Abstract

The effective preparation of two new pyrimidine- and pyridine-based organic crystalline salts with substituted acidic moieties (i.e., (Z)-4-(naphthalen-2-ylamino)-4-oxobut-2-enoic acid (DCNO) and 2-hydroxy-3,5-dinitrobenzoic acid (PCNP)) using methanol as a solvent has been reported. These molecular salts have ionic interactions that are responsible for their structural stabilization in their solid-state assemblies. The crystal structures of DCNO and PCNP were determined by the single-crystal X-ray diffraction (SCXRD) technique. The SCXRD study inferred that cations and anions are strongly packed due to N–H···O, N–H···N, and C–H···O noncovalent interactions in DCNO, whereas in PCNP, N–H···N noncovalent interactions are absent. The noncovalent interactions in both organic crystalline salts were comprehensively investigated by Hirshfeld surface analysis. Further, a detailed density functional theory (DFT) study of both compounds was performed. The optimized structures of both compounds supported the existence of the H-bonding and weak dispersion interactions in the synthesized organic crystalline salt structures. Both compounds were shown to have large and noticeably different HOMO/LUMO energy gaps. The atomic charge analysis results supported the SCXRD and HSA results, showing the formation of intermolecular noncovalent interactions in both organic crystalline salts. The results of the natural bond orbital (NBO) analysis confirmed the existence of (relatively weak) noncovalent interactions between the cation and anion moieties of their organic crystalline salts. The global reactivity parameters (GRPs) analysis showed that both organic crystalline salts’ compounds should be quite thermodynamically stable and that DCNO should be less reactive than PCNP. For both compounds, the molecular electrostatic potential (MEP) analysis results support the existence of intermolecular electrostatic interactions in their organic crystalline salts.
1. Introduction
Infectious diseases like typhus, influenza, typhoid, and malaria are traditionally considered as the most fatal diseases due to the inaccessibility of effective medications.1 Pyrimidine- and pyridine-based synthetic chemical architectures have been globally used effectively as medications for malaria and other infectious diseases to save human lives.2 Pyrimethamine, medication used for the treatment of malaria, is of no more use because of the drug resistance phenomenon.3 The combinations of various compounds such as sulfalene-pyrimethamine, sulfamethoxazole-trimethoprim, and sulfadoxine-pyrimethamine have also lost their effectiveness against malaria due to the same reason.4 Among various synthetic approaches for rapid modification of the existing drugs, co-crystallization procedure is found to be an efficient protocol to obtain new drug combinations with high potential against various harmful parasites.5,6 Co-crystallization could enhance the physical properties like solubility and stability, as can be seen in the case of the increased hydration stability of the co-crystals of isoniazid with 5-fluorocytosine.7 The isoniazid-entacapone co-crystal system was found to have improved permeability, solubility, and diffusion.8 Likewise, the antitubercular compound ethionamide and salicylic acid co-crystal system were found to have increased dissolution rate.9 Pyrimethamine could also be revived via co-crystal preparation using co-partners such as theophyllinen, carbamazepine,10 and benzoic acids.11
Density functional theory has proven to be the most promising tool for investigating the electrochemical features of various compounds via computer-based calculations. The weak interaction forces such as noncovalent interactions (NCIs), can also be investigated via DFT studies,12 particularly it could be employed to examine the NCI aptitude of the co-crystal compounds.13 Moreover, the HOMO/LUMO energy gaps and nonlinear optical (NLO) properties of different classes of organic compounds could also be investigated by means of DFT calculations. Our research group has reported the synthesis of different classes of compounds, accompanied by their computational exploration, such as hydrazones,14,15 peptoids,16 β-hydroxy carbonyl compounds and chalcones,17 piperidone derivatives,18 functionalized esters,19 phosphonates,20 functionalized pyrimidines,21,22 monocarbonyl curcuminoids,23,24 unsymmetrical acyl thioureas,25 functionalized indoles,26 and co-crystal salt systems.27,28 Motivated by the above-provided considerations, herein we are presenting our research findings regarding the preparation of two new pyrimidine- and pyridine-based organic crystalline salts with substituted acidic moieties, i.e., (Z)-4-(naphthalen-2-ylamino)-4-oxobut-2-enoic acid (DCNO) and 2-hydroxy-3,5-dinitrobenzoic acid (PCNP) using methanol as the solvent; their crystalline nature was found via single-crystal X-ray diffraction (SCXRD) study, along with their electronic properties studied using DFT calculations.
2. Experimental Part
2.1. Chemicals and Instrumentation
Solvents and chemicals of the highest possible quality were used with no further purification. In the single-crystal analysis, data for the title salts were recorded using a Bruker designed diffractometer having a graphite monochromator (Kappa APEX-II CCD) at temperature 296 K. The XRD data collection was performed using finely focused Mo Kα X-rays and APEX2 software.29 SHELXT-201430 and SHEXL 2019/231 software were used for the structure solution and refinement, respectively. ORTEP-3,32 PLATON,33 and Mercury, version 4.0,34 were used for the graphical representation of the SCXRD results.
2.2. Preparation Procedure
Both organic crystalline salts were prepared by the following procedure. Accordingly, 1.2 mmol of each reacting species was placed in a 50 mL round-bottom flask containing 10 mL of methanol as a solvent. The mixture was stirred under reflux conditions for 4 h. Subsequently, the reaction mixture was cooled down to room temperature and then was left for 24 h to secure crystals of the title ionic organic crystalline salts, i.e., DCNO and PCNP (Scheme 1).
Scheme 1. Preparation of the 2,4-Diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium(Z)(4-napthalen-2-ylamino)-4-oxobut-2-enoate (1:1) (DCNO) and 2-Amino-3-methylpyridin-1-ium 2-carboxy-4,6-dinitrophenolate (1:1) (PCNP).
2.3. Computational Details
DFT studies were performed using Gaussian 16 software.35 Using the structures from the SCXRD analysis as the starting geometries, we optimized the DCNO and PCNP molecules without any symmetry constraints and then performed frequency calculations to verify that the optimized structures are true energy minima. All calculations were performed with the hybrid density functional B3LYP36 with the D2 version of Grimme’s dispersion37 included and the triple-zeta split-valence polarized basis set 6–311+G*38,39 (one set of polarization functions on heavier atoms). This approach is further referred to as B3LYP-D2/6–311+G*. We did the computational studies and all analyses listed below with the B3LYP-D2/6–311+G* approach and with the implicit effects from methanol (dielectric constant ε = 32.613) taken into account, employing the self-reliable IEF-PCM approach40 with the UFF default model as implemented in the Gaussian 16 software, with the electrostatic scaling factor α = 1.0. Below we compare the calculated structural parameters, natural population analysis (NPA) charges, natural bond orbital (NBO) interactions,41 and frontier molecular orbitals (FMOs) for both compounds. We used the values of the energies of HOMO and LUMO to compute the global reactivity parameters (GRPs)42−44 (see eqs 1–6). Equations 1 and 2 were used to calculate the values of the ionization potential (IP) and electron affinity (EA)
| 1 |
| 2 |
For global hardness η and electronegativity X values, we used eqs 3 and 4
| 3 |
| 4 |
and global electrophilicity ω value was calculated using eq 5
| 5 |
where  is the chemical potential of the system.
is the chemical potential of the system.
Finally, the global softness σ value was computed using eq 6
| 6 |
The open GL version of Molden 5.8.2 visualization program was used for the visualization of the structures and FMOs of the title compounds.45 Avogadro, version 1.1.1, was used to visualize the molecular electrostatic potential (MEP) maps.46,47
3. Results and Discussion
3.1. Single-Crystal XRD of DCNO and PCNP
The Cambridge Structure Database CSD 2022 updated in September 2022 confirmed that the crystal structures of DCNO and PCNP are novel. For both compounds, the SCXRD experimental details are listed in Table 1, whereas important bond lengths and bond angles are given in Table S1. Both compounds are crystallized in the triclinic crystal system with space group P1̅ as in the crystal structure of the literature.48
Table 1. Experimental Details of DCNO and PCNP.
| crystal data | DCNO | PCNP |
|---|---|---|
| CCDC | 2214651 | 2214652 |
| chemical formula | C26H24ClN5O3 | C13H12N4O7 |
| Mr | 489.95 | 336.27 |
| crystal system, space group | triclinic, P1̅ | triclinic, P1̅ |
| temperature (K) | 296 | 296 |
| a, b, c (Å) | 11.0693 (9), 11.6693 (10), 11.9213 (10) | 7.556 (3), 8.689 (3), 11.871 (5) |
| α, β, γ (°) | 101.780 (3), 117.440 (2), 105.139 (3) | 78.39 (2), 83.03 (2), 71.471 (19) |
| V (Å3) | 1220.91 (18) | 722.4 (5) |
| Z | 2 | 2 |
| radiation type | Mo Kα | Mo Kα |
| μ (mm–1) | 0.20 | 0.13 |
| crystal size (mm) | 0.44 × 0.40 × 0.36 | 0.38 × 0.26 × 0.16 |
| data collection | ||
| diffractometer | Bruker Kappa APEXII CCD Diffractometer | Bruker Kappa APEXII CCD Diffractometer |
| absorption correction | Multi-scan (SADABS; Bruker, 2007) | Multi-scan (SADABS; Bruker, 2007) |
| no. of measured, independent and observed [I > 2σ(I)] reflections | 31 330, 5881, 4166 | 10 310, 2822, 1593 |
| Rint | 0.050 | 0.063 |
| (sin θ/λ)max (Å–1) | 0.660 | 0.617 |
| refinement | ||
| R[F2 > 2σ(F2)], wR(F2), S | 0.072, 0.194, 1.04 | 0.068, 0.217, 1.02 |
| no. of reflections | 5881 | 2822 |
| no. of parameters | 259 | 238 |
| H-atom treatment | H-atom parameters constrained | H-atoms treated with a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å–3) | 0.99, −0.53 | 0.40, −0.20 |
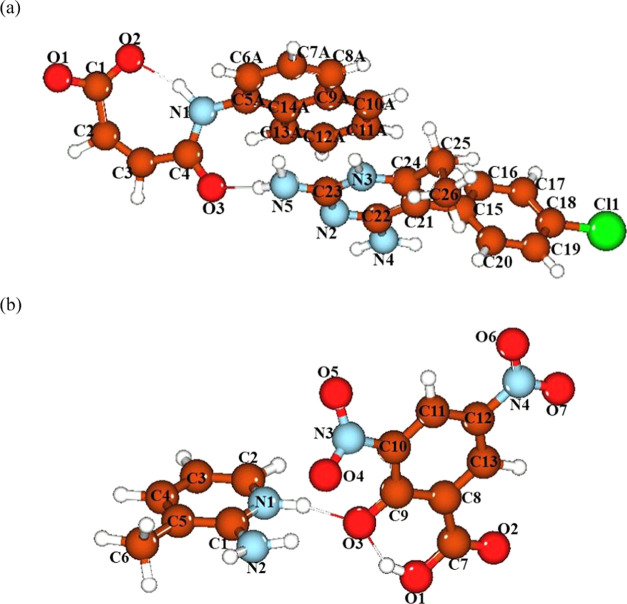
In the asymmetric unit of the organic crystalline salt DCNO (Figure 1 and Table 1), there is a cation named as 2,4-diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium (C1-C4/C5A-C14A/N1/O1-O3) and an anion named as (Z)-4-(naphthalen-2-ylamino)-4-oxobut-2-enoate (C15-C26/N2-N5/Cl1). In the cation, the naphthalene ring is disordered over two sets of sites with an occupancy ratio of 0.606(4):0.394(4). The acetate group A (C1/C2/O1/O2) and acrylamide group B (C3/C4/N1/O3) are planar with root mean square (rms) deviations of 0.0043 and 0.0095 Å, respectively, with a dihedral angle A/B of 25.1(7)°. Group B makes the dihedral angles of 62.9(1) and 60.9(1)° with the major and minor parts of the naphthalene ring, respectively. In the anion, the chlorophenyl ring C (C15-C20/Cl1) and 2,4-diaminopyrimidin-1-ium ring D (C21-C24/N2-N5) are planar with r.m.s. deviations of 0.0044 and 0.0121 Å, respectively, with a dihedral angle C/D of 77.5(7)°. The atoms of the ethyl group E (C25/C26) are at distances of −0.0544(4) and −1.4591(5) Å, respectively, from the root mean square of their parent ring D. The conformation of the anion is stabilized by intramolecular N–H···O bonding to form the S(7) loop. The cations are interlinked in the form of dimers through N–H···N bonding to form the R22 (8) loop, where NH is one of the amino groups and the N-atom of the pyrimidine ring acts as a H-bonding acceptor, whereas the anions are not directly connected with each other through any kind of H-bonding. One of the carboxylate O-atoms acts as the H-bonding acceptor for the NH of the ring and the other carboxylate O-atom acts as the H-bonding acceptor for one of the amino groups to form another R2 (8) loop. The anion and cation are also connected through C–H···O and C–H···Cl bonding in which the carbonyl O-atom of the anion acts as the H-bonding acceptor (Figure S1 and Table S2). A similar C–H···O bonding is found in the related structure.49 The crystal packing is further stabilized by C–H···π and C–O···π interactions to form an infinite chain of molecules that runs along the a-axis50 (Figure 2a and Table S2).
Figure 1.
ORTEP diagram of DCNO drawn at the probability of 30%. H-atoms are shown by small circles of arbitrary radii. Only the major part of the disordered group is shown for clarity.
Figure 2.
Graphical representation of (a) C–H···π and C–O···π interactions in DCNO and (b) C–H···π and N–O···π interactions in PCNP. Only selected H-atoms are shown for clarity. Distances are measured in Å.
In the asymmetric unit of organic crystalline salt PCNP (Figure 3 and Table 1), there is a cation named as amino-3-methylpyridin-1-ium (C1-C6/N1/N2) and an anion named as 2-carboxy-4,6-dinitrophenolate (C7-C13/N3/N4/O1-O7). The cation is planar with an rms deviation of 0.0167 Å, whereas the phenolate ring A (C8-C13/O3) is planar with an r.m.s. deviation of 0.0225 Å. The ring A makes dihedral angles of 5.08(4), 4.82(5), and 7.14(5)° with the carboxylate group B (C7/O1/O2), the first nitro group (N3/O4/O5), and the second nitro group (N4/O6/O7), respectively. These dihedral angles indicate that the anion is also almost planar. The conformation of the anion is stabilized by intramolecular O–H···O bonding to form the S(6) loop. The cations are not directly interlinked through any H-bonding; the same is the case of anions. The cation and anion are interlinked through N–H···O and C–H···O bonding (Figure 4 and Table S2). Both H-atoms of the amino group and the ring NH act as the H-bonding donor. The carboxylate O-atom (O2) acts as a H-bonding acceptor for the NH of the amino group as well as for the ring NH to form the R21 (6) loop. The NH of the amino group acts as a H-bonding donor for one of the O-atoms of the nitro group as well as for the carbonyl O-atom to form the R1 (6) loop. For C–H···O bonding, CH is from the pyrimidinium ring and the acceptor O-atom is from the nitro group that is not involved in N–H···O bonding. As a result of H-bonding, a one-dimensional chain of molecules is formed that runs along the [11̅0] direction. The crystal packing is further stabilized by C–H···π and N–O···π interactions with a H···π distance of 2.87 Å and O···π distances ranging from 3.61 to 3.82 Å (Figure 2b and Table S2). By the combination of by C–H···π and N–O···π interactions, an infinite chain containing cations and anions is formed that runs along the b-axis. The stimulated powder XRD patterns of DCNO and PCNP are shown in Figure S2. The stimulated powder patterns showed the high crystallinity of DCNO and PCNP.
Figure 3.

ORTEP diagram of PCNP drawn at the probability of 30%. H-atoms are shown by small circles of arbitrary radii.
Figure 4.
Packing diagram of the PCNP organic crystalline salt. Only selected H-atoms are shown for clarity.
3.2. Hirshfeld Surface Analysis (HSA)
The intermolecular interactions or, in general, the noncovalent interactions in DCNO and PCNP are further explored by Hirshfeld surface analysis. The analysis is done using Crystal Explorer version 21.5.51 The Hirshfeld surface concept is based on division of the molecular electron density into small fragments for the sake of integration. The Hirshfeld surface plotted over normalized distances (dnorm) provides vital information about the short interatomic contacts or the intermolecular interactions.52−54 Red, white, and blue spots on this surface, respectively, stand for the contacts with interatomic spacing less than, equal to, and greater than the sum of the Van der Waal radii of the atoms. Figure 5 shows the Hirshfeld surface plotted over dnorm for the cation and anion of DCNO and PCNP. The red spots on the surface around the amino groups, N-atom, and NH of the ring in Figure 5a show that these atoms are involved in H-bonding interactions. Similarly, the red spots on the surface around the carboxylate O-atoms and carbonyl O-atom in Figure 5b show that these atoms are engaged in H-bonding. Likewise, the red spots on the Hirshfeld surface plotted over dnorm for the cation and anion of PCNP (Figure 5c,d) show the short contacts or the H-bonding interactions.
Figure 5.
Hirshfeld surface plotted over dnorm for (a) cation of DCNO in the range −0.7012 to 1.4908 a.u., (b) anion of DCNO in the range −0.7003 to 1.3568 a.u., (c) cation of PCNP in the range −0.5145 to 1.2622 a.u., (d) anion of PCNP in the range −0.5154 to 1.1764 a.u.
2D fingerprint plots provide a unique way for the qualitative as well as quantitative exploration of the interatomic contacts in the single crystals.55−57 In this perspective, we are going to explore the important 2D fingerprint plots including the reciprocal contacts for both compounds. di and de signify the distance from the surface to the nearest nucleus interior and exterior to the surface, respectively. The 2D fingerprint plots of the important contacts for both compounds are shown in Figure 8. The contact that has the greatest contribution in the crystal packing is H···H (41.7%, Figure 6a) for DCNO and O···H (54.5%, Figure 6g) for PCNP. Although the NH is involved in H-bonding, the contribution of the N–H contact in the crystal packing of both compounds is small. Although the Cl–H contact is important as it has a large contribution in the crystal packing of DCNO (10.3%, Figure 6d), that contact is absent in PCNP.
Figure 8.
Graphical representation of voids in the crystal packing of (a) DCNO, (b) PCNP.
Figure 6.
Important 2D fingerprint plots for DCNO (a–d) and PCNP (e–h).
The interaction energy between the molecular pairs (in our case, ionic pairs) assists in understanding the supramolecular assembly of the single crystal. One can determine which pairs are strongly interacting with each other as compared to other pairs by calculating the interaction energy. The interaction energy is the sum of electrostatic coulomb, dispersion, polarization, and repulsion energies. The dispersion and polarization energies of the pair are attractive, whereas the repulsion energy is repulsive. The electrostatic coulomb energy can be attractive or repulsive. Crystal Explorer version 21.5 is used for the calculations of interaction energies. Figure 7 shows the interaction energies between ionic pairs in DCNO and PCNP. The two R22 (8) loops formed in the crystal packing of DCNO are represented by L1 and L2, respectively. Similarly, the R2 (6) and R12 (6) loops formed in the crystal packing of PCNP are represented by L3 and L4, respectively. The center of the ions is connected by a cylinder and the width of the cylinder is directly proportional to the strength of the interaction energy. For all of the loops except L4, the electrostatic coulomb energy plays a dominant role in the stabilization of the ionic pair through H-bonding, whereas for L4, the dominating one is the dispersion energy. The values of electrostatic coulomb energy for loops L1-L4 are −72.9, −23.4, −40.8 and −13.8 kJ/mol, respectively, and the corresponding dispersion energy values are −22.3, −17.8, −12.5, and −16.5 kJ/mol. The electrostatic coulomb energy contributes 62.7, 37.3, 64.1, and 41.1% of the total attractive energy for L1-L4, respectively, and the corresponding dispersion energy contribution is 19.2, 28.4, 19.7, and 49.1%.
Figure 7.
Interaction energies between ionic pairs that form H-bonding loops (a) L1 and L2 in DCNO and (g–l) L3 and L4 in PCNP. Energies shown are measured in kJ/mol.
Voids play a vital role in predicting and understanding the physical properties of the single crystals like melting point, mechanical response to stress, etc. We calculate voids in both compounds by using Hartree Fock theory.58,59 Voids are calculated by assuming that all of the atoms are spherically symmetric, and then, the electron densities of all of the atoms are added up (Figure 8). The volume of voids in the unit cell of DCNO and PCNP is 180.20 and 77.23 Å3, respectively. The space occupied by voids is 14.8 and 10.7% in DCNO and PCNP, respectively. The calculations of voids indicated that the cations and anions are strongly packed in both compounds. There is no large cavity in the crystal packing of both compounds.
4. Computational Results
4.1. Structural Features
In Figure 9, the structures of the DCNO (a) and PCNP (b) organic crystalline salt compounds optimized at the B3LYP-D2/6-311+G* level with the implicit effects from CH3OH are presented; in Table S1, the important bond lengths and bond angles in both compounds obtained using the SCXRD analysis and the DFT optimizations are compared, and Table S3 contains more selected structural parameters for the optimized structures: interatomic distances and dihedral angles. Analysis of the structures presented in Figure 9 and the selected structural parameters provided in both tables shows the following. (i) Upon the optimization, the PCNP structure becomes more distorted relative to the experimental structure compared to the DCNO compound (cf. Figures 1, 3, and 9). The distortions upon the optimization can be explained by the lack of the organic crystalline salt structure restrictions.60 Further, more significant distortions of the PCNP structure upon the optimization might be explained by the somewhat more noticeable (weak) interactions between the moieties of the DCNO structure. (ii) Comparison of the important bond distances and bond angles between the SCXRD and DFT structures (Table S1) shows quite good agreement between the experimentally measured and calculated values. (iii) Consideration of the interatomic distances provided in Table S3 shows the presence of both intra- and intermolecular H-bonding in both compounds O2···H(N1) and O3···H(N5), with distances 1.680 and 1.814 Å, respectively, in the DCNO compound (Figure 9a), and O3···H(O1), O3···H(N1), O4···H(N2), and O3···H(N2), with distances 1.658, 1.801, 2.314, and 2.130 Å, respectively, in the PCNP structure (cf. Figure 9b). These results are in qualitative agreement with the H-bonding data obtained by the SCXRD (cf. Table S3) and HS analysis.
Figure 9.
Structures of the DCNO (a) and PCNP (b) organic crystalline salt compounds optimized at the B3LYP-D2/6-311+G* level with the implicit effects from CH3OH. DCNO atom numbering scheme corresponds to Figure 1 and PCNP atom numbering scheme corresponds to Figure 4. Color coding: brown for C, red for O, light gray for H, and bright green for Cl.
Thus, it can be seen that the DFT-optimized structures of both compounds, despite the more significant distortions for PCNP, show quite good agreement for the important bond distances and bond angles with the experimental results and further also support the existence of the H-bonding and weak dispersion interactions in the synthesized organic crystalline salt structures.
4.2. Frontier Molecular Orbitals (FMOs) Analysis
Figure 10 shows the plots of the HOMOs and LUMOs of the compounds DCNO and PCNP and Table 2 provides the energies (eV) of their selected frontier MOs (HOMO – 2–LUMO + 2) along with the HOMO/LUMO gaps (eV). Analysis of these results shows the following. (i) For DCNO, the HOMO is dominated by the naphthalene ring of the (Z)-4-(naphthalen-2-ylamino)-4-oxobut-2-enoate moiety along with smaller contributions from other parts of the (Z)-4-(naphthalen-2-ylamino)-4-oxobut-2-enoate moiety and small contributions of the cation moiety. Thus, the HOMO could be considered as responsible for binding between these moieties. The LUMO is dominated by the (Z)-4-(naphthalen-2-ylamino)-4-oxobut-2-enoate moiety along with smaller contributions from the 2,4-diaminopyrimidin-1-ium ring of the cation moiety (Figure 10a). On the contrary, for the compound PCNP, the HOMO is contributed by both moieties and could be considered as responsible for binding between these moieties, whereas the LUMO is dominated by the contributions of the 2-carboxy-4,6-dinitrophenolate moiety (Figure 10b). This might imply that the HOMO → LUMO transition in this compound would mean the intermolecular charge transfer from the cationic moiety to the anionic moiety. (ii) Next, as can be seen from Table 2, both compounds have significant HOMO/LUMO gap values: 4.40 and 3.84 eV for DCNO and PCNP, respectively. The DCNO HOMO is destabilized by 0.67 eV compared to the PCNP HOMO, whereas the DCNO LUMO is destabilized by 1.23 eV compared to the PCNP LUMO. This implies certain differences in the reactivity between the compounds (see also the global reactivity parameters discussion below). (iii) For the compound DCNO, the energy difference between HOMO and HOMO – 1 is 0.46 eV and that between HOMO – 1 and HOMO – 2 is 0.30 eV, which makes HOMO – 1 and HOMO – 2 unlikely to participate in redox processes. However, the situation is different for LUMO and LUMO + 1; the energy difference between them is smaller, 0.11 eV, and the energy difference between LUMO + 1 and LUMO + 2 is 0.21 eV. This makes LUMO + 1 and LUMO + 2 more probable to participate in the redox reactions. For PCNP, the energy difference between HOMO and HOMO – 1 is merely 0.06 eV, which implies the participation of HOMO – 1 in redox processes; however, HOMO – 2 is by 1.00 eV lower in energy than HOMO – 1 and thus, would not participate in redox processes. The situation is similar for LUMO, LUMO + 1, and LUMO + 2: the energy difference between LUMO and LUMO+1 is 0.14 eV, whereas the energy gap between LUMO + 1 and LUMO + 2 is noticeably larger, 0.94 eV. This again implies that LUMO + 1 of PCNP would participate in redox reactions and LUMO+2 would not. (iv) Consideration of the FMOs HOMO – 2 and LUMO + 2 for DCNO (Figure S3a) shows that HOMO – 1 and HOMO – 2 look quite different from HOMO: HOMO – 1 is contributed only by the part of the anion moiety, whereas HOMO – 2 is dominated by the cation moiety and the naphthalene rings of the anion moiety. As for the unoccupied FMOs, LUMO + 1 has contributions from both moieties and LUMO + 2 is dominated by the anion moiety. The situation is different again for the compound PCNP: its HOMO – 1 is very similar to HOMO, and HOMO – 2 is dominated by contributions from the 2-carboxy-4,6-dinitrophenolate moiety. Both LUMO and LUMO + 1 are dominated by contributions from the 2-carboxy-4,6-dinitrophenolate moiety, and LUMO + 2 is dominated by contributions from the amino-3-methylpyridin-1-ium moiety.
Figure 10.
Plots of the selected FMOs (HOMO and LUMO) of the DCNO (a) and PCNP (b) compounds calculated at the B3LYP-D2/6-311+G* level with the implicit effects from methanol (isosurface value = 0.012). Orbital energies are given in a.u.
Table 2. Energies of the Selected FMOs (HOMO – 2–LUMO + 2) of DCNO and PCNP (eV) along with Their HOMO/LUMO Gaps (eV), Calculated at the B3LYP-D2/6-311+G* Level with the Implicit Effects from CH3OH.
| MOs | energy, eV | ΔE (HOMO/LUMO), eV |
|---|---|---|
| DCNO | ||
| LUMO | –1.86 | 4.40 |
| HOMO | –6.26 | |
| LUMO + 1 | –1.75 | 4.97 |
| HOMO – 1 | –6.72 | |
| LUMO + 2 | –1.54 | 5.48 |
| HOMO – 2 | –7.02 | |
| PCNP | ||
| LUMO | –3.09 | 3.84 |
| HOMO | –6.93 | |
| LUMO + 1 | –2.95 | 4.04 |
| HOMO – 1 | –6.99 | |
| LUMO + 2 | –2.01 | 5.98 |
| HOMO – 2 | –7.99 | |
Thus, DCNO and PCNP have large and noticeably different HOMO/LUMO energy gaps, and both DCNO HOMO and LUMO are destabilized compared to the PCNP HOMO and LUMO, but in different degrees, which implies certain differences in the reactivity between the compounds. For DCNO, HOMO – 1 and HOMO – 2 are unlikely to participate in redox processes, but LUMO + 1 and LUMO + 2 are more likely to participate in redox reactions, whereas for PCNP both HOMO – 1 and LUMO + 1 would participate in redox reactions.
4.3. Natural Population Analysis (NPA)
Analysis of the NPA charges provided in Table 3 shows the following. (i) Cl-atoms of the DCNO compound bear negligible negative charge, −0.004e, and thus would weakly participate in noncovalent intermolecular interactions. (ii) On the contrary, the oxygen atoms of the carboxyl and carbonyl groups of the DCNO anion moiety have quite significant negative charges, ca. −0.72 to −0.8e; therefore, they can participate effectively in both intramolecular and intermolecular H-bonding and electrostatic noncovalent interactions. The nitrogen atoms of the anionic and cationic moieties also bear significant negative charges, ranging within ca. −0.55 to −0.70e, and the hydrogen bond to the nitrogens N1, N3, N4, and N5 bear noticeable positive charges, ca. 0.42–0.45e. This supports the above-considered participation of these nitrogens and hydrogens in both intramolecular and intermolecular H-bonding and electrostatic noncovalent interactions (see the discussion of the DCNO crystal packing and HSA). (iii) In PCNP, the oxygen atoms of the anionic 2-carboxy-4,6-dinitrophenolate moiety bear significant negative charges, ca. −0.41 to −0.70e, and the hydrogen of the carboxyl group has a high positive charge, ca. 0.50e. This implies the formation of a quite strong intramolecular hydrogen bond O3···H(O1)-O1 as well as intermolecular H-bonding within the complex, and the possibility of formation of H-bonding and electrostatic interactions within the organic crystalline salt structure; see the discussion above. (iv) The nitrogens of the PCNP cation moiety, N1 and N2, bear quite noticeable negative charges, ca. −0.50 and −0.74e, respectively, and the hydrogens connected to them have significant positive charges, ca. 0.44–0.45e, which implies the formation of quite strong intermolecular H-bonding within the organic crystalline salt complex as well as intermolecular noncovalent interactions. The nitrogens of the anion moiety, N3 and N4, have quite significant positive charges, ca. 0.50e, and thus might participate in intermolecular noncovalent interactions as well.
Table 3. NPA Charges, e, on the Selected Atoms of the Compounds DCNO and PCNP, Computed at the B3LYP-D2/6-311+G* Level with the Implicit Effects from Methanol.
| atom | charge, e | atom | charge, e |
|---|---|---|---|
| DCNO | PCNP | ||
| Cl1 | –0.004 | N1 | –0.501 |
| O1 | –0.765 | H(N1) | 0.454 |
| O2 | –0.799 | N2 | –0.738 |
| O3 | –0.715 | HA(N2)a | 0.438 |
| N1 | –0.640 | O1 | –0.698 |
| H(N1) | 0.448 | H(O1) | 0.502 |
| N5 | –0.749 | O2 | –0.650 |
| HA(N5)a | 0.445 | O3 | –0.776 |
| N2 | –0.575 | O4 | –0.405 |
| N3 | –0.547 | O5 | –0.408 |
| H(N3) | 0.431 | N3 | 0.494 |
| N4 | –0.700 | O6 | –0.423 |
| HA(N4) | 0.418 | O7 | –0.420 |
| HB(N4) | 0.416 | N4 | 0.489 |
H-atoms participating in hydrogen bonding.
Thus, the charge analysis results support the SCXRD and HSA results showing the formation of various intermolecular noncovalent interactions in both organic crystalline salts.
4.4. Natural Bonding Orbital (NBO) Analysis
The NBO analysis results provide us with knowledge of the orbital interactions of different types, both intra- and intermolecular.41 The NBO analysis is performed by consideration of all possible interactions among the filled, or donor, Lewis-type NBOs and empty, or acceptor, non-Lewis NBOs. Their energetic contributions in the interactions are evaluated using the 2nd-order perturbation theory. These interactions result in the decrease of the localized NBOs’ occupancy in the idealized Lewis structure and corresponding increase of the occupancy of the empty non-Lewis orbitals. As a consequence, they are referred to as ‘delocalization’ corrections to the 0th-order natural Lewis structure. The stronger donor-acceptor interactions are characterized by higher stabilization energies. The 2nd-order stabilization energy E(2) is computed according to eq 7
| 7 |
where εi and εj are off-diagonal elements and Fi·j is the diagonal element of the NBO Fock matrix, qi is the donor orbital possession, and E(2) is the energy of stabilization.41
Representative values of the NBO analysis results for the studied compounds are provided in Table S2. Analysis of these results shows the following. (i) The DCNO compound is characterized by a broad range of stabilization energies within both cationic and anionic moieties (intramolecular stabilization energies), from 5.07 till 90.73 kcal/mol, whereas the intermolecular stabilization interactions are not numerous, with the stabilization energies being within a narrow range, 6.15—11.15 kcal/mol (values underlined in Table S2). The intramolecular donor-acceptor stabilizing interactions are represented by the following types: σ(N–H) → σ*(O–C), σ(C–H) → σ*(C–C), π(C–C) → π*(O–C), π(C–C) → π*(C–C), π(C–C) → LP*(C), LP(O) → σ*(O–C), LP(O) → σ*(C–C), LP(O) → σ*(N–H), LP(O) → σ*(N–C), LP(N) → π*(O–C), LP(N) → σ*(C–C), LP(N) → π*(C–C), LP(N) → π*(N–C), LP(C) → π*(C–C), π(N–C) → π*(N–C), π(N–C) → π*(C–C), σ(N–H) → σ*(N–C), σ(C–C) → σ*(C–C), σ(C–H) → σ*(N–C), and LP(Cl) → π*(C–C). Among them, the following interactions with the most significant stabilization energies, kcal/mol (given in red in Table S2), can be highlighted: LP(O2) → π*(O1-C1), 90.73, responsible for the interaction within the carboxyl group of the anionic moiety; LP(N1) → π*(O3-C4), 80.67, responsible for the interaction within the amido group of the anionic moiety; LP(C9A) → π*(C7A-C8A) and LP(C9A) → π*(C10A-C11A), 56.85 and 55.28, respectively, responsible for the stabilization of the naphthalene ring of the anionic moiety; π(N2-C22) → π*(N3-C23), 53.30, responsible for the stabilization of the 2,4-diaminopyrimidin-1-ium ring of the cationic moiety; and LP(N4) → π*(N2-C22) and LP(N5) → π*(N3-C23), 69.54 and 72.65, respectively, responsible for the stabilization of the 2,4-diaminopyrimidin-1-ium ring of the cationic moiety. The intermolecular stabilizing interactions with the stabilization energies, kcal/mol, are as follows: LP1(O3) → σ*(N5-H(N5)), 6.15, and LP2(O3) → σ*(N5-H(N5)), 11.15, responsible for the H-bonding between the cation and anion moieties, and LP(C9A) → π*(N3-C23), 6.94, responsible for the (weak) interaction between the naphthalene ring of the anion moiety and 2,4-diaminopyrimidin-1-ium ring of the cationic moiety. These results imply that there are (relatively weak) H-bonding and other stabilizing interactions between the moieties of the DCNO organic crystalline salt compound, which further supports the SCXRD results and HSA results. (ii) The PCNP compound is characterized by a much broader range of stabilization energies within both cationic and anionic moieties (intramolecular stabilization energies), 5.23–825.16 kcal/mol, whereas there are only two intermolecular stabilization interactions with the stabilization energies high enough, 6.90 and 8.62 kcal/mol (values underlined in Table S2). The intramolecular donor-acceptor stabilizing interactions are represented by the following types: σ(N–H) → σ*(N–C), π(N–C) → π*(C–C), π(C–C) → π*(N–C), π(C–C) → π*(C–C), LP(N) → σ*(N–C), σ(O–H) → σ*(O–C), π(O–N) → LP*(O), π(O–N) → π*(O–N), π(C–C) → LP*(C), π(C–C) → π*(O–C), π(C–C) → π*(O–N), LP(O) → σ*(C–C), LP(O) → π*(C–C), LP(O) → π*(O–C), LP(O) → σ*(O–H), LP(O) → σ*(N–H), LP(O) → LP*(C), LP(O) → σ*(N–O), LP(O) → π*(N–O), LP(C) → LP*(C), LP(C) → π*(N–O), and LP(C) → π*(C–C). Among them, the following interactions with the most significant stabilization energies, kcal/mol (highlighted in red in Table S2), can be noticed: LP(N2) → σ*(N1-C1), 70.39, responsible for the stabilization in the cationic moiety; π(C8-C13) → LP*(C9), 52.95, responsible for the stabilization of the benzene ring in the anionic moiety; LP(O1) → π*(O2-C7), 50.21, responsible for the stabilization in the carboxyl group of the anionic moiety; LP(O3) → LP*(C9), 147.66, responsible for the carbonyl group interaction with the benzene ring in the anionic moiety; LP(O5) → π*(O4-N3), 146.22, responsible for the stabilization of the nitro group of the anion moiety interacting with the cation moiety by H-bonding; LP(O6) → π*(O7-N4), 153.48, responsible for the stabilization of another nitro group of the anion moiety; LP(C10) → LP*(C9), 825.16, responsible for the stabilization of the benzene ring of the anion moiety; LP(C10) → π*(O4-N3), 477.41, responsible for the interaction between the benzene ring and nitro group of the anion moiety; and LP(C10) → π*(C11-C12), responsible for the stabilization of the benzene ring of the anion moiety. The intermolecular stabilizing interactions with the stabilization energies, kcal/mol, are as follows: LP1(O3) → σ*(N1-H(N1)) and LP2(O3) → σ*(N1-H(N1)), 6.90 and 8.62, respectively, responsible for the H-bonding between two moieties of the crystal. These results imply that there is (relatively weak) H-bonding between the moieties of the PCNP crystalline compound, which further supports the SCXRD results and HSA results. It is also worthwhile to notice that sometimes the NBO approach does not completely correctly describe the aromatic structures, which gives rise to the interactions like LP(C) → LP*(C), π(C–C) → LP*(C), etc.
Thus, as can be seen from the results of the NBO analysis, for both compounds there exist (relatively weak) interactions (H-bonding and other interactions) between the moieties of their organic crystalline salts, which further supports the SCXRD and HSA results.
4.5. Global Reactivity Parameter (GRP) Analysis
The global reactivity parameters, ionization potential (IP), electron affinity (EA), global softness (σ), global electrophilicity (ω), global hardness (η), global electronegativity (X), and chemical potential (μ), were computed using the FMOs energies (Table 2) according to eqs 1–6 (see Section 2.3),42−44 and the computed values in eV are presented in Table 4.
Table 4. Calculated GRPs for the DCNO and PCNP Compounds (eV).
| IP | EA | gap | X | η | μ | σ | ω |
|---|---|---|---|---|---|---|---|
| DCNO | |||||||
| 6.26 | 1.86 | 4.40 | 4.06 | 2.20 | –4.06 | 0.227 | 3.746 |
| PCNP | |||||||
| 6.93 | 3.09 | 3.84 | 5.01 | 1.92 | –5.01 | 0.260 | 6.536 |
Analysis of the values of the GRPs for the two compounds shows the following. (i) Both DCNO and PCNP compounds have high ionization potential values, 6.26 and 6.93 eV, respectively, and the PCNP electron affinity value, 3.09 eV, is noticeably higher than the DCNO electron affinity value, 1.86 eV. This implies that both compounds should be quite stable in oxidation processes, especially PCNP; that is, they would be poor electron donors, but the PCNP compound would be much a better electron acceptor compared to DCNO and thus should be more active in reduction processes. (ii) Judging by their large HOMO/LUMO gap values, 4.40 and 3.84 eV, both compounds should be quite thermodynamically stable, which is also supported by their significant chemical potential values, −4.06 and −5.01 eV. However, in this case, the HOMO/LUMO gap values and chemical potential values show opposite trends, because according to the first value, DCNO should be more stable, and according to the second value, PCNP should be more stable. This implies that other factors should be also taken into account. (iii) Next, global electronegativity X and global electrophilicity ω values for DCNO, 4.06 and 3.746 eV, respectively, are noticeably lower than for PCNP, 5.01 and 6.536 eV, respectively, which is in agreement with the above-considered electron affinity values, implying that PCNP should be more active in reduction processes. (iv) Finally, the global hardness η value for DCNO, 2.20 eV, is larger than for PCNP, 1.92 eV, and the DCNO global softness σ value, 0.227 eV, is smaller than the PCNP global softness value, 0.260 eV, which implies that the compound DCNO should be less reactive than the PCNP compound.
Thus, the GRP analysis shows that both organic crystalline salt compounds should be quite thermodynamically stable. Also, they should be both stable in oxidation processes and DCNO should be quite stable in reduction processes. Finally, the compound DCNO should be less reactive than the PCNP compound.
4.6. Molecular Electrostatic Potential (MEP) Mapping
The MEP plots of the DCNO and PCNP compounds provided in Figure 11 show the following. (i) In the compound DCNO, there is significant accumulation of negative MEP (as indicated by red) at the carboxyl group of the anion moiety along with slight accumulation of negative MEP on the Cl-atom and in the benzene ring of the cation moiety, as well as in the naphthalene ring of the anion moiety. Also, there is significant accumulation of positive MEP (as indicated by blue) on the nitrogen-containing groups of the cation moiety and less significant accumulation of positive MEP on hydrogens of the benzene ring of the cation moiety and on hydrogens of the naphthalene ring of the anion moiety. This implies the possibility of intermolecular electrostatic interactions in the DCNO crystal, in agreement with the SCXRD and HSA results (see above). (ii) In the compound PCNP, there is significant accumulation of negative MEP (as indicated by red) on the nitro group of the anion moiety located on the side from the cation moiety and on the side of the cation moiety benzene ring opposite to the amino group along with smaller accumulation of negative MEP on the oxygens of the carboxyl group of the anion moiety and on the oxygens of its nitro group participating in H-bonding with the cation moiety. Further, there is significant accumulation of positive MEP (as indicated by blue) on the nitrogen-containing groups of the cation moiety and slight accumulation of positive MEP on hydrogens of the benzene ring of the cation moiety and on the hydrogen of the carboxyl group of the anion moiety. This again implies the possibility of intermolecular electrostatic interactions in the PCNP crystal, in agreement with the SCXRD and HSA results (see above).
Figure 11.
Molecular Electrostatic Potential plots for the DCNO (a) and PCNP (b) compounds calculated at the B3LYP-D2/6-311+G* level with the implicit effects from methanol. Bright red color corresponds to the negative MEP, −0.01 a.u., whereas the bright blue color corresponds to the positive MEP, +0.01 a.u.
Thus, for both compounds the MEP analysis results support the existence of intermolecular electrostatic interactions in their organic crystalline salts, in good agreement with the SCXRD and HSA results.
5. Conclusions and Prospectives
Two new organic crystalline salts DCNO and PCNP have been prepared via the acid-base neutralization reaction. The structures of both compounds having crystalline nature were investigated and confirmed via SCXRD studies, which revealed that the crystal packing of both compounds was stabilized by strong as well as comparatively weak noncovalent interactions. The noncovalent interactions were thoroughly explored by the Hirshfeld surface analysis as well. Void analysis conformed that the cation and anions were strongly packed with no large cavity in both compounds. DFT-optimized structures of both compounds show quite good agreement for the important bond distances and bond angles with the experimental results and support the existence of the H-bonding and weak dispersion interactions in the synthesized organic crystalline salt structures. DCNO and PCNP have large and noticeably different HOMO/LUMO energy gaps, and both DCNO HOMO and LUMO are destabilized compared to PCNP HOMO and LUMO, but in different degrees, which implies certain differences in the reactivity between the compounds. For DCNO, HOMO – 1 and HOMO – 2 are unlikely to participate in redox processes, but LUMO + 1 and LUMO + 2 are more likely to participate in redox reactions, whereas for PCNP both HOMO – 1 and LUMO + 1 would participate in redox reactions. The NPA results support the SCXRD and HSA results, showing the formation of various intermolecular noncovalent interactions in both organic crystalline salts. The results of the NBO analysis for both compounds confirm that there exist (relatively weak) interactions (H-bonding and other interactions) between the moieties of their co-crystals, which further supports the SCXRD and HSA results. The GRP analysis shows that both organic crystalline salt compounds should be quite thermodynamically stable and stable in oxidation processes, whereas DCNO should be quite stable in reduction processes. Finally, the compound DCNO should be less reactive than the PCNP compound. For both compounds the MEP analysis results support the existence of intermolecular electrostatic interactions in their organic crystalline salts, in good agreement with the SCXRD and HSA results.
Acknowledgments
The authors express their appreciation to the Deanship of Scientific Research at King Khalid University, Saudi Arabia, for funding this work through research group program under grant no. RGP-2/370/44. Akbar Ali gratefully acknowledges the support of TWAS-CNPq and HEC Pakistan. Aleksey Kuznetsov appreciates the financial support of USM. Powered@NLHPC: this research was partially supported by the supercomputing infrastructure of the NLHPC (ECM-02).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c01659.
Selected bond distances and bond angles for the DNCO and PCNP compounds, SCXRD/B3LYP-D2/6-311+G*, implicit CH3OH (Table S1); hydrogen-bond geometry (Å, °) for DCNO and PCNP (Table S2); important interatomic distances and dihedral angles for the DNCO and PCNP compounds, SCXRD/B3LYP-D2/6-311+G*, implicit CH3OH (Table S3); representative values of the NBO analysis for the studied compounds (Table S4); packing diagram of the DCNO organic crystalline salt, only selected H-atoms are shown for clarity (Figure S1); stimulated powder XRD pattern of (a) DCNO, (b) PCNP (Figure S2); plots of the selected FMOs (HOMO – 2—LUMO + 2) of the DCNO (a) and PCNP (b) compounds calculated at the B3LYP-D2/6-311+G* level with the implicit effects from methanol (isosurface value = 0.012), orbital energies are given in a.u ((Figure S3)) (PDF)
Processing Summary (CIF)
Processing Summary (CIF)
The authors declare no competing financial interest.
Notes
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary Material
References
- Quirke V.; Gaudillière J. P. The era of biomedicine: Science, medicine, and public health in Britain and France after the second world war. Med. Hist. 2008, 52, 441–452. 10.1017/S002572730000017X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan E.; Bhandari S.; Gilbert R. E.; Stanford M. Antibiotics versus no treatment for toxoplasma retinochoroiditis. Cochrane Database Syst. Rev. 2016, 2016, 1–25. 10.1002/14651858.CD002218.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper C.; Pearce R.; Nair S.; Sharp B.; Nosten F.; Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science 2004, 305, 1124. 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Gatton M. L.; Martin L. B.; Cheng Q. Evolution of resistance to sulfadoxine-pyrimethamine in Plasmodium falciparum. Antimicrob. Agents Chemother. 2004, 48, 2116–2123. 10.1128/AAC.48.6.2116-2123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Du S.; Zhang R.; Jia X.; Yang T.; Zhang X. Drug-drug cocrystals: Opportunities and challenges. Asian J. Pharm. Sci. 2021, 16, 307–317. 10.1016/j.ajps.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A.; Kuznetsov A.; Ibrahim M.; Abbas A.; Akram N.; Maqbool T.. Chemistry and modern techniques of characterization of Co-crystals. 2022.
- Bu F.-Z.; Yu Y.-M.; Shen Y.-L.; Wu Z.-Y.; Li Y.-T. Cocrystallization with nutrient ferulic acid towards reducing the dissolubility behaviors of antifungal drug 5-fluorocytosine: An integrated theoretical and experimental case research. J. Mol. Struct. 2023, 1275, 134601 10.1016/j.molstruc.2022.134601. [DOI] [Google Scholar]
- Bommaka M. K.; Mannava M. C.; Suresh K.; Gunnam A.; Nangia A. Entacapone: Improving aqueous solubility, diffusion permeability and cocrystal stability with theophylline. Cryst. Growth Des. 2018, 18, 6061–6069. 10.1021/acs.cgd.8b00921. [DOI] [Google Scholar]
- Bernasconi D.; Bordignon S.; Rossi F.; Priola E.; Nervi C.; Gobetto R.; Voinovich D.; Hasa D.; Duong N. T.; Nishiyama Y. Selective synthesis of a salt and a cocrystal of the ethionamide–salicylic acid system. Cryst. Growth Des. 2019, 20, 906–915. 10.1021/acs.cgd.9b01299. [DOI] [Google Scholar]
- Delori A.; Galek P. T.; Pidcock E.; Patni M.; Jones W. Knowledge-based hydrogen bond prediction and the synthesis of salts and cocrystals of the anti-malarial drug pyrimethamine with various drug and GRAS molecules. CrystEngComm 2013, 15, 2916–2928. 10.1039/c3ce26765b. [DOI] [Google Scholar]
- Ashfaq M.; Ali A.; Kuznetsov A.; Tahir M. N.; Khalid M. DFT and single-crystal investigation of the pyrimethamine-based novel co-crystal salt: 2, 4-diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium-4-methylbenzoate hydrate (1: 1: 1)(DEMH). J. Mol. Struct. 2021, 1228, 129445 10.1016/j.molstruc.2020.129445. [DOI] [Google Scholar]
- Cavallo G.; Metrangolo P.; Pilati T.; Resnati G.; Terraneo G. Naming interactions from the electrophilic site. Cryst. Growth Des. 2014, 14, 2697–2702. 10.1021/cg5001717. [DOI] [Google Scholar]
- Khalid M.; Ali A.; Haq S.; Tahir M. N.; Iqbal J.; Braga A. A. C.; Ashfaq M.; Akhtar S. U. H. O-4-acetylamino-benzenesulfonylated pyrimidine derivatives: synthesis, SC-XRD, DFT analysis and electronic behaviour investigation. J. Mol. Struct. 2020, 1224, 129308 10.1016/j.molstruc.2020.129308. [DOI] [Google Scholar]
- Khalid M.; Ali A.; Abid S.; Tahir M. N.; Khan M. U.; Ashfaq M.; Imran M.; Ahmad A. Facile ultrasound-based synthesis, SC-XRD, DFT exploration of the substituted acyl-hydrazones: An experimental and theoretical slant towards supramolecular chemistry. ChemistrySelect 2020, 5, 14844–14856. 10.1002/slct.202003589. [DOI] [Google Scholar]
- Khalid M.; Ali A.; Asim S.; Tahir M. N.; Khan M. U.; Vieira L. C. C.; de la Torre A. F.; Usman M. Persistent prevalence of supramolecular architectures of novel ultrasonically synthesized hydrazones due to hydrogen bonding [X–H··· O; X = N]: Experimental and density functional theory analyses. J. Phys. Chem. Solids 2021, 148, 109679 10.1016/j.jpcs.2020.109679. [DOI] [Google Scholar]
- Concepcion O.; Ali A.; Khalid M.; F de la Torre A.; Khan M. U.; Raza A. R.; Kamal G. M.; Rehman M. F. U.; Alam M. M.; Imran M.; et al. Facile synthesis of diversely functionalized peptoids, spectroscopic characterization and DFT-based nonlinear optical exploration. ACS Omega 2021, 6, 26016–26025. 10.1021/acsomega.1c02962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A.; Khalid M.; Din Z. U.; Asif H. M.; Imran M.; Tahir M. N.; Ashfaq M.; Rodrigues-Filho E. Exploration of structural, electronic and third order nonlinear optical properties of crystalline chalcone systems: Monoarylidene and unsymmetrical diarylidene cycloalkanones. J. Mol. Struct. 2021, 1241, 130685 10.1016/j.molstruc.2021.130685. [DOI] [Google Scholar]
- Khalid M.; Ali A.; Rehman M. F. U.; Mustaqeem M.; Ali S.; Khan M. U.; Asim S.; Ahmad N.; Saleem M. Exploration of noncovalent interactions, chemical reactivity, and nonlinear optical properties of piperidone derivatives: a concise theoretical approach. ACS Omega 2020, 5, 13236–13249. 10.1021/acsomega.0c01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan B.; Khalid M.; Shah M. R.; Tahir M. N.; Khan M. U.; Ali A.; Muhammad S. Efficient synthesis by mono-carboxy methylation of 4, 4′-biphenol, X-ray diffraction, spectroscopic characterization and computational study of the crystal packing of ethyl 2-((4′-hydroxy-[1, 1′-biphenyl]-4-yl) oxy) acetate. ChemistrySelect 2019, 4, 9274–9284. 10.1002/slct.201901422. [DOI] [Google Scholar]
- Khalid M.; Ali A.; De la Torre A. F.; Marrugo K. P.; Concepcion O.; Kamal G. M.; Muhammad S.; Al-Sehemi A. G. Facile synthesis, spectral (IR, mass, UV– Vis, NMR), linear and nonlinear investigation of the novel phosphonate compounds: A combined experimental and simulation study. ChemistrySelect 2020, 5, 2994–3006. 10.1002/slct.201904224. [DOI] [Google Scholar]
- Khan I.; Khalid M.; Adeel M.; Khan M. U.; Khan M. S.; Ahmad N.; Ali A.; Akram M. Palladium-catalyzed synthesis of pyrimidine substituted diaryl ethers through Suzuki Miyaura coupling reactions: Experimental and DFT studies. Optik 2020, 219, 165285 10.1016/j.ijleo.2020.165285. [DOI] [Google Scholar]
- Ali A.; Khalid M.; Rehman M. F. U.; Haq S.; Ali A.; Tahir M. N.; Ashfaq M.; Rasool F.; Braga A. A. C. Efficient synthesis, SC-XRD, and theoretical studies of O-Benzenesulfonylated pyrimidines: Role of noncovalent interaction influence in their supramolecular network. ACS Omega 2020, 5, 15115–15128. 10.1021/acsomega.0c00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid M.; Ali A.; Adeel M.; Din Z. U.; Tahir M. N.; Rodrigues-Filho E.; Iqbal J.; Khan M. U. Facile preparation, characterization, SC-XRD and DFT/DTDFT study of diversely functionalized unsymmetrical bis-aryl-α, β-unsaturated ketone derivatives. J. Mol. Struct. 2020, 1206, 127755 10.1016/j.molstruc.2020.127755. [DOI] [Google Scholar]
- Ali A.; Din Z. U.; Khalid M.; Tahir M. N.; Rodrigues-Filho E.; Ali B.; Asim S.; Muhammad S. Crystal and quantum chemical exploration of the potent monocarbonyl curcuminoids to unveil their structural and intriguing electronic properties. ChemistrySelect 2020, 5, 3735–3745. 10.1002/slct.201904757. [DOI] [Google Scholar]
- Ashfaq M.; Tahir M. N.; Muhammad S.; Munawar K. S.; Ali A.; Bogdanov G.; Alarfaji S. S. Single-crystal investigation, Hirshfeld surface analysis, and DFT study of third-order NLO properties of unsymmetrical acyl thiourea derivatives. ACS Omega 2021, 6, 31211–31225. 10.1021/acsomega.1c04884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq S.; Raza A. R.; Khalid M.; Rubab S. L.; Khan M. U.; Ali A.; Tahir M. N.; Braga A. A. C. Synthesis and structural analysis of novel indole derivatives by XRD, spectroscopic and DFT studies. J. Mol. Struct. 2020, 1203, 127438 10.1016/j.molstruc.2019.127438. [DOI] [Google Scholar]
- Tahir M. N.; Mirza S. H.; Khalid M.; Ali A.; Khan M. U.; Braga A. A. C. Synthesis, single crystal analysis and DFT based computational studies of 2, 4-diamino-5-(4-chlorophenyl)-6-ethylpyrim idin-1-ium 3, 4, 5-trihydroxybenzoate-methanol (DETM). J. Mol. Struct. 2019, 1180, 119–126. 10.1016/j.molstruc.2018.11.089. [DOI] [Google Scholar]
- Tahir M. N.; Ashfaq M.; de la Torre A. F.; Caballero J.; Hernández-Rodríguez E. W.; Ali A. Rationalizing the stability and interactions of 2, 4-diamino-5-(4-chlorophenyl)-6-ethylpyrimidin-1-ium 2-hydroxy-3, 5-dinitrobenzoate salt. J. Mol. Struct. 2019, 1193, 185–194. 10.1016/j.molstruc.2019.05.003. [DOI] [Google Scholar]
- Shahwar D.; Tahir M. N.; Chohan M. M.; Ahmad N.; Raza M. A. 3-Acetyl-1-(2-methylphenyl) thiourea. Acta Crystallogr. E: Crystallogr. 2012, 68, o1160. 10.1107/S1600536812011658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. SHELXT–Integrated space-group and crystal-structure determination. Acta Crystallogr. A 2015, 71, 3–8. 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. Crystal structure refinement with SHELXL. Acta Crystallogr., Sect. C: Struct. Chem. 2015, 71, 3–8. 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia L. J. WinGX and ORTEP for windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. 10.1107/S0021889812029111. [DOI] [Google Scholar]
- Spek A. L. Structure validation in chemical crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2009, 65, 148–155. 10.1107/S090744490804362X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SJ M. C. S. I. C.; Cottrell S. I.; Galek P. T. A.; McCabe P. Mercury 4.0: from visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. 10.1107/S1600576719014092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M.; Trucks G.; Schlegel H.; Scuseria G.; Robb M.; Cheeseman J.; Scalmani G.; Barone V.; Petersson G.; Nakatsuji H.. Gaussian 16, Revision B. 01; Gaussian. Inc.: Wallingford CT, 2016.
- Becke A. D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. 10.1063/1.464913. [DOI] [Google Scholar]
- Grimme S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. 10.1002/jcc.20495. [DOI] [PubMed] [Google Scholar]
- McLean A.; Chandler G. Contracted Gaussian basis sets for molecular calculations. I. Second row atoms, Z = 11–18. J. Chem. Phys. 1980, 72, 5639–5648. 10.1063/1.438980. [DOI] [Google Scholar]
- Krishnan R.; Binkley J. S.; Seeger R.; Pople J. A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 1980, 72, 650–654. 10.1063/1.438955. [DOI] [Google Scholar]
- Tomasi J.; Mennucci B.; Cammi R. Quantum mechanical continuum solvation models. Chem. Rev. 2005, 105, 2999–3094. 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- Reed A. E.; Curtiss L. A.; Weinhold F. Intermolecular interactions from a natural bond orbital, donor-acceptor viewpoint. Chem. Rev. 1988, 88, 899–926. 10.1021/cr00088a005. [DOI] [Google Scholar]
- Geerlings P.; De Proft F.; Langenaeker W. Conceptual density functional theory. Chem. Rev. 2003, 103, 1793–1874. 10.1021/cr990029p. [DOI] [PubMed] [Google Scholar]
- Chakraborty A.; Pan S.; Chattaraj P. K. Biological activity and toxicity: A conceptual DFT approach. Appl. Density Funct. Theory Biol. Bioinorg. Chem. 2013, 143–179. [Google Scholar]
- Jorio S.; Salah M.; Abou El Makarim H.; Tabyaoui M. Reactivity indices related to DFT theory, the electron localization function (ELF) and non-covalent interactions (NCI) calculations in the formation of the non-halogenated pyruvic esters in solution. Mediterr. J. Chem. 2019, 8, 476–485. 10.13171/mjc8619072612sj. [DOI] [Google Scholar]
- Schaftenaar G.; Noordik J. H. Molden: a pre-and post-processing program for molecular and electronic structures. J. Comput. Aided Mol. Des. 2000, 14, 123–134. 10.1023/A:1008193805436. [DOI] [PubMed] [Google Scholar]
- Hanwell M. D.; Curtis D. E.; Lonie D. C.; Vandermeersch T.; Zurek E.; Hutchison G. R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 2012, 4, 1–17. 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avogadro A.An Open-Source Molecular Builder and Visualization Tool, Version 1.1. 0, 2009.
- Mirzaei M.; Eshtiagh-Hosseini H.; Hassanpoor A. Different behavior of PDA as a preorganized ligand versus PCA ligand in constructing two inorganic-organic hybrid materials based on Keggin-type polyoxometalate. Inorg. Chim. Acta 2019, 484, 332–337. 10.1016/j.ica.2018.09.053. [DOI] [Google Scholar]
- Taghipour F.; Mirzaei M. A survey of interactions in crystal structures of pyrazine-based compounds. Acta Crystallogr., Sect. C: Struct. Chem. 2019, 75, 231–247. 10.1107/S2053229619002225. [DOI] [PubMed] [Google Scholar]
- Mirzaei M.; Nikpour M.; Bauzá A.; Frontera A. On the importance of C—H/π and C—H···H—C interactions in the solid state structure of 15-Lipoxygenase inhibitors based on eugenol derivatives. ChemPhysChem 2015, 16, 2260–2266. 10.1002/cphc.201500287. [DOI] [PubMed] [Google Scholar]
- Spackman P. R.; Turner M. J.; McKinnon J. J.; Wolff S. K.; Grimwood D. J.; Jayatilaka D.; Spackman M. A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. 10.1107/S1600576721002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei M.; Sadeghi F.; Molčanov Ki.; Zarȩba J. K.; Gomila R. M.; Frontera A. Recurrent supramolecular motifs in a series of acid–base adducts based on Pyridine-2, 5-dicarboxylic acid N-oxide and organic bases: Inter-and intramolecular hydrogen bonding. Cryst. Growth Des. 2020, 20, 1738–1751. 10.1021/acs.cgd.9b01475. [DOI] [Google Scholar]
- Spackman M. A.; Jayatilaka D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. 10.1039/B818330A. [DOI] [Google Scholar]
- Madni M.; Ahmed M. N.; Hafeez M.; Ashfaq M.; Tahir M. N.; Gil D. M.; Galmés B.; Hameed S.; Frontera A. Recurrent π–π stacking motifs in three new 4, 5-dihydropyrazolyl–thiazole–coumarin hybrids: X-ray characterization, Hirshfeld surface analysis and DFT calculations. New J. Chem. 2020, 44, 14592–14603. 10.1039/D0NJ02931A. [DOI] [Google Scholar]
- McKinnon J. J.; Jayatilaka D.; Spackman M. A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. commun. 2007, 3814–3816. 10.1039/b704980c. [DOI] [PubMed] [Google Scholar]
- Ashfaq M.; Tahir M. N.; Kuznetsov A.; Mirza S. H.; Khalid M.; Ali A. DFT and single crystal analysis of the pyrimethamine-based novel co-crystal salt: 2, 4-diamino-5-(4-chloro-phenyl)-6-ethylpyrimidin-1-ium: 4-hydroxybenzoate: Methanol: Hydrate (1: 1: 1: 1)(DEHMH). J. Mol. Struct. 2020, 1199, 127041 10.1016/j.molstruc.2019.127041. [DOI] [Google Scholar]
- Ashfaq M.; Ali A.; Tahir M. N.; Khalid M.; Assiri M. A.; Imran M.; Munawar K. S.; Habiba U. Synthetic approach to achieve halo imine units: Solid-state assembly, DFT based electronic and non linear optical behavior. Chem. Phys. Lett. 2022, 803, 139843 10.1016/j.cplett.2022.139843. [DOI] [Google Scholar]
- Turner M. J.; McKinnon J. J.; Jayatilaka D.; Spackman M. A. Visualisation and characterisation of voids in crystalline materials. CrystEngComm 2011, 13, 1804–1813. 10.1039/C0CE00683A. [DOI] [Google Scholar]
- Ashfaq M.; Munawar K. S.; Tahir M. N.; Dege N.; Yaman M.; Muhammad S.; Alarfaji S. S.; Kargar H.; Arshad M. U. Synthesis, crystal structure, Hirshfeld surface analysis, and computational study of a novel organic salt obtained from benzylamine and an acidic component. ACS Omega 2021, 6, 22357–22366. 10.1021/acsomega.1c03078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi S. H.; Eshtiagh-Hosseini H.; Mirzaei M. Computational study of the intramolecular proton transfer reactions of dipicolinic acid (pyridine-2, 6-dicarboxylic acid) and its dimers. Comput. Theor. Chem. 2013, 1004, 69–75. 10.1016/j.comptc.2012.10.024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.