Abstract
Dimethylsulfide (DMS) is the major biosulfur source emitted to the atmosphere with key roles in global sulfur cycling and potentially climate regulation. The main precursor of DMS is thought to be dimethylsulfoniopropionate. However, hydrogen sulfide (H2S), a widely distributed and abundant volatile in natural environments, can be methylated to DMS. The microorganisms and the enzymes that convert H2S to DMS, and their importance in global sulfur cycling were unknown. Here we demonstrate that the bacterial MddA enzyme, previously known as a methanethiol S-methyltransferase, could methylate inorganic H2S to DMS. We determine key residues involved in MddA catalysis and propose the mechanism for H2S S-methylation. These results enabled subsequent identification of functional MddA enzymes in abundant haloarchaea and a diverse range of algae, thus expanding the significance of MddA mediated H2S methylation to other domains of life. Furthermore, we provide evidence for H2S S-methylation being a detoxification strategy in microorganisms. The mddA gene was abundant in diverse environments including marine sediments, lake sediments, hydrothermal vents and soils. Thus, the significance of MddA-driven methylation of inorganic H2S to global DMS production and sulfur cycling has likely been considerably underestimated.
Subject terms: Biogeochemistry, Water microbiology, Biogeochemistry
Introduction
Dimethylsulfide (DMS) is a volatile organic sulfur compound that plays important roles in chemotaxis [1], global sulfur cycling and climate regulation [2–4]. It is the major organosulfur compound emitted to the atmosphere, representing 20 Tg of sulfur annually [5, 6]. Atmospheric DMS oxidation products serve as cloud condensation nuclei and aerosols that influence the global radiation budget and climate [3, 4, 7].
The abundant marine osmolyte dimethylsulfoniopropionate (DMSP), produced by many phytoplankton and bacteria [8], is regarded as the major biosource of DMS [9] through the action of bacterial and algal DMSP lyase enzymes [9–12]. However, there are many DMSP-independent biopathways that generate DMS, for example, microbial cycling of S-methyl-methionine, dimethyl sulfoxide (DMSO) and methoxyaromatic compounds [13–15]. Furthermore, microorganisms in oxic and anoxic terrestrial and marine environments generate DMS from the methylation of methanethiol (MeSH) [16–18], a reactive volatile derived from DMSP demethylation [19] and/or hydrogen sulfide (H2S) methylation [20, 21]. Many diverse bacteria (including cyanobacteria, Proteobacteria and Actinobacteria) which are abundant in sediment environments, contain the MeSH S-methyltransferase MddA [17, 18, 22], but the enzyme(s) responsible for H2S methylation have not been identified.
H2S is one of the Earth’s most common and abundant volatile sulfur compounds [6, 23], often reaching several hundred micromolar and several millimolar concentrations in hydrothermal vents and marine sediment environments [24–26]. However, H2S is toxic to cells because it inhibits cytochrome oxidase activity and blocks respiratory electron transport chains [27, 28]. Various H2S detoxification pathways exist in living organisms [29, 30], including the methylation of H2S to MeSH in higher animals and plants [29, 31]. Recently, a human thiol S-methyltransferase (METTL7B) was shown to catalyze H2S S-methylation to DMS rather than MeSH [32]. Previous studies have also shown that bacteria in anoxic sediments S-methylate H2S to DMS [20, 21], but to the best of our knowledge, H2S methylation by aerobic bacteria has not been reported.
Here we evaluated the methylation of H2S to DMS in various environmental samples, including seawater, marine and lake sediments, and soils, under oxic conditions. Furthermore, using Neptunicoccus sediminis, an aerobic Rhodobacteraceae bacterium isolated from marine sediment, as a model organism which converted H2S to DMS, we established that its mddA gene, formerly only known to S-methylate MeSH [18], was responsible for this H2S-dependent DMS production as a detoxification strategy. Biochemical analysis of the recombinant MddA confirmed its in vitro S-methylation of H2S via MeSH to produce DMS and the kinetic feasibility. Based on sequence alignment, structure prediction, and site-directed mutagenesis, the putative catalytic mechanism of MddA was proposed, which enabled identification of functional MddA enzymes in diverse Archaea and eukaryotic algae, not previously suspected of DMS production. Finally, bioinformatic analysis showed that mddA is widely distributed in diverse organisms and environments. Thus, H2S-dependent DMS production may play an important, yet largely unappreciated role in microbial H2S detoxification, global DMS emissions and sulfur cycling, and chemotaxis.
Materials and methods
Product analyses
Gas chromatography (GC) analyses were performed using a Nexis GC-2030 (Shimadzu, Japan) equipped with a flame photometric detector and a fused silica capillary column (30 m × 0.53 mm × 1 μm). The sample gas was injected into the GC using a purge-and-trap device. The carrier gas was nitrogen. The column temperature was 70 °C and the detector temperature was 250 °C. Standard curves for DMS were generated (using DMS standards from 0.1 nmol to 10 nmol) and used for quantification of DMS produced from sediment samples, bacterial cultures and enzyme reaction mixtures. The detecting lower limit of the method was 0.1 nmol. Volatile organic sulfur compounds were also determined by headspace GC-MS. The GC-MS analyses were performed using a Q Exactive GC Orbitrap GC-MS/MS System (Thermo Fisher Scientific, United States) equipped with a DB-5ms Ultra Inert GC column (Agilent Technologies, United States). All samples were analyzed in triplicate.
High performance liquid chromatography (HPLC) analyses were performed using an UltiMate 3000 (Thermo Fisher Scientific, United States) attached with a SunFire C18 reversed-phase column (4.6 × 250 mm, 5 μm particle size, Waters, United States). The detection wavelength was 260 nm, and injection volume was 10 μl. The samples were eluted with a linear gradient of 1–20% (v/v) acetonitrile in 50 mM ammonium acetate (pH 5.5) over 15 min at a flow rate of 1 ml/min. All samples were analyzed in triplicate. S-adenosyl-L-homocysteine (SAH) standard curves were established using SAH standards from 0.5 μM to 1 mM and used for quantitative detections. The detecting lower limit of the method was 5 pmol.
Assays of H2S methylation by environmental samples and bacteria
Marine sediments were collected using a box corer from the East China Sea (30°00′ N, 124°00′ E and 31°30′ N, 123°30′ E) and the South China Sea (21°00′ N, 117°30′ E). Lake surface sediments were collected from the East Lake (30°33′ N, 114°22′ E). Soil samples were collected from garden soil (36°11′ N, 120°29′ E). Samples in the upper 10 cm were collected from each location and stored at −20 °C. Seawater samples were collected from the North Pacific Ocean (17°24′ N, 153°09′ E) at a depth of 30 meters and was filtered (every 5 L) through a 0.22 μm pore size polyethersulfone membrane (Millipore, United States).
The marine sediment and seawater samples were resuspended with artificial seawater which was prepared using sea salts (Sigma-Aldrich, United States) while the terrestrial samples were resuspended with distilled water. Then 1 ml resuspended mixture was added to a 10 ml sterile glass vial. The saturated H2S aqueous solution (110 mM) was prepared by bubbling H2S gas into water at 25 °C. Then H2S solution was added to the vial to a final concentration of 1 mM. The vial was sealed immediately with aluminum crimp cap (molded polytetrafluoroethylene/butyl septa) using a manual crimper after the addition of H2S, and incubated at 20 °C for 48 h. The same volume of distilled water instead of H2S solution was used as the negative control. All operations were carried out under oxic conditions. GC was used for the detection of gaseous sulfur products as described above.
N. sediminis was cultured in Marine Broth 2216 medium (Becton, Dickinson and Company, United States) at 30 °C to an OD600 of 0.6. The cells were washed twice and resuspended with artificial seawater. Then 1 ml resuspended cells was transferred to a 10 ml sterile glass vial. H2S solution was added to the culture to a final concentration of 1 mM and then the vial was sealed with aluminum crimp cap (molded polytetrafluoroethylene/butyl septa) immediately. The vials were incubated at 20 °C for 24 h without shaking. The products were also detected using GC.
Pseudomonas deceptionensis M1T wild-type and ΔmddA strains cultures on LB medium [33] were adjusted to an OD600 of 0.6. Cells were washed twice with M9 minimal medium [33] and inoculated into fresh M9 medium with 2 mM H2S. After incubation at 30 °C, DMS generated from H2S was quantified by GC as described [18]. DMS production rates are expressed as pmol mg protein−1 min−1. The protein content in the cells was estimated by a Bradford method (BioRad, United States).
RT-qPCR analyses
N. sediminis was cultured in Marine Broth 2216 medium at 30 °C to an OD600 of 0.6. Then H2S solution was added to the culture to a final concentration of 1 mM. The same volume of distilled water was added to the culture as the negative control. Samples were collected 2 h after the addition of different additives. Each sample for qPCR was performed in triplicate. Total RNA extraction was performed using RNeasy Kit (QIAGEN, Germany). Reverse transcription was performed using the PrimeScript RT Reagent Kit (TaKaRa, Japan). The RT-qPCR reaction was performed using LightCycler 480 II (Roche, Switzerland). Data were analyzed by the 2−ΔΔCt method and recA was used was the reference gene. Significance was determined using paired two-tailed t test.
Genetic manipulations of N. sediminis
The knockout mutant ΔmddA of N. sediminis was constructed by homologous recombination. Two 1 kb DNA fragments, one upstream of mddA and one downstream of mddA, were amplified by PCR from its genomic DNA. The two DNA fragments were joined by overlapping PCR and cloned into plasmid pK18mobsacB-Ery [34]. The constructed vector was then conjugated into N. sediminis to generate ΔmddA mutant and ΔmddA mutants were confirmed by PCR and DNA sequencing, as described previously [34]. To complement the ΔmddA mutant, the mddA gene was amplified by PCR from N. sediminis genomic DNA and then cloned into plasmid pBBR1MCS-4 [35]. The constructed plasmid was conjugated into the ΔmddA mutant yielding the complemented strain ΔmddA/pBBRmddA. The primers used in this study are listed in Supplementary Table S1.
Protein expression and purification
All mddA genes were synthesized, codon optimized for expression in E. coli, by BGI Tech (China), and subcloned into the pET-22b vector to allow protein expression work with incorporation of a C-terminal hexahistidine tag. All site-directed mutations were introduced using QuikChange II mutagenesis kit (Agilent Technologies, United States) and were verified by sequencing. The expression plasmids were transformed into E. coli C43 (DE3) cells. Cells were grown in LB medium at 37 °C to an OD600 of 0.8. Then the culture was induced with 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) overnight at 20 °C. The cells were resuspended in a lysis buffer (50 mM Tris-HCl pH 8.0, 200 mM NaCl and 10% glycerol) and lysed using a high-pressure homogenizer. The lysate was centrifuged at 16,000 g for 20 min and the supernatant was then centrifuged at 200,000 g for 60 min to collect the membrane fraction. The membrane pellets were resuspended in lysis buffer containing 1.5% n-dodecyl-β-D-maltoside (DDM). After solubilization overnight at 4 °C, the mixture was centrifuged at 200,000 g for 30 min. The protein was purified using a Ni-nitriloacetic acid (Ni-NTA) agarose column followed by a size-exclusion chromatography column (Superdex 200, Cytiva, United States). The peak fractions were collected and stored at −80 °C.
Enzyme assays in vitro
Reaction mixtures (100 μl) contained 400 mM Tris-HCl (pH 8.0), 4 µg purified protein (or cell extract), 1 mM S-adenosyl-L-methionine (SAM) and 0.5 mM substrate (H2S, MeSH, potassium iodide (KI), potassium thiocyanate (KSCN), dithiothreitol (DTT), captopril or D-penicillamine). For apparent optimum pH assays, H2S was used as the substrate and Tris-HCl was replaced with Britton-Robinson buffer (20 mM final concentration). The reactions were carried out in 10 ml sterile glass vials. The vials were sealed immediately after the addition of the substrate. After incubation at 30 °C for 1 h, hydrochloric acid was injected to the reaction mixture to a final concentration of 0.1 M to terminate the reaction. The detection of gaseous DMS, MeSH and H2S was performed by GC while the detection of non-volatile SAH and SAM was performed using HPLC.
For kinetic parameter assays, the reaction mixtures (100 μl) contained 400 mM Tris-HCl (pH 8.0), 4 µg purified PdMddA, 20 mM SAM and varying concentrations of substrate (H2S, MeSH or DTT). The reactions were performed as described above. The production of SAH by PdMddA when using H2S as the substrate was halved because it is a two-step methylation. Non-linear fitting of the data was performed using the Origin software.
Growth analyses of N. sediminis and E. coli
N. sediminis cells were cultured in Marine Broth 2216 medium at 30 °C for 48 h, washed and diluted 1/50 into fresh media for growth analysis. E. coli cells were cultured in LB medium at 37 °C overnight, washed and diluted 1/100 into fresh LB medium for growth analysis. H2S (in solution) was added to the bacterial suspensions to a final concentration of 1 mM. The same volume of distilled water was added to the bacterial suspensions as negative controls. The bacterial suspensions with different additions were transferred to the wells in the microplate and then were incubated at 20 °C. The turbidity of the bacterial suspensions was measured at 600 nm using an FP-1100-C Automated Microbiology Growth Curve Analysis System (Bioscreen, Finland).
Bioinformatics analysis
The three-dimensional structure of PdMddA was predicted using AlphaFold2 [36]. The structural data can be obtained from the AlphaFold Protein Structure Database [37] with accession code A0A0F6P9C0. The figures of the structure were generated with PyMOL (https://pymol.org/2/).
To explore the distribution of mddA gene in all domains, verified sequences of MddA were used as reference to extract homologs from public databases. For bacteria and archaea, MddA homologs were extracted from NCBI NR database by BLASTp with an e-value of 1e-30 and a minimum identity of 40%. Considering huge amount of hits were detected, these hits were further clustered at 80% amino acid identity and one representative sequence of each cluster was kept using CD-HIT [38]. For Eukaryotes, MddA homologs were extracted from re-assemblies of Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) (10.5281/zenodo.740440) by hmmsearch with an e-value of 1e−30. To reduce false positives, only MddA sequences containing the catalytic histidine and five SAM binding residues were retained for tree building. Sequences alignment and trimming were respectively conducted by MAFFT [39] and TrimAl [40]. The tree was constructed using Maximum likelihood method in Fasttree [41]. Visualization of the tree was performed with iTOL [42].
The distribution of prokaryotic and eukaryotic mddA genes in global ocean was estimated in Tara Oceans datasets OM-RGC-v2 and MATOU, respectively. This analysis was conducted using the online webserver Ocean Gene Atlas [43] with hmmsearch (e-value < 1e−30) as the search method. Briefly, hmm databases based on the amino acid sequences of ratified mddA genes, two reference genes (DMSP lyase gene alma1 for eukaryotic mddA and DMSP lyase gene dddP for prokaryotic mddA), and recA genes for normalization were submitted to Ocean Gene Atlas for searching homologs in OM-RGC-v2 metagenomes/metatranscriptomes and MATOU metagenomes/metatranscriptomes. For MATOU, only samples with a filter size within 0.8–20 μm (picoplankton/nanoplankton) were included in our analyses.
Environmental metagenomes analysis was conducted using the online webserver from the Integrated Microbial Genomes & Microbiomes (IMG/M) system [44]. The MddA, DddP and RecA sequences used to search the metagenomes are summarized in Supplementary Table S2. The metagenomes examined in this study are listed in Supplementary Table S3. The homologs of MddA, DddP and RecA in metagenomes of different environments with an e-value cut-off of 1e−30 and a minimum identity of 30% to reference sequences were extracted using BLASTP from IMG/M analysis system [44]. The abundance of mddA and dddP in metagenomes of different environments were calculated by using the numbers of unique MddA and DddP sequences normalized to the number of unique RecA sequences.
Results
Microorganisms in diverse environments methylate H2S to DMS
H2S methylation to DMS is often prominent in anoxic sediments [20]; however, it is unclear whether H2S is methylated to DMS in oxic environments. Considering H2S is often abundant in marine sediments [24], we conducted oxic incubation experiments on East China Sea surface marine sediment to test for H2S-dependent DMS production. GC analysis showed that DMS production under oxic conditions was stimulated by H2S addition to the sediment, but no DMS was detected from heat-inactivated samples (Fig. 1A, B), suggesting that DMS formation from H2S was mediated by microorganisms. No MeSH was detected in these incubations, indicating that if MeSH was an intermediate in H2S-dependent DMS production it did not accumulate and was likely quickly S-methylated to DMS or lost to biotic and/or abiotic oxidative processes.
Fig. 1. Analyses of H2S S-methylation by environmental samples and microorganisms.
All experiments were conducted in triplicate. A GC analyses of the sulfur gases produced by marine sediment samples. Sediment samples were incubated with H2S solution (1 mM final concentration) or H2O (as the control). Heat-inactivated sediment was used as a microorganism-killed control. The major sulfur gas detected had the same retention time as the DMS standard. H2S was not detected in the heat-inactivated samples, which have resulted from H2S adsorption by the sediment. B GC-MS analysis of the sulfur gases liberated from the marine sediment in A. The fragment peaks showed that the main product was DMS. C H2S-dependent DMS produced from diverse environmental samples. D GC analyses of N. sediminis cells incubated with H2S solution (1 mM final concentration) or H2O (control). E DMS produced from H2S by N. sediminis wild-type and mutant strains. F In vitro SAM-dependent S-methylation of H2S activities of cell extracts of E. coli expressing the NsMddA and PdMddA. H2S was added to the mixture and DMS was produced. Cell extract of E. coli with empty pET-22b vector was used as negative control. G RT-qPCR analyses of NsmddA in N. sediminis. NsmddA gene transcription was significantly enhanced by growth in the presence of H2S, and not MeSH. ns not significant. *p < 0.05 (paired two-tailed t test).
To investigate the potential for H2S-dependent DMS production in more diverse oxic environments, we collected various environmental samples, including marine surface sediment, seawater, lake surface sediment, and soil, and incubated them with H2S under oxic conditions (see methods). All these samples generated DMS at 0.08–1.10 pmol DMS min−1 g−1 wet weight (0.009 pmol DMS min−1 L−1 for the seawater sample) (Fig. 1C), implying that oxic H2S-dependent DMS production occurs in diverse oxic environments when H2S is present. The observed DMS production rates varied in the diverse oxic samples, but the marine sediments produced more DMS than those from terrestrial samples.
N. sediminis S-methylates H2S to DMS
Bacteria isolated from marine sediment samples were screened for H2S-dependent DMS production to identify model bacteria with this ability. Of these isolates, N. sediminis, an aerobic Rhodobacteraceae member originally isolated from Yellow Sea sediment [45], methylated H2S to produce DMS at a rate of 150.8 pmol DMS min−1 g−1 wet weight under oxic conditions (Fig. 1D, E). No MeSH was detected in these incubations of N. sediminis with H2S (Fig. 1D), which was consistent with the sediment incubations described above.
Identification of a key enzyme driving H2S-dependent DMS production
The conversion of H2S to DMS is a methyl transfer reaction, and we postulated that a thiol (R-SH) S-methyltransferase (EC 2.1.1.9) was likely involved. To identify the methyltransferases catalyzing the conversion of H2S to DMS in N. sediminis, its genome was analyzed for candidate methyltransferases homologous to those known to methylate H2S [31, 32, 46] or other thiol S-methyltransferases (Supplementary Table S4). This analysis only identified a candidate MeSH S-methyltransferase MddA protein in N. sediminis (NsMddA, WP_069301345.1) with 46% amino acid identity to P. deceptionensis M1T MddA (PdMddA, WP_048359798.1), which S-methylates MeSH to DMS but was previously reported to lack the ability to S-methylate H2S [18].
We next investigated whether the putative NsMddA was involved in H2S-dependent DMS production. The results showed that E. coli cell extracts expressing cloned NsmddA showed in vitro SAM-dependent DMS production from H2S (Fig. 1F), indicating that NsMddA likely S-methylates H2S to MeSH and then MeSH to DMS. In addition, PdMddA also showed in vitro SAM-dependent DMS production from H2S in E. coli extracts (Fig. 1F) under the conditions used here, which contradicted the previous work done on this protein [18]. To confirm that MddA was responsible for in vivo H2S-dependent methylation, ultimately generating DMS, we constructed a N. sediminis ΔmddA knock out mutant, and analyzed its ability to S-methylate H2S along with the P. deceptionensis ΔmddA mutant [18]. H2S-dependent DMS production was ~2-fold reduced in the N. sediminis ΔmddA strain compared to the wild type N. sediminis strain, with this reduction in activity being fully restored to wild type levels by cloned NsMddA (Fig. 1E). Furthermore, H2S-dependent DMS production was abolished in the P. deceptionensis ΔmddA mutant (Supplementary Table S5). These data indicate that mddA in N. sediminis and P. deceptionensis encode a functional H2S S-methyltransferase that generates DMS via MeSH, but also that N. sediminis likely contains another unidentified H2S S-methyltransferase. This is consistent with previous research, in which diverse isolated bacteria had Mdd activity but lacked mddA in their genomes [17, 22]. The transcription of NsmddA was upregulated 3-fold when N. sediminis was grown with H2S, but not with MeSH (Fig. 1G), likely explaining the induction of DMS production seen in oxic sediment and water incubation experiments when H2S was added.
Characterization of the MddA enzyme
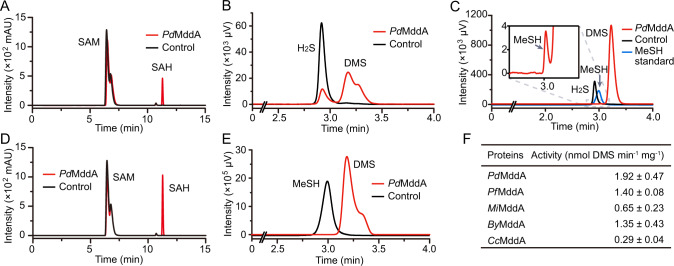
When expressed and purified from E. coli, the NsMddA protein was unstable and readily precipitated. Thus, the PdMddA, which was more stable when purified from E. coli, was used as the model protein to examine the enzymology of MddA. The PdMddA was shown to have in vitro SAM-dependent H2S and MeSH S-methyltransferase activity yielding SAH and DMS (Fig. 2A–E). In addition to PdMddA, several other purified MddA enzymes from the bacteria known to contain it [18] were also shown to have SAM-dependent S-methylation activity on H2S to produce DMS (Fig. 2F), indicating that the activity of MddA towards H2S is likely of universal significance to the MddA enzyme family. We noted that in H2S-dependent DMS production assays of MddA, the levels of MeSH detected were far lower than the DMS product (Fig. 2C). These data support the hypothesis that MddA primarily S-methylates H2S to the intermediate MeSH, which is subsequently S-methylated to DMS.
Fig. 2. Analyses of MddA enzymatic activities.
Enzymatic activities of PdMddA using H2S as the substrate analyzed by HPLC (A) and GC (B). The reaction mixture contained 1 mM SAM, 4 µg purified MddA and 0.5 mM H2S. The same reaction mixture but without the addition of PdMddA was used as the control. C GC analyses of intermediate MeSH in H2S methylation process. The peaks of MeSH are indicated by arrows. Enzymatic activity of PdMddA using MeSH as the substrate analyzed by HPLC (D) and GC (E). The reaction mixture contained 1 mM SAM, 4 µg purified MddA and 0.5 mM MeSH. The same reaction mixture but without the addition of PdMddA was used as the control. F Enzymatic activities of MddA homologs using H2S as the substrate. The MddAs included in the analyses are: PdMddA from P. deceptionensis M1T, PfMddA from P. fragi (Gammaproteobacteria), MiMddA from Mycobacterium intracellulare (Actinobacteria), ByMddA from Bradyrhizobium sp. YR681 (Alphaproteobacteria), CcMddA from Crocosphaera chwakensis (cyanobacteria).
The PdMddA enzyme had an apparent optimum pH of 8.0 and temperature of 30 °C (Supplementary Fig. S1). Thiol S-methyltransferases can catalyze the methylation of diverse substrates, including potassium iodide (KI), potassium thiocyanate (KSCN), dithiothreitol (DTT), captopril and D-penicillamine [31, 32, 46]. These compounds, in addition to several cellular thiols (L-cysteine, L-homocysteine, glutathione) and carboxylate substrates (2-(methylthio)acetic acid and thiodiglycolic acid), were used to test the substrate specificity of PdMddA. Of these potential substrates, PdMddA only S-methylated H2S, MeSH and DTT with Km values of 0.41 mM, 1.99 mM and 1.62 mM, respectively (Table 1 and Supplementary Fig. S2). PdMddA showed notably high kcat and kcat/Km values towards the artificial substrate DTT which does not accumulate in cells under natural conditions. The concentration of H2S in natural environments range from several hundred micromolar to several millimolar [25, 26], while MeSH is nanomolar [20]. Thus, H2S is the only substrate that will likely reach the Km levels for PdMddA in the environment. PdMddA exhibited a ~5-fold lower Km value towards the H2S than for MeSH (Table 1), which was higher than human [32] and rat [29] thiol S-methyltransferases but considerably lower than those in plants (Supplementary Table S6). The kcat and kcat/Km values of PdMddA towards MeSH were higher than for H2S (Table 1), indicating that the consumption rate of MeSH is likely higher than its production rate. This is consistent with the very low MeSH (intermediate) levels detected in comparison to DMS in H2S S-methylation enzyme assays (Fig. 2C). The higher specific activity of PdMddA towards MeSH than H2S, may reflect that the reactive gas MeSH is also toxic to cells if allowed to accumulate [47, 48], whereas DMS is not toxic.
Table 1.
Kinetic parameters of PdMddA with different substrates.
| Substrate | Km (mM) | kcat (s−1) | kcat/Km (M−1s−1) |
|---|---|---|---|
| H2S | 0.41 ± 0.04 | 7.1 × 10–3 | 17.07 |
| MeSH | 1.99 ± 0.11 | 93.0 × 10–3 | 46.73 |
| DTT | 1.62 ± 0.22 | 253.3 × 10–3 | 156.17 |
| KI | NAa | − | − |
| KSCN | NAa | − | − |
| Captopril | NAa | − | − |
| D-penicillamine | NAa | − | − |
| L-cysteine | NAa | − | − |
| L-homocysteine | NAa | − | − |
| Glutathione | NAa | − | − |
| 2-(methylthio)acetic acid | NAa | − | − |
| Thiodiglycolic acid | NAa | − | − |
aNo activity detected.
Key residues of PdMddA in methylation process
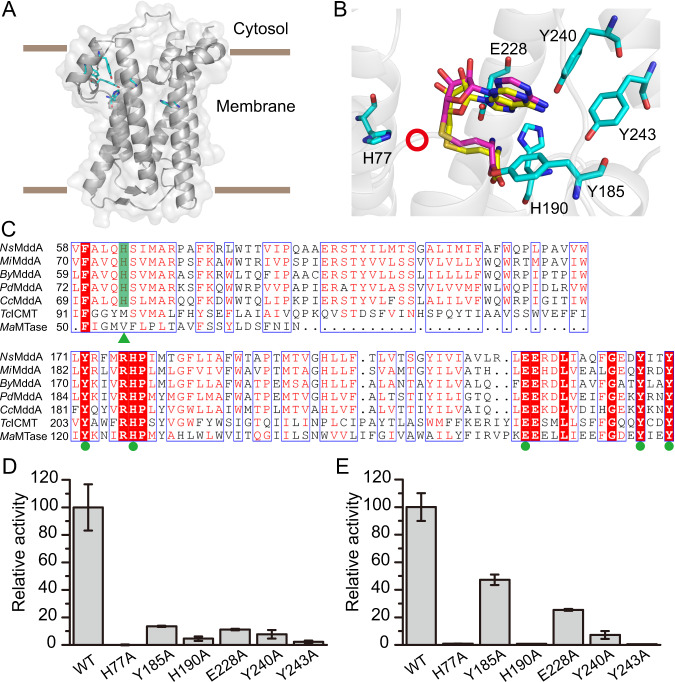
In X-ray crystallography work to elucidate the catalytic mechanism of H2S-dependent DMS production, we obtained PdMddA crystals but their diffractions were poor, and no structures could be solved. We therefore predicted the structure of PdMddA using AlphaFold2 [36], only analyzing residues R10-S253 that had a > 70 confidence score. PdMddA was predicted to comprise eight main transmembrane helices, with several small helices and two antiparallel β-strands (Fig. 3A).
Fig. 3. Important PdMddA residues for enzymatic activity.
A The AlphaFold2 predicted structure of PdMddA, with key residues colored in cyan. B Key residues of the predicted PdMddA structure. The structures of MaMTase, TcICMT and PdMddA are superimposed and the SAH molecules of MaMTase and TcICMT are represented in yellow and magenta, respectively. The residues of PdMddA analyzed in this study are colored in cyan. The potential position of H2S is indicated by a red circle. C Sequence alignment of MddA enzymes and other known methyltransferases. The amino acid sequences included in the alignment are: NsMddA from N. sediminis, MiMddA from M. intracellulare, ByMddA from B. sp. YR681, PdMddA from P. deceptionensis M1T, CcMddA from C. chwakensis, TcICMT from T. castaneum and MaMTase from M. acetivorans. The catalytic histidines are indicated with a green triangle. Residues participate in SAM binding are indicated with green dots. D Relative activity of wild-type PdMddA and its derivatives with the indicated amino acid substitutions, towards H2S. The enzymatic activity of wild-type PdMddA towards H2S is defined as 100%. Data represent the average of three biological replicates with their respective standard deviations. E Relative activity of wild-type PdMddA and derived variants on MeSH.
Analyzing the Protein Data Bank (PDB), PdMddA was most similar to Tribolium castaneum isoprenylcysteine carboxyl methyltransferase (TcICMT, PDB code 5V7P, 27% identity, 33% coverage) and Methanosarcina acetivorans isoprenylcysteine carboxyl methyltransferase (MaMTase, PDB code 4A2N, 30% identity, 27% coverage). Despite these low sequence identities, structural alignment showed that the structures of PdMddA, TcICMT and MaMTase shared four of the eight predicted transmembrane helices (Supplementary Fig. S3). Sequence and structural alignments also showed that five conserved residues (Y185, H190, E228, Y240, Y243) of PdMddA were located near to the SAH molecules of MaMTase and TcICMT (Fig. 3B, C), suggesting that these residues may participate in SAM binding. Substitution of these residues to alanine significantly decreased the enzymatic activity of PdMddA towards H2S and MeSH (Fig. 3D, E). Residue H77 of PdMddA, strictly conserved in MddA homologs but not in TcICMT or MaMTase (Fig. 3C), was located near the sulfur atom of SAH (Fig. 3B). Biochemical analyses showed that substituting H77 to alanine completely abolished the S-methyltransferase activity of PdMddA towards both H2S and MeSH (Fig. 3D, E), implying that H77 could be the catalytic residue.
The PdMddA proposed catalytic mechanism
SAM-dependent methylation is an SN2 nucleophilic replacement reaction [49, 50]. We predicted the PdMddA catalytic mechanism for H2S S-methylation based on biochemical and structural analyses, which is likely common to all MddA proteins. The S-methylation of H2S can be divided into two steps (Fig. 4A). In the first step, PdMddA binds the SAM and H2S molecules. H77 acts as a general base and abstracts a proton from the H2S molecule. The movement of electrons drives the transfer of the methyl group from SAM to the sulfur atom of H2S, resulting in the formation of MeSH. Subsequently, SAH is released from the active site and a new SAM molecule binds to the active site. The second step is similar to the first step. The residue H77 would become uncharged again in solution, deprotonate the MeSH molecule, and facilitate electron transfer, triggering the methyl transfer from SAM to MeSH, thus enabling the formation of DMS (Fig. 4A). In addition to acting as a base, the residue H77 may also help stabilize the binding of substrate to the active site and retain MeSH at the active site by hydrogen bonding for subsequent conversion to DMS.
Fig. 4. The predicted catalytic mechanism of MddA and the enzymatic activities of MddA-like proteins in algae and archaea.
A The proposed catalytic mechanism of H2S methylation by PdMddA. Putative electron transfers are indicated by red arrows. B Enzymatic activities of MddA-like proteins in algae and archaea. The MddA-like proteins included in the analyses are: PpMddA from P. provasolii (chlorophyte algae), NrMddA from N. sp. RCC80, LgMddA from Lotharella globosa CCCM811, CfMddA from C. fragilis CCMP3189, HwMddA from Haladaptatus sp. W1 (halotolerant archaea), HpMddA from Haladaptatus sp. PSR5 (halotolerant archaea). The same reaction mixture without the addition of enzyme was used as negative control. C Distribution of MddA in bacteria, archaea and eukaryotes. Branches are colored according to their taxonomic affiliations at phylum or class level. All ratified MddA sequences are marked in red solid circles. The dark stars are used to mark phylum containing ratified MddA. Tree scale indicates evolutionary distance as rate of substitution per site. The scale bar corresponds to 0.1.
Mechanistic informed extension of the MddA enzyme family
With insight into key residues for MddA-driven H2S and MeSH S-methylation, we re-evaluated the occurrence of this enzyme in more diverse organisms. MddA-like proteins with the predicted catalytic H77 and conserved SAM binding residues were identified in many more diverse organisms than were previously reported in Carrión et al. [18], including some chlorophyte (Pycnococcus provasolii), diatom (Nitzschia sp. RCC80), Chlorarachniophyte (Lotharella globosa CCCM811) and Ochraphyta (Chrysocystis fragilis CCMP3189) algae and halotolerant archaea (Haladaptatus spp.) (Supplementary Fig. S4), not before suspected to produce DMS from H2S or MeSH. Given that MddA proteins were only thought to exist in diverse bacteria and cyanobacteria, these more diverse candidate MddA enzymes from algae (P. provasolii, PpMddA; N. sp. RCC80, NrMddA; L. globose, LgMddA and C. fragilis, CfMddA) and archaea (Haladaptatus sp. W1, HwMddA; Haladaptatus sp. PSR5, HpMddA) were expressed and purified from E. coli and tested for SAM-dependent H2S S-methylation generating DMS. These proteins all showed in-vitro S-methylation activity on H2S and produced DMS (Fig. 4B). By searching homologs of ratified MddA sequences from NCBI NR database and MMETSP (Marine Microbial Eukaryote Transcriptome Sequencing Project) database, we found mddA genes are widely distributed in many bacterium phyla (e.g., proteobacteria, acidobacteria and actinobacteria) and several eukaryotic phyla (Chlorophyta, Bacillariophyta, Cercozoa, and Rhodophyta) (Fig. 4C and Supplementary Table S7). These results showed that some diverse algae and archaea likely S-methylate H2S and generate DMS in a DMSP-independent pathway, thus significantly extending the domains of life that conduct this process.
The potential for H2S-dependent DMS production in diverse environments
Previous research suggested that bacteria with mddA are highly abundant (3–77% relative abundance, RA) in diverse soils, rhizosphere and surface saltmarsh sediment environments, but extremely scarce (0.01% RA) in marine water samples [17, 18, 22]. This led to the prediction that MeSH-, and now H2S-dependent DMS production was likely a significant process in sediment environments but insignificant in marine environments [17, 18, 22]. As this study significantly extended the suite and diversity organisms known to contain a functional mddA gene, we re-examined the importance of mddA in more diverse marine settings in comparison to the most abundant DMSP lyase gene dddP.
Analysis of metagenomes from diverse environments confirmed previous work by showing mddA to be far more abundant in marine or terrestrial sediments than in diverse seawater or freshwater samples (Fig. 5A). In addition, it was clear that mddA in hydrothermal vents was also abundant (Fig. 5A). This was not surprising given H2S is naturally abundant in many sediment and hydrothermal vent environments [24–26]. The previously held prediction that MeSH- and now H2S-dependent DMS production, was an important process in sediment environments is strongly supported here. Compared to dddP, mddA was less abundant in most of our examined environments, except for lake sediment and soil environments (Fig. 5A). Within marine water samples, eukaryotic Alma1 and bacterial dddP DMSP lyase gene sequences and transcripts were more abundant than their mddA equivalents (Supplementary Fig. S5A, B). These data imply that MddA-driven DMS production is a considerable, although may not be the major, source of DMS in H2S-rich environments (Fig. 5B).
Fig. 5. MddA mediated H2S S-methylation in natural environments.
A Relative abundance of MddA and DddP protein-encoding sequences in different environmental metagenomes. The numbers of MddA and DddP sequences were normalized to the number of RecA sequences in each metagenome. B Diagram of H2S methylation mediated by aerobic bacteria. Sulfate is transformed into H2S by sulfate-reducing microorganisms in soil and marine sediments. H2S can also be directly released from oceanic crust through hydrothermal vents. H2S from both terrestrial environments and marine environments can be methylated to MeSH then to DMS, which act as a detoxification process.
It was difficult to resolve the taxonomy of many of the prokaryotic mddA genes beyond the bacterial domain and Proteobacterial phylum in most metagenomic samples (see Supplementary Fig. S5 for marine samples). The marine eukaryotic mddA genes were more easily assigned and were affiliated to Chlorophyta, Cercozoa and Bacillariophyta or, unexpectedly, more than half of these sequences were from Appendicularia, a group of solitary and free-swimming tunicates found throughout the world’s oceans (Supplementary Fig. S5C, D). The Appendicularia had not previously been linked to DMS production whatsoever.
Role for MddA in detoxification of H2S
H2S is a toxic compound, whereas, to our knowledge, DMS is not harmful to organisms. Thus, the MddA-driven conversion of H2S to DMS may be a cellular detoxification strategy. To test this hypothesis, we examined the growth characteristics of wild type N. sediminis ΔmddA strain in the presence of H2S. Growth of the ΔmddA mutant was significantly impaired by the addition of 2 mM H2S compared to the wild type strains and this phenotype was fully complemented by cloned mddA (Fig. 6A, B). In addition to work in N. sediminis, we also performed growth inhibition work in E. coli expressing MddA. The growth of E. coli in the presence of 2 mM H2S was enhanced by the expression of MddA (Fig. 6C, D). These data indicate that MddA plays a role in detoxification of H2S in diverse organisms and environments.
Fig. 6. Growth of N. sediminis and E. coli in response to H2S.
A Growth of N. sediminis strains with H2O (control). B Growth of N. sediminis strains in the presence of H2S. C Growth of E. coli with H2O added (control). D Growth curve of E. coli strains amended with H2S. H2S was used at 1 mM final concentration. Values shown represent the average of three biological replicates with their respective standard deviations.
Discussion
H2S and DMS are abundant and important forms of inorganic and organic sulfur, respectively, in natural environments. The S-methylation of H2S to DMS via MeSH represents a biological route from inorganic to organic sulfur (Fig. 5B). H2S readily reacts with metal ions to produce metal sulfides, can be oxidized to thiosulfate by dissolved organic matter in sediments [51], and is highly toxic to cells by virtue of inhibiting cytochrome c oxidase activity electron transport chains [27, 28]. In contrast, DMS is relatively stable in environments and is non-toxic to cells [52]. The majority of DMS in the environment is degraded by microbial or, to a lesser extent, photochemical processes [52]. Some bacteria utilize DMS as a source of reduced carbon and/or sulfur and/or energy [53]. Therefore, through the methylation of H2S, some bacteria (not necessarily those generating the DMS via MddA) can incorporate sulfur from H2S into organic matter necessary for growth.
Several thiol S-methyltransferase enzymes that catalyze H2S methylation to DMS had been identified in higher animals and plants [29, 31, 32], but to the best of our knowledge, there had been no aerobic bacteria, archaea or algae reported to have this activity. Our data showed that many and diverse organisms including aerobic bacteria and, likely, archaea and photosynthetic bacteria, and algae, S-methylate H2S to produce DMS via the MddA S-methyltransferase for protection against the cellular toxicity of H2S and potentially MeSH.
MddA is located in the cell membrane, which may be important for bacteria to respond to H2S toxicity since it was reported that no channels or facilitators were needed for H2S to permeate cell membranes [54, 55]. Therefore, environmental H2S gas can easily enter the cell. The partition coefficient of H2S between membrane and water is ~2, and thus, H2S concentration in the membrane would be higher than the concentration in cytosol [55, 56]. In addition, terminal oxidases in cellular respiration such as cytochrome c oxidase and cytochrome bo3 oxidase, which H2S inhibits, are also membranous enzymes. In addition to detoxification, S-methylation of H2S to DMS may also play other physiological roles. For example, the DMS produced from H2S S-methylation may act as a sulfur and/or an energy source for some bacteria [57–59]; DMS and/or its oxidation product DMSO [60] may serve as antioxidants to protect against oxidative stress [61]; or DMS could act as chemical signaling molecule to attract or deter grazers [1, 62].
H2S S-methylation to yield DMS was previously observed in anoxic freshwater sediments [20]. Furthermore, previous research showed that grassland and forest soils, and saltmarsh sediment samples displayed MeSH-dependent DMS production [17, 18]. Here, we demonstrate that diverse soil, marine and lake sediments, and seawater samples display significant levels of H2S-dependent DMS production. H2S-dependent DMS production is likely more prominent than suggested from our examination of environmental omics datasets because many organisms exhibiting MeSH and/or H2S-dependent DMS production phenotypes contain unknown enzymes with these activities since their genomes lack mddA [17] or they contain multiple different enzymes with these activities, as was predicted for N. sediminis. It will be important to identify these enzymes in the future to better evaluate the environmental significance of H2S- and MeSH-dependent DMS production.
The data presented here imply that both eukaryotic and prokaryotic MddA-driven H2S and/or MeSH-dependent DMS production pathways are important in aquatic and marine settings, but that their importance vastly increases in marine and terrestrial sediment and hydrothermal vent settings where mddA sequences can be relatively abundant. Considering that H2S is abundant in various environments and our re-evaluation of the abundance of mddA in diverse organisms and environments, the significance of MddA in global DMS production influencing atmospheric chemistry and potentially climate, and sulfur cycling has been previously underestimated.
Supplementary information
Acknowledgements
We thank Terry McGenity from University of Essex for providing the Haladaptatus sp. W1 strain, and Emese Bartha from University of East Anglia for providing advice on how to culture it. This work was supported by the Marine S&T Fund of Shandong Province for Qingdao Marine Science and Technology Center (No. 2022QNLM030004-3), the National Key Research and Development Program of China (2022YFC2807500), the National Science Foundation of China (grants 42276102, 92251303, 42076229, 31961133016), the Fundamental Research Funds for the Central Universities (202172002, 202041011), the Major Scientific and Technological Innovation Project (MSTIP) of Shandong Province (2019JZZY010817), the Program of Shandong for Taishan Scholars (tspd20181203), the Biotechnology and Biological Sciences Research Council, UK, grant (BB/X005968), Natural Environment Research Council, UK, Standard grants (NE/X000990, NE/V000756 and NE/S001352) and the Leverhulme Trust research grant (RPG-2020-413).
Author contributions
CYL and YZZ designed and directed the research. JDT designed some experiments. HYC, CYL, QW, OC, XZ and JM performed the experiments. PW, XZ and XLC helped in data analysis. HYC, CYL, JDT and YZZ wrote the manuscript. XLC edited the manuscript.
Data availability
The predicted structure can be obtained from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/) with accession code A0A0F6P9C0. The sequences of MddA and other proteins can be found in NCBI database as well as Supplementary Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chun-Yang Li, Hai-Yan Cao.
Contributor Information
Hai-Yan Cao, Email: haiyancao92@126.com.
Yu-Zhong Zhang, Email: zhangyz@sdu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01430-z.
References
- 1.Shemi A, Alcolombri U, Schatz D, Farstey V, Vincent F, Rotkopf R, et al. Dimethyl sulfide mediates microbial predator-prey interactions between zooplankton and algae in the ocean. Nat Microbiol. 2021;6:1357–66.. doi: 10.1038/s41564-021-00971-3. [DOI] [PubMed] [Google Scholar]
- 2.De Zwart JMM, Kuenen JG. C1-cycle of sulfur compounds. Biodegradation. 1992;3:37–59. [Google Scholar]
- 3.Charlson RJ, Lovelock JE, Andreae MO, Warren SG. Oceanic phytoplankton, atmospheric sulfur, cloud albedo and climate. Nature. 1987;326:655–61.. [Google Scholar]
- 4.Quinn PK, Bates TS. The case against climate regulation via oceanic phytoplankton sulphur emissions. Nature. 2011;480:51–6. doi: 10.1038/nature10580. [DOI] [PubMed] [Google Scholar]
- 5.Kettle AJ, Andreae MO. Flux of dimethylsulfide from the oceans: a comparison of updated data seas and flux models. J Geophys Res Atmos. 2000;105:26793–808.. [Google Scholar]
- 6.Watts SF. The mass budgets of carbonyl sulfide, dimethyl sulfide, carbon disulfide and hydrogen sulfide. Atmos Environ. 2000;34:761–79.. [Google Scholar]
- 7.Vallina SM, Simo R. Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science. 2007;315:506–8. doi: 10.1126/science.1133680. [DOI] [PubMed] [Google Scholar]
- 8.Curson ARJ, Liu J, Martinez AB, Green RT, Chan YH, Carrion O, et al. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat Microbiol. 2017;2:17009. doi: 10.1038/nmicrobiol.2017.9. [DOI] [PubMed] [Google Scholar]
- 9.Curson AR, Todd JD, Sullivan MJ, Johnston AW. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Microbiol. 2011;9:849–59. doi: 10.1038/nrmicro2653. [DOI] [PubMed] [Google Scholar]
- 10.Yoch DC. Dimethylsulfoniopropionate: Its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl Environ Microbiol. 2002;68:5804–15. doi: 10.1128/AEM.68.12.5804-5815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A. Identification of the algal dimethyl sulfide-releasing enzyme: A missing link in the marine sulfur cycle. Science. 2015;348:1466–9. doi: 10.1126/science.aab1586. [DOI] [PubMed] [Google Scholar]
- 12.Li CY, Wang XJ, Chen XL, Sheng Q, Zhang S, Wang P, et al. A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA. eLife. 2021;10:e64045. doi: 10.7554/eLife.64045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiene RP, Hines ME. Microbial formation of dimethyl sulfide in anoxic sphagnum peat. Appl Environ Microbiol. 1995;61:2720–6. doi: 10.1128/aem.61.7.2720-2726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zinder SH, Brock TD. Dimethyl sulphoxide reduction by micro-organisms. J Gen Microbiol. 1978;105:335–42. doi: 10.1099/00221287-105-2-335. [DOI] [PubMed] [Google Scholar]
- 15.Spiese CE, Kieber DJ, Nomura CT, Kiene RP. Reduction of dimethylsulfoxide to dimethylsulfide by marine phytoplankton. Limnol Oceanogr. 2009;54:560–70.. [Google Scholar]
- 16.Stets EG, Hines ME, Kiene RP. Thiol methylation potential in anoxic, low-pH wetland sediments and its relationship with dimethylsulfide production and organic carbon cycling. FEMS Microbiol Ecol. 2004;47:1–11. doi: 10.1016/S0168-6496(03)00219-8. [DOI] [PubMed] [Google Scholar]
- 17.Carrion O, Pratscher J, Curson ARJ, Williams BT, Rostant WG, Murrell JC, et al. Methanethiol-dependent dimethylsulfide production in soil environments. ISME J. 2017;11:2379–90.. doi: 10.1038/ismej.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrion O, Curson AR, Kumaresan D, Fu Y, Lang AS, Mercade E, et al. A novel pathway producing dimethylsulphide in bacteria is widespread in soil environments. Nat Commun. 2015;6:6579. doi: 10.1038/ncomms7579. [DOI] [PubMed] [Google Scholar]
- 19.Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature. 2011;473:208–11. doi: 10.1038/nature10078. [DOI] [PubMed] [Google Scholar]
- 20.Lomans BP, Smolders AJP, Intven LM, Pol A, denCamp HJMO, vanderDrift C. Formation of dimethyl sulfide and methanethiol in anoxic freshwater sediments. Appl Environ Microbiol. 1997;63:4741–7. doi: 10.1128/aem.63.12.4741-4747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bak F, Finster K, Rothfuss F. Formation of dimethylsulfide and methanethiol from methoxylated aromatic compounds and inorganic sulfide by newly isolated anaerobic bacteria. Arch Microbiol. 1992;157:529–34.. [Google Scholar]
- 22.Carrion O, Pratscher J, Richa K, Rostant WG, Ul Haque MF, Murrell JC, et al. Methanethiol and dimethylsulfide cycling in stiffkey saltmarsh. Front Microbiol. 2019;10:1040. doi: 10.3389/fmicb.2019.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreae MO. Ocean-atmosphere interactions in the global biogeochemical sulfur cycle. Mar Chem. 1990;30:1–29. [Google Scholar]
- 24.Bagarinao T. Sulfide as an environmental factor and toxicant: tolerance and adaptations in aquatic organisms. Aquat Toxicol. 1992;24:21–62. [Google Scholar]
- 25.Johnson KS, Beehler CL, Sakamoto-Arnold CM, Childress JJ. In situ measurements of chemical distributions in a deep-sea hydrothermal vent field. Science. 1986;231:1139–41. doi: 10.1126/science.231.4742.1139. [DOI] [PubMed] [Google Scholar]
- 26.Thompson BE, Bay SM, Anderson JW, Laughlin JD, Greenstein DJ, Tsukada DT. Chronic effects of contaminated sediments on the urchin Lytechinus pictus. Environ Toxicol Chem. 1989;8:629–37.. [Google Scholar]
- 27.Malone Rubright SL, Pearce LL, Peterson J. Environmental toxicology of hydrogen sulfide. Nitric Oxide. 2017;71:1–13. doi: 10.1016/j.niox.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuevasanta E, Moller MN, Alvarez B. Biological chemistry of hydrogen sulfide and persulfides. Arch Biochem Biophys. 2017;617:9–25. doi: 10.1016/j.abb.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Weisiger RA, Pinkus LM, Jakoby WB. Thiol S-methyltransferase: suggested role in detoxication of intestinal hydrogen sulfide. Biochem Pharm. 1980;29:2885–7. doi: 10.1016/0006-2952(80)90029-5. [DOI] [PubMed] [Google Scholar]
- 30.Sun Y, Wang M, Zhong Z, Chen H, Wang H, Zhou L, et al. Adaption to hydrogen sulfide-rich environments: Strategies for active detoxification in deep-sea symbiotic mussels, Gigantidas platifrons. Sci Total Environ. 2022;804:150054. doi: 10.1016/j.scitotenv.2021.150054. [DOI] [PubMed] [Google Scholar]
- 31.Itoh N, Toda H, Matsuda M, Negishi T, Taniguchi T, Ohsawa N. Involvement of S-adenosylmethionine-dependent halide/thiol methyltransferase (HTMT) in methyl halide emissions from agricultural plants: isolation and characterization of an HTMT-coding gene from Raphanus sativus (daikon radish) BMC Plant Biol. 2009;9:116. doi: 10.1186/1471-2229-9-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maldonato BJ, Russell DA, Totah RA. Human METTL7B is an alkyl thiol methyltransferase that metabolizes hydrogen sulfide and captopril. Sci Rep. 2021;11:4857. doi: 10.1038/s41598-021-84218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Russell DW. Molecular cloning, a laboratory manual. 3rd ed. New York, USA: Cold Spring Harbor Laboratory Press; 2001.
- 34.Wang PX, Yu ZC, Li BY, Cai XS, Zeng ZS, Chen XL, et al. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Micro Cell Fact. 2015;14:11. doi: 10.1186/s12934-015-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–6. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 36.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D44.. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–6. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W6.. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vernette C, Lecubin J, Sanchez P, Tara Oceans C, Sunagawa S, Delmont TO, et al. The Ocean Gene Atlas v2.0: online exploration of the biogeography and phylogeny of plankton genes. Nucleic Acids Res. 2022;50:W516–26. doi: 10.1093/nar/gkac420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen IA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, et al. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res. 2021;49:D751–D63.. doi: 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang YJ, Liu XF, Kuang BZ, Zhang XY, Zhou MY, Chen S. Neptunicoccus sediminis gen. nov., sp. nov., a member of the family Rhodobacteraceae isolated from the Yellow Sea. Int J Syst Evol Microbiol. 2018;68:1702–6. doi: 10.1099/ijsem.0.002728. [DOI] [PubMed] [Google Scholar]
- 46.Toda H, Itoh N. Isolation and characterization of a gene encoding a S-adenosyl-L-methionine-dependent halide/thiol methyltransferase (HTMT) from the marine diatom Phaeodactylum tricornutum: Biogenic mechanism of CH3I emissions in oceans. Phytochemistry. 2011;72:337–43. doi: 10.1016/j.phytochem.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Furne J, Springfield J, Koenig T, DeMaster E, Levitt MD. Oxidation of hydrogen sulfide and methanethiol to thiosulfate by rat tissues: a specialized function of the colonic mucosa. Biochem Pharm. 2001;62:255–9. doi: 10.1016/s0006-2952(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 48.Wirth JS, Wang T, Huang QY, White RH, Whitman WB. Dimethylsulfoniopropionate sulfur and methyl carbon assimilation in Ruegeria species. mBio. 2020;11:e00329-20. [DOI] [PMC free article] [PubMed]
- 49.Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat Prod Rep. 2012;29:1238–50. doi: 10.1039/c2np20029e. [DOI] [PubMed] [Google Scholar]
- 50.Sun Q, Huang MY, Wei YQ. Diversity of the reaction mechanisms of SAM-dependent enzymes. Acta Pharmaceutica Sin B. 2021;11:632–50.. doi: 10.1016/j.apsb.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tobias H, Christian B. Oxidation and incorporation of hydrogen sulfide by dissolved organic matter. Chem Geol. 2006;235:12–20. [Google Scholar]
- 52.Kiene RP, Bates TS. Biological removal of dimethyl sulfide from sea-water. Nature. 1990;345:702–5. [Google Scholar]
- 53.Kappler U, Schafer H. Transformations of dimethylsulfide. Met Ions Life Sci. 2014;14:279–313. doi: 10.1007/978-94-017-9269-1_11. [DOI] [PubMed] [Google Scholar]
- 54.Mathai JC, Missner A, Kugler P, Saparov SM, Zeidel ML, Lee JK, et al. No facilitator required for membrane transport of hydrogen sulfide. Proc Natl Acad Sci USA. 2009;106:16633–8. doi: 10.1073/pnas.0902952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riahi S, Rowley CN. Why can hydrogen sulfide permeate cell membranes? J Am Chem Soc. 2014;136:15111–3. doi: 10.1021/ja508063s. [DOI] [PubMed] [Google Scholar]
- 56.Cuevasanta E, Denicola A, Alvarez B, Moller MN. Solubility and permeation of hydrogen sulfide in lipid membranes. PLoS One. 2012;7:e34562. doi: 10.1371/journal.pone.0034562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horinouchi M, Kasuga K, Nojiri H, Yamane H, Omori T. Cloning and characterization of genes encoding an enzyme which oxidizes dimethyl sulfide in Acinetobacter sp. strain 20B. FEMS Microbiol Lett. 1997;155:99–105. doi: 10.1111/j.1574-6968.1997.tb12692.x. [DOI] [PubMed] [Google Scholar]
- 58.Fuse H, Takimura O, Murakami K, Yamaoka Y, Omori T. Utilization of dimethyl sulfide as a sulfur source with the aid of light by Marinobacterium sp. strain DMS-S1. Appl Environ Microbiol. 2000;66:5527–32. doi: 10.1128/aem.66.12.5527-5532.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boden R, Murrell JC, Schafer H. Dimethylsulfide is an energy source for the heterotrophic marine bacterium Sagittula stellata. FEMS Microbiol Lett. 2011;322:188–93. doi: 10.1111/j.1574-6968.2011.02349.x. [DOI] [PubMed] [Google Scholar]
- 60.Lidbury I, Krober E, Zhang ZD, Zhu YJ, Murrell JC, Chen Y, et al. A mechanism for bacterial transformation of dimethylsulfide to dimethylsulfoxide: a missing link in the marine organic sulfur cycle. Environ Microbiol. 2016;18:2754–66. doi: 10.1111/1462-2920.13354. [DOI] [PubMed] [Google Scholar]
- 61.Sunda W, Kieber DJ, Kiene RP, Huntsman S. An antioxidant function for DMSP and DMS in marine algae. Nature. 2002;418:317–20. doi: 10.1038/nature00851. [DOI] [PubMed] [Google Scholar]
- 62.Teng ZJ, Wang P, Chen XL, Guillonneau R, Li CY, Zou SB, et al. Acrylate protects a marine bacterium from grazing by a ciliate predator. Nat Microbiol. 2021;6:1351–6. doi: 10.1038/s41564-021-00981-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The predicted structure can be obtained from the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/) with accession code A0A0F6P9C0. The sequences of MddA and other proteins can be found in NCBI database as well as Supplementary Information files.