AN OVERVIEW OF IRON METABOLISM
Iron is a prerequisite for in vitro growth of mycobacteria.
Iron is an obligate cofactor for at least 40 different enzymes encoded in the Mycobacterium tuberculosis genome (8). It is required for the cytochromes involved in electron transport and other hemoproteins involved in oxygen metabolism, such as catalase-peroxidase KatG. Most of the intracellular iron in mycobacteria, however, is in the form of nonheme iron (48). One important form of nonheme iron is the iron-sulfur clusters that are cofactors of proteins involved in amino acid and pyrimidine biogenesis, as well as in enzymes involved in the tricarboxylic acid cycle and electron transport. Iron is also required for DNA synthesis in ribonucleotide reductase, superoxide dismutase, and 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (Rv2178c; DAHP synthase [Every gene in the genome of M. tuberculosis H37Rv has been assigned a gene identifier beginning with Rv which can be used to retrieve information on line; see reference 30a]). It has been estimated that 7 to 64 μg of Fe/g of mycobacterial cell mass is required to support growth (39, 48). Iron limitation in vitro to levels below these can result in growth restriction in many species of mycobacteria, including pathogens such as M. tuberculosis. Since iron is the fourth most abundant element, difficulty in its acquisition may seem counterintuitive. However, the extremely low solubility of ferric ion in aqueous solution makes obtaining it difficult and so microorganisms have evolved many strategies for acquiring sufficient soluble iron for aerobic growth.
Iron sequestration represents a formidable challenge to in vivo growth of pathogenic mycobacteria.
An important component of the mammalian host defense against bacterial pathogens involves restricting access of such organisms to iron (35). M. tuberculosis is a highly specialized pathogen of humans and must contend with iron sequestration in order to survive in the human lung. Pulmonary tuberculosis patients are often anemic, suggesting sequestration of available iron by the host (5). The serum of many mammals, including humans, is tuberculostatic because of its ability to sequester iron from the bacilli (32, 34). The addition of iron to such sera relieves the bacteriostatic effect completely. In a tragic attempt to rectify what was perceived as a debilitating iron deficiency in infected patients in Somalia, iron supplementation was found to actually promote the development of active tuberculosis (54). Other clinical studies in Africa have also established a strong correlation between dietary iron overload and an enhanced risk of death from tuberculosis (20, 52). Purified transferrin inhibits the growth of M. tuberculosis in vitro (33), and haptoglobin may play a role in restricting access to heme iron (29, 57). Thus, iron availability or sequestration and the bacterium’s efforts to circumvent this restriction are critical determinants of the outcome of infection with M. tuberculosis.
Infections with mycobacteria of the M. avium complex (MAC) occur predominantly in human immunodeficiency virus-infected patients and are a major source of mortality for such patients. Iron storage in macrophages is known to be increased in patients with AIDS (although overall iron levels in serum are lower than those of patients without AIDS), and this observation has been proposed to underlie the predisposition of such patients to MAC infections (11, 12, 22). Animal model experiments support this hypothesis in that mice fed iron-rich diets more rapidly develop disease from MAC (11). In cultured human macrophages, the rate of MAC replication has been shown to be directly correlated with the concentration and iron loading of transferrin (12).
Mycobacterial iron acquisition is mediated by siderophores.
The problem of iron acquisition has been solved by many microorganisms, including mycobacteria, by producing small, soluble iron chelators known as siderophores. These substances are generally secreted by the organism to compete with environmental iron binding molecules for the small amount of available iron. Siderophore synthesis is typically controlled by extracellular iron abundance, and these molecules display a high affinity for ferric iron. Four types of structurally distinct molecules are thought to be involved in iron acquisition in mycobacteria, salicylic acid and citric acid being the simplest, followed by two classes of more complex siderophores. The first, the mycobactins (MBs), are distinguished primarily by the presence of a phenyloxazolidine ring, while the second class, the exochelins, are peptidic siderophores whose iron-chelating ability is associated with ornithine-derived hydroxamates. MBs and exochelins are complex molecules with very high affinities for iron and are integral to the bacterial strategy for obtaining iron from the environment.
Early work focused on the dramatic accumulation of salicylic acid secreted into the growth medium coincident with iron starvation of either slow-growing or fast-growing mycobacterial species (60). Salicylate was postulated to be an important extracellular siderophore, but the stability of the salicylate-iron complex appears to be too low to support this hypothesis. However, the amount of salicylate secreted into growth media upon iron starvation reaches as much as 2.28 mg/100 ml of medium and this concentration suggests a secondary physiological role, perhaps in facilitating iron solubilization (60). Citric acid has also been proposed to be involved in iron acquisition in M. smegmatis on the basis of cell association of an iron-citrate complex (51). However, the very high concentrations of citric acid required, the lack of inducibility of citrate synthesis upon iron starvation, and the lack of an energy-dependent specific uptake system for such complexes all argue against such a role.
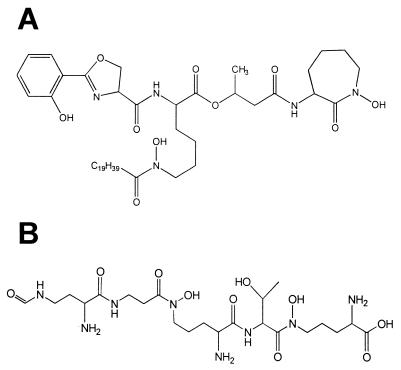
The MBs and exochelins (Fig. 1A and B) are produced specifically in response to iron starvation and are known to have a high affinity for iron(III). The increase in MB production in iron-limiting medium has been recognized since the very first isolation work (15). Iron sufficiency has been reported to decrease MB content by as much as 10,000 times (45, 59), although a much smaller decrease is generally reported (58). Mycobacterial species fall into four groups based upon the expression of these molecules. M. tuberculosis produces only the MB type of siderophore, M. vaccae produces only the exochelin type, M. smegmatis produces both types, and some strains of M. paratuberculosis produce none (50, 60). The real distribution of organisms among these groups is difficult to assess precisely because of substantial confusion over the use of the term exochelin (when what is meant is a water-soluble MB or a mixture of MBs and exochelins in species which produce both). Many authors equate exochelin with water-soluble MB in spite of the fact that exochelin was first proposed to represent the non-salicylate-containing peptidic siderophores of M. smegmatis (Fig. 1B) (70, 71). Use of the term exochelin should be limited to those molecules structurally related to the peptidohydroxamate siderophores typified by the structure of exochelin MS (Fig. 1B). Molecules related to the salicylate-containing siderophores which contain chemical functionality rendering them more water soluble should be more properly referred to as MBs (or water-soluble MBs, also referred to as exomycobactins [84]). Some strains of mycobacteria, in particular, of M. paratuberculosis, that do not produce any siderophores are dependent for growth upon the inclusion of such molecules in the medium (4, 24, 79). Species, such as M. bovis, which produce only MBs are not able to utilize water-soluble peptidic exochelins for uptake of iron (77). In vitro growth of an MB-deficient mutant of M. tuberculosis H37Rv was significantly impaired under low-iron conditions compared to the wild-type strain, supporting a critical role for this molecule in iron acquisition and transport (10).
FIG. 1.
Siderophores of mycobacterial species. (A) Structure of the major MB of M. tuberculosis (MB T). (B) Structure of the major exochelin of M. smegmatis (exochelin MS). Peptidic siderophores such as exochelin MS have only been identified from fast-growing saprophytic strains, while MBs such as that shown in panel A have been found widely distributed among mycobacterial species.
The interplay of cell-associated and secreted siderophores in iron acquisition.
In many species of mycobacteria, it has been noted that two forms of siderophore are often present, one primarily cell associated and one secreted or medium associated. This has given rise to a hypothesis that there are two distinct roles for such molecules. The extracellular form competes directly for environmental iron, while the cell-associated form acts as an ionophore, facilitating transport into the cell cytoplasm (43, 64). The water-soluble form of MB from M. bovis BCG (confusingly labeled exochelin MB-2) is capable of supporting ferritin-dependent growth of M. bovis BCG even when the iron source is separated from the growing cells by a dialysis membrane (43). The restriction of growth which accompanies culture of the tubercle bacillus in serum can be reversed by addition of MB and transferrin or ferritin but not by that of either molecule alone (19). More recently, the water-soluble MBs of M. tuberculosis (again confusingly mislabeled exochelins) were shown to acquire iron from 40% iron-saturated transferrin and to transfer this iron to the cell-associated MBs (17). Transferrin receptors, and presumably therefore transferrin, are present in the phagosomal membrane surrounding living M. tuberculosis within human monocytes (7). These experiments support the hypothesis that both soluble and cell-associated forms of siderophore are required for iron acquisition from the mammalian host in vivo for mycobacterial pathogenesis. Finally, and definitively, the availability of a knockout in the biosynthetic pathway which produces both forms of MB in M. tuberculosis has allowed us to demonstrate that a mutant incapable of producing both types of MBs has lost the ability to grow within macrophages (10). The definitive experiments regarding the hypothetical specific role for cell-associated compared to secreted siderophores must await the identification and specific modification of the acyl transferases which attach the R5 chain. These bifurcate the biosynthetic pathway and determine the ultimate ratio of MB to water-soluble MB produced by M. tuberculosis.
Storage of iron in mycobacterial species.
Mycobacterial siderophores are thought to be taken up and transported across the cell membrane by specific receptors and transporters, by analogy to other siderophore uptake mechanisms. Siderophores are transient reservoirs of iron which bind to ferric iron during transport into the cell cytoplasm. Recent experimental evidence shows that very little intracellular iron is contained within these molecules on a long-term basis (31). In situ Mössbauer spectroscopy and electron paramagnetic resonance spectroscopy have been used to confirm these results, as signals associated with siderophore-bound iron are only observed upon short-term labeling of iron pools with 57Fe (46, 47). Long-term labeling experiments have revealed that under iron-sufficient conditions, iron is accumulated in a form with spectral parameters consistent with bacterioferritin (46). Bacterioferritin proteins have been purified from M. leprae (55), and a bacterioferritin gene has been cloned from M. avium (30). On the chromosome of M. tuberculosis, there are apparently two bacterioferritin homologs, bfrA (Rv1876) and bfrB (Rv3841) (2).
STRUCTURE OF MYCOBACTERIAL SIDEROPHORES
An invariant “core” MB structure.
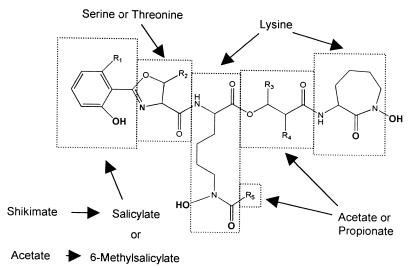
The structure of MB P, isolated from M. phlei, was initially established through chemical degradation by Snow (74). Since that time, a large number of MBs have been isolated but their core structure displays remarkably little variation (Fig. 2) (73). They all have a 2-hydroxyphenyloxazoline moiety linked via an amide bond to an acylated ɛ-N-hydroxylysine residue. This lysine is, in turn, esterified at the α-carboxyl with a β-hydroxy acid that forms an amide link with a second ɛ-N-hydroxylysine which has cyclized to give a seven-membered lactam. The absolute configurations of the stereogenic centers derived from an amino acid (where known) all appear to arise from the naturally occurring L series. This basic structure serves to chelate ferric iron very effectively via the two hydroxamic acids of the ɛ-N-hydroxylysines, the N of the oxazoline ring, and the phenolate oxygen atom (25). Although a formation constant of 1030 M−1 is widely quoted, the only direct measurements have monitored the dissociation of 59Fe-labeled ferri-MB S under strongly acidic conditions. From this data, one can extrapolate a formation constant of approximately 4 × 1026 M−1 at pH 7 (41). Coordination to ferrous iron by MB is much less effective, allowing release of iron from a ferri-MB complex by reduction. Computational work suggests that the reason for ferric ion selectivity lies in the confluence of the size of the ferric cation and the cavity created by a sterically favorable organization of the ligand (40).
FIG. 2.
General structure of MBs. Iron-chelating groups are in boldface. Letters and R groups correspond to the structures listed in Fig. 3.
Within this core of the MB, a methyl group may or may not be present at the position 6 of the phenolic ring (R1) and/or at the 5′ position of the oxazoline (R2) (Fig. 3). The presence of both types of oxazoline in M. tuberculosis has been reported (18), and MBs containing both aromatic moieties are found in M. fortuitum (75). One final core variation seen in some of the MBs from Nocardia sp. (the nocobactins) is an aromatic oxazole in place of the oxazoline ring (53, 65).
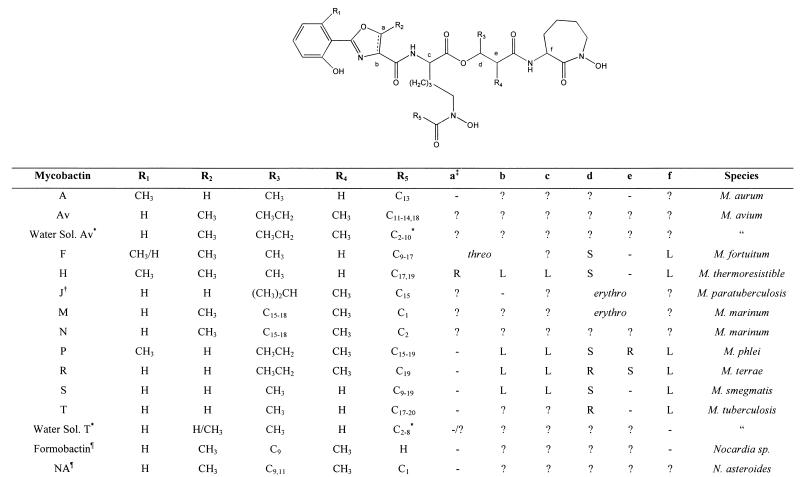
FIG. 3.
General structures of known MBs. Symbols: ∗, the terminal carbon in this chain of this water-soluble MB is either a carboxylic acid or its methyl ester; †, the structure as originally reported (49); §, MB that contains an oxazole moiety instead of the usual oxazoline and is found in Nocardia sp; ‡, letters refer to the stereocenters shown in the structure above, − indicates that the center is not chiral in this compound, and ? indicates that the stereochemistry is unknown.
Variation in alkyl substitution patterns of MBs.
In contrast to the relatively subtle variation seen in the nuclei of MBs, the alkyl substituents of the hydroxy acid (R3 and R4) and the acyl moiety R5 are highly variable (Fig. 3) (73). The β-hydroxy acid is usually β-hydroxybutyrate or 2-methyl-3-hydroxypentanoic acid. Both absolute configurations of the β-hydroxybutyrate have been reported from different MBs (75). Similarly, the absolute stereochemistry of the 2-methyl-3-hydroxypentanoic acid has been reported, and although the relative stereochemistry is always erythro, both absolute configurations are seen in different MBs. There are also reports of more complex β-hydroxy acids with branched chains (R3 = i-Pr, R4 = Me) (49), long-chain alkyl substituents (R3 = C15–18, R4 = Me) (3, 75), or both (R3 = C9, R4 = Me2) (53) being employed.
The acyl moiety (R5) is the source of most of the observed structural variation in the MBs (73). In general, the acyl group is a long-chain fatty acid (C10 to C21) that is often unsaturated with an unusual cis double bond conjugated to the carbonyl group (73). However, enormous variation on this theme is observed. Ratledge and Ewing detailed 17 different MBs from a single strain of M. smegmatis that differ only at R5; 7 of these compounds were present in significant amounts (61)! Thin-layer chromatography and high-performance liquid chromatography have been employed extensively, with some success, to identify strains of mycobacteria to the species level; however, they give no information on the exact structures of the MBs isolated (38). At least three different structures for MB J (the major MB from M. paratuberculosis) have appeared, the final one without comment in the context of other work (4, 18, 49). In addition, recent work on the carboxy-MBs or the chloroform-extractable exochelins has added to the complexity of the situation (18, 36, 37, 81). These compounds are clearly MBs with a modified acyl group (R5) derived from a dicarboxylic acid. This may be present as the free acid, the methyl ester, or a mixture of the two. Once again, a number of analogues are observed, with 20 different compounds identified from M. avium that differ only at R5 (81).
Nomenclature of extracellular MBs.
The small structural difference between the prototypical MBs and the extracellular compounds, i.e., the presence of an additional carboxyl or ester functionality on R5, leads to a profound increase in water solubility. This prompted the two groups of workers involved to each suggest a new name for this class of compounds, i.e., the carboxy-MBs (37) or the exochelins (18). Unfortunately, both are problematic. The name exochelin has already been applied to structurally unrelated, extracellular, peptidic siderophores from M. smegmatis (71) and M. neoaurum (70) and belies the structural similarity of these exochelins and the MBs. The name carboxy-MB, while addressing the latter concern, is inappropriate, as both the methyl esters and the free acids of these compounds have been reported (81). It seems best simply to refer to this class of compounds as water-soluble MBs, a term that is indicative of both their structure and their distinguishing physical characteristic. (The actual situation is even more complex. Ratledge and coworkers discovered water-soluble, mycobacterial siderophores and collectively termed them exochelins [42]. It was known that there were at least two types based on solubilities [43], and the first to be structurally characterized were peptidic compounds unrelated to MBs and named exochelins [70, 71]. Subsequently, the water-soluble MBs were identified [18, 37].) The more insightful term exo-MB, which evokes both the putative functional difference of these compounds from and the structural similarity of these compounds to cell-associated MBs, has also been used to describe these extracellular MB analogs (84).
As might be expected for such a complex array of structures, systematic chemical nomenclature is too unwieldy and trivial names are generally employed. The best system still appears to be that suggested by Snow in his excellent review in 1970 (73). Families of MBs are identified by their core structures, each of which is indicated by a letter referring to a previously identified MB. The letter historically indicated the species from which the compound was isolated, e.g., MB P from M. phlei.
A second structural class of siderophores: exochelins.
The existence of a second family of mycobacterial siderophores has been recognized for some time (42), but only recently have some of its members been characterized (70, 71). Two structures are known, both highly modified peptides with iron coordination sites provided by hydroxamic acids derived from ornithine (71), and in one case, a novel β-hydroxyhistidine (70). Exochelin MS (Fig. 1B), the major exochelin from M. smegmatis, reportedly representing 25 to 35% of the total of these compounds in this organism, was shown to be a formylated pentapeptide derived from three molecules of δ-N-hydroxyornithine, a β-alanine, and a threonine. Hydroxamic acids derived from the three ornithine moieties provide the iron coordination sites. Exochelin MN, one of the exochelins from M. neoaurum, is a hexapeptide consisting of two δ-N-hydroxyornithine residues and a β-hydroxyhistidine, which provide the iron coordination sites; two β-alanines; and an ornithine. The differences between exochelin MS and exochelin MN implies that there is a greater variation in structure and iron coordination strategy in this class of siderophore than is seen in the MBs. Thus far, such peptidic siderophores have only been reported from rapidly growing saprophytic mycobacterial species.
BIOSYNTHESIS OF MYCOBACTERIAL SIDEROPHORES
The basic building blocks.
Soon after the first structure of an MB was reported, the identities of the basic biosynthetic building blocks were elucidated (Fig. 2). It was shown that l-lysine was the precursor of both ɛ-N-hydroxylysines through radiolabeling studies (1, 78). The β-hydroxy acid moiety was shown to be derived from the condensation of two propionate molecules in the case of the 2-methyl-3-hydroxypentanoic acid of MB P (78). Results obtained with [1-14C]acetate were consistent with the β-hydroxybutyrate and the acyl moiety at R5 of MB S being derived from acetate (78). The aromatic moiety of MB S from M. smegmatis has been shown to arise from salicylate formed via the shikimic acid pathway (27, 62, 63). The aromatic moiety of MB P, with its extra methyl group, is believed to arise from 6-methylsalicylate instead of salicylate. Once again, this acid is a known extracellular metabolite in mycobacteria that forms MBs bearing the methylated aromatic moiety (73). However, the biosynthetic origins of these two aromatic acids are very different. In contrast to salicylate, 6-methylsalicylate is not a shikimate metabolite but rather a polyketide formed from the condensation of four acetate units (28). This provides an interesting example of two very different biosynthetic pathways being utilized to provide the required structural unit, a hydroxyaromatic acid. Even more surprising is the co-occurrence of MBs in M. fortuitum (75) that have both a salicylate- and a 6-methylsalicylate-derived aromatic moiety. This implies that both pathways must apparently be employed by this organism (73). No studies have been carried out on the origins of the oxazoline, but serine and threonine are presumed to be the precursors of the unsubstituted and methyl-bearing rings, respectively (73).
The enzymes which assemble MBs belong to the family of nonribosomal peptide synthases.
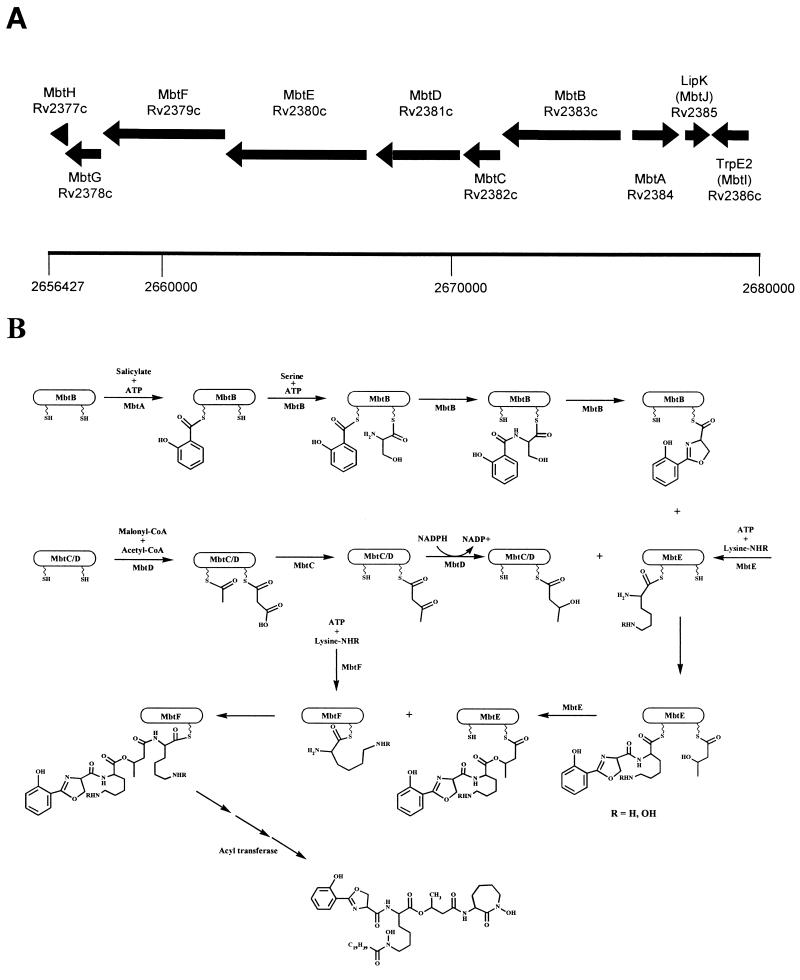
Studies of MB biosynthesis have languished for the last 20 years, only to be reinvigorated by the recent sequencing of the M. tuberculosis genome. A gene cluster designated the mbt genes was recognized by Cole et al. (8) and by Quadri et al. (56) as having the potential to encode the appropriate enzymes for MB formation (Fig. 4A). Several of the encoded proteins are highly homologous to nonribosomal peptide synthases (NRPSs), which have been implicated in the biosynthesis of other siderophores, as well as a variety of peptide-derived secondary metabolites (44). These NRPSs are large modular enzymes that activate amino acids as their acyl adenylates through an activating domain and covalently link them to the enzyme via a phosphopantetheinyl thioester attached to a peptide carrier protein domain. Two such activated amino acids are then condensed to form a peptide bond in a reaction catalyzed by a condensation domain. The activation domains specify the amino acids incorporated, and their order encodes the sequence of the final peptide. Numerous other domains or activities are also encountered in these proteins, i.e., epimerization domains that convert l amino acids to d amino acids, cyclization domains that form either macrocyclic esters or amides or smaller rings such as oxazolines and thiazolines, and thioesterase domains that cleave the final peptide product from the enzyme.
FIG. 4.
Proposed biosynthetic scheme for MB T. (A) Region of the genome of M. tuberculosis H37Rv containing the putative MB biosynthetic enzymes. Numbering refers to chromosomal positions reported by Cole et al. (8). (B) Proposed biosynthetic cascade catalyzed by the Mbt locus. See the text for details of individual enzymatic predictions. There are significant areas of uncertainty in this model, as suggested by the following questions. (i) Does the ketoacyl synthase domain of MbtC act upon the MbtD-bound acyl carrier protein domains as shown? (ii) Does the second peptidyl/acyl carrier protein domain of MbtE function to attach the β-hydroxybutyrate as shown, or does it transfer the acyl group to the first lysine (the R5) position in Fig. 2? (iii) Do MbtE and MbtF act in the order in which they appear in the genomic locus, or are they reversed? (iv) Is MbtG involved in N-hydroxylation as shown? (v) What is the nature of the acyltransferase, and how is the specificity determined? When does this act and upon what substrate? Since this determines the ratio of water-soluble to cell-associated siderophore, this is a critical question. (vi) What is the role played by MbtH and MbtJ?
A unified scheme for the production of MBs.
The proposed biosynthetic scheme based upon the known precursor incorporation results and the sequence homologies of the deduced mbt-encoded proteins is as follows (Fig. 4B). Salicylate is activated as its acyl adenylate by MbtA (Rv2384) and is then covalently attached to a phosphopantetheine prosthetic group of MbtB (Rv2383c). This activation-attachment appears to be a common motif in the biosynthesis of siderophores which contain a phenolate moiety. Proteins homologous to MbtA have been observed in the biosynthetic pathways of enterobactin (67) from Escherichia coli and yersiniabactin from Yersinia pestis (16). MtbB is an NRPS that is believed to activate serine, condense it with the salicylate moiety, and cyclize this product to a hydroxyphenyloxazoline. Once again, MtbB has homologues with analogous functions in both the pyochelin (from Pseudomonas aeruginosa) (66) and yersiniabactin (16) pathways. There are two other NRPSs, encoded by mbtE (Rv2380c) and mbtF (Rv2379c), that have the appropriate activation, condensation, and peptide carrier domains for donation of the two lysine-derived moieties of MB. MbtF has a terminal domain that was assigned a role as either an epimerization domain or as a thioesterase responsible for releasing the MB from the enzyme by lactamization of the terminal hydroxylysine residue. The latter function appears much more likely, as all amino acid-derived centers with known absolute configurations in the MBs arise from the naturally occurring L series (25, 73, 75, 76). It should be noted, however, that the absolute stereochemistries of the oxazoline and the acylated lysine (i.e., b and c in Fig. 3) have not been determined unambiguously for MB T, the MB from M. tuberculosis. Also in the gene cluster are mbtC (Rv2382c) and mbtD (Rv2381c), which encode proteins that are homologous to polyketide synthases. The encoded proteins appear to contain the appropriate modules to produce the required β-hydroxybutyrate. Additionally, mbtG (Rv2378c) encodes a protein that is homologous to known ornithine and lysine oxygenases, an activity required for the production of the ɛ-N-hydroxylysines. Thus, these seven genes, mbtA to mbtG, appear to encode sufficient activities for the biosynthesis of the core of the MBs (Fig. 4). There are two other proposed gene products, MbtH (Rv2377c) and MbtJ (Rv2385), to which no clear biochemical role has been assigned.
The genes involved in the formation of salicylate are not clearly defined. Cole et al. (8) suggested that a distinct operon with two genes (entC and entD [Rv3215 and Rv3214, respectively]) homologous to those implicated in salicylate formation in P. aeruginosa (69) is responsible for its formation. Walsh et al. (56) concluded that the product of another gene, mbtI (Rv2386c), located at the 5′ end of the mbt gene cluster and homologous to salicylate- and anthranilate-forming enzymes was most likely to be responsible for salicylate formation. This gene was originally called trpE2 and assigned the encoding of an anthranilate synthase (8). Given the similarity between the different biochemical functions postulated for these various enzymes (anthranilate is the amino analog of salicylate), certain assignment will await the expression and characterization of these proteins. It is interesting, however, that the DNA sequence upstream of the mbtI gene contains a very convincing match for an IdeR binding consensus sequence, which suggests repression of the expression of this gene under high-iron conditions, consistent with a predicted role in salicylate synthesis (13). It should also be noted that a gene (acpS [Rv2523c]) which encodes a phosphopantetheinyl transferase, necessary for the formation of the holoenzyme of the NRPSs, has been identified in the M. tuberculosis genome (56).
What proof exists for the function of the above enzymes (MbtA to MbtG), postulated on the basis of homology alone? The protein encoded by mbtA has been expressed and shown to form the acyl adenylate of salicylate (56). This, in turn, acylates a fragment of MbtB which was predicted to encode the necessary phosphopantetheinyl carrier domain. This only occurred after treatment of the MbtB protein fragment with the appropriate phosphopantetheinyl transferase (56). We have constructed an mbtB mutant strain of M. tuberculosis through homologous recombination with a DNA fragment in which a portion of the mbtB gene was replaced with a hygromycin resistance marker (10). The resulting ΔmbtB::hyg strain was shown to be incapable of siderophore production, thus conclusively linking this gene cluster with MB biosynthesis. The absence of siderophore production in this mutant furthermore demonstrates that all of the MBs, including the water-soluble ones, arise through a common biogenic pathway. This latter work (10) also was the first to demonstrate the long-suspected importance of siderophore production to virulence in M. tuberculosis, as the mbtB mutant strain was avirulent in macrophage infections. Thus, the importance of this operon in MB biosynthesis has been established. The roles of MbtA and MbtB (in part) are as predicted, but the completion of much of the MB biosynthetic story awaits the functional characterization of the remaining enzymes.
The biosynthesis of exochelins.
Recently, some results concerning the biosynthesis of the other mycobacterial siderophore, the exochelins, have been reported (14, 83, 84). Complementation studies carried out with M. smegmatis mutants defective in exochelin production have identified a cluster of three genes believed to be involved in the biosynthesis of exochelin MS (the exochelin from M. smegmatis). Two of these genes encode large NRPSs (83, 84), while the third encodes a protein with homology to phosphoribosylglycineamide formyltransferases (14). This latter enzyme was believed to be responsible for the production of the formylornithine moiety of exochelin MS, while the NRPSs catalyzed the assembly of the peptide backbone. However, the identification of a total of six amino acid activation domains for the production of this pentapeptide indicates that, as with the MBs, much remains to be understood about exochelin biosynthesis. Part of the explanation for this discrepancy may lie in the fact that exochelin MS is only one of the family of exochelins produced by M. smegmatis.
THE REGULATION OF IRON METABOLISM
Iron-responsive alterations in protein synthesis.
Combination of sophisticated mass spectrometry techniques with the genome sequence to analyze protein patterns on two-dimensional gels has been a useful tool in the identification of proteins that are upregulated or downregulated in response to iron concentration (82). Fifteen proteins from M. tuberculosis were upregulated and 12 proteins were downregulated under low-iron conditions. In another study of protein profiles of M. tuberculosis grown in low and high concentrations of iron, several iron-regulated proteins were isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to N-terminal sequencing (6). The gene encoding Irp10, a 10-kDa protein prominent in iron-deficient cell extracts but virtually absent from extracts of iron-replete cultures, was designated irpA (Rv3269). This gene is similar to those that encode heat shock proteins or chaperonins and lies immediately upstream of a gene that encodes a putative cation-transporting ATPase named CtpC (Rv3270). This protein is a member of the P-type cation-transporting ATPase family. Both irpA and ctpC appear to be induced by low iron, although no consensus binding sites for known iron-dependent regulators were identified upstream of either gene. If further studies reveal that Irp10 and CtpC do, in fact, transport iron, it will be the first example of iron transport by a P-type ATPase, rather than an ABC transporter ATPase.
The N-terminal sequences of two other iron-regulated proteins that were upregulated under low-iron conditions were also reported (6). By searching the M. tuberculosis genome database, it is now possible to identify these proteins. The N-terminal sequence of the upregulated Irp13 protein corresponds to Acr (Rv2031c), the same protein previously shown by these authors to be downregulated by iron starvation (82). Irp28 contains the sequence found in HupB (Rv2986c), an HU-histone fusion. Interestingly, the same N-terminal sequence was found for both Irp28 and Irp29. While Irp28 is upregulated by iron starvation, Irp29 is downregulated by iron starvation, suggesting that the same histone-like protein is modified differently under the two conditions.
Phenotypic responses to iron deprivation.
The effect of iron starvation on stress response-related protein synthesis in M. smegmatis was examined (39). Killing by hydrogen peroxide was increased in cultures grown in 0.2 μM or a lower iron concentration. The simplest interpretation is that during iron starvation, iron-containing enzymes, such as catalase, which are required for protection from hydrogen peroxide are not produced in their active, iron-containing form. However, the situation may be more complicated. Patterns of stress-induced proteins were examined by placing M. smegmatis under heat, ethanol, or hydrogen peroxide stress while labeling the induced proteins with 35S. When the labeling experiments were performed with iron-sufficient cells, all three stress conditions induced expression of GroEL1 (Rv3417c) and DnaK (Rv0350). Surprisingly, when the labeling experiments were done with iron-starved cells, GroEL and DnaK were not induced by hydrogen peroxide but were induced by heat shock or ethanol, suggesting that the regulatory networks involved in iron metabolism and stress responses are interrelated. The response of iron-starved M. tuberculosis to hydrogen peroxide stress may differ from that described for M. smegmatis, since M. smegmatis displays a broader phenotypic response to oxidative stress than does M. tuberculosis (72).
Control of gene expression by iron in mycobacteria.
The best-studied iron-responsive transcriptional regulator is the ferric uptake regulator Fur (9). Encoded by the fur gene, this protein binds its corepressor, ferric iron, and then binds to operator sequences known as Fur boxes, thereby repressing transcription. Under iron starvation, Fur no longer binds to the Fur box and the corresponding genes become derepressed. M. tuberculosis contains as many as four such iron-dependent regulators. IdeR (Rv2711) is the only protein for which experimental evidence of a role in iron binding and DNA binding exists (68). It contains extensive similarity to the DtxR family. This family, named after the diphtheria toxin repressor, is only distantly related to the gram-negative iron-dependent regulator Fur and binds a different DNA operator sequence. Interestingly, M. tuberculosis contains two genes, furA and furB, that encode proteins more similar to E. coli Fur than to IdeR/DtxR. FurA (Rv1909c) clusters more tightly with Fur proteins found in other mycobacteria and FurS from Streptomyces reticuli. In each case, immediately downstream of the fur homolog is a gene encoding a catalase peroxidase. In the mycobacteria, this gene is katG; in S. reticuli, it is called cpeB. In S. reticuli, FurS has been shown to negatively regulate the expression of cpeB in the presence of iron (85). The sequence similarity of both the regulator and catalase-peroxidase, as well as the conserved gene arrangement, suggests functional conservation between S. reticuli and M. tuberculosis. Even more similar to E. coli Fur is FurB (Rv2359), for which no other mycobacterial homologs have been described. Finally, there is SirR (Rv2788), putatively described as an iron-dependent regulator based on similarity to SirR from Staphylococcus epidermidis (23), TroR from Treponema pallidum (21), and the IdeR/DtxR family.
The iron-responsive regulatory protein encoded by ideR, which is homologous to the dtxR gene from Corynebacterium diphtheriae, is biochemically the best characterized. The mycobacterial protein has been analyzed both in vitro and in vivo for function. The recombinant protein overproduced in E. coli was shown by band shift analysis to bind the C. diphtheriae tox promoter-operator sequence in a divalent metal-dependent manner (68). The role of IdeR in the repression of siderophore production was shown with the construction of an ideR mutant of M. smegmatis (13). This mutant produces siderophore when grown in high- or low-iron media, demonstrating the requirement for IdeR to repress siderophore production under high-iron conditions. Interestingly, but perhaps not surprisingly in light of the presence of furA, furB, and sirR in M. tuberculosis, the mutant was still capable of upregulating siderophore production under low-iron conditions, suggesting the presence of a second iron-sensing regulator in M. smegmatis. Another phenotype associated with the ideR mutant was increased sensitivity to hydrogen peroxide. Native gel activity assays demonstrated a reduction in the activities of the catalase-peroxidase KatG and the superoxide dismutase activity of Mn superoxide dismutase. However, the activity of another catalase, KatE, was unaffected by the ideR mutation. These results again demonstrate the link between the responses to iron starvation and oxidative stress in M. smegmatis.
CONCLUSIONS
Iron is an essential nutrient whose concentration is critical in determining the outcome of infection with pathogenic mycobacteria. These organisms have evolved a complex web of biosynthetic and metabolic pathways by which to obtain it, store it, and regulate its use. Unraveling of the intricacies of this system may reveal an Achilles heel that can be used to guide and inform the development of new antibiotics. As a precedent for such antibiotics, p-aminosalicylic acid is a clinically useful second-line antitubercular agent, the target of which may lie in the enzymes which biosynthesize MBs (80). MB structural analogs have inhibitory potential as well, attracting the interest of synthetic chemists. M. tuberculosis, which biosynthesizes and utilizes MB T, is inhibited by synthetic MB S, which only appears to differ from MB T in the absolute configuration of one to three of its stereogenic centers (26, 73). Further progress toward new therapies based upon a rational understanding of iron acquisition and metabolism by mycobacteria requires an even greater understanding of the complexities of the structure, biosynthesis, and functions of siderophores; their uptake into bacterial cells; and the long-term storage and retrieval of iron.
ACKNOWLEDGMENT
This work was supported in part by NHMRC grant 961249 to J.J.D.V.
REFERENCES
- 1.Allen M, Birch A J, Jones A R. Studies in relation to biosynthesis. XLIII. Incorporation of l-lysine into mycobactin-P. Aust J Chem. 1970;23:427–429. [Google Scholar]
- 2.Andrews S C. Iron storage in bacteria. Adv Microb Physiol. 1998;40:281–351. doi: 10.1016/s0065-2911(08)60134-4. [DOI] [PubMed] [Google Scholar]
- 3.Barclay R, Ewing D F, Ratledge C. Isolation, identification, and structural analysis of the mycobactins of Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium scrofulaceum, and Mycobacterium paratuberculosis. J Bacteriol. 1985;164:896–903. doi: 10.1128/jb.164.2.896-903.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barclay R, Ratledge C. Iron-binding compounds of Mycobacterium avium, M. intracellulare, M. scrofulaceum, and mycobactin-dependent M. paratuberculosis and M. avium. J Bacteriol. 1983;153:1138–1146. doi: 10.1128/jb.153.3.1138-1146.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baynes R D, Flax H, Bothwell T H, Bezwoda W R, MacPhail A P, Atkinson P, Lewis D. Haematological and iron-related measurements in active pulmonary tuberculosis. Scand J Haematol. 1986;36:280–287. doi: 10.1111/j.1600-0609.1986.tb01735.x. [DOI] [PubMed] [Google Scholar]
- 6.Calder K M, Horwitz M A. Identification of iron-regulated proteins of Mycobacterium tuberculosis and cloning of tandem genes encoding a low iron-induced protein and a metal transporting ATPase with similarities to two-component metal transport systems. Microb Pathog. 1998;24:133–143. doi: 10.1006/mpat.1997.9999. [DOI] [PubMed] [Google Scholar]
- 7.Clemens D L, Horwitz M A. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. . (Erratum, 396:190, 1998.) [DOI] [PubMed] [Google Scholar]
- 9.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Voss, J. J., K. Rutter, Y. Zhu, B. G. Schroeder, and C. E. Barry III. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 11.Dhople A M, Ibanez M A, Poirier T C. Role of iron in the pathogenesis of Mycobacterium avium infection in mice. Microbios. 1996;87:77–87. [PubMed] [Google Scholar]
- 12.Douvas G S, May M H, Crowle A J. Transferrin, iron, and serum lipids enhance or inhibit Mycobacterium avium replication in human macrophages. J Infect Dis. 1993;167:857–864. doi: 10.1093/infdis/167.4.857. [DOI] [PubMed] [Google Scholar]
- 13.Dussurget O, Rodriguez M, Smith I. An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol Microbiol. 1996;22:535–544. doi: 10.1046/j.1365-2958.1996.1461511.x. [DOI] [PubMed] [Google Scholar]
- 14.Fiss E H, Yu S, Jacobs W R., Jr Identification of genes involved in the sequestration of iron in mycobacteria: the ferric exochelin biosynthetic and uptake pathways. Mol Microbiol. 1994;14:557–569. doi: 10.1111/j.1365-2958.1994.tb02189.x. [DOI] [PubMed] [Google Scholar]
- 15.Francis J, Macturk H M, Madinaveitia J, Snow G A. Mycobactin, a growth factor for Mycobacterium johnei. I. Isolation from M. phlei. Biochem J. 1953;55:596–607. doi: 10.1042/bj0550596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehring A M, Mori I, Perry R D, Walsh C T. The nonribosomal peptide synthetase HMWP2 forms a thiazoline ring during biogenesis of yersiniabactin, an iron-chelating virulence factor of Yersinia pestis. Biochemistry. 1998;37:11637–11650. doi: 10.1021/bi9812571. [DOI] [PubMed] [Google Scholar]
- 17.Gobin J, Horwitz M A. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall. J Exp Med. 1996;183:1527–1532. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gobin J, Moore C H, Reeve J R, Jr, Wong D K, Gibson B W, Horwitz M A. Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci USA. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golden C A, Kochan I, Spriggs D R. Role of mycobactin in the growth and virulence of tubercle bacilli. Infect Immun. 1974;9:34–40. doi: 10.1128/iai.9.1.34-40.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordeuk V R, McLaren C E, MacPhail A P, Deichsel G, Bothwell T H. Associations of iron overload in Africa with hepatocellular carcinoma and tuberculosis: Strachan’s 1929 thesis revisited. Blood. 1996;87:3470–3476. [PubMed] [Google Scholar]
- 21.Hardham J M, Stamm L V, Porcella S F, Frye J G, Barnes N Y, Howell J K, Mueller S L, Radolf J D, Weinstock G M, Norris S J. Identification and transcriptional analysis of a Treponema pallidum operon encoding a putative ABC transport system, an iron-activated repressor protein homolog, and a glycolytic pathway enzyme homolog. Gene. 1997;197:47–64. doi: 10.1016/s0378-1119(97)00234-5. [DOI] [PubMed] [Google Scholar]
- 22.Harris C E, Biggs J C, Concannon A J, Dodds A J. Peripheral blood and bone marrow findings in patients with acquired immune deficiency syndrome. Pathology. 1990;22:206–211. doi: 10.3109/00313029009086664. [DOI] [PubMed] [Google Scholar]
- 23.Hill P J, Cockayne A, Landers P, Morrissey J A, Sims C M, Williams P. SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect Immun. 1998;66:4123–4129. doi: 10.1128/iai.66.9.4123-4129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Homuth M, Valentin-Weigand P, Rohde M, Gerlach G F. Identification and characterization of a novel extracellular ferric reductase from Mycobacterium paratuberculosis. Infect Immun. 1998;66:710–716. doi: 10.1128/iai.66.2.710-716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hough E, Rogers D. Crystal structure of ferrimycobactin P, a growth factor for the mycobacteria. Biochem Biophys Res Commun. 1974;57:73–77. doi: 10.1016/s0006-291x(74)80358-x. [DOI] [PubMed] [Google Scholar]
- 26.Hu J, Miller M J. Total synthesis of a mycobactin S, a siderophore and growth promoter of Mycobacterium smegmatis, and determination of its growth inhibitory activity against Mycobacterium tuberculosis. J Am Chem Soc. 1997;119:3462–3468. [Google Scholar]
- 27.Hudson A T, Bentley R. Utilization of shikimic acid for the formation of mycobactin S and salicyclic acid by Mycobacterium smegmatis. Biochemistry. 1970;9:3984–3987. doi: 10.1021/bi00822a017. [DOI] [PubMed] [Google Scholar]
- 28.Hudson A T, Campbell I M, Bentley R. Biosynthesis of 6-methylsalicylic acid by Mycobacterium phlei. Biochemistry. 1970;9:3988–3992. doi: 10.1021/bi00822a018. [DOI] [PubMed] [Google Scholar]
- 29.Immanuel C, Acharyulu G S, Kannapiran M, Segaran R, Sarma G R. Acute phase proteins in tuberculous patients. Indian J Chest Dis Allied Sci. 1990;32:15–23. [PubMed] [Google Scholar]
- 30.Inglis N F, Stevenson K, Hosie A H, Sharp J M. Complete sequence of the gene encoding the bacterioferritin subunit of Mycobacterium avium subspecies silvaticum. Gene. 1994;150:205–206. doi: 10.1016/0378-1119(94)90889-3. [DOI] [PubMed] [Google Scholar]
- 31.Kikuchi S, Fukumoto M, Takahashi H. Iron storage in Mycobacterium smegmatis grown under iron-sufficient and iron-overload conditions. Biosci Biotechnol Biochem. 1994;58:885–888. [Google Scholar]
- 32.Kochan I. The role of iron in bacterial infections, with special consideration of host-tubercle bacillus interaction. Curr Top Microbiol Immunol. 1973;60:1–30. doi: 10.1007/978-3-642-65502-9_1. [DOI] [PubMed] [Google Scholar]
- 33.Kochan I, Ishak K, Said M, Stotts J. Study on the tuberculostatic factor of mammalian serum. Am Rev Respir Dis. 1963;88:818–826. doi: 10.1164/arrd.1963.88.6.818. [DOI] [PubMed] [Google Scholar]
- 34.Kochan I, Patton C, Ishak K. Tuberculostatic activity of normal sera. J Immunol. 1963;90:711–719. [PubMed] [Google Scholar]
- 35.Kontoghiorghes G J, Weinberg E D. Iron: mammalian defense systems, mechanisms of disease, and chelation therapy approaches. Blood Rev. 1995;9:33–45. doi: 10.1016/0268-960x(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 36.Lane S J, Marshall P S, Upton R J, Ratledge C. Isolation and characterization of carboxymycobactins as the second extracellular siderophores in Mycobacterium smegmatis. BioMetals. 1998;11:13–20. [Google Scholar]
- 37.Lane S J, Marshall P S, Upton R J, Ratledge C, Ewing M. Novel extracellular mycobactins, the carboxymycobactins from Mycobacterium avium. Tetrahedron Lett. 1995;36:4129–4132. [Google Scholar]
- 38.Leite C Q F, Barreto A M W, Leite S R A. Thin-layer chromatography of mycobactins and mycolic acids for the identification of clinical mycobacteria. Rev Microbiol. 1995;26:192–199. [Google Scholar]
- 39.Lundrigan M D, Arceneaux J E L, Zhu W, Byers B R. Enhanced hydrogen peroxide sensitivity and altered stress protein expression in iron-starved Mycobacterium smegmatis. BioMetals. 1997;10:215–225. doi: 10.1023/a:1018355928990. [DOI] [PubMed] [Google Scholar]
- 40.MacCordick H J. Restrictive spatial modeling in structure and stability concepts of metallo-mycobactins. Nouv J Chim. 1985;9:535–538. [Google Scholar]
- 41.MacCordick H J, Schleiffer J J, Duplatre G. Radiochemical studies of iron binding and stability in ferrimycobactin S. Radiochim Acta. 1985;38:43–47. [Google Scholar]
- 42.Macham L P, Ratledge C. New group of water-soluble iron-binding compounds from mycobacteria. Exochelins. J Gen Microbiol. 1975;89:379–382. doi: 10.1099/00221287-89-2-379. [DOI] [PubMed] [Google Scholar]
- 43.Macham L P, Ratledge C, Nocton J C. Extracellular iron acquisition by mycobacteria: role of the exochelins and evidence against the participation of mycobactin. Infect Immun. 1975;12:1242–1251. doi: 10.1128/iai.12.6.1242-1251.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marahiel M A, Stachelhaus T, Mootz H D. Modular peptide synthetases involved in non-ribosomal peptide synthesis. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 45.Marshall B J, Ratledge C. Salicylic acid biosynthesis and its control in Mycobacterium smegmatis. Biochim Biophys Acta. 1972;264:106–116. doi: 10.1016/0304-4165(72)90122-5. [DOI] [PubMed] [Google Scholar]
- 46.Matzanke B F, Boehnke R, Moellmann U, Reissbrodt R, Schuenemann V, Trautwein A X. Iron uptake and intracellular metal transfer in mycobacteria mediated by xenosiderophores. BioMetals. 1997;10:193–203. doi: 10.1023/a:1018351728081. [DOI] [PubMed] [Google Scholar]
- 47.Matzanke B F, Mollmann U, Reissbrodt R, Schunemann V, Trautwein A X. Siderophore transport in mycobacteria analyzed by Mössbauer spectroscopy: possible routes to novel antibiotics against these organisms? Hyperfine Interact. 1998;112:123–128. [Google Scholar]
- 48.McCready K A, Ratledge C. Amounts of iron, heme and related compounds in Mycobacterium smegmatis grown in various concentrations of iron. Biochem Soc Trans. 1978;6:421–423. doi: 10.1042/bst0060421. [DOI] [PubMed] [Google Scholar]
- 49.McCullough W G, Merkal R S. Structure of mycobactin J. Curr Microbiol. 1982;7:337–341. [Google Scholar]
- 50.Messenger A J M, Hall R M, Ratledge C. Iron uptake processes in Mycobacterium vaccae R877R, a mycobacterium lacking mycobactin. J Gen Microbiol. 1986;132:845–852. doi: 10.1099/00221287-132-3-845. [DOI] [PubMed] [Google Scholar]
- 51.Messenger A J M, Ratledge C. Iron transport in Mycobacterium smegmatis: uptake of iron from ferric citrate. J Bacteriol. 1982;149:131–135. doi: 10.1128/jb.149.1.131-135.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moyo V M, Gangaidzo I T, Gordeuk V R, Kiire C F, Macphail A P. Tuberculosis and iron overload in Africa: a review. Cent Afr J Med. 1997;43:334–339. [PubMed] [Google Scholar]
- 53.Murakami Y, Kato S, Nakajima M, Matsuoka M, Kawai H, Shin-Ya K, Seto H. Formobactin, a novel free radical scavenging and neuronal cell protecting substance from Nocardia sp. J Antibiot. 1996;49:839–845. doi: 10.7164/antibiotics.49.839. [DOI] [PubMed] [Google Scholar]
- 54.Murray M J, Murray A B, Murray M B, Murray C J. The adverse effect of iron repletion on the course of certain infections. Br Med J. 1978;2:1113–1115. doi: 10.1136/bmj.2.6145.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pessolani M C V, Smith D R, Rivoire B, McCormick J, Hefta S A, Cole S T, Brennan P J. Purification, characterization, gene sequence, and significance of a bacterioferritin from Mycobacterium leprae. J Exp Med. 1994;180:319–327. doi: 10.1084/jem.180.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quadri L E, Sello J, Keating T A, Weinreb P H, Walsh C T. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem Biol. 1998;5:631–645. doi: 10.1016/s1074-5521(98)90291-5. [DOI] [PubMed] [Google Scholar]
- 57.Raghu B, Sarma G R, Venkatesan P. Effect of haptoglobin on hemoglobin-supported growth and siderophore production of mycobacteria. Med Sci Res. 1994;22:99–100. [Google Scholar]
- 58.Raghu B, Sarma G R, Venkatesan P. Effect of iron on the growth and siderophore production of mycobacteria. Biochem Mol Biol Int. 1993;31:341–348. [PubMed] [Google Scholar]
- 59.Ratledge C. Metabolism of iron and other metals by mycobacteria. Microbiol Ser. 1984;15:603–627. [Google Scholar]
- 60.Ratledge C. Nutrition, growth and metabolism. In: Ratledge C, Stanford J, editors. The biology of the mycobacteria. Vol. 1. San Diego, Calif: Academic Press Limited; 1982. pp. 185–271. [Google Scholar]
- 61.Ratledge C, Ewing D F. The separation of the mycobactins from Mycobacterium smegmatis by using high-pressure liquid chromatography. Biochem J. 1978;175:853–857. doi: 10.1042/bj1750853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ratledge C, Hall M J. Isolation and properties of auxotrophic mutants of Mycobacterium smegmatis requiring either salicylic acid or mycobactin. J Gen Microbiol. 1972;72:143–150. doi: 10.1099/00221287-72-1-143. [DOI] [PubMed] [Google Scholar]
- 63.Ratledge C, Hall M J. Uptake of salicylic acid into mycobactin S by growing cells of Mycobacterium smegmatis. FEBS Microbiol Lett. 1970;10:309–312. doi: 10.1016/0014-5793(70)80460-4. [DOI] [PubMed] [Google Scholar]
- 64.Ratledge C, Marshall B J. Iron transport in Mycobacterium smegmatis. Role of mycobactin. Biochim Biophys Acta. 1972;279:58–74. doi: 10.1016/0304-4165(72)90241-3. [DOI] [PubMed] [Google Scholar]
- 65.Ratledge C, Patel P V. Lipid-soluble, iron-binding compounds in Nocardia and related organisms. In: Goodfellow M, Brownell G H, Serrano J A, editors. The biology of the Nocardiae. London, England: Academic Press; 1976. pp. 372–385. [Google Scholar]
- 66.Reimmann C, Serino L, Beyeler M, Haas D. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology. 1998;144:3135–3148. doi: 10.1099/00221287-144-11-3135. [DOI] [PubMed] [Google Scholar]
- 67.Rusnak F, Faraci W S, Walsh C T. Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl)adenylate product. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 68.Schmitt M P, Predich M, Doukhan L, Smith I, Holmes R K. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect Immun. 1995;63:4284–4289. doi: 10.1128/iai.63.11.4284-4289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Serino L, Reimmann C, Baur H, Beyeler M, Visca P, Haas D. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol Gen Genet. 1995;249:217–228. doi: 10.1007/BF00290369. [DOI] [PubMed] [Google Scholar]
- 70.Sharman G J, Williams D H, Ewing D F, Ratledge C. Determination of the structure of exochelin MN, the extracellular siderophore from Mycobacterium neoaurum. Chem Biol. 1995;2:553–561. doi: 10.1016/1074-5521(95)90189-2. [DOI] [PubMed] [Google Scholar]
- 71.Sharman G J, Williams D H, Ewing D F, Ratledge C. Isolation, purification and structure of exochelin MS, the extracellular siderophore from Mycobacterium smegmatis. Biochem J. 1995;305:187–196. doi: 10.1042/bj3050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sherman D R, Sabo P J, Hickey M J, Arain T M, Mahairas G G, Yuan Y, Barry III C E, Stover C K. Disparate responses to oxidative stress in saprophytic and pathogenic mycobacteria. Proc Natl Acad Sci USA. 1995;92:6625–6629. doi: 10.1073/pnas.92.14.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Snow G A. Mycobactins: iron-chelating growth factors from mycobacteria. Bacteriol Rev. 1970;34:99–125. doi: 10.1128/br.34.2.99-125.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Snow G A. The structure of mycobactin P, a growth factor for Mycobacterium johnei, and the significance of its iron complex. Biochem J. 1965;94:160–165. doi: 10.1042/bj0940160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Snow G A, White A J. Chemical and biological properties of mycobactins isolated from various mycobacteria. Biochem J. 1969;115:1031–1045. doi: 10.1042/bj1151031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Snow G A, White A J. Isolation of mycobactins from various mycobacteria. Biochem J. 1969;111:785–792. doi: 10.1042/bj1110785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stephenson M C, Ratledge C. Specificity of exochelins for iron transport in three species of mycobacteria. J Gen Microbiol. 1980;116:521–523. doi: 10.1099/00221287-116-2-521. [DOI] [PubMed] [Google Scholar]
- 78.Tateson J E. Early steps in the biosynthesis of mycobactins P and S. Biochem J. 1970;118:747–753. doi: 10.1042/bj1180747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78a.TubercuList Web Server. 1999, copyright date. [Online.] Institut Pasteur, Paris, France. http://www.pasteur.fr/Bio/TubercuList/. [7 June 1999, last date accessed.]
- 79.Wheeler P R, Ratledge C. Metabolism of Mycobacterium tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C: American Society for Microbiology; 1994. pp. 353–385. [Google Scholar]
- 80.Winder F G. Mode of action of antimycobacterial agents and associated aspects of the molecular biology of the mycobacteria. In: Ratledge C, Standford J, editors. The biology of the mycobacteria. Vol. 1. London, England: Academic Press; 1982. pp. 353–438. [Google Scholar]
- 81.Wong D K, Gobin J, Horwitz M A, Gibson B W. Characterization of exochelins of Mycobacterium avium: evidence for saturated and unsaturated and for acid and ester forms. J Bacteriol. 1996;178:6394–6398. doi: 10.1128/jb.178.21.6394-6398.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wong D K, Lee B-Y, Horwitz M A, Gibson B W. Identification of Fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect Immun. 1999;67:327–336. doi: 10.1128/iai.67.1.327-336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu S, Fiss E, Jacobs W R., Jr Analysis of the exochelin locus in Mycobacterium smegmatis: biosynthesis genes have homology with genes of the peptide synthetase family. J Bacteriol. 1998;180:4676–4685. doi: 10.1128/jb.180.17.4676-4685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu W, Arceneaux J E L, Beggs M L, Byers B R, Eisenach K D, Lundrigan M D. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two non-ribosomal peptide synthetase genes. Mol Microbiol. 1998;29:629–639. doi: 10.1046/j.1365-2958.1998.00961.x. [DOI] [PubMed] [Google Scholar]
- 85.Zou P, Borovok I, Lucana D, Muller D, Schrempf H. The mycelium-associated Streptomyces reticuli catalase-peroxidase, its gene and regulation by FurS. Microbiology. 1999;145:549–559. doi: 10.1099/13500872-145-3-549. [DOI] [PubMed] [Google Scholar]