Abstract
A Bacillus subtilis promoter, Px, that functions in a convergent manner with the sigA operon promoter P3 has been found in the sigA operon. Promoter Px is turned on at the same time as promoter P3 during early sporulation. The transcript from promoter Px codes for a small protein with partial homology to the OmpR protein from Escherichia coli and also carries an untranslated sequence at its 3′ end that is complementary to the 5′ end of the P3 transcript, which codes for the ribosome binding site of dnaE. The gene controlled by Px has been called antE. The expression of antE does not require ςB, ςE, or ςH. Px was transcribed in vitro by the ςA holoenzyme and is the seventh promoter to be recognized in the ςA operon. A possible role for the antE gene during early sporulation is proposed.
Promoter switching observed in Bacillus subtilis resulted in transcription of the sigA operon from two ςA promoters (P1 and P2) (19) during growth and from a ςH promoter (P3) (2) located in the 5′ end of the P23 gene of the operon during early sporulation. This promoter switching allowed transcription of the sigA operon during early sporulation and the expression of the 3′ end of P23, the dnaE gene, and the sigA gene (19, 20).
The second gene of the sigA operon, dnaE, codes for DNA primase (21), which is not required during sporulation after the final round of DNA replication preceding forespore formation. The requirement for the product of the third gene of the operon, sigA, during early sporulation has been demonstrated (9, 14). We report here a novel promoter (Px) and a gene (antE) that is transcribed convergently to promoter P3 and is located in dnaE. This is the seventh promoter to be recognized in the sigA operon (see reference 16 for promoters P1 through P6).
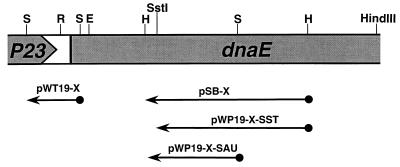
During a systematic search for promoters controlling the expression of the sigA operon, a convergent transcribing promoter activity was found on a 436-bp HaeIII fragment located within the dnaE coding region (17, 21). The Px promoter was further mapped between the SstI and Sau3A sites, which are 338 bp apart (Fig. 1). By using transcription terminator probe vector pWT19 (19) and shotgun cloning of Sau3A fragments generated from the EcoRI-SstI fragment at the 5′ end of the sigA operon (17, 18, 21), a 129-bp fragment showing strong termination activity was isolated. Sequence analysis of the 129-bp Sau3A fragment revealed a stem-and-loop structure followed by a stretch of T (U in RNA) residues (see Fig. 2 for the sequence), indicative of a prokaryotic rho-independent transcription terminator, which functions in the same orientation as Px, i.e., antisense to the sigA coding strand.
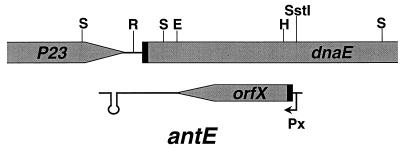
FIG. 1.
Mapping of promoter and terminator for antE gene. The shaded regions represent the truncated P23 gene and dnaE genes transcribing from left to right. Promoter probe vectors pSB and pWP19 and terminator probe vector pWT19 were constructed by using the subtilisin gene as a reporter gene and have been previously described by us (19). Restriction fragments showing positive activities when inserted into proper probe vectors are indicated by solid lines with arrowheads pointing to the direction of transcription of the reporter gene in the probe vectors. Clone designations are given above the solid lines, and relevant restriction sites used for subcloning are shown at the top of the figure. Abbreviations for restriction enzymes: S, Sau3A; R, RsaI; E, EcoRV; H, HaeIII.
FIG. 2.
DNA sequence of the antE region and the deduced amino acid sequence of the AntE protein. The DNA sequence shown is the antisense strand of the Sau3A (nt 1278)-HaeIII (nt 2239) fragment from the rpoD sequence published previously (17, 21). The numbering of DNA begins with the first base of the HaeIII site. Relevant restriction sites are shown. The deduced amino acid sequence of AntE is given underneath, numbered from the first Met (GTG) residue. The putative EςA promoter sequence and the RBS for AntE are indicated by single and double underlines, respectively. The putative transcription terminator sequence is indicated by arrows. The region of AntE homologous to E. coli OmpR is shown by dots.
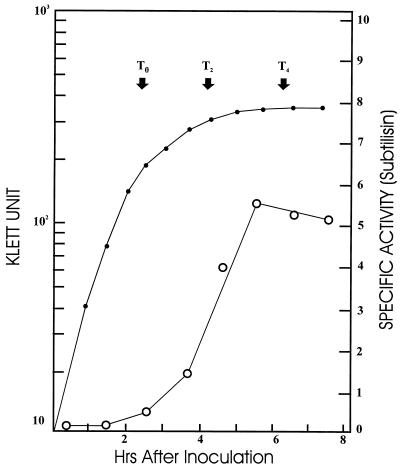
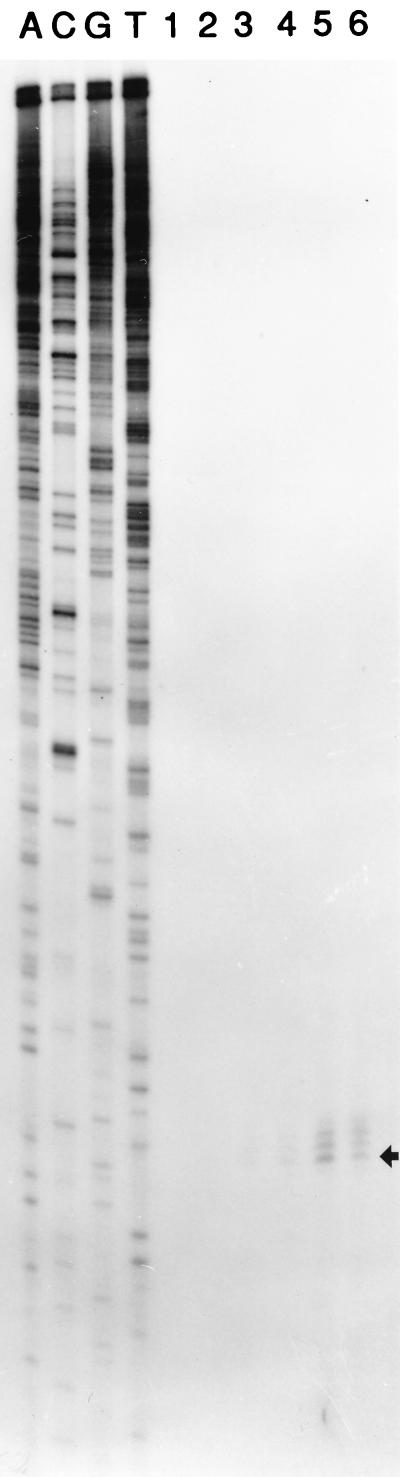
When DB104(pSB-X) (Table 1), a strain containing the subtilisin gene under the control of promoter Px, was subjected to time course studies under sporulation conditions, an expression pattern strongly resembling that of promoter P3 of the sigA operon was obtained (Fig. 3). To further confirm this pattern of expression and to exclude the possibility that Px in a multicopy plasmid (pSB-X) might have an expression profile different from that of the chromosomal single-copy gene, quantitative S1 nuclease mapping was conducted by using the 5′-end-labeled HaeIII fragment as a probe. These results, shown in Fig. 4, illustrated several important features of promoter Px: (i) the transcription initiation site (+1) was mapped at a position 47 bp away from the HaeIII site, which was consistent with the previous mapping and Bal 31 deletion data; (ii) levels of RNA transcripts derived from Px at different growth stages matched the previously observed temporal expression pattern (Fig. 3); and (iii) by defining the +1 site, we were able to find a ςA RNA polymerase holoenzyme (EςA)-type promoter sequence at the −10 (TAAAAT) and −35 (TTGTAT) regions with a spacer of 17 bp (Fig. 2).
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| DB2 | trpC | Lab stock |
| DB42 | dnaE20 | 15 |
| DB104 | hisH nprR2 nprE18 ΔaprE | 8 |
| IS233 | trpC2 phe-1 spo0HΔHindIII | 23 |
| ML1 | trpC2 sigB::cat | 7 |
| EU8701 | trpC2 phe-1 ΔsigE::erm | 9 |
| WB29 | hisH nrpR2 nrpE18 ΔaprE sigB::cat | This work |
| WB71 | hisH nrpR2 nrpE18 ΔaprE ΔsigE::erm | This work |
| WB72 | nprR2 nprE18 ΔaprE spo0HΔHindIII | This work |
| DB104(pSB) | DB104 containing plasmid pSB | This work |
| DB104(pSB-X) | DB104 containing plasmid pSB-X | This work |
| WB29(pSB-X) | WB29 containing plasmid pSB-X | This work |
| WB71(pSB-X) | WB71 containing plasmid pSB-X | This work |
| WB72(pSB-X) | WB72 containing plasmid pSB-X | This work |
FIG. 3.
Expression time course of promoter Px. The solid circles represent Klett units (for the growth curve), and the open circles represent subtilisin activities (for the expression curve), of DB104(pSB-X) culture in sporulation medium 2× SG (11). The specific activity of subtilisin was measured and defined as described previously (19). A parallel culture of DB104(pSB) was used as a negative control for enzyme assays. T0, onset of sporulation.
FIG. 4.
Quantitative S1 nuclease mapping of Px transcription start point. Fifty micrograms of yeast tRNA or B. subtilis total RNA was annealed to the 5′-end-labeled single-stranded probe (the sense strand of the 436-bp HaeIII fragment) with 20,000 to 50,000 cpm by procedures described previously (19). The protected DNA probe fragments after S1 digestion were resolved on a 6% sequencing gel together with an M13-derived sequencing ladder as size markers. Lanes A, C, G, and T, ladders generated from M13mp9 with universal sequencing primer; lane 1, yeast tRNA; lanes 2 to 6, total RNA from DB104 at T0 to T4, respectively. The arrow on the right indicates the position of the +1 nucleotide, which is located 47 bp from the end of the HaeIII fragment (see Fig. 2 for the relevant position in the sequence).
Although the sequence at the −10 and −35 regions suggested that Px was a ςA-type promoter, the fact that its expression was extremely similar to that of P3 made it difficult to exclude the possibility that Px might be controlled by EςH or by more than one species of RNA polymerase holoenzyme. To test this, we introduced Px into the sigH (spo0H) mutant strain WB72 and compared the expression patterns of Px in wild-type DB104 and WB72. In contrast to the drastic difference observed previously in a similar test for P3 of sigA (2), no significant difference was found in this case (data not shown). This suggested that although Px and P3 had similar patterns of expression, they were controlled by different types of RNA polymerases in vivo. In similar tests with the sigB and sigE null mutation strains WB29 and WB71, respectively, no difference of expression was observed when Px was present in either DB104 or any of the null mutants (data not shown).
To test whether EςA could utilize the promoter, the EcoRI-digested plasmid DNA containing Px was used as a template for in vitro transcription by EςA and the RNA synthesized was examined by the method of Goldfarb et al. (4). An RNA runoff band in the range of 50 to 60 nucleotides (nt) was observed (data not shown), which was close to what had been expected from the S1 mapping data, i.e., 47 bp from the +1 site to the HaeIII site, plus 10 bp from the vector polylinker region (19). Given the negative effects of sigH, sigB, and sigE null mutations on Px expression in vivo and the positive results of EςA runoff transcription in vitro, we propose that Px is recognized in vivo by EςA in a temporally regulated fashion different from that for other ςA promoters.
Sequence analysis indicated that there was an open reading frame (orfX) coding for a putative protein product of 98 amino acid residues (11 kDa) within the antE transcript (Fig. 2). A GTG codon very close to the 5′ end of the transcript was preceded by a putative ribosome binding site (RBS) sequence (AGGAG 5 bp upstream, which is a proper spacer for most B. subtilis genes [1, 6, 13]). As shown in Fig. 2, there is a stretch of 9 amino acid residues with high sequence homology to the N-terminal domain of the Escherichia coli transcription regulatory protein OmpR (3, 25), which has been shown to be a member of the highly conserved two-component sensor-regulator systems found in E. coli, B. subtilis, and other organisms (10). One other interesting observation is that this highly conserved sequence coding for CRpLASQSN in the antisense strand also codes for a very highly conserved sequence HCFCGCGAhGN (residues 60 to 69) of DNA primase in the sense strand (21); the variable residues (shown as lowercase letters in the two sequences above) happen to be located in the same position. This might be the first example of a highly conserved region under natural selection for sequences on both strands. Whether this sort of homology indicates any functional significance for the AntE protein has to be experimentally proven.
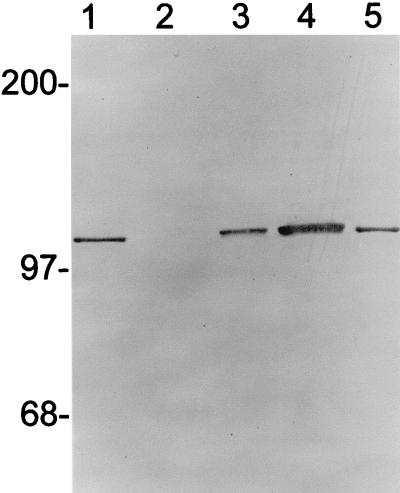
Although the function of the AntE protein remains unclear at present, we were able to show its in vivo expression by fusion of the gene to lacZ. To accomplish this, the 436-bp HaeIII fragment from pSB-X was cut out as a SmaI cassette and subcloned into the unique SmaI site in the lacZ translational fusion vector pMC1871 (20) to form pMC-X so that the expression of the lacZ gene was under the control of Px and the putative translational initiation signals of orfX. The correct orientation and in-frame fusion between two genes in pMC-X were confirmed by sequencing through the fusion junction region (data not shown). For expression in B. subtilis, the orfX-lacZ fusion gene from pMC-X was moved, as a PstI cassette, to pUB18, forming pUBZ-X. The fusion gene thus constructed was demonstrated to be expressed in both E. coli and B. subtilis. Figure 5 is a Western blot profile of the AntE-LacZ proteins expressed from the two organisms. The sizes of the fusion proteins expressed from B. subtilis (Fig. 5, lane 3) and E. coli (lane 4) were essentially the same, indicating that the same translational initiation site was probably used. The sizes of the fusion proteins were also very close to that of the authentic β-galactosidase (Fig. 5, lanes 1 and 5), as one would have expected if the putative GTG codon was used for translational initiation. We are currently trying to assess the possible in vivo function of AntE by carrying out site-directed mutagenesis of the orfX coding region on the antisense strand, at the same time maintaining the dnaE codons on the sense strand.
FIG. 5.
Western blot profile of AntE-LacZ fusion proteins expressed in B. subtilis and E. coli. Lanes 1 and 5, 0.1 μg (each) of authentic E. coli β-galactosidase (Boehringer); lane 2, 3 mg of total cell lysate from DB104 culture at T3 as a negative control; lane 3, 3 mg of total cell lysate from DB104(pUBZ-X) at T3; lane 4, 1.5 mg of total lysate from MC1061(pMC-X) at mid-log growth. Cell lysate preparation and Western blot analysis were carried out as described previously (20, 22).
Figure 6 summarizes the structural features of the antE transcription unit in relation to its complementary part in the sense strand of the sigA operon. The overall gene organization of antE could be described as compact or economical, considering the facts that the entire transcript is only about 440 nt long and that the RBS for orfX is located at the very 5′ end of the RNA transcript. The relatively long nontranslated tail at the 3′ end is complementary to the intercistronic region between P23 and dnaE and to the RBS and the translation initiation site at the N terminus of the dnaE gene. This unique organization did not occur, we believe, by chance, but rather suggests the potential role of the “naked” 3′ tail as a messenger-interfering complementary RNA (micRNA), a class of small regulatory RNA molecules first described by Green and colleagues for the regulatory micF RNA of E. coli (5). Since DNA synthesis ceases around 1 h after the onset of sporulation (T1) in B. subtilis (12, 24), it would not be a surprise to find that the expression of dnaE, the gene coding for DNA primase involved in DNA replication, begins to slow down at a similar developmental stage. The expression pattern of Px (see Fig. 3) fits very well with the postulation that the 3′ nontranslated tail of the antE transcript negatively regulates the expression of dnaE at the initiation stages of sporulation by means of complementarity to the dnaE translational initiation region. We are currently testing this hypothesis using different experimental strategies.
FIG. 6.
Gene organization of antE in relation to the sigA operon. The upper diagram represents part of the sigA operon, showing the truncated P23 and dnaE genes transcribing from left to right. The lower diagram represents the antE gene transcribing from right to left. Shaded areas, coding regions; solid rectangle at the beginning of the coding regions, RBS. The locations of the promoter and terminator are indicated by the arrow and stem-loop, respectively.
From these analyses, it seems possible that the antE transcript has a dual function, with its 5′ portion coding for AntE and its 3′ portion acting as a regulatory micRNA. If this is the case, antE would be the first example, to our knowledge, of a dual-function gene of this type.
Acknowledgments
We thank I. Smith, R. Losick, and C. Moran, Jr., for providing sigma factor null mutants IS233, ML1, and EU8701, respectively.
This research was supported in part by National Institute of General Medical Sciences grant GM 19673.
REFERENCES
- 1.Band L, Henner D J. Bacillus subtilis requires a “stringent” Shine-Dalgarno region for gene expression. DNA. 1984;3:17–21. doi: 10.1089/dna.1.1984.3.17. [DOI] [PubMed] [Google Scholar]
- 2.Carter H L, III, Wang L-F, Doi R H, Moran C P., Jr Promoter in the rpoD operon used by sigma-H RNA polymerase in Bacillus subtilis. J Bacteriol. 1988;170:1617–1621. doi: 10.1128/jb.170.4.1617-1621.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comeau D E, Ikenaka K, Tsung K L, Inouye M. Primary characterization of the protein products of the Escherichia coli ompB locus: structure and regulation of synthesis of the OmpR and EnvZ proteins. J Bacteriol. 1985;164:578–584. doi: 10.1128/jb.164.2.578-584.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldfarb D S, Wong S L, Kudo T, Doi R H. A temporally regulated promoter from Bacillus subtilis is transcribed only by an RNA polymerase with a 37,000-dalton sigma factor. Mol Gen Genet. 1983;191:319–325. doi: 10.1007/BF00334833. [DOI] [PubMed] [Google Scholar]
- 5.Green P J, Pines O, Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:560–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- 6.Hager P W, Rabinowitz J C. Translational specificity in Bacillus subtilis. In: Dubnau D A, editor. The molecular biology of Bacilli. Vol. 2. New York, N.Y: Academic Press; 1985. pp. 1–32. [Google Scholar]
- 7.Igo M, Lampe M, Ray C, Schafer W, Moran C P, Jr, Losick R. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3464–3469. doi: 10.1128/jb.169.8.3464-3469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawamura F, Doi R H. Construction of a Bacillus subtilis double mutant deficient in both extracellular alkaline and neutral proteases. J Bacteriol. 1984;160:442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenney T J, Moran C P., Jr Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3329–3339. doi: 10.1128/jb.169.7.3329-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunst F, Debarbouille M, Msadek T, Young M, Mauel C, Karamata D, Klier A, Rapoport G, Dedonder R. Deduced polypeptides encoded by the Bacillus subtilis sacU locus share homology with two-component sensor-regulator systems. J Bacteriol. 1988;170:5093–5101. doi: 10.1128/jb.170.11.5093-5101.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leighton T J, Doi R H. The stability of messenger ribonucleic acid during sporulation of Bacillus subtilis. J Biol Chem. 1971;246:3189–3195. [PubMed] [Google Scholar]
- 12.Mandelstam J, Higgs S A. Induction of sporulation during synchronized chromosome replication in Bacillus subtilis. J Bacteriol. 1974;120:38–42. doi: 10.1128/jb.120.1.38-42.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran C P, Jr, Lang N, Legrice S F J, Lee G, Stephens M, Sonenshein A L, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 14.Park S-S, Wong S-L, Wang L-F, Doi R H. Bacillus subtilis subtilisin gene (aprE) is expressed from a ςA (ς43) promoter in vitro and in vivo. J Bacteriol. 1989;171:2657–2665. doi: 10.1128/jb.171.5.2657-2665.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price C W, Doi R H. Genetic mapping of rpoD implicates the major sigma factor of Bacillus subtilis RNA polymerase in sporulation initiation. Mol Gen Genet. 1985;201:88–95. doi: 10.1007/BF00397991. [DOI] [PubMed] [Google Scholar]
- 16.Qi F-X, He X-S, Doi R H. Localization of a new promoter, P5, in the sigA operon of Bacillus subtilis and its regulation in some spo mutant strains. J Bacteriol. 1991;173:7050–7054. doi: 10.1128/jb.173.21.7050-7054.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L-F, Doi R H. Nucleotide sequence and organization of Bacillus subtilis RNA polymerase major sigma 43 operon. Nucleic Acids Res. 1986;14:4293–4307. doi: 10.1093/nar/14.10.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L-F, Doi R H. Organization of the major sigma operons of Bacillus subtilis and Escherichia coli. In: Ganesan A, Hoch J A, editors. Genetics and biology of Bacilli. Vol. 2. New York, N.Y: Academic Press; 1986. pp. 367–376. [Google Scholar]
- 19.Wang L-F, Doi R H. Promoter switching during development and termination site of the sigma 43 operon of Bacillus subtilis. Mol Gen Genet. 1987;207:114–119. doi: 10.1007/BF00331498. [DOI] [PubMed] [Google Scholar]
- 20.Wang L-F, Doi R H. Developmental expression of three proteins from the first gene of the RNA polymerase ς43 operon of Bacillus subtilis. J Bacteriol. 1987;169:4190–4195. doi: 10.1128/jb.169.9.4190-4195.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L-F, Price C W, Doi R H. Bacillus subtilis dnaE encodes a protein homologous to DNA primase of Escherichia coli. J Biol Chem. 1985;260:3368–3372. [PubMed] [Google Scholar]
- 22.Wang L-F, Wong S-L, Lee S-G, Kalyan N K, Hung P P, Hilliker S, Doi R H. Expression and secretion of human atrial natriuretic alpha-factor in Bacillus subtilis using the subtilisin signal peptide. Gene. 1988;69:39–47. doi: 10.1016/0378-1119(88)90376-9. [DOI] [PubMed] [Google Scholar]
- 23.Weir J, Dubnau E, Ramakrishna N, Smith I. Bacillus subtilis spo0H gene. J Bacteriol. 1984;157:405–412. doi: 10.1128/jb.157.2.405-412.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winston S, Sueoka N. DNA replication in Bacillus subtilis. In: Dubnau D A, editor. The molecular biology of the Bacilli. Vol. 1. New York, N.Y: Academic Press; 1982. pp. 36–71. [Google Scholar]
- 25.Wurtzel E T, Chou M Y, Inouye M. Osmoregulation of gene expression. I. DNA sequence of the ompR gene of the ompB operon of Escherichia coli and characterization of its gene product. J Biol Chem. 1982;257:13685–13691. [PubMed] [Google Scholar]