Summary
Previous studies suggested that severe epilepsies, e.g., developmental and epileptic encephalopathies (DEEs), are mainly caused by ultra-rare de novo genetic variants. For milder disease, rare genetic variants could contribute to the phenotype. To determine the importance of rare variants for different epilepsy types, we analyzed a whole-exome sequencing cohort of 9,170 epilepsy-affected individuals and 8,436 control individuals. Here, we separately analyzed three different groups of epilepsies: severe DEEs, genetic generalized epilepsy (GGE), and non-acquired focal epilepsy (NAFE). We required qualifying rare variants (QRVs) to occur in control individuals with an allele count ≥ 1 and a minor allele frequency ≤ 1:1,000, to be predicted as deleterious (CADD ≥ 20), and to have an odds ratio in individuals with epilepsy ≥ 2. We identified genes enriched with QRVs primarily in NAFE (n = 72), followed by GGE (n = 32) and DEE (n = 21). This suggests that rare variants may play a more important role for causality of NAFE than for DEE. Moreover, we found that genes harboring QRVs, e.g., HSGP2, FLNA, or TNC, encode proteins that are involved in structuring the brain extracellular matrix. The present study confirms an involvement of rare variants for NAFE that occur also in the general population, while in DEE and GGE, the contribution of such variants appears more limited.
Keywords: rare genetic variants, epilepsy, NAFE
Rare genetic variants that also occur in the general population play an important role in milder epilepsy types such as non-acquired focal epilepsy (NAFE). Severe epilepsies such as developmental and epileptic encephalopathies (DEEs) show limited contribution from these variants, while their importance for genetic generalized epilepsies (GGEs) is still unclear.
Introduction
Epilepsy is one of the most common neurological diseases worldwide, affecting almost 1% of the population in the United States.1 Early pedigree studies showed a high genetic component and a heritability of up to 70%.2,3 With the help of next-generation sequencing, there was a significant advance in gene discovery. Currently, hundreds of genes are established as monogenic causes for epilepsy,4 while recent studies have identified a few genes associated with polygenic forms of epilepsy.5 Yet, the biggest leap in diagnostic yield happened mainly for the most severe type of epilepsies, developmental and epileptic encephalopathy (DEE).6 For this type of epilepsy, the heritability is very low, since such diseases are often caused by deleterious de novo variants, as the severely affected individuals usually do not reproduce.
The role of common7,8,9 and ultra-rare de novo genetic variants10,11 for epilepsy has been extensively researched. Epidemiological studies accounting for the similar prevalence across populations and the increased risk of individuals in more densely affected families suggested that polygenic predisposition should have a predominant role over the monogenic etiology.12 This has been addressed by genome-wide association studies and polygenic risk scores,7,8,9 which identified mostly non-coding variants with individually small effects—median odds ratio (OR) generally lower than 1.3—but with a high aggregate effect explaining in part the missing heritability. Conversely, the ultra-rare de novo genetic variants have much larger effects on individual risk, but they make only a small contribution to the overall heritability in the population because of their rarity.13,14
Our understanding of the underlying genetic architecture leading to increased susceptibility to epilepsy due to a middle tier of variants that are rare (neither ultra-rare de novo nor common, i.e., allele frequency of >1%) is still very limited. Based on evolutionary theory, forces of negative natural selection will keep large-effect risk variants at much lower frequencies in the population, especially for a disorder such as epilepsy, which results in reduced fitness, i.e., reproduction. Analysis of rare variants’ contribution to the disease could be a useful tool for a better understanding of the heritability and disease pathomechanism15 as it was shown in some other conditions, e.g., autism.16
In this study, we focused only on rare variants (allele count in control individuals ≥ 1 from the same cohort, minor allele frequency ≤ 1:1,000) predicted to be deleterious with an excess in affected individuals (i.e., OR ≥ 2 compared to control individuals).15 Finally, to understand the underlying pathomechanism, we performed a combined analysis of the identified genes and their interacting partners aimed at identifying molecular pathways that are potentially disrupted.
Material and methods
Cohort and data description
Genetic and phenotype information were obtained from the Epi25 Collaborative11 (http://epi-25.org/). Phenotyping procedures, case definitions, and ancestry of the participating individuals are reported in a previous Epi25 Collaborative study.11 To account for differences in ancestry and exome capture technologies among individuals, the data has undergone previously described thorough quality check procedures and we analyzed only individuals of European descent.11 Briefly, variant calling was performed with GATK17 and only variants with a genotype quality > 20 were kept. Variants called heterozygous were required to have an allele frequency of 0.2–0.8. To control for kit enrichment artifacts, we retained only variants where 80% of both Agilent- and Illumina-sequenced samples show at least 10× coverage. We previously ruled out ancestry stratification by using principal-component analyses to identify ancestral backgrounds and only analyzed individuals of European ancestry classified by random forest with 1,000 Genomes data further.11 Annotation of variants was performed with Ensembl’s Variant Effect Predictor18 for human genome assembly GRCh37 (RefSeq: GCF_000001405.13).
To understand differences in genetic susceptibility across different types of epilepsy, we analyzed 1,021 individuals with developmental and epileptic encephalopathy (DEE), 3,108 individuals with genetic generalized epilepsy (GGE), and 3,597 individuals with non-acquired focal epilepsy (NAFE). Each cohort was compared to 8,436 matched-ancestry, unrelated control individuals.11
Ethical declaration and informed consent
This study was approved by the ethics committee of the University of Leipzig, Germany (224/16-ek and 402/16-ek) and by the Epi25 Strategy Committee (approval from 16.10.2019). The availability of written informed consent from the tested individuals was checked as part of Epi2511 sample inclusion criteria. Because data analysis was performed across multiple centers, we used only aggregated data to assure participant anonymity.
Qualifying rare variants and variant set enrichment analysis
Qualifying rare variants (QVRs) are variants that we hypothesize potentially influence the susceptibility to epilepsy. A QRV is any variant that meets the following criteria.
-
•
The variant is present in control individuals, i.e., AC_CTRL ≥ 1 from our cohort of 8,436 unaffected individuals.
-
•
The minor allele frequency ≤ 1:1,000.
-
•
OR ≥ 2, p value ≤ 0.05 in individuals with epilepsy per epilepsy group.
All variants regardless of their variant type—synonymous and non-synonymous (frameshift, missense, splice, etc.)—can qualify as QRVs. For the background set, variants coming from the synonymous and non-synonymous group are tested separately, as we expect that enrichment in the QRVs occurs only in the non-synonymous variant set.
To account for gene length and different mutation rate across genes, we counted the total number of observed variants per gene and the number of variants with allele frequency (AF) ≤ 0.001. Using these two parameters, we modeled a simple linear regression to characterize the relationship of the number of rare variants and total variants per gene. On the basis of the linear regression, we estimated the expected number of rare variants with OR ≥ 2 for each gene and compared this to the observed. Only genes with an excess of QRVs were considered in the QRV filtering (Figure S2).
We estimated the susceptibility and risk burden for each gene by testing for enrichment of QRVs. To this end, we assigned an empirical enrichment score (ES) to each gene. The generation of the ES was inspired by the significance and functional enrichment (SAFE) framework19 and gene set enrichment analysis (GSEA),20 hence termed as variant set enrichment analysis (VSEA).
VSEA allows us to score the genes on the basis of the number of QRVs they have across the control population and individuals with epilepsy. We defined following lists of variants.
-
•
Ranked variant list (L)—list of variants per gene with a CADD21 score ≥ 20 ordered (descending) by their corresponding OR in each of the epilepsy types.
-
•
Background set (S)—list of QRVs grouped into synonymous or nonsynonymous variants per gene and per epilepsy type. The background set comes from the same cohort as the ranked variant list. These variants are required to be rare (AF in control individuals ≤ 1:1,000), present in control individuals, and with higher frequency in individuals with epilepsy (OR ≥ 2). However, these variants are not required to have a CADD21 score ≥ 20.
Both list L and S were generated for each gene and each different epilepsy subtype. The enrichment analysis is designed to check whether predicted deleterious variants in L are randomly distributed throughout S or skewed to a side where OR is higher or lower; the skewness will be associated to the enrichment of predicted deleterious variants meeting the QRV criteria. With this method, we can also identify the variants in each gene that contribute most to the enrichment, referred to as leading edge. We intended grouping and generating background sets for synonymous and non-synonymous separately to check whether the signal is different for the synonymous variants, which are generally considered to be neutral, i.e., not deleterious.
The ES assigned to the genes is calculated with the maximum deviation observed among cumulative ranked sum19 of both hits and misses across the variants (Figure S3) normalized by the number of observed hits and misses as seen from Equation 2. The vector of either hits or misses can be represented by vector y and ES as the maximum difference between the two vectors, defined as follows:
| (Equation 1) |
From Equation 1, y is a vector of the running cumulative ranked sum of either hits or misses vector. Each element y[i] in the vector y is calculated as the cumulative sum of the elements in the vector x from index 1 to index i. So, for example, y[1] is the first element of y and is equal to x[1], while y[2] is the second element of y and is equal to x[1] + x[2], and so on up to y[n], which is equal to the sum of all elements in the vector x.
| (Equation 2) |
The ES is calculated as the maximum difference between the normalized cumulative sum of hits and misses. We use the maximum difference to capture the most significant enrichment or depletion of the variant set (S) in the ranked list (L) of variants.
A high ES of a gene indicates an enrichment of variants having higher OR in comparison to the variants found in the lower rank. After calculating the ES, to determine the significance, we performed n = 1,000 permutations where a set of variants having the length n(L) were randomly selected from set S. We used this to calculate an empirical distribution of ES. We counted the number of times the permuted ES (ESp) exceeds the observed ES (ESobs) and divided this by the number of permutations (n) to calculate the p values (pval).
| (Equation 3) |
For multiple testing correction, we determined false discovery rate (FDR) by using the ratio of the normalized enrichment score (NES) – observed and permuted ES. The steps and equations are as follow.
| (Equation 4) |
NESobs is the score calculated by rescaling the observed ES (ESobs) of a gene resulted from Equation 2 and dividing it by the mean of all positive permuted ES (ESp), i.e., ESp > 0, from the permutation (n = 1,000) previously described.
| (Equation 5) |
NESp is a vector of score calculated from all positive permuted ES (ESp) per gene normalized by its mean.
| (Equation 6) |
where NESp is the vector of permuted NES calculated from Equation 5; NESobs is the NES observed per gene across the dataset analyzed, as seen from Equation 4; and NESglobal is the vector of all NES observed across the dataset (a whole set of NESobs). Notation n refers to the length of a vector defined, i.e., n(x) = length of vector x.
The FDR is then calculated by dividing the proportion of NESp that exceeds the value of NESobs with the proportion of NESglobal exceeding NESobs. The threshold is set to 0.05 to be more conservative because setting a threshold of FDR ≤ 0.25 implies that 1 out of 4 is a false positive. Unlike GSEA, here, we do not identify biological pathways, which could be more tolerant to error. Here, we aim to identify genes that we infer to have impaired functionality, which is why a very conservative threshold was adopted, keeping in mind that we may not detect the whole spectrum of genes bearing QRVs. To test for a potential inflation of NES as a result of population stratification, we performed quantile-quantile (QQ) plots for each epilepsy type (Figure S4).
Functional module analysis and protein-protein interactions
To identify the functional pathways in which genes bearing an excess of QRVs play a role, we performed enrichment analysis across multiple gene sets including but not limited to Gene Ontology (GO), Allen Brain Atlas, Reactome, and KEGG pathways.
Allen Brain Atlas enrichment
To identify gene sets that are specific to certain brain areas and/or developmental stages, we used the Allen Human Brain Atlas. This resource delivers information about gene expression levels in various parts of the human brain during the course of brain development.22,23 We used the R package ABAEnrichment to test whether genes with excess QRVs show significant enrichment in specific brain regions or brain developmental stages.24 The package integrates human brain expression datasets provided by the Allen Brain Atlas in both the prenatal and adult stage. The expression data is analyzed over 47 brain regions and 20 age time points. The gene expression is evaluated during development from prenatal stage to adult. If the change is high in a specific region, the gene is annotated to that region and the score mirrors the deviation from prenatal to adult stage.24
Overrepresentation analysis (ORA)
The gene lists were also subjected to functional class analysis via ORA with MsigDb Reactome database for gene set collections C2 (Reactome) and C5 (GO)20,25 and GOFuncR (https://github.com/sgrote/GOfuncR). The method uses a hypergeometric test26 to assess the probability of observing at least k genes from the list across the pathway database.25,27
| (Equation 7) |
For the customized gene set ORA, we used the datasets from MSigDB – C5 for sets associated with ion channels, neurotransmitter, glutamatergic and GABAergic signaling, nervous system development, and synaptic functions.20,25 For GoFuncR overrepresentation, we used C5 (GO) of MSigDB for restricting the background genes to those that are found expressed in the brain.28
Analysis of distance in the human protein interacting network (PIN)
To elucidate the putative roles of the genes significantly enriched with QRVs in epilepsy, we investigated the distance between them and the known epilepsy-related genes in the human PIN. The source of the human PIN data is InBio Map29 and epilepsy-related genes were acquired from the consolidated list of epi-25.org. The distance is defined as the shortest path length between genes u and v in the human PIN. All paired shortest path lengths between genes are calculated by the Dijkstra algorithm. We identified which genes with QRVs are significantly more closely located to the known epilepsy-related genes within the PIN compared to the distance of all other genes.
Results
Enrichment of QRVs in three subtypes of epilepsies
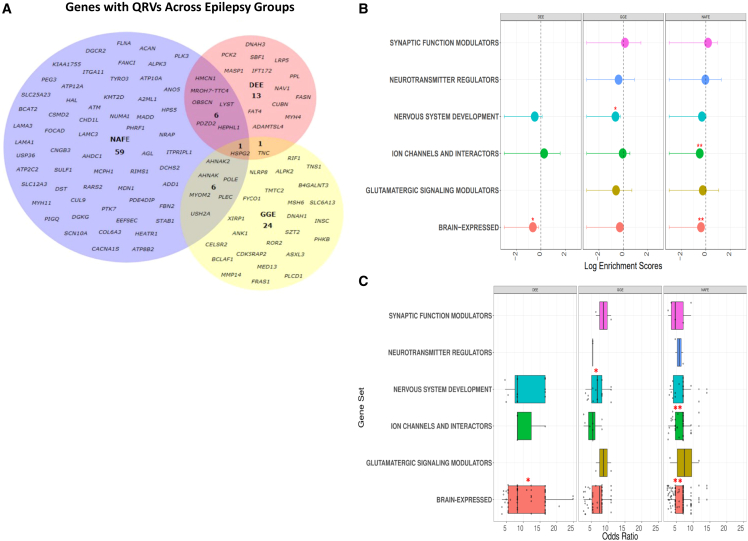
We tested the burden of QRVs per gene in each epilepsy group: DEE, NAFE, and GGE. The only significant gene that was found to be enriched across all types of epilepsies was HSPG2 (p value = 0.0001, from 10,000 permutations to test the significance of the overlap among three different epilepsy groups in this gene). HSPG2 (heparan sulfate proteoglycan 2) encodes perlecan, which belongs to the glycosaminoglycans family, which are major components of the brain extracellular matrix (ECM). Although the gene was common to all epilepsy groups, there were different variants in this gene, which contributed to the increased enrichment score of the gene for each group (Table S1).
Further, consistent with the presumed de novo origin of pathogenic variants and a highly penetrant phenotype, DEE showed the lowest number of genes enriched with QRVs (Figure 1A); this supports DEE’s mainly monogenic origin. In other words, for DEE, the most severe of the epilepsy phenotypes, it is less likely that variants present in control individuals contribute as causal risk factors. The largest number of genes with a significantly high enrichment score was retrieved for NAFE (Figure 1A, Table S1). This result contrasts the previous findings on ultra-rare de novo genetic variants, which show mainly no significant burden in the NAFE-affected individuals11 and may suggest that rare variants also present in control individuals could contribute more to the NAFE pathophysiology compared to highly damaging de novo variation.
Figure 1.
QRV-enriched genes across different epilepsy groups
(A) For each epilepsy group, a set of genes were identified to be enriched with QRVs (FDR ≤ 0.05, Z score ≥ 1.96). The number of genes found for NAFE are greater than the two other groups, seemingly implying that rare variants in this particular group could have a larger contribution to the etiology compared to DEE and GGE.
(B) Overrepresentation of QRV-enriched genes. The graph shows the log enrichment scores and intervals with confidence interval = 0.95 for the overrepresentation of the QRV-enriched genes. The genes enriched with QRVs were found to be overrepresented across highly expressed genes in the brain (DEE, NAFE), nervous system development (GGE), and ion channel and interactors (NAFE), suggesting a role in the brain and brain-related processes.
(C) Odds ratio distribution of QRV-enriched genes across different gene sets. The graph shows the odds ratio distribution of the variants per genes across different gene sets. (∗FDR ≤ 0.05, ∗∗FDR ≤ 0.01).
To gain insight into the functionality of the genes, we classified them into six categories, which are considered to play a role in the epileptic pathomechanism: modulators of synaptic functions, neurotransmitter regulators, nervous system development, ion channels and their interacting partners, modulators of glutamatergic signaling, and genes with high expression in brain (Figures 1B and 1C, Table 1). Our results showed for DEE (FDR = 0.03) and NAFE (FDR = 0.004) a significant enrichment especially for genes highly expressed in brain. GGE showed an enrichment for genes annotated to nervous system development (FDR = 0.026). In NAFE, we identified genes enriched for ion channels and their interactors (FDR = 0.004) (Figure 1B). Variants in these genes generally have median ORs > 5 (Figure 1C), suggesting they may have a large effect size and could contribute to the underlying pathomechanism.
Table 1.
QRV-enriched genes across gene sets annotated to brain processes and molecular functions
| Gene set | DEE | NAFE | GGE |
|---|---|---|---|
| Ion channel and interactors | HEPHL1 | ADD1, AHNAK, ANO5, ATM, ATP10A, ATP12A, ATP2C2, ATP8B2, CACNA1S, CNGB3, FLNA, HEPHL1, RIMS1, SCN10A, SLC12A3, SLC25A23 | AHNAK, SLC6A13, TMTC2 |
| Neurotransmitter regulators | – | RIMS1 | SLC6A13 |
| Nervous system development | FAT4, HSPG2, IFT172 | ACAN, ATM, HSPG2, LAMC3, MCPH1, PLEC, TYRO3 | CDK5RAP2, CELSR2, HSPG2, PLEC, ROR2, SZT2 |
| Synaptic function modulators | – | FLNA, RIMS1 | ROR2 |
| Glutamatergic signaling modulators | – | HAL | ROR2 |
| Brain-expressed | ADAMTSL4, CUBN, FASN, FAT4, HMCN1, HSPG2, IFT172, LRP5, LYST, MASP1, NAV1, OBSCN, PCK2, PDZD2, PPL, SBF1, TNC | A2ML1, ACAN, ADD1, AGL, AHDC1, AHNAK, AHNAK2, ALPK3, ANO5, ATM, ATP10A, ATP8B2, BCAT2, CHD1L, COL6A3, CSMD2, CUL9, DGCR2, DGKG, DST, FANCI, FLNA, FOCAD, HEATR1, HMCN1, HPS5, HSPG2, KIAA1755, KMT2D, LAMA1, LAMA3, LAMC3, LYST, MADD, MCPH1, MDN1, MYH11, NUMA1, OBSCN, PDE4DIP, PDZD2, PEG3, PHRF1, PIGQ, PLEC, PLK3, PTK7, RARS2, RIMS1, SLC25A23, STAB1, SULF1, TYRO3, USP36 | AHNAK, AHNAK2, ANK1, BCLAF1, CDK5RAP2, CELSR2, FRAS1, FYCO1, HSPG2, MED13, MMP14, MSH6, PHKB, PLCD1, PLEC, RIF1, SLC6A13, SZT2, TMTC2, TNC, TNS1 |
The table contains the QRVs-enriched genes per epilepsy groups, developmental and epileptic encephalopathy (DEE), non-acquired focal epilepsy (NAFE), and genetic generalized epilepsy (GGE), which are found to be enriched in gene sets derived from molecular functions in the GO database and brain gene expression level from Protein Atlas and GTEx. For each gene set, we selected processes associated with ion channels, GABAergic- and glutamatergic-related pathways, synaptic functions, neurotransmission, nervous system development, and genes with nTPM ≥ 1 in brain.28
Variant set enrichment captures genes encoding for ion channels or their interactors in NAFE
Ion channels play a major role in genetic epilepsies.30,31 Although, since the beginning of the epilepsy-related gene discoveries, many other gene classes and biological pathways have been revealed to play a role, it is still a significant proportion (∼25%) of the epilepsy genes known to date that encode for ion channels.30 Since we identified the ion-channel-related molecular function pathway to be enriched in NAFE (Figure 1B), we performed ORA based on KEGG (https://www.genome.jp/kegg/) and Reactome.32 For KEGG, the ion channel pathway is annotated only with respect to drug development, and thus, we could not identify an over-representation based on the QRV-enriched genes in NAFE. Using the Reactome annotation, we identified the ion channel transport category to be significantly enriched. NAFE-related genes annotated to the Reactome category are ATP2C2, ATP12A, ATP8B2, ATP10A, and ANO5. While, the first four genes encode for ATPases involved in ions transport, such as Ca2+ or H+/K+, ANO5 encodes for an anoctamin, which belongs to a protein family of Ca2+-activated chloride channels and phospholipid scramblases.33 A founder mutation in ANO5 has been implicated in muscular dystrophy.33 Despite the high expression in brain and the controversial muscle phenotype in the mouse knockout models,34,35 its function in the brain remains unknown.
Using the GO annotation, we identified a few ion channel genes and multiple genes encoding proteins that interact with ion channels, which showed QRV enrichment in NAFE (Table 1), e.g., CACNA1S and SCN10A. Another identified gene is ADD1, coding for adducin. Although adducin is primarily responsible for the assembly of spectrin-actin, which provides functional support to the cytoskeleton, the GO also annotates the gene to ion transport and synaptic functions. Variants in ADD1 have also been recently identified in intellectual disability, corpus callosum dysgenesis, and ventriculomegaly in humans.36 Similarly, ATM, another gene with QRVs for NAFE, was recently shown to be involved in hippocampal and cortical development, as well as synaptic functions.37
On the basis of our analysis, we did not retrieve genes encoding for ion channels that have already been associated with monogenic epilepsy in reference to curated lists of epilepsy genes in the Seizure-Associated Genes Across Species (SAGAS)5 database and Genes4Epilepsy (https://github.com/bahlolab/Genes4Epilepsy). Those genes generally bear ultra-rare variants that do not occur in control individuals, which does not comply with our definition of QRVs. Some of the genes we identified to be significantly enriched in NAFE have already been associated with Mendelian disorders on the basis of OMIM,38 e.g., HSPG2 and FLNA, yet their phenotype does not appear to be severe or highly penetrant, which meets our hypothesis that variants in these genes could confer an increased risk for epilepsy.
Involvement in brain development
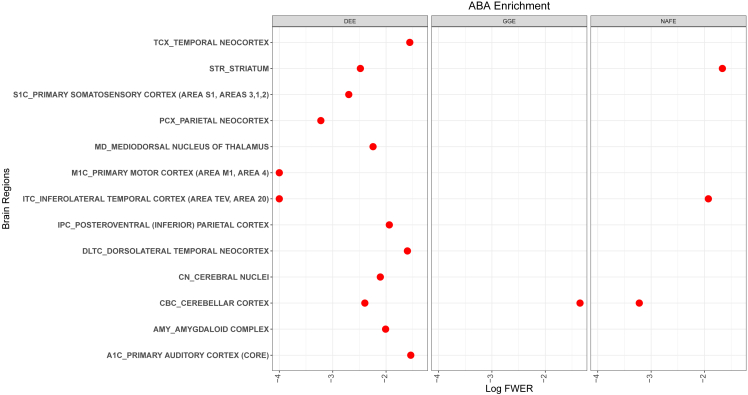
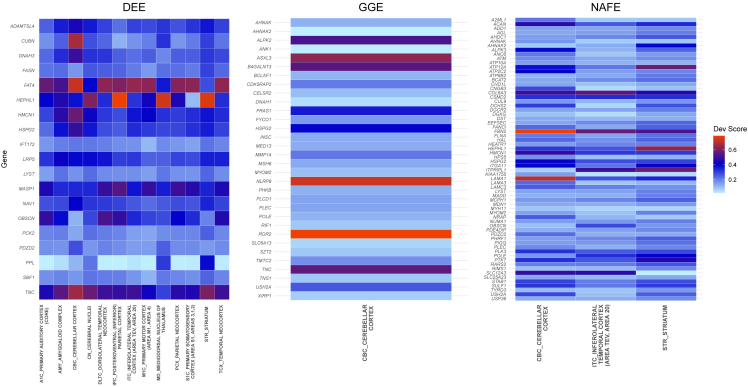
Using the ABAEnrichment package,24 we tested whether the genes identified in the different types of epilepsy play a role during development in any brain region. From the results, we identified genes from DEE to show the highest involvement during development with significant enrichment over 13 brain regions (Figure 2). For GGE, only the cerebellar cortex showed a signal (family-wise error rate, FWER = 0.045), while for NAFE, we identified a signal in this region and two additional ones in the striatum and inferolateral temporal cortex. When we examined the list of genes with the largest developmental scores based on expression differences between prenatal and adult stages, we identified genes encoding for ECM proteins (LAMA1, FBN2, COL6A3) to contribute to the enrichment in the different brain regions for NAFE (Figure 3). Similarly, TNC, which encodes for the ECM protein tenascin C, showed a high developmental score contributing to the brain region enrichment in both DEE and GGE. For DEE, FAT4, a gene encoding for a protocadherin, a calcium-dependent cell adhesion protein, showed the highest developmental score (Figure 3). FAT4, which was previously related to epilepsy,39 plays a role in the maintenance of planar cell polarity as well as in neuroprogenitor proliferation.40 For GGE, ROR2 is the leading gene in respect to the developmental score. ROR2 encodes a tyrosine-protein kinase transmembrane receptor also known as the neurotrophic tyrosine kinase, receptor-related 2, which also appears to play a role in the maintenance of neuroprogenitor cells in the developing neocortex.41 On the basis of the observed functions for the genes with the highest developmental scores, we were further prompted to perform pathway analyses and understand in which molecular processes genes with QRVs are involved.
Figure 2.
ABA enrichment of QRV-enriched genes across all epilepsy types
The graph shows brain regions with QRV-enriched genes with respect to their developmental score and the log-transformed FWER (family-wise error rate) associated with the significance of the enrichment. The smaller the value of log FWER (to the left) means that it is more significant.
Figure 3.
ABA developmental score of QRV-enriched genes
Heatmap with developmental scores of QRV-enriched genes significantly associated with the different brain regions during development. The listed regions were identified to be overrepresented with QRV-enriched genes (Figure 2). Note: it should be noted that some brain regions, which include but are not limited to PCx_parietal neocortex, CN_cerebral nuclei, DLTC_dorsolateral temporal neocortex, and TCx_temporal neocortex in the ABAEnrichment database, have no gene expression annotation and the resulting enrichment was generated from equivalent subregions; in this case the average developmental scores of the subregions were used.
Pathway and network analyses
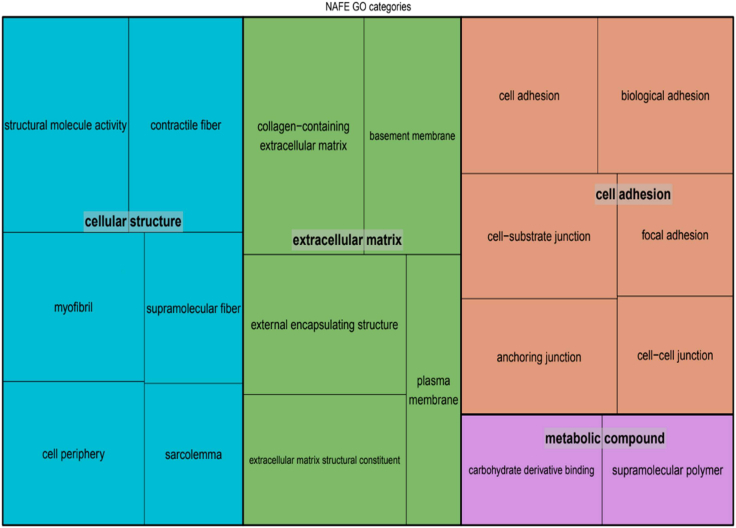
We performed pathway enrichment analyses to determine whether the identified genes cluster within specific functions. For DEE, we could not identify any significant categories after multiple testing correction. NAFE showed enrichment of many GO categories that clustered mainly within the ECM or cell adhesion (Figure 4). For GGE, there were only three significant categories, all related to cellular junctions or adhesion (Table S2).
Figure 4.
Overrepresentation of QRV-enriched genes from NAFE
The GO terms with enrichment in NAFE cluster into a more general category to represent their functional class. The overrepresentation analysis was run with GoFuncR with genes expressed in brain as background. Note: the size of the squares correspond to the number of genes on that GO category.
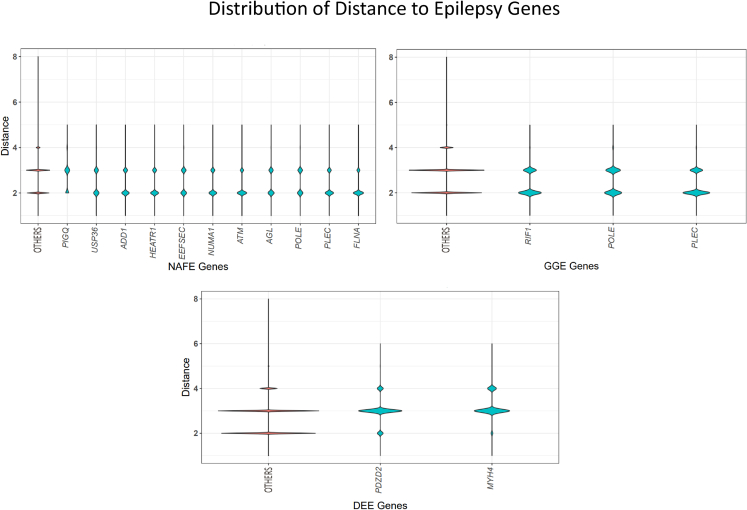
To further test how the identified genes with QRVs are connected to already known epilepsy genes, we determined the distance between the identified gene and the epilepsy genes from Epi25 (http://epi-25.org/). To this end, we assessed within the protein-protein interaction (PPI) network how many nodes represented by protein interacting partners lie between the identified gene and any known epilepsy gene. While for NAFE, the mean distance between QRV-enriched genes and epilepsy genes was at 2.57, significantly smaller than the mean distance of between the non-QRV-enriched genes and epilepsy genes (2.66, p value < 0.001), the effect size is too small to conclude that overall, the identified genes with QRVs are closer to known epilepsy genes. We thus further determined the genes with a shorter distance to known epilepsy genes within the PIN (Figure 5). For GGE and DEE, the overall mean distance did not reach significance, and only three and two genes, respectively, showed a significantly closer distance to epilepsy genes.
Figure 5.
QRV-enriched genes and their distance to known epilepsy genes
The distance (y axis) of the QRV-enriched genes (x axis) to known epilepsy genes from Epi25 (http://epi-25.org/) was determined by counting the nodes within the protein-protein interaction network. We show genes with significantly shorter distance to known epilepsy genes compared to the distance of randomly selected genes. A larger distribution assigned to the lower distance in the violin plot implies a higher number of genes with short distance to known epilepsy genes in the PIN.
Discussion
Unlike common and de novo ultra-rare variants, rare variants in epilepsy have not been researched extensively. On the basis of the “common disease, rare variant” hypothesis, multiple rare sequence variants, with relatively high penetrance, confer an increased genetic susceptibility to a common disease.42 While the identification of such variants is paramount for understanding their role, their detection is statistically more challenging because they are present at low frequencies in the general population. To understand how rare variants contribute to the etiology and pathomechanism of epilepsy, we applied a method to estimate the risk burden of a gene on the basis of the enrichment of rare variants posing a relatively high risk in the Epi25 cohort, i.e., OR ≥ 2. Using the proportion of QRVs in affected individuals and control individuals, we calculated a score for each gene. A high score of a gene implies a higher disease probability due to the presence of the rare variants with higher ORs in affected individuals. By this approach we identified sets of QRV-enriched genes: DEE (n = 21), NAFE (n = 72), and GGE (n = 32) (Figure 1). In a previous study that analyzed the same cohort in respect to the enrichment of deleterious ultra-rare variants, individuals with DEE and GGE had significantly more such variants compared to those diagnosed with NAFE,11 while for our study on the enrichment of rare variants, we identified the lowest number of genes for DEE and the highest for NAFE. Our results and the results of the previous study11 could suggest that, while for DEE the pathomechanism relies on highly deleterious and penetrant variants, NAFE may result from an enrichment of more frequent and less penetrant rare variants, identified also in control individuals, albeit at lower frequency than in affected individuals.
To further analyze the gene set and its relevance to specific molecular pathways, we performed a GO enrichment analysis. This revealed a significant overrepresentation of the NAFE genes across ECM and structural related pathways (Figure 4).41 ECM represents approximately 10%–20% of the brain’s volume. The constitutive ECM molecules are synthesized and secreted by neural and glial cells.43,44,45 These molecules organize themselves in structures surrounding axons and synapses and participate in developmental processes and nervous system plasticity.43,46 ECM components participating in plasticity, mostly perineuronal nets (PNs),47 are involved in the regulation of neural excitation and inhibition. PNs can be found surrounding GABAergic interneurons,45,47,48 where they limit the neuronal remodeling, which in turn prohibits epileptogenesis. They are embedded in a basal lamina that includes collagen, laminin, heparan sulfate proteoglycans (HSPGs), and other glycoproteins, such as aggrecans.49 We identified genes encoding for these proteins (LAMA1, LAMAC3, COL6A3, ACAN) to be enriched in QRVs in NAFE. Interestingly, changes in ECM have been directly implicated in the pathophysiology of temporal lobe epilepsy,50,51 the most common form of NAFE. HSPG2, the only gene found to be enriched across all types of epilepsy in this study, encodes for perlecan—a heparan sulfate proteoglycan—also an important constituent of the basal lamina. Little is known about HSPG2 and its association to epilepsy, but some studies have revealed its role in acetylcholinesterase clustering at the synapse, which has the capability to interfere in synaptic transmission.52 HSPG2 is, however, ubiquitously expressed, and pathogenic variants have been implicated in the Schwartz-Jampel syndrome type I, a rare autosomal-recessive disease with cardinal symptoms consisting of skeletal dysplasia and neuromuscular hyperactivity.53 Some of the affected individuals also show impaired neurologic development, consistent with perlecan’s neuroprotective effect and its involvement in neurogenesis and normalization of neocortical excitability after insult events.54 In further support of our finding, previous studies have also considered HSPG2 to be an epilepsy-associated gene, although the underlying mechanism is still not clear.39
While among the identified genes there was an enrichment of QRV-enriched genes with high brain expression, both in the DEE and NAFE groups, for NAFE, we additionally identified variants in ion channels and their interactors to play a role (Figures 1B and 1C). CACNA1S is one of the genes with an excess of QRVs in NAFE-affected individuals. The gene has low expression in the brain and is highly expressed in the muscles being implicated in the hypokalemic periodic paralysis. However, we observed outliers in respect to brain expression (Figure S5),55 suggesting that Cav1.1, the L-type voltage-gated calcium channel encoded by CACNA1S, could play a role in the calcium influx in response to large depolarizing shifts in membrane potential for some individuals. In support of the potential involvement of CACNA1S in brain functions, rare variants in this gene have been associated with schizophrenia.56 Similarly, SCN10A is not highly expressed in the brain but shows an enrichment signal in our dataset. Bi-allelic variants in this gene have been potentially linked to epilepsy-related phenotypes.57
In an additional analysis to understand whether the genes we identified are closer in the PIN to already known epilepsy genes, we calculated the PPI distance (Figure 5). Most of the genes with significantly shorter PPI paths are connected to epilepsy genes for NAFE. A gene with significantly shorter distance to known epilepsy genes is PDZD2, enriched for both NAFE (FDR = 0.005) and DEE (FDR = 0.006). This gene is expressed mainly in the basal ganglia and cerebral cortex with high specificity among oligodendrocytes precursor cells and excitatory neurons.26,49 PDZD2 also contributes to the functional expression of Nav1.8 ion channel, which is encoded by SCN10A, a QRV-enriched gene in NAFE.58 Another gene with shorter distance to known epilepsy genes, FLNA, has itself been found to have variants associated with epilepsy and seizure disorders.5 FLNA is also known to interact with channels encoded by HCN1 during neuronal excitability modulation in the mature brain.59 Additionally, FLNA also controls ECM remodeling by regulating metalloproteinase activity and hence ECM degradation.60 On the basis of the enrichment of ECM genes, we suggest that especially for NAFE, genetic variants that may impact ECM morphology could lead to imbalance in excitatory and inhibitory signals44 and hence underlie epileptogenesis.
Consistent with the assumed pathomechanisms, we identified a significant enrichment of QRVs in genes related to brain development only in DEE (Figure 2) and GGE (Figure 1). Interestingly, most genes with high developmental scores are assigned to DEE (Figure 3), suggesting that rare variants in these genes may contribute to the disease development. TNC, which encodes an ECM protein, was identified in both DEE and GGE (Figure 1). It controls neurite growth and axon guidance61 and it is highly active during early brain development, which is mirrored by a high developmental score (Figure 3). Intriguingly, TNC is higher expressed by both neurons and glias after seizures, which can lead to ECM remodeling and induce additional epileptic events.61,62 For NAFE, genes with high developmental scores cluster in the inferolateral temporal cortex (Figure 2), a signal that may be triggered by a high number of individuals with temporal lobe epilepsy within the NAFE cohort.
Aside from the established list of epilepsy-associated genes from the Epi25 cohort, there are a number of curated lists for epilepsy genes, for instance the SAGAS database, containing candidate genes with possible polygenic and monogenic causal tendencies,5 and Genes4Epilepsy (https://github.com/bahlolab/Genes4Epilepsy). We identified a significant overlap of the QRV-enriched genes from our study with both SAGAS (DEE: pval = 0.001, NAFE: pval = 1.30e−8, GGE: pval = 0.02) and Genes4Epilepsy (NAFE: pval = 0.001), which lends additional support to our findings.
A previous study of the Epi25 Collaborative suggested that clinical presentations of GGE and NAFE are influenced by common and rare variants, as opposed to DEE, which is mainly caused by de novo ultra-rare highly deleterious variants.11 Here, we focused on rare variants, present in control individuals, but at higher frequency in individuals with epilepsy. Our results support the hypothesis that rare variants could be important in the NAFE pathomechanism. Moreover, ECM appears to play a central role in NAFE. For DEE, we retrieved genes that are highly expressed during development, which meets the expected pathomechanism; however, the number of identified genes is rather low. On the basis of the genes identified for GGE, we cannot infer which pathways play an important role in the pathomechanism. It is possible that either enlarged cohorts of individuals with epilepsy or a focus on common variants will shed more light on GGE63 pathophysiology.
Consortia
The following persons are participating members and/or collaborators of the Epi25 Consortium: Yen-Chen Anne Feng, Daniel P. Howrigan, Liam E. Abbott, Katherine Tashman, Felecia Cerrato, Tarjinder Singh, Henrike Heyne, Andrea Byrnes, Claire Churchhouse, Nick Watts, Matthew Solomonson, Dennis Lal, Erin L. Heinzen, Ryan S. Dhindsa, Kate E. Stanley, Gianpiero L. Cavalleri, Hakon Hakonarson, Ingo Helbig, Roland Krause, Patrick May, Sarah Weckhuysen, Slave′ Petrovski, Sitharthan Kamalakaran, Sanjay M. Sisodiya, Patrick Cossette, Chris Cotsapas, Peter De Jonghe, Tracy Dixon-Salazar, Renzo Guerrini, Patrick Kwan, Anthony G. Marson, Randy Stewart, Chantal Depondt, Dennis J. Dlugos, Ingrid E. Scheffer, Pasquale Striano, Catharine Freyer, Kevin McKenna, Brigid M. Regan, Susannah T. Bellows, Costin Leu, Caitlin A. Bennett, Esther M.C. Johns, Alexandra Macdonald, Hannah Shilling, Rosemary Burgess, Dorien Weckhuysen, Melanie Bahlo, Terence J. O’Brien, Marian Todaro, Hannah Stamberger, Danielle M. Andrade, Tara R. Sadoway, Kelly Mo, Heinz Krestel, Sabina Gallati, Savvas S. Papacostas, Ioanna Kousiappa, George A. Tanteles, Katalinerbova′, Marke′ta Vl ckova′, Lucie Sedla′ckova′, Petra La s suthova′, Karl Martin Klein, Felix Rosenow, Philipp S. Reif, Susanne Knake, Wolfram S. Kunz, Ga′bor Zsurka, Christian E. Elger, Jü rgen Bauer, Michael Rademacher, Manuela Pendziwiat, Hiltrud Muhle, Annika Rademacher, Andreas van Baalen, Sarah von Spiczak, Ulrich Stephani, Zaid Afawi, Amos D. Korczyn, Moien Kanaan, Christina Canavati, Gerhard Kurlemann, Karen Mü ller-Schlü ter, Gerhard Kluger, Martin Häusler, Ilan Blatt, Johannes R. Lemke, Ilona Krey, Yvonne G. Weber, Stefan Wolking, Felicitas Becker, Christian Hengsbach, Sarah Rau, Ana F. Maisch, Bernhard J. Steinhoff, Andreas Schulze-Bonhage, Susanne Schubert-Bast, Herbert Schreiber, Ingo Borggräfe, Christoph J. Schankin, Thomas Mayer, Rudolf Korinthenberg, Knut Brockmann, Gerhard Kurlemann, Dieter Dennig, Rene Madeleyn, Reetta Kälviäinen, Pia Auvinen, Anni Saarela, Tarja Linnankivi, Anna-Elina Lehesjoki, Mark I. Rees, Seo-Kyung Chung, William O. Pickrell, Robert Powell, Natascha Schneider, Simona Balestrini, Sara Zagaglia, Vera Braatz, Michael R. Johnson, Pauls Auce, Graeme J. Sills, Larry W. Baum, Pak C. Sham, Stacey S. Cherny, Colin H.T. Lui, Nina Bari si c, Norman Delanty, Colin P. Doherty, Arif Shukralla, Mark McCormack, Hany El-Naggar, Laura Canafoglia, Silvana Franceschetti, Barbara Castellotti, Tiziana Granata, Federico Zara, Michele Iacomino, Francesca Madia, Maria Stella Vari, Maria Margherita Mancardi, Vincenzo Salpietro, Francesca Bisulli, Paolo Tinuper, Laura Licchetta, Tommaso Pippucci, Carlotta Stipa, Raffaella Minardi, Antonio Gambardella, Angelo Labate, Grazia Annesi, Lorella Manna, Monica Gagliardi, Elena Parrini, Davide Mei, Annalisa Vetro, Claudia Bianchini, Martino Montomoli, Viola Doccini, Carla Marini, Toshimitsu Suzuki, Yushi Inoue, Kazuhiro Yamakawa, Birute Tumiene, Lynette G. Sadleir, Chontelle King, Emily Mountier, S. Hande Caglayan, Mutluay Ar- slan, Zuhal Yapıcı, Uluc Yis, Pınar Topaloglu, Bulent Kara, Dilsad Turkdogan, Aslı Gundogdu-Eken, Nerses Bebek, Sibel Ug ureri, Betü l Baykan, Barış Salman, Garen Haryanyan, Emrah Yü cesan, Yeşim Kesim, Ç ig dem Ö zkara, Annapurna Poduri, Beth R. Shiedley, Catherine Shain, Russell J. Buono, Thomas N. Ferraro, Michael R. Sperling, Warren Lo, Michael Privitera, Jacqueline A. French, Steven Schachter, Ruben I. Kuzniecky, Orrin Devinsky, Manu Hegde, Pouya Khankhanian, Katherine L. Helbig, Colin A. Ellis, Gianfranco Spalletta, Fabrizio Piras, Federica Piras, Tommaso Gili, Valentina Ciullo, Andreas Reif, Andrew McQuillin, Nick Bass, Andrew McIntosh, Douglas Blackwood, Mandy Johnstone, Aarno Palotie, Michele T. Pato, Carlos N. Pato, Evelyn J. Bromet, Celia Barreto Carvalho, Eric D. Achtyes, Maria Helena Azevedo, Roman Kotov, Douglas S. Lehrer, Dolores Malaspina, Stephen R. Marder, Helena Medeiros, Christopher P. Morley, Diana O. Perkins, Janet L. Sobell, Peter F. Buckley, Fabio Macciardi, Mark H. Rapaport, James A. Knowles, Genomic Psychiatry Cohort (GPC) Consortium, Ayman H. Fanous, Steven A. McCarroll, Namrata Gupta, Stacey B. Gabriel, Mark J. Daly, Eric S. Lander, Daniel H. Lowenstein, David B. Goldstein, Holger Lerche, Samuel F. Berkovic, and Benjamin M. Neale.
Acknowledgments
This study is funded by the Else Kröner-Fresenius-Stiftung 2020_EKEA.42 to D.L.D. and the German Research Foundation SFB 1052 project B10 to D.L.D. and A.G. D.L.D. is funded through the “Clinician Scientist Programm, Medizinische Fakultät, Universität Leipzig.” We thank the Epi25 principal investigators, local staff from individual cohorts, and all of the individuals with epilepsy who participated in the study for making possible this global collaboration and resource to advance epilepsy genetics research. This work is part of the Centers for Common Disease Genomics (CCDG) program, funded by the National Human Genome Research Institute (NHGRI) and the National Heart, Lung, and Blood Institute (NHLBI). CCDG-funded Epi25 research activities at the Broad Institute, including genomic data generation in the Broad Genomics Platform, are supported by NHGRI grant UM1 HG008895 (PIs: Eric Lander, Stacey Gabriel, Mark Daly, Sekar Kathiresan). The Genome Sequencing Program efforts were also supported by NHGRI grant 5U01HG009088-02. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the Stanley Center for Psychiatric Research at the Broad Institute for supporting the genomic data generation efforts.
Author contributions
L.B., conceptualization; writing – original draft; formal analysis; investigation; methodology. D.L.D., investigation; methodology. S.C., A.V., F.B., D.L., and H.O.H., methodology; validation; writing – original draft. Y.-Y.S. and C.-C.L., methodology; formal analysis; writing – original draft. A.-S.K. and A.G., conceptualization; writing – original draft. J.R.L. and D.L.D., conceptualization; writing – original draft; supervision; funding acquisition.
Declaration of interests
The authors declare no competing interests.
Published: June 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.06.004.
Contributor Information
Linnaeus Bundalian, Email: linnaeusbundalian@gmail.com.
Diana Le Duc, Email: diana_leduc@eva.mpg.de.
Web resources
Epi25 Collaborative, " \o "http://epi-25.org/http://epi-25.org/
Epi25 WES results browser, http://epi25.broadinstitute.org/
GoFuncR, " \o "https://github.com/sgrote/GOfuncRhttps://github.com/sgrote/GOfuncR
Genes4Epilepsy, https://github.com/bahlolab/Genes4Epilepsy
Supplemental information
The table contains all the results of the Variant Set Enrichment Analysis for non-synonymous and synonymous variants across 3 epilepsy subtypes, DEE, NAFE, and GEE. A gene is considered to be QRV-enriched if it has Z score ≥ 1.96 and FDR ≤ 0.05
The table contains the resulting overrepresented GO terms for the QRV-enriched genes across 3 epilepsy subtypes in the study. Note: The analysis was done using GoFuncR with a 95% confidence level. The returned result does not include the confidence interval
The table contains the variants that cause the enrichment for the QRV-enriched genes. The annotation of variants was done using the Ensembl’s Variant Effect Predictor for human genome assembly GRCh37
Data and code availability
The data that support this study are available in https://epi25.broadinstitute.org/downloads. This dataset consists of WES analysis for three primary epilepsy types—developmental epileptic encephalopathy (DEE), genetic generalized epilepsy (GGE), and non-acquired focal epilepsy (NAFE) compared against controls from independent sources. The code used in the analysis is deposited under https://github.com/lbundalian/EPI25_VSEA.
References
- 1.Zack M.M., Kobau R. National and State Estimates of the Numbers of Adults and Children with Active Epilepsy — United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 2017;66:821–825. doi: 10.15585/mmwr.mm6631a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T., Giri M., Xia Z., Subedi Y.N., Li Y. Genetic and epigenetic mechanisms of epilepsy: A review. Neuropsychiatr. Dis. Treat. 2017;13:1841–1859. doi: 10.2147/NDT.S142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annegers J.F., Hauser W.A., Anderson V.E., Kurland L.T. The risks of seizure disorders among relatives of patients with childhood onset epilepsy. Neurology. 1982;32:174–179. doi: 10.1212/wnl.32.2.174. [DOI] [PubMed] [Google Scholar]
- 4.Perucca P., Bahlo M., Berkovic S.F. The Genetics of Epilepsy. Annu. Rev. Genomics Hum. Genet. 2020;21:205–230. doi: 10.1146/annurev-genom-120219-074937. [DOI] [PubMed] [Google Scholar]
- 5.Gracie L., Rostami-Hochaghan D., Taweel B., Mirza N., SAGAS Scientists' Collaborative. Ahmed A., Al-Bahri T., Alexander M., Ali L., Ashton J., et al. The Seizure-Associated Genes Across Species (SAGAS) database offers insights into epilepsy genes, pathways and treatments. Epilepsia. 2022;63:2403–2412. doi: 10.1111/epi.17352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerrini R., Balestrini S., Wirrell E.C., Walker M.C. Monogenic Epilepsies: Disease Mechanisms, Clinical Phenotypes, and Targeted Therapies. Neurology. 2021;97:817–831. doi: 10.1212/WNL.0000000000012744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell C., Leu C., Feng Y.C.A., Wolking S., Moreau C., Ellis C., Ganesan S., Martins H., Oliver K., Boothman I., et al. The role of common genetic variation in presumed monogenic epilepsies. EBioMedicine. 2022;81:104098. doi: 10.1016/j.ebiom.2022.104098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leu C., Stevelink R., Smith A.W., Goleva S.B., Kanai M., Ferguson L., Campbell C., Kamatani Y., Okada Y., Sisodiya S.M., et al. Polygenic burden in focal and generalized epilepsies. Brain. 2019;142:3473–3481. doi: 10.1093/brain/awz292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International League Against Epilepsy Consortium on Complex Epilepsies. Auce P., Avbersek A., Bahlo M., Balding D.J., Bast T., Baum L., Becker A.J., Becker F., Berghuis B., et al. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat. Commun. 2018;9:5269. doi: 10.1038/s41467-018-07524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epi4K consortium. Epilepsy Phenome/Genome Project. Berkovic S.F., Bridgers J., Burgess R., Cavalleri G., Chung S.K., Cossette P., Delanty N., Dlugos D., et al. Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol. 2017;16:135–143. doi: 10.1016/S1474-4422(16)30359-3. [DOI] [PubMed] [Google Scholar]
- 11.Epi25 Collaborative Electronic address sberkovic@unimelbeduau. Epi25 Collaborative. Abbott L.E., Tashman K., Cerrato F., Singh T., Heyne H., Byrnes A., Churchhouse C., Watts N., et al. Ultra-Rare Genetic Variation in the Epilepsies: A Whole-Exome Sequencing Study of 17,606 Individuals. Am. J. Hum. Genet. 2019;105:267–282. doi: 10.1016/j.ajhg.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peljto A.L., Barker-Cummings C., Vasoli V.M., Leibson C.L., Hauser W.A., Buchhalter J.R., Ottman R. Familial risk of epilepsy: A population-based study. Brain. 2014;137:795–805. doi: 10.1093/brain/awt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandez R.D., Uricchio L.H., Hartman K., Ye C., Dahl A., Zaitlen N. Ultrarare variants drive substantial cis heritability of human gene expression. Nat. Genet. 2019;51:1349–1355. doi: 10.1038/s41588-019-0487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuk O., Schaffner S.F., Samocha K., Do R., Hechter E., Kathiresan S., Daly M.J., Neale B.M., Sunyaev S.R., Lander E.S. Searching for missing heritability: Designing rare variant association studies. Proc. Natl. Acad. Sci. USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu J.M., Satterstrom F.K., Peng M., Brand H., Collins R.L., Dong S., Wamsley B., Klei L., Wang L., Hao S.P., et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022;54:1320–1331. doi: 10.1038/s41588-022-01104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Auwera G.A., O’Connor B.D. O’Reilly Media; 2020. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. [Google Scholar]
- 18.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barry W.T., Nobel A.B., Wright F.A. Significance analysis of functional categories in gene expression studies: A structured permutation approach. Bioinformatics. 2005;21:1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 20.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kircher M., Witten D.M., Jain P., O’roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Ebbert A., Riley Z.L., Royall J.J., Aiona K., et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., Van De Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L., et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grote S., Prüfer K., Kelso J., Dannemann M. ABAEnrichment: An R package to test for gene set expression enrichment in the adult and developing human brain. Bioinformatics. 2016;32:3201–3203. doi: 10.1093/bioinformatics/btw392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Draghici S., Khatri P., Tarca A.L., Amin K., Done A., Voichita C., Georgescu C., Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wieder C., Frainay C., Poupin N., Rodríguez-Mier P., Vinson F., Cooke J., Lai R.P., Bundy J.G., Jourdan F., Ebbels T. Pathway analysis in metabolomics: Recommendations for the use of over-representation analysis. PLoS Comput. Biol. 2021;17:e1009105. doi: 10.1371/journal.pcbi.1009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sjöstedt E., Zhong W., Fagerberg L., Karlsson M., Mitsios N., Adori C., Oksvold P., Edfors F., Limiszewska A., Hikmet F., et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science. 2020;367:eaay5947. doi: 10.1126/SCIENCE.AAY5947. [DOI] [PubMed] [Google Scholar]
- 29.Li T., Wernersson R., Hansen R.B., Horn H., Mercer J., Slodkowicz G., Workman C.T., Rigina O., Rapacki K., Stærfeldt H.H., et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods. 2017;14:61–64. doi: 10.1038/nmeth.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oyrer J., Maljevic S., Scheffer I.E., Berkovic S.F., Petrou S., Reid C.A. Ion channels in genetic epilepsy: From genes and mechanisms to disease-targeted therapies. Pharmacol. Rev. 2018;70:142–173. doi: 10.1124/pr.117.014456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid C.A., Berkovic S.F., Petrou S. Mechanisms of human inherited epilepsies. Prog. Neurobiol. 2009;87:41–57. doi: 10.1016/j.pneurobio.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Ge S.X., Yao R., Jung D., Yao R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penttilä S., Palmio J., Udd B. University of Washington, Seattle; 1993. ANO5-Related Muscle Diseases. [PubMed] [Google Scholar]
- 34.Xu J., El Refaey M., Xu L., Zhao L., Gao Y., Floyd K., Karaze T., Janssen P.M.L., Han R. Genetic disruption of Ano5 in mice does not recapitulate human ANO5-deficient muscular dystrophy. Skelet. Muscle. 2015;5 doi: 10.1186/s13395-015-0069-z. 43–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiruvengadam G., Sreetama S.C., Charton K., Hogarth M., Novak J.S., Suel-Petat L., Chandra G., Allard B., Richard I., Jaiswal J.K. Anoctamin 5 Knockout Mouse Model Recapitulates LGMD2L Muscle Pathology and Offers Insight Into in vivo Functional Deficits. J. Neuromuscul. Dis. 2021;8:S243–S255. doi: 10.3233/JND-210720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi C., Feng I., Costa A.R., Pinto-Costa R., Neil J.E., Caluseriu O., Li D., Ganetzky R.D., Brasch-Andersen C., Fagerberg C., et al. Variants in ADD1 cause intellectual disability, corpus callosum dysgenesis, and ventriculomegaly in humans. Genet. Med. 2022;24:319–331. doi: 10.1016/j.gim.2021.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Focchi E., Cambria C., Pizzamiglio L., Murru L., Pelucchi S., D’Andrea L., Piazza S., Mattioni L., Passafaro M., Marcello E., et al. ATM rules neurodevelopment and glutamatergic transmission in the hippocampus but not in the cortex. Cell Death Dis. 2022;13:616–713. doi: 10.1038/s41419-022-05038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amberger J.S., Bocchini C.A., Scott A.F., Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019;47:D1038–D1043. doi: 10.1093/nar/gky1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Lin Z.J., Liu L., Xu H.Q., Shi Y.W., Yi Y.H., He N., Liao W.P. Epilepsy-associated genes. Seizure. 2017;44:11–20. doi: 10.1016/j.seizure.2016.11.030. [DOI] [PubMed] [Google Scholar]
- 40.Badouel C., Zander M.A., Liscio N., Bagherie-Lachidan M., Sopko R., Coyaud E., Raught B., Miller F.D., McNeill H. Fat1 interacts with Fat4 to regulate neural tube closure, neural progenitor proliferation and apical constriction during mouse brain development. Development (Camb.) 2015;142:2781–2791. doi: 10.1242/dev.123539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Endo M., Doi R., Nishita M., Minami Y. Ror family receptor tyrosine kinases regulate the maintenance of neural progenitor cells in the developing neocortex. J. Cell Sci. 2012;125:2017–2029. doi: 10.1242/jcs.097782. [DOI] [PubMed] [Google Scholar]
- 42.Iyengar S.K., Elston R.C. Methods in molecular biology. Methods Mol Biol; 2007. The Genetic Basis of Complex Traits; pp. 71–84. [DOI] [PubMed] [Google Scholar]
- 43.Song I., Dityatev A. Crosstalk between glia, extracellular matrix and neurons. Brain Res. Bull. 2018;136:101–108. doi: 10.1016/j.brainresbull.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Dityatev A., Fellin T. Extracellular matrix in plasticity and epileptogenesis. Neuron Glia Biol. 2008;4:235–247. doi: 10.1017/S1740925X09000118. [DOI] [PubMed] [Google Scholar]
- 45.Dityatev A. Remodeling of extracellular matrix and epileptogenesis. Epilepsia. 2010;51(Suppl 3):61–65. doi: 10.1111/j.1528-1167.2010.02612.x. [DOI] [PubMed] [Google Scholar]
- 46.Wong M. Degrading epilepsy: the role of extracellular proteases and the extracellular matrix. Epilepsy Curr. 2012;12:118–120. doi: 10.5698/1535-7511-12.3.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pitkänen A., Ndode-Ekane X.E., Łukasiuk K., Wilczynski G.M., Dityatev A., Walker M.C., Chabrol E., Dedeurwaerdere S., Vazquez N., Powell E.M. Progress in Brain Research. Prog Brain Res; 2014. Neural ECM and epilepsy; pp. 229–262. [DOI] [PubMed] [Google Scholar]
- 48.McRae P.A., Porter B.E. The perineuronal net component of the extracellular matrix in plasticity and epilepsy. Neurochem. Int. 2012;61:963–972. doi: 10.1016/j.neuint.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eskici N.F., Erdem-Ozdamar S., Dayangac-Erden D. The altered expression of perineuronal net elements during neural differentiation. Cell. Mol. Biol. Lett. 2018;23:5. doi: 10.1186/s11658-018-0073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Devinsky O., Vezzani A., O’Brien T.J., Jette N., Scheffer I.E., De Curtis M., Perucca P. Epilepsy. Nat. Rev. Dis. Primers. 2018;4:18024. doi: 10.1038/nrdp.2018.24. [DOI] [PubMed] [Google Scholar]
- 51.Heck N., Garwood J., Loeffler J.P., Larmet Y., Faissner A. Differential upregulation of extracellular matrix molecules associated with the appearance of granule cell dispersion and mossy fiber sprouting during epileptogenesis in a murine model of temporal lobe epilepsy. Neuroscience. 2004;129:309–324. doi: 10.1016/j.neuroscience.2004.06.078. [DOI] [PubMed] [Google Scholar]
- 52.Zai C.C., Lee F.H., Tiwari A.K., Lu J.Y., De Luca V., Maes M.S., Herbert D., Shahmirian A., Cheema S.Y., Zai G.C., et al. Investigation of the HSPG2Gene in tardive dyskinesia-new data and meta-analysis. Front. Pharmacol. 2018;9:974. doi: 10.3389/fphar.2018.00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin P.Y., Hung J.H., Hsu C.K., Chang Y.T., Sun Y.T. A Novel Pathogenic HSPG2 Mutation in Schwartz–Jampel Syndrome. Front. Neurol. 2021;12:632336. doi: 10.3389/fneur.2021.632336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trout A.L., Kahle M.P., Roberts J.M., Marcelo A., de Hoog L., Boychuk J.A., Grupke S.L., Berretta A., Gowing E.K., Boychuk C.R., et al. Perlecan Domain-V Enhances Neurogenic Brain Repair After Stroke in Mice. Transl. Stroke Res. 2021;12:72–86. doi: 10.1007/s12975-020-00800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Velluva A., Radtke M., Horn S., Popp B., Platzer K., Gjermeni E., Lin C.-C., Lemke J.R., Garten A., Schöneberg T., et al. Phenotype-tissue expression and exploration (PTEE) resource facilitates the choice of tissue for RNA-seq-based clinical genetics studies. BMC Genom. 2021;22:802. doi: 10.1186/s12864-021-08125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heyes S., Pratt W.S., Rees E., Dahimene S., Ferron L., Owen M.J., Dolphin A.C. Genetic disruption of voltage-gated calcium channels in psychiatric and neurological disorders. Prog. Neurobiol. 2015;134:36–54. doi: 10.1016/J.PNEUROBIO.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kambouris M., Thevenon J., Soldatos A., Cox A., Stephen J., Ben-Omran T., Al-Sarraj Y., Boulos H., Bone W., Mullikin J.C., et al. Biallelic SCN10A mutations in neuromuscular disease and epileptic encephalopathy. Ann. Clin. Transl. Neurol. 2017;4:26–35. doi: 10.1002/acn3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao D., Baker M.D., Abrahamsen B., Rugiero F., Malik-Hall M., Poon W.Y.L., Cheah K.S.E., Yao K.M., Wood J.N., Okuse K. A multi PDZ-domain protein Pdzd2 contributes to functional expression of sensory neuron-specific sodium channel NaV1.8. Mol. Cell. Neurosci. 2009;42:219–225. doi: 10.1016/j.mcn.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hiromoto Y., Azuma Y., Suzuki Y., Hoshina M., Uchiyama Y., Mitsuhashi S., Miyatake S., Mizuguchi T., Takata A., Miyake N., et al. Hemizygous FLNA variant in West syndrome without periventricular nodular heterotopia. Hum. Genome Var. 2020;7:43–45. doi: 10.1038/s41439-020-00131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baldassarre M., Razinia Z., Brahme N.N., Buccione R., Calderwood D.A. Filamin A controls matrix metalloproteinase activity and regulates cell invasion in human fibrosarcoma cells. J. Cell Sci. 2012;125:3858–3869. doi: 10.1242/jcs.104018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chelyshev Y.A., Kabdesh I.M., Mukhamedshina Y.O. Extracellular Matrix in Neural Plasticity and Regeneration. Cell. Mol. Neurobiol. 2022;42:647–664. doi: 10.1007/s10571-020-00986-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferhat L., Chevassus-Au-Louis N., Khrestchatisky M., Ben-Ari Y., Represa A. Seizures induce tenascin-C mRNA expression in neurons. J. Neurocytol. 1996;25:535–546. doi: 10.1007/bf02284821. [DOI] [PubMed] [Google Scholar]
- 63.International League Against Epilepsy Consortium on Complex Epilepsies Electronic address epilepsy-austin@unimelbeduau Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. Lancet Neurol. 2014;13:893–903. doi: 10.1016/S1474-4422(14)70171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The table contains all the results of the Variant Set Enrichment Analysis for non-synonymous and synonymous variants across 3 epilepsy subtypes, DEE, NAFE, and GEE. A gene is considered to be QRV-enriched if it has Z score ≥ 1.96 and FDR ≤ 0.05
The table contains the resulting overrepresented GO terms for the QRV-enriched genes across 3 epilepsy subtypes in the study. Note: The analysis was done using GoFuncR with a 95% confidence level. The returned result does not include the confidence interval
The table contains the variants that cause the enrichment for the QRV-enriched genes. The annotation of variants was done using the Ensembl’s Variant Effect Predictor for human genome assembly GRCh37
Data Availability Statement
The data that support this study are available in https://epi25.broadinstitute.org/downloads. This dataset consists of WES analysis for three primary epilepsy types—developmental epileptic encephalopathy (DEE), genetic generalized epilepsy (GGE), and non-acquired focal epilepsy (NAFE) compared against controls from independent sources. The code used in the analysis is deposited under https://github.com/lbundalian/EPI25_VSEA.