Summary
Newborn genomic sequencing (NBSeq) to screen for medically important genetic information is of considerable interest but data characterizing the actionability of such findings, and the downstream medical efforts in response to discovery of unanticipated genetic risk variants, are lacking. From a clinical trial of comprehensive exome sequencing in 127 apparently healthy infants and 32 infants in intensive care, we previously identified 17 infants (10.7%) with unanticipated monogenic disease risks (uMDRs). In this analysis, we assessed actionability for each of these uMDRs with a modified ClinGen actionability semiquantitative metric (CASQM) and created radar plots representing degrees of penetrance of the condition, severity of the condition, effectiveness of intervention, and tolerability of intervention. In addition, we followed each of these infants for 3–5 years after disclosure and tracked the medical actions prompted by these findings. All 17 uMDR findings were scored as moderately or highly actionable on the CASQM (mean 9, range: 7–11 on a 0–12 scale) and several distinctive visual patterns emerged on the radar plots. In three infants, uMDRs revealed unsuspected genetic etiologies for existing phenotypes, and in the remaining 14 infants, uMDRs provided risk stratification for future medical surveillance. In 13 infants, uMDRs prompted screening for at-risk family members, three of whom underwent cancer-risk-reducing surgeries. Although assessments of clinical utility and cost-effectiveness will require larger datasets, these findings suggest that large-scale comprehensive sequencing of newborns will reveal numerous actionable uMDRs and precipitate substantial, and in some cases lifesaving, downstream medical care in newborns and their family members.
Keywords: newborn sequencing, newborn screening, genomic screening, population health, precision medicine
Among 17 infants found to have unanticipated monogenic disease risks through genome screening to date in the BabySeq Project, all of the genomic findings were actionable. After 3–5 years, there had been interventions in the families of 13 of these infants, including three risk-reducing surgeries among mothers of infants sequenced.
Introduction
Recent advances in the clinical deployment of genome-scale sequencing now make it possible to determine an infant’s complete genome sequence shortly after birth, enabling the identification of deleterious variants associated with monogenic disorders.1 Genomic sequencing as a screening tool in newborns is currently being explored2,3,4 and supplementary genomic screening panels are offered by several commercial laboratories,5 but there are a number of evidentiary, ethical, and cost concerns.1,6,7,8,9
The BabySeq Project is a series of NIH-funded clinical trials of newborn screening by genomic sequencing (GS) that has generated empirical data on mechanisms of consent, gene curation, variant interpretation, and disclosure methods as well as medical, behavioral, and economic outcomes.10,11,12,13,14,15 In the first phase of the project, we recruited apparently healthy infants from a newborn nursery (NBN) and sick infants from intensive care units (ICUs) who were randomized to receive either standard of care newborn screening (NBS) or NBS plus GS.10 Participants randomized to GS underwent whole-exome sequencing with clinical reporting of pathogenic or likely pathogenic variants (PLPVs) for any genetic condition that was childhood onset and highly penetrant or childhood actionable and at least moderately penetrant, such that 954 genes were included as previously described.11,16 A limited subset of actionable adult-onset-only conditions from the American College of Medical Genetics and Genomics (ACMG) secondary findings list version 217 were also included in the analysis of the newborn’s genome and offered separately to families as previously described.14 Results were disclosed to participants’ parents in a counseling session and a disclosure letter was delivered to the family and the newborn’s clinician(s). Among the 325 newborns enrolled in the initial phase of the BabySeq Project, 159 were randomized to the GS arm. We previously reported that 18 (11.3%) of these were found to have a PLPV in one of the genes evaluated, and in one of these infants, the monogenic disease risk was recognized retrospectively to be associated with their clinical presentation.16 Thus, 17 of these 159 infants had PLPVs characterized as unanticipated monogenic disease risks (uMDRs), in that the GS was not performed to uncover genetic etiology for any presenting indication or family history in either the apparently healthy infants or the ICU infants. Among the 17 infants with uMDRs, we identified PLPVs in 13 unique genes, all of which were inherited.
In this report, we examined the medical conditions associated with each of these 17 uMDRs, mapping them onto a standardized semiquantitative measure of potential actionability, and we created a visual representation of these scores for each infant. We then tracked the actual downstream medical appointments, tests, imaging studies, and procedures that were ordered by the pediatricians and specialists who followed these infants and their families to ascertain the follow-up conducted in the 3–5 years following disclosure.
Material and methods
The BabySeq Project is a series of NIH-funded randomized controlled trials of GS in newborns. The initial phase of the study enrolled families from the NBN or ICU who were randomized to conventional care plus family history assessment and genetic counseling alone or with GS. Detailed methodology for the study design and recruitment has been previously reported.10,15 In brief, parents and newborns from the NBN at Brigham and Women’s Hospital and parents and sick newborns from the neonatal ICU and other ICUs at Brigham and Women’s Hospital, Boston Children’s Hospital, and Massachusetts General Hospital were approached, consented, and enrolled into the study. Infants from ICUs were not selected for suspicion of genetic disease. A blood sample was obtained from each newborn for DNA isolation and analysis and parent(s) provided saliva samples. Within each cohort, healthy and sick, the families were randomized to receive family history assessment and standard newborn screening (NBS) [the control arm] or to receive family history assessment and standard NBS plus GS [the GS arm]. Both arms had a three-generation family history collected and evaluated by a study genetic counselor, with genetic counseling provided if genetic risk was evident on the basis of family history. Additionally, parental surveys at enrollment (baseline), immediately post-disclosure, and 3 and 10 months after disclosure were administered.
Gene selection and variant analysis
Detailed description of the rationale for gene selection and variant analysis in the BabySeq Project have been previously published.11,16 In brief, we annotated and filtered the exomes and analyzed the results to identify PLPVs that met criteria for return, including variants found in genes with definitive or strong disease-gene association and high penetrance regardless of actionability or variants found in genes with moderate evidence or moderate penetrance but high actionability. GS was performed at the CLIA/CAP-accredited Clinical Research Sequencing Platform of the Broad Institute of MIT and Harvard, and Sanger confirmation was performed at the CLIA-accredited Mass General Brigham Laboratory for Molecular Medicine as previously described. All GS results were returned to parents and placed into the medical record via a BabySeq report, which included an indication-based analysis (IBA) for any additional diagnostic assessment related to a clinical indication, if requested.
Disclosure protocol
Parents attended a disclosure session facilitated by a study physician and genetic counselor, on average 4 months after enrollment (range: 1.2 to 10.2 months), where they were informed of their randomization status and given their family history report. Parents in the GS arm also received the BabySeq report, and if an IBA was requested, received those results. At disclosure, the study physicians (who are all trained in either clinical genetics, neurology, and/or neonatology) performed detailed physical examination to identify features that might have been previously missed. After disclosure of results, the genetic counselor and physician prepared a note summarizing the visit. These notes, along with the family history report, NBS report, and, for those in the sequencing arm, the BabySeq report/IBA, were then mailed to the parents and faxed to the infant’s pediatrician and other providers, and all these documents were uploaded to the infant’s medical record.
Actionability analysis
Clinical severity of potential conditions identified and available interventions were graded with a modification of the ClinGen actionability semiquantitative metric (CASQM).18 The CASQM evaluates outcome-intervention pairs on four axes: severity (the threat to health for an individual carrying the deleterious variant), likelihood (the chance that a serious outcome will occur, similar to penetrance), effectiveness (how effective is the proposed intervention for preventing or diminishing the risk of harm), and nature of the intervention (how medically burdensome or dangerous is the intervention). We did not modify the domains themselves or their scoring weights, i.e., the criteria upon which each domain was assigned a 0–3 score. However, instead of limiting the assessment to actionability specifically during childhood or adulthood (as is the case with the unmodified CASQM), we assessed potential actionability throughout the lifetime in order to determine the value of returning this result. We shortened and clarified the domain labels to improve readability. When more than one outcome was possible on a domain within the CASQM, we scored outcome-intervention pairs for outcomes anticipated to have the highest penetrance. The scoring was carried out by one author (N.S.) with review by all co-authors.
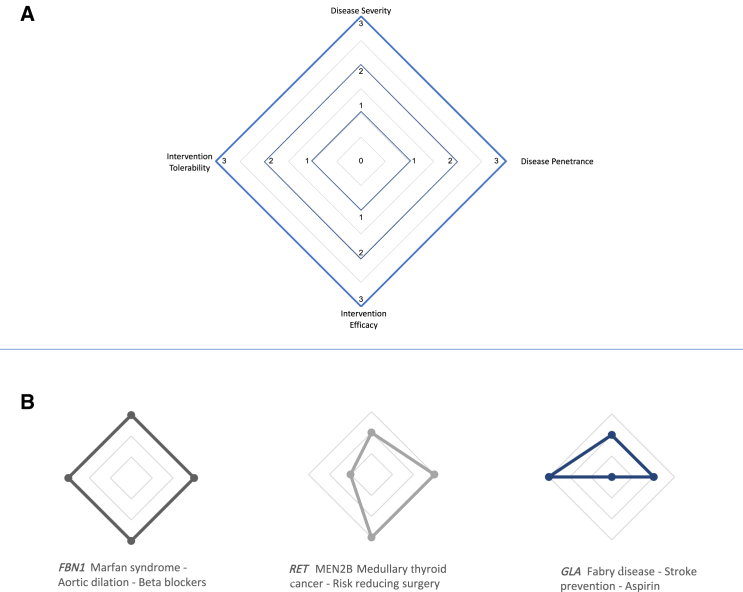
We then generated radar plots based on the modified CASQM to more intuitively explore the data visually; a perfect diamond shape represents the ideal actionable condition with high penetrance as well as higher morbidity/mortality that had a highly effective yet low-burden intervention. We applied this visual representation to several conditions associated with genes on the ACMG secondary findings list as well as to the uMDRs in BabySeq participants on the basis of their modified CASQM scores.
Healthcare utilization
For this analysis, we reviewed all available medical records to ascertain downstream medical care provided both for the proband and family members as a direct result of the uMDR identified. We also conducted phone interviews with the participant families at 37.6 to 60.6 months after the conclusion of the study to identify any relevant information that may not have been captured in the medical records. We collected pre-specified measures, such as downstream healthcare use attributable to the genomic results both for primary care as well as specialty care. We also collected outcomes that were not pre-specified, such as family member cascade testing and family member healthcare utilization, which included primary or specialty care initiated as a result of the uMDR finding in the parent from whom the uMDR was inherited as well as other family members at risk. We tracked both initial interventions based on recommendations made by the disclosing medical team/primary care physician as well as subsequent interventions over the follow-up period for both the proband as well as family members at risk. These interventions included primary care and specialty care consultations and diagnostic imaging and laboratory studies as well as therapeutic interventions such as medications and risk-reducing surgical procedures in adult relatives of the infants.
Results
Clinical actionability of uMDRs was classified with the CASQM adapted in modified form from the ClinGen Actionability Working Group (Table 1) and visualized with the radar plots (Figures 1 and 2).18,19 The uMDRs discovered in our study had a mean score of 9 out of a maximum possible score of 12 (range: 7–11) on the CASQM. On the basis of criteria defined by the CASQM, potential interventions were available for all of the 13 conditions associated with uMDRs in the 17 infants, ranging from the initiation of surveillance for cancer risk, hearing loss, and cardiac abnormalities to biotin supplementation in the case of partial biotinidase deficiency.
Table 1.
Clinical actionability of specific genes in which pathogenic and likely pathogenic variants (PLPVs) were identified in infants with uMDR
| Infant | Presentation | Gene | Disease intervention | Severity | Likelihood of disease | Efficacy of intervention | Burden of intervention | Total score | Knowledge base strength | ClinGen actionability score (where available) | NC NEXUS ASQM score (ASQM knowledge base score) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | well baby | ELN | supravalvular aortic stenosis, AD; screening cardiac ultrasound | 2 | 2 | 3 | 3 | 10 | 3 | – | 11 (2) |
| 2 | well baby | BTD | biotinidase deficiency, AR; biotin supplementation | 2 | 3 | 3 | 3 | 11 | 3 | 11CC | 10 (3) |
| 3 | ICU admission for tetralogy of Fallot, pulmonic stenosis, cryptorchidism | GLMN | glomuvenous malformations, AD; monitoring for lesions | 0 | 3 | 2 | 3 | 8 | 1 | – | – |
| 4 | ICU admission for aortic coarctation | G6PD | glucose-6-phosphate dehydrogenase (G6PD) deficiency, XLR; drug safety interventions | 0 | 3 | 3 | 3 | 9 | 3 | – | 9 (3) |
| 5 | well baby | TTN | dilated cardiomyopathy, AD; screening cardiac ultrasound | 2 | 1 | 2 | 3 | 8 | 2 | – | 9 (2) |
| 6 | well baby | TTN | dilated cardiomyopathy, AD; screening cardiac ultrasound | 2 | 1 | 2 | 3 | 8 | 2 | – | 9 (2) |
| 7 | well baby | TTN | dilated cardiomyopathy, AD; screening cardiac ultrasound | 2 | 1 | 2 | 3 | 8 | 2 | – | 9 (2) |
| 8 | well baby | TTN | dilated cardiomyopathy, AD; screening cardiac ultrasound | 2 | 1 | 2 | 3 | 8 | 2 | – | 9 (2) |
| 9 | ICU admission for hypoplastic left heart syndrome | BRCA2 | hereditary breast and ovarian cancer syndrome, AD; screening mammography | 2 | 2 | 3 | 3 | 10 | 3 | 8-10AA | 10 (3) |
| 10 | well baby | BRCA2 | hereditary breast and ovarian cancer syndrome, AD; screening mammography | 2 | 2 | 3 | 3 | 10 | 3 | 8-10AA | 10 (3) |
| 11 | ICU admission for neonatal pneumonia | SLC7A9 | cystinuria, AD; hydration, urinary alkalization, thiol medications | 1 | 1 | 3 | 3 | 8 | 2 | – | 8 (2) |
| 12 | well baby | KCNQ4 | non-syndromic hearing loss, AD; audiologic screening, hearing aids/implants | 1 | 3 | 3 | 3 | 10 | 2 | – | 10 (2) |
| 13 | well baby | VCL | dilated cardiomyopathy, AD; screening cardiac ultrasound | 2 | 2 | 2 | 3 | 9 | 2 | – | 10 (1) |
| 14 | well baby | CD46 | atypical hemolytic-uremic syndrome, AD; screening, plasma exchange, anti-C5 monoclonal antibody treatment | 2 | 2 | 2 | 1 | 7 | 2 | – | – |
| 15 | well baby | MYBPC3 | hypertrophic cardiomyopathy, AD; screening cardiac ultrasound | 3 | 2 | 3 | 2 | 10 | 3 | 10NN | 10 (3) |
| 16 | ICU admission for respiratory distress | MSH2 | Lynch syndrome, AD; screening colonoscopy | 2 | 3 | 3 | 2 | 10 | 3 | 8AA-9AB-10AA | 10 (3) |
| 17 | ICU admission for extreme prematurity | CYP21A2 | congenital adrenal hyperplasia due to 21-hydroxylase deficiency, AR; anticipatory guidance, glucocorticoid therapy | 1 | 3 | 3 | 3 | 10 | 3 | – | 11(3) |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; XLR, X-linked recessive. The table illustrates the score assignment based on the modified semiquantitative metric adapted from ClinGen as described in the material and methods. Where available, the ClinGen actionability scores as well as age-related semiquantitative metric actionability scores from the NC NEXUS study (with NC NEXUS knowledge base scores in brackets) are also shown to facilitate comparison.
Figure 1.
Clinical actionability radar plots
(A and B) Radar plots that illustrate visualization of clinical actionability. The plots utilize a modified semi-quantitative metric adapted from ClinGen as described in the material and methods. As shown in (A), the four points of the diamond on the radar graph (starting from the top and moving clockwise with each figure) represent severity of the fully expressed genetic condition, the penetrance or likelihood that the condition will manifest over an individual’s lifetime, the effectiveness of the specific intervention shown in the figure, and the tolerability of the intervention, i.e., its burden and acceptability to patients. A radar plot with maximum area within the diamond would represent a severe genetic condition that has high penetrance with a highly effective intervention that is particularly acceptable to patients because it is minimally invasive or dangerous. (B) shows sample clinical actionability radar plots for three genes from the ACMG secondary findings list.
Figure 2.
Clinical actionability of the specific uMDR genes
Radar plots that illustrate the pattern of clinical actionability in the 13 specific genes in which pathogenic and likely pathogenic variants (PLPVs) were identified in the 17 infants with an unanticipated monogenic disease risk. Axes labels correspond to reference diamond plot in Figure 1.
Having defined potential interventions, we examined the actual interventions that followed in these infants. We tracked immediate and long-term medical outcomes for these infants and their families over 3–5 years, as shown in Table 2. In three infants, genomic findings prompted discovery of a subclinical phenotype that had not been previously recognized (ELN [MIM: 130160], BTD [MIM: 60919], and GLMN [MIM: 601749], Table 2); for example, echocardiogram revealed mild but abnormal aortic stenosis in the infant with a pathogenic variant in ELN (infant 1). In the remainder of the infants (14/17) molecular findings were associated with future disease risk with PLPVs in the following genes: TTN (MIM: 188840) in four infants; BRCA2 (MIM: 600185) in two infants; and G6PD (MIM: 305900), SLC7A9 (MIM:604144), KCNQ4 (MIM:603537), CD46 (MIM:120920), VCL (MIM:193065), MYBPC3 (MIM:600958), MSH2 (MIM:609309), and CYP21A2 (MIM: 613815) in one infant each. In eight of those 14 infants, post-disclosure review of the newborn’s family history raised the possibility of additional previously unsuspected and undiagnosed family members with the condition. For example, the maternal grandfather of an infant with a maternally inherited likely pathogenic variant in TTN (infant 5) had previously been clinically diagnosed with dilated cardiomyopathy and, as a result of the finding in his grandchild (the BabySeq infant-participant), was subsequently confirmed to carry the same variant. Consequently, the infant’s mother is now routinely followed by echocardiography.
Table 2.
Demographic and clinical characteristics (including phenotype and family history information) of 17 infants with unanticipated monogenic disease risks (uMDRs) identified along with medical interventions in infants as well as their family members
| Infant | Presentation (sex) | Gene; Variant | Classification (Origin) | Disease | Infant phenotype related to uMDR? | Previously known or subsequently recognized family history related to uMDR? | uMDR-prompted Initial Interventions (Proband) | uMDR-prompted initial interventions (family) | Months of follow-up | uMDR-prompted subsequent interventions (proband) | Results of interventions (proband) | uMDR-prompted subsequent interventions (family) | Results of interventions (family) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | well baby (M) | ELN, c.1957G>T (p.Gly653∗) (GenBank: NM_000501.3) | P (Pat) | supravalvular aortic stenosis, AD | yes, mild supravalvular hypoplasia detected on follow-up ECG | no | specialist consultation [cardiology]; imaging studies [ECG] | father: imaging studies [ECG]; paternal grandmother: specialist consultation [cardiology] | 60.1 | regular cardiology follow up | routine surveillance for mild supravalvar hypoplasia detected on initial echo | father follows yearly with PCP with focus on cardiac risk | father, PGM asymptomatic from cardiac perspective with normal echocardiograms |
| 2 | well baby (F) | BTD, c.44+1G>A (p.?) (GenBank: NM_000060.2); c.1612C>T (p. Arg538Cys) | LP (Mat), P (Pat) | biotinidase deficiency, AR | yes, partial biotinidase deficiency detected on follow-up biochemical testing | no | specialist consultation [genetics]; treatment intervention [biotin supplementation] | brother: specialist consultation [genetics] | 55.5 | regular metabolism follow-up for ongoing supplementation | initial evaluation detected low biotin levels, continues ongoing biotin supplementation | none | brother found to have normal biotin levels |
| 3 | ICU admission for tetralogy of Fallot, pulmonic stenosis, crypt-orchidism (M) | GLMN, c.554_558delinsG (p.Lys185Serfs∗19) (GenBank: NM_053274.2) | LP (Pat) | glomuvenous malformations, AD | yes, vascular skin lesions | yes, recognized after infant sequencing | specialist consultation [dermatology, vascular surgery]; diagnostic procedure [endoscopy to look for vascular anomalies] | father: specialist consultation [dermatology] | 42.2 | routine surveillance, targeted evaluation for concerning symptoms (fatiguability) | vascular lesions identified on physical exam | cardiology consultation due to abnormal finding on echocardiogram (possible mass) | glomuvenous malformations identified in father |
| 4 | ICU admission for aortic coarctation (M) | G6PD, c.961G>A (p.Val321Met) (GenBank: NM_000402.3) | LP (Mat) | glucose-6-phosphate dehydrogenase (G6PD) deficiency, XLR | no, predicts risk of future disease | no | uMDR alert in EMR | none, mother informed of reproductive risk | 41.8 | not available | not available | not available | not available (family is lost to follow-up) |
| 5 | well baby (F) | TTN, c.12344delC (p.Pro4115Glnfs∗14) (GenBank: NM_133432.3) | LP (Mat) | dilated cardiomyopathy, AD | no, predicts risk of future disease | yes, known prior to infant sequencing | specialist consultation [cardiology]; imaging studies [ECG] | mother, multiple maternal first-degree relatives: imaging studies [ECG] | 45 | regular cardiology follow-up | no evidence of dilated cardiomyopathy in proband to date | mother follows with echo every 5 years; maternal relatives tested | no evidence of dilated cardiomyopathy in mother and maternal uncle confirmed to carry variant, maternal grandfather confirmed to have variant and known dilated cardiomyopathy |
| 6 | well baby (M) | TTN, c.54172C>T (p.Arg18058∗) (GenBank: NM_133378.4) | LP (Pat) | dilated cardiomyopathy, AD | no, predicts risk of future disease | yes, recognized after infant sequencing | specialist consultation [cardiology]; imaging studies [ECG] | father: imaging studies [ECG] | 46.1 | regular cardiology follow-up | no evidence of dilated cardiomyopathy in proband to date | none | borderline dilated ventricle on echo in father |
| 7 | well baby (M) | TTN, c.34894_34895insG (p.Met11632Serfs∗8) (GenBank: NM_133378.4) | LP (Mat) | dilated cardiomyopathy, AD | no, predicts risk of future disease | possible, recognized after infant sequencing | specialist consultation [cardiology]; imaging studies [ECG] | mother, multiple maternal first-degree relatives: imaging studies [ECG] | 52.7 | regular cardiology follow-up | no evidence of dilated cardiomyopathy in proband to date | none | no evidence of dilated cardiomyopathy in mother, maternal grandfather with known dilated cardiomyopathy (not tested for variant) |
| 8 | well baby (F) | TTN, c.64276_64282delinsTA (p.Ala21426∗) (GenBank: NM_133378.4) | P (Pat) | dilated cardiomyopathy, AD | no, predicts risk of future disease | no | specialist consultation [cardiology]; imaging studies [ECG] | father: imaging studies [ECG]; paternal grandmother: specialist consultation [cardiology, genetics] | 41 | regular cardiology follow-up | no evidence of dilated cardiomyopathy in proband to date | not available | no evidence of dilated cardiomyopathy in father, paternal grandmother tested and found negative for variant |
| 9 | ICU admission for hypoplastic left heart syndrome (M) | BRCA2, c.3545_3546del (p.Phe1182∗) (GenBank: NM_000059.3) | P (Mat) | hereditary breast and ovarian cancer syndrome, AD | no, predicts risk of future disease | yes, recognized after infant sequencing | (passed away from complications of primary presenting disease) | mother: specialist consultation [oncology] | N/A | not applicable | not applicable | ongoing surveillance | mother underwent prophylactic risk-reducing surgery and undergoing ongoing surveillance |
| 10 | well baby (M) | BRCA2, c.8297delC (p.Thr2766Asnfs∗11) (GenBank: NM_000059.3) | P (Mat) | hereditary breast and ovarian cancer syndrome, AD | no, predicts risk of future disease | yes, known prior to infant sequencing | specialist consultation in future [oncology] | mother: specialist consultation [oncology, was previously followed because of family history] | 60.6 | not applicable | not applicable | ongoing surveillance | mother underwent prophylactic risk-reducing surgery and undergoing ongoing surveillance |
| 11 | ICU admission for neonatal pneumonia (M) | SLC7A9, c.614dupA (p.Asn206Glufs∗3) (GenBank: NM_014270.4) | P (Mat) | cystinuria, AD | no, predicts risk of future disease | yes, recognized after infant sequencing | specialist consultation in future [nephrology] | none, mother informed of risk | 42.5 | not applicable | not applicable | none | mother asymptomatic |
| 12 | well baby (F) | KCNQ4, c.1671_1672insACGAC (p.Val558Thrfs∗3) (GenBank: NM_004700.3) | LP (Pat) | non-syndromic hearing loss, AD | no, predicts risk of future disease | possible, recognized after infant sequencing | specialist consultation [audiology, otorhinolaryngology] | none, father reported normal hearing | 46.2 | none | no evidence of hearing loss in proband on initial evaluation, no further follow-up planned | none | father asymptomatic and paternal grandfather was known to have hearing loss |
| 13 | well baby (F) | VCL, c.1713delA (p. Ala573Hisfs∗8) (GenBank: NM_014000.2) | LP (Mat) | dilated cardiomyopathy, AD | no, predicts risk of future disease | no | specialist consultation [cardiology]; imaging studies [ECG] | mother: imaging studies [ECG] | 60.3 | none | no evidence of dilated cardiomyopathy in proband to date | none | no evidence of dilated cardiomyopathy in mother |
| 14 | well baby (F) | CD46, c.286+2T>G (p.?) (GenBank: NM_002389.4) | LP (Mat) | atypical hemolytic-uremic syndrome, AD | no, predicts risk of future disease | no | specialist consultation [nephrology] | mother: specialist consultation [nephrology] | 44.5 | regular nephrology follow-up | no evidence of hemolytic uremic syndrome in proband; family provided with risk information | none | mother asymptomatic |
| 15 | well baby (M) | MYBPC3, c.1624G>C (p.Glu542Gln) (GenBank: NM_000256.3) | P (Mat) | hypertrophic cardiomyopathy, AD | no, predicts risk of future disease | no | specialist consultation [cardiology]; imaging studies [ECG] | mother: imaging studies [ECG, cardiac MRI] | 37.6 | none | no evidence of hypertrophic cardiomyopathy in proband to date | none | not available (family is lost to follow-up) |
| 16 | ICU admission for espiratory distress (F) | MSH2, c.1637_1638insA (p.Asn547Glufs∗4) (GenBank: NM_000251.2) | P (Mat) | Lynch syndrome, AD | no, predicts risk of future disease | yes, known prior to infant sequencing | specialist consultation in future [oncology] | mother: specialist consultation [oncology] | 44.7 | not applicable | not applicable | ongoing surveillance | mother underwent prophylactic risk-reducing surgery and undergoing ongoing surveillance |
| 17 | ICU admission for extreme prematurity (F) | CYP21A2, c.844G>T (p.Val282Leu) (GenBank: NM_000500.7); c.1447C>T (p.Pro483Ser) | P (Unk), P (Pat) | congenital adrenal hyperplasia due to 21-hydroxylase deficiency, AR | unclear, infant deceased after premature birth | no | specialist consultation [endocrinology] | none, parents informed of reproductive risk | 37.9 | not available | not available | not applicable (recessive) | none |
Abbreviations: uMDR, unanticipated monogenic disease risk; M, male; F, female; AD, autosomal dominant; AR, autosomal recessive; XLR, X-linked recessive; P, pathogenic variant; LP, likely pathogenic variant; Pat, paternal; Mat, maternal; Unk, unknown; EMR, electronic medical record; echocardiogram, ECG.
The infants with uMDRs, and their families, were followed for a median of 44.7 months following disclosure (range: 37.6 to 60.6 months). We tracked all medical interventions taken on behalf of the infant, as well as medical interventions taken by other family members as a result of the uMDR findings in the sequenced infant (Table 2). In all but one infant (G6PD deficiency), the disclosure of uMDR findings generated recommendations for specialist consultation and follow-up testing for the child and/or at least one of the parents. Nearly two-thirds (11/17; 64.7%) of the infants received immediate referrals for specialist care as part of the results disclosure, of which ten completed an initial referral and eight of the 10 continued to receive ongoing specialty care. In three of these 17 infants (17.6%), an exclusively cancer-related adult-onset uMDR was identified in the infant, and parents were encouraged to alert their child when older to seek appropriate screening and care in adulthood. Confirmatory parental testing was performed for all infants with a uMDR, and in 13 of 17 (76.5%) of the infants, genomic findings prompted specialist evaluations and/or diagnostic procedures for one or more family members. To date, at least four of these family member evaluations (see data associated with infants 3, 5, 6 and 7 in Table 2) have led to medically significant findings. Of the at-risk parents of the six infants with cardiovascular uMDRs, all six parents carrying the PLPV of interest had screening workups, and two continue to receive ongoing surveillance and care. Of the three parents discovered to have hereditary cancer risk through their infant’s uMDR findings, all three have undergone risk-reducing surgery.
Discussion
Among 159 infants sequenced, with interpretation of 954 genes, there were 17 infants (10.7%) with unanticipated monogenic disease risks, and all of these met standardized criteria for medical actionability. In three infants, the genomic findings led to the discovery of a phenotype that had not been previously detected or correctly diagnosed, and in eight infants, the genomic findings led to the discovery of previously unknown at-risk family members. In the 37–60 months following disclosure of the unanticipated genetic findings, two-thirds of the infants and all of their at-risk first-degree family members with uMDRs were referred for specialty consultation, surveillance, or treatment.
In the three infants with previously unrecognized phenotypes, uMDRs were no longer considered to be “risk variants” but were discovered to be penetrant—albeit with mild or subclinical features. This finding highlights how difficult it is to determine the true penetrance of most monogenic conditions and the importance of considering differences in expressivity over time. While it is well recognized that estimations of penetrance for many genetic conditions are biased toward higher estimates because clinicians are more likely to recognize genetic disease in families with robust inheritance patterns, our findings suggest that penetrance may also be underestimated when genetic diseases present with milder or subclinical symptoms or in families without obvious family history. Indeed, the very concept of penetrance depends on which particular phenotype is being measured and over what time period. For example, in an epidemiological study where penetrance of hereditary cardiomyopathy was defined as asymptomatic thickening of the cardiac septum or as one of several electrophysiological abnormalities, penetrance could be recognized far earlier than if penetrance were designated as the appearance of exercise intolerance or sudden cardiac death.20 This phenomenon highlights the importance of describing uMDRs detected through screening in terms of “risk stratification” rather than in terms of “diagnosis” and suggests that the use of terms such as “false positive” to describe the discovery of pathogenic variants in apparently healthy newborns is inappropriate, since the clinical syndrome in question may require additional testing to detect or years of follow-up to manifest.
In the BabySeq Project, we only reported variants from genes that were selected for definitive and strong disease-gene association and high penetrance regardless of actionability or moderate evidence or moderate penetrance but high actionability in childhood or adolescence.11 In addition, the parents of infants enrolled in BabySeq were offered the opportunity to have their infants’ DNA analyzed for additional genes related to actionable adult-onset conditions such as hereditary breast and ovarian cancer. Since the majority of the 954 genes ultimately interpreted were selected without regard for actionability, it was surprising to find that all of the uMDRs discovered in sequenced infants were associated with conditions that were scored as actionable on a modified version of the CASQM. This suggests that the differential in results disclosed between “parents who just want actionable information” and “parents who want any information that is of medical significance to their infant” may not be large.
The concept of actionability is difficult to discuss because for some the ability to provide enhanced surveillance or even knowledgeable anticipation justify a broader definition, while for others this concept only includes conditions where a treatment can slow or stop progression or unequivocally improve the individual’s prognosis. Figure 1 illustrates how the modified CASQM domains can be represented visually to facilitate interpretation through pattern recognition. A full diamond with a score of 3 in each domain represents the most actionable manifestation of a condition whereas alternative shapes can signal variations in which, for example, penetrance is expected to be lower or treatment is expected to be more burdensome. This or other types of visual dashboards could allow policy-makers, clinicians, and parents to intuitively understand the specific pros and cons around the actionability of the genetic information they are choosing or are receiving by reducing complex information into digestible features. There is a wide range of opinion around the specific disease-gene pairs that are recommended for newborn sequencing by rare disease experts, but in a recent survey of over 200 such experts, 87.9% agreed that NBSeq for monogenic treatable disorders should be made available to all newborns, 58.5% agreed that NBSeq should include genes associated with treatable disorders even if those conditions were low penetrance, 37.2% agreed that actionable adult-onset conditions should be sequenced in newborns to facilitate cascade testing in parents, and 27.9% agreed that NBSeq should include screening for conditions for which there were no established therapies or management guidelines.21
Of the infants identified with uMDRs, 64.7% were seen by specialists knowledgeable about the genetic condition and 76.5% had findings that prompted specialist evaluations and/or follow-up procedures for one or more family members, including life-saving risk-reducing surgeries. This speaks to the benefits of alerting family members to a genotype carried by one individual and to the powerful advantages of cascade testing, a phenomenon that has been recognized as far more efficient and cost-effective than population screening22,23 but that is implemented inconsistently and with variable success in clinical genetics today.24
Recommendations for best practices from most expert bodies caution against testing children for adult-onset conditions,25,26,27,28 but several contain language that recognizes the possibility of mitigating circumstances within specific families, and the need for expanded considerations in research studies such as ours in order to collect outcomes data on this point.27 In this project, we elected to screen a limited number of genes associated with adult-onset conditions deemed actionable by the ACMG secondary findings recommendations17,29,30 under the logic that identification of such infants could permit the further identification of parents or other adult relatives who were carrying the same variant and that efforts to preserve the health and life of the parent and extended family of the infant were very much in the best interests of the child.14,31 It is striking that among only 159 infants sequenced and only 17 with uMDRs, we discovered two infants carrying pathogenic variants in BRCA2 and one carrying a pathogenic variant in MSH2 that together have stimulated three risk-reducing surgeries among the mothers of these infants (Table 2). Our experience illustrates the real-world value of disclosing unanticipated findings of adult-onset risk variants discovered in minors. A number of recent ethical analyses have also indicated that the current consensus position on genetic testing in children is evolving.31,32,33
The BabySeq Project is a rigorous examination of uMDR findings in an unselected cohort of newborns, yet it has several limitations. Participation in the first phase of this project was offered to the parents of infants born in a single urban tertiary care medical center, and those who enrolled were more educated and of higher socio-economic status than the general population.12,15 The sample size of infants carrying uMDRs is small and the constellation of unanticipated genetic findings will not be representative of what might be found in population-level sequencing. In five instances of uMDR disclosure, the physical examination, subsequent testing, and the family history did not reveal any new or useful information, leaving these five families with risk information that may be less medically useful at this point in their lives and therefore more emotionally burdensome. A larger study with a more diverse population will be needed to ascertain whether the distress to parents and cost to the healthcare system associated with specialty consultations and surveillance around these risks is justified. Our most recently NIH-funded second phase of the BabySeq Project has begun recruitment to perform similar screening and examine downstream outcomes including healthcare utilization in a larger, more diverse population. Moreover, it is reassuring that in a separate report we recently demonstrated that neither sequencing nor receiving uMDRs was associated with greater parental distress or disruption of the parent-child bond and that parents felt empowered by both positive and negative results.12 In assessing the reactions of parents, it is important to keep in mind that parents self-select to enroll, and their self-selection reflects a desire to receive risk information and may be biased toward greater optimism around the results.
In summary, these early data on medical utilization from the BabySeq Project suggest that over 10% of infants may carry unanticipated monogenic risks for actionable conditions that over 3–5 years will result in important medical consequences for those infants and their families.
Acknowledgments
This work was supported by the following NIH awards: HD077671, TR003201 (R.C.G., B.Z., A.L.M., I.A.H., K.D.C., H.L.R., A.H.B., M.S.L.); HG011798 (P.B.A.); HG009173 (K.D.C.); HD090019, HG009922, HL143295, HG008685 (R.C.G.).
Declaration of interests
R.C.G. has received compensation for advising Allelica, Atria, Fabric, Genome Web, Genomic Life, and VinBigData and is co-founder of Genome Medical and Nurture Genomics. N.S. is a member of the scientific advisory board for Neuberg Center for Genomic Medicine. C.A.G. has received compensation for consulting for Kate Therapeutics. T.W.Y. has consulted and received compensation or honoraria from Eisai, BioMarin, GeneTx, Takeda, and Alnylam and serves on the scientific advisory boards for several not-for-profit rare disease foundations. B.Z. has received compensation for consulting for Novartis Gene Therapies. M.S.L. is employed by a not-for-profit, fee-for-service clinical laboratory at Mass General Brigham offering genomic screening. P.B.A. is a member of the scientific advisory boards for GeneDx and Illumina, Inc. A.L.M. is a member of the board of the Greenwall Foundation and is on the scientific advisory boards for Nurture Genomics, Geisinger Research, and the Morgridge Institute for Research. H.L.R. is employed as the medical and clinical laboratory director for a fee-for-service laboratory offering genomic sequencing at the Broad Institute of MIT and Harvard. I.A.H. is a member of the scientific advisory board for Biomarin for vosoritide. A.H.B. has received funding from Muscular Dystrophy Association (USA), Chan Zuckerberg Initiative, Alexion Pharmaceuticals Inc, Avidity Biosciences, Dynacure SAS, Kate Therapeutics, and Pfizer Inc; has consulted and received compensation or honoraria from Audentes Therapeutics, F. Hoffman-LaRoche AG, GLG Inc, Guidepoint Global LLC, and Kate Therapeutics Inc; and holds equity in Kinea Bio and Kate Therapeutics Inc.
Published: June 5, 2023
Data and code availability
The genomic data/analyses reported in this paper have been deposited in the NBSTRN LPDR (https://nbstrn.org/tools/lpdr) under accession identifier nbs000002.v1.p1. Data access is restricted; for information on how to request access, please contact the corresponding author.
References
- 1.Downie L., Halliday J., Lewis S., Amor D.J. Principles of genomic newborn screening programs: A systematic review. JAMA Netw. Open. 2021;4:e2114336. doi: 10.1001/jamanetworkopen.2021.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg J.S., Agrawal P.B., Bailey D.B., Jr., Beggs A.H., Brenner S.E., Brower A.M., Cakici J.A., Ceyhan-Birsoy O., Chan K., Chen F., et al. Newborn sequencing in genomic medicine and public health. Pediatrics. 2017;139:e20162252. doi: 10.1542/peds.2016-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kingsmore S.F., Smith L.D., Kunard C.M., Bainbridge M., Batalov S., Benson W., Blincow E., Caylor S., Chambers C., Del Angel G., et al. A genome sequencing system for universal newborn screening, diagnosis, and precision medicine for severe genetic diseases. Am. J. Hum. Genet. 2022;109:1605–1619. doi: 10.1016/j.ajhg.2022.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bick D., Ahmed A., Deen D., Ferlini A., Garnier N., Kasperaviciute D., Leblond M., Pichini A., Rendon A., Satija A., et al. Newborn Screening by Genomic Sequencing: Opportunities and Challenges. Int. J. Neonatal Screen. 2022;8:40. doi: 10.3390/ijns8030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeCristo D.M., Milko L.V., O'Daniel J.M., Foreman A.K.M., Mollison L.F., Powell B.C., Powell C.M., Berg J.S. Actionability of commercial laboratory sequencing panels for newborn screening and the importance of transparency for parental decision-making. Genome Med. 2021;13:50. doi: 10.1186/s13073-021-00867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnston J., Lantos J.D., Goldenberg A., Chen F., Parens E., Koenig B.A., members of the NSIGHT Ethics and Policy Advisory Board Sequencing newborns: A call for nuanced use of genomic technologies. Hastings Cent. Rep. 2018;48:S2–S6. doi: 10.1002/hast.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston J., Juengst E. Are parents really obligated to learn as much as possible about their children's genomics? Hastings Cent. Rep. 2018;48:S14–S15. doi: 10.1002/hast.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pichini A., Ahmed A., Patch C., Bick D., Leblond M., Kasperaviciute D., Deen D., Wilde S., Garcia Noriega S., Matoko C., et al. Developing a National Newborn Genomes Program: An Approach Driven by Ethics, Engagement and Co-design. Front. Genet. 2022;13:866168. doi: 10.3389/fgene.2022.866168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armstrong B., Christensen K.D., Genetti C.A., Parad R.B., Robinson J.O., Blout Zawatsky C.L., Zettler B., Beggs A.H., Holm I.A., Green R.C., et al. Parental Attitudes Toward Standard Newborn Screening and Newborn Genomic Sequencing: Findings From the BabySeq Study. Front. Genet. 2022;13:867371. doi: 10.3389/fgene.2022.867371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm I.A., Agrawal P.B., Ceyhan- Birsoy O., Christensen K.D., Fayer S., Frankel L.A., Genetti C.A., Krier J.B., LaMay R.C., Levy H.L., et al. The BabySeq Project: Implementing genomic sequencing in newborns. BMC Pediatr. 2018;18:225. doi: 10.1186/s12887-018-1200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceyhan-Birsoy O., Machini K., Lebo M.S., Yu T.W., Agrawal P.B., Parad R.B., Holm I.A., McGuire A., Green R.C., Beggs A.H., Rehm H.L. A curated gene list for reporting results in newborn genomic sequencing. Gen. Med. 2017;19:809–818. doi: 10.1038/gim.2016.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira S., Smith H.S., Frankel L.A., Christensen K.D., Islam R., Robinson J.O., Genetti C.A., Blout Zawatsky C.L., Zettler B., Parad R.B., et al. The psychosocial impact of newborn genomic sequencing on families in the BabySeq Project: A randomized clinical trial. JAMA Peds. 2021;175:1132–1141. doi: 10.1001/jamapediatrics.2021.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira S., Robinson J.O., Gutierrez A.M., Petersen D.K., Hsu R.L., Lee C.H., Schwartz T.S., Holm I.A., Beggs A.H., Green R.C., et al. Perceived benefits, risks, and utility of newborn genomic sequencing in the BabySeq Project. Pediatrics. 2019;143:S6–S13. doi: 10.1542/peds.2018-1099C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holm I.A., McGuire A., Pereira S., Rehm H., Green R.C., Beggs A.H., BabySeq Project Team Returning a genomic result for an adult-onset condition to the parents of a newborn: Insights from the BabySeq Project. Pediatrics. 2019;143:S37–S43. doi: 10.1542/peds.2018-1099H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genetti C.A., Schwartz T.S., Robinson J.O., VanNoy G.E., Petersen D., Pereira S., Fayer S., Peoples H.A., Agrawal P.B., Betting W.N., et al. Parental interest in genomic sequencing of newborns: enrollment experience from the BabySeq Project. Genet. Med. 2019;21:622–630. doi: 10.1038/s41436-018-0105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceyhan-Birsoy O., Murry J.B., Machini K., Lebo M.S., Yu T.W., Fayer S., Genetti C.A., Schwartz T.S., Agrawal P.B., Parad R.B., et al. Interpretation of genomic sequencing results in healthy and ill newborns: Results from the BabySeq Project. Am. J. Hum. Genet. 2019;104:76–93. doi: 10.1016/j.ajhg.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P., Herman G.E., Hufnagel S.B., Klein T.E., Korf B.R., et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 18.Hunter J.E., Irving S.A., Biesecker L.G., Buchanan A., Jensen B., Lee K., Martin C.L., Milko L., Muessig K., Niehaus A.D., et al. A standardized, evidence-based protocol to assess clinical actionability of genetic disorders associated with genomic variation. Genet. Med. 2016;18:1258–1268. doi: 10.1038/gim.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg J.S., Foreman A.K.M., O'Daniel J.M., Booker J.K., Boshe L., Carey T., Crooks K.R., Jensen B.C., Juengst E.T., Lee K., et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet. Med. 2016;18:467–475. doi: 10.1038/gim.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah R.A., Asatryan B., Sharaf Dabbagh G., Aung N., Khanji M.Y., Lopes L.R., van Duijvenboden S., Holmes A., Muser D., Landstrom A.P., et al. Frequency, Penetrance, and Variable Expressivity of Dilated Cardiomyopathy-Associated Putative Pathogenic Gene Variants in UK Biobank Participants. Circulation. 2022;146:110–124. doi: 10.1161/CIRCULATIONAHA.121.058143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gold N.B., Adelson S.M., Shah N., Williams S., Bick S.L., Zoltick E.S., Gold J.I., Strong A., Ganetzky R., Roberts A.E., et al. Perspectives of rare disease experts on newborn genetic sequencing. JAMA Netw. Open. 2023;6:e2312231. doi: 10.1001/jamanetworkopen.2023.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Offit K., Tkachuk K.A., Stadler Z.K., Walsh M.F., Diaz-Zabala H., Levin J.D., Steinsnyder Z., Ravichandran V., Sharaf R.N., Frey M.K., et al. Cascading after peridiagnostic cancer genetic testing: An alternative to population-based screening. J. Clin. Oncol. 2020;38:1398–1408. doi: 10.1200/JCO.19.02010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cernat A., Hayeems R.Z., Prosser L.A., Ungar W.J. Incorporating cascade effects of genetic testing in economic evaluation: A scoping review of methodological challenges. Children. 2021;8:346. doi: 10.3390/children8050346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srinivasan S., Won N.Y., Dotson W.D., Wright S.T., Roberts M.C. Barriers and facilitators for cascade testing in genetic conditions: A systematic review. Eur. J. Hum. Genet. 2020;28:1631–1644. doi: 10.1038/s41431-020-00725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borry P., Stultiens L., Nys H., Cassiman J.J., Dierickx K. Presymptomatic and predictive genetic testing in minors: A systematic review of guidelines and position papers. Clin. Genet. 2006;70:374–381. doi: 10.1111/j.1399-0004.2006.00692.x. [DOI] [PubMed] [Google Scholar]
- 26.Ross L.F., Saal H.M., David K.L., Anderson R.R., American Academy of Pediatrics. American College of Medical Genetics and Genomics Technical report: Ethical and policy issues in genetic testing and screening of children. Genet. Med. 2013;15:234–245. doi: 10.1038/gim.2012.176. [DOI] [PubMed] [Google Scholar]
- 27.Botkin J.R., Belmont J.W., Berg J.S., Berkman B.E., Bombard Y., Holm I.A., Levy H.P., Ormond K.E., Saal H.M., Spinner N.B., et al. Points to consider: ethical, legal, and psychosocial implications of genetic testing in children and adolescents. Am. J. Hum. Genet. 2015;97:6–21. doi: 10.1016/j.ajhg.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Society of Genetic Counselors . 2018. Genetic Testing of Minors for Adult-Onset Conditions.https://www.nsgc.org/Policy-Research-and-Publications/Position-Statements/Position-Statements/Post/genetic-testing-of-minors-for-adult-onset-conditions [Google Scholar]
- 29.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O’Daniel J.M., Ormond K.E., et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller D.T., Lee K., Chung W.K., Gordon A.S., Herman G.E., Klein T.E., Stewart D.R., Amendola L.M., Adelman K., Bale S.J., et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2021 update: A policy statement of the Amercian College of Medical Genetics and Genomics (ACMG) Genet. Med. 2021;23:1381–1390. doi: 10.1038/s41436-021-01172-3. [DOI] [PubMed] [Google Scholar]
- 31.Wilfond B.S., Fernandez C.V., Green R.C. Disclosing secondary findings from pediatric sequencing to families: considering the “benefit to families”. J. Law Med. Ethics. 2015;43:552–558. doi: 10.1111/jlme.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrett J.R., Lantos J.D., Biesecker L.G., Childerhose J.E., Chung W.K., Holm I.A., Koenig B.A., McEwen J.E., Wilfond B.S., Brothers K., Clinical Sequencing Exploratory Research CSER Consortium Pediatrics Working Group Rethinking the "open future" argument against predictive genetic testing of children. Genet. Med. 2019;21:2190–2198. doi: 10.1038/s41436-019-0483-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miner S.A., Similuk M., Jamal L., Sapp J., Berkman B.E. Genomic tools for health: Secondary findings as findings to be shared. Genet. Med. 2022;24:2220–2227. doi: 10.1016/j.gim.2022.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genomic data/analyses reported in this paper have been deposited in the NBSTRN LPDR (https://nbstrn.org/tools/lpdr) under accession identifier nbs000002.v1.p1. Data access is restricted; for information on how to request access, please contact the corresponding author.