region common to animal and bacterial phosphoribosyltransferases that have a variable intron/exon arrangement. Proc Natl Acad Sci USA. 1985;82:2731–2735. doi: 10.1073/pnas.82.9.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stotz A, Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and vectors. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 30.Tuttle J V, Krenitsky J V. Purine phosphoribosyltransferases from Leishmania donovani. J Biol Chem. 1980;255:909–916. [PubMed] [Google Scholar]
- 31.Wolfe K H, Sheilds D C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 32.Yuryev A, Corden J L. A Saccharomyces cerevisiae gene encoding a potential adenine phosphoribosyltransferase. Yeast. 1994;10:659–662. doi: 10.1002/yea.320100510. [DOI] [PubMed] [Google Scholar]