Abstract
Background
Diet, a key component of type 1 diabetes (T1D) management, modulates the intestinal microbiota and its metabolically active byproducts—including SCFA—through fermentation of dietary carbohydrates such as fiber. However, the diet–microbiome relationship remains largely unexplored in longstanding T1D.
Objectives
We evaluated whether increased carbohydrate intake, including fiber, is associated with increased SCFA-producing gut microbes, SCFA, and intestinal microbial diversity among young adults with longstanding T1D and overweight or obesity.
Methods
Young adult men and women with T1D for ≥1 y, aged 19–30 y, and BMI of 27.0–39.9 kg/m2 at baseline provided stool samples at baseline and 3, 6, and 9 mo of a randomized dietary weight loss trial. Diet was assessed by 1–2 24-h recalls. The abundance of SCFA-producing microbes was measured using 16S rRNA gene sequencing. GC-MS measured fecal SCFA (acetate, butyrate, propionate, and total) concentrations. Adjusted and Bonferroni-corrected generalized estimating equations modeled associations of dietary fiber (total, soluble, and pectins) and carbohydrate (available carbohydrate, and fructose) with microbiome-related outcomes. Primary analyses were restricted to data collected before COVID-19 interruptions.
Results
Fiber (total and soluble) and carbohydrates (available and fructose) were positively associated with total SCFA and acetate concentrations (n = 40 participants, 52 visits). Each 10 g/d of total and soluble fiber intake was associated with an additional 8.8 μmol/g (95% CI: 4.5, 12.8 μmol/g; P = 0.006) and 24.0 μmol/g (95% CI: 12.9, 35.1 μmol/g; P = 0.003) of fecal acetate, respectively. Available carbohydrate intake was positively associated with SCFA producers Roseburia and Ruminococcus gnavus. All diet variables except pectin were inversely associated with normalized abundance of Bacteroides and Alistipes. Fructose was inversely associated with Akkermansia abundance.
Conclusions
In young adults with longstanding T1D, fiber and carbohydrate intake were associated positively with fecal SCFA but had variable associations with SCFA-producing gut microbes. Controlled feeding studies should determine whether gut microbes and SCFA can be directly manipulated in T1D.
Keywords: type 1 diabetes, fiber, gut microbiome, short-chain fatty acids, obesity
Introduction
Diet is a key contributor to weight and glycemia—cardiometabolic risk factors that are paramount to co-manage in people with type 1 diabetes (T1D) [1]. The American Diabetes Association provides general healthy eating guidelines for people with diabetes [2], but few people with T1D meet weight and glycemic targets [3, 4]. Numerous diet trials conducted in people without T1D demonstrate substantial heterogeneity in the metabolic response to diets of equivalent macronutrient composition [5]. This suggests the need to look beyond the known direct links between macronutrients and metabolic responses to identify novel modifiers of therapeutic diet efficacy.
One potential source of variability in the metabolic response to diet is how an individual’s gut microbiota—the complex community of microbes residing in the intestinal tract—and its fermentative metabolites shift in response to dietary change [6, 7]. A broad literature demonstrates potential biological mechanisms between diet, the gut microbiota and SCFA—metabolites generated by microbial fiber fermentation, and energy balance and glucose homeostasis in animal models [[8], [9], [10], [11], [12], [13]]. A much more limited body of evidence has demonstrated such links in humans [[14], [15], [16]] and none, to our knowledge, in people with T1D. Furthermore, although diet has a measurable effect on the composition and diversity of the intestinal microbiota, the effect size may be small in comparison with interpersonal variability in the gut microbiome [17]. Nonetheless, it is critical to determine whether augmenting the gut microbiota—whether through diet or supplementation—can have synergistic health benefits to the human host above and beyond those conferred by diet alone. This is particularly important to understand in people with T1D, who have an altered gut microbial ecosystem and reduced plasma SCFA compared with that of control individuals without T1D [18].
Studies conducted in individuals without diabetes have shown that dietary interventions change the composition of the intestinal microbiota and SCFA primarily through modulation of nondigestible carbohydrates such as fiber [particularly, soluble fiber [19, 20]] and, to a lesser extent, of simple carbohydrates [21] [although fructose may contribute appreciably to acetate production [22]]. The diet–gut microbiome relationship is critical to understand because dietary fiber may augment gut and systemic health through promoting gut microbial diversity (that is, the number of unique species in one’s intestinal microbiome) and intestinal barrier integrity [partly through SCFA production [23]], which may protect against opportunistic infection by otherwise harmless gut pathogens [24] and dampen low-grade, chronic inflammation. In addition, SCFA reduce adiposity in mice [9] and are associated with increased insulin sensitivity in adults with metabolic syndrome [16]. A limited number of observational studies have assessed the relationship between diet and the intestinal microbiota in people with T1D: 2 among youth near the time of the development of islet autoimmunity or T1D diagnosis [25, 26] and 1 that developed a predictive algorithm for glycemic response to standardized meals in adults with T1D, of which the composition of the intestinal microbiota was an important component and outperformed a model that emulated standard of care [7].
To bolster the very limited evidence base describing the diet–microbiome relationship in adults with longstanding T1D, we conducted an ancillary, hypothesis-generating pilot study to evaluate whether increased intake of dietary fiber (total, soluble, and pectins) and carbohydrate (available carbohydrate and fructose) was associated with an increased abundance of SCFA-producing gut microbes, fecal SCFA, and intestinal microbial diversity among young adults with longstanding T1D and overweight or obesity who participated in a randomized dietary weight loss pilot [27].
Methods
The design and main results of the Advancing Care for Type 1 Diabetes and Obesity Network (ACT1ON) pilot study has been described elsewhere [28, 29]. In brief, ACT1ON was a 9-mo pilot feasibility and acceptability pilot conducted at the University of North Carolina (UNC) and Stanford University to identify acceptable and effective dietary strategies (hypocaloric low carbohydrate, hypocaloric Look AHEAD, or Mediterranean diet without calorie restriction) to co-optimize weight (weight loss or weight maintenance if a BMI <25 kg/m2 was achieved during the study) and glycemia among adults with T1D aged 19–30 y (1DP3DK113358, NCT03651622). Registered dietitians provided guidelines about general healthy eating and specific dietary recommendations tailored to each diet assignment using motivational interviewing and problem-solving skills training aimed at overcoming barriers to adherence [30, 31]. The intervention consisted of 8 full-length counseling and education sessions and 15 shorter “check-in” sessions. Counseling strategies related to carbohydrate counting for insulin dosing and encouragement of usual physical activity were consistent across diets.
Primary outcomes were change in weight, hemoglobin A1c (HbA1c) concentration, and percentage of time in level 1 and level 2 hypoglycemia (<70 mg/dL) [32] as assessed by continuous glucose monitoring (CGM). Measurements were obtained at baseline and after each of 3 3-mo dietary periods. Secondary outcomes were change in percentage body fat as assessed by DXA, percent time in target glucose range (70–180 mg/dL), and percent time in level 2 hypoglycemia (<54 mg/dL). After an initial diet randomization, the sequential multiple assignment randomized trial design adapted dynamically to participant responses by rerandomizing those for whom the assigned diet was not acceptable or not effective [33]. This was performed based on a priori decision rules at intervention months 3 and 6. The rerandomization criteria included clinical outcomes [<2% weight reduction (unless weight loss resulted in a BMI <25 kg/m2), HbA1c increase of ≥0.5%, and self-reported increased or problematic hypoglycemia] and self-reported diet unacceptability. Measurement visits occurred at baseline and at the end of each diet period.
Study sample
This study was approved by the UNC and Stanford University institutional review boards, and study participation began after participants signed informed consent. Participants were adults aged 19–30 y at baseline with T1D for ≥1 y, with literacy in English, an HbA1c of <13.0% (<119 mmol/mol), and a BMI of 27.0–39.9 kg/m2. Participants were excluded if they presented with a history of a diagnosed eating disorder, gastrointestinal or bowel disorder, dietary restrictions that precluded following the study diets, were pregnant or lactating, had experienced any episode of diabetic ketoacidosis or level 2 hypoglycemia requiring third-party assistance in the last 6 mo, or were weight unstable [change of ±10 lb (kg) in the last 6 mo].
Young adults meeting all eligibility criteria according to medical record data were enrolled beginning on 12 November, 2018. The COVID-19 pandemic caused closure of the research clinics at both sites as of 27 March, 2020; thus, enrollment ceased with the last participant enrolled on 6 March, 2020 (68 of the anticipated 72 participants were enrolled). The study transitioned to a virtual format using a HIPAA-secure video conferencing platform (Zoom Video Communications), including dietary counseling and data collection, and recruitment ceased. This created the need to alter the measurement methods for the primary outcomes of weight and glycemia during COVID-19. Using standardized protocols with support from study staff, weight and waist measurements and HbA1c samples were collected at home, participants inserted CGM sensors at home, and DXA was discontinued. The final study visit was completed on 22 February, 2021. Given the alteration of measurement methods for the primary parent study outcomes, the changes to intervention delivery, and the delay in study visits owing to establishing these protocol changes, results of the COVID-19–influenced periods of the study are reported in Supplemental Material. In this study we primarily present data collected under the original study protocol before COVID-19 research clinic closures.
Ancillary gut microbiome pilot study
This analysis used data from the ACT1ON ancillary gut microbiome pilot study. ACT1ON participants who had not taken antibiotics in the last month were invited to provide fecal samples through home collection using provided collection kits during the 2-wk timeframe in which all other measurement visit data were collected (that is, before each diet period was initiated). Stool sample collection was added to measurement visits 3 and 4 during COVID-19 to consider an anticipated diminished sample size owing to COVID-19 dropouts. Thus, the primary analysis of pre–COVID-19 data includes data from the baseline (that is, preintervention) visit and measurement visit 2 (that is, after 3 mo of the diet intervention).
Of the 68 total enrolled parent ACT1ON Study participants, 6 were excluded from the ancillary gut microbiome pilot study for antibiotic use in the past 4 wk, 2 did not return a stool sample before randomization owing to shipment issues, 2 had difficulty with producing a sample, 4 declined to participate, and 9 agreed to participate but did not return the sample before diet randomization. Thus, 45 individuals participated in the ancillary gut microbiome study and provided 112 stool samples across 4 timepoints (approximately, at baseline and 3, 6, and 9 mo, corresponding to the parent study measurement visits). The final sample size in the pre–COVID-19 period consisted of 40 participants with 52 and 48 samples for SCFA and gut microbiome analysis, respectively. A CONSORT with the full sample size derivation, including for the supplemental analysis of all available data (both pre– and during COVID-19), is shown in Supplemental Figure 1.
Measurements
Dietary intake
At each measurement visit, 24-h dietary recalls were administered through telephone by trained UNC NIH/NIDDK Nutrition Obesity Research Center (NORC, P30DK056350) staff using a multipass method [34]. Given that the objective was to estimate usual intake [35], recalls deemed to be unreliable by NORC staff or for which participants indicated the amount consumed was “a lot more” or “a lot less” than usual were excluded from the analysis. Nutrition Data System for Research (NDSR) (version 2019; University of Minnesota, MN) was used to derive nutrients associated with recalled foods and beverages.
When 2 dietary recalls per participant were available for a given measurement visit, nutrient values were averaged per participant across the 2 d. Daily intake of fiber (total, soluble, and pectins), available carbohydrates (starch and sugars, that is, total carbohydrates minus total fiber), and fructose were used as exposure variables (all in grams per day) for all analyses. We focused on soluble fiber and pectins—a type of soluble fiber—given that they are the ideal fermentative substrates for the intestinal microbiota, with insoluble fiber contributing much less to microbial fermentation [36]. We included available carbohydrate as a “control” nutrient (that is, a nutrient whose relationships with gut microbes and fecal SCFA are likely to be weaker because available carbohydrate is largely absorbed by the human host and thereby rendered less available for microbial fermentation); and fructose, given preclinical evidence that it is a substrate for acetate production by the gut microbiota [22].
Stool collection and processing
Per the parent study protocol, all data were collected during a 14-d measurement period at baseline and at the beginning of each 3-mo diet period (that is, at time 0 and 3, 6, and 9 mo). This was done to allow for a 14-d CGM wear period. Participants were asked to return stool samples at any point during this 14-d time frame, to enhance the feasibility of collecting stool samples on a voluntary basis for this ancillary pilot study.
Participants were instructed to store stool samples on ice in a 4°C fridge prior to shipping the sample priority overnight to UNC. Given evidence that storage medium has a small effect on the composition of the gut microbiota, samples were collected in a sterile collection cup and stored without any added reagents [37]. Immediately on receipt (or within 1 h if first stored at 4°C), samples were homogenized, aliquoted, and frozen at −80°C. Given rolling study recruitment, aliquots were stored between 1 y and 2.5 y before the analysis. To prevent batch effects, samples were randomized before the laboratory analysis.
Gut microbiota characterization
Genomic microbial DNA from human fecal samples was isolated by a phenol–chloroform extraction method combined with a bead beating step using 0.1-mm glass beads (Bio Spec) to physically disrupt bacterial cells and using a DNA clean-up kit (Qiagen DNeasy Blood and Tissue extraction kit), as previously described [38]. Fecal microbiotas were characterized by creating sequencing libraries from the variable 4 region of the 16S rRNA gene using polymerase chain reaction and sequencing on the Illumina MiSeq platform (Illumina) at the High-Throughput Sequencing Facility in the Carolina Center for Genome Sciences at the UNC School of Medicine, as previously described [39].
The 16S rRNA gene sequences generated by the Illumina MiSeq platform were managed using the Quantitative Insights into Microbial Ecology 2 platform, including demultiplexing and denoizing via the Divisive Amplicon Denoizing Algorithm (DADA2) [40]. The DADA2 pipeline was used to generate sequence variants at 100% identity threshold. The total number of sequence reads was 11,105,926 [98,558.5 (IQR: 78,072.0, 129,325.8) per sample] and total number of sequence variants generated by DADA2 was 2339. After consideration of alternate normalization approaches [41, 42], we used a previously published method to normalize read counts using the following formula [43]:
Taxonomic classification was performed using the DADA2-formatted reference database Silva version 132 (>99% sequence identity) [44]. QIIME2 was used to derive rarefied intestinal microbial diversity—reported as the number of unique sequence variants (that is, the number of unique taxa) per sample. To reduce bias stemming from imbalanced replication of reads during polymerase chain reaction steps, we performed alpha-rarefaction (that is, normalization of sequencing depth) in the estimation of intestinal microbial diversity [45]. Based on the alpha-rarefaction curves, we normalized intestinal microbial diversity to a depth of 3000 sequence reads per sample. Per published methods, only taxa that were present in ≥25% of samples (that is, “nonrare” taxa) were retained [43].
We identified both genus- and species-level SCFA-producing taxa through a rigorous literature review [9, 27, [46], [47], [48]]. We detected the following 24 SCFA-producing taxa in the stool of our study participants: Akkermansia, Alistipes, Anaerostipes, Bacteroides, Bifidobacterium, 2 members of the Clostridium genus (Clostridium sensu stricto cluster 1 and Clostridium innocuum), Dialister, 3 members of the Eubacterium genus (Eubacterium eligens, Eubacterium hallii, and Eubacterium ventriosum), Faecalibacterium, Intestinimonas, Lachnospira, 2 members of the Prevotella genus (Prevotella clusters 7 and 9), Roseburia, 4 members of the Ruminococcus genus (Ruminococcus gnavus, Ruminococcus torques, and Ruminococcus clusters 1 and 2), Sutterella, Streptococcus, and Veillonella. After removal of 3 taxa that were present in <25% of samples (Sutterella and Prevotella clusters 7 and 9), 21 taxa were used in the analysis.
SCFA analysis
Total and specific (acetate, propionate, and butyrate) fecal SCFA were analyzed using GC-MS (Agilent 7820), as previously described, and converted to micromoles per gram [49]. Samples were analyzed on a wet matter basis. Because SCFA are volatile, we conducted a sensitivity analysis to ascertain whether results for fecal SCFA changed when excluding the 5 samples that were received >1 d after collection. Doing so did not substantively change the results of the crude models, so we retained all samples in the analysis of SCFA.
Demographic and clinical covariates
Standardized questionnaires collected self-reported demographic data such as age, gender, race and ethnicity, and insulin regimen (twice daily, 3 times daily, >3 times daily injections, or insulin pump). Self-reported race categories included African American, American Indian/Alaska Native, Asian, Native Hawaiian/Other Pacific Islander, other race, or White. Ethnicity was classified as Spanish/Hispanic/Latino or not. Owing to sample size limitations, we collapsed race and ethnicity into a single indicator variable denoting other race and ethnicity or non-Hispanic White race and ethnicity. Missing data for insulin regimen (n = 3 observations) were imputed forward or backward from the closest visit in time.
Design covariates
Design covariates included diet assignment at each visit and study site.
Statistical analysis
We assessed representativeness by comparing the baseline characteristics of ACT1ON participants included and excluded from this analysis.
Effect size and power
After correction for multiple comparisons, we were powered to detect an R2 of 0.14 with 80% power and an R2 of 0.18 with 90% power given our sample size of 52 stool samples. The magnitude of these effect sizes is smaller than those found in a previous study on diet and the intestinal microbiota (R2 = 0.2–0.4) with a smaller sample size than this study (n = 10), suggesting we were powered to detect observable effects [50].
Modeled analysis.
Using data from each of the 4 measurement time points occurring at time 0 and roughly at 3, 6, and 9 mo, we fit separate generalized estimating equation (GEE) models predicting outcomes [the abundance of each of 21 SCFA-producing taxa, fecal SCFA (butyrate, propionate, acetate, and total) concentrations, and intestinal microbial diversity (number of unique taxa per sample)] from each dietary exposure variable.
We selected GEEs because they consider nonindependence of repeated measures using a repeated statement (PROC GEE in SAS) and are robust to nonnormally distributed data [51], which is particularly relevant to zero-inflated gut microbiome data [52]. We specified a compound symmetric covariance structure, which assumes that observations within a participant are equally correlated. We report the β estimate and 95% CIs for changes in the gut microbiota and fecal SCFA that were associated with a 10-g/d increase in each of the dietary exposure variables given that at baseline, the 40 study participants included in the primary analysis reported consuming 12.6 g/d total fiber (IQR: 9.5, 21.3 g), which is >10 g lower than adequate intake [25 g/d for women and 38 g/d for men according to the Institute of Medicine’s Dietary Reference Intakes [53]]. Therefore, a 10-g/d increase in total fiber intake would have facilitated near-achievement of this recommendation.
Model 1 was unadjusted but considered within-subject correlations of repeated measures. To maximize utility of the sample, we evaluated which of our nondesign adjustment covariates (age, gender, race and ethnicity, BMI, and insulin regimen) should be retained in adjusted model 2 based on stepdown approaches in which the “least significant” covariate was removed from the model sequentially until only covariates with a P value of <0.1 remained [54]. The informative variables (P <.1) that were retained in model 2 were age and race and ethnicity. Moreover, model 2 included the design covariates of diet assignment, study site, and diet period. For each association, we computed standardized β coefficients by dividing each β estimate from GEE models by its standard error. We report these unitless standardized coefficients in the figures rather than the raw β estimates, allowing for comparability of coefficients across models [51]. Given that the relationship between diet and the composition of the gut microbiota could either be confounded or mediated (or both) by BMI [55], we included BMI separately in the fully adjusted model as part of an exploratory analysis.
For 2 microbes that were associated inversely with available carbohydrate and fiber intake, we conducted post hoc adjustments of model 2 by protein and fat intake (g/d) to explore the possibility that residual confounding by these nutrients may have led to spurious associations. We did not adjust for total calorie intake given that we were primarily interested in the absolute grams of fiber (total, soluble, and pectins) or carbohydrate (available, fructose) that could lead to a particular response in the fecal microbiota or SCFA.
Bonferroni-corrected [56] P values were considered to be statistically significant at an α level of P < 0.1. We selected Bonferroni correction given that the false discovery rate (FDR) assumes independence of tested comparisons, which is generally not true of correlated gut microbiota data; however, given that FDR is commonly used in microbiome research, we present FDR-adjusted results in Supplemental Material when q < 0.05. Descriptive statistics comparing the baseline characteristics of ACT1ON Study participants included and excluded from the analysis used an α threshold of P < 0.05. Power calculations were estimated with R software (version 4.1.1). All other analyses were conducted using SAS, version 9.4.
Results
At enrollment, the 40 participants included in the analysis had a mean age of 25.5 ± 3.2 y, a mean BMI (in kg/m2) of 29.8 (range, 28.1, 33.9), a HbA1c of 7.8% ± 1.3%, and a diabetes duration of 15.6 ± 6.2 y and were similar to excluded ACT1ON participants in all baseline characteristics (Table 1).
TABLE 1.
Baseline characteristics of ACT1ON participants included and excluded from the primary analysis (n = 68)
| Included (n = 40) | Excluded (n = 28) | P | |
|---|---|---|---|
| Age (y)1 | 25.5 ± 3.2 | 25.4 ± 3.0 | 0.77 |
| Female gender2 | 28 (70.0) | 21 (75.0) | 0.79 |
| Non-Hispanic White race and ethnicity2,3 | 30 (75.0) | 15 (53.6) | 0.08 |
| UNC site2 | 25 (37.5) | 14 (50.0) | 0.33 |
| BMI (kg/m2)1 | 29.8 (28.1, 33.9) | 30.8 (27.3, 33.8) | 0.80 |
| Hemoglobin A1c (%)1 | 7.8 ± 1.3 | 8.0 ± 1.5 | 0.31 |
| Diabetes duration (y)1 | 15.6 ± 6.2 | 11.8 ± 6.0 | 0.84 |
| Insulin pump use2 | 23 (57.5) | 17 (60.7) | 0.81 |
| Diet4 | Included (n = 36) | Excluded (n = 22) | |
| Total fiber intake (g/d)1 | 12.6 (9.5, 21.3) | 14.3 (10.5, 19.3) | 0.68 |
ACT1ON, Advancing Care for Type 1 Diabetes and Obesity Network; UNC, University of North Carolina. 1Continuous variables are presented as mean ± SD if normally distributed or as median (IQR) if nonnormally distributed. Group differences were tested with the independent t test and Mann-Whitney U test for normally and nonnormally distributed continuous variables, respectively. 2Categorical variables are presented as n (%). The χ2 test of equal proportions was used to test for group differences in categorical variables. 3Race and ethnicity were collapsed into other or non-Hispanic White race and ethnicity. To avoid participant identification, we expressed frequencies with fewer than 3 individuals as “n < 3.” Included participants: African American, n = 3 (7.9%); Asian, n < 3; >1 race, n < 3; and White, n = 32 (84.2%). Excluded participants: African American, n = 3 (12.5%); Native Hawaiian/Other Pacific Islander, n < 3; >1 race, n = 4 (14.3%); other race, n < 3; Asian, n < 3; and White, n = 17 (70.8%); 6 (15.0%) included and 5 (17.9%) excluded participants were identified as Spanish/Hispanic/Latino ethnicity. Three participants (2 included and 1 excluded) were missing race data but identified with a Spanish/Hispanic/Latino ethnicity. 4Sample sizes differed for dietary intake owing to missing data.
The time interval between collection of diet recalls and stool samples (n = 52 samples) was −2 d (IQR: −4, 2 d; range: −14, 13 d). Twenty-four (46.2%) stool samples were collected within 5 d of both dietary recalls. Forty-seven (90.4%) samples were received within 1 d of collection. Three samples were received 2 d after collection, and 1 sample each was received 3 and 4 d after collection.
Intake (in grams per day) of fiber (total, soluble, and pectins) and carbohydrate (available carbohydrates and fructose) and fecal SCFA concentrations, are presented in Table 2. At baseline, 23 participants had 2 diet recalls and 13 participants 1 diet recall. At measurement visit 2, 13 participants had 2 diet recalls and 3 participants 1 recall. At baseline, the measured values for fecal SCFA were 52.2 ± 18.8 μmol/g for acetate, 7.0 ± 1.8 μmol/g for butyrate, and 16.1 ± 5.7 μmol/g for propionate.
TABLE 2.
Repeated measures of dietary intake and fecal SCFA among ACT1ON participants (n = 40) using pre–COVID-19 data1
| Baseline (n = 36) | Measurement visit 2 (n = 16) | |
|---|---|---|
| Dietary intake (g/d)2 | ||
| Total fiber | 12.6 (9.5, 21.3) | 16.5 (11.4, 23.5) |
| Soluble fiber | 4.3 (2.9, 6.7) | 4.7 (4.1, 6.8) |
| Pectins | 1.5 (0.79, 2.7) | 2.6 (1.1, 4.5) |
| Fructose | 7.6 (5.0, 11.2) | 11.6 (6.6, 21.0) |
| Available carbohydrate | 155 ± 70.3 | 150 ± 90.2 |
| SCFA (μmol/g)2 | ||
| Acetate | 52.2 ± 18.8 | 54.0 ± 23.8 |
| Butyrate | 7.0 ± 1.8 | 6.2 ± 1.8 |
| Propionate | 16.1 ± 5.7 | 15.1 ± 7.8 |
| Total | 75.3 ± 22.3 | 75.3 ± 30.4 |
ACT1ON, Advancing Care for Type 1 Diabetes and Obesity Network. 1Data are from the 40 participants with available data at either baseline or measurement visit 2, or both. At baseline, 23 participants had 2 diet recalls and 13 participants had 1 diet recall. At measurement visit 2, 13 participants had 2 diet recalls and 3 participants had 1 recall. If 2 dietary recalls were available, averages were computed for each nutrient. 2Continuous variables are presented as mean ± SD if normally distributed or as median (IQR) if nonnormally distributed.
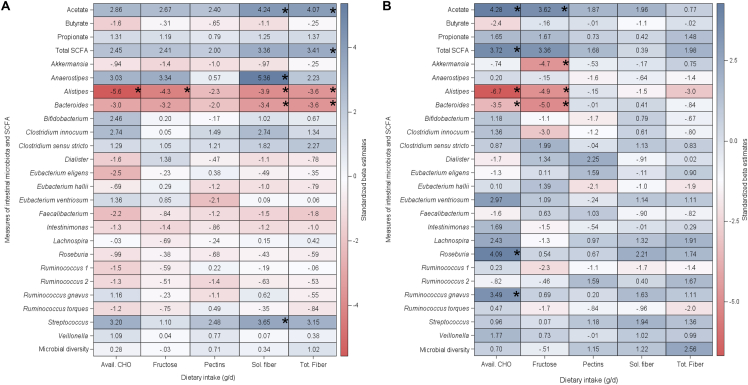
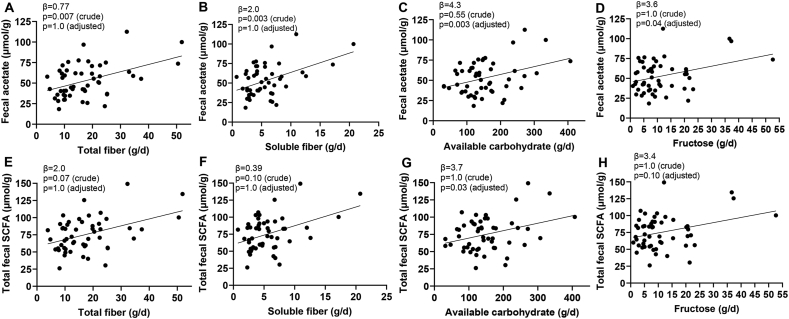
According to the crude (unadjusted) Bonferroni-corrected results, a 10-g/d increase in available carbohydrate, fructose, soluble fiber, and total fiber was associated inversely with the normalized abundance of the SCFA-producing genus Alistipes (Figure 1). A 10-g/d increase in total and soluble fiber was associated positively with fecal acetate (unstandardized β estimates: 8.8 μmol/g; 95% CI: 4.5, 12.8 μmol/g; P = 0.006; unstandardized β estimates: 24.0 μmol/g; 95% CI: 12.9, 35.1 μmol/g; P = 0.003, respectively) (Figure 2).
FIGURE 1.
Associations of diet with SCFA-producing gut microbes, fecal SCFA, and intestinal microbial diversity among ACT1ON participants (n = 40). The heatmaps show standardized β estimates from crude (A) and covariate-adjusted (B) GEE models with a repeated statement to consider multiple measurements within participants over time, for the primary pre–COVID-19 analysis. P values were Bonferroni-corrected and statistically significant at P < 0.1 (asterisks). Results are from 40 participants (52 visits) for SCFA and from 40 participants (48 visits) for abundance of intestinal microbes and intestinal microbial diversity. ACT1ON, Advancing Care for Type 1 Diabetes and Obesity Network; avail., available; GEE, generalized estimating equation; sol., soluble; tot., total.
FIGURE 2.
Raw visualizations of dietary intake in association with fecal SCFA among ACT1ON participants (n = 40). Scatterplots of raw pre–COVID-19 data show associations of total fiber, soluble fiber, available carbohydrate, and fructose intake (all g/d) with fecal acetate (A, B, C, D) and total fecal SCFA (E, F, G, H, both μmol/g), respectively. All associations were statistically significant after Bonferroni correction of crude or adjusted modeled results (P < 0.1), except for the associations of soluble fiber and fructose intake with total fecal SCFA (F, G, P = 0.1). Data are from 40 participants (52 visits) and were modeled using generalized estimating equations, which included a repeated statement to account for multiple measurements within participants over time. ACT1ON, Advancing Care for Type 1 Diabetes and Obesity Network.
After covariate adjustment and Bonferroni correction, a 10-g/d increase in available carbohydrate and fructose intake was associated inversely with the normalized abundance of the SCFA-producing genera Bacteroides and Alistipes. Post hoc adjustment by protein and fat intake (in grams per day) did not attenuate associations with these 2 gut microbe genera. Each covariate-adjusted 10-g/d increase in available carbohydrate and fructose intake was associated with a 2.5-μmol/g (95% CI: 1.4, 3.7 μmol/g; P = 0.003) and an 8.1-μmol/g (95% CI: 3.7, 12.5 μmol/g; P = 0.04) increase in fecal acetate, respectively (Supplemental Table 1). A 10-g/d increase in fructose intake was associated with a reduced normalized abundance of the SCFA-producing genus Akkermansia. The relationships between available carbohydrate intake and the normalized abundance of SCFA-producing genus Roseburia and R. gnavus showed positive associations. No relationships with total or soluble fiber intake were statistically significant after covariate adjustment.
Supplemental results for crude unadjusted estimates using pre–COVID-19 data that were statistically significant after FDR but not after Bonferroni correction are summarized in Supplemental Table 1 and Supplemental Figure 2. The results of the supplemental analysis of all available data are presented in Supplemental Table 2, Supplemental Table 3, and Supplemental Figure 3 and are described in the Supplemental Methods. Supplemental Table 4 summarizes the results of the exploratory adjustment by BMI for the primary analysis restricted to pre–COVID-19 data.
Discussion
In our hypothesis-generating gut microbiome pilot study, we found evidence to support our conjecture that an increased intake of dietary fiber (before adjustment) and carbohydrate (after adjustment), irrespective of the specific type, is associated with increased fecal SCFA (total and acetate) among adults with longstanding T1D. Echoing our results, a study of 42 children with islet autoimmunity or T1D showed that a 10-g/d increase in carbohydrate intake was associated with a 5.2-μmol/L (95% CI: 1.1, 9.2 μmol/L; P = 0.01) increase in plasma acetate [25]. The study authors also detected a reduced gut microbial diversity in association with a 5-g/d increase in “junk food intake.” Given the complexity of dietary intake, future studies could consider assessing dietary patterns or food groups, in addition to other nutrients such as protein and fat, in association with the structure, diversity, and functional genomic potential [57] of the gut microbiome in adults with T1D.
The compositional nature of both the gut microbiota and diet should be considered when interpreting the variable directionality of our diet–gut microbiome associations. Although all dietary measures (except for pectin, which generally constitutes a small proportion of dietary fiber and showed no associations) were associated negatively with the SCFA-producing genera Bacteroides and Alistipes, soluble fiber and available carbohydrate intake were associated positively with the SCFA-producing genera Anaerostipes and Streptococcus per crude results. Gut microbial abundances are intercorrelated, which reflects that gut microbes must compete for limited dietary substrates: as one gut microbe occupies a dietary niche, the abundance of another gut microbe that relies on the same dietary substrate may decline [58]. Thus, it is possible that members of Bacteroides and Alistipes were competitively inhibited by other fiber and carbohydrate fermenters [59]. We ruled out the possibility that the inverse associations of total and soluble fiber (crude) and available carbohydrate and fructose intake (adjusted) with abundances of the SCFA-producing taxa Bacteroides and Alistipes were confounded by fat and protein intake [that is, that these microbes were competitively excluded by fat and protein fermenters [21, 24]. The intake of fructose—which can be fermented to acetate—was inversely associated with the abundance of the acetate producer Akkermansia after covariate adjustment [60]. Future studies should investigate dietary substitution effects: perhaps increased fructose intake displaced dietary fiber [61], leading to a relative reduction in Akkermansia abundance.
We did not find associations of our dietary exposures with fecal butyrate or propionate, nor with gut microbial diversity. Butyrate is rapidly used as an energy source by colonocytes [48]. More direct measures of SCFA production, including in vivo tracer techniques, can provide more direct measures of SCFA production in response to acute dietary intake [62]. Intake of total dietary fiber was generally low in the r study sample, which may underlie the lack of associations between reported fiber (total and soluble) intake with gut microbial diversity.
Although it is initially surprising that available carbohydrate intake was associated positively with fecal SCFA and the abundance of several SCFA-producing gut microbes, this may be because available carbohydrates may include resistant starch—a substrate for microbial fermentation [63] that is not available as a standalone nutrient in the NDSR database given the lack of a standardized method for its quantification in foods [64]. Intake of available carbohydrate was associated positively with the normalized abundance of genus Roseburia and R. gnavus, whose beneficial functions include inhibition of proinflammatory cytokines through butyrate production [65, 66]. However, the abundance of Roseburia intestinalis may be elevated among individuals who are overweight or obese compared with those who are lean [67, 68]. This raises the often-cited conundrum about whether the metabolic benefits of SCFA outweigh their potential harms [for example, through their contribution to positive energy balance through energy extraction from nondigestible carbohydrates [69]]. This is especially relevant to T1D, in which the comanagement of glycemia and weight is necessary for cardiometabolic risk reduction but is complicated by the dysregulation of multiple hormones governing energy homeostasis and appetite (for example, insulin, glucagon, amylin, and incretin hormones) [[17], [18], [19]]. Furthermore, individuals with T1D should strive for consumption of complex (fiber) rather than simple (that is, available carbohydrate or fructose) sources of carbohydrate to promote achievement of glucose targets [70]. The mean fiber intake among our sample of young adults with T1D and overweight or obesity was far below recommendations [53, 71]. Thus, the multitude of health benefits conferred by dietary fiber, whether or not they are mediated by the gut microbiota, (for example, cholesterol lowering, glucose and appetite regulation) should continue to be emphasized in the clinical management of T1D [72, 73].
The results of this study should be interpreted in the context of its limitations. The most notable limitation is the duration of time between the measurement of dietary intake and the collection of certain stool samples. Approximately half of our stool samples were collected >5 d before or after the corresponding dietary recall(s); although not available when we designed or implemented this study, more recent guidance suggests that it is optimal to collect diet recalls ≤5 d before stool sample collection for assessment of the diet–gut microbiome relationship [74]. We avoided restricting the study sample to observations for which diet and stool were collected more proximally to one another owing to sample size considerations and the potential for inducing missing data bias (that is, a longer duration of time between diet and stool sample collection may have been an indicator of nonadherence to diet recommendations in the parent trial, so exclusion could introduce systematic bias related to the diet exposures at hand) [75]. Furthermore, the within-sample variation in the gut microbiome over time is much smaller than the between-sample variation [76], which minimizes the possibility that intraindividual changes in gut microbial composition over a short duration of time (that is, within the 2 wk collection period) biased our interindividual analysis. Another limitation is that home stool collection poses challenges for measurement of fecal SCFA, which are volatile [77]. Studies that collect and snap-freeze feces on-site provide a means to better preserve SCFA. As with all fecal microbiome studies, we randomly sampled the larger gut microbial ecosystem. Thus, our fecal measurements are proxies for the true underlying data distribution of gut microbes and SCFA [78].
Given the hypothesis-generating nature of this observational pilot study with a modest sample size and the small effect sizes that are characteristic of associations of diet with the intestinal microbiota, our results should be confirmed in additional observational and controlled feeding studies [21]. We were not powered to test the relationships between dietary intake and all gut microbial metabolites. Future larger studies should consider other diet-derived microbial metabolites [including but not limited to bile acids [79], branched-chain fatty acids [80], and urolithin A [81]] that are metabolically relevant. We focused on SCFA, given the large body of evidence in preclinical models and in clinical studies of people without T1D that these metabolites are measurably affected by diet and hold promise for augmenting metabolism [9, 16, 27]. Although stepdown variable selection is preferable to step-forward selection [54], such procedures generally may not result in the optimal set of adjustment covariates. We considered variable selection to be a necessary step in constructing statistical models, given our sample size. Given that the effect of our primary exposure variable, diet, is likely to affect the gut microbiome more appreciably than any of the confounders considered in the full set of potential adjustment covariates (age, gender, race and ethnicity, and insulin regimen) [17], we believe the effect of erroneously excluding 1 or more of these variables is likely to be modest. We considered BMI as a potential confounder in a supplemental analysis, but avoid overinterpreting this result because it is unclear biologically whether BMI mediates or confounds the relationship between diet and the composition of the gut microbiota [55]. Model misspecification may have biased estimates of effect toward or away from the null. Although data missingness was likely to be completely at random for some participants, for others, it may have been related to the study exposure of dietary intake or gut microbiota–related outcomes; therefore, we cannot rule out the possibility that this introduced bias in the GEE estimates. Future studies designed specifically to assess the diet–microbiome relationship would likely reduce data missingness. The 16S rRNA gene sequencing is limited to taxonomic identification. Future whole genome sequencing studies can identify dietary effects on functional gut microbial gene pathways in T1D and map specific taxa to enzymatic pathways necessary for SCFA production. Because there is a great deal of overlap in the genes that gut microbes harbor (that is, functional redundancy), understanding the relationship between diet (a selective pressure) and microbial gene pathways in longitudinal data sets can reveal how gut microbes and their genomes evolve in response to changes in the host environment—which may be unique in T1D [82]. As a free-living diet study, our measure of self-reported dietary intake was subject to dietary recall bias [83]. However, differential misreporting of the dietary exposures relative to outcomes under study is unlikely because the participants did not know their gut microbiome (outcome) status. Generalizability of our findings to individuals with T1D who are not overweight or obese, racially or ethnically more diverse than our study participants, or recently diagnosed with T1D may be limited.
This study also includes several strengths. Previous studies investigating the intestinal microbiota and SCFA in T1D have largely focused on the development of autoimmunity near the time of T1D diagnosis [25, [84], [85], [86]]. Although this topic is one that is of great importance, we focused on the links among diet and the intestinal microbiota in longstanding T1D, about which very little is known. Furthermore, no studies, to our knowledge, have assessed the intestinal microbiota among individuals with T1D and overweight or obesity at multiple time points throughout a weight management intervention. If gut microbes affect clinical outcomes or can serve as biomarkers of diet efficacy in people with T1D, their role in diet interventions may be clinically relevant. We used rigorous methods of statistical analysis and adjusted our models for potential confounding variables.
Much work remains to be performed to fully elucidate the role of the gut microbiota and its response to diet in T1D management. Future studies can focus on the following: 1) the feasibility of inducing durable changes in the composition and metabolic byproducts of the intestinal microbiota through diet; 2) clarifying whether total and specific SCFA are truly beneficial to people with T1D because they may contribute to positive energy balance [69] and gut mucosal inflammation [87]; and 3) the relationships among diet, inflammation, and the gut microbiota in the context of longstanding T1D autoimmunity. Such studies can inform whether future research and ultimately clinical practitioners should target dietary modifications that promote a specific milieu of intestinal microbes and metabolites to improve glycemia, adiposity [88], and downstream microvascular and cardiovascular outcomes in people with T1D.
Author disclosures
DMM has had research support from the NIH, JDRF, NSF, and the Helmsley Charitable Trust and his institution has had research support from Medtronic, Dexcom, Insulet, Bigfoot Biomedical, Tandem, and Roche; and has consulted for Abbott, Aditxt, the Helmsley Charitable Trust, Lifescan, Mannkind, Sanofi, Novo Nordisk, Eli Lilly, Medtronic, Insulet, Dompe, and Biospex. EJM-D has consulted for Helmsley Charitable Trust. CMB reports the following: Shire (grant recipient, Scientific Advisory Board member); Idorsia (consultant); Lundbeckfonden (grant recipient); Pearson (author, royalty recipient); Equip Health, Inc. (Clinical Advisory Board); and is supported by NIMH (R01MH120170; R01MH124871; R01MH119084; R01MH118278; R01 MH124871); Brain and Behavior Research Foundation Distinguished Investigator Grant; Swedish Research Council (Vetenskapsrådet, award: 538-2013-8864); Lundbeck Foundation (Grant no. R276-2018-4581). IMC is a consultant for Vivilex; is a former consultant for Salix Pharmaceuticals, and receives funding from NIH (R21-AI125800–01-02) and NIMH (R01-MN105684–03). DI was supported by the Global Cardiometabolic Disease training grant (National Heart, Lung, and Blood Institute of the National Institutes of Health) awarded to the Department of Nutrition at the University of North Carolina at Chapel Hill under Award Number HL129969. DI, JC, KDC, JH, JMT, BWP, REP, and MRK declare no conflict of interest.
Acknowledgments
The ACT1ON Study is indebted to the young adults whose participation made this study possible. The 24-h recalls were obtained by the UNC Nutrition Obesity Research Center staff, funded by the NIDDK of the NIH under Award Number 1DP3DK113358; MPIs were EJM-D, DMM, REPP. The analysis of SCFA was performed by Yifei Yang and Dr. Yunjia Lai under the supervision of Dr. Kun Lu at the University of North Carolina at Chapel Hill. We thank Dr. Anthony Fodor for his provision of technical advice related to analysis of the intestinal microbiota. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tjnut.2022.12.017.
Author contribution
The authors’ responsibilities were as follows—DI, IMC, and EJM-D: designed the research; DI, IMC, EJM-D, JMT, DMM, KDC, RP, and JH: conducted the research; IMC: provided essential materials; DI: wrote the initial manuscript, conducted the analyses and holds responsibility for the accuracy of the data analysis and the final content; IMC, JC, and MK: supervised the analyses; DI, IMC, MK, JC, and EJM-D: designed the analyses; CMB and BP: reviewed the manuscript; and all authors: read and approved the final version of the manuscript.
Funding
This study was supported by NIH/NIDDK (1DP3DK113358-01, PNC DK056350, and P30 DK034987).
Data Availability
Data will be shared as appropriate following the submission of a formal application to the ACT1ON Publications and Presentations Committee. If necessary, a data use or other data sharing agreement will be established.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Purnell J.Q., Braffett B.H., Zinman B., Gubitosi-Klug R.A., Sivitz W., Bantle J.P., et al. Impact of excessive weight gain on cardiovascular outcomes in type 1 diabetes: results from the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Study. Diabetes Care. 2017;40(12):1756–1762. doi: 10.2337/dc16-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Diabetes Association Cardiovascular disease and risk management: Standards of Medical Care in Diabetes—2021. Diabetes Care. 2021;44(Supplement 1):S125–S150. doi: 10.2337/dc21-S010. 10. [DOI] [PubMed] [Google Scholar]
- 3.Miller K.M., Foster N.C., Beck R.W., Bergenstal R.M., DuBose S.N., DiMeglio L.A., et al. Current state of type 1 diabetes treatment in the U.S.: updated data from the T1D Exchange clinic registry. Diabetes Care. 2015;38(6):971–978. doi: 10.2337/dc15-0078. [DOI] [PubMed] [Google Scholar]
- 4.Wallace A.S., Chang A.R., Shin J.I., Reider J., Echouffo-Tcheugui J.B., Grams M.E., et al. Obesity and chronic kidney disease in US adults with type 1 and type 2 diabetes mellitus. J Clin Endocrinol Metab. 2022;107(5):1247–1256. doi: 10.1210/clinem/dgab927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez J.A., Navas-Carretero S., Saris W.H., Astrup A. Personalized weight loss strategies—the role of macronutrient distribution. Nat Rev Endocrinol. 2014;10(12):749–760. doi: 10.1038/nrendo.2014.175. [DOI] [PubMed] [Google Scholar]
- 6.Zeevi D., Korem T., Zmora N., Israeli D., Rothschild D., Weinberger A., et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Shilo S., Godneva A., Rachmiel M., Korem T., Kolobkov D., Karady T., et al. Prediction of personal glycemic responses to food for individuals with type 1 diabetes through integration of clinical and microbial data. Diabetes Care. 2022;45(3):502–511. doi: 10.2337/dc21-1048. [DOI] [PubMed] [Google Scholar]
- 8.den Besten G., Bleeker A., Gerding A., van Eunen K., Havinga R., van Dijk T.H., et al. Short-chain fatty acids protect against high-fat diet–induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes. 2015;64(7):2398–2408. doi: 10.2337/db14-1213. [DOI] [PubMed] [Google Scholar]
- 9.Kimura I., Ozawa K., Inoue D., Imamura T., Kimura K., Maeda T., et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4(1):1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weitkunat K., Stuhlmann C., Postel A., Rumberger S., Fankhänel M., Woting A., et al. Short-chain fatty acids and inulin, but not guar gum, prevent diet-induced obesity and insulin resistance through differential mechanisms in mice. Sci Rep. 2017;7(1):1. doi: 10.1038/s41598-017-06447-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita H., Fujisawa K., Ito E., Idei S., Kawaguchi N., Kimoto M., et al. Improvement of obesity and glucose tolerance by acetate in Type 2 diabetic Otsuka Long-Evans Tokushima Fatty (OLETF) rats. Biosci Biotechnol Biochem. 2007;71(5):1236–1243. doi: 10.1271/bbb.60668. [DOI] [PubMed] [Google Scholar]
- 12.Xiong Y., Miyamoto N., Shibata K., Valasek M.A., Motoike T., Kedzierski R.M., et al. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci U S A. 2004;101(4):1045–1050. doi: 10.1073/pnas.2637002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montague C.T., Farooqi I.S., Whitehead J.P., Soos M.A., Rau H., Wareham N.J., et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387(6636):903–908. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- 14.Canfora E.E., van der Beek C.M., Jocken J.W., Goossens G.H., Holst J.J., Damink S.W.O., et al. Colonic infusions of short-chain fatty acid mixtures promote energy metabolism in overweight/obese men: a randomized crossover trial. Sci Rep. 2017;7(1):2360. doi: 10.1038/s41598-017-02546-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vrieze A., Van Nood E., Holleman F., Salojärvi J., Kootte R.S., Bartelsman J.F., et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913. doi: 10.1053/j.gastro.2012.06.031. 6.e7. [DOI] [PubMed] [Google Scholar]
- 16.Kootte R.S., Levin E., Salojärvi J., Smits L.P., Hartstra A.V., Udayappan S.D., et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26(4):611. doi: 10.1016/j.cmet.2017.09.008. 9.e6. [DOI] [PubMed] [Google Scholar]
- 17.Falony G., Joossens M., Vieira-Silva S., Wang J., Darzi Y., Faust K., et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 18.de Groot P.F., Belzer C., Aydin Ö., Levin E., Levels J.H., Aalvink S., et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLOS ONE. 2017;12(12) doi: 10.1371/journal.pone.0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James S.L., Muir J.G., Curtis S.L., Gibson P.R. Dietary fibre: a roughage guide. Intern Med J. 2003;33(7):291–296. doi: 10.1046/j.1445-5994.2003.00404.x. [DOI] [PubMed] [Google Scholar]
- 20.Rideout T.C., Harding S.V., Jones P.J., Fan M.Z. Guar gum and similar soluble fibers in the regulation of cholesterol metabolism: current understandings and future research priorities. Vasc Health Risk Manag. 2008;4(5):1023–1033. doi: 10.2147/vhrm.s3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao S., Jang C., Liu J., Uehara K., Gilbert M., Izzo L., et al. Dietary fructose feeds hepatic lipogenesis via microbiota-derived acetate. Nature. 2020;579(7800):586–591. doi: 10.1038/s41586-020-2101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki T., Yoshida S., Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr. 2008;100(2):297–305. doi: 10.1017/S0007114508888733. [DOI] [PubMed] [Google Scholar]
- 24.De Filippo C., Di Paola M., Ramazzotti M., Albanese D., Pieraccini G., Banci E., et al. Diet, environments, and gut microbiota. A preliminary investigation in children living in rural and urban Burkina Faso and Italy. Front Microbiol. 2017;8:1979. doi: 10.3389/fmicb.2017.01979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harbison J.E., Thomson R.L., Wentworth J.M., Louise J., Roth-Schulze A., Battersby R.J., et al. Associations between diet, the gut microbiome and short chain fatty acids in youth with islet autoimmunity and type 1 diabetes. Pediatr Diabetes. 2021;22(3):425–433. doi: 10.1111/pedi.13178. [DOI] [PubMed] [Google Scholar]
- 26.Stewart C.J., Ajami N.J., O’Brien J.L., Hutchinson D.S., Smith D.P., Wong M.C., et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 28.Corbin K.D., Igudesman D., Addala A., Casu A., Crandell J., Kosorok M.R., et al. Design of the Advancing Care for Type 1 Diabetes and Obesity Network energy metabolism and sequential multiple assignment randomized trial nutrition pilot studies: an integrated approach to develop weight management solutions for individuals with type 1 diabetes. Contemp Clin Trials. 2022;117 doi: 10.1016/j.cct.2022.106765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Igudesman D., Crandell J., Corbin K.D., Zaharieva D.P., Addala A., Thomas J.M., et al. Weight management in young adults with type 1 diabetes: the advancing care for type 1 diabetes and obesity network sequential multiple assignment randomized trial pilot results. Diabetes Obes Metab. 2022 doi: 10.1111/dom.14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hettema J., Steele J., Miller W.R. Motivational interviewing. Annu Rev Clin Psychol. 2005;1:91–111. doi: 10.1146/annurev.clinpsy.1.102803.143833. [DOI] [PubMed] [Google Scholar]
- 31.Nezu A.M., D’Zurilla T.J. Springer Publishing Company; 2006. Problem-solving therapy: a positive approach to clinical intervention. [Google Scholar]
- 32.Battelino T., Danne T., Bergenstal R.M., Amiel S.A., Beck R., Biester T., et al. Clinical targets for continuous glucose monitoring data interpretation: recommendations from the International Consensus on Time in Range. Diabetes Care. 2019;42(8):1593–1603. doi: 10.2337/dci19-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy S.A. An experimental design for the development of adaptive treatment strategies. Stat Med. 2005;24(10):1455–1481. doi: 10.1002/sim.2022. [DOI] [PubMed] [Google Scholar]
- 34.Beaton G.H., Milner J., Corey P., McGuire V., Cousins M., Stewart E., et al. Sources of variance in 24-hour dietary recall data: implications for nutrition study design and interpretation. Am J Clin Nutr. 1979;32(12):2546–2559. doi: 10.1093/ajcn/32.12.2546. [DOI] [PubMed] [Google Scholar]
- 35.Dodd K.W., Guenther P.M., Freedman L.S., Subar A.F., Kipnis V., Midthune D., et al. Statistical methods for estimating usual intake of nutrients and foods: a review of the theory. J Am Diet Assoc. 2006;106(10):1640–1650. doi: 10.1016/j.jada.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Stark A.H., Madar Z. In vitro production of short-chain fatty acids by bacterial fermentation of dietary fiber compared with effects of those fibers on hepatic sterol synthesis in rats. J Nutr. 1993;123(12):2166–2173. doi: 10.1093/jn/123.12.2166. [DOI] [PubMed] [Google Scholar]
- 37.Song S.J., Amir A., Metcalf J.L., Amato K.R., Xu Z.Z., Humphrey G., et al. Preservation methods differ in fecal microbiome stability, affecting suitability for field studies. mSystems. 2016;1(3):e00021. doi: 10.1128/mSystems.00021-16. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fouladi F., Glenny E.M., Bulik-Sullivan E.C., Tsilimigras M.C.B., Sioda M., Thomas S.A., et al. Sequence variant analysis reveals poor correlations in microbial taxonomic abundance between humans and mice after gnotobiotic transfer. ISME J. 2020;14(7):1809–1820. doi: 10.1038/s41396-020-0645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleiman S.C., Glenny E.M., Bulik-Sullivan E.C., Huh E.Y., Tsilimigras M.C.B., Fodor A.A., et al. Daily changes in composition and diversity of the intestinal microbiota in patients with anorexia nervosa: a series of three cases. Eur Eat Disord Rev. 2017;25(5):423–427. doi: 10.1002/erv.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin H., Peddada S.D. Analysis of microbial compositions: a review of normalization and differential abundance analysis. NPJ Biofilms Microbiomes. 2020;6(1):60. doi: 10.1038/s41522-020-00160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van den Berge K., Perraudeau F., Soneson C., Love M.I., Risso D., Vert J.-P., et al. Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications. Genome Biol. 2018;19(1):24. doi: 10.1186/s13059-018-1406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noble E.E., Hsu T.M., Jones R.B., Fodor A.A., Goran M.I., Kanoski S.E. Early-life sugar consumption affects the rat microbiome independently of obesity. J Nutr. 2017;147(1):20–28. doi: 10.3945/jn.116.238816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willis A.D. Rarefaction, alpha diversity, and statistics. Front Microbiol. 2019;10:2407. doi: 10.3389/fmicb.2019.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Wang H., Howard A.G., Meyer K.A., Tsilimigras M.C.B., Avery C.L., et al. Circulating short-chain fatty acids are positively associated with adiposity measures in Chinese adults. Nutrients. 2020;12(7):2127. doi: 10.3390/nu12072127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Remely M., Aumueller E., Merold C., Dworzak S., Hippe B., Zanner J., et al. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene. 2014;537(1):85–92. doi: 10.1016/j.gene.2013.11.081. [DOI] [PubMed] [Google Scholar]
- 48.Louis P., Young P., Holtrop G., Flint H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA: acetate CoA-transferase gene. Environ Microbiol. 2010;12(2):304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 49.van der Lelie D., Oka A., Taghavi S., Umeno J., Fan T.-J., Merrell K.E., et al. Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat Commun. 2021;12(1):3105. doi: 10.1038/s41467-021-23460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.-Y., Keilbaugh S.A., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanley J.A., Negassa A., Edwardes M.D., Forrester J.E. Statistical analysis of correlated data using generalized estimating equations: an orientation. Am J Epidemiol. 2003;157(4):364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- 52.Weiss S., Xu Z.Z., Peddada S., Amir A., Bittinger K., Gonzalez A., et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5(1):27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyers L.D., Hellwig J.P., Otten J.J. National Academies Press; 2006. Dietary reference intakes: the essential guide to nutrient requirements. [Google Scholar]
- 54.Mantel N. Why stepdown procedures in variable selection. Technometrics. 1970;12(3):621–625. [Google Scholar]
- 55.Maruvada P., Leone V., Kaplan L.M., Chang E.B. The human microbiome and obesity: moving beyond associations. Cell Host Microbe. 2017;22(5):589–599. doi: 10.1016/j.chom.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Bland J.M., Altman D.G. Multiple significance tests: the Bonferroni method. BMJ. 1995;310(6973):170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deehan E.C., Duar R.M., Armet A.M., Perez-Muñoz M.E., Jin M., Walter J. Modulation of the gastrointestinal microbiome with nondigestible fermentable carbohydrates to improve human health. Microbiol Spectr. 2017;5(5) doi: 10.1128/microbiolspec.bad-0019-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foster K.R., Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22(19):1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 59.Chen T., Long W., Zhang C., Liu S., Zhao L., Hamaker B.R. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci Rep. 2017;7(1):2594. doi: 10.1038/s41598-017-02995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopetuso L.R., Quagliariello A., Schiavoni M., Petito V., Russo A., Reddel S., et al. Towards a disease-associated common trait of gut microbiota dysbiosis: the pivotal role of Akkermansia muciniphila. Dig Liver Dis. 2020;52(9):1002–1010. doi: 10.1016/j.dld.2020.05.020. [DOI] [PubMed] [Google Scholar]
- 61.Boeing H. Nutritional epidemiology: new perspectives for understanding the diet-disease relationship? Eur J Clin Nutr. 2013;67(5):424–429. doi: 10.1038/ejcn.2013.47. [DOI] [PubMed] [Google Scholar]
- 62.Boets E., Gomand S.V., Deroover L., Preston T., Vermeulen K., De Preter V., et al. Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: a stable isotope study. J Physiol. 2017;595(2):541–555. doi: 10.1113/JP272613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DeMartino P., Cockburn D.W. Resistant starch: impact on the gut microbiome and health. Curr Opin Biotechnol. 2020;61:66–71. doi: 10.1016/j.copbio.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 64.Slavin J.L. Carbohydrates, dietary fiber, and resistant starch in white vegetables: links to health outcomes. Adv Nutr. 2013;4(3):351S. doi: 10.3945/an.112.003491. 5S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamanai-Shacoori Z., Smida I., Bousarghin L., Loreal O., Meuric V., Fong S.B., et al. Roseburia spp.: a marker of health? Future Microbiol. 2017;12(2):157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 66.Nilsen M., Madelen Saunders C., Leena Angell I., Arntzen M.Ø., Lødrup Carlsen K.C., Carlsen K.H., et al. Butyrate levels in the transition from an infant- to an adult-like gut microbiota correlate with bacterial networks associated with Eubacterium rectale and Ruminococcus gnavus. Genes (Basel). 2020;11(11):1245. doi: 10.3390/genes11111245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kasai C., Sugimoto K., Moritani I., Tanaka J., Oya Y., Inoue H., et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15(1):100. doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tims S., Derom C., Jonkers D.M., Vlietinck R., Saris W.H., Kleerebezem M., et al. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013;7(4):707–717. doi: 10.1038/ismej.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cummings J.H., Rombeau J.L., Sakata T. Cambridge University Press; 1995. Physiological and clinical aspects of short-chain fatty acids. [Google Scholar]
- 70.American Diabetes Association 5. Facilitating behavior change and well-being to improve health outcomes: standards of medical care in diabetes—2021. Diabetes Care. 2021;44(Supplement 1):S53–72. doi: 10.2337/dc21-S005. [DOI] [PubMed] [Google Scholar]
- 71.Dietary guidelines for Americans. U.S. Department of Agriculture and U.S. Department of Health and Human Services; 2020. 2020–2025 [Internet] [date updated December 2020, date cited 11/4/2022] [Google Scholar]
- 72.Brown L., Rosner B., Willett W.W., Sacks F.M. Cholesterol-lowering effects of dietary fiber: a meta-analysis. Am J Clin Nutr. 1999;69(1):30–42. doi: 10.1093/ajcn/69.1.30. [DOI] [PubMed] [Google Scholar]
- 73.Pereira M.A., Ludwig D.S. Dietary fiber and body-weight regulation: observations and mechanisms. Pediatr Clin North Am. 2001;48(4):969–980. doi: 10.1016/s0031-3955(05)70351-5. [DOI] [PubMed] [Google Scholar]
- 74.Johnson A.J., Vangay P., Al-Ghalith G.A., Hillmann B.M., Ward T.L., Shields-Cutler R.R., et al. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe. 2019;25(6):789–802.e5. doi: 10.1016/j.chom.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 75.Heitjan D.F., Basu S. Distinguishing “missing at random” and “missing completely at random. Am Stat. 1996;50(3):207–213. [Google Scholar]
- 76.Faith J.J., Guruge J.L., Charbonneau M., Subramanian S., Seedorf H., Goodman A.L., et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141) doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang S., Wang H., Zhu M.J. A sensitive GC/MS detection method for analyzing microbial metabolites short chain fatty acids in fecal and serum samples. Talanta. 2019;196:249–254. doi: 10.1016/j.talanta.2018.12.049. [DOI] [PubMed] [Google Scholar]
- 78.Zoetendal E.G., von Wright A., Vilpponen-Salmela T., Ben-Amor K., Akkermans A.D., de Vos W.M. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68(7):3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ridlon J.M., Kang D.J., Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 80.Rasmussen H.S., Holtug K., Mortensen P.B. Degradation of amino acids to short-chain fatty acids in humans. An in vitro study. Scand J Gastroenterol. 1988;23(2):178–182. doi: 10.3109/00365528809103964. [DOI] [PubMed] [Google Scholar]
- 81.Espín J.C., Larrosa M., García-Conesa M.T., Tomás-Barberán F. Biological significance of urolithins, the gut microbial ellagic acid-derived metabolites: the evidence so far. Evid Based Complement Alternat Med. 2013 doi: 10.1155/2013/270418. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tian L., Wang X.W., Wu A.K., Fan Y., Friedman J., Dahlin A., et al. Deciphering functional redundancy in the human microbiome. Nat Commun. 2020;11(1):6217. doi: 10.1038/s41467-020-19940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hebert J.R., Clemow L., Pbert L., Ockene I.S., Ockene J.K. Social desirability bias in dietary self-report may compromise the validity of dietary intake measures. Int J Epidemiol. 1995;24(2):389–398. doi: 10.1093/ije/24.2.389. [DOI] [PubMed] [Google Scholar]
- 84.Davis-Richardson A.G., Ardissone A.N., Dias R., Simell V., Leonard M.T., Kemppainen K.M., et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown C.T., Davis-Richardson A.G., Giongo A., Gano K.A., Crabb D.B., Mukherjee N., et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLOS ONE. 2011;6(10) doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mejía-León M.E., Petrosino J.F., Ajami N.J., Domínguez-Bello M.G., De La Barca A.M.C. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep. 2014;4(1):3814. doi: 10.1038/srep03814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim M.H., Kang S.G., Park J.H., Yanagisawa M., Kim C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145(2):396–406.e1. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 88.Boulangé C.L., Neves A.L., Chilloux J., Nicholson J.K., Dumas M.E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8(1):42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be shared as appropriate following the submission of a formal application to the ACT1ON Publications and Presentations Committee. If necessary, a data use or other data sharing agreement will be established.