ABSTRACT
The transmembrane proteins cdon and boc are implicated in regulating hedgehog signaling during vertebrate development. Recent work showing roles for these genes in axon guidance and neural crest cell migration suggest that cdon and boc may play additional functions in regulating directed cell movements. We use newly generated and existing mutants to investigate a role for cdon and boc in zebrafish neural crest cell migration. We find that single mutant embryos exhibit normal neural crest phenotypes, but that neural crest migration is strikingly disrupted in double cdon;boc mutant embryos. We further show that this migration phenotype is associated with defects in the differentiation of slow-twitch muscle cells, and the loss of a Col1a1a-containing extracellular matrix, suggesting that neural crest defects may be a secondary consequence to defects in mesoderm development. Combined, our data add to a growing literature showing that cdon and boc act synergistically to promote hedgehog signaling during vertebrate development, and suggest that the zebrafish can be used to study the function of hedgehog receptor paralogs.
Keywords: Adaxial cell, Cell guidance, Cell migration, Extracellular matrix, Hedgehog signaling, Neural crest cell
Summary: Hedgehog co-receptors Cdon and Boc regulate neural crest cell migration and differentiation of slow-twitch muscle in the zebrafish trunk.
INTRODUCTION
During vertebrate development, neural crest cells (NCCs) migrate ventrally from the dorsal neural tube to contribute to numerous traits, such as the craniofacial skeleton, peripheral nervous system and pigmentation (Theveneau and Mayor, 2012). Defects in NCC migration result in multiple hereditary disorders (Tobin et al., 2008; Vega-Lopez et al., 2018), and modifications to NCC migration are associated with the evolution of morphological diversity among some vertebrate taxa (Powder et al., 2014). Diseases, such as metastatic melanoma, progress through a reactivation of a NCC-like migratory genetic program (Diener and Sommer, 2021; Kaufman et al., 2016). Understanding the genetic regulation of NCC migration is fundamental to understanding the etiology of many human diseases and is a major morphogenetic event that occurs in every vertebrate embryo.
NCC migration is coordinated along conserved routes in the embryo (Mayor and Etienne-Manneville, 2016) and is regulated by both NCC intrinsic factors, such as genes regulating cell adhesion and actin cytoskeleton (Clay and Halloran, 2011, 2013, 2014; Williams et al., 2018), and NCC extrinsic factors, such as proteins in the extracellular matrix (Perris and Perissinott, 2000; Szabo et al., 2016), tissue stiffness (Barriga et al., 2018; Shellard and Mayor, 2021) and chemical gradients (Barriga et al., 2013; Olesnicky Killian et al., 2009; Shellard et al., 2018). Mediating cell-intrinsic and -extrinsic regulation of NCC migration are cell signaling interactions (Carmona-Fontaine et al., 2008; Mayor and Etienne-Manneville, 2016; Scarpa et al., 2015; Szabo et al., 2016; Theveneau and Mayor, 2012). These include the chemical gradients that attract and inhibit migrating NCCs, and adhesion molecules that adhere NCCs to each other and mediate interactions such as contact inhibition of locomotion (Carmona-Fontaine et al., 2008; Mayor and Etienne-Manneville, 2016; Scarpa et al., 2015; Szabo et al., 2016; Theveneau and Mayor, 2012; Vega-Lopez et al., 2017).
The immunoglobin superfamily member cell adhesion associated, oncogene regulated (cdon) is a cell surface protein implicated in trunk NCC (tNCC) migration in zebrafish (Powell et al., 2015). Morpholino knockdown of cdon results in arrested tNCC migration, with tNCCs mis-localizing N-cadherin. cdon and its paralog boc (brother of cell adhesion associated, oncogene regulated), are vertebrate members of a larger gene family with identifiable orthologs in most metazoan taxa, including the Drosophila paralogs ihog and boi. Across taxa, these proteins activate hedgehog signaling. cdon and boc act as co-receptors that bind to hedgehog ligand and patched receptor to promote smoothened activity. cdon and boc also interact with Wnt and Nodal signaling pathways (Hong et al., 2020; Jeong et al., 2017). cdon forms a complex with N-cadherin/Cdc42/Bnip-2 in a hedgehog-independent manner to regulate actin cytoskeletal dynamics during myogenic differentiation in cell culture (Kang et al., 2008; Lu and Krauss, 2010). boc plays a role in axon guidance and neuron growth (Connor et al., 2005; Okada et al., 2006). Both proteins localize to the ends of long membrane extensions (e.g. cytonemes), suggesting that these receptors help cells sense the external environment and communicate over long distances (Bilioni et al., 2013; Ferent et al., 2019; Hall et al., 2021). We hypothesize that these transmembrane proteins are reiteratively used as cell surface proteins to integrate multiple signals during cell migration and axon guidance. Although a role for cdon in tNCC migration has been suggested (Powell et al., 2015), the mechanism by which cdon and boc regulate NCC movements is not fully understood.
In zebrafish, hedgehog signaling is thought to affect tNCC migration in a non-cell autonomous manner by regulating the differentiation of adaxial mesoderm (Honjo and Eisen, 2005). Adaxial cells form at the border of the notochord during gastrulation and differentiate into slow-twitch muscle that migrates laterally through the somite (Elworthy et al., 2008; Wolff et al., 2003). A non-migratory population of adaxial cells, called muscle pioneers, remains at the body midline. In hedgehog mutant embryos, adaxial cells fail to differentiate into slow-twitch muscle and muscle pioneer derivatives (Elworthy et al., 2008; Roy et al., 2001; Yin et al., 2018). Zebrafish embryos with slow-twitch muscle defects exhibit tNCC migration defects. Although the mechanisms of how these two tissues interact is not fully understood, it is suggested to depend on extracellular matrix deposition and/or modification (Banerjee et al., 2011, 2013; Guillon et al., 2016; Honjo and Eisen, 2005). Whether cdon and boc affect slow-twitch muscle differentiation is unknown (but see Bergeron et al., 2011), and it is possible that cdon and boc affect NCC development through autonomous mechanism (Powell et al., 2015).
We use previously unreported CRISPR/Cas9-generated mutations and existing mutants to investigate the role of cdon and boc in tNCC migration and slow-twitch muscle differentiation. We show that cdon and boc are expressed by tNCCs and slow-twitch muscle. Loss of either cdon or boc individually has no observable effect on tNCC migration and has minimal effects on slow-twitch muscle differentiation. However, embryos that are double mutant for cdon and boc exhibit both tNCC migratory defects and loss of slow-twitch muscle. tNCC motility is not impaired by loss of cdon or boc, suggesting that the tNCC migration defect may be a non-cell autonomous consequence of loss of the migratory path. We provide data suggesting that cdon and boc modify hedgehog signal reception during mesoderm differentiation. These data contribute to a growing literature on the crucial role of cdon and boc in modulating hedgehog signaling across tissues during embryogenesis.
RESULTS
cdon and boc exhibit shared sequence conservation in extracellular domains
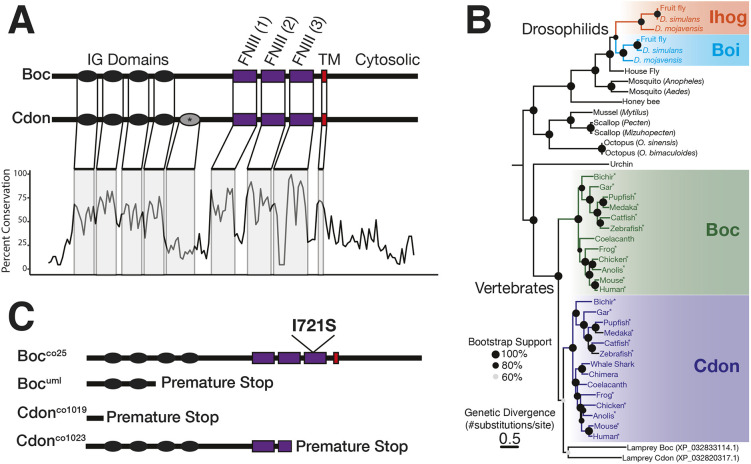
cdon and boc share a similar genomic domain architecture that includes four immunoglobin (IG) domains, three fibronectin type 3 repeat (FNIII) domains and a cytosolic C-terminal tail (Fig. 1A). A fifth IG domain is present in cdon orthologs of some tetrapod taxa. The FNIII domains bind hedgehog ligand and patched receptor (Izzi et al., 2011; Song et al., 2015), and the C-terminal region is needed for protein localization in the cell (Okumura et al., 2021).
Fig. 1.
cdon and boc are transmembrane receptors and share conserved domain structure. (A) cdon and boc share a domain structure of four or five immunoglobin (IG) domains, three fibronectin (FNIII) domains, a transmembrane (TM) region and a cytosolic C-terminal tail. A fifth cdon IG domain (*) is found in some tetrapod taxa. Percent conservation (20 amino acid sliding window) is shown for an alignment of cdon and boc amino acid sequences from 11 tetrapod and ray-finned fish taxa. The extracellular domains exhibit high sequence conservation, while the cytosolic region is less conserved. (B) Maximum likelihood tree (phyml) for curated cdon and boc ortholog amino acid sequences from multiple bilaterian taxa. There are independent duplications of Ihog/Boi in drosophilids and of Cdon/Boc in vertebrates. Node support was calculated based on 1000 bootstraps; nodes with less than 60% bootstrap support are shown in gray. Asterisks indicate taxa used to generate conservation in A. Lamprey cdon and boc sequences are uncolored due to topological position as an outgroup with respect to vertebrate cdon sequences. (C) Schematic of cdon and boc mutant alleles characterized in the current study.
As functional conservation is reflected by sequence conservation across evolutionary time, we investigated the molecular evolution of cdon and boc orthologs across vertebrate taxa. An alignment of cdon and boc amino acid sequences from 11 select jawed vertebrate taxa shows that extracellular domains exhibit high conservation across taxa and paralogs, suggesting that these extracellular domains are crucial for gene function (Fig. 1A). In contrast, the C-terminal cytosolic tail exhibits relatively low sequence conservation (Kang et al., 2002), suggesting that cytosolic region is evolutionarily labile.
Orthologs to cdon and boc are identifiable in non-vertebrate bilaterians. These include Drosophila paralogs interference hedgehog (ihog) and brother of ihog (boi), the functions of which in hedgehog signaling have been well characterized (Bilioni et al., 2013; Camp et al., 2010; Yao et al., 2006). However, the orthology relationships between cdon and boc and non-vertebrate orthologs, such as ihog and boi, are less well known. To explore the molecular evolution of this gene family, we built a maximum likelihood tree (phyml) using 44 amino acid sequences from taxa representative of major bilaterian clades (Fig. 1B). These data identify cdon and boc as shared vertebrate-specific paralogs that emerge from a duplication event at either the base of the vertebrate phylogeny or along the gnathostome lineage after splitting from cyclostomes (hagfish and lamprey). Both lamprey cdon and boc paralogs group with other vertebrate cdon sequences in our tree; however, bootstrap node support is relatively low in support of this topology. Regardless of the true position of the lamprey cdon and boc genes, these data are consistent with a hypothesis that cdon and boc originated during one of the two vertebrate-specific genome duplications (Dehal and Boore, 2005). Analyses suggest that an independent gene duplication event led to the origins of ihog and boi paralogs in drosophilids (Fig. 1B) (Allen et al., 2011; Izzi et al., 2011; Sanchez-Arrones et al., 2012), a finding concordant with gene trees made publicly available by the TreeFam consortium (Li et al., 2006; Ruan et al., 2008).
To explore the function of cdon and boc we used mutations that target the conserved functional extracellular domains (Fig. 1C, see Materials and Methods). Two mutants for each gene were initially used to confirm an association between phenotypes and gene mutations. As we observed similar phenotypes for both cdon mutations and both boc mutations, we focused our analyses on two mutations in particular, cdonco1019 and bocco25, and refer to these mutants simply as cdon and boc, except where stated.
cdon and boc transcripts are expressed in tNCCs and slow-twitch muscle cells
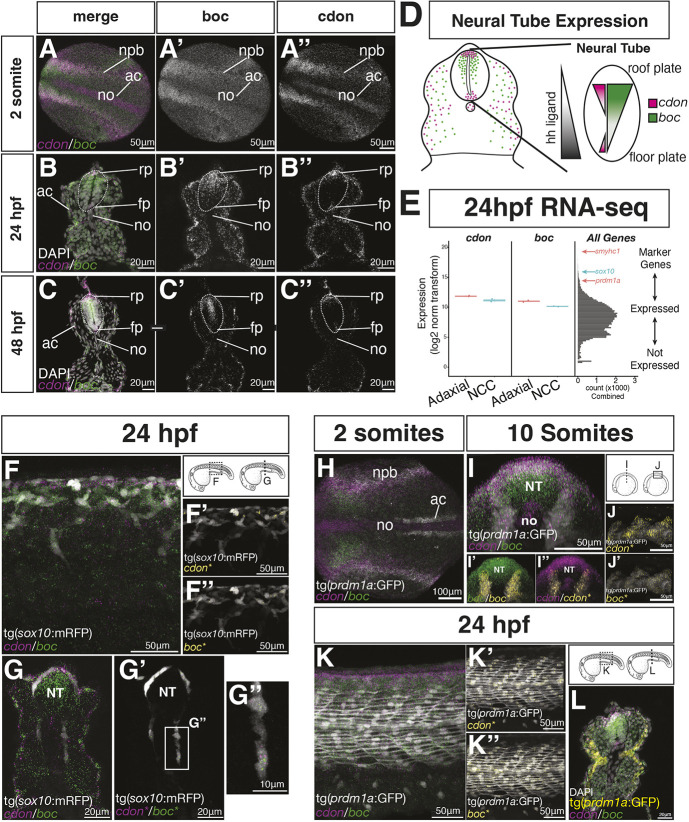
To determine where endogenous cdon and boc transcripts are expressed, we used multiplexed hybrid chain reaction in situ hybridization (HCR), focusing on tNCC migration stages. We found that cdon and boc transcripts were abundant in the dorsal central nervous system (CNS) of 24-48 hpf zebrafish embryos (Fig. 2A-D), consistent with data from previous studies (Bergeron et al., 2011; Cardozo et al., 2014; Kearns et al., 2021). This pattern is established early in development. cdon and boc are initially expressed along the border of the neural plate in cells that contribute to the dorsal neural tube and NCC (Fig. 2A). Whereas boc transcripts in the CNS form a broad dorsal-ventral gradient (Kearns et al., 2021), cdon transcripts are restricted to the dorsal-most roofplate cells and the ventral-most floorplate cells of the CNS. As such, cdon creates a dorsal sub-domain of boc expression (Fig. 2D). Floorplate cells express hedgehog ligand. This CNS expression at 24 hpf is similar to expression patterns reported in mouse embryos (Izzi et al., 2011; Kang et al., 2002; Mulieri et al., 2000, 2002; Tenzen et al., 2006).
Fig. 2.
cdon and boc are expressed by NCCs and adaxial mesoderm cells. (A-C) Hybrid chain reaction in situ hybridization (HCR) shows cdon (magenta) and boc (green) expression in the developing neural tube of two somites shown in a dorsal view (A), and in 24 hpf (B) and 48 hpf (C) cross-sectioned embryos (20 µm sections at the level of the yolk extension). (D) Schematic of a cross-section at yolk extension summarizing cdon and boc expression in the neural tube. The gradient of hedgehog ligand is indicated in gray. (E) Bulk RNA-seq from two replicate samples of FAC-sorted tNCCs (blue) and slow-twitch muscle (red) suggest both tissues express cdon and boc. Right panel is a histogram of gene expression levels for all samples for all genes. Location of genes known to mark NCCs (blue) or slow-twitch muscle (red) are indicated. (F) Whole-mount lateral view of 24 hpf embryo showing expression of cdon and boc with NCCs (gray) labeled by the tg(sox10:mRFP) transgene at level of yolk extension. (F′,F″) HCR puncta overlapping the sox10 transgene (*) are shown in yellow. (G-G″) Cross-section shows tNCCs expressing cdon and boc transcripts. (H-L) HCR expression of cdon and boc in adaxial mesoderm labeled with the tg(prdm1a:eGFP) transgene (gray). (H) Dorsal view of a two-somite embryo. (I-J′) Optical cross-sections (I-I″) and lateral views (J,J′) of a 10-somite embryo. (I′,I″,J,J′) HCR puncta overlapping the tg(prdm1a:eGFP) transgene are shown in yellow. Only HCR puncta overlapping the tg(prdm1a:eGFP) transgene are shown in J and J′. (K) Lateral view of 24 hpf embryo showing cdon and boc transcripts in slow-twitch muscle. K′ and K″ show overlapping puncta labeled in yellow. (L) Cross-section of a 24 hpf embryo shows tg(prdm1a:eGFP) in yellow, cdon/boc puncta in slow-twitch muscle along with the nuclear marker DAPI in white. ac, adaxial cells; fp, floor plate; no, notochord; rp, roof plate.
boc is known to be expressed by slow-twitch muscle at 24 hpf (Bergeron et al., 2011); however, detailed investigation of cdon and boc expression in NCCs and slow-twitch muscle have not been undertaken. To further characterize expression, we used the tg(sox10:mRFP) and tg(prdm1a:eGFP) transgenic reporter lines to isolate both tissues from the trunks of 24 hpf zebrafish embryos, and sequenced the transcriptome of these tissues using bulk RNA-seq (see Materials and Methods). These data identify cdon and boc transcripts in both tissues, supporting the hypothesis that tNCCs and adaxial mesoderm express cdon and boc at low to moderate levels (Fig. 2E).
We asked whether this expression is observable by HCR. Using the tg(sox10:mRFP) transgene to label NCCs, we identified cdon and boc HCR puncta that overlap the sox10 transgene, confirming the presence of cdon and boc transcripts in tNCCs at 24 hpf (Fig. 2F,G), albeit at levels lower than surrounding tissues.
To confirm expression in slow-twitch muscle and adaxial cell mesoderm, we used the tg(prdm1a:eGFP) (Elworthy et al., 2008). At the two-somite stage, adaxial cells are labeled with the tg(prdm1a:eGFP) transgene, but we did not observe significant expression of cdon/boc in these cells (Fig. 2H). By the 10-somite stage, cdon and boc transcripts were observed throughout the mesoderm (Fig. 2I,J). At 24 hpf, slow-twitch muscle cells continue to express cdon and boc (Fig. 2K,L), consistent with findings from our bulk RNA-seq data (Fig. 2E).
tNCC migration and slow-twitch muscle development are affected in cdon;boc mutants
In wild-type embryos, tNCCs collectively migrate during the period when slow-twitch muscle cells are migrating (Fig. 3A,B). To visualize the interaction of tNCCs with adaxial cells, we live imaged double transgenic tg(sox10:mRFP);tg(prdm1a:gfp) embryos from 10 somites to 24 hpf (Movie 1). These data show tNCCs and slow-twitch muscle migrating at similar times and potentially interacting. tNCCs initially migrate to the dorsal extent of the slow-twitch muscle, and then come into contact with the muscle pioneer cells at the body midline before continuing to migrate ventrally.
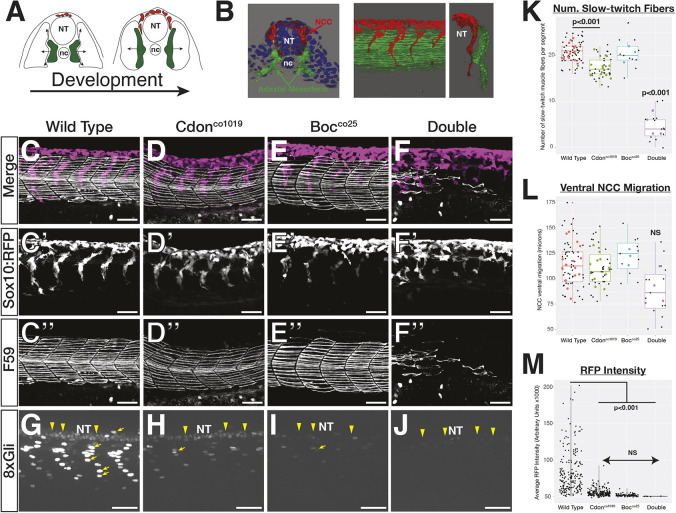
Fig. 3.
cdon and boc affect NCC migration and slow-twitch muscle differentiation. (A) Schematic cross-sections of a 10-somite to 24 hpf embryo show NCC (red) and adaxial mesoderm (green) migration. Adaxial cells in green are specified adjacent to the notochord and migrate laterally to differentiate into slow-twitch muscle. NCCs (red) migrate ventrally from the dorsal neural tube. (B) 3D renderings of a 10 μm cross-section from the trunk of a 15-somite (left) and a whole-mount 24 hpf embryo (right) showing association between migrating adaxial mesoderm (green) and NCCs (red). (C-F) Lateral views of maximum projection images show NCC streams, tg(sox10:mRFP) and immunostaining for slow-twitch muscle (F59; white) in 24 hpf wild-type embryos at the level of yolk extension. Single fluorescent images are shown in black and white below for tNCC (C′-F′) and adaxial cells (C″-F″). (G-J) Maximum projection images of tg(8×Gli:mCherry) transgene in 24 hpf embryos. There are mCherry-positive nuclei in slow-twitch muscle (yellow arrows) and neural tube (yellow arrowheads). (K-M) Quantification of (K) the number of F59 stained fibers per segment in each genotype, (L) ventral migration of the NCC stream, and (M) mCherry intensity. For K and L, black dots represent individual segments and colored dots represent individual averages. Horizontal line in box plot indicates the median. Lower and upper edges of boxes indicate the first and third quartiles, respectively. Whiskers indicate minimum and maximum values. NT, neural tube; nc, notochord.
To determine whether cdon and boc mutations affect tNCC migration and slow-twitch muscle development, we used the tg(sox10:mRFP) line to label migrating tNCCs and immunostaining of a slow-twitch muscle-specific myosin (DSHB F59) to label adaxial mesoderm derivatives in 24 hpf embryos. We analyzed two different alleles of each, cdonco1019 and cdonco1023, and two existing alleles of bocty54z and bocco25 (Bergeron et al., 2011; Kearns et al., 2021).
tNCC migration was normal in single cdon and boc mutant embryos (Fig. 3C-E; Fig. S1). These data from homozygous single mutant embryos are in contrast to those reported by morpholino knockdown of cdon (Powell et al., 2015). We hypothesized that this may be due to compensation in mutant lines. To test this, we created double cdon;boc mutant embryos and determined that tNCCs failed to form distinguishable streams. In double mutant embryos, tNCC migration arrested at the body midline, similar to what was observed from cdon knockdown (Fig. 3F,L; Fig. S1) (Powell et al., 2015). These analyses confirm that the bocco25 allele is phenotypically similar to the bocty54 in both adaxial cell formation and tNCC migration, and that both cdonco1019 and cdonco1023 alleles behave similarly (also see Kearns et al., 2021).
Similar to our findings of tNCC migration, slow-twitch muscle differentiation and migration was normal in cdon and boc single mutants (Bergeron et al., 2011). However, double mutants exhibited defects in slow-twitch muscle differentiation (Fig. 3C-F,K; Fig. S1). In double mutant embryos, slow-twitch muscle fibers appeared dysmorphic, with loosely packed myosin bundles relative to wild-type embryos (e.g. compare myosin bundle width and packing in Fig. 3; Fig. S1). Loss of slow-twitch muscle cell differentiation was not complete and posterior body segments tended to have fewer slow-twitch muscle fibers than anterior segments.
To determine whether this slow-twitch muscle defect can be observed at earlier stages, we performed in situ hybridization and HCR for myod1 and prdm1a at the 10-somite stage (Fig. S2). In situ hybridization with myod1 of wild type when compared with the double mutant shows that loss of cdon;boc results is a severe reduction of myod1-expressing adaxial cells (Fig. S2A), whereas the somitic mesoderm is not affected. We next assayed both myod1 and prdm1a expression at 10 somite stage by HCR (Fig. S2B). Here, we determined that while single mutants have normal adaxial cells, cdon;boc double mutants display absent myod1 expression in the adaxial domain and severely reduced expression of prdm1a.
cdon and boc affect hedgehog signal transduction in slow-twitch muscle cells
As cdon and boc play a role in hedgehog signaling (Allen et al., 2011; Zhang et al., 2006), we hypothesized that defects in tNCC migration and slow-twitch muscle differentiation were due to loss of hedgehog signaling. To test this, we measured hedgehog signal reception by using the transgenic, tg(8xGliBS:mCherry-NLS-Odc1) reporter line (hereafter 8xGli:mCherry) that drives nuclear localized mCherry under the control of eight tandem Gli-binding sites (Mich et al., 2014). The mCherry fluorophore contains an Odc1 destabilization domain that facilitates proteasomal degradation to promote rapid fluorophore turnover.
At 24 hpf, we observed mCherry-positive nuclei in ventral neural tube cells and in slow-twitch muscle nuclei (Fig. 3G) (Kearns et al., 2021; Mich et al., 2014). We never observed mCherry-positive nuclei in tNCCs. In cdon and boc mutants, the number and signal intensity of mCherry-positive nuclei was lower when compared with wild-type siblings (Fig. 3G-J,M). Although not statistically significant, 8×Gli:mCherry intensity in slow-twitch muscle nuclei qualitatively varied across mutant genotypes, with boc mutants displaying lower 8×Gli:mCherry signal than cdon mutants, and double mutants exhibiting lower signal than all other genotypes. This results from a reduction in the number of high mCherry-expressing cells (Fig. 3M). These data suggest that cdon and boc mutations decrease hedgehog activity in slow-twitch muscle cells, which is consistent with a hypothesis that hedgehog signaling is responsible for the slow-twitch muscle differentiation phenotype in mutant embryos (Barresi et al., 2000; Devoto et al., 1996).
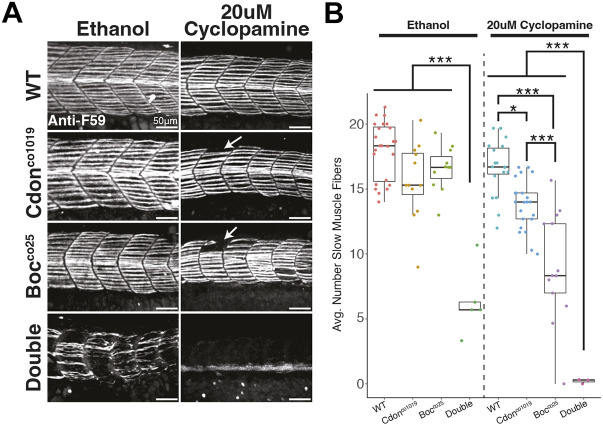
Interestingly, the tg(8×Gli:mCherry) transgene reported substantially lower hedgehog signal reception in single cdon and boc mutant embryos that exhibit normal slow-twitch muscle differentiation. These data suggest that slow-twitch muscle development is robust to moderate perturbations in hedgehog signals. If this hypothesis is correct, we reasoned that cdon and boc mutations should sensitize embryos to further pharmacological manipulation of smoothened signaling with cyclopamine. To test this, we in-crossed double heterozygous cdon;boc parents and treated embryos with a subthreshold dose of 20µM cyclopamine, or ethanol as a control, from 50% epiboly to 24 hpf. We then quantified the number of slow-twitch muscle fibers (F59) at 24 hpf as a readout.
Although single mutants treated with ethanol had a similar number of slow-twitch muscle fibers to their wild-type treated siblings, single mutants treated with 20µM cyclopamine had significantly fewer slow-twitch muscle fibers than their similarly cyclopamine-treated wild-type siblings (Fig. 4). This pattern was even more striking for double mutants. Whereas double mutants treated with ethanol have an average of approximately five to six slow-twitch muscle fibers per segment, double mutants lost all slow-twitch muscle fibers when treated with 20µM cyclopamine. We also noted qualitative changes to slow-twitch muscle morphology. Mutants treated with 20µM cyclopamine often exhibited gaps or missing slow-twitch muscle fibers in body segments, and muscle fibers were more likely to have loosely packed myosin. We interpret these results to indicate that cdon and boc are modifying hedgehog signals reception by slow-twitch muscles, and that loss of cdon or boc sensitize embryos to further perturbations of hedgehog signals.
Fig. 4.
cdon and boc sensitize zebrafish embryos to a reduction in hedgehog signal by cyclopamine treatment. (A) Lateral views of representative images show slow-twitch muscle fiber morphology in 24 hpf embryos treated with either 20µM cyclopamine or an equivalent volume of ethanol as a control. cdon and boc mutants exhibit more severe phenotypes when treated with cyclopamine, as evidenced by gaps in segments (arrows), and complete loss of slow-twitch muscles in the double mutants. (B) Quantification of average number of slow-twitch muscle fibers per segment shows that cdon and boc mutations are sensitizing embryos to cyclopamine treatment. All images and data are taken from segments at the level of the yolk extension. Horizontal line in box plot indicates the median. Lower and upper edges of boxes indicate the first and third quartiles, respectively. Whiskers indicate minimum and maximum values. Significance values are from Tukey pairwise post-hoc tests (*P<0.05, ***P<0.001).
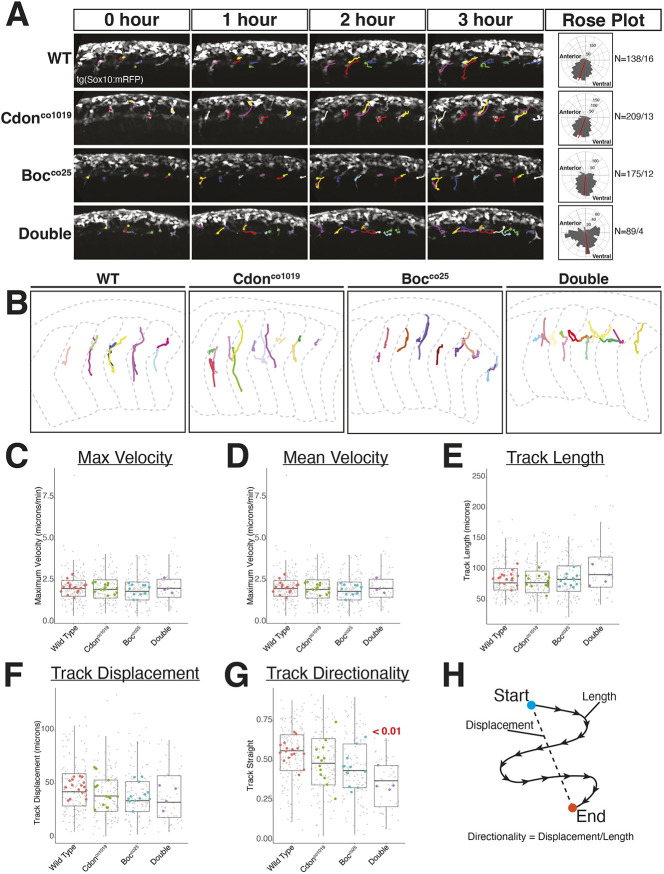
tNCCs lose directionality in cdon;boc double mutants
The arrested tNCC migratory phenotype in double mutants is striking and could result from either a loss of NCC migratory ability or a loss of directionality. To investigate this further, we used the tg(sox10:mRFP) transgene to label migrating tNCCs and live imaged NCC migration at the level of the yolk extension from 22 hpf over a 6 h time frame (Fig. 5; Movie 2). We chose this stage to focus on imaging of active tNCC migration and not the initiation of migration. We found that across all genotypes, tNCCs were qualitatively migratory, and general measures of motility (e.g. velocity, etc.) in mutants were similar to that observed in wild-type controls (Fig. 5). In contrast, we observed a significant loss of directional tNCC movement in double cdon;boc mutant embryos. In double mutants, tNCCs migrate to the body midline and then move along the anterior-posterior axis, which is in contrast to the directed ventral movement typical of wild-type embryos. This is seen quantitatively as a lower ability to migrate directionally and an increased proportion of movements in lateral directions (Fig. 5A,G). Single cdon or boc mutants did not display migratory phenotypes that were different from that of wild-type controls. We interpret these results to indicate that loss of cdon and boc affects tNCC directionality and path-finding ability, but that cdon and boc are not required for overall tNCC motility.
Fig. 5.
NCC directionality, but not motility, is disrupted in cdon;boc mutants. (A) Maximum projection images show migrating tNCCs in representative wild-type, cdon, boc and double mutant embryos over a 6 h time period at the level of yolk extension. Colored lines are tracks of individual cells. There are horizontal tracks at body midline in double mutant embryos. Matched rose plots (right panels) show directional movement of NCCs in each genotype. Samples sizes reflect number of tracks/number of embryos. (B) Migration tracks overlaid onto outlines of somites (gray dashed lines) for representative embryos. Somite segments in the double mutant are wider and dysmorphic. (C-G) Quantification of NCC migration for maximum velocity (C), mean velocity (D), track length (E), track displacement (F) and track directionality (G). Small gray points are individual track values; large colored points are means for individuals; boxplots reflect variability across tracks. (H) Schematic of track straightness calculated as track displacement divided by track length. All images and data are taken at yolk extension. Horizontal line in box plot indicates the median. Lower and upper edges of boxes indicate the first and third quartiles, respectively. Whiskers indicate minimum and maximum values.
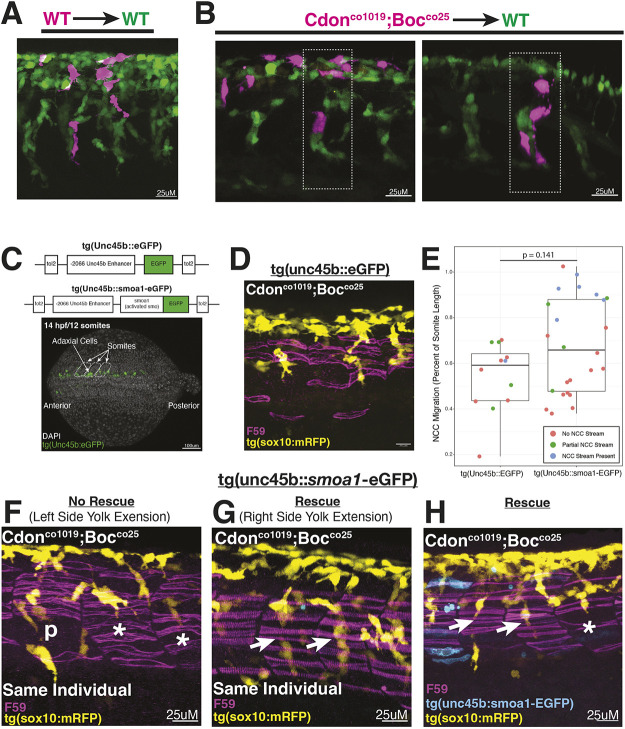
We therefore hypothesized that tNCC migratory defects in cdon;boc mutants are a secondary consequence of failed slow-twitch muscle differentiation. To test cell autonomy versus non-cell autonomy, we attempted to rescue tNCC migration in double cdon;boc mutant embryos using two complementary methods.
First, we created genetic mosaics by transplanting double mutant tNCCs into wild-type hosts (Fig. 6A-C). The rationale for these experiments is that if the observed tNCC migration defects are a non-cell autonomous consequence of slow-twitch muscle differentiation, then we would expect transplanted mutant tNCCs to migrate normally in wild-type hosts with normal muscle differentiation.
Fig. 6.
Transplant and smo overexpression experiments in cdon/boc double mutants suggest a possible non-autonomous role. (A,B) Wild-type (A) and mutant (B) tNCCs transplanted into wild-type hosts. Double mutant cdon;boc tNCCs migrate normally in wild-type hosts when transplanted into the trunk (B, 3/3 embryos). (C, top) tol2 constructs used for rescues. Dorsal view of a 10-somite embryo shows mosaic expression in adaxial cells. (C, bottom) Mosaic expression of smoa1-eGFP in adaxial cell progenitors. (D) Lateral view of slow-twitch muscle morphology (F59; magenta) and tNCC streams (yellow) in a double mutant embryo injected with control eGFP construct. (E) Quantification of NCC ventral migration (as percent of somite length) in control- and smoa1-eGFP-injected embryos. Colors indicate qualitative rescue of the NCC stream. Control injected, n=4 embryos; smoa1a-eGFP injected, n=8 embryos. (F-H) Representative images of smoa1a-eGFP-injected embryos. (F) Left side of an embryo where tNCC migration (yellow) was not rescued. (G) Right side of same embryo where tNCC migration was rescued. Note the slow-twitch muscle fiber morphology (magenta). (H) Mosaic expression of smoa1a-eGFP (cyan). Arrows indicate segments with qualitatively rescued tNCC streams. Asterisks label segments with non-rescued streams, ‘p’ labels a representative ‘partial rescue’.
When wild-type tNCCs are transplanted into wild-type hosts, we observed transplanted tNCCs migrated in well-organized streams alongside host tNCCs (Fig. 6A). Similarly, in three out of three cases (out of 490 transplanted embryos) where double mutant tNCCs were transplanted into wild-type hosts, we observed mutant tNCCs migrating alongside host tNCCs in organized streams (Fig. 6B,C). Each embryo contained between one and three transplanted tNCCs. Although a low sample size, these data suggest that double cdon;boc mutant tNCCs migrate normally in wild-type hosts with normal mesoderm development, which is consistent with a hypothesis of a non-autonomous role for cdon and boc in tNCC migration (but see Discussion).
The second approach used tol2 transgenesis and the −2.066 kb fragment of the unc45b enhancer (Berger and Currie, 2013) to drive expression of a constitutively active smoothened, smoa1-eGFP (Ju et al., 2014, 2009), in adaxial mesoderm at early segmentation stages to rescue cdon;boc mutations in mesoderm. Importantly, this early adaxial cell expression turns on before slow-twitch muscle cell differentiation (Fig. 6D). We reasoned that if cdon;boc are affecting hedgehog signaling then driving smoa1-eGFP in the slow-twitch muscle progenitor population should rescue both slow-twitch muscle morphology and tNCC migration (Fig. 6D).
Out of eight double mutant embryos injected with the smoa1a-eGFP construct, four exhibited at least one tNCC stream that was qualitatively normal. This is in comparison with four double mutant embryos injected with a control eGFP construct where only one exhibited a tNCC stream that was considered a wild-type phenotype. However, tNCCs did not migrate significantly further (as a percentage of total somite length) in rescue injected embryos when compared with control injected embryos (Fig. 6F). We note that we observed low transgenesis efficiency in the smoa1-eGFP plasmid relative to the control plasmid, which we hypothesize is due to downregulation of the transgene (see Materials and Methods).
Importantly, these are F0-injected embryos, tol2 transgenesis is mosaic and the rescue phenotype is expected to be mosaic. In support of this, in one embryo injected with the smoa1-egfp construct, the left side of the embryos exhibited a typical mutant phenotype, whereas the right side of the embryo exhibited tNCC streams that appeared wild-type like (Fig. 6G,H). The slow-twitch muscle morphology on the right side of this embryo was similarly wild-type like, whereas the non-rescued left side had loosely packed myosin bundles. Across all embryos, body segments with qualitatively rescued tNCC streams (see methods) tended to have more slow-twitch muscle fibers than either non-rescued or partially rescued segments (Fig. S3A). We observed that slow-twitch muscle fiber morphology appeared normal in these ‘rescued’ segments (note the tightly packed slow-twitch muscle fibers in Fig. 6H,I compared with loosely packed fibers in Fig. 6E,G).
We noticed that some control injected embryos had similar numbers of slow-twitch muscle fibers to those observed in the smoa1-eGFP injected embryos (Fig. S3A). In these cases, control injected embryos also exhibited a qualitative ‘partial rescue’ or, in one case, a ‘wild-type like’ tNCC stream. To quantify this, we measured the ventral extent of the tNCC stream and related this to the number of F59 fibers in a segment (Fig. S3B). However, a correlation between number of F59 fibers per somite and ventral migration of the tNCC stream was not significant (Fig. S3B). We hypothesize that there is likely some degree of incomplete penetrance in the cdon;boc mutants, whereby occasional segments in some embryos exhibit wild-type-like phenotypes.
Deposition of a Col1a1a extracellular matrix is affected in cdon;boc mutants
Previous research has suggested that slow-twitch muscle cells express or modify extracellular matrix proteins and genes, including Col15a1b (Guillon et al., 2016), tenascin (Schweitzer et al., 2005), col18a1 (Banerjee et al., 2013; Schneider and Granato, 2006) and the collagen-modifying enzyme plod3 (also known as lh3) (Schneider and Granato, 2006). Trunk NCCs are known to migrate on collagen substrates (Lallier et al., 1992), and we hypothesized that changes to a collagen extracellular matrix (ECM) may be responsible for mediating the effects of cdon and boc on tNCC migration (Banerjee et al., 2013).
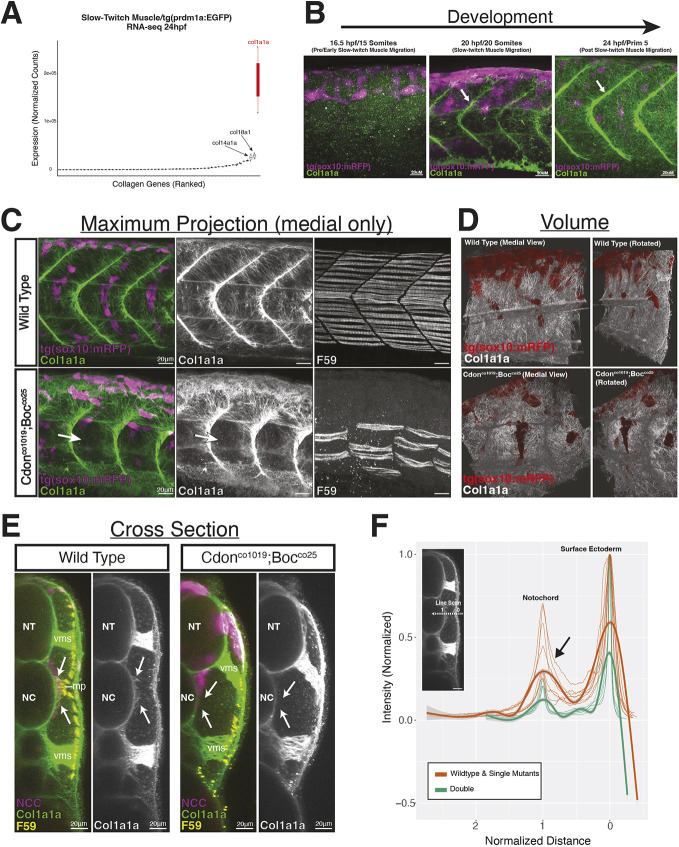
To investigate this, we identified collagen genes expressed by slow-twitch muscle in our RNAseq dataset on FAC sorted tg(prdm1a:eGFP) cells. We found many collagen genes expressed by slow-twitch muscle cells. In particular, col1a1a is highly expressed by slow-twitch muscle, with more than twice the number of estimated transcripts than the next two most highly expressed collagen genes (Fig. 7A).
Fig. 7.
Col1a1a matrix deposition in cdon;boc mutants. (A) RNA-seq from FAC sorted tg(prdm1a:eGFP) cells at 24 hpf identify expression of collagen genes. (B) 3D projections of Col1a1a deposition at the level of yolk extension. At 20 hpf, Col1a1a expression is strongest in the vertical myosepta (arrows). (C) Medial view shows Col1a1a deposition along the somite-notochord boundary in wild-type and double mutant embryos at 24 hpf. Col1a1a signal in skin ectoderm was removed for images in C. The loss of Col1a1a in the double mutant is associated with loss of slow-twitch muscle differentiation (arrows). Retention of Col1a1a around dorsal neural tube (arrows) coincides with the location of tNCC migration arrest. (D) Volume rendering shows medial deposition of Col1a1a in wild-type (top) and double mutant (bottom) embryos at 24 hpf. (E) Optical cross-section shows loss of medial Col1a1a deposition along the NCC migratory path (arrows) in double mutant embryos at 24 hpf. (F) Line scan quantification showing reduced Col1a1a intensity in double mutant embryos. Line scans taken on optically sectioned images and normalized for length and intensity. NC, notochord; NT, neural tube; mp, muscle pioneers; vms, vertical myosepta.
We looked at Col1a1a protein deposition using an antibody raised against the zebrafish Col1a1a protein (GeneTex). At 16 h, we observed punctate expression, suggesting that this ECM component is deposited after this time period (Fig. 7B; Fig. S4). A control experiment with secondary antibody only indicates that punctate Col1a1a staining at 16 hpf is not background at this stage (Fig. S4). Interestingly, we observed premature Col1a1a fibers in some double mutant embryos at 16 hpf located on the surface ectoderm (Fig. S4). From 20 to 24 hpf, Col1a1a was observed in the presumptive skin ectoderm, vertical myosepta and medial border of the mesoderm adjacent to the neural tube and notochord (Fig. 7B,C; Fig. S4). This period between 16 hpf and 24 hpf coincides with slow-twitch muscle migration (Elworthy et al., 2008).
Importantly, we observed Col1a1a fibers deposited along notochord/somite boundary that medial path tNCCs migrate on (Fig. 7C-E; Movie 3). In wild-type embryos, Col1a1a fibers extend into the region where tNCC streams are observed along the medial somite border. In double cdon;boc mutants, Col1a1a deposition in along this tNCC path is disrupted. Optical cross-sections at 24 hpf show few collagen fibers in this medial region of double mutant embryos where tNCCs tend to stall (arrows in Fig. 7E). We further quantified these data by drawing line scans from the skin through the notochord, normalizing each line scan to intensity and skin to notochord distance (Fig. 7F). These data show reduced Col1a1a intensity in double mutant embryos. We note a steep change in Col1a1a intensity values between the notochord peak and skin ectoderm peak in double mutant embryos, suggesting absence of Col1a1a that is consistent with our observations of collagen deposition and fiber formation being disrupted along the tNCC migratory path in cdon;boc double mutants.
We interpret these data to suggest that tNCC migratory defects may be a consequence of loss of the Col1a1a ECM along the migratory path (Fig. 7). Supporting this hypothesis is the observation that Col1a1a is still expressed dorsally in the double mutants, but stops at the position where tNCC migration often arrests.
DISCUSSION
We show that cdon and boc affect tNCC migration and differentiation of slow-twitch muscle cells. Despite our previous knockdown studies suggesting that cdon alone functions in tNCC migration (Powell et al., 2015), we find that only in double cdon;boc mutants do we observe noticeable tNCC migratory defects associated with loss of slow-twitch muscle. We hypothesize compensation between cdon and boc in homozygous mutant embryos, as double mutants have a similar tNCC migration phenotype to that seen in knockdown embryos. Earlier research suggests that slow-twitch muscle differentiation requires hedgehog signaling (Elworthy et al., 2008), and that tNCC migration requires proper slow-twitch muscle differentiation (Honjo and Eisen, 2005). Our data are consistent with these results and suggest that the tNCC migratory defect in cdon;boc mutants is a consequence of defects in mesodermal differentiation (Banerjee et al., 2011, 2013; Honjo and Eisen, 2005).
Although mesodermal tissues are appreciated as being crucial for neural crest migration (Krull, 2010; Kulesa and Gammill, 2010; Theveneau and Mayor, 2012), the mechanisms underlying how the mesoderm affects NCC migration vary among both taxa and different NCC subpopulations. In tetrapod taxa, mesoderm restricts cranial and trunk NCC migration along conserved paths through neuropilin, semaphorin and ephrin ligand-receptor interactions (Barriga et al., 2018; Davy and Soriano, 2007; Gammill et al., 2007, 2006; Krull, 2010; Krull et al., 1997; Santiago and Erickson, 2002). Localization of receptors and ligands within tissues creates permissive/non-permissive zones that restrict NCC migratory paths. Mesodermal tissues can also act as sources of chemoattractant molecules, such as chemokine signaling (Kasemeier-Kulesa et al., 2010; Olesnicky Killian et al., 2009). In zebrafish, semaphorin and neuropilin interactions mediate cranial NCC migration (Yu and Moens, 2005), although the role of this pathway in zebrafish tNCC migration is less established. We searched our RNA-seq data for complementary ligand-receptor pairs in FAC sorted tNCCs and slow-twitch muscle cells. Although some possible interacting molecules could be found, there were no clear candidate signaling pathways, leading us to hypothesize that there is not a direct ligand-receptor interaction between tNCCs and slow-twitch muscle mesoderm (not shown; data have ben deposited in GEO under accession number GSE213728).
Instead, we build on earlier research in both tNCC migration (Banerjee et al., 2011, 2013) and motor axon guidance (Guillon et al., 2016; Schneider and Granato, 2006; Zeller and Granato, 1999; Zhang et al., 2004) to suggest that slow-twitch muscle cells affect tNCC stream formation through ECM deposition. Adaxial derived slow-twitch muscle cells express Col15a1b (Guillon et al., 2016), col18a1 (Schneider and Granato, 2006) and chondroitin sulfate proteoglycans associated with collagen-rich ECMs (Zhang et al., 2004). Morpholino knockdown of col18a1 leads to tNCC migratory defects that are similar to those we observe in cdon;boc mutants (Banerjee et al., 2013). Our data add to this literature, showing that slow-twitch muscle expresses other collagen genes in addition to those previously studied, including col1a1a at exceptionally high levels. Deposition of col1a1a along the medial somitic boundary is disrupted in cdon;boc mutants, consistent with a model in which slow-twitch muscle cells are responsible for depositing a collagen-rich ECM along the tNCC migratory path.
How the ECM regulates NCC migration is an active area of research (Leonard and Taneyhill, 2020; Vega-Lopez et al., 2017). One hypothesis is that ECM components create a permissive zone that bias NCCs to move on specific migratory routes (Banerjee et al., 2013; Leonard and Taneyhill, 2020). Our hypothesis of a collagen ECM creating a permissive migratory route is supported by chick neural tube explant studies, which show that trunk, but not cranial, NCCs use collagen ECMs as preferred migratory substrates (Lallier et al., 1992). Thus, slow-twitch muscle cells may be depositing a complex collagen-rich ECM that is crucial for tNCC stream formation. However, other mechanisms may also be at play. For example, cranial NCCs use durotaxis to migrate along a stiffness gradient of pre-chordal mesoderm in Xenopus (Shellard and Mayor, 2021). Muscle segment organization is disrupted in different slow-twitch muscle mutants (Henry and Amacher, 2004; Snow and Henry, 2009) and mesoderm stiffness may be affected in the cdon;boc mutants that exhibit defects in slow-twitch muscle differentiation. Whether tNCCs use durotaxis has not been studied to our knowledge.
Cell-autonomous or non-autonomous function for cdon/boc?
Because of the nature of the transplantation and rescue experiments performed, we avoid interpreting our results to suggest a non-cell autonomous function for cdon and boc. Although our data are consistent with a hypothesis whereby cdon and boc affect tNCC migration non-cell autonomously through slow-twitch muscle differentiation, cdon and boc may also play cell autonomous roles in NCC migration. Indeed, NCC autonomous and non-autonomous function are not mutually exclusive hypotheses. Transplantation experiments using cdon knockdown embryos from our previous study suggest a cell-autonomous function in migration (Powell et al., 2015). It is further possible that cdon and boc function differently in cranial neural crest than in the tNCC, on which we have focused. Mouse Cdon and Boc mutants have specific craniofacial phenotypes (Allen et al., 2011). We also refer to data showing cdon and boc expression in both the neural plate border, migrating tNCCs and mesoderm to suggest multiple roles for these genes. Recent work in zebrafish showed that notch signaling among tNCC pre-migratory progenitors establishes a unique ‘leader cell’ identity that is required for stream formation (Alhashem et al., 2022; McLennan et al., 2015; Morrison et al., 2017; Richardson et al., 2016). Intriguingly, notch mutant embryos that lack leader cell identity have normal slow-twitch muscle differentiation and tNCC migratory phenotypes that are remarkably similar to what we observe in cdon;boc mutants (Alhashem et al., 2022). Whether cdon and boc also play a role in establishing a ‘leader cell’ identity, or function in ‘leader cell’ guidance is unknown. It is worth noting that our transplant rescue experiments cannot differentiate between a role for mesoderm or ‘leader cell’ identity, as leader cells will be present in the wild-type hosts.
cdon and boc as modifiers of hedgehog signaling
Our data add to a growing literature showing that cdon and boc act synergistically during zebrafish development to modify hedgehog signal reception and regulate the development of multiple traits (Echevarria-Andino and Allen, 2020; Song et al., 2015; Zhang et al., 2011). Interestingly, we found that cdon single mutants do not have noticeable phenotypes in zebrafish, and we can recover adult homozygous mutant zebrafish (not shown). This is contrast to mouse, where loss of CDON results in mild holoprosencephaly (Allen et al., 2011, 2007; Echevarria-Andino and Allen, 2020; Zhang et al., 2011). Although it is possible that our cdon mutations do not result in full loss of function, we find this possibility unlikely. Both cdon mutations lead to a predicted early stop before the transmembrane domain, but we cannot fully rule out the possibility that a late start rescues cdon protein, resulting in a hypomorphic but functional receptor, or creates a dominant negative. Future research could investigate these possibilities.
An alternative possibility is that cdon and boc interact with other hedgehog pathway members, resulting in species-specific phenotypic responses. In zebrafish, other genes may provide robustness against cdon mutations. The cyclopamine pharmacological experiments support this hypothesis. Intriguingly, cdon expression is often nested within boc expression, suggesting that cdon and boc act together to modify hedgehog morphogen gradients. Kearns et al. (2021) found that patched-2 (ptch2) expression in the dorsal neural tube is expanded in bocco25 mutant embryos, indicating that loss of boc leads to more dorsal hedgehog reception (ptch2 is a readout for hedgehog activity). These data were interpreted to suggest that boc sequesters hedgehog ligand, which drives hedgehog signal reception to ventral tissues. Expanding on this hypothesis, we note that cdon transcripts are restricted dorsally in the neural tube. We hypothesize that boc and cdon gradients may act as strong sequestering agents to limit hedgehog signal activation and inhibit ligand activity in tissues (Appel and Eisen, 1998; Cardozo et al., 2014).
Summary
We show that cdon and boc act synergistically in zebrafish to regulate slow-twitch muscle cell differentiation and affect tNCC migration. Our data add to a growing literature in zebrafish showing the crucial role of the slow-twitch muscle in regulating the migration of cells. Intriguingly, homologous adaxial/slow-twitch muscle tissues in other taxa are less understood (Grimaldi et al., 2004; Hammond et al., 2009), and to our knowledge there is no known tissue in amniotes that is homologous to adaxial cells. Despite this, cdon and boc are known to be crucial for myogenesis in amniotes (Jeong et al., 2017; Lu and Krauss, 2010), and genes expressed by slow-twitch muscle in zebrafish regulate tNCC migration in mouse (Banerjee et al., 2011). Furthermore, cdon and boc are expressed in a broad range of tissues. cdon, but not boc, transcripts are co-expressed in hedgehog ligand-producing cells, and cdon plays a role in presenting hedgehog ligand to responsive cells (Hall et al., 2021). Thus, it is likely that cdon and boc play different roles in different tissues, and that cellular context, expression level and distance from ligand source may be crucial for determining the biological function of these receptors.
MATERIALS AND METHODS
Fish stocks, husbandry and generation of mutants
Adult fish and embryos were maintained at the University of Colorado Anschutz Medical school according to standard zebrafish rearing protocols and guidelines for the rearing and maintenance of vertebrate animals. Two boc mutations were investigated. The bocco25 mutant was previously recovered from an ENU screen (Kearns et al., 2021) and results in an non-synonymous isoleucine-to-serine substitution in the third FNIII domain (Fig. 1C). The bocuml/ty54z mutant was acquired from the Zebrafish International Resource Center and results in a predicted early stop (Bergeron et al., 2011). Both mutations were previously characterized as loss of function (Bergeron et al., 2011; Kearns et al., 2021).
We used CRISPR/Cas9 to generate previously unreported mutations in cdon. Two sets of guide RNAs (sgRNA) were designed to target either the 2nd and 3rd exons (ggcagUUgaggagcagagUa, UggUaggagccUgUgagagc, UgcUUcagaaUcgcUgagag) or to target the 11th exon (UcgccaUUgagaUUgUgggg, cUccacgcgaaaUgcagUga) of cdon. DNA oligos were annealed to a common backbone scaffold, which was used to generate sgRNAs for injections (Williams et al., 2018). For each sgRNA set, our final CRISPR injection solution consisted of sgRNAs diluted to 20 ng/µl, 1 µg Cas9 protein (Invitrogen, TrueCut V2) and 5 µg rhodamine dextran in a total volume of 10 µl. Approximately 2-4 nl were injected into the cell of a one-cell stage embryo, and F0-injected embryos were reared to adulthood.
We recovered a single allele for each sgRNA set. These include the cdonco1019 allele, which is a 229 bp deletion that spans the second to third exon of cdon, removes the intronic sequence between these exons and is predicted to result in an early stop (Fig. 1C). The cdonco1023 allele is a 5 bp deletion resulting in a predicted early stop in the 11th exon that codes for the second FNIII domain. F0-injected embryos were outcrossed for a minimum of two generations to minimize effects of off-target cutting. Sequence information for cdon mutations is available in Fig. S5.
Zebrafish boc and cdon mutants were crossed into existing transgenic reporter lines, including the tg(-7.2kbsox10:mRFP)vu234 that labels NCCs with cytosolic mRFP (Kucenas et al., 2008) and the tg(8xgli-Xla.Cryaa:NLS-d1mCherry) line that has eight gli-binding sites driving a destabilized nuclear-localized mCherry (Mich et al., 2014). The tgPAC(prdm1a:eGFP) line was used to label adaxial mesoderm for reference (Elworthy et al., 2008).
cdon and boc molecular conservation and gene tree
Amino acid-coding sequences for select bilaterian taxa were downloaded from NCBI GenBank. Orthologous sequences were searched directly or identified through BLAST searches and manual (Table S1) curation. The final set of sequences was chosen to include taxonomic representatives of the major bilaterian taxa for which high-confidence sequences are available. Effort was made to identify two paralogs for each taxon. Amino acid sequences were aligned using MUSCLE (v3.8.31) with default parameters (Edgar, 2004). Amino acid conservation (Fig. 1A) was calculated by pairwise comparisons of cdon and boc homologues from 11 taxa (human, mouse, chicken, Anolis, frog, bichir, Gar, sheepshead minnow, medaka, catfish and zebrafish), and plotted in R by smoothing conservation values across 20 amino acid windows. Substitution models were selected using ProtTest3 by AIC (Darriba et al., 2011). A maximum likelihood gene tree was built using phyml (v3.3) and a JTT+I+G+F model with 1000 bootstraps (Guindon and Gascuel, 2003). We recovered similar results from filtered alignments that removed poorly aligned regions and regions with gaps.
Immunohistochemistry, in situ hybridization and stains
Adaxial mesoderm derivatives were stained by fluorescent immunohistochemistry against a slow-twitch muscle-specific myosin (anti-MYH1A, F59, DSHB) at a concentration of 1:20, as previously described (Elworthy et al., 2008; Nguyen-Chi et al., 2012). Col1a1a was labeled using the GeneTex antibody raised against the zebrafish Col1a1a protein (GTX133063; RRID AB_2886813) at a concentration of 1:100. Standard in situ hybridization of myod1 was performed as previously described (Hsu et al., 2022). Hybrid chain reaction (HCR) in situ hybridizations were performed as previously described (Choi et al., 2018; Lencer et al., 2021). Unlike traditional enzymatic methods for in situ hybridization, HCR allows single molecule measurement of gene expression at cellular resolution, which enables identifying cells and cell types that express transcripts at relatively low levels (Kearns et al., 2021). Custom HCR probes for zebrafish cdon, boc, prdm1a and myod1 were acquired from Molecular Instruments and used at a concentration of 2 pM/500 μl in hybridization buffer.
RNA-seq of tNCCs and adaxial cell mesoderm
We used bulk RNA-seq to characterize the transcriptome of tNCCs and adaxial cell mesoderm at 24 hpf. To do this, we took advantage of embryos that were doubly transgenic for the tg(sox10:mRFP) transgene that labels NCCs and for the tg(prdm1a:eGFP) transgene that labels adaxial mesoderm. Trunks from 80 embryos were dissected and cells were dissociated by incubating in Accumax (Stem Cell Technologies) at 37°C for 30 min with intermittent agitation. Cells were cleaned of debris by passing through a 40 µM mesh filter, and washed in Dulbecco's phosphate-buffered saline plus DNaseI (a final concentration of 7500 U/ml). NCCs and adaxial mesoderm cells were isolated by FACS using a MoFlo XDP100 at the CU Anschutz Flow Cytometry Core.
Total RNA was extracted from FAC-sorted samples of tNCCs and adaxial mesoderm using the RNAqueous-Micro Isolation Kit (ThermoFisher). Library preparation and Illumina sequencing (PE 150 bp) were performed by the CU Anschutz Genomics core on a NovaSeq6000. Reads were aligned to the zebrafish genome (GRCz11, ensemble annotations) using STAR aligner (Dobin et al., 2013) and gene counts were calculated using featurecounts (Liao et al., 2014) with default parameters. DEseq2 (Love et al., 2014) was used to estimate normalized expression levels using the normTransform function.
Image analysis
Image analysis was performed in ImageJ. Some images for presentation were generated using napari (Sofroniew et al., 2019). 3D surface renderings of Col1a1a were generated using Leica Las X software. Images are presented as either maximum projection images or as 3D volume renderings. Quantification of slow-twitch fiber number was performed by manually counting the number of fibers in each segment. Ventral tNCC migration was measured as the straight-line distance from the dorsal-most neural crest to the ventral-most medial tNCC at a given somite location. Intensity quantification of tg(8×GliBS::mCherry-NLS-ODC) nuclei was performed on sum projecting images by manually tracing slow-twitch muscle nuclei in ImageJ. Mean intensity is the raw intensity value/area of the region of interest. For Col1a1a intensity, measurements were taken on optically sectioned z-stacks using ImageJ. For these analyses, the z position of the approximate center of a segment was identified manually from lateral projection images. This approximate center was used to generate 15 µm (50 slices) optically sectioned z-stacks. Line intensity scans were taken in ImageJ from sum-projected images generated from these 15 µM optical sections centered at the approximate middle of the segment. Line scans were normalized by length such that the position of the surface ectoderm and notochord were at the relative positions of 0 and 1, respectively. For presentation, Col1a1a intensity was normalized to the surface ectoderm intensity. All quantification and images shown in figures were taken at the level of the yolk extension (somite numbers 8-16), unless otherwise noted.
Live imaging and tracking of migrating cells
For live-imaging and tracking, neural crest cells were labelled using the tg(sox10:mRFP) line. Embryos were mounted in 0.4% low melt agarose and neural crest cells were imaged over the yolk extension every 5 min from 22 hpf to 30 hpf on an Andor spinning disk confocal microscope at 28°C. Neural crest cells were manually tracked along the xy axis from maximum projection images using the ImageJ mTrackJ plug-in (Meijering et al., 2012). Drift was accounted for using the Correct_3D_Drift.py plugin, as implemented in ImageJ (Parslow et al., 2014). As we only observed a tNCC migratory defect in the medial migrating streams, we limited our tracking of cells to tNCCs migrating along this route. Quantitative analysis of track motility was performed in R. A small number of short tracks with fewer than 30 points (2.5 h) were excluded. As we also noted that NCCs often stopped migrating upon reaching a ventral position, we further limited tracks to a maximum of 6 h to capture cell migration. Limiting all tracks to the same time (e.g. number of points) produces similar results. Rose plots depict cell movements with a velocity greater than 0.3 µM/min, which was empirically determined as a cutoff for capturing cell movements while excluding noise from drift and changes in cell shape. Using different cutoffs produced similar results. Linear models were built in R by including track length as a fixed effect, and embryo identity and date of imaging as random effects, with the packages lme4 (Bates et al., 2015), lmerTest (Kuznetsova et al., 2017) and emmeans.
Transplant rescue experiments
Transplant rescues were performed by transplanting cells from gastrula stage embryos produced by in crossing double heterozygous cdonco1019;bocco25 parents into gastrula stage wild-type hosts. All donor embryos were transgenic for the tg(sox10:mRFP) transgene, whereas hosts were transgenic for the tg(sox10:eGFP) transgene (Carney et al., 2006), allowing us to differentiate transplanted NCCs (red) from host NCCs (green). Embryos were fixed at 24 hpf, genotyped and imaged for tNCC migration. For all transplants, donor embryos were derived by in-crossing double heterozygous cdon;boc parents where 1/16 will be double mutants. All donor embryos were transgenic for the tg(sox10:mRFP) transgene (magenta), and wild-type embryos carry the tg(sox10:eGFP) transgene (green).
Tol2 rescue experiments
Tol2 rescue experiments were performed following standard gateway cloning and tol2 transgenic methods (Kwan et al., 2007). Briefly, the primers 5′-GCGTAAAACTGTGGCGCGTAAAAC-3′ and 5′-CATTGAAGTCAATTCAGCTTCGTC-3′ were used to clone the Unc45b promoter from wild-type genomic DNA following Berger and Currie (2013). This produced a 2.066 kb fragment of the 5′ upstream region to the unc45b gene. This fragment was placed into a tol2kit p5Entry vector using TA cloning (Invitrogen). A smoa1-eGFP construct was a generous gift from Michael Taylor (University of Wisconsin, Madison, USA) and was subcloned into a tol2 middle entry vector. Expression constructs using the −2.066Unc45b enhancer to drive either smoa1-eGFP or eGFP, as a control, were used to rescue hedgehog signal in adaxial cell progenitors.
For rescues, double heterozygous cdonco1019;bocco25 parents were in-crossed and embryos were injected at the one-cell stage with tol2 constructs and tol2 mRNA at a total injection volume of ∼2 nl. Embryos were reared to 24 hpf and then stained for F59 by immunofluorescence (see above) to assess tNCC stream formation and slow-twitch muscle cell morphology. Wild-type and double mutant embryos were imaged over the yolk extension.
To account for mosaic rescue, we analyzed three body segments over the yolk extension for each embryo by qualitatively categorizing tNCC streams into rescue, partial rescue or no-rescue categories based on tNCC stream formation. Rescue was defined as a body segment with a defined single stream of multiple migratory tNCCs. Partial rescue was defined as streams of migratory cells that had gaps between cells (in wild-type conditions, migrating tNCCs maintain contact during migration). No rescue was defined as body segments where no migratory stream was apparent. These were instances of singly migrating cells or cells arrested at the body midline. Sick embryos were not used for analyses.
We observed high transgenesis efficiency in the GFP control, but much lower GFP signal in the smoa1-eGFP rescues. Indeed, very few cells expressed smoa1-eGFP in our analyses. We suspect that cells are downregulating the smoa1-eGFP construct (pre or post transcriptionally) in order to compensate for smoothened overexpression (C. Mosimann, personal communication). An example of this could be silencing of the transgene or trafficking of the protein to the lysosomal pathway.
Pharmacological manipulation
For pharmacological manipulation, embryos were manually dechorionated and treated with 20 µM cyclopamine in ethanol (5 µl/ml) from 50% epiboly to 24 hpf. Embryos that appeared developmentally delayed were excluded.
Imaging, statistical analysis, and biological replication
All imaging was performed on either a Leica Sp8 LSM or Andor spinning disk confocal microscope. Image analysis and presentation was performed in ImageJ or napari. All statistical analyses were performed in R. Experiments were repeated at least three times and biological variation was accounted for in models and through inclusion of biological replicates.
Supplementary Material
Acknowledgements
We thank members of the Artinger and Prekeris laboratories for comments on this manuscript and project. Special thanks to Caleb Doll, Bruce Appel and Christina Kearns for providing tools and advice. We thank Michael Taylor for his generous gift of the smoa1-eGFP construct. The comments of three anonymous reviewers greatly improved this manuscript. We thank the CU Anschutz Flow Cytometry Core and Genomics Core for their help with cell sorting and sequencing performed as part of this study.
Footnotes
Author contributions
Conceptualization: E.L., R.P., K.B.A.; Methodology: E.L., K.B.A.; Validation: E.L.; Formal analysis: E.L.; Investigation: E.L., A.R., E.B., K.B.A.; Resources: E.L., K.B.A.; Data curation: E.L.; Writing - original draft: E.L.; Writing - review & editing: E.L., R.P., K.B.A.; Visualization: E.L., A.R., E.B.; Supervision: R.P., K.B.A.; Project administration: E.L., R.P., K.B.A.; Funding acquisition: E.L., R.P., K.B.A.
Funding
This work was in part supported by the National Institutes of Health (GM122768 to R.P.) and a University of Colorado RNA Biosciences Initiative award to E.L. and K.A. E.L. was supported by National Institutes of Health Ruth L. Kirschstein National Research Service Awards (T32CA17468 and F32HD103406). Deposited in PMC for release after 12 months.
Data availability
Software used is publicly available and includes ImageJ, napari, R, and associated R and ImageJ packages. RNA-seq data have been deposited in GEO under accession number GSE213728.
Contributor Information
Ezra Lencer, Email: lencere@lafayette.edu.
Kristin B. Artinger, Email: kristin.artinger@cuanschutz.edu.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.201304.reviewer-comments.pdf
References
- Alhashem, Z., Feldner-Busztin, D., Revell, C., Alvarez-Garcillan Portillo, M., Camargo-Sosa, K., Richardson, J., Rocha, M., Gauert, A., Corbeaux, T., Milanetto, M.et al. (2022). Notch controls the cell cycle to define leader versus follower identities during collective cell migration. Elife 11, e73550. 10.7554/eLife.73550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, B. L., Tenzen, T. and McMahon, A. P. (2007). The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes Dev. 21, 1244-1257. 10.1101/gad.1543607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, B. L., Song, J. Y., Izzi, L., Althaus, I. W., Kang, J. S., Charron, F., Krauss, R. S. and McMahon, A. P. (2011). Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev. Cell 20, 775-787. 10.1016/j.devcel.2011.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel, B. and Eisen, J. S. (1998). Regulation of neuronal specification in the zebrafish spinal cord by Delta function. Development 125, 371-380. 10.1242/dev.125.3.371 [DOI] [PubMed] [Google Scholar]
- Banerjee, S., Gordon, L., Donn, T. M., Berti, C., Moens, C. B., Burden, S. J. and Granato, M. (2011). A novel role for MuSK and non-canonical Wnt signaling during segmental neural crest cell migration. Development 138, 3287-3296. 10.1242/dev.067306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, S., Isaacman-Beck, J., Schneider, V. A. and Granato, M. (2013). A novel role for Lh3 dependent ECM modifications during neural crest cell migration in zebrafish. PLoS One 8, e54609. 10.1371/journal.pone.0054609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi, M. J. F., Stickney, H. L. and Devoto, S. H. (2000). The zebrafish slow-muscle-omitted gene product is required fro Hedgehog signal transduction and the development of slow muscle identity. Development 127, 2189-2199. 10.1242/dev.127.10.2189 [DOI] [PubMed] [Google Scholar]
- Barriga, E. H., Franze, K., Charras, G. and Mayor, R. (2018). Tissue stiffening coordinates morphogenesis by triggering collective cell migration in vivo. Nature 554, 523-527. 10.1038/nature25742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barriga, E. H., Maxwell, P. H., Reyes, A. E. and Mayor, R. (2013). The hypoxia factor Hif-1alpha controls neural crest chemotaxis and epithelial to mesenchymal transition. J. Cell Biol. 201, 759-776. 10.1083/jcb.201212100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, D., Mächler, M., Bolker, B. and Walker, S. (2015). Fitting linear mixed-effects models Usinglme4. J. Stat. Softw. 67, 1-48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Berger, J. and Currie, P. D. (2013). 503unc, a small and muscle-specific zebrafish promoter. Genesis 51, 443-447. 10.1002/dvg.22385 [DOI] [PubMed] [Google Scholar]
- Bergeron, S. A., Tyurina, O. V., Miller, E., Bagas, A. and Karlstrom, R. O. (2011). Brother of cdo (umleitung) is cell-autonomously required for Hedgehog-mediated ventral CNS patterning in the zebrafish. Development 138, 75-85. 10.1242/dev.057950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilioni, A., Sanchez-Hernandez, D., Callejo, A., Gradilla, A. C., Ibanez, C., Mollica, E., Carmen Rodriguez-Navas, M., Simon, E. and Guerrero, I. (2013). Balancing Hedgehog, a retention and release equilibrium given by Dally, Ihog, Boi and shifted/DmWif. Dev. Biol. 376, 198-212. 10.1016/j.ydbio.2012.12.013 [DOI] [PubMed] [Google Scholar]
- Camp, D., Currie, K., Labbé, A., van Meyel, D. J. and Charron, F. (2010). Ihog and Boi are essential for Hedgehog signaling in Drosophila. Neural Dev. 5, 28. 10.1186/1749-8104-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo, M. J., Sanchez-Arrones, L., Sandonis, A., Sanchez-Camacho, C., Gestri, G., Wilson, S. W., Guerrero, I. and Bovolenta, P. (2014). Cdon acts as a Hedgehog decoy receptor during proximal-distal patterning of the optic vesicle. Nat. Commun. 5, 4272. 10.1038/ncomms5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine, C., Matthews, H. K., Kuriyama, S., Moreno, M., Dunn, G. A., Parsons, M., Stern, C. D. and Mayor, R. (2008). Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature 456, 957-961. 10.1038/nature07441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney, T. J., Dutton, K. A., Greenhill, E., Delfino-Machin, M., Dufourcq, P., Blader, P. and Kelsh, R. N. (2006). A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development 133, 4619-4630. 10.1242/dev.02668 [DOI] [PubMed] [Google Scholar]
- Choi, H. M. T., Schwarzkopf, M., Fornace, M. E., Acharya, A., Artavanis, G., Stegmaier, J., Cunha, A. and Pierce, N. A. (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753. 10.1242/dev.165753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay, M. R. and Halloran, M. C. (2011). Regulation of cell adhesions and motility during initiation of neural crest migration. Curr. Opin. Neurobiol. 21, 17-22. 10.1016/j.conb.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay, M. R. and Halloran, M. C. (2013). Rho activation is apically restricted by Arhgap1 in neural crest cells and drives epithelial-to-mesenchymal transition. Development 140, 3198-3209. 10.1242/dev.095448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay, M. R. and Halloran, M. C. (2014). Cadherin 6 promotes neural crest cell detachment via F-actin regulation and influences active Rho distribution during epithelial-to-mesenchymal transition. Development 141, 2506-2515. 10.1242/dev.105551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor, R. M., Allen, C. L., Devine, C. A., Claxton, C. and Key, B. (2005). BOC, brother of CDO, is a dorsoventral axon-guidance molecule in the embryonic vertebrate brain. J. Comp. Neurol. 485, 32-42. 10.1002/cne.20503 [DOI] [PubMed] [Google Scholar]
- Darriba, D., Taboada, G. L., Doallo, R. and Posada, D. (2011). ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164-1165. 10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy, A. and Soriano, P. (2007). Ephrin-B2 forward signaling regulates somite patterning and neural crest cell development. Dev. Biol. 304, 182-193. 10.1016/j.ydbio.2006.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal, P. and Boore, J. L. (2005). Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3, e314. 10.1371/journal.pbio.0030314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto, S. H., Melancon, E., Eisen, J. S. and Westerfield, M. (1996). Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 122, 3371-3380. 10.1242/dev.122.11.3371 [DOI] [PubMed] [Google Scholar]
- Diener, J. and Sommer, L. (2021). Reemergence of neural crest stem cell-like states in melanoma during disease progression and treatment. Stem. Cells Transl. Med. 10, 522-533. 10.1002/sctm.20-0351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevarria-Andino, M. L. and Allen, B. L. (2020). The hedgehog co-receptor BOC differentially regulates SHH signaling during craniofacial development. Development 147, dev189076. 10.1242/dev.189076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792-1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elworthy, S., Hargrave, M., Knight, R., Mebus, K. and Ingham, P. W. (2008). Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development 135, 2115-2126. 10.1242/dev.015719 [DOI] [PubMed] [Google Scholar]
- Ferent, J., Giguere, F., Jolicoeur, C., Morin, S., Michaud, J. F., Makihara, S., Yam, P. T., Cayouette, M. and Charron, F. (2019). Boc acts via numb as a Shh-dependent endocytic platform for Ptch1 internalization and Shh-mediated axon guidance. Neuron 102, 1157-1171.e1155. 10.1016/j.neuron.2019.04.003 [DOI] [PubMed] [Google Scholar]
- Gammill, L. S., Gonzalez, C., Gu, C. and Bronner-Fraser, M. (2006). Guidance of trunk neural crest migration requires neuropilin 2/semaphorin 3F signaling. Development 133, 99-106. 10.1242/dev.02187 [DOI] [PubMed] [Google Scholar]
- Gammill, L. S., Gonzalez, C. and Bronner-Fraser, M. (2007). Neuropilin 2/semaphorin 3F signaling is essential for cranial neural crest migration and trigeminal ganglion condensation. J. Neurobiol. 67, 47-56. 10.1002/neu.20326 [DOI] [PubMed] [Google Scholar]
- Grimaldi, A., Tettamanti, G., Martin, B. L., Gaffield, W., Pownall, M. E. and Hughes, S. M. (2004). Hedgehog regulation of superficial slow muscle fibres in Xenopus and the evolution of tetrapod trunk myogenesis. Development 131, 3249-3262. 10.1242/dev.01194 [DOI] [PubMed] [Google Scholar]
- Guillon, E., Bretaud, S. and Ruggiero, F. (2016). Slow Muscle Precursors Lay Down a Collagen XV Matrix Fingerprint to Guide Motor Axon Navigation. J. Neurosci. 36, 2663-2676. 10.1523/JNEUROSCI.2847-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon, S. and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696-704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hall, E. T., Dillard, M. E., Stewart, D. P., Zhang, Y., Wagner, B., Levine, R. M., Pruett-Miller, S. M., Sykes, A., Temirov, J., Cheney, R. E.et al. (2021). Cytoneme delivery of Sonic Hedgehog from ligand-producing cells requires Myosin 10 and a Dispatched-BOC/CDON co-receptor complex. Elife 10, e61432. 10.7554/eLife.61432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, K. L., Baxendale, S., McCauley, D. W., Ingham, P. W. and Whitfield, T. T. (2009). Expression of patched, prdm1 and engrailed in the lamprey somite reveals conserved responses to Hedgehog signaling. Evol. Dev. 11, 27-40. 10.1111/j.1525-142X.2008.00300.x [DOI] [PubMed] [Google Scholar]
- Henry, C. A. and Amacher, S. L. (2004). Zebrafish slow muscle cell migration induces a wave of fast muscle morphogenesis. Dev. Cell 7, 917-923. 10.1016/j.devcel.2004.09.017 [DOI] [PubMed] [Google Scholar]
- Hong, M., Christ, A., Christa, A., Willnow, T. and Krauss, R. S. (2020). Cdon mutation and fetal alcohol converge on nodal signaling in a gene-environment interaction model of holoprosencephaly. eLife 9, e60351. 10.7554/eLife.60351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo, Y. and Eisen, J. S. (2005). Slow muscle regulates the pattern of trunk neural crest migration in zebrafish. Development 132, 4461-4470. 10.1242/dev.02026 [DOI] [PubMed] [Google Scholar]
- Hsu, J. Y., Danis, E. P., Nance, S., O'Brien, J. H., Gustafson, A. L., Wessells, V. M., Goodspeed, A. E., Talbot, J. C., Amacher, S. L., Jedlicka, P.et al. (2022). SIX1 reprograms myogenic transcription factors to maintain the rhabdomyosarcoma undifferentiated state. Cell Rep. 38, 110323. 10.1016/j.celrep.2022.110323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi, L., Levesque, M., Morin, S., Laniel, D., Wilkes, B. C., Mille, F., Krauss, R. S., McMahon, A. P., Allen, B. L. and Charron, F. (2011). Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev. Cell 20, 788-801. 10.1016/j.devcel.2011.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, M. H., Kim, H. J., Pyun, J. H., Choi, K. S., Lee, D. I., Solhjoo, S., O'Rourke, B., Tomaselli, G. F., Jeong, D. S., Cho, H.et al. (2017). Cdon deficiency causes cardiac remodeling through hyperactivation of WNT/beta-catenin signaling. Proc. Natl. Acad. Sci. USA 114, E1345-E1354. 10.1073/pnas.1615105114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, B., Spitsbergen, J., Eden, C. J., Taylor, M. R. and Chen, W. (2009). Co-activation of hedgehog and AKT pathways promote tumorigenesis in zebrafish. Mol. Cancer 8, 40. 10.1186/1476-4598-8-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, B., Chen, W., Spitsbergen, J. M., Lu, J., Vogel, P., Peters, J. L., Wang, Y. D., Orr, B. A., Wu, J., Henson, H. E.et al. (2014). Activation of Sonic hedgehog signaling in neural progenitor cells promotes glioma development in the zebrafish optic pathway. Oncogenesis 3, e96. 10.1038/oncsis.2014.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.-S., Mulieri, P. J., Hu, Y., Taliana, L. and Krauss, R. S. (2002). BOC, an Ig superfamily member, associates with CDO to positively regulate myogenic differentiation. EMBO J. 21, 114-124. 10.1093/emboj/21.1.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J. S., Bae, G. U., Yi, M. J., Yang, Y. J., Oh, J. E., Takaesu, G., Zhou, Y. T., Low, B. C. and Krauss, R. S. (2008). A Cdo-Bnip-2-Cdc42 signaling pathway regulates p38alpha/beta MAPK activity and myogenic differentiation. J. Cell Biol. 182, 497-507. 10.1083/jcb.200801119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasemeier-Kulesa, J. C., McLennan, R., Romine, M. H., Kulesa, P. M. and Lefcort, F. (2010). CXCR4 controls ventral migration of sympathetic precursor cells. J. Neurosci. 30, 13078-13088. 10.1523/JNEUROSCI.0892-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, C. K., Mosimann, C., Fan, Z. P., Yang, S., Thomas, A. J., Ablain, J., Tan, J. L., Fogley, R. D., van Rooijen, E., Hagedorn, E. J.et al. (2016). A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197. 10.1126/science.aad2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, C. A., Walker, M., Ravanelli, A. M., Scott, K., Arzbecker, M. R. and Appel, B. (2021). Zebrafish spinal cord oligodendrocyte formation requires boc function. Genetics 218, iyab082. 10.1093/genetics/iyab082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull, C. E. (2010). Neural crest cells and motor axons in avians: Common and distinct migratory molecules. Cell Adh. Migr. 4, 631-634. 10.4161/cam.4.4.13594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull, C. E., Lansford, R., Gale, N. W., Collazo, A., Marcelle, C., Yancopoulos, G. D., Fraser, S. E. and Bronner-Fraser, M. (1997). Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Curr. Biol. 7, 571-580. 10.1016/S0960-9822(06)00256-9 [DOI] [PubMed] [Google Scholar]
- Kucenas, S., Snell, H. and Appel, B. (2008). nkx2.2a promotes specification and differentiation of a myelinating subset of oligodendrocyte lineage cells in zebrafish. Neuron Glia Biol. 4, 71-81. 10.1017/S1740925X09990123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa, P. M. and Gammill, L. S. (2010). Neural crest migration: patterns, phases and signals. Dev. Biol. 344, 566-568. 10.1016/j.ydbio.2010.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova, A., Brockhoff, P. B. and Christensen, R. H. B. (2017). lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 82, 1-26. 10.18637/jss.v082.i13 [DOI] [Google Scholar]
- Kwan, K. M., Fujimoto, E., Grabher, C., Mangum, B. D., Hardy, M. E., Campbell, D. S., Parant, J. M., Yost, H. J., Kanki, J. P. and Chien, C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Lallier, T., LeBlanc, G., Artinger, K. B. and Bronner-Fraser, M. (1992). Cranial and trunk neural crest cells use different mechanisms for attachment to extracellular matrices. Development 116, 531-541. 10.1242/dev.116.3.531 [DOI] [PubMed] [Google Scholar]
- Lencer, E., Prekeris, R. and Artinger, K. B. (2021). Single-cell RNA analysis identifies pre-migratory neural crest cells expressing markers of differentiated derivatives. Elife 10, e66078. 10.7554/eLife.66078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, C. E. and Taneyhill, L. A. (2020). The road best traveled: Neural crest migration upon the extracellular matrix. Semin. Cell Dev. Biol. 100, 177-185. 10.1016/j.semcdb.2019.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Coghlan, A., Ruan, J., Coin, L. J., Heriche, J. K., Osmotherly, L., Li, R., Liu, T., Zhang, Z., Bolund, L.et al. (2006). TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34, D572-D580. 10.1093/nar/gkj118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth, G. K. and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Love, M. I., Huber, W. and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M. and Krauss, R. S. (2010). N-cadherin ligation, but not Sonic hedgehog binding, initiates Cdo-dependent p38alpha/beta MAPK signaling in skeletal myoblasts. Proc. Natl. Acad. Sci. USA 107, 4212-4217. 10.1073/pnas.0908883107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, R. and Etienne-Manneville, S. (2016). The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 17, 97-109. 10.1038/nrm.2015.14 [DOI] [PubMed] [Google Scholar]
- McLennan, R., Schumacher, L. J., Morrison, J. A., Teddy, J. M., Ridenour, D. A., Box, A. C., Semerad, C. L., Li, H., McDowell, W., Kay, D.et al. (2015). Neural crest migration is driven by a few trailblazer cells with a unique molecular signature narrowly confined to the invasive front. Development 142, 2014-2025. 10.1242/dev.117507 [DOI] [PubMed] [Google Scholar]
- Meijering, E., D'zyubachyk, O. and Smal, I. (2012). Methods for cell and particle tracking. Methods Enzymol. 504, 183-200. 10.1016/B978-0-12-391857-4.00009-4 [DOI] [PubMed] [Google Scholar]
- Mich, J. K., Payumo, A. Y., Rack, P. G. and Chen, J. K. (2014). In vivo imaging of Hedgehog pathway activation with a nuclear fluorescent reporter. PLoS One 9, e103661. 10.1371/journal.pone.0103661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, J. A., McLennan, R., Wolfe, L. A., Gogol, M. M., Meier, S., McKinney, M. C., Teddy, J. M., Holmes, L., Semerad, C. L., Box, A. C.et al. (2017). Single-cell transcriptome analysis of avian neural crest migration reveals signatures of invasion and molecular transitions. Elife 6, e28415. 10.7554/eLife.28415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulieri, P. J., Okada, A., Sassoon, D. A., Mcconnell, S. K. and Krauss, A. R. S. (2000). Developmental expression pattern of the cdo gene. Dev. Dyn. 219, 40-49. [DOI] [PubMed] [Google Scholar]
- Mulieri, P. J., Kang, J. S., Sassoon, D. A. and Krauss, R. S. (2002). Expression of the boc gene during murine embryogenesis. Dev. Dyn. 223, 379-388. 10.1002/dvdy.10063 [DOI] [PubMed] [Google Scholar]
- Nguyen-Chi, M. E., Bryson-Richardson, R., Sonntag, C., Hall, T. E., Gibson, A., Sztal, T., Chua, W., Schilling, T. F. and Currie, P. D. (2012). Morphogenesis and cell fate determination within the adaxial cell equivalence group of the zebrafish myotome. PLoS Genet. 8, e1003014. 10.1371/journal.pgen.1003014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, A., Charron, F., Morin, S., Shin, D. S., Wong, K., Fabre, P. J., Tessier-Lavigne, M. and McConnell, S. K. (2006). Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature 444, 369-373. 10.1038/nature05246 [DOI] [PubMed] [Google Scholar]
- Okumura, F., Oki, N., Fujiki, Y., Ikuta, R., Osaki, K., Hamada, S., Nakatsukasa, K., Hisamoto, N., Hara, T. and Kamura, T. (2021). ZSWIM8 is a myogenic protein that partly prevents C2C12 differentiation. Sci. Rep. 11, 20880. 10.1038/s41598-021-00306-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky Killian, E. C., Birkholz, D. A. and Artinger, K. B. (2009). A role for chemokine signaling in neural crest cell migration and craniofacial development. Dev. Biol. 333, 161-172. 10.1016/j.ydbio.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parslow, A., Cardona, A. and Bryson-Richardson, R. (2014). Sample drift correction following 4D confocal time-lapse imaging. J. Vis. Exp. 12, 51086. 10.3791/51086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perris, R. and Perissinott, D. (2000). Role of the extracellular matrix during neural crest cell migration. Mech. Dev. 95, 3-21. 10.1016/S0925-4773(00)00365-8 [DOI] [PubMed] [Google Scholar]
- Powder, K. E., Cousin, H., McLinden, G. P. and Craig Albertson, R. (2014). A nonsynonymous mutation in the transcriptional regulator lbh is associated with cichlid craniofacial adaptation and neural crest cell development. Mol. Biol. Evol. 31, 3113-3124. 10.1093/molbev/msu267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, D. R., Williams, J. S., Hernandez-Lagunas, L., Salcedo, E., O'Brien, J. H. and Artinger, K. B. (2015). Cdon promotes neural crest migration by regulating N-cadherin localization. Dev. Biol. 407, 289-299. 10.1016/j.ydbio.2015.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, J., Gauert, A., Briones Montecinos, L., Fanlo, L., Alhashem, Z. M., Assar, R., Marti, E., Kabla, A., Hartel, S. and Linker, C. (2016). Leader cells define directionality of trunk, but not cranial, neural crest cell migration. Cell Rep. 15, 2076-2088. 10.1016/j.celrep.2016.04.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., Wolff, C. and Ingham, P. W. (2001). The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes Dev. 15, 1563-1576. 10.1101/gad.195801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, J., Li, H., Chen, Z., Coghlan, A., Coin, L. J., Guo, Y., Heriche, J. K., Hu, Y., Kristiansen, K., Li, R.et al. (2008). TreeFam: 2008 Update. Nucleic Acids Res. 36, D735-D740. 10.1093/nar/gkm1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Arrones, L., Cardozo, M., Nieto-Lopez, F. and Bovolenta, P. (2012). Cdon and Boc: Two transmembrane proteins implicated in cell-cell communication. Int. J. Biochem. Cell Biol. 44, 698-702. 10.1016/j.biocel.2012.01.019 [DOI] [PubMed] [Google Scholar]
- Santiago, A. and Erickson, C. A. (2002). Ephrin-B ligands play a dual role in the control of neural crest cell migration. Development 129, 3621-3632. 10.1242/dev.129.15.3621 [DOI] [PubMed] [Google Scholar]
- Scarpa, E., Szabo, A., Bibonne, A., Theveneau, E., Parsons, M. and Mayor, R. (2015). Cadherin switch during EMT in neural crest cells leads to contact inhibition of locomotion via repolarization of forces. Dev. Cell 34, 421-434. 10.1016/j.devcel.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, V. A. and Granato, M. (2006). The myotomal diwanka (lh3) glycosyltransferase and type XVIII collagen are critical for motor growth cone migration. Neuron 50, 683-695. 10.1016/j.neuron.2006.04.024 [DOI] [PubMed] [Google Scholar]
- Schweitzer, J., Becker, T., Lefebvre, J., Granato, M., Schachner, M. and Becker, C. G. (2005). Tenascin-C is involved in motor axon outgrowth in the trunk of developing zebrafish. Dev. Dyn. 234, 550-566. 10.1002/dvdy.20525 [DOI] [PubMed] [Google Scholar]
- Shellard, A. and Mayor, R. (2021). Collective durotaxis along a self-generated stiffness gradient in vivo. Nature 600, 690-694. 10.1038/s41586-021-04210-x [DOI] [PubMed] [Google Scholar]
- Shellard, A., Szabo, A., Trepat, X. and Mayor, R. (2018). Supracellular contraction at the rear of neural crest cell groups drives collective chemotaxis. Science 362, 339-343. 10.1126/science.aau3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow, C. J. and Henry, C. A. (2009). Dynamic formation of microenvironments at the myotendinous junction correlates with muscle fiber morphogenesis in zebrafish. Gene Expr. Patterns 9, 37-42. 10.1016/j.gep.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew, N., Lambert, T., Evans, K., Nunez-Iglesias, J., Bokota, G., Peña-Castellanos, G., Winston, P., Yamauchi, K., Bussonnier, M., Pop, D. D.et al. (2019). napari: a multi-dimensional image viewer for python. 10.5281/zenodo.6598542 [DOI]
- Song, J. Y., Holtz, A. M., Pinskey, J. M. and Allen, B. L. (2015). Distinct structural requirements for CDON and BOC in the promotion of Hedgehog signaling. Dev. Biol. 402, 239-252. 10.1016/j.ydbio.2015.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, A., Melchionda, M., Nastasi, G., Woods, M. L., Campo, S., Perris, R. and Mayor, R. (2016). In vivo confinement promotes collective migration of neural crest cells. J. Cell Biol. 213, 543-555. 10.1083/jcb.201602083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen, T., Allen, B. L., Cole, F., Kang, J. S., Krauss, R. S. and McMahon, A. P. (2006). The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev. Cell 10, 647-656. 10.1016/j.devcel.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Theveneau, E. and Mayor, R. (2012). Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev. Biol. 366, 34-54. 10.1016/j.ydbio.2011.12.041 [DOI] [PubMed] [Google Scholar]
- Tobin, J. L., Di Franco, M., Eichers, E., May-Simera, H., Garcia, M., Yan, J., Quinlan, R., Justice, M. J., Hennekam, R. C., Briscoe, J.et al. (2008). Inhibition of neural crest migration underlies craniofacial dysmorphology and Hirschsprung's disease in Bardet-Biedl syndrome. Proc. Natl. Acad. Sci. USA 105, 6714-6719. 10.1073/pnas.0707057105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Lopez, G. A., Cerrizuela, S. and Aybar, M. J. (2017). Trunk neural crest cells: formation, migration and beyond. Int. J. Dev. Biol. 61, 5-15. 10.1387/ijdb.160408gv [DOI] [PubMed] [Google Scholar]
- Vega-Lopez, G. A., Cerrizuela, S., Tribulo, C. and Aybar, M. J. (2018). Neurocristopathies: New insights 150 years after the neural crest discovery. Dev. Biol. 444 Suppl. 1, S110-S143. 10.1016/j.ydbio.2018.05.013 [DOI] [PubMed] [Google Scholar]
- Williams, J. S., Hsu, J. Y., Rossi, C. C. and Artinger, K. B. (2018). Requirement of zebrafish pcdh10a and pcdh10b in melanocyte precursor migration. Dev. Biol. 444 Suppl. 1, S274-S286. 10.1016/j.ydbio.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, C., Roy, S. and Ingham, P. W. (2003). Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the Zebrafish embryo. Curr. Biol. 13, 1169-1181. 10.1016/S0960-9822(03)00461-5 [DOI] [PubMed] [Google Scholar]
- Yao, S., Lum, L. and Beachy, P. (2006). The ihog cell-surface proteins bind Hedgehog and mediate pathway activation. Cell 125, 343-357. 10.1016/j.cell.2006.02.040 [DOI] [PubMed] [Google Scholar]
- Yin, J., Lee, R., Ono, Y., Ingham, P. W. and Saunders, T. E. (2018). Spatiotemporal coordination of FGF and Shh signaling underlies the specification of myoblasts in the zebrafish embryo. Dev. Cell 46, 735-750.e734. 10.1016/j.devcel.2018.08.024 [DOI] [PubMed] [Google Scholar]
- Yu, H. H. and Moens, C. B. (2005). Semaphorin signaling guides cranial neural crest cell migration in zebrafish. Dev. Biol. 280, 373-385. 10.1016/j.ydbio.2005.01.029 [DOI] [PubMed] [Google Scholar]
- Zeller, J. R. and Granato, M. (1999). The zebrafish diwanka gene controls an early step of motor growth cone migration. Development 126, 3461-3472. 10.1242/dev.126.15.3461 [DOI] [PubMed] [Google Scholar]
- Zhang, J., Lefebvre, J. L., Zhao, S. and Granato, M. (2004). Zebrafish unplugged reveals a role for muscle-specific kinase homologs in axonal pathway choice. Nat. Neurosci. 7, 1303-1309. 10.1038/nn1350 [DOI] [PubMed] [Google Scholar]
- Zhang, W., Kang, J. S., Cole, F., Yi, M. J. and Krauss, R. S. (2006). Cdo functions at multiple points in the Sonic Hedgehog pathway, and Cdo-deficient mice accurately model human holoprosencephaly. Dev. Cell 10, 657-665. 10.1016/j.devcel.2006.04.005 [DOI] [PubMed] [Google Scholar]
- Zhang, W., Hong, M., Bae, G. U., Kang, J. S. and Krauss, R. S. (2011). Boc modifies the holoprosencephaly spectrum of Cdo mutant mice. Dis. Model. Mech. 4, 368-380. 10.1242/dmm.005744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.