Summary
Although the best-known spinocerebellar ataxias (SCAs) are triplet repeat diseases, many SCAs are not caused by repeat expansions. The rarity of individual non-expansion SCAs, however, has made it difficult to discern genotype-phenotype correlations. We therefore screened individuals who had been found to bear variants in a non-expansion SCA-associated gene through genetic testing, and after we eliminated genetic groups that had fewer than 30 subjects, there were 756 subjects bearing single-nucleotide variants or deletions in one of seven genes: CACNA1A (239 subjects), PRKCG (175), AFG3L2 (101), ITPR1 (91), STUB1 (77), SPTBN2 (39), or KCNC3 (34). We compared age at onset, disease features, and progression by gene and variant. There were no features that reliably distinguished one of these SCAs from another, and several genes—CACNA1A, ITPR1, SPTBN2, and KCNC3—were associated with both adult-onset and infantile-onset forms of disease, which also differed in presentation. Nevertheless, progression was overall very slow, and STUB1-associated disease was the fastest. Several variants in CACNA1A showed particularly wide ranges in age at onset: one variant produced anything from infantile developmental delay to ataxia onset at 64 years of age within the same family. For CACNA1A, ITPR1, and SPTBN2, the type of variant and charge change on the protein greatly affected the phenotype, defying pathogenicity prediction algorithms. Even with next-generation sequencing, accurate diagnosis requires dialogue between the clinician and the geneticist.
Keywords: Spinocerebellar Ataxia, SCA, CACNA1A, PRKCG, AFG3L2, ITPR1, STUB1, SPTBN2, KCNC3, onset
This study describes 756 non-expansion ataxia-affected individuals bearing variants in CACNA1A, PRKCG, AFG3L2, ITPR1, STUB1, SPTBN2, or KCNC3. Ages at onset can range over six decades, in some cases for the very same variant. Moreover, some variants predicted to exert little effect can actually be quite deleterious.
Introduction
The autosomal-dominant spinocerebellar ataxias (SCAs) are a clinically and genetically heterogeneous group of progressive neurological diseases characterized by cerebellar atrophy that can be accompanied by dysfunction in the brainstem, basal ganglia, cerebral cortex, spinal cord, or peripheral nervous system.1 There are three major classes of SCA, defined by molecular mechanism. The most common are the polyglutamine SCAs, which are caused by the expansion of a translated CAG repeat;2 together these account for ∼60% of SCAs worldwide, with geographical variations.3 The two remaining classes are non-coding repeat expansions and non-expansion SCAs caused by single-nucleotide variants (SNVs) or genomic rearrangements such as microdeletions, duplications, or insertions.4 CACNA1A (MIM: 601011) is the exception to the rule: it contains CAG repeats that can undergo expansion, causing SCA6 (MIM: 183086) but it can also carry SNVs.5 Nonetheless, the molecular basis of many SCAs has yet to be identified, and it seems likely that new mutational classes will emerge, as suggested by the recent discovery of splice variants in PNPT1 (MIM: 610316) (SCA25 [MIM: 608703]).6
One of the great challenges of the ataxias is their phenotypic heterogeneity, which has been easiest to appreciate amongst the polyglutamine SCAs. Because the CAG repeat tends to expand upon intergenerational transmission, each subsequent generation develops a more severe phenotype that appears at earlier ages, a phenomenon known as anticipation.7 In some cases, the presentation in the child is very different from the ataxia that afflicted the parent in middle age; the child of an adult with SCA7 (MIM: 164500), for example, might first present with vision loss or mood changes.8 Not only is there phenotypic heterogeneity within families, but the same repeat length can cause age of onset to vary by decades in different families. Some heterogeneity has been observed in the non-expansion SCAs, but their rarity has made it difficult to assess genotype-phenotype correlations.
To better understand the clinical and genetic features of non-expansion SCAs, we formed an international collaboration to gather the greatest number of affected individuals that we could. The phenotypic heterogeneity we observed among non-expansion SCAs far exceeds even that seen in the polyglutamine SCAs: in some cases, the very same variant can cause either a late-onset pure ataxia or a complex infantile syndrome with intellectual disability.
Subjects and methods
Subjects
This multicenter cross-sectional study was initiated (by P.C., A.B., and A.D.) in October 2020 with neurologists, clinical geneticists, and pediatricians (Figure S1) under the following institutional review board approvals: Paris Brain Institute (SPATAX RBM01-29/BIOMOV APH210069 Sud-est IV. 2021-A00989-32); Fondazione IRCCS Istituto Neurologico Carlo Besta (Care4NeuroRare CP 20/2018); University of Brescia (SCA OBSERVE NP 4506); University of Tubingen (598/2011BO1); University of Bonn (ATAXIE/HSP 176/16); University of Düsseldorf (ATAXIE 2020-981); University of Cincinnati (2017-5985). Anonymized information on affected individuals diagnosed with non-expansion mutations in known SCA-associated genes was obtained from genetic centers in Europe, the US, Israel, and the French West Indies (Table S1).
We used REDCap electronic data capture tools to collect and manage data, which were hosted and insured by the Paris Brain Institute (Institut du Cerveau, ICM).9 Collaborators were granted database access and an internal ID by formal request. As required by the respective institutional review boards of all centers, all person information in the database is anonymized (no name, date or place of birth, or ethnicity information). We performed quality control for missing or aberrant data. Of 1,316 potential subjects, 353 had incomplete data, which left 963 subjects in November 2021.
Classification of genetic variants
The 963 subjects carried either a missense, null, frameshift, or splice site variant or an in-frame deletion or a large deletion in one of 22 SCA-associated genes (Table S2). To increase the robustness of our phenotypic characterization, we excluded genes that were represented by fewer than 30 affected individuals. For the remaining seven genes, investigators met regularly via video conference to perform data complementation with quality control to ensure correct classification of variants (Table S3). We adopted American College of Medical Genetics and Genomics (ACMG) guidelines10 and performed manual curation by using the gnomAD database to refine the public frequency threshold (https://gnomad.broadinstitute.org; <0.01% for heterozygous variants). We used multiple algorithms to predict pathogenicity: BayesDel_addAF, DANN, DEOGEN2, EIGEN, SIFT, FATHMM-MKL, LIST-S2, M-CAP, MVP, MutationAssessor, MutationTaster, and PrimateAI.
Among the remaining 963 affected individuals, there were 297 pathogenic variants (189 missense, 37 large deletions, 24 frameshift, 21 nonsense and 16 splice site variants and ten in-frame deletions), of which 291 were initially classified as ACMG class V or IV. There were eight class I/II variants and 92 variants of unknown significance (VUSs, class III). Six VUSs were then reclassified as pathogenic or likely pathogenic following functional testing for two variants in AFG3L2 (MIM: 604581), protein modeling for three variants in PRKCG (MIM: 176980) by SwissModel webserver,11 and recurrence among different index cases with phenotypes compatible with one previously published variant in STUB1 (MIM: 607207).12 Carriers of the remaining 86 VUSs were not included in this study, leaving 756 subjects in the final cohort, of which 449 are familial cases (164 in the CACNA1A group; 83 in PRKCG; 51 in AFG3L2; 48 in STUB1; 63 in ITPR1 [MIM: 147265]; 20 in SPTBN2 [MIM: 604985]; and 20 in KCNC3 [MIM: 176264]).
Phenotypic characterization
We perused the clinical information associated with each subject to gather information pertaining to phenotypes. We defined age at onset (AO) as the age of ataxia onset reported by the affected individual, parent, or clinician, even though ataxia was not always the first abnormality detected. We assessed cerebellar signs by the scale for the assessment and rating of ataxia (SARA),13 other neurological signs by the Inventory of Non-Ataxia Signs (INAS),14 and functional impairment by the spinocerebellar degeneration functional score (SDFS).15 Pyramidal signs include hyperreflexia in four limbs and/or Babinski and/or Hoffmann signs. Intellectual disability (ID) refers to developmental cognitive delay, and cognitive deterioration refers to the loss of acquired cognitive skills. Psychiatric symptoms included anxiety, depression, obsessive-compulsive behavior, and psychotic episodes. Epilepsy in this study was defined as at least two unprovoked seizures and therefore excluded febrile seizures in infancy.
Statistical analysis
Demographic data and phenotypic features were summarized as frequencies with percentages for qualitative variables and medians with interquartile ranges ([IQR Q1–Q3]) for quantitative variables. Genetic groups were compared with chi-square tests for qualitative variables and Kruskal-Wallis or Wilcoxon tests for quantitative variables, with a conventional two-tailed type I error of 0.05.
Disease duration was defined as the number of years between ataxia onset and age at examination. For affected individuals with at least 3 years of disease, we calculated annual functional and SARA progression as the SDFS and the SARA scores, respectively, divided by the disease duration. We observed very large age at onset heterogeneity for each gene and aimed to identify significant age groups. We first tested for the presence of multimodality by using the R multimode package, with the excess mass test.16 If there was a significant multimodality, we estimated the Gaussian mixtures by using the expectation maximization algorithm for up to ten groups with the mixtools R package. We then selected the best fit on the basis of Bayesian information criteria.
We attempted to develop an AI decision tree to classify genes according to clinical features by using the R rpart package, which enables the use of data with missing values. All variables described in Table S4 were included in the model. We determined the best “splits” (separation of affected individuals according to clinical features) in the tree with the Gini index and pruned the tree by using the optimal complexity parameter with 10-fold cross-validation. Data were analyzed with R version 3.6.2.
Results
Cohort characteristics
The most salient feature of these seven genetic groups is that the ages at onset (AOs) ranged widely (Figure 1A), but the overall progression is slow (Figure S2). There was no detectable influence of sex on AO (Figure S3) or any other aspect of the clinical presentation (p > 0.50 after correction for multiple comparisons). Most disease-related features occurred in less than a quarter of the affected individuals in each group, meaning that it would be hard to distinguish one of these SCAs from another on a clinical basis alone (Figure 1B). Clinical diagnosis would be made even more challenging by the substantial phenotypic differences within certain SCAs (see below) (Table S4). Below we outline the main findings by genetic group, in descending order of cohort size.
Figure 1.
Age at onset distribution and disease features beyond ataxia in seven non-expansion SCAs
(A) Age at onset distribution for each of the seven genetic groups in our cohort, in descending size of group from top to bottom. The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles).
(B) The percentage of affected individuals in each group with signs beyond cerebellar ataxia. The majority of disease features affect fewer than one in four members of a given genetic group, indicating considerable heterogeneity.
CACNA1A
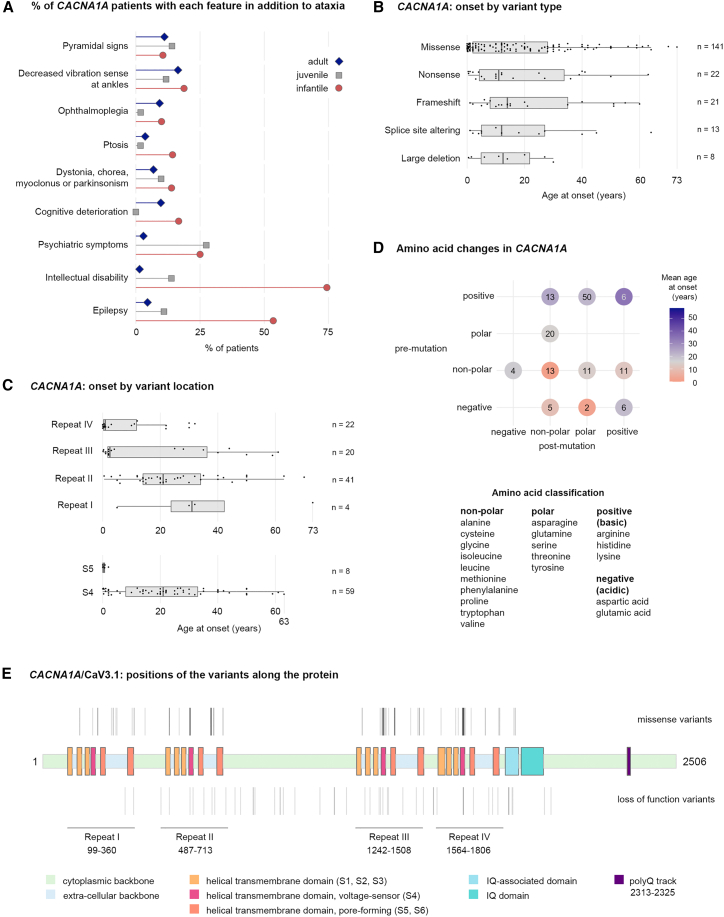
Different mutations in CACNA1A, which encodes the voltage-dependent P/Q-type calcium channel subunit alpha-1A (Cav2.1), have already been associated with a variety of phenotypes (familial hemiplegic migraine, episodic ataxia, and several types of epilepsy in addition to polyglutamine-expanded SCA6).17 The phenotypic heterogeneity in our cohort became more coherent when we grouped subjects by AO, which fell into three peaks determined by multimodality testing at 0.8 (±0.7), 9.2 (±5.2), and 33.7 (±16.1) years (for infantile, juvenile, and adult onset, respectively) (Figure 2A). The infantile form was most strongly associated with epilepsy and intellectual disability (Table S5). The juvenile form was the mildest in terms of SARA, the least likely to involve ophthalmoplegia or cognitive deterioration, but the most likely to include psychiatric symptoms. Adult affected individuals typically had pure cerebellar disease, although pyramidal signs developed with longer disease duration (Table S6). SARA and SDFS progression rates were the slowest in the whole cohort (Figure S2).
Figure 2.
Infantile, juvenile, and adult-onset CACNA1A phenotypes and their correlations with variant location and amino acid properties
(A) Infantile, juvenile, and adult-onset phenotypes differed in their clinical presentations, but no single feature was exclusive to one category.
(B) Distributions of ages at onset for each of the variant type observed in CACNA1A. The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles) for all boxplots in the figure.
(C) Distributions of ages at onset for each CACNA1A missense variant according to their location in the protein by repeat region. Variants within transmembrane segment S4 were found in all the repeats, but in segment S5 they were located only in repeats IV (n = 7) and III (n = 1) and exclusively associated with very-early-onset disease.
(D) Amino acid change properties also influence age at onset. On the left are the initial amino acid properties and at the bottom are the amino acid properties post-mutation. The majority of mutations affected positive residues.
(E) Diagram of CACNA1A shows the locations of missense and loss-of-function variants (above and below the gene, respectively) in this cohort.
AO did not depend much on variant type, although large deletions and altered splicing tended to produce symptoms before midlife (Figure 2B). Three variants presented a particularly wide range of onsets: c.1082G>A (p.Gly361Glu) (GenBank: NM_001127221.2) (0–64 years in six affected individuals from one family); c.1748G>A (p.Arg583Gln) (GenBank: NM_001127221.2) (5–63 years in 37 affected individuals from ten families); and c.5251C>T (p.Arg1751Trp) (GenBank: NM_001127221.2) (1–59 years in seven affected individuals from two families) (Table S3). This led us to examine variant types and location more closely (Figures 2C–2E). The variants in our cohort clustered in areas most pertinent to Cav2.1’s function as a calcium channel: four homologous repeat domains (I–IV) each contain six helical transmembrane segments (S1–S6), of which segment S4 serves as a voltage sensor, while the S5 and S6 segments and the S5–S6 loops form the channel pore that governs the passage of ions.18 Most variants in our cohort were located in segment S4 of repeat II (Figure 2C), which showed a six-decade range of AOs. Most infantile presentations were associated with variants in transmembrane segment S5 of repeat IV that affected non-polar amino acids without causing a change in charge. A large proportion of mutations caused a change of polarity (Figure 2D) and were associated with later onset.
PRKCG (SCA14)
SCA14 (MIM: 605361) had a wide range of AO (3–66 years) and showed such phenotypic heterogeneity that no single disease feature manifested in even one-fifth of affected individuals (Figure 1B; Table S4). The most frequent signs were psychiatric symptoms, decreased ankle vibration sense, cognitive deterioration, and the cluster of dystonia, chorea, myoclonus, or parkinsonism. This cluster also tended to correlate with pyramidal signs, whereas cognitive deterioration often co-occurred with ophthalmoplegia (Table S7).
PRKCG encodes protein kinase C gamma, whose phosphorylation function is activated by calcium. Most pathogenic variants (54/59) were missense mutations; there were three in-frame deletions and two frameshift mutations. No correlation was found between clinical features and variant type or location, although most variants altered non-polar amino acids and clustered in the phorbol-ester/diacylglycerol-binding region (Figure 3A, bottom).
Figure 3.
Amino acid properties and variant characteristics in PRKCG (SCA14) and AFG3L2 (SCA28)
(A) Top: distributions of ages at onset for each type of mutation observed in PRKCG. The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles) in all the boxplots in the figure. Frameshifts and indels seem to have a narrower range of onset, but the number of subjects is too small to judge with certainty. Middle: the majority of variants in PRKCG (70%) affected non-polar residues. Bottom: diagram of PRKCG showing the locations of variants in this cohort.
(B) Top: AFG3L2 missense variants produce a wide range of ages at onset. Middle: nearly half the AFG3L2 mutations affected non-polar residues. Bottom: diagram of AFG3L2 showing the locations of variants in our cohort.
AFG3L2 (SCA28)
SCA28 (MIM: 610246) presented a large range in AO (0–74 years), but only one affected individual had infantile onset. The two most prevalent features were ophthalmoplegia (50%) and ptosis (31%) (Figure 1B; Table S4), which could develop before ataxia but were typically associated with 7–10 years longer disease duration (Table S6). Pyramidal symptoms were also seen in affected individuals with greater (7.6 years) disease duration. Cognitive deterioration tended to co-occur with psychiatric symptoms and dystonia, chorea, myoclonus, and parkinsonism (Table S8). Only two affected individuals had ID.
AFG3L2 encodes AFG3-like matrix AAA peptidase subunit 2, part of an ATP-dependent complex of the mitochondrial inner membrane that regulates ribosome assembly and degrades misfolded proteins. Loss of function impairs respiratory chain activity and mitochondrial gene expression.19 Homozygous mutations in AFG3L2 can cause spastic ataxia 5 (SPAX5 [MIM: 147265]), which has similar features to SCA28.20 Thirty of the 31 variants were missense; one was a nonsense variant. There was no clear correlation between mutation type and polarity or charge, but most mutations occurred in the proteolytic domain (exons 15 and 16), while two were located in the exon 10 catalytic domain. (Figure 3B, bottom).
ITPR1 (SCA15/29)
Although only a handful of infantile SCA15/29 (MIM: 606658)/(MIM: 117360) cases have been reported in the literature,21 in our cohort ITPR1 mutations were associated with a one in three chance of infantile presentation (at 0.5 ± 0.5 years). Even the adult onset was relatively early (33.4 ± 14.9 years). Ophthalmoplegia and the cluster of dystonia, chorea, myoclonus, or parkinsonism were more likely to occur with infantile onset and missense mutations (Figure 4A, top; Table S9). Depression or anxiety, which developed with longer disease duration (11.5 years), were more likely with deletions. AO strongly correlated with the variant type: deletions were associated only with adult onset, but more than three-quarters of missense carriers had infantile disease (Figure 4A, middle; Table S9).
Figure 4.
Missense variants dominate infantile onset in ITPR1 (SCA15/29), but no pattern is discernible with STUB1 (SCA48)
(A) Top: infantile- and adult-onset ITPR1-affected individuals were differentiated primarily by the presence of intellectual disability. Middle: missense mutations in ITPR1 were strongly associated with infantile cases. Bottom: diagram of variant locations in ITPR1.
(B) Top: age at onset in STUB1-associated disease did not correlate with variant type. Middle: two-thirds of variants affected non-polar residues. Bottom: diagram of variant locations in STUB1.
ITPR1 encodes the inositol 1,4,5-triphosphate receptor 1 (also known as IP3R1), which mediates calcium release from the endoplasmic reticulum. This in turn leads to cytosolic and mitochondrial calcium spikes that stimulate mitochondrial energetics22; indeed, too much ITPR1 activity can cause mitochondrial Ca2+ toxicity.23 We found both deletions (>5 Mb) and missense variants distributed throughout the gene (Table S3, Figure 4A, bottom). One in-frame deletion (c.7786_7788del [p.Lys2596del] [GenBank: NM_001168272.2]) and two missense variants (c.7727A>T [p.Asn2576Ile] [GenBank: NM_001168272.2]; c.7616G>T [p.Gly2539Val]) presented a Gillespie syndrome phenotype (congenital hypotonia, bilateral aniridia, ataxia, and ID [MIM: 206700]) and were located in the transmembrane domain, like all Gillespie missense variants published so far.24
STUB1 (SCA48)
SCA48 (MIM: 618093) had a median age at onset of 40 years; there were no cases of infantile or juvenile onset or ID in our cohort. The STUB1 group were the most likely out of the entire cohort, however, to suffer cognitive deterioration (65%) and psychiatric symptoms (44%), which tended to co-occur (Table S10). These features could precede the ataxia by a decade or more, but it was the rapid disease progression that best predicted that an affected individual had a mutation in STUB1 (Table S4).
STUB1 (STIP1 homology and U-box containing protein 1) encodes the CHIP protein, an E3 ubiquitin ligase that influences autophagy and mitochondrial biogenesis.25 The protein contains tetratricopeptide repeats and a U-box, but the 33 heterozygous variants were distributed throughout the protein, with no discernible hotspots or correlations with particular disease features (Table S3, Figure 4B).
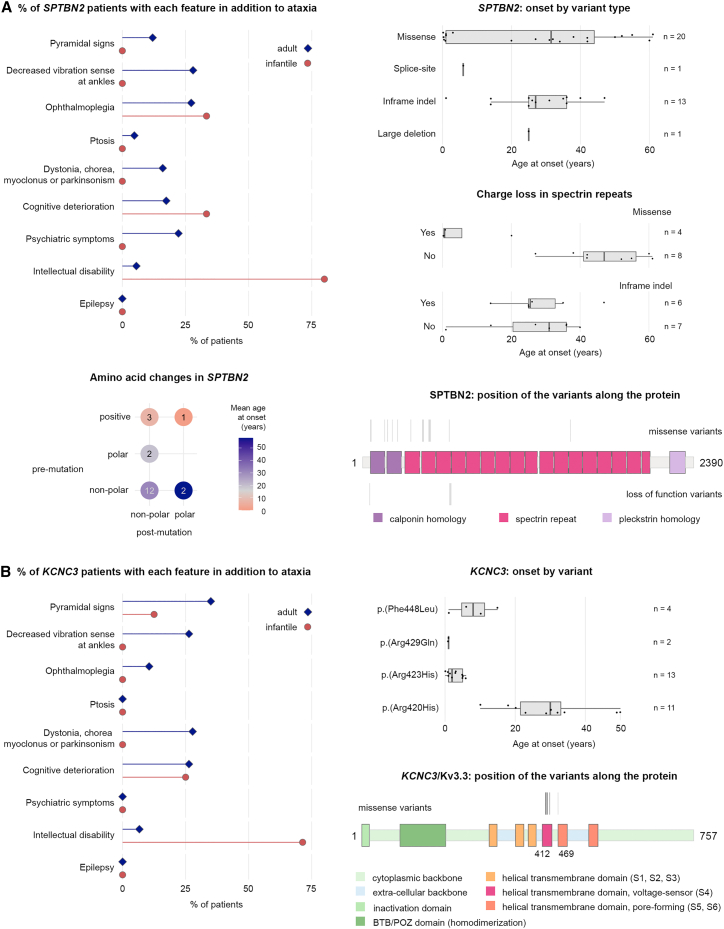
SPTBN2 (SCA5)
SCA5 (MIM: 600224) showed a bimodal distribution in AOs, with the multimodality distributions peaking at 0.7 ± 0.3 years (20%) and 33.3 ± 14.7 years (80%). Infantile-onset cases were associated with ID (80%), but 6% of the adult-onset group also showed ID (recall that we defined AO by ataxia) (Figure 5A, Table S11). Ophthalmoplegia was equally prevalent among infantile- and adult-onset individuals (24%) and associated with longer disease duration (Table S6). Oculomotor symptoms were the initial feature in the few persons that did not present with ataxia.
Figure 5.
Correlations between variant type and amino acid changes in SPTBN2 (SCA5) and KCNC3 (SCA13) with early- and later-onset phenotypes
(A) Left: the majority of infantile-onset SCA5-affected individuals have intellectual disability, which is strongly associated with missense variants and charge loss in the spectrin repeats. The majority of mutations changed the residue from one non-polar amino acid to another. Right: missense mutations were associated with the earliest age at onset (top) and with charge loss in a spectrin repeat (middle panel). The bottom right shows the location of variants, which favored the calponin homology domain and early spectrin repeats.
(B) Intellectual disability is also prominent in SCA13 infantile-onset disease (left), which is strongly associated with mutations affecting amino acids in the region following c.1259G>A (p.Arg420His) (GenBank: NM_004977.3) (right). Mutations clustered in the S4 and S5 domains (right, bottom).
SPTBN2 (spectrin beta non-erythrocytic 2) encodes the beta-III spectrin protein that stabilizes the glutamate transporter EAAT4 at the surface of the plasma membrane. Beta-III spectrin contains a calponin-homology domain, a series of spectrin repeats, and a pleckstrin homology domain. Heterozygous intragenic deletions and missense variants have been associated with infantile-onset disease (homozygosity leads to SCAR14 [MIM: 615386]).26 Most of the previously published variants were present in our cohort.27 We found no correlation between disease features and variant types but noted a strong correlation between loss of charge and earlier onset and a tendency for mutations to occur in the calponin homology domain or the first three spectrin repeats (Figure 5A). A change from a positive to a non-polar or polar amino acid in the helical spectrin repeats nearly always led to infantile onset.
KCNC3 (SCA13)
Overall, the SCA13 (MIM: 605259) group had the earliest median age at onset (5.5 years) (Table S4). Infantile onset was associated with intellectual disability (71% versus 6% of adult-onset individuals) and further cognitive deterioration (25%). Adult-onset individuals were more likely to present pyramidal signs (35%) as well as dystonia, chorea, myoclonus, and parkinsonism (28%) or decreased ankle vibration sense (26%) (Figure 5B, Table S12). Overall progression was slow (median 0.11 point of SDFS and 0.66 point of SARA per year).
KCNC3 encodes the potassium voltage-gated channel sub-family C member 3 (Kv3.3), which consists of six transmembrane segments and a pore re-entrant loop. There were only four missense pathogenic variants, all in exon 2, leading to amino acid substitutions in highly conserved domains in transmembrane segments S4 (the main voltage-sensing element) and S5 (ion-selective pore) (Figure 5B). Although these two variants are only three residues apart, the age at onset for c.1268G>A (p.Arg423His) (GenBank: NM_004977.3) is much earlier than for c.1259G>A (p.Arg420His) (GenBank: NM_004977.3), which is consistent with a previous report.28
Decision tree
We used a machine learning classifier to create a decision tree to aid diagnostic discrimination among these seven genes. The algorithm had a genotype-phenotype correlation accuracy of 48%, as opposed to 20% for random selection, based on five variables: onset before 16 years of age, presence of ophthalmoplegia, intellectual disability, cognitive deterioration, and either slow (<0.08/year) or fast (>0.13/year) disease progression, as measured by the SDFS (Figure S2). Onset before age 16 suggested CACNA1A, because it accounts for one-third of our cohort and the majority of early-onset individuals. Among the 71% of our sample with onset after 16 years, ophthalmoplegia suggested AFG3L2. When ophthalmoplegia was absent, cognitive deterioration suggested STUB1. The absence of a distinctive feature led to heterogeneous groups populated by PRKCG, which was the second-most numerous group after CACNA1A. To improve the result, we tried using a random forest classifier, but this required imputing many missing values; even so, the model achieved only ∼54% accuracy.
Discussion
This study has uncovered two crucial features of the non-expansion SCAs. First, these diseases are remarkably heterogeneous in presentation. The AFG3L2 variants in our cohort together presented a six-decade range in AOs, and three specific variants in CACNA1A achieved a comparably wide range. Infantile-, juvenile-, and adult-onset phenotypes for the same gene can differ considerably. This variability is particularly noteworthy given that neither imaging nor neuropathological studies in these SCAs have yet been able to discern structural abnormalities beyond the cerebellum. This distinguishes non-expansion SCAs from polyglutamine disorders and also raises the intriguing possibility that cerebellar dysfunction may be sufficient to cause cognitive decline or even intellectual disability.
Second, we cannot predict pathogenicity solely on the basis of the amino acid change. We were quite surprised that infantile-onset individuals in the CACNA1A group had variants that did not even change the charge of the residues. All these variants were located within the pore, which is the most sensitive region of the protein29,30; here, even seemingly inconsequential change leads to infantile-onset disease. Only electrophysiological studies can determine the functional consequences of amino acid changes. There were similarly conservative changes in SPTBN2. Protein prediction algorithms therefore cannot be relied on for clinical decision-making, even when multiple algorithms agree: the clinical presentation must inform our interpretation of variants of uncertain pathogenicity.21
Although certain variant types or locations were strongly associated with earlier-onset disease, the extreme heterogeneity we observed foiled our attempts to derive a diagnostic algorithm. We expect that genetic modifiers or at least genetic background effects explain much of this variability. For example, an interaction between STUB1 (SCA48) and TBP [MIM: 600075] (SCA17 [MIM: 607136]) has recently been identified in two studies. In STUB1-affected individuals, the cognitive phenotype and overall survival are driven by the size of the intermediate TBP allele (41–46 repeats, which are not pathogenic on their own).31,32 In this study, we were unable to test whether TBP was responsible for the more rapid progression of STUB1-affected individuals, but this question is relevant not only for nosology but for genetic counseling. We were also unable to analyze geographic or ethnic specificities because we did not include this information; future studies should shed light on genetic background effects.
Notwithstanding the overall clinical heterogeneity, the phenotype of these seven non-expansion SCAs can resemble that of mitochondrial ataxias, which are usually slowly progressive and often include loss of reflexes, ophthalmoplegia, ptosis, and sometimes cognitive impairment and seizures. Interestingly, AFG3L2, ITPR1, and STUB1 all influence mitochondrial function (AFG3L2, ITPR1, and STUB1), while the other proteins involved in these SCAs are involved in neurotransmission through transport of or response to calcium (CACNA1A, PRKCG), potassium (KCNC3), or glutamate (SPTBN2). It is conceivable that polymorphisms in mitochondrial or neurotransmission-related genes could act as genetic modifiers influencing the tremendous range in AOs, as has been found in subjects with polyglutamine SCAs33 and particularly in SCA7.34 The clear dominance of CACNA1A variants in our cohort certainly confirms the prevalence of channelopathies among the SCAs35 and the exquisite sensitivity of channels to even slight alterations in structure.
Last but not least, given the extreme heterogeneity and the difficulty of ascribing pathogenicity to variants via existing prediction tools, it may be that these non-expansion SCAs are not as rare as we imagine. It is conceivable that many individuals with isolated disease features or presentations that are non-standard for ataxia would be overlooked or not tested with an ataxia panel. Thus, despite being by far the largest study on non-expansion SCAs, there are likely many more affected individuals and disease features to be discovered in these diseases. The diagnostic reality is that affected individuals who do undergo testing will be missed by typical commercial ataxia panels. (Most of our subjects were tested in research settings: approximately 35% in our cohort were diagnosed by Sanger sequencing, 24% by panel screening, and 31% by whole-exome or whole-genome sequencing.) The results of the current study argue that sequencing should become standard practice if we are to improve diagnosis for ataxias. Even so, we must be alert to the fact that some variants that one would expect to be benign, such as those that do not alter the charge of the affected amino acid, can be pathogenic. The specific context of the affected domain needs to be taken into account when interpreting pathogenicity, as well as the possible presence of an additional modifying variant in another gene.
Acknowledgments
We thank the individuals and their families for participating in this study. We thank the following colleagues for their interest in the initiative and their support: Mathieu Barbier, Magali Barth, Nicole Bigot, Ariane Choumert, Alberto Espay, Lucile Gleyze, Nigel G. Laing, Mathilde Nizon, Olivier Patat, Violaine Planté-Bordeneuve, Caroline Roos, Richard Roxborough, Lyse Ruaud, Jeremy Schmahmann, and Stephan Zuchner. A special thanks to Vicky Brandt for substantive contributions to the manuscript. This project was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) No. 441409627 (to M.S., A.D., B.v.d.W., and F.M.S.) as part of the PROSPAX consortium under the frame of the European Joint Programme on Rare Diseases, under the EJP RD COFUND-EJP No. 825575; the US Department of Veterans Affairs, Seattle, Washington; the Fondazione Regionale per la Ricerca Biomedica (CP 20/2018 [Care4NeuroRare]) to F.T. and M.T.B.; and the Italian Ministry of Health (RRC) to F.T., M.T.B., C.M., and F.M.S. (RC5X1000). Many authors of this publication are members of the European Reference Network for Rare Neurological Diseases - Project ID No. 739510.
Author contributions
All coauthors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Concept and design: A.D. and A.B. Acquisition, analysis, or interpretation of data: all coauthors, with specific responsibilities for different genetic groups as follows. AFG3L2: C.G., D.D.B., F.T., P.C., S.M., and S.B. CACNA1A: E.I., F.R., G.C., L.B., M.S., P.C., P.P., and S.B. ITPR1: F.T., G.Z., J.S., and J.O. KCNC3: L.S. and C.R. PKRCG: A.G., B.V.d.W., E.B., M.M., P.C., and T.S.-H. SPTBN2: C.M., L.N., and P.C. STUB1: A.F., M.B., and M.S. Drafting of the manuscript: P.C., A.D., E.P., and M.C. Critical revision of the manuscript for important intellectual content: A.D. and A.B. Statistical analysis: E.P., M.C., and P.C. Administrative, technical, or material support: A.D. Supervision: A.D.
Declaration of interests
The authors declare no competing interest.
Published: June 9, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.05.009.
Web resources
Mixtools package, https://CRAN.R-project.org/package=mixtools
Rpart package, https://CRAN.R-project.org/package=rpart
Supplemental information
In cases where a feature was significantly more prominent in one group only, the percentage of affected individuals displaying that feature is in bold. Percentages were rounded to the nearest integer
Data and code availability
The dataset supporting the current study has not been deposited in a public repository but is available from the corresponding author on reasonable request.
References
- 1.Klockgether T., Mariotti C., Paulson H.L. Spinocerebellar ataxia. Nat. Rev. Dis. Primers. 2019;5:24. doi: 10.1038/s41572-019-0074-3. [DOI] [PubMed] [Google Scholar]
- 2.Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 2010;9:885–894. doi: 10.1016/S1474-4422(10)70183-6. [DOI] [PubMed] [Google Scholar]
- 3.Ruano L., Melo C., Silva M.C., Coutinho P. The Global Epidemiology of Hereditary Ataxia and Spastic Paraplegia: A Systematic Review of Prevalence Studies. Neuroepidemiology. 2014;42:174–183. doi: 10.1159/000358801. [DOI] [PubMed] [Google Scholar]
- 4.Nibbeling E.A.R., Duarri A., Verschuuren-Bemelmans C.C., Fokkens M.R., Karjalainen J.M., Smeets C.J.L.M., de Boer-Bergsma J.J., van der Vries G., Dooijes D., Bampi G.B., et al. Exome sequencing and network analysis identifies shared mechanisms underlying spinocerebellar ataxia. Brain. 2017;140:2860–2878. doi: 10.1093/brain/awx251. [DOI] [PubMed] [Google Scholar]
- 5.Jodice C., Mantuano E., Veneziano L., Trettel F., Sabbadini G., Calandriello L., Francia A., Spadaro M., Pierelli F., Salvi F., et al. Episodic ataxia type 2 (EA2) and spinocerebellar ataxia type 6 (SCA6) due to CAG repeat expansion in the CACNA1A gene on chromosome 19p. Hum. Mol. Genet. 1997;6:1973–1978. doi: 10.1093/hmg/6.11.1973. [DOI] [PubMed] [Google Scholar]
- 6.Barbier M., Bahlo M., Pennisi A., Jacoupy M., Tankard R.M., Ewenczyk C., Davies K.C., Lino-Coulon P., Colace C., Rafehi H., et al. Heterozygous PNPT1 Variants Cause Spinocerebellar Ataxia Type 25. Ann. Neurol. 2022;92:122–137. doi: 10.1002/ana.26366. [DOI] [PubMed] [Google Scholar]
- 7.Schöls L., Bauer P., Schmidt T., Schulte T., Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3:291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- 8.Bah M.G., Rodriguez D., Cazeneuve C., Mochel F., Devos D., Suppiej A., Roubertie A., Meunier I., Gitiaux C., Curie A., et al. Deciphering the natural history of SCA7 in children. Eur. J. Neurol. 2020;27:2267–2276. doi: 10.1111/ene.14405. [DOI] [PubMed] [Google Scholar]
- 9.Harris P.A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J.G. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roux T., Barbier M., Papin M., Davoine C.S., Sayah S., Coarelli G., Charles P., Marelli C., Parodi L., Tranchant C., et al. Clinical, neuropathological, and genetic characterization of STUB1 variants in cerebellar ataxias: a frequent cause of predominant cognitive impairment. Genet. Med. 2020;22:1851–1862. doi: 10.1038/s41436-020-0899-x. [DOI] [PubMed] [Google Scholar]
- 13.Schmitz-Hübsch T., du Montcel S.T., Baliko L., Berciano J., Boesch S., Depondt C., Giunti P., Globas C., Infante J., Kang J.S., et al. Scale for the assessment and rating of ataxia: Development of a new clinical scale. Neurology. 2006;66:1717–1720. doi: 10.1212/01.wnl.0000219042.60538.92. [DOI] [PubMed] [Google Scholar]
- 14.Jacobi H., Rakowicz M., Rola R., Fancellu R., Mariotti C., Charles P., Dürr A., Küper M., Timmann D., Linnemann C., et al. Inventory of Non-Ataxia Signs (INAS): Validation of a New Clinical Assessment Instrument. Cerebellum. 2013;12:418–428. doi: 10.1007/s12311-012-0421-3. [DOI] [PubMed] [Google Scholar]
- 15.Anheim M., Monga B., Fleury M., Charles P., Barbot C., Salih M., Delaunoy J.P., Fritsch M., Arning L., Synofzik M., et al. Ataxia with oculomotor apraxia type 2: clinical, biological and genotype/phenotype correlation study of a cohort of 90 patients. Brain. 2009;132:2688–2698. doi: 10.1093/brain/awp211. [DOI] [PubMed] [Google Scholar]
- 16.Ameijeiras-Alonso J., Crujeiras R.M., Rodriguez-Casal A. An R Package for Mode Assessment. J. Stat. Softw. 2021;97 [Google Scholar]
- 17.Lipman A.R., Fan X., Shen Y., Chung W.K. Clinical and genetic characterization of CACNA1A -related disease. Clin. Genet. 2022;102:288–295. doi: 10.1111/cge.14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards K.S., Swensen A.M., Lipscombe D., Bommert K. Novel CaV2.1 clone replicates many properties of Purkinje cell CaV2.1 current: P-type current dynamics. Eur. J. Neurosci. 2007;26:2950–2961. doi: 10.1111/j.1460-9568.2007.05912.x. [DOI] [PubMed] [Google Scholar]
- 19.Pareek G., Pallanck L.J. Inactivation of the mitochondrial protease Afg3l2 results in severely diminished respiratory chain activity and widespread defects in mitochondrial gene expression. PLoS Genet. 2020;16:e1009118. doi: 10.1371/journal.pgen.1009118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tunc S., Dulovic-Mahlow M., Baumann H., Baaske M.K., Jahn M., Junker J., Münchau A., Brüggemann N., Lohmann K. Spinocerebellar Ataxia Type 28—Phenotypic and Molecular Characterization of a Family with Heterozygous and Compound-Heterozygous Mutations in AFG3L2. Cerebellum. 2019;18:817–822. doi: 10.1007/s12311-019-01036-2. [DOI] [PubMed] [Google Scholar]
- 21.Synofzik M., Helbig K.L., Harmuth F., Deconinck T., Tanpaiboon P., Sun B., Guo W., Wang R., Palmaer E., Tang S., et al. De novo ITPR1 variants are a recurrent cause of early-onset ataxia, acting via loss of channel function. Eur. J. Hum. Genet. 2018;26:1623–1634. doi: 10.1038/s41431-018-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao X., Chen J., Li D., Xie P., Xu M., Lin W., Li S., Pan G., Tang Y., Xu J., et al. ORP4L couples IP 3 to ITPR1 in control of endoplasmic reticulum calcium release. FASEB J. 2019;33:13852–13865. doi: 10.1096/fj.201900933RR. [DOI] [PubMed] [Google Scholar]
- 23.Tiscione S.A., Casas M., Horvath J.D., Lam V., Hino K., Ory D.S., Santana L.F., Simó S., Dixon R.E., Dickson E.J. IP 3 R-driven increases in mitochondrial Ca 2+ promote neuronal death in NPC disease. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2110629118. e2110629118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall H.N., Williamson K.A., FitzPatrick D.R. The genetic architecture of aniridia and Gillespie syndrome. Hum. Genet. 2019;138:881–898. doi: 10.1007/s00439-018-1934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao L., Sha Y., Eissa N.T. The E3 ubiquitin ligase STUB1 regulates autophagy and mitochondrial biogenesis by modulating TFEB activity. Mol. Cell. Oncol. 2017;4:e1372867. doi: 10.1080/23723556.2017.1372867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romaniello R., Citterio A., Panzeri E., Arrigoni F., De Rinaldis M., Trabacca A., Bassi M.T. Novel SPTBN2 gene mutation and first intragenic deletion in early onset spinocerebellar ataxia type 5. Ann. Clin. Transl. Neurol. 2021;8:956–963. doi: 10.1002/acn3.51345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Accogli A., St-Onge J., Addour-Boudrahem N., Lafond-Lapalme J., Laporte A.D., Rouleau G.A., Rivière J.B., Srour M. Heterozygous Missense Pathogenic Variants Within the Second Spectrin Repeat of SPTBN2 Lead to Infantile-Onset Cerebellar Ataxia. J. Child Neurol. 2020;35:106–110. doi: 10.1177/0883073819878917. [DOI] [PubMed] [Google Scholar]
- 28.Figueroa K.P., Minassian N.A., Stevanin G., Waters M., Garibyan V., Forlani S., Strzelczyk A., Bürk K., Brice A., Dürr A., et al. KCNC3: phenotype, mutations, channel biophysics-a study of 260 familial ataxia patients. Hum. Mutat. 2010;31:191–196. doi: 10.1002/humu.21165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker D., De Waard M. Subunit interaction sites in voltage-dependent Ca2+ channels: role in channel function. Trends Neurosci. 1998;21:148–154. doi: 10.1016/s0166-2236(97)01200-9. [DOI] [PubMed] [Google Scholar]
- 30.Gillard S.E., Volsen S.G., Smith W., Beattie R.E., Bleakman D., Lodge D. Identification of Pore-forming Subunit of P-type Calcium Channels: an Antisense Study on Rat Cerebellar Purkinje Cells in Culture. Neuropharmacology. 1997;36:405–409. doi: 10.1016/s0028-3908(97)00046-4. [DOI] [PubMed] [Google Scholar]
- 31.Magri S., Nanetti L., Gellera C., Sarto E., Rizzo E., Mongelli A., Ricci B., Fancellu R., Sambati L., Cortelli P., et al. Digenic inheritance of STUB1 variants and TBP polyglutamine expansions explains the incomplete penetrance of SCA17 and SCA48. Genet. Med. 2022;24:29–40. doi: 10.1016/j.gim.2021.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Barbier M., Davoine C.S., Petit E., Porché M., Guillot-Noel L., Sayah S., Fauret A.L., Neau J.P., Guyant-Maréchal L., Deffond D., et al. Intermediate repeat expansions of TBP and STUB1: Genetic modifier or pure digenic inheritance in spinocerebellar ataxias? Genet. Med. 2023;25:100327. doi: 10.1016/j.gim.2022.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Safaei S., Houshmand M., Banoei M.M., Panahi M.S.S., Nafisi S., Parivar K., Rostami M., Shariati P. Mitochondrial tRNALeu/Lys and ATPase 6/8 Gene Variations in Spinocerebellar Ataxias. Neurodegener. Dis. 2009;6:16–22. doi: 10.1159/000170885. [DOI] [PubMed] [Google Scholar]
- 34.Ward J.M., Stoyas C.A., Switonski P.M., Ichou F., Fan W., Collins B., Wall C.E., Adanyeguh I., Niu C., Sopher B.L., et al. Metabolic and Organelle Morphology Defects in Mice and Human Patients Define Spinocerebellar Ataxia Type 7 as a Mitochondrial Disease. Cell Rep. 2019;26:1189–1202.e6. doi: 10.1016/j.celrep.2019.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coutelier M., Coarelli G., Monin M.L., Konop J., Davoine C.S., Tesson C., Valter R., Anheim M., Behin A., Castelnovo G., et al. A panel study on patients with dominant cerebellar ataxia highlights the frequency of channelopathies. Brain. 2017;140:1579–1594. doi: 10.1093/brain/awx081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In cases where a feature was significantly more prominent in one group only, the percentage of affected individuals displaying that feature is in bold. Percentages were rounded to the nearest integer
Data Availability Statement
The dataset supporting the current study has not been deposited in a public repository but is available from the corresponding author on reasonable request.