Abstract
We have cloned, sequenced, and characterized the genes encoding the lytic system of the unique Staphylococcus aureus phage 187. The endolysin gene ply187 encodes a large cell wall-lytic enzyme (71.6 kDa). The catalytic site, responsible for the hydrolysis of staphylococcal peptidoglycan, was mapped to the N-terminal domain of the protein by the expression of defined ply187 domains. This enzymatically active N terminus showed convincing amino acid sequence homology to an N-acetylmuramoyl-l-alanine amidase, whereas the C-terminal part, whose function is unknown, revealed striking relatedness to major staphylococcal autolysins. An additional reading frame was identified entirely embedded out of frame (+1) within the 5′ region of ply187 and was shown to encode a small, hydrophobic protein of holin-like function. The hol187 gene features a dual-start motif, possibly enabling the synthesis of two products of different lengths (57 and 55 amino acids, respectively). Overproduction of Hol187 in Escherichia coli resulted in growth retardation, leakiness of the cytoplasmic membrane, and loss of de novo ATP synthesis. Compared to other holins identified to date, Hol187 completely lacks the highly charged C terminus. The secondary structure of the polypeptide is predicted to consist of two small, antiparallel, hydrophobic, transmembrane helices. These are supposed to be essential for integration into the membrane, since site-specific introduction of negatively charged amino acids into the first transmembrane domain (V7D G8D) completely abolished the function of the Hol187 polypeptide. With antibodies raised against a synthetic 18-mer peptide representing a central part of the protein, it was possible to detect Hol187 in the cytoplasmic membrane of phage-infected S. aureus cells. An important indication that the protein actually functions as a holin in vivo was that the gene (but not the V7D G8D mutation) was able to complement a phage λ Sam mutation in a nonsuppressing E. coli HB101 background. Plaque formation by λgt11::hol187 indicated that both phage genes have analogous functions. The data presented here indicate that a putative holin is encoded on a different reading frame within the enzymatically active domain of ply187 and that the holin is synthesized during the late stage of phage infection and found in the cytoplasmic membrane, where it causes membrane lesions which are thought to enable access of Ply187 to the peptidoglycan of phage-infected Staphylococcus cells.
Among the tailed phages, newly synthesized virus particles are usually released from bacterial host cells following the synergistic action of a two-component lysis system: a hydrophobic membrane protein, termed holin, forms nonspecific pores or lesions in the cell membrane to promote access of a cell wall-hydrolyzing enzyme (endolysin) to the peptidoglycan substrate (44, 45). Such dual-component lysis systems, of which the phage λ S and R gene products represent the best studied prototypes, have recently been discovered in several phages of both gram-negative and gram-positive hosts. The system appears to be extremely heterogeneous, since at least 11 apparently unrelated holin gene families have been identified (8) which are associated with one or more of at least five endolysin enzyme functions, such as the hydrolysis of glycosidic linkages (by muramidases and glucosaminidases) and the hydrolysis of amide bonds (by amidases and peptidases) (24, 44).
Putative holin proteins may be identified by a number of characteristic properties: (i) the presence of encoding genes, usually located immediately upstream of the endolysin genes; (ii) the presence of at least two hydrophobic transmembrane domains with no net charge, separated by a short beta-turn linker; (iii) a highly charged, hydrophilic, C-terminal domain; and (iv) a dual-start motif [5′-ATG-(NNN)1 or 2-ATG-…-3′], not always present, permitting the synthesis of two products of different length, which are thought to regulate the pore-forming process (8, 45). Holin proteins range in size between 60 and 185 amino acids (22, 45).
At least for the phages of gram-positive hosts, however, the dual-component lysis system may not be universal. Although the presence of holins has been shown or suggested for several phages (2, 6, 10, 17, 22, 24, 27, 37, 41, 43), no genes encoding putative holins have yet been found to be associated with the endolysin genes of Listeria phage A511 (24) and Bacillus cereus phages (23). It is interesting that the N terminus of the endolysin of one of the Bacillus phages (TP21) shows extensive sequence homology to a signal sequence (including a cleavage site) from related Bacillus cell wall autolysins.
Staphylococcus aureus bacteriophage 187 is of special interest because it is the only member of staphylococcal bacteriophage species 187 and the sole representative of serogroup L phages (1). It differs from all other S. aureus phages by its host range, DNA restriction enzyme profiles (12), and distinctive set of virion proteins (21). Strains lysed by this phage have never been found to be lysed by any other phage (3), probably due to specific teichoic acids which are required for phage reception (29). We describe here the cloning and functional analysis of the unusual lysis system of this virus, which is comprised of a very small, putative holin (Hol187), whose coding sequence is fully embedded into the genetic module containing the enzymatically active domain of the large endolysin gene. We also provide evidence that the peptidoglycan hydrolase is related to major staphylococcal autolytic enzymes, and confirm the nonspecific membrane lesion-forming, holin-like nature of Hol187 by the complementation of a defective λ S allele.
MATERIALS AND METHODS
Organisms, plasmids, and culture conditions.
All bacterial strains, phages, and plasmids used throughout this study are listed in Table 1. Escherichia coli JM109(DE3) was used in initial detection assays and for the overexpression of the ply187 endolysin gene cloned in pSP72. JM109 and W3110 were used for the expression of hol187 and its derivatives, and LE392 and HB101 were hosts for the propagation and assay of λ phages. S. aureus 187 and Micrococcus luteus were grown in brain heart infusion broth or tryptose media at 37°C. Phage 187 was propagated on double-layer agar plates. Virus particles were concentrated from the lysates by polyethylene glycol 8000 precipitation and purified by ultracentrifugation on stepped CsCl gradients as previously described (33, 46). E. coli was grown in standard Luria-Bertani media (33) at 37°C. For the selection of plasmid-bearing cells, ampicillin was added at 100 μg/ml.
TABLE 1.
Bacterial strains, bacteriophages, and plasmids used throughout this study
| Strain, phage, or plasmid | Genotype or relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus 187 | Host for phage 187; assay for lytic activity | H. W. Ackermann |
| M. luteus WS2331 | Assay for lytic activity | Laboratory stock |
| E. coli | ||
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Laboratory stock |
| JM109(DE3) | ::DE3, T7 RNA polymerase under lacUV5 control | Promega |
| LE392 | F−hsdR514 supE44 supF58 lacY1 galK2 galT22 metB1 trpR55 (phage host; permissive for λgt11) | Promega |
| HB101 | F−hsdS20 supE44 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 (phage host; nonpermissive for λgt11) | Promega |
| W3110 | F− λ− | ATCC 27325 |
| Phages | ||
| 187 | Wild type, B1 siphovirus; approx. 45-kbp double-stranded DNA genome | H. W. Ackermann |
| λ | Wild type | ATCC 23724-B2 |
| λgt11 | cIts857; Sam100; lac promoter for expression of cloned genes | Promega |
| λgt11::hol187 | hol187 inserted into EcoRI site of λgt11 | This work |
| λgt11::hol187-S | Modified hol187 (encoding Hol187 ΔM1L2) inserted into EcoRI site of λgt11 | This work |
| λgt11::hol187-L | Modified hol187 (encoding Hol187 M3L) inserted into EcoRI site of λgt11 | This work |
| λgt11::hol187-Ch | Modified hol187 (encoding Hol187 V7D G8D) inserted into EcoRI site of λgt11 | This work |
| Plasmids | ||
| pSP72 | 2.4-kb cloning and expression vector; T7 promoter; Ampr | Promega |
| pBluescript II (SK−) | 3.0-kb cloning and expression vector; lac promoter; Ampr | Stratagene |
| pPL187 | 1,112-bp AluI fragment from phage 187 ligated into SmaI site of pSP72 | This work |
| pPL187-F1 | 471-bp PCR product (N-terminal domain of Ply187) inserted into BamHI site of pSP72 | This work |
| pPL187-F2 | 891-bp PCR product (C-terminally truncated Ply187) inserted into BamHI site of pSP72 | This work |
| pPL187-F3 | 1,887-bp PCR product (complete Ply187) inserted into BamHI site of pSP72 | This work |
| pPL187-F4 | 421-bp PCR product (central portion of Ply187) inserted into BamHI site of pSP72 | This work |
| pPL187-F5 | 1,416-bp PCR product (N-terminally truncated Ply187) inserted into BamHI site of pSP72 | This work |
| pHOL187 | 188-bp PCR product (encoding Hol187) (Fig. 1) inserted into EcoRI site of pBluescript | This work |
| pHOL187-S | 182-bp PCR product (encoding Hol187 ΔM1L2) inserted into EcoRI site of pBluescript | This work |
| pHOL187-L | 188-bp PCR product (encoding Hol187 M3L) inserted into EcoRI site of pBluescript | This work |
| pHOL187-Ch | 188-bp PCR product (encoding Hol187 V7D G8D) inserted into EcoRI site of pBluescript | This work |
Cloning and identification of the lysis genes.
Phage DNA was extracted and purified according to standard methods (33). For the construction of an expression library in E. coli, DNA was partially digested with AluI. Following electrophoresis in low-melting-point agarose, fragments in the range of 1,500 to 3,000 bp were recovered by β-Agarase digestion (Boehringer) and concentrated by ultrafiltration (Microcon 100; Amicon). DNA fragments were then ligated into pSP72, which had been linearized with SmaI.
Ligation reactions were electroporated (Gene-Pulser; Bio-Rad) into E. coli JM109(DE3), followed by the selection of plasmid-bearing cells on antibiotic plates. Lysin-expressing colonies were identified as previously described (24) by replica-plating of colonies on IPTG (isopropyl-β-d-thiogalactopyranoside)-containing agar plates, followed by incubation for 4 to 5 h at 37°C, chloroform vapor treatment, and overlay of the colonies with a concentrated suspension of S. aureus cells in 0.4% water-agar. After approximately 1 h of incubation at room temperature, clear zones of lysis could be observed around clones releasing a staphylolytic activity.
DNA sequencing and computer analysis.
The nucleotide sequence of the plasmid insert was determined on both strands by primer walking with synthetic oligonucleotides. The sequence downstream of the central AluI site in ply187 (see Fig. 1) was determined by direct sequencing of phage 187 DNA, by using primers as sequences became available. Sequenase, version 2.0 (U.S. Biochemicals), and α-35S-dATP (Amersham) was used in all sequencing reactions. The program DNAsis for Windows, version 2.01 (Hitachi), was used for the analysis of nucleotide and amino acid sequences.
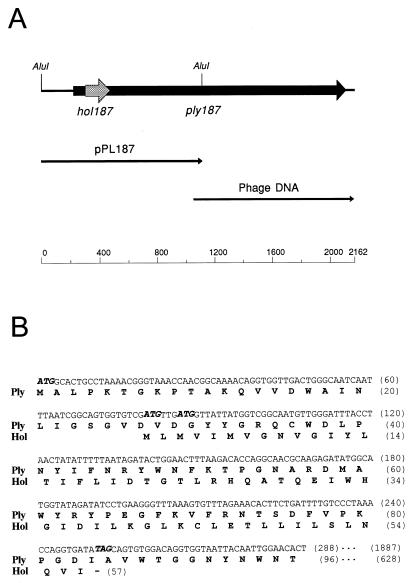
FIG. 1.
(A) Genetic map of the S. aureus phage 187 lysis gene region. The indicated AluI fragment was initially cloned and sequenced on plasmid pPL187, and the remaining nucleotide sequence was determined directly from phage DNA. (B) The holin gene hol187 is fully embedded in the 5′-terminal part of the endolysin gene ply187, in a different reading frame (+1). Deduced amino acid sequences of both gene products (Hol and Ply [partial]) are shown below the nucleotide sequence.
Cloning and expression of truncated Ply187 in E. coli.
To determine the enzymatically active domain(s) of Ply187, specific fragments of ply187 (see Fig. 3) were derived by PCR. For optimal expression, a consensus ribosomal binding site (RBS) and spacer to the ATG start codon was provided on the forward (Fwd) primers. The following forward and reverse (Rev) primers were used (RBSs are underlined and start codons and stop codons are shown in boldface): 1-Fwd, 5′-ACTTGGATCCGAGGAGAAATTACTATGGCACTGCCTAAAACGGG TAAACC-3′; 471-Fwd, 5′-ACTTGGATCCGAGGAGAAATTACTATGGCACAAAACAATCCTGCACCTAAAGAC-3′; 471-Rev, 5′-ACTTGGATCCTTATGGTGGTGTAGGTTTCGGTTCTGC-3′; 892-Rev, 5′-ACTTGGATCCTTAAGCTAATGACAAACATTCGATTTCATT-3′; and 1887-Rev, 5′-ACTTGGATCCTTATTTTTTATATTGATCGTATATAAAAT-3′. Purified phage 187 DNA (20 ng) was used as a template in the amplification reaction with Taq polymerase (Qiagen). PCR products were digested with BamHI and ligated into the BamHI site of pSP72. Transformation of plasmids, screening for clones expressing a lytic activity, and confirmation of correct insertion were done as described above. Finally, the lytic activity of the individual recombinant proteins was scored by comparing the sizes of the clearing zones, which appeared 0.5 to 1 h after overlaying with the indicator cells. For the possible differentiation of amidase from glucosaminidase activity, M. luteus cells were also tested here (38).
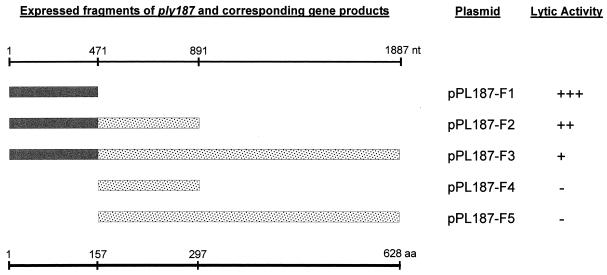
FIG. 3.
Determination of the enzymatically active domain of Ply187. PCR cloning was used to express the native ply187 gene and four fragments derived therefrom in E. coli. The solid bars represent the enzymatically active amidase domains, and the dotted bars indicate the C-terminal domains. The insert in pPL187-F2 corresponds to the DNA fragment initially cloned on plasmid pPL187.
Cloning and expression of hol187 in E. coli.
To investigate the effect of heterologous expression of hol187 in E. coli and to study specific alterations in Hol187, PCR was used for the construction of mutated hol187 genes by modified forward primers (Hol187, 5′-ATCAGAATTCGAGGAGAAATTAATATGTTGATGGTTATTATGGTCGGCAATGTTGGGATT-3′; Hol187-S, 5′-ATCAGAATTCGAGGAGAAATTAATATGGTTATTATGGTCGGCAATGTTGGG ATT-3′; Hol187-L, 5′-ATCAGAATTCGAGGAGAAATTAATATGTTGCTGGTTATTATGGTCGGCAATGTTGGGATT-3′; and Hol187-Ch, 5′-ATCAGAATTCGAGGAGAAATTAATATGTTGATGGTTATTATGGACGACAATG TTGGGATT-3′). The reverse primer was 5′-ATCAGAATTCTTATATCACCTGGTTTAGGGACAAAAT-3′ (see Table 1 for the individual products). The DNA fragments resulting from the amplification of phage 187 DNA were digested with EcoRI (Boehringer) and ligated into pBluescript II, followed by electroporation into JM109. This strain was selected because it features the laqIq mutation, which more efficiently controls the expression of genes cloned under control of the lac promoter. The nucleotide sequences and orientations of the cloned genes were verified by using primers complementary to the T7 and T3 promoters flanking the multiple cloning site in this vector. To investigate the effects of the four gene products on growth (i.e., cell viability) of plasmid-bearing E. coli, log-phase cultures (50-ml volume; optical density at 600 nm [OD600], approximately 0.1) were induced for gene expression by the addition of 1 mM IPTG, and growth (measured as optical density) was monitored over time.
Determination of total and released ATP during hol187 expression.
To determine the effect of Hol187 on the integrity (i.e., leakiness) of the E. coli cytoplasmic membrane, the release of ATP into the medium was determined following the overproduction of Hol187 in JM109(pHOL187). Cells were diluted (in a volume of 50 ml) to approximately 107 cells/ml, and hol187 expression was induced with 1 mM IPTG. Samples (1 ml) were taken at the indicated time points (see Fig. 6). The ATP assay was the highly sensitive firefly luciferase type and was carried out according to the instructions of the manufacturer (bioluminescence assay HS II; Boehringer). The emitted light was quantified in a photon-counting tube luminometer (Lumat 9501/16; Berthold), with a delay of 0.5 s and integration of the signal over the following 10 s.
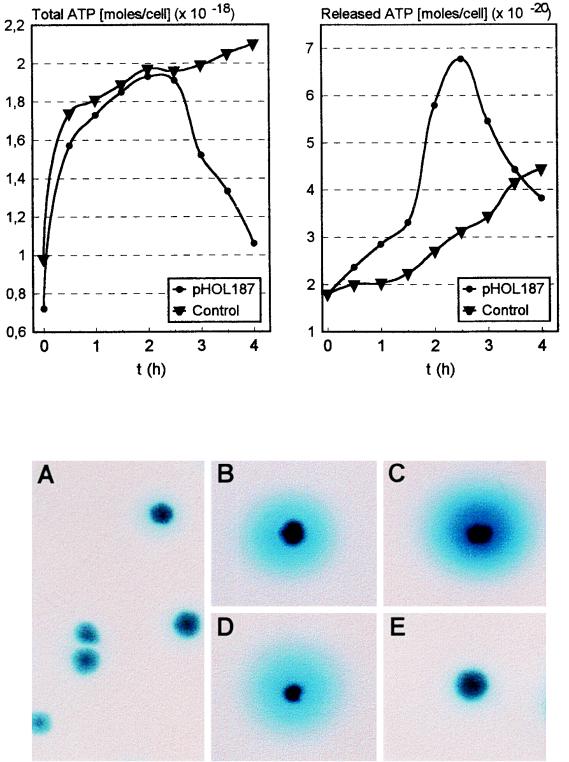
FIG. 6.
(Upper panels) Effect of hol187 expression on total cellular ATP content (left), and ATP release into growth medium (right), by E. coli cells harboring either pBluescript (control) or pHOL187. Gene expression was induced at time zero, and samples were taken and analyzed at the indicated time points. (Lower panels) Effect of the synthesis of Hol187 and mutated proteins on the release of β-galactosidase from E. coli W3110 cells into the medium (indicated by the blue zones around colonies) during growth on agar plates containing the chromogenic substrate X-Gal. (A) Control (W3110 without plasmid); (B) W3110(pHOL187); (C) W3110(pHOL187-S); (D) W3110(pHOL187-L); (E) W3110(pHOL187-Ch).
For the determination of total ATP, a cell lysis reagent (part of the HS-II kit) was added to the cell suspension in a 1:1 ratio before measurement. For the determination of released ATP, cells were pelleted by centrifugation (15,000 × g for 60 s) in a microcentrifuge (Eppendorf). An aliquot of the clear supernatant was then used for immediate measurement. The concentration of ATP was determined from comparison with a standard curve, which was prepared for the working range of the kit (10−12 to 10−16 mol of ATP). In order to calculate the ATP content or release in moles per cell, it was also necessary to determine the number of cells per milliliter at each timepoint. This was done by duplicate surface plating of the appropriate dilutions of the cultures under noninducing conditions (without IPTG), followed by incubation for 16 h and counting of the colonies.
Release of β-galactosidase.
The four plasmids specifying native Hol187 and mutated proteins (pHOL187, pHOL187-S, pHOL187-L, and pHOL187-Ch) were electroporated into E. coli W3110. This strain was used here because it produces native β-galactosidase. Transformants were plated directly onto agar plates containing the chromogenic substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma), and incubated for 24 h. W3110 control cells were plated on medium without antibiotic.
Immunological detection of Hol187.
To verify that Hol187 is actually made in phage-infected S. aureus cells, antibodies were raised in rabbits against a synthetic 18-mer peptide derived from the Hol187 amino acid sequence DTGTLRHQATQEIWHGID. A cysteine residue was added to the N-terminal end, and the peptide was subsequently conjugated to keyhole limpet hemocyanin. The conjugate was used to immunize rabbits, and the specific antibodies from the final bleeding were purified from the serum by affinity chromatography to the immunizing peptide, which had been coupled to an activated Sepharose column (Genosys Biotechnologies, Cambridge, United Kingdom). The resulting purified antibody fraction reacted with the immunizing peptide but showed no cross-reaction with S. aureus proteins, as determined in preliminary control blots. The preimmune serum obtained from the rabbits was tested in a control assay and yielded no signal (results not shown).
Infection of log-phase S. aureus cells (OD600, approximately 0.25) with phage 187 was done at a multiplicity of infection of 5 in a total volume of 200 ml of broth. Phages were allowed to adsorb for 15 min with continuous shaking at 37°C. Both phage-infected cells and the control cells (without phage) were then collected by centrifugation (6,000 × g for 8 min), resuspended in 300 ml of fresh, prewarmed broth, and further incubated for a total of 150 min. Small aliquots (1 ml) were taken every 15 min to monitor the time course of lysis by optical density (OD600) determinations. Cytoplasmic-membrane protein samples from the cells were prepared as described by Chang et al. (9), with modifications. Twenty-milliliter samples were taken from both cultures at time zero (just after infection) and every 15 min thereafter (see Fig. 6) and immediately cooled on ice. Cells were treated by sonication with an HD 2200 ultrasound sonicator (Sonopuls; Bandelin, Germany) equipped with an MS-72 titanium microtip, set for a pulsed mode for 5 min at a 25% duty cycle. After cell disruption, membranes were collected by centrifugation at 100,000 × g for 1 h, and pellets were extracted with 400 μl of ME buffer (10 mM Tris Cl [pH 8.0], 35 mM MgCl2, 1% Triton X-100) for 2 h at room temperature with shaking. Samples were then centrifuged again at 100,000 × g for 30 min to pellet the Triton X-100-insoluble material. Finally, membrane extracts were mixed with equal volumes of 2X sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and boiled for 3 min. In our initial Western blotting approach, small samples (10 μl) were electrophoresed on precast horizontal 15% polyacrylamide gels (ETC, Freiburg, Germany), in a Tris-Tricine-sodium dodecyl sulfate buffer system (34). This was followed by transfer of the proteins onto polyvinylidene difluoride membranes and immunological detection with various dilutions (from 1:100 to 1:10,000) of the purified Hol187 peptide antibody and an anti-rabbit immunoglobulin horseradish peroxidase-labeled secondary antibody (chemiluminescent Western blotting kit; Boehringer). However, the experiments failed to produce a signal of a protein of the desired size reacting with the purified antibody (results not shown). This may have been due to the very small size, extremely hydrophobic character, and low abundance of Hol187. We then used dot blotting in order to apply much larger volumes (100 μl) of each of the samples onto a polyvinylidene difluoride membrane, followed by immunoassay and chemiluminescent detection.
Complementation of the S gene lysis defect in λgt11.
E. coli host cells LE392 and HB101 were grown in Luria-Bertani media supplemented with 0.2% maltose and 10 mM MgSO4 (33). The PCR-generated hol187 genes were ligated (out of frame with lacZ) into the EcoRI site of λgt11 (5, 16, 22) under the control of Plac. Phage genomes were packaged according to the instructions of the manufacturer (Packagene system; Promega), and aliquots containing the recombinant virus particles were plated on soft-agar plates with LE392 or HB101 as hosts. A negative control (λgt11) and a positive control (λ wild type) were also plated. Following incubation for 16 h at 40°C, 10 individual plaques of each of the recombinant phages from the HB101 lawns were picked. The phages were eluted in 200 μl of SM buffer (33) and treated with a drop of chloroform, and the supernatant was replated on HB101. Again, single plaques were picked and the phages were eluted. The presence of the desired hol187 genes and mutations in the phage clones was verified by PCR amplification of the insert and subsequent nucleotide sequencing (results not shown).
Nucleotide sequence accession number.
The DNA sequence reported here appears in the EMBL, GenBank, and DDBJ databases under accession no. Y07740.
RESULTS
Cloning and sequencing of phage 187 lysis genes.
DNA purified from S. aureus phage 187 was shotgun-cloned into the inducible expression vector pSP72. Colonies of E. coli expressing a lytic activity could be readily identified by the clearing zones when overlaid with a lawn of S. aureus cells. Nucleotide sequencing of the plasmid insert and subsequent computer analysis allowed the identification of a putative open reading frame. However, we found that the sequence was incomplete, since no stop codon was present on the cloned fragment. Therefore, the remaining nucleotide sequence was determined directly from phage DNA (Fig. 1A). The ply187 gene (1,887 nucleotides) is preceded by an RBS (5′-GAAGTGAT-3′) with high sequence similarity to that of the 3′ end of the S. aureus 16S rRNA gene (5′-GAGGTGAT-3′ [26]). An inverted sequence repeat (stem-loop structure) is present at the immediate 3′ end of the gene (ΔG = −35 kJ/mol), which includes the TAA stop codon and probably functions as a transcriptional terminator.
Figure 1B shows the position and the deduced amino acid sequence of a second reading frame entirely embedded within the ply187 coding sequence in a different reading frame (+1), which was designated hol187 (see below). It is also preceded by a putative RBS (5′-AGTGGTG-3′).
Ply187 is related to major staphylococcal autolysins.
The complete ply gene product consists of 628 amino acids, has a calculated molecular mass of 71.6 kDa, and has a predicted pI of 9.9. Computer analysis showed no potential signal peptide in Ply187. Comparison of its amino acid sequence to those of other proteins included in some of the current databases (EMBL and GenBank) is presented in Fig. 2. The amino terminus of Ply187 shows 31% identity and 73% similarity over 113 amino acids (aa) to the N terminus of LytA from prophage φ11 of the lysogenic S. aureus strain NCTC 8325 (42), which is an N-acetylmuramoyl-l-alanine amidase (EC 3.5.1.28). Extensive homology was observed between the central-to-C-terminal domain of Ply187 and the C termini of the major autolysins Atl-A of S. aureus (11, 30) and Atl-E of Staphylococcus epidermidis (15). Both proteins show significant sequence identity (46 and 51%, respectively) and similarity (80 and 78%, respectively) to Ply187. Both Atl enzymes are first synthesized as single polypeptides equipped with signal peptides and propeptides of various sizes. However, the Atl enzymes are then processed to yield two functional lytic enzymes: a 60-kDa protein with amidase activity and a 51-kDa portion representing the C-terminal portion of the initial gene product with probable glucosaminidase activity (11, 15, 30). By deducing the sequence homologies to LytA and especially to the Atl enzymes, two domains were tentatively assigned to Ply187: the N terminus represents a probable amidase (17.8 kDa), and the C-terminal portion (53.7 kDa), whose function is unknown, might bear glucosaminidase activity (see Fig. 3). However, as reported below, no staphylolytic activity could be observed when the latter Ply187 domain was separately expressed and tested in the overlay assay.
FIG. 2.
(A) Alignment of the N-terminal amino acid sequences of Ply187 and LytA, an amidase endolysin from the S. aureus phage φ11. Identical amino acid residues and conservative replacements are indicated by vertical lines and colons, respectively. Homologous regions are shown in boldface. (B) Homology of the C-terminal domains of Ply187 and the major autolysins of S. aureus (Atl-A) and S. epidermidis (Atl-E). A few gaps have been introduced to optimize alignment.
Enzymatic activity of Ply187 is located in the N terminus.
The initially observed lytic activity resulted from the expression of only a fragment of ply187 on plasmid pPL187. The cloned fragment consists of 891 bp and enabled the synthesis of a polypeptide corresponding to the N-terminal 297 aa (33.9 kDa) of Ply187, which represents only 47% of the actual enzyme. This result strongly suggested that the catalytic site is located in the N terminus of Ply187. To further support this hypothesis, individual lytic activities of different fragments of Ply187 were determined by overproduction in E. coli (Fig. 3). Expression of the complete enzyme from plasmid pPL187-F3 yielded only relatively weak activity (small lysis zones), compared with the larger lytic zones observed around colonies expressing a fragment corresponding to the initially cloned 891-bp fragment (pPL187-F2). Surprisingly, the strongest activity resulted from the expression of the smallest fragment, i.e., the N-terminal 157 aa of Ply187 (pPL187-F1). In contrast, the polypeptides corresponding to amino acids 158 to 297 of the native enzyme (pPL187-F4) and amino acids 158 to 628 (pPL187-F5) produced no visible lysis in the overlay assays with S. aureus or M. luteus. These results indicated that the N terminus contains the only peptidoglycan-hydrolyzing activity of Ply187.
A holin-like gene is fully embedded in ply187.
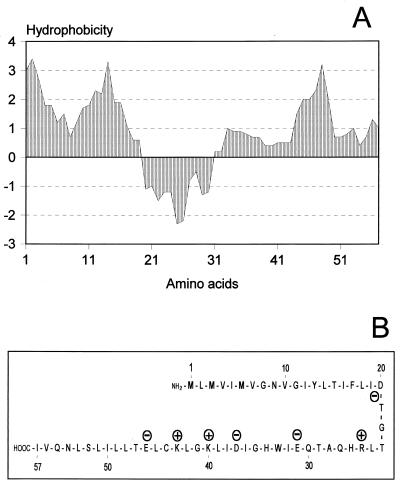
The small reading frame found within the part of the endolysin gene corresponding to the N terminus translates into a putative protein of 6.4 kDa (Fig. 1B and 4), with a predicted pI of 6.1. Its sequence reveals several properties characteristic of the holin protein family (45): (i) two stretches of generally hydrophobic amino acids with a neutral net charge (putative transmembrane helices); (ii) a hydrophilic beta-turn linker separating the membrane-spanning domains; and (iii) a dual-start motif (5′-ATG-TTG-ATG-…-3′), permitting the possible expression of two polypeptides of different lengths (57 and 55 amino acids, respectively). The distinct domains evident from the hydrophobicity plot (Fig. 4A) correlate well with the proposed secondary structure (Fig. 4B). Interestingly, the hol187 gene product lacks a highly charged C-terminal domain, present in most other holins identified to date (45). Moreover, its coding sequence is fully embedded (on the same strand) in a different reading frame (+1) within the endolysin gene, which has not previously been found for any of the known or suspected holin genes.
FIG. 4.
(A) Hydrophobicity analysis graph of Hol187, according to the algorithm of Kyte and Doolittle (20). Hydrophobic regions are in the area above zero, and the central hydrophilic domain is indicated in the area below zero. The mean hydrophobicity index is 0.97, which strongly indicates a membrane location. (B) Simplified schematic representation of the predicted secondary structure of Hol187 (antiparallel helices). Charged amino acids are indicated with plus and minus signs, respectively.
Hol187 is found in the cytoplasmic membrane of phage-infected cells.
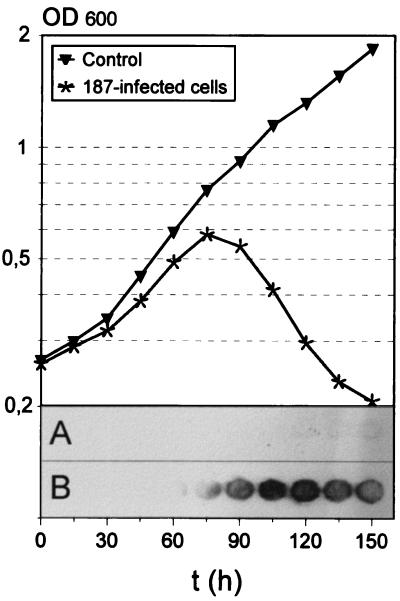
The lysis of S. aureus host cells infected by phage 187 is relatively slow (Fig. 5); cultures start to decrease in optical density at approximately 75 min postinfection and continue to clear during the following hour. Immunoblotting with specific peptide antibodies directed against Hol187 indicated that the protein is synthesized in S. aureus cells upon infection with phage 187 and enabled the detection of the protein in membrane fractions of these cells, whereas uninfected control cells yielded no signal. Since relatively large amounts of membrane extracts were necessary to produce clear signals, it appears that the protein is present in the cells in very small amounts.
FIG. 5.
(Upper panel) Time course of lysis of S. aureus cells infected with phage 187 at time zero, presented as optical density of the cultures. (Lower panel) Spatial appearance of Hol187 in membrane fractions of phage infected cells as revealed by immuno-dot blotting in control cells (A) and phage-infected cells (B). Numbers indicate the time in hours at which the individual samples were taken.
Synthesis of Hol187 in E. coli causes growth retardation and membrane damage.
The expression of hol187 from pHOL187 in E. coli JM109 following induction of the gene caused severe growth impairment, i.e., no further increase in the optical density of the cultures compared to the control. The same effect was observed with cells carrying a mutated hol187 gene (pHOL187-S), where the first two amino acids were deleted. Expression of only the longer, 57-aa product from plasmid pHOL187-L had a somewhat less detrimental effect. The introduction of two negatively charged amino acids into the first putative transmembrane domain of the protein (pHOL187-Ch) completely abolished the protein function; the culture afterward grew normally (results not shown).
To support the hypothesis that the retardation effect is due to membrane damage, we decided to quantify the amount of ATP synthesized and released from hol187-expressing E. coli cells (Fig. 6). The hol187 gene product led to the increased release of ATP from cells into the culture medium, compared to the control. At approximately 2.5 h postinduction, the total cellular ATP content showed a sharp, about twofold, decline whereas ATP synthesis in the control cells remained in a steady state. At the same time, the amount of ATP released from hol187-expressing cells suddenly dropped, from 2.5 h after induction up to the final measurement at 4 h, also approximately twofold. These data taken together indicate that intracellular synthesis and accumulation of Hol187 causes increasing leakiness of the cytoplasmic membrane and that once a critical concentration is reached (here, at about 2.5 h postinduction), de novo ATP synthesis collapses.
Our hypothesis that Hol187 has a membrane-damaging effect was further supported by the observation that the expression of the gene in E. coli W3110 led to release of the enzyme β-galactosidase into the periplasm (and eventually into the surrounding medium), as visualized by the formation of blue zones around colonies growing on X-Gal-containing agar plates (Fig. 6A through E). These results are in agreement with the above-mentioned growth inhibition, i.e., both variants (ΔM1L2 and M3L) of Hol187 have slightly different membrane-disturbing effects, whereas the V7D G8D mutation resulted in loss of activity.
hol187 can substitute for λ S.
Propagation (i.e., plaque formation) of λgt11 requires a host featuring supF, to compensate for the Sam100 mutation. To determine whether Hol187 could cause release of the R gene product into the periplasm, we tested the ability of recombinant λgt11 to form plaques on the nonpermissive host HB101. Results are presented in Table 2 and clearly show that complementation of the defective S allele with functional Hol187 (native, ΔM1L2, and M3L) allowed plaque formation. Again, λgt11::hol187-S produced somewhat larger plaques than λgt11::hol187-L, whereas the V7D G8D mutation in λgt11::hol187-Ch did not support R-mediated cell lysis. These data strongly suggest that Hol187 does, in fact, function as a holin.
TABLE 2.
Ability of Hol187 to complement the lysis defect (Sam100) of λgt11
| Phage | Plaque formation on host:
|
|
|---|---|---|
| LE392 | HB101 | |
| λgt11 | +++ | − |
| λ (wild type) | +++ | +++ |
| λgt11::hol187 | +++ | + |
| λgt11::hol187-S | +++ | ++ |
| λgt11::hol187-L | +++ | + |
| λgt11::hol187-Ch | +++ | − |
+++, large plaques; ++, medium plaques; +, small plaques; −, no plaques.
DISCUSSION
Many peptidoglycan-hydrolyzing (lytic) enzymes are composites of specifically adapted modules, encoding substrate recognition and hydrolysis domains. Such a modular organization was shown or proposed for several investigated lysins from phages or bacteria, where the catalytic activity is (almost always) located in the N-terminal region, while the C-terminal part contains the target-specific binding domains (4, 18, 22, 23, 24, 25, 35, 40, 41). It is shown here that the N-terminal domain (25%) of Ply187 contains the enzymatically active site, which, as deduced from its convincing homology to LytA, may act as an N-acetylmuramoyl-l-alanine amidase. Although the large C-terminal domain of Ply187 shows homology to the proposed glucosaminidase domains of the Atl proteins, the respective Ply187 fragments revealed no lytic activity. Moreover, C-terminal deletion of the enzyme (up to 75%) strongly increased lytic activity in the overlay assays. Thus, our results suggest that the C terminus does not represent an essential substrate recognition and/or binding domain, since it seems to be dispensable for staphylolytic activity. Therefore, the actual in vivo function of this large domain of Ply187 remains to be investigated. In this context, it would be interesting to clarify whether Ply187 exerts its in vivo activity as a full-length polypeptide or, as do the Atl enzymes, might also be posttranslationally processed to yield two polypeptides. A site similar to the one which is proteolytically cleaved in AtlA (Ala776 [30]) can also be tentatively identified in Ply187 (Ala158) by amino acid sequence alignment (results not shown). This would separate the proposed amidase domain from the large C-terminal part but would still leave open the hypothetical function of the latter.
In several cases, lytic phage enzymes are thought to be closely related to the lytic enzymes harbored by their individual host bacteria (autolysins and others), since extensive sequence similarities were found in specific domains (functional modules) of the respective enzymes (13, 22, 23, 35). With Ply187, we found striking homology to major staphylococcal cell wall hydrolases, i.e., the Atl autolysins. Therefore, it is tempting to speculate that the different domains (i.e., genetic modules) of Ply187 could have been acquired or exchanged through horizontal gene transfer between phage DNA and the bacterial genome.
We identified the small hol187 coding sequence, translated from a different reading frame, fully embedded within ply187. Holin genes are generally found to be closely associated with the endolysin sequences on the phage genomes (i.e., directly upstream of the peptidoglycan hydrolase gene), and they may overlap the latter by a few base pairs to allow translational coupling to occur (22, 44). The observation that hol187 is fully embedded out of frame within the endolysin gene on the same strand is an unusual finding. In fact, coding sequences entirely encompassed within other genes seem to be very rare among life forms having double-stranded DNA as genetic material. A relevant example is λ Rz, which contains another small gene (Rz1) translatable from a different reading frame (14). Although Rz1, a small lipoprotein localized in the outer membrane of phage infected E. coli cells, may somehow be involved in the lysis event (19), it is clearly not a holin.
We have shown that Hol187 is synthesized in phage-infected S. aureus cells prior to cell lysis as a late gene product and that it is present in the cytoplasmic membrane. The best indication, however, that Hol187 functions as a holin is that it is able to complement a defective λ S allele and allows propagation and plaque formation by recombinant phages in the nonsuppressive HB101 background. Moreover, the gene product shows all properties characteristic of the group of small, hydrophobic proteins termed holins (44), except that it is the first example described that lacks the highly charged C terminus common to other holins (45). It was previously thought that this region is crucial for correct orientation and/or oligomerization in the cytoplasmic membrane. However, the results of a recent study on C-terminally truncated λ S holin indicated that the hydrophilic C terminus is actually nonessential (32). Hol187 belongs to the class II holins (with only two possible membrane-spanning domains) and is the smallest holin protein described to date. The introduction of a negative charge into the first proposed transmembrane domain of Hol187 resulted in loss of activity. This supports the present model, which suggests that the overall net charge of the membrane-spanning domains must be neutral.
The hol187 gene reveals two possible translational start codons (ATG), separated by a TTG encoding a leucine residue. It is interesting that the latter codon itself may also serve as a translational start, which is not uncommon in staphylococcal genes (16, 28, 31, 39) and has recently been found for the (entirely different) holin gene of S. aureus phage Twort (22). However, it seems unlikely that the dual ATG codons of hol187 are coincidental, because this motif is a frequent and characteristic feature of many holin genes (8). When the mutated hol187 variants were expressed in an E. coli background, we observed that the full-length polypeptide (Hol187-L; 57 aa) may be somewhat less active than the shorter product translated from the second possible start codon (Hol187-S; 55 aa). In the best studied model of holin function (phage λ), this difference seems to be more pronounced: the longer S-107 polypeptide inhibits membrane hole formation and was reported not to cause lysis by itself (7, 9). However, its independent overexpression from a high-copy-number plasmid under the control of the lac promoter also resulted in cell lysis (36). While the opposing nature of the two S variants is explained by different N-terminal net charges, the two possible hol187 products are separated by an uncharged leucine residue. At present, it is entirely unclear how Hol187 function may be regulated in its natural host.
Compared to λ S, Hol187 is relatively slow acting. Low-level background expression from plasmid vectors is slightly inhibitory but not strictly lethal for the cells, whereas promoter induction results in membrane lesions and cell starvation. In phage-infected S. aureus cells, lysis begins at approximately 75 min and continues up to 150 min. Compared to λ-infected E. coli cells, this is more than twice the time needed to complete an infective cycle. This may simply reflect the different timing of lysis, which should not terminate the infective cycle before particle morphogenesis is complete. Nevertheless, we have shown here that Hol187 functions as a holin: it causes release of intracellular molecules into the periplasm and, most notably, is able to substitute for λ S, i.e., it promotes access of the heterologous R-gene product to the E. coli murein.
In conclusion, we found compelling evidence that the embedded hol187 gene encodes a holin-like protein, whose probable function is to permeabilize the staphylococcal cytoplasmic membrane to permit access of the Ply187 amidase endolysin to the cell wall. Hol187 is novel in more than one respect and joins the 11 apparently unrelated holin families currently known (45). These findings indicate that there is an even greater heterogeneity of holin genes (i.e., in gene families) than previously established. Even though the primary amino acid sequences of holins are totally dissimilar, their functional characteristics are well conserved.
ACKNOWLEDGMENTS
We are grateful to Hans Ackermann for supplying phage 187 and the host strain, to Patrick Schiwek for his excellent technical assistance, to Ingo Krause for his valuable advice in performing the affinity purification of Hol187 antibodies, and to Nataša Vukov for help in performing some of the gt11 experiments. We also thank Ry Young for critically reading a first draft of the manuscript.
REFERENCES
- 1.Ackermann H-W, Dubow M S, editors. Viruses of procaryotes. II. Natural groups of bacteriophages. Boca Raton, Fla: CRC Press, Inc.; 1987. [Google Scholar]
- 2.Arendt E K, Daly C, Fitzgerald G F, van de Guchte M. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of genes encoding lysin, a putative holin, and two structural proteins. Appl Environ Microbiol. 1994;60:1875–1883. doi: 10.1128/aem.60.6.1875-1883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asheshov E A, Jevons M P. The effect of heat on the ability of a host strain to support the growth of a Staphylococcus phage. J Gen Microbiol. 1963;31:97–107. doi: 10.1099/00221287-31-1-97. [DOI] [PubMed] [Google Scholar]
- 4.Baba T, Schneewind O. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 1996;15:4789–4797. [PMC free article] [PubMed] [Google Scholar]
- 5.Berkmen M, Benedik M J, Bläsi U. The Serratia marcescens NucE protein functions as a holin in Escherichia coli. J Bacteriol. 1997;179:6522–6524. doi: 10.1128/jb.179.20.6522-6524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birkeland N-K. Cloning, molecular characterization, and expression of the genes encoding lytic functions of lactococcal bacteriophage φLC3: a dual lysis system of modular design. Can J Microbiol. 1994;40:658–665. doi: 10.1139/m94-104. [DOI] [PubMed] [Google Scholar]
- 7.Bläsi U, Chang C-Y, Zagotta M T, Nam K, Young R. The lethal lambda S gene encodes its own inhibitor. EMBO J. 1990;9:981–989. doi: 10.1002/j.1460-2075.1990.tb08200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bläsi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 9.Chang C-Y, Nam K, Young R. S gene expression and timing of lysis by bacteriophage λ. J Bacteriol. 1995;177:3283–3294. doi: 10.1128/jb.177.11.3283-3294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz E, Munthali M, Lünsdorf H, Höltje J-V, Timmis K N. The two-step lysis system of pneumococcal bacteriophage EJ-1 is functional in gram negative bacteria: triggering of the major pneumococcal autolysin in E. coli. Mol Microbiol. 1996;19:667–681. doi: 10.1046/j.1365-2958.1996.399929.x. [DOI] [PubMed] [Google Scholar]
- 11.Foster S J. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J Bacteriol. 1995;177:5723–5725. doi: 10.1128/jb.177.19.5723-5725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaeng S. Identifizierung, Klonierung und Sequenzierung der Endolysin Gene aus zwei Staphylococcus aureus Bakteriophagen. Diploma thesis. Tübingen and Munich, Germany: Universität Tübingen and Technische Universität München; 1995. [Google Scholar]
- 13.García P, García J L, García J M, Sánchez-Puelles J M, López R. Modular organization of the lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Gene. 1990;86:137–142. doi: 10.1016/0378-1119(90)90116-9. [DOI] [PubMed] [Google Scholar]
- 14.Hanych B, Kedzierska S, Walderich B, Uznanski B, Taylor A. Expression of the Rz gene and the overlapping Rz1 reading frame present at the right end of the bacteriophage lambda genome. Gene. 1993;129:1–8. doi: 10.1016/0378-1119(93)90689-z. [DOI] [PubMed] [Google Scholar]
- 15.Heilmann C, Hussain M, Peters G, Götz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 16.Heinrich P, Rosenstein R, Böhmer M, Sonner P, Götz F. The molecular organization of the lysostaphin gene and its sequences repeated in tandem. Mol Gen Genet. 1987;209:563–569. doi: 10.1007/BF00331163. [DOI] [PubMed] [Google Scholar]
- 17.Henrich B, Binishofer B, Bläsi U. Primary structure and functional analysis of the lysis genes of Lactobacillus gasseri bacteriophage φadh. J Bacteriol. 1995;177:723–732. doi: 10.1128/jb.177.3.723-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joris B, Englebert S, Chu C-P, Kariyama R, Daneo-Moore L, Shockman G D, Ghuysen J-M. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEMS Microbiol Lett. 1992;70:257–264. doi: 10.1016/0378-1097(92)90707-u. [DOI] [PubMed] [Google Scholar]
- 19.Kedzierska S, Wawrzynow A, Taylor A. The Rz gene product of bacteriophage lambda is a lipoprotein localized in the outer membrane of Escherichia coli. Gene. 1996;168:1–8. doi: 10.1016/0378-1119(95)00712-1. [DOI] [PubMed] [Google Scholar]
- 20.Kyte J, Doolitle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Lee J S, Stewart P R. The virion proteins and ultrastructure of Staphylococcus aureus bacteriophages. J Gen Virol. 1985;66:2017–2027. doi: 10.1099/0022-1317-66-9-2017. [DOI] [PubMed] [Google Scholar]
- 22.Loessner M J, Gaeng S, Wendlinger G, Maier S K, Scherer S. The two-component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol Lett. 1998;162:265–274. doi: 10.1111/j.1574-6968.1998.tb13008.x. [DOI] [PubMed] [Google Scholar]
- 23.Loessner M J, Maier S K, Daubek-Puza H, Wendlinger G, Scherer S. Three Bacillus cereus bacteriophage endolysins are unrelated but reveal high homology to cell wall hydrolases from different bacilli. J Bacteriol. 1997;179:2845–2851. doi: 10.1128/jb.179.9.2845-2851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loessner M J, Wendlinger G, Scherer S. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol Microbiol. 1995;16:1231–1241. doi: 10.1111/j.1365-2958.1995.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 25.Longchamp P F, Mauël C, Karamata D. Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-l-alanine amidase gene of prophage PBSX. Microbiology. 1994;140:1855–1867. doi: 10.1099/13500872-140-8-1855. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig W, Kirchof G, Klugbauer N, Weizenegger M, Betzl D, Ehrmann M, Hertel C, Jilg S, Tatzel R, Zitzelsberger H, Liebl S, Hochberger M, Shah J, Lane D, Wallnoef P R. Complete 23S ribosomal RNA sequences of gram-positive bacteria with a low G+C content. Syst Appl Microbiol. 1992;15:487–501. [Google Scholar]
- 27.Martín A C, Löpez R, García P. Functional analysis of the two-gene lysis system of the pneumococcal phage Cp-1 in homologous and heterologous host cells. J Bacteriol. 1998;180:210–217. doi: 10.1128/jb.180.2.210-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLaughlin J R, Murray C L, Rabinowitz J C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus β-lactamase gene. J Biol Chem. 1981;256:11283–11291. [PubMed] [Google Scholar]
- 29.Oeding P. Cellular antigens of staphylococci. Ann NY Acad Sci. 1974;236:15–21. doi: 10.1111/j.1749-6632.1974.tb41479.x. [DOI] [PubMed] [Google Scholar]
- 30.Oshida T, Sugai M, Komatsuzawa H, Hong Y-M, Suginaka H, Tomasz A. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence analysis and characterization. Proc Natl Acad Sci USA. 1995;92:285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recsei P A, Gruss A D, Novick R P. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc Natl Acad Sci USA. 1987;84:1127–1131. doi: 10.1073/pnas.84.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rietsch A, Fraisl P, Grasschopf A, Bläsi U. The hydrophilic C-terminal part of the lambda S holin is non-essential for intermolecular interactions. FEMS Microbiol Lett. 1997;153:393–398. doi: 10.1111/j.1574-6968.1997.tb12601.x. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Schägger H, von Jagow G. Tricine sodium dodecylsulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–397. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 35.Sheehan M M, Garcia J L, López R, Garcia P. The lytic enzyme of the pneumococcal phage Dp-1: a chimeric lysin of intergeneric origin. Mol Microbiol. 1997;25:717–725. doi: 10.1046/j.1365-2958.1997.5101880.x. [DOI] [PubMed] [Google Scholar]
- 36.Steiner M, Bläsi U. Charged amino-terminal amino acids affect the lethal capacity of lambda lysis proteins S107 and S105. Mol Microbiol. 1993;8:525–533. doi: 10.1111/j.1365-2958.1993.tb01597.x. [DOI] [PubMed] [Google Scholar]
- 37.Steiner M, Lubitz W, Bläsi U. The missing link in phage lysis of gram-positive bacteria: gene 14 of Bacillus subtilis phage φ29 encodes the functional homolog of lambda S protein. J Bacteriol. 1993;175:1038–1042. doi: 10.1128/jb.175.4.1038-1042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugai M, Komatsuzawa H, Akiyama T, Hong Y-M, Oshida T, Miyake Y, Yamaguchi T, Suginaka H. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J Bacteriol. 1995;177:1491–1496. doi: 10.1128/jb.177.6.1491-1496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlen M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. Complete sequence of the staphylococcal gene encoding protein A. J Biol Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]
- 40.Usobiaga P, Medrano F J, Gasset M, Garcia J L, Saiz J L, Rivas G, Laynez J, Menendez M. Structural organization of the major autolysin from Streptococcus pneumoniae. J Biol Chem. 1996;271:6832–6838. doi: 10.1074/jbc.271.12.6832. [DOI] [PubMed] [Google Scholar]
- 41.Vasala A, Välkkila M, Caldentey J, Alatossava T. Genetic and biochemical characterization of the Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H lysin. Appl Environ Microbiol. 1995;61:4004–4011. doi: 10.1128/aem.61.11.4004-4011.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Wilkinson B J, Jayaswal R K. Sequence analysis of a Staphylococcus aureus gene encoding a peptidoglycan hydrolase activity. Gene. 1991;102:105–109. doi: 10.1016/0378-1119(91)90547-o. [DOI] [PubMed] [Google Scholar]
- 43.Weerakoon L K, Jayaswal R K. Sequence analysis of the region upstream of a peptidoglycan hydrolase-encoding gene from bacteriophage φ11 of Staphylococcus aureus. FEMS Microbiol Lett. 1995;133:9–15. doi: 10.1111/j.1574-6968.1995.tb07853.x. [DOI] [PubMed] [Google Scholar]
- 44.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young R, Bläsi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 46.Zink R, Loessner M J. Classification of virulent and temperate bacteriophages of Listeria spp. on the basis of morphology and protein analysis. Appl Environ Microbiol. 1992;58:296–302. doi: 10.1128/aem.58.1.296-302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]