Abstract
• Context.—
Pancreatic ductal adenocarcinoma has some of the worst prognostic outcomes among various cancer types. Detection of histologic patterns of pancreatic tumors is essential to predict prognosis and decide the treatment for patients. This histologic classification can have a large degree of variability even among expert pathologists.
Objective.—
To detect aggressive adenocarcinoma and less aggressive pancreatic tumors from nonneoplasm cases using a graph convolutional network–based deep learning model.
Design.—
Our model uses a convolutional neural network to extract detailed information from every small region in a whole slide image. Then, we use a graph architecture to aggregate the extracted features from these regions and their positional information to capture the whole slide–level structure and make the final prediction.
Results.—
We evaluated our model on an independent test set and achieved an F1 score of 0.85 for detecting neoplastic cells and ductal adenocarcinoma, significantly outperforming other baseline methods.
Conclusions.—
If validated in prospective studies, this approach has a great potential to assist pathologists in identifying adenocarcinoma and other types of pancreatic tumors in clinical settings.
Pancreatic ductal adenocarcinoma (PDAC) is an aggressive type of cancer derived from the epithelial cells that make up the ducts of the pancreas. PDAC ranks firmly last among all cancer types in terms of worst prognostic outcomes,1 and its incidence and mortality rates have continued to increase for decades in the United States.2–5 According to one study,6 from 1990 to 2017, the numbers of cases and deaths worldwide identified as related to pancreatic carcinoma have doubled. It is estimated that there will be 60 430 pancreatic cancer cases and 48 220 deaths caused by pancreatic cancer in the United States in 2021.7 Furthermore, PDAC is expected to become the second leading cause of cancer death by 2030.8 Therefore, preventive measures, screening, and surveillance are becoming increasingly important for pancreatic cancer.
PDAC makes up the overwhelming majority of pancreatic malignant tumors and is derived from the ductules of the exocrine pancreas, which carry digestive enzymes and other secretions from the exocrine pancreas to the lumen of the small bowel. PDAC has a variable histologic appearance, ranging from high-grade lesions with necrosis and marked cellular atypia to bland, “foamy” infiltrative glands in a highly fibrotic stroma. Inflammation may be prominent, subtle, or essentially absent. The islet cells of the pancreas also have a neoplastic counterpart, which is the pancreatic neuroendocrine tumor (PanNET). PanNET, while prognostically much more favorable, is also histologically diverse and may mimic a benign inflammatory condition (eg, islet aggregation in chronic pancreatitis) or PDAC.
Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) and EUS-guided fine-needle biopsy (EUS-FNB)9 have become the primary diagnostic methodologies used in the evaluation of pancreatic mass lesions. These methods are the least invasive means of procuring tissue for diagnosis currently available, as they can be performed via endoscopy. However, they have significant limitations in terms of tissue fragmentation, crush artifact, and the overall quantity of tissue procured for diagnosis. Considering these limitations, the severity of pancreatic carcinoma, and the enormous morbidity of pancreatectomy, it is imperative that the diagnostic utility of EUS-FNA/FNB be maximized. PDAC is a histologically diverse malignant neoplasm with numerous known patterns, including several that can mimic neuroendocrine tumors (which typically have a much better prognosis) and various nonneoplastic pancreatic lesions. Although there exists a set of guidelines for the classification of pancreatic tumors from the Papanicolaou Society of Cytopathology10 (see Table 1), some cases can be ambiguous even to the trained eyes of pathologists. The low tissue volume of FNA/FNB procedures may increase the frequency of cases that do not receive a definitive diagnosis (ie, cases labeled as atypical or suspicious). Therefore, not only radiologists and oncologists but also pathologists may incorrectly identify some benign tumors or pseudotumor cells for malignant tumors.11–13
Table 1.
Classification Criteria Modified From Papanicolaou Society of Cytopathology System for Reporting Pancreaticobiliary Cytologya
| I. Nondiagnostic |
| Preparation/obscuring artifact precluding evaluation of the cellular component |
| Gastrointestinal contaminant only |
| Normal pancreatic tissue elements (with a solid or cystic mass by imaging study) |
| Acellular aspirate of a solid mass |
| Acellular aspirate of a cyst without evidence of a mucinous etiology (ie, thick colloidlike mucin, elevated CEA or KRAS or GNAS mutation) |
| II. Negative (for malignancy) |
| Benign pancreatic tissue (in the appropriate clinical setting) |
| Acute pancreatitis |
| Chronic and autoimmune pancreatitis |
| Pseudocyst |
| Lymphoepithelial cyst |
| Splenule/accessory spleen |
| III. Atypical |
| Atypical cytologic and/or architectural features not consistent with normal or reactive changes, yet insufficient to be categorized as either neoplastic or suspicious for malignancy |
| IV. Neoplastic |
| Benign |
| Serous cystadenoma |
| Neuroendocrine microadenoma |
| Lymphangioma |
| Other |
| Well-differentiated neuroendocrine tumor (PanNET) |
| Intraductal papillary mucinous neoplasm, all grades of dysplasia |
| Mucinous cystic neoplasm, all grades of dysplasia |
| Solid-pseudopapillary neoplasm |
| V. Suspicious (for malignancy) |
| Significant cytologic and/or architectural atypia suggestive of malignancy though qualitatively and/or quantitatively insufficient for a definite diagnosis |
| VI. Positive or malignant |
| Pancreatic ductal adenocarcinoma and its variants |
| Cholangiocarcinoma |
| Acinar cell carcinoma |
| Poorly differentiated (small or large cell) neuroendocrine carcinoma |
| Pancreatoblastoma |
| Lymphoma |
| Metastatic malignancy |
Data derived from Papanicolaou Society of Cytopathology System for Reporting Pancreaticobiliary Cytology.10 The bold cases are those classes included in this study.
Our study proposes an automatic and accurate method based on graph convolutional neural networks to detect PanNETs and PDAC on digitized histology slides. Furthermore, such an approach can assist pathologists with reviewing the slides by generating additional diagnostic information for consideration, such as the location of cells suspicious for malignancy or neoplasia in a pancreatic tissue specimen.
DATA COLLECTION AND ANNOTATION
In this study, we focused on classifying the 2 most common pancreatic tumors, PDAC and PanNET, in combination with a benign control group. To develop and evaluate our model to classify these pancreatic tumor patterns, we collected 143 digitized formalin-fixed, paraffin-embedded, hematoxylin and eosin–stained whole slide images from Dartmouth-Hitchcock Medical Center in Lebanon, New Hampshire. The EUS-FNA/EUS-FNB cell block slides were digitized using an Aperio AT2 scanner (Leica Biosystems, Nussloch, Germany) at ×20 resolution (0.5 μm/pixel) at Dartmouth-Hitchcock Medical Center. These slides were identified using structured cytopathology diagnosis data from the laboratory information management system. In addition, a full-text pathology report search was performed for further disambiguation of cases within each class. To assure the quality of the slides and their labels in our data set, they were independently reviewed by the expert pathologists involved in our study (X.L. and R.H.) for concordance with the identified diagnosis. Our pathologists assessed all slides identified as negative, positive, and those with neoplastic cells present based on the laboratory information management system structured data and pathology reports. A manual review of report text was used for cases of unusual histologic appearance or any other irregularity concerning the identified diagnosis versus the slide’s appearance.
Our classification criteria for this data set are described in Table 1. Of note, cases without viable tumors (eg, entirely necrotic) were excluded from the data set. The positive cases in our data set include only PDAC. At the same time, lymphomas, acinar cell carcinoma, neuroendocrine carcinoma, and other malignancies were excluded because of the unavailability of a sufficient number of those cases for training at our institution. The neoplastic cell class in our data set is represented by neuroendocrine tumors, which excludes neuroendocrine carcinomas. Also, rarer tumors such as solid pseudopapillary tumors were excluded from this class. In addition, we opted to exclude cystic lesions, such as mucinous cystic neoplasm and intraductal papillary mucinous neoplasm, from the neoplastic category because the diagnosis of these cases often relies heavily on cyst fluid chemistry studies and clinical information, which our proposed neural network does not consider. The negative class contains normal cells as well as blood, fibrin, mild atypia associated with inflammation, leukocytes, and benign gastric or duodenal epithelium due to procedural artifact. Atypical and suspicious cases were also excluded because of lack of definitive diagnosis and interobserver variability. Rarely, some cell blocks from such cases were included in the negative category if they met 1 of 2 criteria: (1) the original pathology report described them as negative or unremarkable; or (2) they were cleared in blinded review by our senior expert cytopathologist (X.L.).
The digitized slides in our data set were partially annotated by our domain expert pathologists (X.L. and R.H.) to indicate the pancreatic cancer subtypes and their locations on the slides. As such, the neoplastic and positive regions were annotated using the polygon annotation feature in the Automated Slide Analysis Platform or ASAP (Radboud University Medical Center, Nijmegen, The Netherlands),14 a fast viewer and annotation tool for high-resolution histology images. These annotations are used as reference standards for developing and evaluating our patch classification models. The distribution of the annotated images is shown in Table 2. Any disagreements in annotations were resolved through joint discussions among annotators and further consultation with our senior expert pathologist (X.L.). We randomly partitioned 91 unannotated slides and the 52 annotated slides into a training set, a validation set, and a test set for the patch-level classification.
Table 2.
Distribution of Our Data Set and Its Annotations
| Annotated Slides | All Slides (Annotated + Unannotated) | ||||
|---|---|---|---|---|---|
| Neoplastic | Positive | Negative | Neoplastic | Positive | |
| Training | 13 | 18 | 32 | 28 | 30 |
| Validation | 2 | 3 | 5 | 4 | 5 |
| Test | 7 | 9 | 16 | 11 | 12 |
| Total | 22 | 30 | 53 | 43 | 47 |
Of note, all whole slides (annotated and unannotated) were reviewed and classified by our expert pathologists (X.L. and R.H.), and whole slide labels were established according to consensus opinions between X.L. and R.H. and the original diagnosis in the clinical report. Slides on which agreement could not be reached were excluded from our data set. All whole slide images were randomly partitioned into the training, validation, and test sets and used for whole slide inference.
METHODS
Given the large size of high-resolution whole slide histology images and the memory capacity of currently available computer hardware, it was not feasible to directly train a model on whole slide images. Therefore, we used a sliding window strategy to extract small fixed-size (224 × 224-pixel) patches from the whole slide images. To analyze and classify a whole slide image, our pipeline has 2 parts: (1) a deep convolutional neural network to extract high-dimensional features from patches extracted from a whole slide image and (2) a graph convolutional neural network to aggregate patches’ high-dimensional features and their positional information to make the whole slide inference. The deep convolutional neural network model is implemented to recognize local features at the patch level, whereas the graph convolutional neural network model is used to capture structural and global patterns at the slide level. As a result, this pipeline allows us to analyze high-resolution images with feasible memory and computational resources while capturing global and structural information of whole slide images.
Deep Convolutional Neural Network
Deep neural networks have been proved a powerful tool in computer vision and are increasingly applied in medical image analysis.15–17 In the histologic image inference domain, deep convolutional neural networks are applied as a backbone for whole slide image analysis and classification.18,19 This study uses a residual neural network20 to extract the image features. The whole slide images are usually high resolution, from 0.25 to 1 μm/pixel. Because of this high resolution and hardware memory limitations, it is not feasible to directly extract features from whole slide images without downsampling. However, by downsampling whole slide images, we may lose critical histologic features for classification. Therefore, to train our deep residual neural network with achievable memory and computational resource requirements, we use a sliding-window strategy to generate small, fixed-size patches from the whole slide images. The labels of these small patches depend on whether they include the annotated regions of interest by pathologists. Using this sliding-window approach, we generated 3091 neoplastic patches, 6275 positive patches, and 94 633 negative patches in the training set. We then trained multiple ResNet models with different numbers of layers, including 18, 50, and 101 layers. Among these, the ResNet-18 model performed the best in the patch-level classification on our validation set. Therefore, we used the ResNet-18 model trained on the augmented annotated training set as our feature extractor. In the training process of the feature extractor, the ResNet-18 model used the extracted tissue patches as inputs and outputted the predicted class probabilities for each patch. All the layers in this model are initialized with He initialization.21 We trained the ResNet-18 model for 60 epochs with an initial learning rate of 0.001 and decayed the learning rate by a factor of 0.9 to the power of the number of epochs.
Graph Neural Network
Using the ResNet-18 model, we can extract features from patches and get the predictions at the patch level. To infer the whole slide image labels, we developed a novel method based on a graph neural network to aggregate the patch-level information extracted by our patch-level ResNet-18 model for the whole slide–level inference. Graphs’ unique ability to capture structural relationships among data points allows for the extraction of more insights and information than analyzing data points in isolation. In recent years, graph neural networks, such as graph convolutional networks (GCNs), have gained massive success in analyzing data with nonregular structures, such as social networks and protein networks.22,23 GCNs use graph convolutional layers to aggregate the neighbor nodes’ information and have achieved state-of-the-art performance on graph classification benchmarks such as Citeseer, Cora, PubMed, and NELL.24–27 Some recent work has proposed using GCNs to make the whole slide inference.28 This method leverages the pretrained ResNet-50 model on ImageNet29 to extract features from patches in a whole slide image. Although this approach’s overall architecture is similar to ours, our approach is different in constructing the graph and extracting the patch-level features. Of note, instead of the ResNet-50 model pretrained on ImageNet, our feature extractor uses ResNet-18 architecture and is trained on labeled pancreas patches from annotations. In our approach, the whole slide images are viewed as graphs. Fixed-size patches and their extracted features are considered nodes and node features, respectively. We use the patches’ positional information and features extracted by our patch classification model to construct graphs from whole slide images; we have named this the Slide2Graph method and describe it below.
Slide2Graph for Whole Slide Inference
Graph Construction.—
An overview of our graph construction pipeline is shown in Figure 1. To construct the computational graphs for whole slide images, we first use a framework developed by our group for slide preprocessing to automatically remove background and extract tissue segments at a ×10 (1 μm/pixel) magnification level.30 Then, tissue images are divided into 224 × 224-pixel fixed-size patches, and the coordinates of patches are saved. We removed the last fully connected and the SoftMax layers from our patch-level ResNet-18 classifier and used the rest of our trained ResNet-18 to extract 512-dimensional feature vectors from each fixed-size patch. Each 224 × 224-pixel fixed-size patch image extracted from a whole slide image is viewed as a node in the computational graph, and its 512-dimensional feature is used as the node feature. We used the previously saved positional information of patches or nodes to add edges in the computation graphs. For each node, we used the KD-Tree algorithm31 to search its 4 nearest nodes in the Euclidean space and then add edges between nodes that were weighted by the nodes’ Euclidean distance. As a result, we converted the whole slide images into computational graphs. Through the constructed graphs, our approach keeps track of the distances between extracted patches from a whole slide image. Figure 2 shows our proposed pipeline for whole slide image classification. We used GCNs, which leverage all patches’ spatial and positional information to aggregate the local patch features and make the whole slide image–level inference. Of note, in comparison with previous aggregation methods, this method incorporates the positional information of each patch and the global structure of the whole slide image.
Figure 1.
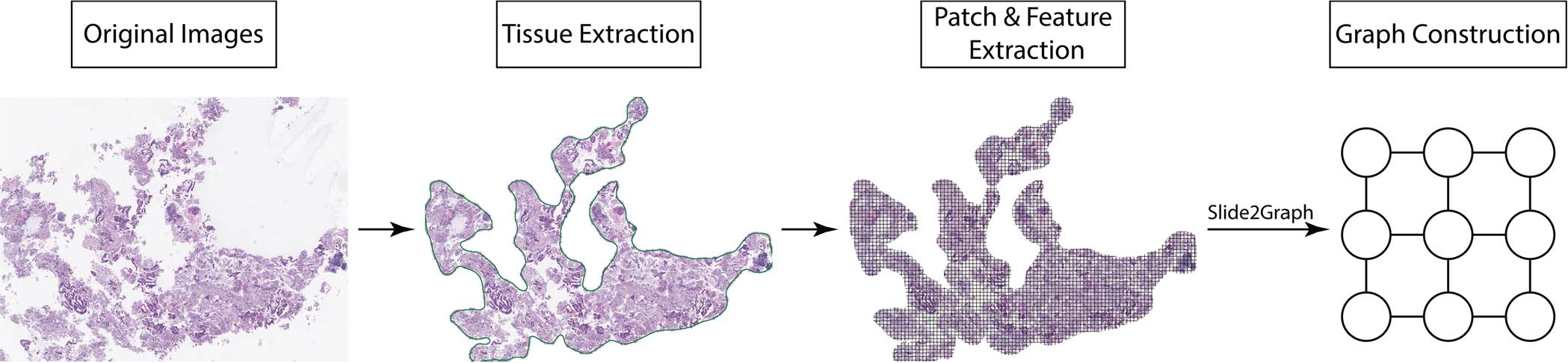
Overview of our Slide2Graph preprocessing pipeline. First, the tissue regions are identified, and the background is removed. Then, a sliding-window method is used to generate small fixed-size patches from each whole slide image, and the corresponding features for each patch are extracted using a convolutional neural network model. Finally, a graph is constructed by considering each patch as a node and connecting each node with its 4 nearest neighbors.
Figure 2.
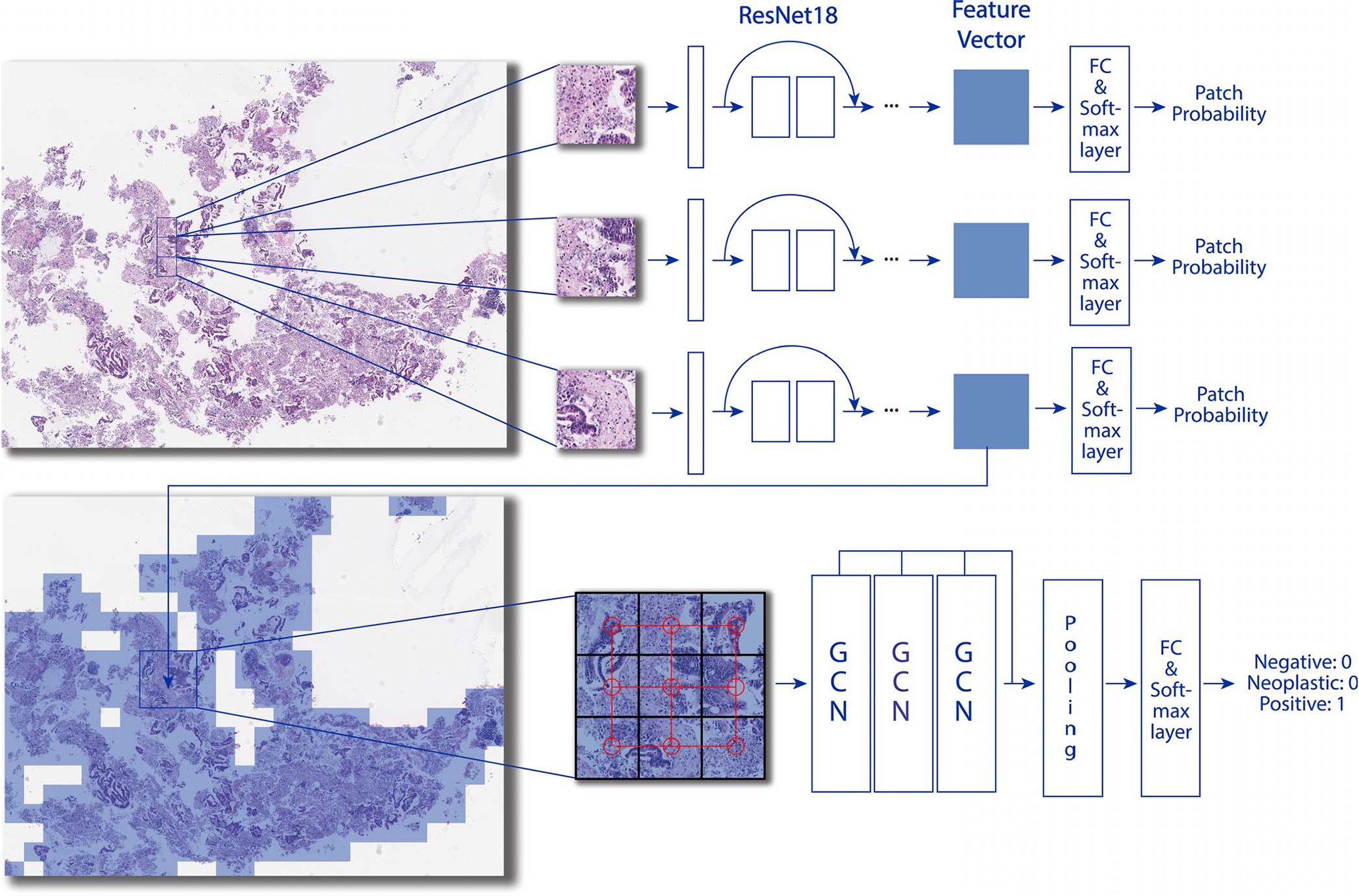
Overview of our Slide2Graph classification pipeline. After background removal, fixed-size patches were extracted from whole slide images using the sliding-window method. A ResNet-18 model was trained on the extracted patches from annotated whole slide images and then used to extract histology features of patches. The features and positional information of patches were used to construct a computational graph for whole slide inferencing. Abbreviations: FC, fully connected; GCN, graph convolutional network.
Graph Model Architecture.—
We modify the graph model proposed by Zhang et al32 and use self-attention global pooling layers33 as our graph model architecture. The overall model structure is shown in Figure 3. After constructing the graph, we apply 3 graph convolutional or GCN layers to update the node features. Each GCN layer generates a new node representation by aggregating features from the node itself and its neighboring nodes in the graph structure. Therefore, every node contains information of its surrounding neighborhood after the 3 feed-forward GCN layers in Slide2Graph architecture. We concatenate the outputs of every GCN layer and then use the self-attention pooling layer to select the top 50% highly weighted nodes that determine the class of a graph. We run a global mean pooling and maximum pooling on these top 50% nodes and concatenate them. Finally, a fully connected layer and a SoftMax layer take the feature matrices and output the predicted whole slide class probabilities. We trained this model for 200 epochs with an initial learning rate of 0.001 and learning rate annealing. Slide2Graph code is publicly available at https://github.com/BMIRDS/Slide2Graph.
Figure 3.
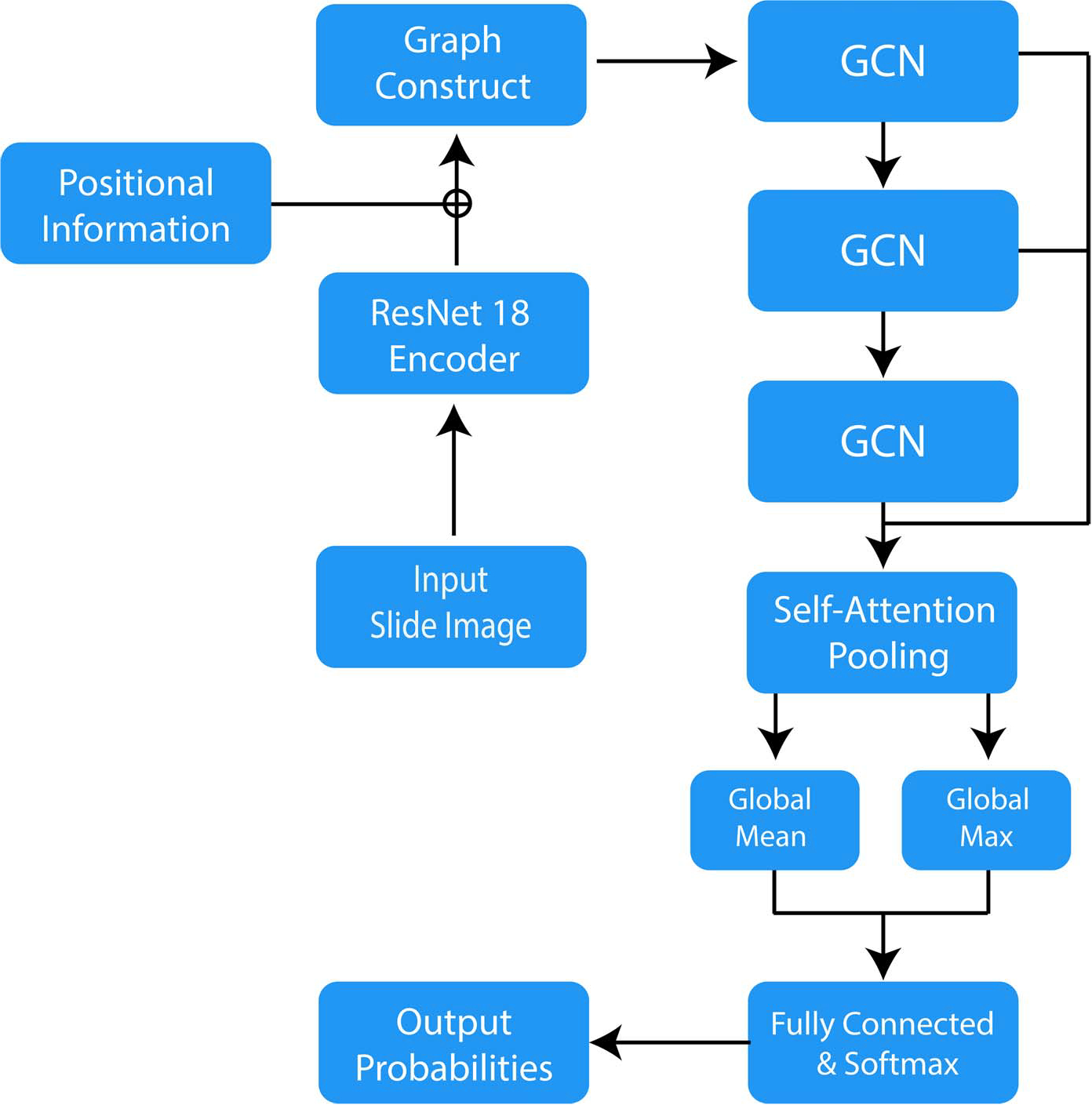
Slide2Graph architecture. Abbreviation: GCN, graph convolutional network.
RESULTS
We evaluated our model’s performance on our holdout test set, which was not used during the model training. Table 3 summarizes the precision, recall, F1 score, and area under the relative operating characteristic curve metrics for each class and overall. In addition, we calculated the 95% CIs for all the metrics using a bootstrapping method with 10 000 iterations. For error analysis, the confusion matrix of our model is shown in Figure 4.
Table 3.
Slide2Graph’s Performance Metrics and 95% CIs on the Test Set
| Precision | Recall | F1 score | AUC | |
|---|---|---|---|---|
| Negative | 0.90 (0.67, 1.00) | 0.56 (0.31, 0.80) | 0.69 (0.44, 0.87) | 0.80 (0.66, 0.93) |
| Neoplastic | 0.73 (0.55, 1.00) | 1.00 (1.00, 1.00) | 0.85 (0.71, 1.00) | 0.94 (0.86, 1.00) |
| Positive | 0.79 (0.50, 0.93) | 0.92 (0.73, 1.00) | 0.85 (0.62, 0.95) | 0.90 (0.73, 0.98) |
| Average | 0.81 (0.57, 0.98) | 0.83 (0.68, 0.93) | 0.80 (0.59, 0.94) | 0.88 (0.75, 0.97) |
Abbreviation: AUC, area under the relative operating characteristic curve.
Figure 4.
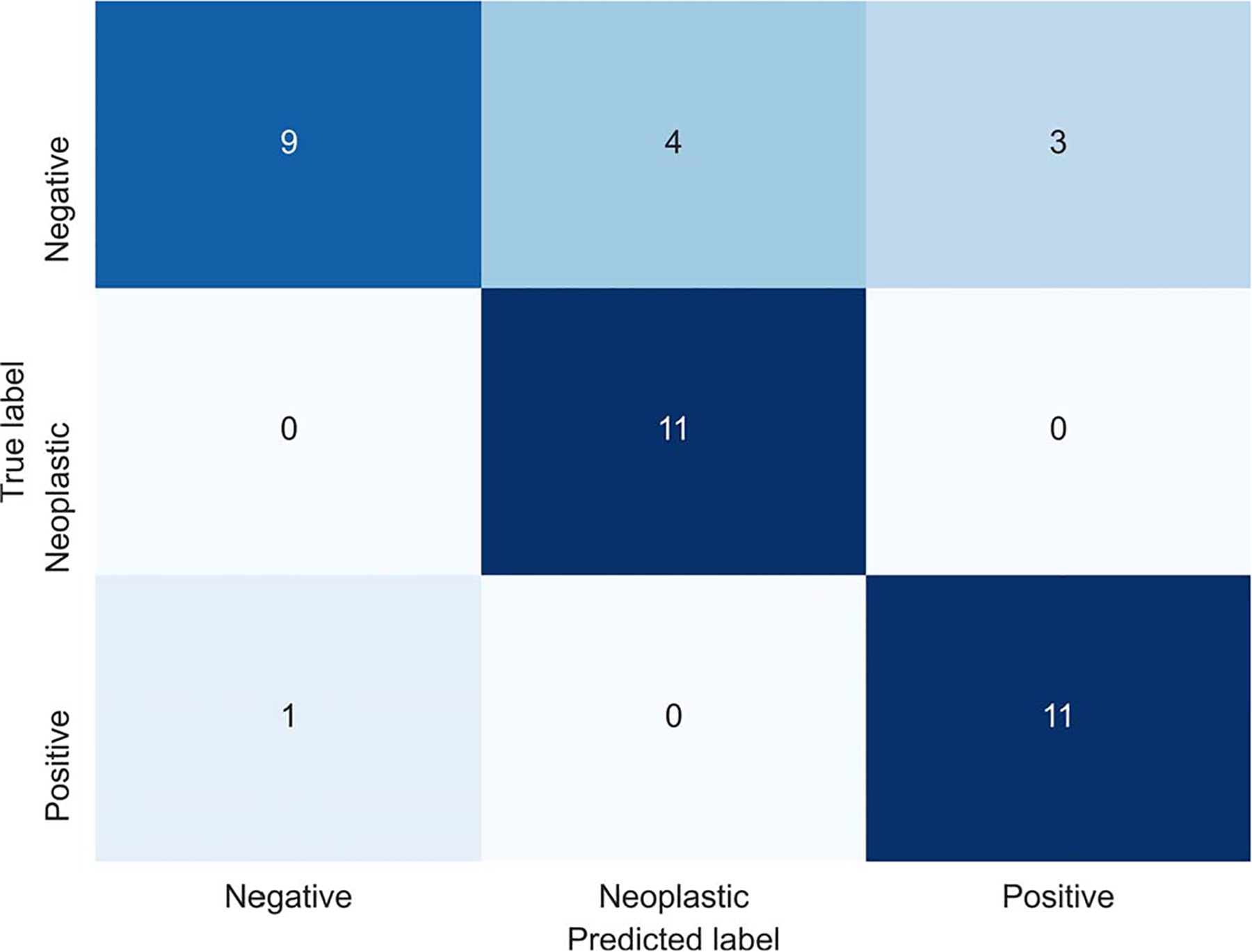
Slide2Graph’s confusion matrix on the test set.
We also implemented other models, including DeepSlide, a decision tree, a random forest, and Adaboost,18,34–36 to aggregate patch information for comparison, and showed the efficacy of Slide2Graph. In DeepSlide,18,34–36 we ran systematic grid searches to find the best thresholds for patch-level confidence score and the required percentages of predicted patches in one slide for developing whole slide inferencing rules using our training and validation sets. In the DeepSlide approach, only patches with a confidence score greater than 0.75 were considered for whole slide inference. In DeepSlide’s whole slide inferencing rule, if the percentage of neoplastic or positive patches in a slide exceeded 20% of the entire patches extracted from the whole slide, the slide was classified as neoplastic or positive, respectively. If both the percentages of neoplastic and positive patches exceeded 20%, then the class with the larger percentage was considered as the class for the slide. Otherwise, this slide was deemed to be negative. In addition, we used the percentages of neoplastic and positive patches extracted from a slide as the independent variables to predict the whole slide label in the other machine learning models, that is, decision tree, random forest, and Adaboost. Sixfold cross-validation and grid search were used to find the best hyperparameters for the random forest and the Adaboost models. As shown in Figure 5, our proposed Slide2Graph graph model performs the best among all 5 whole slide inference models.
Figure 5.
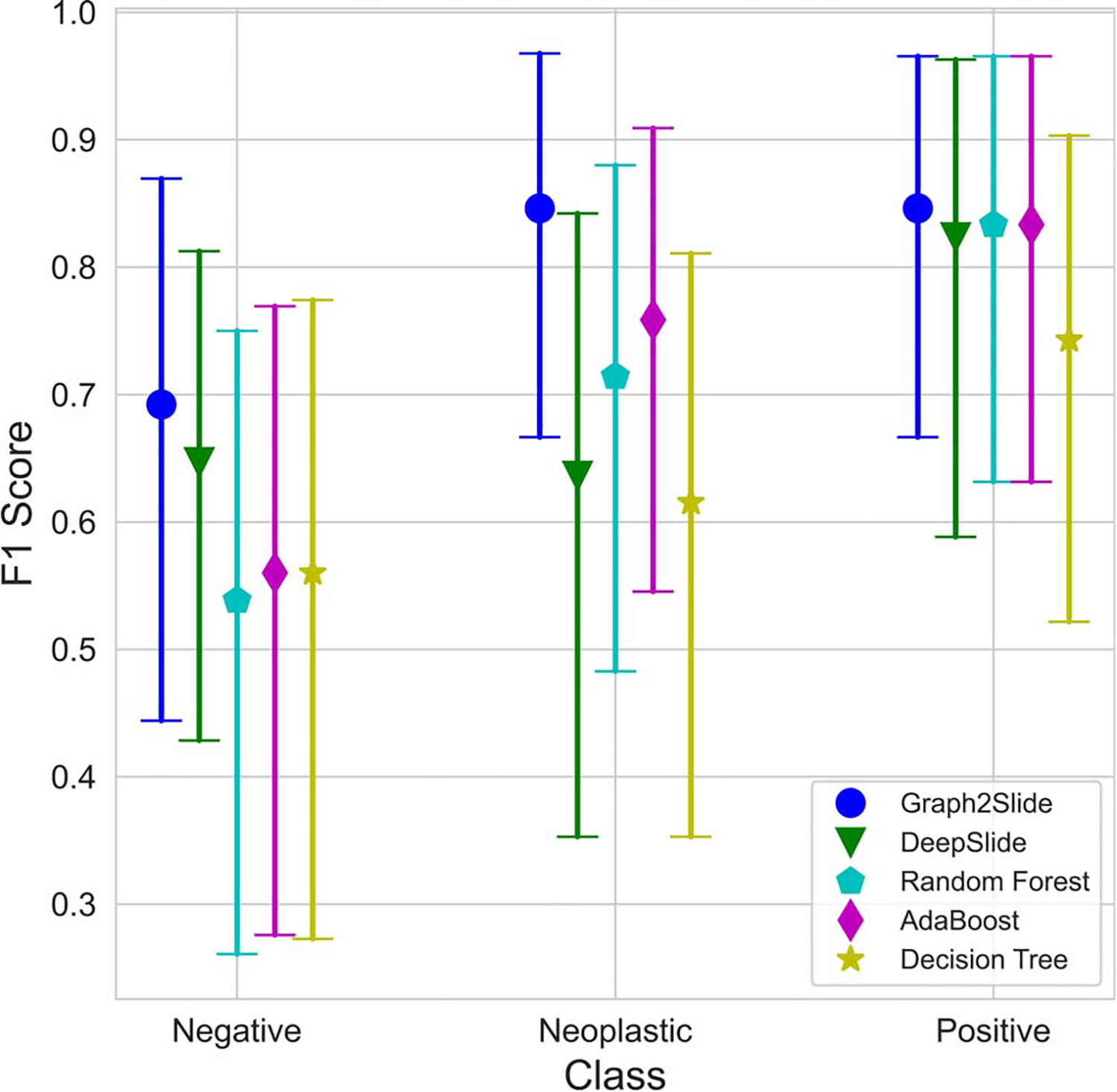
The F1 scores and 95% CIs of different models on the test set stratified by class.
Of note, because of the small size of the testing set, there are overlaps among all models’ F1 score CIs. Therefore, to investigate the statistical significance of the performance differences among different models, we used a bootstrapping approach to randomly sample 50 subsets from our test set. Then, we used the 2-tailed Student t test to examine the statistical significance of difference among F1 scores from various methods. As Table 4 shows, the F1 score of our Slide2Graph model outperformed the other models with a statistical significance level of P < .001, whereas Adaboost was the strongest competitor among the baselines.
Table 4.
Comparisons Between Slide2Graph and Other Baseline Models Based on Bootstrapping and F1 Scoresa
| Slide2Graph | DeepSlide | Decision Tree | Random Forest | Adaboost | |
|---|---|---|---|---|---|
| F1 score | 0.79 | 0.70 | 0.64 | 0.70 | 0.72 |
All P values were less than .001.
DISCUSSION
The approach proposed in this study can automatically and accurately detect pancreas tumor patterns on the whole slide images. Our proposed approach achieved the best performance on our test set compared with other baseline methods. Unlike other studies in this domain, which do not consider the benign class in whole slide inference or rely on detailed benign tissue annotations, we considered tissues outside the region of interest annotations as negative or benign. We used these regions for the training of our patch-level classifier. Therefore, these negative regions could contain noise and findings that may be visually similar to those seen in positive or neoplastic cases. For example, Figure 6 is an image from a case diagnosed with benign pancreatitis. Albeit the slide is correctly labeled as negative or benign, this region does appear atypical because it contains inflammation and fibrosis surrounding residual atrophic, mild atypical ductular structures (ie, reactive atypia). This region bears a striking resemblance to a well-differentiated adenocarcinoma with infiltrating glands.37 Although the cells in this region are still benign, the tissue overall is architecturally more similar to a well-differentiated adenocarcinoma than to normal pancreatic acini/parenchyma. Likewise, Figure 7, A through D, depicts several situations in which benign pancreas might mimic a neuroendocrine tumor; contrariwise, a neuroendocrine tumor may mimic lymphocytes (if discohesive) or carcinoma (if glandular or organoid).38 This phenomenon of mimicry is likely one of the reasons why our patch-level classifier does not perform perfectly, as tissue findings of some reactive and malignant processes are known to demonstrate considerable morphologic overlap.37,38 Because we do not explicitly annotate benign (including reactive atypia) or normal regions in our data set, the developed algorithm is subject to these ambiguities.
Figure 6.
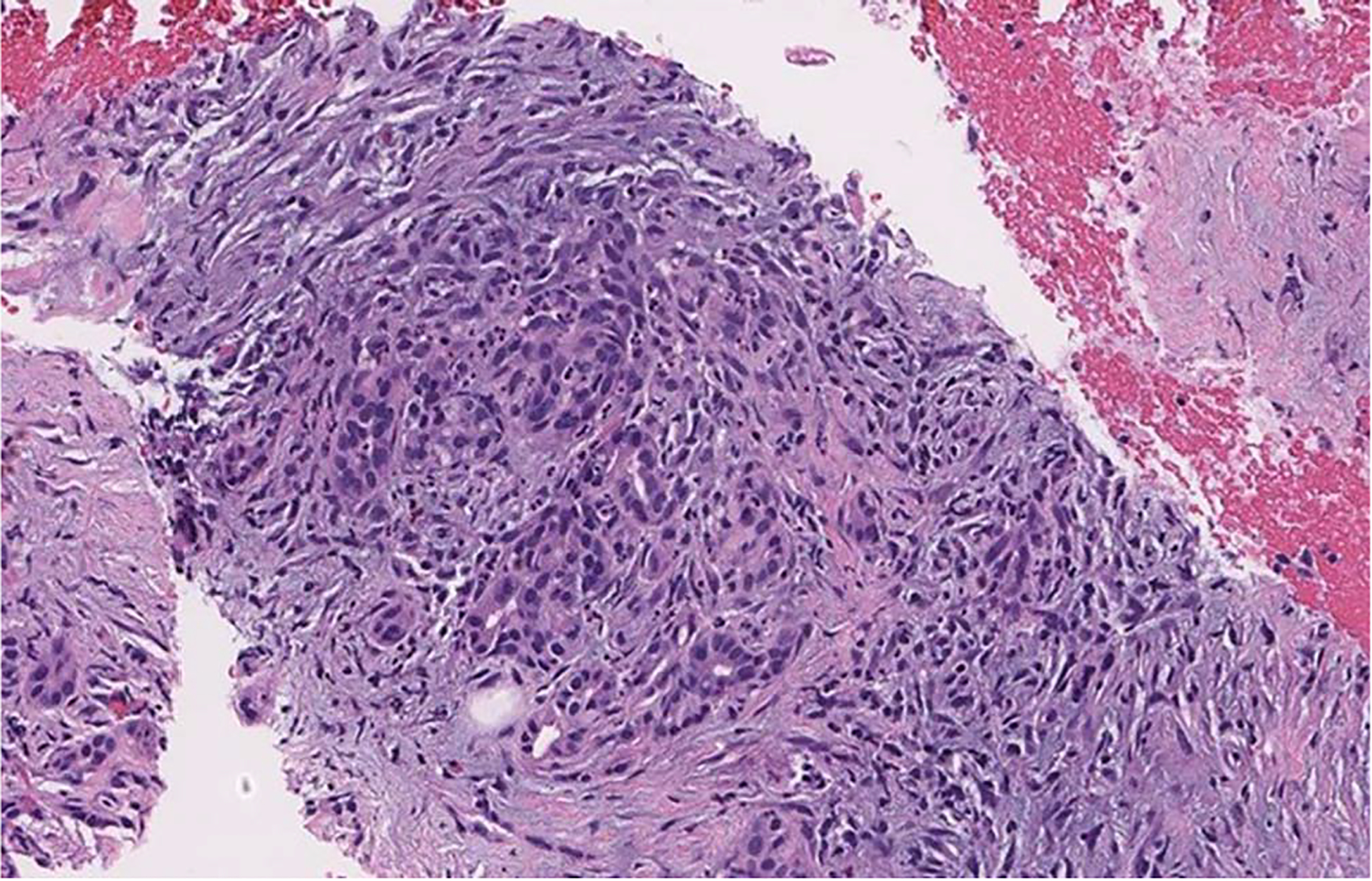
Differential survival of benign pancreatic ducts in chronic pancreatitis can create the illusion of a neoplasm (hematoxylin-eosin, original magnification ×20).
Figure 7.
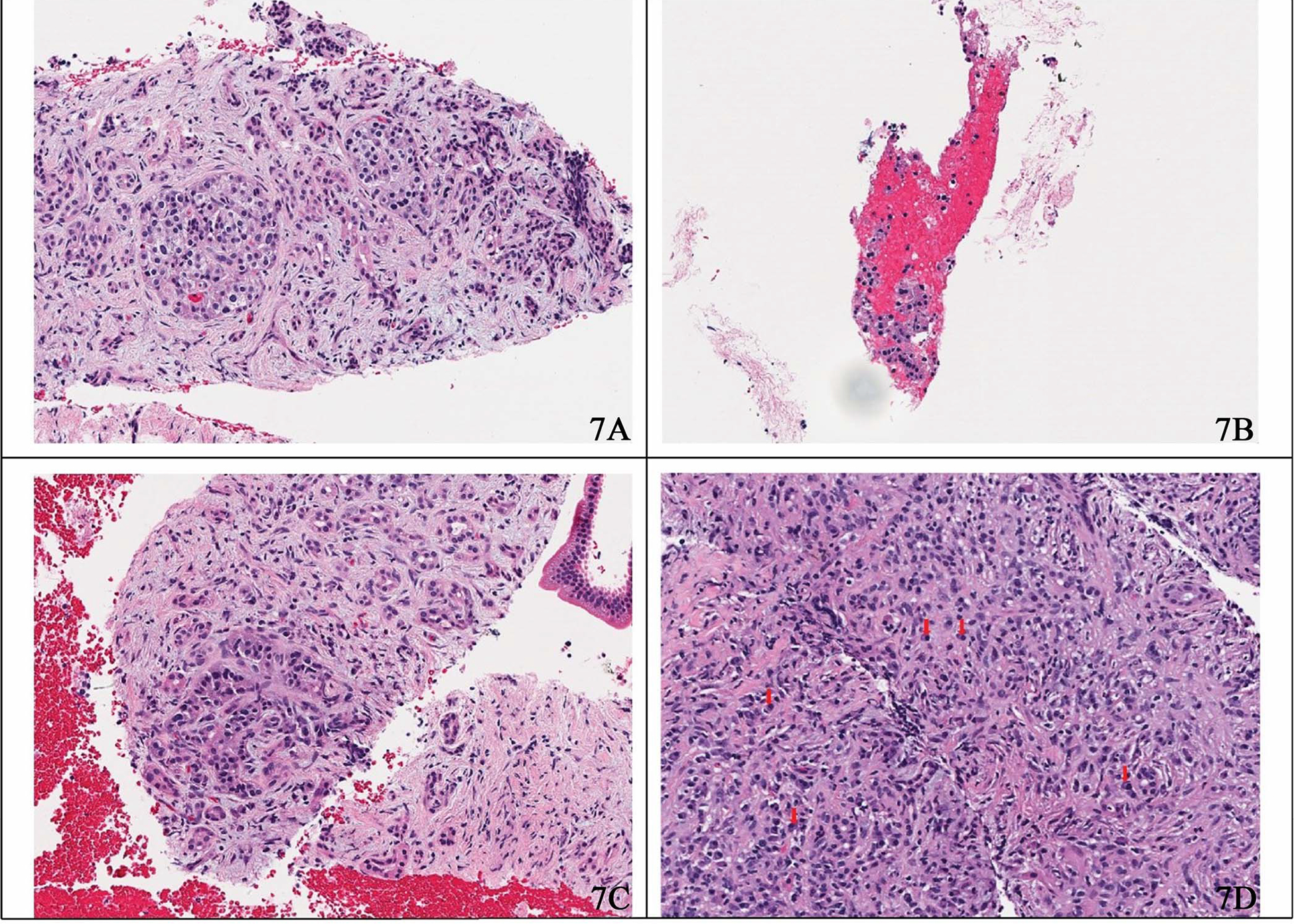
Sample hematoxylin-eosin–stained images. A, Prominent islet cell aggregates in chronic pancreatitis mimicking neoplastic cells. B, Detached and degenerated acinar cells may mimic the detached cells of a neuroendocrine neoplasm. C, Surviving ducts in this case of chronic pancreatitis demonstrate an organoid pattern, an architecture frequently associated with neuroendocrine tumors. D, Plasma cells may resemble single neoplastic neuroendocrine cells, as seen here in immunoglobulin G4–related autoimmune pancreatitis (original magnification ×20).
Notably, although our patch classifier does not achieve the perfect performance at the patch level, our whole slide inference model still showed a high performance in detecting neoplastic and positive patterns. Reviewing the model’s mistakes by expert pathologists shows that our algorithm’s major error type is overcalling of negative cases—that is, labeling them as either neoplastic or positive. Although this error is diagnostically irritating and would be cumbersome in the full-scale clinical use of the algorithm, it does not create the same concern for patient safety as frequent misclassifications of positive or neoplastic cases as negative.
Why the Graph Model Performs Better
It is challenging to classify pancreatic tissue in small patches because of significant histologic overlap among low-grade malignancies, reactive atypia, pancreatitis, etc. For example, although Figure 6 appears benign to trained eyes, it is impossible to exclude the possibility that it might have come from a tissue specimen that contains a tumor; the inflammation and fibrosis seen could easily be found in a case of pancreatitis or at the edge of a malignancy. The ductules seen are minimally atypical and unlikely to be malignant. However, if more atypical epithelium were present elsewhere in the specimen, that evaluation would need to be reconsidered. Neuroendocrine tumors also often exhibit morphologic overlap with other conditions; Figure 7, A through D, depicts several situations where this may occur. Of note, at our institution, in recent memory, we have received at least 1 consultation in which prominent islet cell aggregates deceived the initial reviewer into diagnosing a neuroendocrine tumor. However, the evaluation of the subsequent surgical specimen revealed benign chronic pancreatitis as the source of the neoplastic-appearing cells.
Therefore, the patch-level classifier essentially performs as a weak classifier because of these overlaps. However, our proposed graph model can consider those patch-level predictions and additional positional data to come to a more reliable prediction for classifying a whole slide, particularly for the neoplastic and positive classes.
Several improvements in our approach facilitate this performance. Unlike the models previously used for whole slide–level inferencing, which treat every patch equally and use the percentages of predicted patches for each class to make the whole slide predictions, our graph-based model takes the patches’ positions and the global structure of the whole slide into consideration. As we applied the self-attention pooling layer to aggregate the node features, we can obtain the associated attention map for a whole slide. This attention map can provide insights into the results of our Slide2Graph model by highlighting the regions that significantly influence the whole slide inference. Figure 8, A and B, shows the most important regions by Slide2Graph highlighted in red, which contributed the most to the whole slide–level classification compared with regions of interest annotated by an expert pathologist. The regions highlighted in darker shades of red have a higher impact on the classification results. These visualizations can draw attention to important regions of whole slide images for their classification and provide insights into our approach’s reasoning that pathologists can review and confirm. Therefore, the proposed approach in this study can potentially assist pathologists in reviewing whole slide images and improve their accuracy and efficiency in this diagnostic task.
Figure 8.
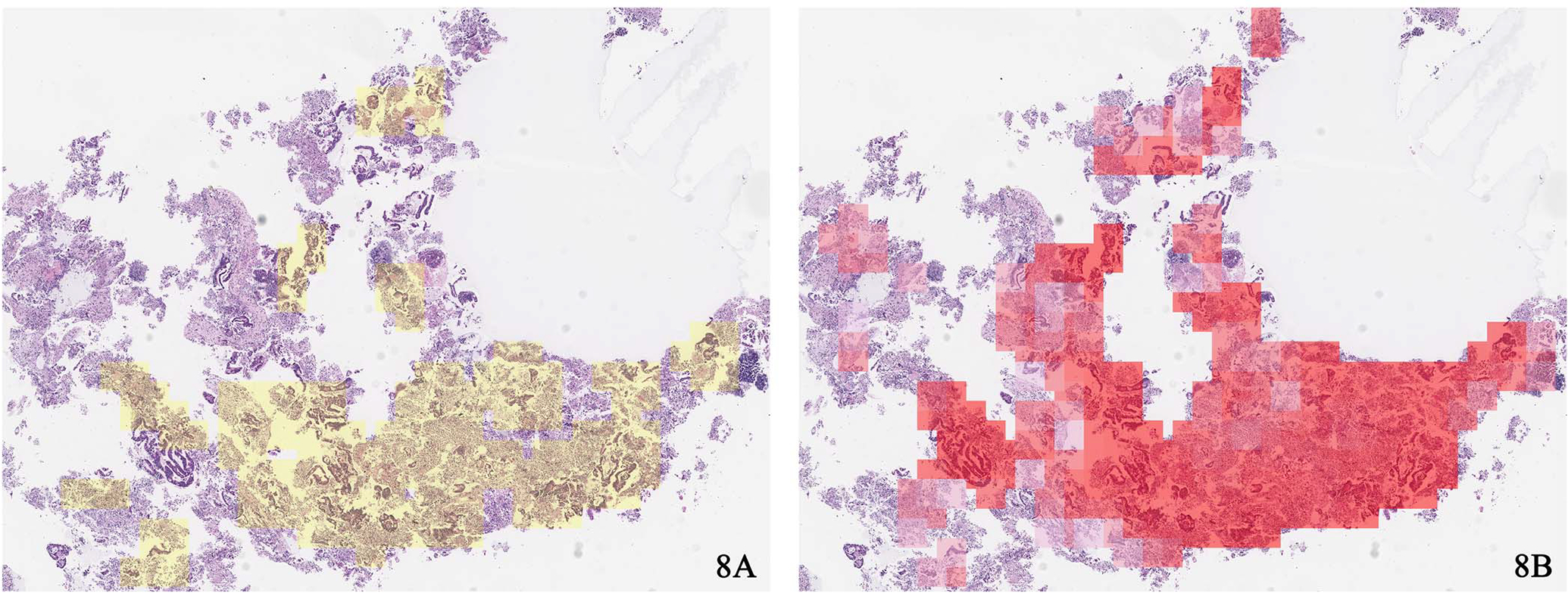
A, Regions of interest annotated by pathologists. B, Important regions by Slide2Graph for whole slide inferencing (hematoxylin and eosin stain, original magnification ×20 [A and B]).
Clinical Utility
We envision that a key function of our model will be to reduce the amount of time pathologists spend reviewing slides and to enable an alternative workflow that improves diagnostic efficacy. In such a workflow, slides are digitized upfront, and the algorithm can be used to highlight potentially malignant cases in the queue and encourage the pathologist to review them as priority cases. This would reduce the turnaround time associated with frankly malignant diagnoses. In addition, up-front augmentation and highlighting of high-risk regions can help pathologists to come to a decision more rapidly or even occasionally prevent misdiagnoses. The ability to rapidly review the most diagnostic fields of a slide could also speed the ordering of ancillary studies, such as immunostains, molecular genetic testing, and studies for microsatellite instability and programmed death ligand-1 expression. Such an alternative workflow would allow a case to undergo full review at the microscope once, concurrently with relevant immunostains, and be signed out on the spot. Future iterations of the algorithm, optimized for specificity, can even order relevant ancillary studies automatically.
This study is the first step toward the goal of deploying and evaluating our model in clinical settings. To start, such methodologies need to be developed and rigorously tested by retrospective evaluations before they can be deployed and evaluated in prospective studies. For this purpose, our research team has worked closely with expert pathologist collaborators who have provided guidance and advice on the design, development, and evaluation of the proposed technology in a retrospective evaluation before the potential deployment in clinical settings. As the next step, our team plans to conduct a prospective clinical trial to measure the impact of this model on pathologist performance in clinical settings.
Limitations and Future Directions
The proposed method in this paper has some limitations. First, the size of our data set in this study is small. Even though we use a partially annotated data set, our sample size, especially in the test and validation sets, is still slim, which resulted in wide 95% CIs in model evaluation. Moreover, when annotating the whole slide images, instead of annotating specific negative regions, we annotated only positive and neoplastic regions. Therefore, mimicries were not differentiated from normal tissues in our annotations and were considered as negative regions in the current setup. As a result, the negative regions used to train our model contain many different types of cells other than positive and neoplastic cells, such as benign epithelium, acinar tissue, normal cells, and inflammation. This lack of annotations likely led to the observed mimicry phenomenon in the patch-level classifier discussed above. The variance and diversity of cells in negative regions introduce noise in our negative class and some morphologic overlap with the positive and neoplastic classes. The inclusion of cell blocks from cases overall labeled atypical in the negative class may have introduced a low level of additional noise as well. Although our reviewers considered atypia in the cell blocks not significant, there is some interobserver variability in the threshold at which specimens are called atypical. The broad scope of the negative class, although important from the perspective of clinical utility (because it reflects the diversity of negative clinical findings), makes the model harder to train. That is likely why our patch-level classifier did not perform ideally in terms of precision.
Besides the noisy negative class, our current model was trained only on a relatively small data set, which did not include several uncommon neoplasms, such as neuroendocrine carcinoma, acinar cell carcinoma, solid pseudopapillary tumor, and lymphoma. Therefore, these neoplasms are not included in the current version of our model. However, our model architecture can be extended to include more classes if we have sufficient training samples from them. In future work, we plan to use a larger data set with a more specific breakdown of annotated negative findings and uncommon neoplasms to train our model to distinguish these neoplasms and atypical cases.
In addition, our model is composed of 2 parts: a patch-level feature extractor and a whole slide inference model. These parts are trained separately. Our future work will use small fixed-size patches as nodes directly to construct computational graphs for whole slide images, so the model can be trained end to end. Also, our approach has the potential to perform multitask learning. We will explore different ways of aggregating the loss function from the patch-level and whole slide–level classifiers to decide whether the information from these 2 classifiers can benefit each other. Through this process, our model can output the predictions for both patches and whole slide images simultaneously. In addition, we plan to explore various graph convolutional and global pooling layers and different approaches to construct graphs for whole slide images to further improve the graph-based model’s performance. Finally, our proposed approach was tested only by a retrospective evaluation. In the following steps, we plan to conduct a prospective clinical trial to measure the impact of the proposed tool in clinical settings and its translational value. In this prospective evaluation, we will engage with expert pathologists to deploy and validate our approach as a decision support system in clinical settings.
Acknowledgments
This research was supported in part by grants from the US National Library of Medicine (R01LM012837 and R01LM013833) and the US National Cancer Institute (R01CA249758).
Footnotes
The authors have no relevant financial interest in the products or companies described in this article.
References
- 1.McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24(43):4846–4861. doi: 10.3748/wjg.v24.i43.4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali H, Pamarthy R, Vallabhaneni M, Sarfraz S, Ali H, Rafique H. Pancreatic cancer incidence trends in the United States from 2000–2017: analysis of Surveillance, Epidemiology and End Results (SEER) database. F1000Res. 2021;10: 529. doi: 10.12688/f1000research.54390.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saad AM, Turk T, Al-Husseini MJ, Abdel-Rahman O. Trends in pancreatic adenocarcinoma incidence and mortality in the United States in the last four decades; a SEER-based study. BMC Cancer. 2018;18(1):688. doi: 10.1186/s12885-018-4610-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu W, He X, Yang L, et al. Rising trends in pancreatic cancer incidence and mortality in 2000–2014. Clin Epidemiol. 2018;10:789–797. doi: 10.2147/CLEP.S160018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Costa WL, Oluyomi AO, Thrift AP. Trends in the incidence of pancreatic adenocarcinoma in all 50 United States examined through an age-period-cohort analysis. JNCI Cancer Spectr. 2020;4(4):1–7. doi: 10.1093/JNCICS/PKAA033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pourshams A, Sepanlou SG, Ikuta KS, et al. The global, regional, and national burden of pancreatic cancer and its attributable risk factors in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2019;4(12):934–947. doi: 10.1016/S2468-1253(19)30347-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 8.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014; 74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155 [DOI] [PubMed] [Google Scholar]
- 9.Levine I, Trindade AJ. Endoscopic ultrasound fine-needle aspiration vs fine needle biopsy for pancreatic masses, subepithelial lesions, and lymph nodes. World J Gastroenterol. 2021;27(26):4194–4202. doi: 10.3748/wjg.v27.i26.4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitman MB, Centeno BA, Ali SZ, et al. Standardized terminology and nomenclature for pancreatobiliary cytology: the Papanicolaou Society of Cytopathology Guidelines. Cytojournal. 2014;11(suppl 1):3. doi: 10.4103/1742-6413.133343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basturk O, Askan G. Benign tumors and tumorlike lesions of the pancreas. Surg Pathol Clin. 2016;9(4):619–641. doi: 10.1016/j.path.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren B, Liu X, Suriawinata AA. Pancreatic ductal adenocarcinoma and its precursor lesions: histopathology, cytopathology, and molecular pathology. Am J Pathol. 2019;189(1):9–21. doi: 10.1016/j.ajpath.2018.10.004 [DOI] [PubMed] [Google Scholar]
- 13.Putra J, Liu X. Autoimmune pancreatitis: a succinct overview. J Pancreas. 2015;16(3):239–243. doi: 10.6092/1590-8577/2989 [DOI] [Google Scholar]
- 14.Litjens G Computation Pathology Group at the Radboud University Medical Center. Automated Slide Analysis Platform (ASAP). https://computationalpathologygroup.github.io/ASAP. Published 2015. Accessed August 25, 2022.
- 15.Tomita N, Cheung YY, Hassanpour S. Deep neural networks for automatic detection of osteoporotic vertebral fractures on CT scans. Comput Biol Med. 2018;98:8–15. doi: 10.1016/j.compbiomed.2018.05.011 [DOI] [PubMed] [Google Scholar]
- 16.Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH. Patch-based convolutional neural network for whole slide tissue image classification. In: 2016. IEEE Conference on Computer Vision and Pattern Recognition (CVPR); June 27–30, 2016; Las Vegas, NV. 2016:2424–2433. doi:10.1109/CVPR.2016.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasir-Moin M, Suriawinata AA, Ren B, et al. Evaluation of an artificial intelligence-augmented digital system for histologic classification of colorectal polyps. JAMA Netw Open. 2021;4(11):1–12. doi: 10.1001/jamanetworkopen.2021.35271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei JW, Tafe LJ, Linnik YA, Vaickus LJ, Tomita N, Hassanpour S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci Rep. 2019;9(1):3358. doi: 10.1038/s41598-019-40041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu M, Ren B, Richards R, Suriawinata M, Tomita N, Hassanpour S. Development and evaluation of a deep neural network for histologic classification of renal cell carcinoma on biopsy and surgical resection slides. Sci Rep. 2021;11(1):7080. doi: 10.1038/s41598-021-86540-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); June 27–30, 2016; Las Vegas, NV. 2016:770–778. doi:10.1109/CVPR.2016.90 [Google Scholar]
- 21.He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: surpassing human-level performance on ImageNet Classification. In: 2015 IEEE International Conference on Computer Vision (ICCV); December 7–13, 2015; Santiago, Chile. 2015:1026–1034. doi:10.1109/ICCV.2015.123 [Google Scholar]
- 22.Monti F, Frasca F, Eynard D, Mannion D, Bronstein MM. Fake news detection on social media using geometric deep learning. Preprint posted online February 10, 2019. ArXiv. doi: 10.48550/ARXIV.1902.06673 [DOI] [Google Scholar]
- 23.Fout A, Byrd J, Shariat B, Ben-Hur A. Protein interface prediction using graph convolutional networks. In: Guyon I, Von Luxburg U, Bengio S, et al. , eds. Advances in Neural Information Processing Systems. Vol 30. Long Beach, CA: Curran Associates Inc; 2017:6533–6542. https://proceedings.neurips.cc/paper/2017/file/f507783927f2ec2737ba40afbd17efb5-Paper.pdf. Accessed January 11, 2022. [Google Scholar]
- 24.Namata G, London B, Getoor L, Huang B. Query-driven active surveying for collective classification. In: Proceedings of the Workshop on Mining and Learning with Graphs (MLG-2012). 2012. http://linqs.cs.umd.edu/basilic/web/Publications/2012/namata:mlg12-wkshp/namata-mlg12.pdf. Accessed March 29, 2022. [Google Scholar]
- 25.London B, Getoor L. Collective classification of network data. In: Aggarwal CC, ed. Data Classification: Algorithms and Applications. Boca Raton, FL: Chapman and Hall/CRC Press; 2014:399–416. doi: 10.1201/b17320 [DOI] [Google Scholar]
- 26.Bhattacharya I, Getoor L. Collective entity resolution in relational data. ACM Trans Knowl Discov Data. 2007;1(1):5–es. doi: 10.1145/1217299.1217304 [DOI] [Google Scholar]
- 27.Kipf TN, Welling M. Semi-supervised classification with graph convolutional networks. Paper presented at: 5th International Conference on Learning Representations, ICLR 2017; April 24–26, 2017; Toulon, France. [Google Scholar]
- 28.Chen RJ, Lu MY, Shaban M, et al. Whole slide images are 2D point clouds: context-aware survival prediction using patch-based graph convolutional networks. In: de Bruijne M, Cattin PC, Cotin S, et al, eds. Medical Image Computing and Computer Assisted Intervention—MICCAI 2021. Cham, Switzerland: Springer International Publishing; 2021:339–349. [Google Scholar]
- 29.Deng J, Dong W, Socher R, Li L-J, Li K, Li F-F. ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Miami, FL. 2009:248–255. doi:10.1109/CVPR.2009.5206848 [Google Scholar]
- 30.Wei JW, Suriawinata AA, Vaickus LJ, et al. Evaluation of a deep neural network for automated classification of colorectal polyps on histopathologic slides. JAMA Netw Open. 2020;3(4):e203398. doi: 10.1001/jamanetworkopen.2020.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bentley JL. Multidimensional binary search trees used for associative searching. Commun ACM. 1975;18(9):509–517. doi: 10.1145/361002.361007 [DOI] [Google Scholar]
- 32.Zhang M, Cui Z, Neumann M, Chen Y. An end-to-end deep learning architecture for graph classification. In: Proceedings of the AAAI Conference on Artificial Intelligence. 2018;32(1). https://ojs.aaai.org/index.php/AAAI/article/view/11782. Accessed March 29, 2022. [Google Scholar]
- 33.Lee J, Lee I, Kang J. Self-attention graph pooling. In: Chaudhuri K, Salakhutdinov R, eds. Proceedings of the 36th International Conference on Machine Learning. Vol 97. Long Beach, CA: Proceedings of Machine Learning Research; 2019:3734–3743. [Google Scholar]
- 34.Freund Y, Schapire RE. A decision-theoretic generalization of on-line learning and an application to boosting. J Comput Syst Sci. 1997;55(1):119–139. doi: 10.1006/jcss.1997.1504 [DOI] [Google Scholar]
- 35.Ho TK. Random decision forests. In: Proceedings of 3rd International Conference on Document Analysis and Recognition. Montreal, Canada: IEEE; 1995:278–282. doi:10.1109/ICDAR.1995.598994 [Google Scholar]
- 36.Breiman L, Friedman J, Stone CJ, Olshen RA. Classification and regression trees. Boca Raton, FL: Taylor & Francis; 1984. [Google Scholar]
- 37.Odze R, Goldblum J. Tumors of the pancreas. In: Odze and Goldblum Surgical Pathology of the GI Tract, Liver, Biliary Tract and Pancreas. 3rd ed. Philadelphia, PA: Saunders; 2014:1081–1119.e8. [Google Scholar]
- 38.Odze R, Goldblum J. Neuroendocrine tumors of the gastrointestinal and pancreatobiliary tracts. In: Odze and Goldblum Surgical Pathology of the GI Tract, Liver, Biliary Tract and Pancreas. 3rd ed. Philadelphia, PA: Saunders; 2014: 803–820.e2. [Google Scholar]


