Abstract
The Tol-PAL system of Escherichia coli is a multiprotein system involved in maintaining the cell envelope integrity and is necessary for the import of some colicins and phage DNA into the bacterium. It is organized into two complexes, one near the outer membrane between TolB and PAL and one in the cytoplasmic membrane between TolA, TolQ, and TolR. In the cytoplasmic membrane, all of the Tol proteins have been shown to interact with each other. Cross-linking experiments have shown that the TolA transmembrane domain interacts with TolQ and TolR. Suppressor mutant analyses have localized the TolQ-TolA interaction to the first transmembrane domain of TolQ and have shown that the third transmembrane domain of TolQ interacts with the transmembrane domain of TolR. To get insights on the composition of the cytoplasmic membrane complex and its possible contacts with the outer membrane complex, we focused our attention on TolR. Cross-linking and immunoprecipitation experiments allowed the identification of Tol proteins interacting with TolR. The interactions of TolR with TolA and TolQ were confirmed, TolR was shown to dimerize, and the resulting dimer was shown to interact with TolQ. Deletion mutants of TolR were constructed, and they allowed us to determine the TolR domains involved in each interaction. The TolR transmembrane domain was shown to be involved in the TolA-TolR and TolQ-TolR interactions, while TolR central and C-terminal domains appeared to be involved in TolR dimerization. The role of the TolR C-terminal domain in the TolA-TolR interaction and its association with the membranes was also demonstrated. Furthermore, phenotypic studies clearly showed that the three TolR domains (N terminal, central, and C terminal) and the level of TolR production are important for colicin A import and for the maintenance of cell envelope integrity.
The Tol-PAL system of Escherichia coli is composed of six proteins localized in the cell envelope (TolQ, TolR, TolA, TolB, PAL, and Orf2) and one cytoplasmic protein, Orf1. These proteins are encoded by clustered genes organized in two operons (54). One operon encodes Orf1, TolQ, TolR, and TolA proteins, which, except Orf1, are localized in the inner membrane. The other operon encodes TolB and Orf2, two periplasmic proteins, and PAL, a lipoprotein of the outer membrane (for reviews, see references 2 and 37). A series of experiments, including cross-linking with formaldehyde, coimmunoprecipitations, and suppressor mutant analysis, have shown that the Tol-PAL complex is organized into two complexes: one in the inner membrane between TolA, TolQ, and TolR and one associated with the outer membrane between TolB, PAL, Orf2, and two proteins of the outer membrane, Lpp and OmpA. In the inner membrane complex, the three Tol proteins appear to interact with each other via their transmembrane domains. The TolA transmembrane domain interacts with the first transmembrane domain of TolQ (18, 22) and with TolR (18), while the third transmembrane domain of TolQ interacts with the transmembrane domain of TolR (39). The C-terminal domain of TolR also seems to play a role in the TolQ-TolR interaction, and the first and third transmembrane domains of TolQ seem to be in close contact (39). At the level of the outer membrane, TolB interacts with PAL (5) but also with Lpp and OmpA (14). PAL also interacts with OmpA (14). There is no direct evidence of interactions between the inner membrane and outer membrane complexes; however, the localization of Tol proteins in contact sites between the inner and outer membranes (25) supports this hypothesis. The Tol-PAL system appears to be conserved in many gram-negative bacteria, including Haemophilus influenzae (17), Brucella abortus (52), Pseudomonas putida (46), and Pseudomonas aeruginosa (41), suggesting that its physiological role is important. Furthermore, tol-pal mutants have been shown to be hypersensitive to drugs and detergents (16), to release periplasmic proteins (20), and to form vesicules (3). Although the diversity of these phenotypes supports a role for the Tol-PAL system in maintaining cell envelope integrity, it does not allow us to precisely define its physiological function. Recently, the finding that TolA and TolB interact with porins in vitro and in the presence of sodium dodecyl sulfate (SDS) has suggested that the Tol-PAL system might play a role in porin biogenesis or in porin activity (19, 45). On the other hand, the existence of an interaction network among TolB, PAL, OmpA, Lpp, and peptidoglycan favors a structural role of the Tol-PAL system in anchoring the inner and outer membranes to peptidoglycan (14). Tol proteins have been parasitized to permit group A colicins (A, E, and K) and single-stranded phage DNA (M13, fd, and f1) to be transported through the outer membrane (2, 16). Another system, composed of TonB, ExbB, and ExbD, is involved in the import of group B colicins (B, D, I, and M) and phage DNA (T1 and Φ80) (2, 15). Its physiological role consists of the active transport of iron siderophores and vitamin B12 across the outer membrane (for a review, see reference 32). The TonB system is thought to couple the proton motive force of the inner membrane to these transport processes (8, 44) by gating the siderophores and vitamin B12 receptors located in the outer membrane (31, 47). This gating may be induced by TonB, which interacts with siderophores and vitamin B12 receptors. ExbB and ExbD are homologous to TolQ and TolR, respectively, and share some functional reactivity (9, 10, 34). TonB and TolA transmembrane domains also exhibit some homology, while the remaining parts of both proteins have an extended conformation in the periplasm but no sequence homology. Like the Tol proteins of the inner membrane, the proteins of the TonB system interact with each other. TonB interacts with ExbB and ExbD via its transmembrane domain (30, 33, 36, 50), and ExbB and ExbD interact with each other (11, 33). TonB has been shown to oscillate between a high-affinity association with the outer membrane (probably via interactions with the receptors) and a high-affinity association with the inner membrane (probably via interactions with ExbB and -D) (40). This cycling might be the key to the mechanism of energy transduction, allowing the gating of the receptors and the pumping of the ligand in the periplasm. Recently, Higgs et al. (26) have shown that ExbB and ExbD form trimers in the inner membrane, suggesting that these proteins might form a heterohexamer, which would have implications for the mechanism of energy transduction.
Since the transmembrane domains of the inner membrane Tol proteins are homologous to those of the proteins of the TonB system, the organizations of the two systems in the inner membrane might be similar. We also wondered if we could demonstrate an interaction linking the inner membrane Tol complex to the outer membrane-associated Tol complex. Therefore, we focused our attention on TolR to answer these questions. Cross-linking and immunoprecipitation experiments were performed, and cross-linked complexes containing TolR were identified. The regions of TolR involved in the formation of these complexes were also determined.
(Some of these results were first presented at the Colicins and Other Bacteriocins European Molecular Biology Organization workshop (University of East Anglia) in April 1998.)
MATERIALS AND METHODS
Strains and media.
The strains used are listed in Table 1. They were grown at 37°C on Luria-Bertani agar plates, in liquid Luria-Bertani medium (42), or in M9 minimal medium (42) containing all amino acids except Cys and Met for specific radiolabelling experiments. The plasmids were maintained with ampicillin (100 μg · ml−1), tetracycline (12.5 μg · ml−1), kanamycin (50 μg · ml−1), or chloramphenicol (30 μg · ml−1).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genetic description | Source or reference |
|---|---|---|
| Strains | ||
| W3110 | Wild type | 1 |
| BL21(DE3) | E. coli B F−ompT (lon) hsdSB (rB− mB−) λT7RNApol | Novagen |
| GM1 | E. coli K-12 ara thi Δlac-pro F′ (lac-pro) | 51 |
| TPS300 | GM1 tolR::Cm | 51 |
| TPS13 | GM1 tolQ1 | 51 |
| 1292 | supE hsdS met gal lacY tonA | W. Wood |
| JC7782 | 1292 tolA | J. C. Lazzaroni |
| JC7752 | 1292 tolB-pal | J. C. Lazzaroni |
| JC3417 | 1292 tolB236 | J. C. Lazzaroni |
| Plasmids | ||
| pOQRAB | Tetrorf1 tolQ tolR tolA tolB | 29 |
| pQA | Amprorf1 tolQ tolA | 18 |
| pOQRA | Amprorf1 tolQ tolR tolA | 18 |
| pOQ925R207A | Amprorf1 tolQ925 tolR207 tolA | 39 |
| pEpTolQ | AmprtolQ (+30 N-terminal amino acids of ColA) | 4 |
| pBSK− | Ampr | Stratagene |
| pIN-III-ompA-2 | Ampr | 23 |
| pART3 | Cmr | 43 |
| pET-22 b(+) | Ampr | Novagen |
| pACY177 | Ampr Kanr | Biolabs |
| pBPAII | Tetrorf1 tolQ tolR tolA tolB pal | This study |
| pARTR | Cmr; pART3 vector; tolR (+3)a | This study |
| pARTR207 | Cmr; pART3 vector; tolR207 (+3) | This study |
| pBSKR | Ampr; pBSK− vector; tolR (+3) | This study |
| pBSKR207 | Ampr; pBSK− vector; tolR207 (+3) | This study |
| pINRII-III | Ampr; pIN-III-ompA-2 vector; tolRII-III | This study |
| pINRII-IIIM | Ampr Kanr; pACY177 vector; tolRII-III | This study |
| pETRII-III | Ampr; pET-22 b(+) vector; tolRII-III-His6 | This study |
| pARTRI-II | Cmr; pART3 vector; tolRI-II (+3) | This study |
| pBSKRI-II | Ampr; pBSK− vector; tolRI-II (+3) | This study |
| pINRII | Ampr; pIN-III-ompA-2 vector; tolRII | This study |
| pETRII | Ampr; pET-22 b(+) vector; tolRII-His6 | This study |
| pETRIIΔ108-117 | Ampr; pET-22 b(+) vector; tolRIIΔ108-117-His6 | This study |
| pETRIII | Ampr; pET-22 b(+) vector; tolRIII-His6 | This study |
Three unrelated residues are added at the N terminus.
Plasmids.
All the plasmids used are listed in Table 1. pBPAII was constructed as pBP (5), except that the PCR fragment encoding PAL, which was amplified from total cells and digested by AseI and BsaI, was introduced into the pOQRAB plasmid opened with the same sites.
The DNA fragments encoding the different TolR derivatives were amplified from the pBPAII plasmid by PCR. For each TolR derivative, the oligonucleotides used were designed so as to place restriction sites upstream and downstream of the amplified fragment in order to allow the in-frame fusions of the resulting fragments in the different vectors. Oligonucleotides TolR1 (5′GCCATGGAATTCGAGGTCGATCTGCCAGAC3′) and TolR2 (5′CTTAAGCTTGATAGGCTGCGTCAT3′) allowed us to obtain a DNA fragment encoding domains II and III of TolR and flanked on the 5′ side by the NcoI and EcoRI sites and on the 3′ side by the HindIII and AflII sites. This fragment was digested with NcoI/HindIII and was ligated between the same sites in pET-22 b(+) to create pETRII-III. To obtain pINRII-III, the same fragment was digested by EcoRI/HindIII and ligated in the same sites of pIN-III-ompA-2 (23). Oligonucleotides TolR3 (5′TGCTGCAGGCGCCAGAGCGCGT3′) and TolR4 (5′TGCTGCAGGCTTAGATAGGCTGCGT3′) allowed the amplification of a fragment encoding the entire TolR (except its start codon) and placed a PstI site upstream and downstream of the coding sequence. After digestion with PstI, the fragment was ligated into the pART3 plasmid (18) opened with the same site to create the pARTR plasmid. The pARTR207 plasmid was constructed along the same lines, but the matrix used for amplification was the pOQ925R207A plasmid instead of pBPAII. The DNA fragment encoding TolRI-II was amplified with oligonucleotides TolR3 and TolR5.1 (5′CACTGCAGAAGCTTTTAGTAAGGCACATCTTT3′). TolR5.1 introduced a stop codon after residue 117 and the HindIII and PstI sites 3′ of the coding sequence. pARTRI-II was obtained by ligating this fragment digested with PstI into the pART3 plasmid opened with the same site. The DNA fragment encoding TolRII was amplified by using oligonucleotides TolR1 and TolR5.1. The fragment obtained was then digested with EcoRI/HindIII and ligated into pIN-III-ompA-2 opened with the same sites to create pINRII. Other fragments encoding TolRII and TolRIIΔ108-117 were amplified by using oligonucleotides TolR1 and TolR5.2 (5′CACTGCAGAAGCTTGTAAGGCACATCTTT3′) and oligonucleotides TolR1 and TolR6 (5′GTAAGCTTAAAGACCGTTTTCGG3′), respectively. TolR5.2 and TolR6 introduced the HindIII site 3′ of the TolRII and TolRIIΔ108-117 coding sequence, respectively. The fragments obtained were then digested with NcoI/HindIII and ligated into pET-22 b(+) opened with the same sites to create pETRII and pETRIIΔ108-117, respectively. pINRII-IIIM was obtained by ligating the BamHI/BglII fragment of pINRII-III, encoding TolRII-III, into the pACY177 vector opened with the same sites. A DNA fragment encoding domain III of TolR was obtained by using the oligonucleotides TolR7 (5′GCCATGGAAGTGGACGGAATTCTGATCGGTGGCGCAAA3′) and TolR2.
TolR7 allows the amplification of a fragment that encodes domain III of TolR but also the first three residues (residues 44 to 46) and the last nine residues (residues 108 to 117) of TolR domain II separated by two residues (Gly and Ile) encoded by an EcoRI site introduced for easier screening. TolR7 also introduces an NcoI site 5′ of the coding sequence. The resulting fragment was digested by NcoI/HindIII and ligated into pET-22 b(+) opened with the same sites to create pETRIII. pBSKR and pBSKR207 were obtained by inserting the EcoRI/HindIII fragment of pARTR and pARTR207 (encoding TolR and TolR207) into the pBSK− plasmid opened with the same sites. PBSK− and pART3 have compatible replication origins, and pBSK− is a higher-copy-number plasmid than pART3.
Antibodies.
The antiserum directed against TolR (AbTolR) was raised in a rabbit by using purified TolRII-III–His protein. Three injections of purified TolRII-III–His protein (300 μg) in phosphate-buffered saline mixed with Freund’s adjuvant were given over a period of 40 days. Using defined amounts of purified TolRII-His and TolRII-III–His, we could determine that for the same amount of both proteins, AbTolR gives a three-times-stronger signal for TolRII-III than for TolRII. To lower the background of the AbTolR antiserum, it was absorbed against crude tolR cell extracts (TPS300) as previously described (5). Antisera directed against TolA (AbTolA) (18), TolB (45), PAL (5), and the monoclonal antibodies (MAbs) directed against colicin A (MAb 1C11) (13) have been described previously. The antiserum directed against LexA was a gift of R. Lloubès. All of the antisera were used at 1/500 dilution except MAb 1C11 (1/1,000).
Colicin sensitivity tests.
Cell sensitivity to colicin A was tested in liquid medium as described previously (6, 12, 21). Before their sensitivity to colicin A was tested, W3110 cells transformed with pINRII-III, pINRII, and pIN-III-ompA-2 (pIN2) were induced for 30 min with 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside) and W3110 cells transformed with pBSKR, pBSKRI-II, and pART3 were induced for 2 h with 0.5 mg of arabinose/ml.
Assays of outer membrane leakage.
The leakiness of wild-type cells overproducing TolR and TolR derivatives was assessed by testing on plates, as described by Lazzaroni and Portalier (38), the release of RNase in the extracellular medium. W3110(pINRII-III), W3110(pINRII), and W3110(pIN2) cells were induced with IPTG (10 μM), while W3110(pBSKR), W3110(pBSKRI-II), and W3110(pART3) were induced with arabinose (0.2 mg/ml). As a control for cell lysis, the release of a cytoplasmic protein marker was checked on these strains by measuring the β-galactosidase activity in the extracellular medium (42) upon culture in liquid medium and after the same inductions as for the colicin sensitivity tests.
Purification of TolR derivatives tagged with six histidines.
A 1-liter culture of BL21(DE3) cells transformed with pETRII-III, pETRII, or pETRIII plasmids (optical density at 600 nm [OD600], 1) was induced for 1.5 h with IPTG (50 μM). The cells were harvested and resuspended in 20 ml of 50 mM sodium phosphate buffer, pH 8, containing 50 mM NaCl. The purification of TolRII-III–His, TolRII-His, and TolRIII-His was then performed as previously described for TolB-His (45). TolRIII-His became undetectable after purification even when it was performed in the presence of protease inhibitor cocktail (Sigma) at 4°C. TolRII-III–His and TolRII-His were about 90% pure, but TolRII-III–His was detected as two bands of similar molecular mass. Both preparations were concentrated at 1 mg/ml in 50 mM sodium phosphate buffer, pH 8.
In vivo and in vitro cross-linking with formaldehyde.
In vivo cross-linking experiments with formaldehyde were performed directly on whole cells for 20 min as previously described by Bouveret et al. (5). The production of TolR derivatives or EpTolQ was induced (as described in the figure legends) before the cross-linkings were carried out. In vitro cross-linking experiments were carried out with 10 μg of TolRII-III–His and 10 μg of TolRII-His in a 50-μl volume. After 20 min at room temperature in the presence of 1% formaldehyde, cross-linking was stopped by adding 10 mM Tris (pH 6.8) and incubating the mixture for 15 min. Sample buffer was added, and the samples were heated for 30 min at 37°C before being analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Immunoprecipitation of cells producing EpTolQ and TolR derivatives with the 1C11 MAbs.
A 100-ml culture of TPS13(pEpTolQ) cells transformed with pINRII-IIIM, pARTR, or pARTRI-II plasmids was induced for 2.5 h with mitomycin C (300 μg/ml) (OD600, 0.7). The cells transformed with pINRII-IIIM were also induced for 30 min with IPTG (50 μM), and the cells transformed with pARTR and pARTRI-II were induced for 2.5 h with arabinose (0.5 mg/ml). Half of the cultures were treated for 20 min with formaldehyde at room temperature. The cells, treated with formaldehyde or untreated, were then converted into spheroplasts and lysed by four freeze-thaw cycles, and 2% Triton X-100 was added to solubilize the membranes (under vigorous agitation for 2 h at 4°C) as described previously (5). After centrifugation at 105,000 × g for 30 min, the soluble material was immunoprecipitated by adding Affi-gel 10 beads (Bio-Rad) coupled to 1C11 antibodies in phosphate-buffered saline buffer (10 μl for 2 OD600 units). After overnight incubation at 4°C and two washes with 150 mM phosphate buffer (pH 7.0) containing 1% Triton X-100 and 0.5% sodium deoxycholate and then with 10 mM Tris, pH 7.5, the immunoprecipitates obtained were dissociated from the beads by adding sample buffer (15 μl per 10 μl of beads) and heating for 2 min at 96°C. Samples were then analyzed by SDS-PAGE and Western blotting.
Specific radiolabelling of TolRIII-His.
BL21(pETRIII) cells were grown in M9 minimal medium at 37°C (OD600, 0.3) and were then induced for 15 min with 100 μM IPTG. Rifampin (0.2 mg/ml) was then added, and after 15 min of incubation, 1 μl of [35S]methionine/ml of culture (10 μCi/μl) was added. After a 2-min pulse, a chase was performed with cold methionine (0.5 mg/ml) for 3 min, and protein synthesis was stopped by cooling the mixture at 4°C and harvesting the cells.
Fractionations.
TolRII-III, TolRII, and TolRIII-His (after labelling) localization was assessed by cell fractionation as described previously (6, 7). Briefly, a pellet of exponentially growing cells was resuspended in 10 mM Tris (pH 6.8)–20% sucrose and incubated for 10 min at 0°C after 100 μg of lysozyme per ml and 0.5 mM EDTA were added. Spheroplasts were pelleted and washed with 10 mM Tris–20% sucrose and subjected to five freeze-thaw cycles. Membranes and cytoplasm were separated after 30 min of centrifugation at 105 × g. Periplasmic, cytoplasmic, and membrane fractions were precipitated with 20% trichloroacetic acid and analyzed by Western blotting with the AbTolR antiserum (for TolRII-III and TolRII) and autoradiography (for TolRIII-His). Protein markers of different compartments (TolB for the periplasm and membranes, LexA for the cytoplasm, and TolA for the membranes) were detected in the corresponding fractions.
A sucrose gradient fractionation was performed on W3110(pINRII-III) and W3110(pINRII) cells. A 100-ml culture of these cells was induced for 30 min with 50 and 100 μM IPTG, respectively. After being harvested, the cells were resuspended at an OD600 of 50 in 1.3 ml of 10 mM HEPES (pH 7.4)–20% sucrose with RNase and DNase (10 μg/ml each) and broken in a French pressure cell. The membranes were collected after a passage on a two-step sucrose gradient (SG0) and were then subjected to a multistep sucrose gradient (SG1) (28). The fractions were analyzed by Western blotting with the AbTolR antiserum and the antisera directed against TolA, PAL, and TolB. It should be noted that TolA, TolR, and TolB proteins have been shown to be enriched in contact sites (25).
Miscellaneous.
Standard methods were used for DNA manipulations (48). PCR amplifications were performed as described by Ho et al. (27), and DNA sequences were determined by the method of Sanger et al. (49) with the Sequenase 2.0 sequencing kit (U.S. Biochemicals). SDS-PAGE, electrotransfer onto nitrocellulose membranes, and Western blot analysis were performed as previously described (24, 35, 53).
RESULTS
TolR and TolQ can be cross-linked in vivo.
In an attempt to characterize all the interactions between TolR and the other Tol proteins, an antiserum directed against TolR was raised and in vivo cross-linking experiments with formaldehyde were performed. The antiserum was raised against a TolR derivative protein with its cytoplasmic and transmembrane domains (amino acids 1 to 43) deleted and tagged with a six-His motif for easier purification (TolRII-III–His). The antiserum obtained (AbTolR) revealed two nonspecific bands at 30 and 75 kDa in total bacterial extracts. After adsorption of the antiserum on tolR cell extracts (TPS300), the 30-kDa band was no longer recognized. Upon cross-linking of wild-type cells overproducing Orf1 (with a theoretical molecular mass of 14.7 kDa), TolQ (25.5 kDa), TolR (15.5 kDa but migrating at 19 kDa), TolA (44.2 kDa), TolB (47.3 kDa), and PAL (19 kDa) proteins [GM1(pBPAII)], four major cross-linked products were revealed by the adsorbed AbTolR antiserum after Western blotting (Fig. 1). Two complexes had very similar molecular masses at 38 and 38.5 kDa; the other two migrate at 65 and 70 kDa.
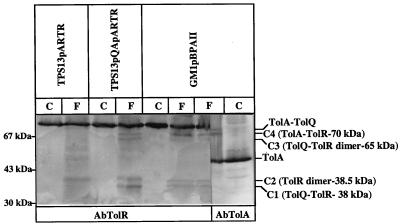
FIG. 1.
TolR is involved in four specific cross-linked complexes. Strain GM1 carrying plasmid pBPAII (which encodes Orf1, TolQ, TolR, TolA, TolB, and PAL), strain TPS13 carrying the pARTR plasmid (encoding a TolR derivative with three unrelated residues), or carrying pARTR and pQA plasmids (the latter encoding TolQ and TolA) were cross-linked for 20 min in the presence of 1% formaldehyde as described in Materials and Methods. TolR production from pARTR was induced for 2 h with 0.5 mg of arabinose/ml prior to cross-linking. Extracts of cells treated (F) or not treated (C) with formaldehyde were analyzed by immunoblotting of an SDS–7.5% PAGE gel with the AbTolR adsorbed antiserum and the AbTolA antiserum. The Western blot was visualized by the peroxidase colorimetric method. A total of 0.3 OD600 units of each cell extract was loaded. The positions of the four complexes (C1 to C4) and their molecular masses are indicated on the right, and those of the molecular mass markers are indicated on the left.
The 70-kDa complex (C4) was revealed by the antiserum directed against TolA (AbTolA) (Fig. 1) and was shifted towards lower molecular masses in tolA cells overproducing Orf1, TolQ, TolR, and TolA with the central domain of TolA deleted [JC7782(pOQRAΔh)] (data not shown); therefore, the C4 complex corresponds to the previously described TolA-TolR complex (18).
The cross-linking pattern was checked in the absence of the TolQ protein. However, the amount of TolR (and to a lesser extent of TolA) is drastically reduced in tolQ cells (TPS13). To overproduce TolR in tolQ mutant cells, TPS13 cells were transformed with a plasmid allowing the inducible overproduction of TolR (pARTR). pARTR encodes a TolR protein with three unrelated residues at its N terminus (two Ala and a Gly). pARTR, which confers resistance to chloramphenicol, cannot be maintained in TPS300 cells (tolR), which are already chloramphenicol resistant; however, it complements other cells with a tolR mutant phenotype [TPS13(pQA)]. In TPS13(pQA, pARTR) cross-linked cells, the migration of the C1 complex was not affected but that of the C2 complex was slightly shifted towards a higher molecular mass (Fig. 1). Therefore, the three additional residues of the TolR derivative encoded by pARTR allow a better separation of C1 and C2 complexes. In TPS13(pARTR) cross-linked cells, due to the low TolA production, C4 could only be detected when the chemiluminescence-sensitive revelation method was used; furthermore, both C1 and C3 complexes disappeared (Fig. 1). This suggests that TolQ might be a component of the C1 and C3 complexes. C1, with a molecular mass of 38 kDa, might correspond to a TolQ-TolR complex. In contrast, C3 might contain TolQ, TolR, and another protein. TolQ has been tagged with an epitope localized in the N-terminal domain of colicin A and recognized by the 1C11 MAbs (4). We could not directly detect any EpTolQ-TolR complex upon cross-linking of tolQ cells producing EpTolQ and TolR [TPS13(pEpTolQ, pARTR)] by using the AbTolR antiserum or MAb 1C11 because EpTolQ is produced in insufficient amounts. However, immunoprecipitation experiments with MAb 1C11 on extracts of TPS13(pARTR) and TPS13(pEpTolQ, pARTR) cells with or without cross-linking by formaldehyde clearly showed that TolR was specifically coimmunoprecipitated with EpTolQ, even without cross-linking (Fig. 2). Upon cross-linking of the cells, MAb 1C11 immunoprecipitated two other bands (around 38 and 45 kDa) which were specific for the production of TolR (Fig. 2). One of them, at 45 kDa, has a molecular mass consistent with an EpTolQ-TolR complex. Indeed, it corresponds to such a complex, as revealed by MAb 1C11 and AbTolR antisera (Fig. 2). These results, therefore, strongly suggest that TolQ and TolR interact and that the C1 complex corresponds to a TolQ-TolR complex.
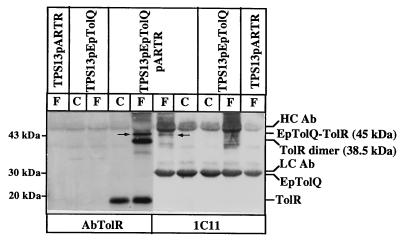
FIG. 2.
TolR is coimmunoprecipitated with EpTolQ by MAb 1C11. Extracts of TPS13(pARTR) cells (producing TolR with three unrelated residues) and TPS13(pARTR, pEpTolQ) cells (producing TolR with three unrelated residues and EpTolQ) cross-linked (F) or not (C) with 1% formaldehyde were immunoprecipitated with MAb 1C11. The immunoprecipitates were analyzed by immunoblotting with AbTolR and MAb 1C11 after separation by SDS–15% PAGE. Visualization was performed as for Fig. 1. HCAb and LCAb indicate the heavy chain and the light chain of MAb 1C11, respectively. The positions of TolR, EpTolQ, and EpTolQ-TolR (labelled by arrows) are indicated on the right; the positions of the molecular mass markers are indicated on the left.
TolR dimerizes in vivo.
In TPS13(pQA, pARTR) cells treated with formaldehyde, the C2 complex migrates slightly higher than the C2 complex of cells producing wild-type TolR (Fig. 1). The three-residue difference in TolR cannot account for such a difference. However, the difference in C2 migration might be explained if the complex corresponded to a TolR dimer.
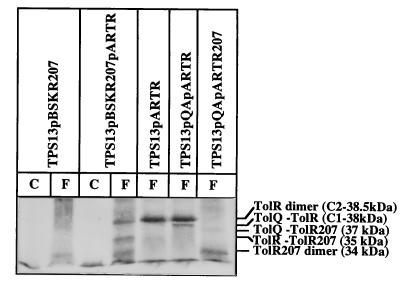
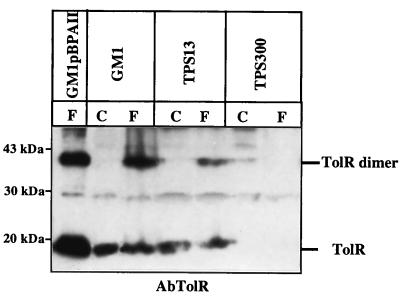
TolR207 mutant protein has its Pro in position 37 replaced by Ala (P37A), and it has been shown to suppress the tolQ925 mutation (A177V) (39). The migration of TolR207 is modified compared to wild-type TolR (18 instead of 19 kDa) (39). pARTR207 encodes a TolR (P37A) protein with three unrelated residues at its N terminus. In TPS13(pQA, pARTR207) cross-linked cells, the migration of C1 and C2 complexes was drastically affected. Indeed, they were replaced by two complexes of 37 and 34 kDa (Fig. 3). To obtain further information on the components of these complexes, cross-linking experiments were performed in tolQ cells producing TolR207 [TPS13(pBSKR207)]. In the absence of TolQ, the 37-kDa band disappeared, indicating that it might correspond to the C1 (TolQ-TolR207) complex. Therefore, the 34-kDa band might correspond to the C2 complex. The shift in the molecular mass of the C2 complex from 38.5 to 34 kDa might again be explained if the complex corresponded to a TolR dimer. This hypothesis is unambiguously confirmed by the fact that TPS13 cells producing both TolR207 and the wild-type TolR (pARTR is compatible with pBSKR207) exhibit three cross-linked products: one at 37 kDa at the level of C2 (TolR dimer) in TPS13(pARTR) cells, one at 34 kDa at the level of C2 (TolR207 dimer) in TPS13(pBSKR207) cells, and one with an intermediate migration at 35 kDa that might correspond to a TolR-TolR207 complex (Fig. 3). These different results argue in favor of a dimerization of TolR. This dimerization is not an artifact of TolR overproduction. Indeed, the dimer can be immunodetected by AbTolR when TPS13 cell extracts (which only produce a small residual amount of TolR) are overloaded on SDS-PAGE and revealed with sensitive methods (Fig. 4).
FIG. 3.
C2 complex is a TolR dimer. Strain TPS13 carrying different plasmids (pARTR, which encodes TolR with three unrelated residues at the N terminus; pARTR207 and pBSKR207, which encode the TolR207 mutant with three unrelated residues at the N terminus; and pQA, which encodes TolQ and TolA) was cross-linked for 20 min in the presence of 1% formaldehyde as described in Materials and Methods. TolR and TolR207 production from pARTR and from pARTR207 or pBSKR207, respectively, was induced for 2 h with 0.5 mg of arabinose/ml prior to cross-linking. Extracts of cells treated (F) or not treated (C) with formaldehyde were analyzed by immunoblotting of an SDS–7.5% PAGE gel with the AbTolR antiserum. Visualization was performed as for Fig. 1. A total of 0.3 OD600 units of each cell extract was loaded. The positions of TolR dimer, TolR207 dimer, TolR-TolR207, TolR-TolQ, and TolR207-TolQ complexes and their molecular masses are indicated on the right.
FIG. 4.
TolR dimerizes even without overproduction. Strains GM1 (wild-type cells), TPS13 (tolQ), TPS300 (tolR), and GM1(pBPAII) were cross-linked for 20 min in the presence of 1% formaldehyde as described in Materials and Methods. Extracts of these cells treated (F) or not treated (C) with formaldehyde were analyzed by immunoblotting with the AbTolR antiserum. Twice as much cell extract of TPS13 and TPS300 (0.6 OD600 units) as of GM1 (0.3 OD600 units) was loaded on an SDS–12.5% PAGE gel. A total of 0.2 OD600 units of GM1(pBPAII) cross-linked cells were loaded. The Western blot was visualized by secondary antibodies coupled to peroxidase and by chemoluminescence, which is a more sensitive method than the colorimetric one used for the other Western blots. The positions of TolR and TolR dimer are indicated on the right, and the positions of the molecular mass markers are indicated on the left.
In the absence of TolQ [TPS13(pARTR cells)], the C3 complex was not replaced by a lower-molecular-mass complex. Two hypotheses might account for such an observation. One is that C3 is composed of TolQ, TolR, and another protein, but TolR does not interact with the other protein in the absence of TolQ. The other possibility is that C3 is composed of only TolQ and TolR and that one of them dimerizes in the complex. Along this line, C3 might correspond to a TolQ-TolR dimer complex, since TolR has been shown to dimerize. The latter hypothesis is strengthened by the fact that MAb 1C11 coimmunoprecipitates a 38.5-kDa band that is recognized by AbTolR (Fig. 2) and migrates at exactly the same position as the TolR dimer in TPS13(pARTR) cross-linked cells (data not shown). Furthermore, the shifts in the migration of the C3 complex in cross-linked TPS13(pQA, pARTR207) and TPS13(pQA, pARTR) cells compared to that in bacteria producing wild-type TolR [TPS13(pOQRA)] are consistent with a TolQ-TolR dimer complex (data not shown).
C-terminal domain of TolR cofractionates with membranes.
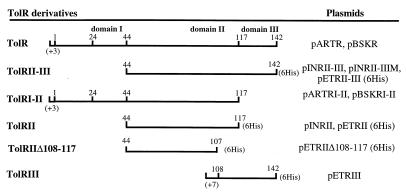
To determine the regions of TolR protein implicated in the interactions with itself and with TolQ and TolA, four TolR deletion derivatives were constructed (Fig. 5). Initially, their cellular locations were determined.
FIG. 5.
The TolR domains and the truncated TolR protein constructs. The names of TolR derivatives are indicated on the left. The corresponding vectors encoding these constructs (with or without the addition of six C-terminal His, as indicated in parentheses) are indicated on the right. The positions of the different domain boundaries are indicated (1, 44, 117, and 142); 24 corresponds to the limit of the transmembrane domain, and 108 corresponds to the first TolR residue of the TolRIII derivative; (+3) and (+7) indicate that three and seven unrelated residues, respectively, are added to the N terminus of some TolR derivatives for the constructs.
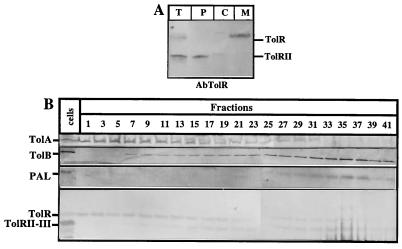
Fractionation experiments were performed on cells producing the different TolR derivatives. As expected, due to the presence of the transmembrane domain, TolRI-II was found in the membrane fractions of TPS13(pQA, pARTRI-II) cells (data not shown). TolRII was found exclusively in the periplasmic fraction (Fig. 6A), while TolRII-III was detected in the periplasmic and the membrane fractions (data not shown). The localization of TolRIII-His protein was also assessed by fractionating BL21(DE3)(pETRIII) cells. The AbTolR antiserum still recognizes TolRIII-His but with a lower affinity than the other TolR deletion mutants. To avoid any detection problems due to its low molecular mass (6 kDa), TolRIII-His protein was specifically labelled with [35S]methionine by using the T7 polymerase system, and the resulting BL21(DE3)(pETRIII) cell extracts were fractionated. TolRIII-His appeared to be partially associated with the membrane fraction (data not shown). To rule out the possibility of an aggregation problem for TolRII-III membrane localization, a sucrose gradient fractionation was performed. Fractions corresponding to the inner membrane, the contact sites, and the outer membrane were determined after the localization of different protein markers (TolA, TolB, TolR, and PAL). TolRII-III appeared to be enriched in fractions with a sucrose density intermediate between the fractions corresponding to the inner membrane and the outer membrane (Fig. 6B). TolRII-III was also present in fractions corresponding to the outer membrane. As expected, TolRII was not detected in any of the gradient fractions, confirming that it was not membrane associated (data not shown). Again, these results argue in favor of an interaction of the TolR C-terminal domain with the membranes, but they go further by suggesting a localization of this domain in the contact sites between the inner and outer membranes.
FIG. 6.
Fractionation of cells producing TolRII-III and TolRII proteins. (A) W3110(pINRII) cells were induced for 45 min with 100 μM IPTG and were fractionated as described in Materials and Methods into periplasmic (P), cytoplasmic (C), and membrane (M) fractions. Total cells is the total fraction before cell fractionation (T). Equal amounts of each cell fraction (OD600, 0.3) were loaded on SDS–12.5% PAGE gels, transferred onto nitrocellulose, and analyzed by AbTolR. (B) W3110(pINRII-III) cells were induced for 45 min with 50 μM IPTG. They were broken in a French pressure cell, and the membranes were subjected to a sucrose gradient. Fractions (100 μl) were collected, and 20 μl of each of the fractions indicated by numbers was analyzed by Western blotting with different antibodies. AbTolA, AbTolB, and AbPAL antisera allowed the determination of the fractions corresponding to the inner membrane (TolA), the contact sites (TolA and TolB), and the outer membrane (TolB and PAL). AbTolR revealed the localization of TolR and TolRII-III. Total cell extracts (cells) were also analyzed with the different antisera.
Role of TolR domains in its interaction with TolA and TolQ and in its dimerization.
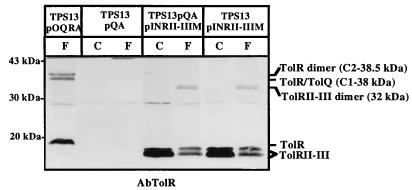
To evaluate the role of the TolR domains in its interactions with TolA and TolQ and in its dimerization, in vivo cross-linking experiments were performed on cells producing different TolR deletion derivatives. The experiments were carried out in tolQ cells (TPS13) transformed with the pQA plasmid (to restore the synthesis of TolQ and TolA) and with a compatible plasmid encoding the TolR derivatives. pINRII-IIIM, encoding TolRII-III and compatible with pQA, was constructed, and cross-linking experiments were performed on TPS13(pQA, pINRII-IIIM) cells [with TPS13(pQA) and TPS13(pINRII-IIIM) cells as controls]. After detection with the AbTolR antiserum, a single band migrating at 32 kDa appeared in TPS13(pINRII-IIIM) and TPS13(pQA, pINRII-IIIM) cells (Fig. 7). This band probably corresponds to the TolRII-III dimer complex, since its presence is correlated to TolRII-III production. No band that may correspond to TolRII-III–TolQ or TolRII-III–TolA complexes was detected with AbTolR or AbTolA antiserum (data not shown). Therefore, when its domain I has been deleted, TolR seems to still be able to dimerize but does not seem to interact with TolA and TolQ any longer. The ability of TolRII-III to dimerize was confirmed in vitro with the purified protein tagged with six His (TolRII-III–His) (data not shown).
FIG. 7.
TolRII-III dimerizes and does not interact with TolA and TolQ. TPS13(pQA), TPS13(pINRII-IIIM), and TPS13(pQA, pINRII-IIIM) cells were cross-linked for 20 min in the presence of 1% formaldehyde as described in Materials and Methods. TolRII-III production from pINRII-IIIM was induced for 30 min with 50 μM IPTG prior to cross-linking. Extracts of these cells, treated (F) or not treated (C) with formaldehyde, were analyzed by immunoblotting on an SDS–12.5% PAGE gel with the AbTolR antiserum, as described in the legend to Fig. 1. The positions of TolRII-III and TolRII-III dimer and their molecular masses are indicated on the right, and the positions of the molecular mass markers are indicated on the left.
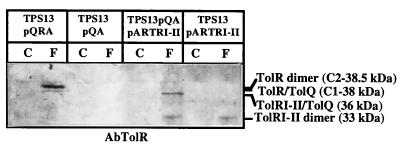
To assess the role of TolR domain III, the cross-linking patterns of TPS13(pQA, pARTRI-II), TPS13(pARTRI-II), and TPS13(pQA) cells were compared. The AbTolR antiserum revealed a unique band at 33 kDa in TPS13(pARTRI-II) cells and two bands, one migrating at 33 kDa and the other migrating at 36 kDa, in TPS13(pQA, pARTRI-II) cells (Fig. 8). The 33-kDa complex may correspond to the TolRI-II dimer because its detection is linked to TolRI-II production. In view of its molecular mass, the 36-kDa complex may correspond to the TolRI-II–TolQ complex. No band corresponding to the TolRI-II–TolA complex was revealed by the AbTolR antiserum. Furthermore, TPS13(pQA) and TPS13(pQA, pARTRI-II) cells exhibited no difference when revealed with the AbTolA antiserum (data not shown). Therefore, TolR domain III is not essential for TolR dimerization, and it plays a role in the TolA-TolR interaction. Cross-linking experiments performed on the purified TolRII-His protein (Fig. 9) showed that TolRII-His forms a 30-kDa dimer (and even a trimer). These experiments, therefore, confirm that domains I and III of TolR are not required for TolR dimerization. The exact role of TolR domain III was also evaluated by performing cross-linking experiments on BL21(DE3)(pETRIII) cells. Strikingly, TolRIII-His appeared to dimerize, but no TolA-TolRIII-His nor TolQ-TolRIII-His complexes were detected with AbTolA and AbTolR antisera, respectively (data not shown). Since our TolRIII-His mutant shares nine residues with TolRII, we wondered whether these residues were involved in TolR dimerization. A new TolRII deletion mutant with residues 108 to 117 deleted still dimerized [BL21(pETRIIΔ108-117) cells were cross-linked with formaldehyde] (data not shown). Therefore, TolR domains II and III have the intrinsic capacity to dimerize.
FIG. 8.
TolRI-II dimerizes and interacts with TolQ, but not with TolA. TPS13(pARTRI-II), TPS13(pQA), and TPS13(pQA, pARTRI-II) cells were cross-linked for 20 min in the presence of 1% formaldehyde, as described in Materials and Methods. TolRI-II production from pARTRI-II was induced for 2 h with 0.5 mg of arabinose/ml prior to cross-linking. Extracts of these cells, treated (F) or not treated (C) with formaldehyde, were analyzed by immunoblotting on an SDS–10% PAGE gel with the AbTolR antiserum as described in the legend to Fig. 1. The positions of TolRI-II dimer and TolRI-II–TolQ complex and their molecular masses are indicated on the right.
FIG. 9.
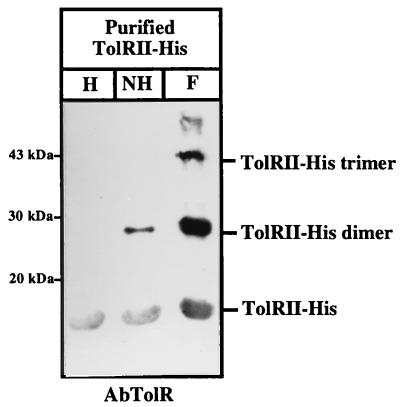
TolRII-His dimerizes in vitro. Purified TolRII-His (10 μl; 1 mg/ml) was treated (F) or not treated with formaldehyde. The samples not treated with formaldehyde were heated (H) or not heated (NH) for 20 min at 96°C before being loaded on SDS-PAGE gels. After transfer to nitrocellulose, bands were revealed with the AbTolR antiserum as described in the legend to Fig. 1. The positions of TolRII-His, TolRII-His dimer, and TolRII-His trimer are indicated on the right. The positions of the molecular mass markers are indicated on the left.
TolR, TolRII, and, to a lesser extent, TolRII-III overproduction induce a tolerant phenotype.
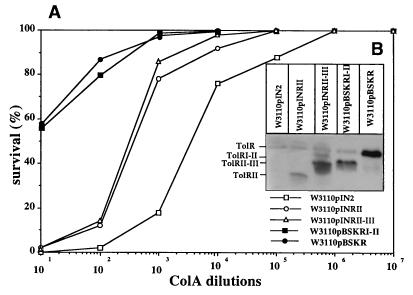
When produced in tolR cells, TolRI-II and periplasmic TolRII-III and TolRII could not restore the sensitivity of the cells to colicin A nor complement their envelope integrity defects (data not shown). However, wild-type cells producing TolRI-II [W3110(pBSKRI-II)] or TolRII-III [W3110(pINRII-III)] and TolRII [W3110(pINRII)] in their periplasms became tolerant to colicin A (Fig. 10A). Surprisingly, wild-type cells overproducing wild-type TolR [W3110(pBSKR)] also became tolerant to colicin A (Fig. 10A). All of the experiments were done in duplicate or triplicate. In parallel, the production levels of TolR and TolR derivatives were compared (Fig. 10B). TolR and TolR derivative overproduction also caused the leakage of a periplasmic marker protein (RNase I) but no significant cell lysis (data not shown), indicating that the cell envelope integrity might be affected.
FIG. 10.
Protective effect of TolR and TolR derivative overproduction against colicin A. (A) W3110 cells overproducing TolRII (pINRII) or TolRII-III (pINRII-III), or transformed with the pIN-III-OmpA-2 plasmid (pIN2) as a control, were induced for 30 min with 100 μM IPTG, and their sensitivity to colicin A was tested as described in Materials and Methods. Similarly, the colicin A sensitivity of W3110 cells overproducing TolRI-II (pBSKRI-II) and TolR (pBSKR) or transformed with the pART3 plasmid was tested after 2 h of induction with 0.5 mg of arabinose/ml. The dilution factors and the cells tested are indicated. Since the sensitivity curves of the two controls [W3110(pART3) and W3110(pIN2)] are superimposable, only one curve is presented. (B) Comparison of the amount of TolRII, TolRII-III, TolRI-II, and TolR production to that of the chromosomally encoded TolR in W3110. A total of 0.3 OD600 units of W3110(pIN2)-, W3110(pINRII)-, and W3110(pINRII-III)-induced cells (100 μM IPTG) or W3110(pBSKR)- and W3110(pBSKRI-II)-induced cells (0.5 mg/ml of arabinose), used for the sensitivity tests to colicin A, were analyzed by Western blotting with AbTolR as described in the legend to Fig. 1.
DISCUSSION
TolA, TolQ, and TolR interact with each other inside the inner membrane. TolR interacts with the transmembrane domain of TolA (18) and with the third transmembrane domain of TolQ (39); however, it is not known whether these interactions are simultaneous, since no trimeric TolA-TolR-TolQ complex has ever been detected. One of the objectives of this work was to identify other proteins of the Tol-PAL system interacting with TolR. Using formaldehyde as a cross-linker, antisera directed against many proteins of the Tol-PAL system, and mutants in the tol-pal genes, we biochemically confirmed the TolQ-TolR interaction and unambiguously showed that TolR dimerizes in vivo, without TolR overproduction. This dimerization was fully confirmed by the double-hybrid technique in Saccharomyces cerevisiae in a systematic screening of Tol protein interactions (14a). Surprisingly, in our hands, the TolR dimer (with a theoretical molecular mass in accordance with the one observed around 38.5 kDa) migrates slightly more slowly than the TolQ-TolR complex (which has an apparent molecular mass around 38 kDa but a theoretical molecular mass of 44 kDa). This might be due to the use of a different SDS-PAGE system than that used by Germon et al. (22). Indeed, the three unrelated residues of the TolR protein used to identify the two complexes induce a slight displacement of TolR dimer towards a higher molecular mass, but by no means could it be responsible for the inversion of the bands corresponding to TolR dimer and TolQ-TolR complexes. Higgs et al. (26) have recently shown that ExbB and ExbD form homodimers and homotrimers in vivo. We could not demonstrate the existence of TolR trimers in vivo, but purified forms of six-histidine-tagged TolR deletion mutants were shown to trimerize in vitro. Interestingly, a TolQ-TolR dimer complex was also detected. This means that at least one TolR molecule of the TolR dimer is able to interact with TolQ. It suggests that even in a dimerized form, TolR might still be involved in other complexes with Tol proteins of the inner membrane. If each TolR molecule of the dimer interacts with TolQ and TolA, and if these interactions are simultaneous, the Tol complex in the inner membrane would be a hexamer composed of 10 transmembrane domains. This hypothesis is consistent with what Higgs et al. (26) have proposed for the TonB system. Another alternative is that there might be a subtle equilibrium between TolQ-TolA, TolR-TolA, and TolQ-TolR dimer interactions. Despite our efforts, no TolA-TolR dimer complex or TolQ dimer-TolR dimer complex could be detected. However, the low cross-linking efficiency, the requirement for the close proximity of formaldehyde-reactive residues, and the limits of detection of the antibodies do not allow us to exclude the existence of these two complexes. Along the same line, the absence of other cross-linked products between TolR and other proteins of the Tol-PAL system does not mean that such interactions do not exist.
TolR can be divided into three distinct domains. One domain, extending from residues 1 to 43 and called domain I, includes the transmembrane domain (amino acids 23 to 43), which exhibits a high percentage of conserved residues among the different TolR and ExbD proteins sequenced so far. It is separated from another conserved domain at the C terminus (extending from residues 117 to 142 and called domain III) by the central region of the protein (domain II), which is, in contrast, poorly conserved. The conserved C-terminal domain has a residue distribution suggesting that it may form an amphiphilic α-helix (39). This suggested that it may be in contact with the inner membrane and may play a role in the TolQ-TolR interaction. TolR deletion mutants were constructed, and cross-linking experiments were performed to determine the TolR domains involved in its dimerization and its interaction with TolA and TolQ. Our results directly confirm the role of the TolR transmembrane domain in its interaction with TolA and TolQ, since no cross-linked complexes were detected between TolR with its domain I deleted and TolA or TolQ. Our results also demonstrate that both domains II and III of TolR have the intrinsic capacity to dimerize. We cannot exclude, however, the possibility that TolR domain I dimerizes. Nevertheless, this seems unlikely because it already interacts with TolQ and TolA, unless these interactions are not simultaneous. Lazzaroni et al. (39) showed that TolR domain III participates in the TolQ-TolR interaction; however, TolR with its domain III deleted still cross-linked to TolQ. Therefore, the interaction might be affected in a way that does not lower or disrupt the cross-linking. Such a phenomenon has also been observed for the tolQ925 mutant. This tolQ mutant [TolQ(A177V)], which harbors a tolR phenotype and has been shown to be affected in its interaction with TolR (39), exhibits a TolQ(A177V)-TolR cross-linked complex (data not shown). Therefore, the two proteins still interact, but TolR function is altered because the characteristics of the interaction have changed or because TolR conformation is modified. In any case, the equilibrium of TolR interactions with other proteins might be disturbed. Since TolR dimerizes and interacts with TolA, it might be that these two interactions are indirectly affected by the tolQ925 mutation. This was tested, but no difference was detected in the formation of TolR dimer and TolA-TolR complexes compared to that in bacteria expressing wild-type TolQ (data not shown). Therefore, even if the tolQ925 mutation has an indirect effect on the TolQ(A177V)-TolR, TolR dimer, and TolA-TolR interactions, this is not detectable by cross-linking. The deletion of domain III resulted in the disappearance of the TolA-TolR complex, demonstrating that the C-terminal domain of TolR also plays a role in the TolA-TolR interaction. Fractionation experiments on TolR deletion mutants have shown that TolR domain III partially cofractionates with the membranes. Furthermore, this domain is responsible for TolRII-III localization in contact sites and in the outer membrane upon sucrose gradient fractionation. Rather than a direct interaction with the inner membrane, these results suggest that TolRIII interacts with proteins localized in the contact sites and the outer membrane. These results are consistant with those of Lazzaroni et al. (39), who showed that the C-terminal domain of TolR was not accessible to carboxypeptidase. In the case of ExbD, a role for its C-terminal domain in the interaction with ExbB and TonB has also been suggested. Indeed, a deletion in ExbD domain III suppresses the negative-complementation phenotype induced by a point mutation in its transmembrane domain (11). TolRII-III and TolRI-II are unable to complement tolR cells. Therefore, domains I and III play a role in TolR function. Since these two domains are involved in the interactions with TolA and TolQ, it seems likely that these interactions are important for TolR function. Two point mutations localized in the transmembrane domain and the C-terminal domain of ExbD (D25N and L132Q, respectively) also pointed out the importance of these two domains in ExbD function (11). We could not demonstrate a direct interaction of TolR domain III with TolA or TolQ (data not shown); therefore, it might be that domain III affects the TolQ-TolR and TolA-TolR interactions by affecting domain I conformation. If we consider that TolA, TolQ, and TolR proteins do not form a stable complex but cycle between different interactions, we could imagine that TolR domain III plays a role in specific complex formation in response to different physiological states of the cells. TolR domain II also seems to play an important role in TolR function because TolRII production in the periplasms of wild-type cells induces a partial inhibition of TolR function. Its role might be to allow the formation of a TolR dimer or to allow an interaction of TolR with another protein of the Tol-PAL system. However, these interactions, if they exist, would not result in TolRII association with the membranes, as is the case for TolRIII. When produced in wild-type cells, the TolRII-III mutant also induced a negative complementation. However, the same level of negative complementation is induced by much higher overproduction of TolRII-III compared to TolRII. It might be that the presence of domain III somehow inhibits the competitive effect of domain II by affecting its conformation or by hindering its interaction with other envelope proteins.
When overproduced, wild-type TolR also induces a negative complementation. Different explanations might account for this result. First, the excess of TolR molecules might sequester TolA and TolQ, preventing them from forming active TolA-TolQ-TolR complexes in the inner membrane. Second, the excess of TolR molecules which are not involved in the TolA-TolQ-TolR inner membrane complex might interact with another protein of the Tol-PAL system, preventing it from functionally interacting with the TolA-TolQ-TolR complex. These explanations are not mutually exclusive, and this might explain why the competition induced by TolR overproduction is stronger than that induced by the TolR central domain. When TolR has its domain III deleted, it still induces a negative complementation. This result was unexpected because when ExbD (D25N) had its domain III deleted, its negative complementation was abolished (11). However, the effects induced by wild-type TolR and ExbD (D25N), although both result in a modification of the phenotype, are probably different, and this might account for this discrepancy.
It is noteworthy that the two conserved domains of TolR are involved in interactions with other Tol proteins of the inner membrane. It appears that the regions of TolA and TolQ in interaction with TolR (transmembrane domains) are also conserved among TolA, TonB, TolQ, and ExbB proteins sequenced so far in other gram-negative bacteria. These domains might be conserved because they are all involved in a specific function which is similar in the Tol and TonB systems.
Our study has allowed us to demonstrate for the first time that TolR interacts with another partner, itself, and that both its transmembrane and C-terminal domains are involved in the interactions with TolA and TolQ. We do not know if the Tol system function needs the formation of a stable multimer of a definite stoichiometry between TolA, TolQ, and TolR or results from a subtle equilibrium between different interactions. Cross-linking experiments alone cannot completely answer this question. They should be combined with suppression mutant analyses, double-hybrid system analysis, and purification experiments in conditions preserving the interactions in wild-type or in tol mutant cells and with information on the Tol-PAL system function to elucidate the stoichiometry and the dynamics of the inner membrane Tol complex.
ACKNOWLEDGMENTS
We are grateful to R. Lloubès for helpful discussions, critical reading of the manuscript, and the gift of the anti-LexA antiserum. We also thank E. Bouveret for helpful advice and L. M. Olivera for careful reading of the manuscript.
This work was supported by the Life Science department and the Mission Physique et Chimie du Vivant of the CNRS, the INSERM Programme Environnement Santé 96 (EN96C3), and the European Community (BIO4-CT97-2313).
REFERENCES
- 1.Bachman B J. Derivatives and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1190–1219. [Google Scholar]
- 2.Bénédetti H, Géli V. Colicin transport, channel formation and inhibition. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Vol. 2. New York, N.Y: Elsevier Science; 1996. pp. 665–691. [Google Scholar]
- 3.Bernadac A, Gavioli M, Lazzaroni J C, Raina S, Lloubès R. Escherichia coli tol/pal mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourdineaud J P, Howard S P, Lazdunski C. Localization and assembly onto the Escherichia coli envelope of a protein required for entry of colicin A. J Bacteriol. 1989;171:2458–2465. doi: 10.1128/jb.171.5.2458-2465.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouveret E, Derouiche R, Rigal A, Lloubès R, Lazdunski C, Bénédetti H. Peptidoglycan-associated lipoprotein-TolB interaction. J Biol Chem. 1995;270:11071–11077. doi: 10.1074/jbc.270.19.11071. [DOI] [PubMed] [Google Scholar]
- 6.Bouveret E, Rigal A, Lazdunski C, Bénédetti H. The N-terminal domain of colicin E3 interacts with TolB which is involved in the colicin translocation step. Mol Microbiol. 1997;23:909–920. doi: 10.1046/j.1365-2958.1997.2751640.x. [DOI] [PubMed] [Google Scholar]
- 7.Bouveret E, Rigal A, Lazdunski C, Bénédetti H. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol Microbiol. 1998;27:143–157. doi: 10.1046/j.1365-2958.1998.00667.x. [DOI] [PubMed] [Google Scholar]
- 8.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun V. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J Bacteriol. 1989;171:6387–6390. doi: 10.1128/jb.171.11.6387-6390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braun V, Hermann C. Evolutionary relation of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol Microbiol. 1993;8:261–268. doi: 10.1111/j.1365-2958.1993.tb01570.x. [DOI] [PubMed] [Google Scholar]
- 11.Braun V, Gaisser S, Herrmann C, Kampfenkel K, Killmann H, Traub I. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J Bacteriol. 1996;178:2836–2845. doi: 10.1128/jb.178.10.2836-2845.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavard D, Bernadac A, Pagès J M, Lazdunski C. Colicins are not transiently accumulated in the periplasmic space before release from colicinogenic cells. Biol Cell. 1984;51:79–86. doi: 10.1111/j.1768-322x.1984.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 13.Cavard D, Crozel V, Gorvel J P, Pattus F, Baty D, Lazdunski C. A molecular, genetic and immunological approach to the functioning of colicin A, a pore-forming protein. J Mol Biol. 1986;187:449–459. doi: 10.1016/0022-2836(86)90445-6. [DOI] [PubMed] [Google Scholar]
- 14.Clavel T, Germon P, Vianney A, Portalier R, Lazzaroni J C. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol Microbiol. 1998;29:359–367. doi: 10.1046/j.1365-2958.1998.00945.x. [DOI] [PubMed] [Google Scholar]
- 14a.Corda, Y., et al. Unpublished data.
- 15.Davies J K, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12. Cross-resistance among colicins of group B. J Bacteriol. 1975;123:96–101. doi: 10.1128/jb.123.1.96-101.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies J K, Reeves P. Genetics of resistance to colicins in Escherichia coli K-12. Cross-resistance among colicins of group A. J Bacteriol. 1975;123:102–117. doi: 10.1128/jb.123.1.102-117.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deich R A, Metcalf B N J, Finn C W, Farley J E, Green B A. Cloning of genes encoding a 15,000-dalton peptidoglycan-associated outer membrane lipoprotein and an antigenically related 15,000-dalton protein from Haemophilus influenzae. J Bacteriol. 1988;170:489–498. doi: 10.1128/jb.170.2.489-498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derouiche R, Bénédetti H, Lazzaroni J C, Lazdunski C, Lloubès R. Protein complex within Escherichia coli inner membrane—TolA N-terminal domain interacts with TolQ and TolR proteins. J Biol Chem. 1995;270:11078–11084. doi: 10.1074/jbc.270.19.11078. [DOI] [PubMed] [Google Scholar]
- 19.Derouiche R, Gavioli M, Bénédetti H, Prilipov A, Lazdunski C, Lloubès R. TolA central domain interacts with Escherichia coli porins. EMBO J. 1997;15:6408–6415. [PMC free article] [PubMed] [Google Scholar]
- 20.Fognini-Lefebvre N, Lazzaroni J C, Portalier R. tolA, tolB and excC, three cistrons involved in the control of pleiotropic release of periplasmic proteins by Escherichia coli K12. Mol Gen Genet. 1987;209:391–395. doi: 10.1007/BF00329670. [DOI] [PubMed] [Google Scholar]
- 21.Géli V, Baty D, Lazdunski C. Use of a foreign epitope as a tag for the localization of minor proteins: the case of the immunity protein to colicin A. Proc Natl Acad Sci USA. 1988;85:689–693. doi: 10.1073/pnas.85.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Germon P, Clavel T, Vianney A, Portalier R, Lazzaroni J C. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J Bacteriol. 1998;180:6433–6439. doi: 10.1128/jb.180.24.6433-6439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghrayeb J, Kimura H, Takahara M, Hsiung H, Masui Y, Inouye M. Secretion cloning vectors in Escherichia coli. EMBO J. 1984;3:2437–2442. doi: 10.1002/j.1460-2075.1984.tb02151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorvel J P, Rigal A, Sarles J, Maroux S. Aminopeptidase N and human blood group A-antigenicity along the digestive tract and associated glands in the rabbit. Cell Tissue Res. 1985;239:241–248. doi: 10.1007/BF00214925. [DOI] [PubMed] [Google Scholar]
- 25.Guihard G, Boulanger P, Bénédetti H, Lloubès R, Besnard M, Letellier L. Colicin A and the Tol proteins involved in its translocation are preferentially located in the contact sites between the inner and outer membranes of Escherichia coli cells. J Biol Chem. 1994;269:5874–5880. [PubMed] [Google Scholar]
- 26.Higgs P I, Myers P S, Postle K. Interactions in the TonB-dependent energy transduction complex: ExbB and ExbD form homomultimers. J Bacteriol. 1998;180:6031–6038. doi: 10.1128/jb.180.22.6031-6038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extinction using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 28.Ishidate K, Creeger E S, Zike J, Deb S, Glauner B, MacAlister T J, Rothfield L I. Isolation and differentiated membrane domains from Escherichia coli and Salmonella typhimurium, including a fraction containing attachment sites between the inner and outer membranes and the murein skeleton of the cell envelope. J Biol Chem. 1986;261:428–443. [PubMed] [Google Scholar]
- 29.Isnard M, Rigal A, Lazzaroni J C, Lazdunski C, Lloubès R. Maturation and localization of the TolB protein required for colicin import. J Bacteriol. 1994;176:6392–6396. doi: 10.1128/jb.176.20.6392-6396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaskula J C, Letain T E, Roof S K, Skare J T, Postle K. Role of the TonB amino terminus in energy transduction between membranes. J Bacteriol. 1994;175:2326–2338. doi: 10.1128/jb.176.8.2326-2338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X, Payne M A, Cao Z, Foster S B, Feix J B, Newton M C, Klebba P E. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science. 1997;276:1261–1264. doi: 10.1126/science.276.5316.1261. [DOI] [PubMed] [Google Scholar]
- 32.Kadner R J, Franklund C V, Lathrop J T. Communication between membranes in TonB-dependent transport across the bacterial outer membrane. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biological physics. Vol. 2. New York, N.Y: Elsevier Science; 1996. pp. 637–664. [Google Scholar]
- 33.Karlsson M, Hannavy K, Higgins C F. A sequence-specific function for the N-terminal signal-like sequence of the TonB protein. Mol Microbiol. 1993;8:379–388. doi: 10.1111/j.1365-2958.1993.tb01581.x. [DOI] [PubMed] [Google Scholar]
- 34.Koebnik R. The molecular interaction between components of the TonB-ExbBD-dependent and the TolQRA-dependent bacterial uptake systems. Mol Microbiol. 1993;9:219. doi: 10.1111/j.1365-2958.1993.tb01683.x. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Larsen R A, Thomas M G, Wood G E, Postle K. Partial suppression of an Escherichia coli TonB transmembrane domain mutation (D17V) by a missense mutation in ExbB. Mol Microbiol. 1994;13:627–640. doi: 10.1111/j.1365-2958.1994.tb00457.x. [DOI] [PubMed] [Google Scholar]
- 37.Lazdunski C J, Bouveret E, Rigal A, Journet L, Lloubes R, Bénédetti H. Colicin import into Escherichia coli cells. J Bacteriol. 1998;180:4993–5002. doi: 10.1128/jb.180.19.4993-5002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazzaroni J C, Portalier R. Genetic and biochemical characterization of periplasmic-leaky mutants of Escherichia coli K-12. J Bacteriol. 1981;145:1351–1358. doi: 10.1128/jb.145.3.1351-1358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazzaroni J C, Vianney A, Popot J L, Benedetti H, Samatey F, Lazdunski C, Portalier R, Geli V. Transmembrane alpha-helix interactions are required for the functional assembly of the Escherichia coli Tol complex. J Mol Biol. 1995;246:1–7. doi: 10.1006/jmbi.1994.0058. [DOI] [PubMed] [Google Scholar]
- 40.Letain T E, Postle K. TonB protein appears to transduce energy by shuttling between the cytoplasmic membrane and the outer membrane in Escherichia coli. Mol Microbiol. 1997;24:271–283. doi: 10.1046/j.1365-2958.1997.3331703.x. [DOI] [PubMed] [Google Scholar]
- 41.Lim A, Jr, De Vos D, Brauns M, Mossialos D, Gaballa A, Quing D, Cornelis P. Molecular and immunological characterization of Opr1, the 18 kDa outer membrane peptidoglycan-associated lipoprotein (PAL) of Pseudomonas aeruginosa. Microbiology. 1997;143:1709–1711. doi: 10.1099/00221287-143-5-1709. [DOI] [PubMed] [Google Scholar]
- 42.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 435–447. [Google Scholar]
- 43.Pérez-Pérez J, Gutierrez J. An arabinose-inducible expression vector, pART3, compatible with ColE1-derived plasmids. Gene. 1995;158:141–142. doi: 10.1016/0378-1119(95)00127-r. [DOI] [PubMed] [Google Scholar]
- 44.Postle K, Skare J T. Escherichia coli TonB protein is exported from the cytoplasm without proteolytic cleavage of its amino terminus. J Biol Chem. 1988;263:11000–11007. [PubMed] [Google Scholar]
- 45.Rigal A, Bouveret E, Lloubes R, Lazdunski C, Bénédetti H. The TolB protein interacts with the porins of Escherichia coli. J Bacteriol. 1997;179:7274–7279. doi: 10.1128/jb.179.23.7274-7279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodríguez-Herva J J, Ramos-González M I, Ramos J L. The Pseudomonas putida peptidoglycan-associated outer membrane lipoprotein is involved in maintenance of the integrity of the cell envelope. J Bacteriol. 1996;178:1699–1706. doi: 10.1128/jb.178.6.1699-1706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutz J M, Liu J, Lyons J A, Gotanson J, Armstrong S K, McIntosh M A, Feix J B, Klebba P E. Formation of a gated channel by a ligand-specific transport protein in the bacterial membrane. Science. 1992;258:471–475. doi: 10.1126/science.1411544. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skare J T, Ahmer B M M, Seachord C L, Darveau R P, Postle K. Energy transduction between membranes. TonB, a cytoplasmic membrane protein, can be chemically cross-linked in vitro to the outer membrane receptor FepA. J Biol Chem. 1993;268:16302–16308. [PubMed] [Google Scholar]
- 51.Sun T P, Webster R E. fii, a bacterial locus required for filamentous phage infection, and its relation to colicin-tolerant tolA and tolB. J Bacteriol. 1986;165:107–115. doi: 10.1128/jb.165.1.107-115.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tibor A, Weynants V, Denoel P, Lichtfouse B, de Bolle X, Saman E, Limet J N, Letesson J J. Molecular cloning, nucleotide sequence, and occurrence of a 16.5-kilodalton outer membrane protein of Brucella abortus with similarity to PAL lipoproteins. Infect Immun. 1994;62:3633–3639. doi: 10.1128/iai.62.9.3633-3639.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vianney A, Muller M M, Clavel T, Lazzaroni J C, Portalier R, Webster R E. Characterization of the tol-pal region of Escherichia coli K-12: translational control of tolR expression by TolQ and identification of a new open reading frame downstream of pal encoding a periplasmic protein. J Bacteriol. 1996;178:4031–4038. doi: 10.1128/jb.178.14.4031-4038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]