Abstract
Protective antigen (PA) is an important component of the edema and lethal toxins produced by Bacillus anthracis. PA is essential for binding the toxins to the target cell receptor and for facilitating translocation of the enzymatic toxin components, edema factor and lethal factor, across the target cell membrane. The structural gene for PA, pagA (previously known as pag), is located on the 182-kb virulence plasmid pXO1 at a locus distinct from the edema factor and lethal factor genes. Here we show that a 300-bp gene located downstream of pagA is cotranscribed with pagA and represses expression of the operon. We have designated this gene pagR (for protective antigen repressor). Two pagA mRNA transcripts were detected in cells producing PA: a short, 2.7-kb transcript corresponding to the pagA gene, and a longer, 4.2-kb transcript representing a bicistronic message derived from pagA and pagR. The 3′ end of the short transcript mapped adjacent to an inverted repeat sequence, suggesting that the sequence can act as a transcription terminator. Attenuation of termination at this site results in transcription of pagR. A pagR mutant exhibited increased steady-state levels of pagA mRNA, indicating that pagR negatively controls expression of the operon. Autogenous control of the operon may involve atxA, a trans-acting positive regulator of pagA. The steady-state level of atxA mRNA was also increased in the pagR mutant. The mutant phenotype was complemented by addition of pagR in trans on a multicopy plasmid.
Bacillus anthracis is the etiological agent of anthrax, a disease that affects all mammals, including humans. Key virulence factors of the bacterium include the proteins that comprise the edema and lethal toxins. These binary toxins are composed of distinct enzymatic proteins and a common protein that mediates entry into target cells. Protective antigen (PA) and edema factor (EF) comprise edema toxin, while PA plus lethal factor (LF) comprise lethal toxin. PA binds to a specific receptor on target cells and following endocytosis of the toxin-receptor complex, facilitates translocation of EF and LF across the cell membrane such that the enzymatic proteins can contact their cytosolic substrates (15, 19, 34).
The genes encoding the toxin proteins, pagA (previously known as pag) (which encodes PA), cya (which encodes EF), and lef (which encodes LF), are located noncontiguously within a 30-kb region of the 182-kb plasmid, pXO1 (12, 17, 22, 23, 29, 32). As might be expected due to the pivotal role of PA in intoxication, the pagA gene is highly expressed. When B. anthracis is cultured under optimal conditions, culture supernatants contain up to 20 mg of PA per liter, while LF and EF are present at 5 and 1 mg per liter, respectively (16). Analyses of toxin gene expression employing reporter gene fusions suggest that the steady-state level of mRNA of pagA is 4-fold higher than that of lef and 14-fold higher than that of cya (25).
Two host-related cues, CO2-bicarbonate and temperature, are important signals for expression of all three toxin genes. During in vitro growth, optimal expression of pagA, cya, and lef is achieved when B. anthracis is cultured in buffered R medium, a defined medium containing glucose, salts, and all amino acids, or CA medium, a medium containing Casamino Acids and glucose (16, 21, 27). Toxin synthesis is greatest when cultures are incubated in elevated (5% or greater) atmospheric CO2 or when bicarbonate is added to culture medium in a closed vessel. Toxin synthesis is also increased when cultures are incubated at 37°C compared to when they are incubated at 28°C. CO2-bicarbonate- and temperature-controlled gene expression is at the level of transcription (2, 4, 12, 25, 31).
The atxA gene, located within the toxin gene region of pXO1, positively controls transcription of all three toxin genes and at least one gene required for capsule synthesis, capB, which is located on the 93-kb plasmid pXO2 (9, 12, 29, 30). Under all growth conditions tested, atxA is essential for transcription from the unique start sites of cya and lef and for transcription from the major start site, P1, of the pagA gene (7, 12). The mechanism by which atxA activates expression of virulence genes and the relationship(s) between host-related cues and atxA function are not known.
In an effort to identify regulatory genes that act downstream of atxA to activate toxin gene expression, we screened for CO2-enhanced atxA-dependent loci on pXO1. We created random transcriptional lacZ fusions using transposon Tn917-pLTV3 and tested insertion mutants for CO2-enhanced atxA-dependent β-galactosidase activity. In addition to mutants harboring transposon insertions in the toxin genes, our screen yielded a number of mutants harboring apparent CO2-enhanced atxA-regulated fusions at distinct loci on pXO1 (10). Here we characterize a 300-bp open reading frame (ORF), pagR, identified in our screen. We show that the pagR gene is cotranscribed with the toxin gene pagA and that a pagR mutant shows increased levels of pagA and atxA transcripts.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Escherichia coli JM109 was used as a host for cloning. B. anthracis strains and their relevant characteristics are listed in Table 1. All B. anthracis strains are derivatives of the Weybridge strain, a noncapsulated toxigenic isolate originally obtained from the Microbiological Research Establishment, Porton Down, England. For DNA extractions, the B. anthracis strains were grown in brain heart infusion medium (Difco, Detroit, Mich.) containing 10% horse serum. For electroporation experiments, B. anthracis strains were grown in brain heart infusion medium containing 0.5% glycerol. For RNA extractions, B. anthracis strains were grown in CA broth (27) buffered with 100 mM HEPES (pH 8.0). CA medium contained 0.8% sodium bicarbonate for cultures incubated in 5% CO2.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant characteristic(s)a | Source and/or reference |
|---|---|---|
| Plasmids | ||
| B. anthracis | ||
| pXO1 | Tox+ | 26 |
| E. coli | ||
| pBSIIKS+ | Apr | Stratagene |
| pG+host5 | Emr | Appligene |
| pGEM-T | Apr | Promega |
| pSL301 | Apr | Invitrogen |
| pUC18::Ωkm-2 | pUC18 carrying the Ωkm-2 cassette; Apr Kmr | 18 |
| pUTE40 | 3.0-kb HpaI-BamHI pXO1 fragment, containing pagA, in pMK4 | 12 |
| pUTE41 | 3.0-kb HpaI-BamHI pXO1 fragment, containing pagA, in pBSIIKS+ | This work |
| pUTE103 | 4.3-kb PstI-XbaI pXO1 fragment containing pagR and flanking sequences in pG+host5 | This work |
| pUTE308 | BamHI-KpnI PCR product containing 0.9 kb upstream of pagR and the 5′ end of pagR in pSL301 | This work |
| pUTE331 | 1.1-kb PCR product containing the 3′ end of pagA, the pagA attenuator, and the 5′ end of pagR in pGEM-T | This work |
| pUTE333 | 2.6-kb BamHI-SacI fragment from pUTE103, containing pagR, in pUTE41 | This work |
| Bifunctional | ||
| pUTE29 | Apr in E. coli, Tcr in B. anthracis | 12 |
| pUTE34 | 2.7-kb SnaBI-EcoRI pXO1 fragment containing atxA in pUTE29 | 12 |
| pUTE314 | 4.0-kb SacI-PstI pXO1 fragment containing pagR and flanking sequences in pUTE29 | This work |
| pUTE315 | Ωkm-2 cloned into BamHI site between pagA and pagR in pUTE314 | This work |
| pUTE334 | 5.6-kb SacI-KpnI fragment from pUTE333, containing pagA and pagR, in pUTE29 | This work |
| pUTE366 | pUTE334 containing +1 frameshift within pagR | This work |
| Strains | ||
| B. anthracis | ||
| UM44b | Tox+ Ind− | 26 |
| UT53 | atxA-Null derivative of UM44, atxA is replaced by Ωkm-2; Ind− Kmr | 7 |
| UT62 | P1 start site of pagA disrupted with the Ωkm-2, derivative of UM44; Ind− Kmr | 7 |
| UT82 | Tn917-LTV3 insertion mutant of UM44; Cmr MLSr, CO2-atxA-regulated lacZ expression | 10 |
| UT119 | pagR Mutant derivative of UM44, Ωkm-2 inserted at BamHI site between pagA and pagR; Ind− Kmr | This work |
| E. coli | ||
| GM1684 | F′ F-lacIqΔM15 pro+/dam-4 Δ(lac-pro)X111 thi-1 glnV44 (relA1) | R. Kolter |
| JM109 | F′ traD36 proA+ proB+ lacIqlacZΔM15/recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) mcrA | 35 |
| BMH 71-18 | thi supE Δ(lac-proAB) [mutS::Tn10][F′ proAB lacIqZΔM15] | Clontech |
Abbreviations: Apr, ampicillin resistant; Emr, erythromycin resistant; Ind, indole; Kmr, kanamycin resistant; MLSr, macrolide resistant; Tcr, tetracycline resistant; Tox, anthrax toxin proteins.
UM44 was derived from the Weybridge strain.
All antibiotics were purchased from Sigma (St. Louis, Mo.) or Fisher Scientific (Pittsburgh, Pa.) and were added to media at the following concentrations when appropriate: for E. coli, ampicillin, 100 μg/ml; kanamycin, 20 μg/ml, and tetracycline, 10 μg/ml; for B. anthracis, erythromycin, 1 μg/ml; lincomycin, 25 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 5 μg/ml.
Plasmid and strain constructions.
Plasmid DNA was extracted from B. anthracis by the method of Green et al. (8). Preparation of plasmid DNA from E. coli, transformation of E. coli, and recombinant DNA techniques were carried out by standard procedures (1). B. anthracis was electroporated with plasmid DNA from E. coli GM1684, as described previously (12). Restriction enzymes, T4 ligase, and Taq polymerase were purchased from Promega (Madison, Wis.) or New England Biolabs (Beverly, Mass.).
Plasmids and their relevant characteristics are shown in Table 1. Plasmid pUTE308 contains DNA downstream of pagA, including the 5′ end of pagR. To construct pUTE308, a 946-bp PCR product was generated by using oligonucleotides 5′-GTAAGAAATACAAGGAGAGTATG-3′ and 5′-CGCATAGGAGGTACCATTGTTTTT-3′. The former oligonucleotide is complementary to a region 62 to 85 bp downstream of the translational stop of pagA and just upstream of a BamHI site (see Fig. 1). The latter oligonucleotide is complementary to a region 62 to 86 bp downstream of the predicted translational start of pagR and contains an engineered KpnI site (shown in boldface type). The PCR product was digested with BamHI and KpnI and ligated into BamHI-KpnI-digested pSL301.
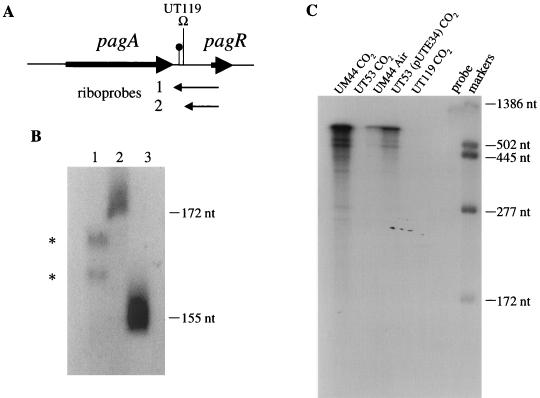
FIG. 1.
pXO1 DNA sequence extending from the 3′ end of pagA through pagR. Predicted amino acid sequences are indicated under the pagA and pagR DNA sequences. Straight arrows indicate an inverted repeat sequence, located downstream of pagA, that functions as a weak transcription terminator. A potential ribosome binding site upstream of pagR is underlined. A tyrosine residue predicted to be a tyrosine kinase phosphorylation site is boxed. Locations of a BamHI site (the site of the Ωkm-2 insertion in UT119) and a Tn917-LTV3 insertion in UT82 are as indicated.
Plasmid pUTE331 contains DNA from the 3′ end of pagA through the 5′ end of pagR. To construct pUTE331, a 1.1-kb PCR product was generated with oligonucleotides 5′-GGGGATACTTAGTACCAACGGG-3′ and 5′-GTTCAGCATCATCTTCTAAACTC-3′. The former oligonucleotide is complementary to a region 45 to 68 bp upstream of the translational stop codon of pagA. The latter oligonucleotide is complementary to a region 36 to 58 bp downstream of the predicted translational start of pagR. The PCR product was ligated into pGEM-T.
Plasmid pUTE334 carries pagA and pagR. It was created by first ligating a BamHI-SacI fragment (containing pagR) from pUTE103 into BamHI-SacI-digested pUTE41 (containing pagA), resulting in pUTE333. The 5.6-kb SacI-KpnI fragment from pUTE333 was ligated into SacI-KpnI-digested pUTE29, resulting in pUTE334. The frameshift mutation within pagR (pUTE366) was generated by using the Transformer Site-Directed Mutagenesis kit from Clontech (Palo Alto, Calif.). The oligonucleotide, 5′-GACAGTATTTGTACGATCATAAAATTG-3′, was used as a mutagenic primer to add a C residue (underlined) 15 bases downstream of the ATG start codon of pagR. The frameshift mutation was confirmed by DNA sequencing.
UT119 carries the Ωkm-2 element inserted in the BamHI site between pagA and pagR (see Fig. 1). Insertion of the Ωkm-2 element into the B. anthracis genome was accomplished as described previously (7). To create UT119, pUTE315 (Table 1) was electroporated into UM44 with selection for kanamycin resistance. UT119 was isolated following a screen for tetracycline-sensitive kanamycin-resistant mutants. The location of the Ωkm-2 element was confirmed by using PCR.
RNA analysis.
Methods for RNA extraction, primer extension reactions, and RNase T2 protection assays have been described (12, 28). RNA was extracted from B. anthracis cultures grown to late log phase (optical density at 600 nm of 0.8 to 1.0) in CA medium. RNA was quantified spectrophotometrically and by visualization on 1.2% formaldehyde gels. Oligonucleotides used for atxA and pagA primer extensions were described previously (6). Oligonucleotides were labeled with [γ-32P]ATP (6,000 Ci/mmol) (Amersham Corp., Arlington Heights, Ill.), hybridized to 20 μg of RNA, and extended by using avian myeloblastosis virus reverse transcriptase (Promega). The 5′ ends of the atxA and pagA genes were sequenced by the dideoxy-chain termination method (1), using the appropriate primers and a Sequenase version 2.0 DNA sequencing kit purchased from United States Biochemical Corp. (Cleveland, Ohio). The [α-35S]dATP (>1,000 Ci/mmol) for sequencing was purchased from Amersham Corp. Primer extension and sequencing reaction mixtures were subjected to electrophoresis on 6% polyacrylamide and 42% urea gels. Primer extension products were quantified by using a Packard Instant Imager.
Antisense RNA probes (riboprobes) 1 and 2 (for illustrations see Fig. 3A) were used for RNase T2 protection assays. RNA polymerases and RNase T2 were purchased from Promega. To make riboprobe 1, pUTE331 was linearized with NcoI and antisense RNA was generated by using Sp6 RNA polymerase and [α-32P]CTP (>800 Ci/mmol) (Amersham). To make riboprobe 2, pUTE308 was linearized with BamHI and antisense RNA was generated by using T7 RNA polymerase and [α-32P]CTP. Riboprobes were hybridized to 20 μg of B. anthracis RNA prior to digestion with RNase T2.
FIG. 3.
Mapping the 3′ ends of the monocistronic pagA transcript and determining the effects of CO2 and atxA on pagR expression. RNase T2 protection assays were performed with 20 μg of RNA. (A) Locations of antisense riboprobes, the Ωkm-2 insertion in UT119, and the transcription attenuator are shown. (B) Lane 1: riboprobe 1 was hybridized to RNA from UM44 grown in 5% CO2. RNase T2-protected fragments are indicated. Lanes 2 and 3: nucleotide size markers, as indicated. (C) Riboprobe 2 was hybridized to RNA from cells grown in 5% CO2 or in air, as indicated. RNase T2-protected fragments are shown. Size markers are as indicated. The probe is 1,167 nt (including 222 nt of vector-derived sequences).
For Northern hybridizations, biotinylated DNA probes complementary to the internal DNA sequences of pagA and pagR were created by using a Phototope kit (New England Biolabs). A 1.7-kb XmnI-HincII fragment from pUTE40 was biotinylated to make the pagA probe. The pagR probe was made from a 242-bp PCR product generated by the following oligonucleotides: 5′-GAGTTTAGAAGATGATGCTGAAC-3′ and 5′-TAATAATCCCTTCAACTTTTGG-3′. The oligonucleotide primers were complementary to regions 35 to 58 and 264 to 286 bp downstream of the predicted translational start codon of pagR. Twenty micrograms of RNA was run on a 1.2% agarose gel (containing formaldehyde) and capillary blotted to Maximum Strength Nytran Plus membranes (Schleicher & Schuell, Keene, N.H.). Membranes were hybridized with probes overnight at 45°C. Blots were developed by using a Phototope-Star detection kit (New England Biolabs) and exposed to film.
Immunoblotting.
Relative levels of PA produced by parent and mutant strains were determined by assaying cell lysates for protein that reacted with polyclonal anti-PA antiserum. PA in culture supernatants is subject to varying levels of degradation by proteases secreted by B. anthracis. Therefore, for monitoring small differences in PA levels, more-reproducible data can be obtained by examining cell lysates rather than culture supernatants. B. anthracis cultures were grown in 25 ml of CA medium to an optical density at 600 nm of 0.8 to 1.0. Cells were collected on cellulose acetate membranes (pore size, 0.45 μm) (Nalge, Rochester, N.Y.), resuspended in 1 ml of buffer (1% sodium dodecyl sulfate, 200 mM dithiothreitol, 28 mM Tris HCl, 22 mM Tris OH, 2 mM phenylmethylsulfonyl fluoride), and passed through a French press minicell three times at 20,000 lb/in2. The soluble fraction was obtained following centrifugation at 16,000 × g for 5 min, and protein concentrations were determined by using a Bio-Rad protein assay reagent (Bio-Rad, Hercules, Calif.). Samples (5 μg) were subjected to electrophoresis on sodium dodecyl sulfate–7.5% polyacrylamide gels.
Following electrophoresis, proteins were transferred to nitrocellulose membranes by electroblotting. Membranes were reacted with rabbit anti-PA serum (diluted 1:6,000 in TBS-T (20 mM Tris OH, 137 mM NaCl, 0.1% Tween 20 [pH 7.6]) containing 5% milk for 2 h at room temperature. Membranes were washed in TBS-T and finally reacted with donkey anti-rabbit immunoglobulin G antibody conjugated to horseradish peroxidase (1:4,000 in TBS-T with 5% milk) for 1 h at room temperature. Cross-reactive material was visualized on autoradiographs by using an ECL immunodetection kit purchased from Amersham (Little Chalfont, Buckinghamshire, England).
Nucleotide sequence accession number.
The complete nucleotide sequence of the pagR gene and flanking regions has been deposited in the GenBank database under accession no. AF031382.
RESULTS
Identification of pagR.
In experiments reported previously, we used the transposon Tn917-pLTV3 to identify B. anthracis promoters that exhibited increased expression during growth in elevated CO2 and were atxA-dependent (10). Tn917-LTV3 carries a promoterless lacZ gene at one end and generates a transcriptional fusion when inserted downstream of an active promoter (3). In this investigation, we studied one Tn917-pLTV3 insertion mutant that showed CO2- and atxA-dependent β-galactosidase activity.
Restriction analysis and Southern hybridization experiments revealed that the mutant UT82 contained a Tn917-pLTV3 insertion in the approximately 2.8-kb region between the pagA and lef genes on pXO1 (data not shown). We cloned and sequenced this intergenic region from the parent strain. To determine the exact insertion site and orientation of Tn917-pLTV3 in UT82, we cloned and sequenced the B. anthracis DNA flanking the 5′ end of the transposon insertion. As shown in Fig. 1, sequence analysis indicated that the transposon insertion was 1,339 nucleotides (nt) downstream of pagA. A 300-nt ORF was noted between pagA and the Tn917-LTV3 insertion. This ORF lies in the same orientation as the pagA gene and the promoterless lacZ gene associated with the transposon. The 3′ end of the ORF maps to 122 bp upstream of the transposon insertion. For reasons made evident below, we have designated the downstream ORF pagR (for protective antigen repressor). A potential ribosome binding site beginning 12 bp upstream of the translational start codon of pagR was noted.
The pagR gene is predicted to encode a 99-amino-acid protein with sequence similarity to two proteins that regulate transcription in other species. The predicted pagR gene product is 26% identical and 52% similar to CadC (13.5 kDa), a predicted regulator of cadmium resistance in Listeria monocytogenes (14). The putative PagR protein is 33% identical and 57% similar to NolR (13.3 kDa), a trans-acting negative regulator of the nod regulon of Rhizobium meliloti (13). The NolR protein has been shown to bind specifically to promoter regions of several nod genes. The sequence similarity of PagR to both NolR and CadC is found throughout the predicted amino acid sequences of the proteins. Analysis of the PagR amino acid sequence by using the motifs program from the University of Wisconsin Genetics Computer Group (GCG) software package identified a potential tyrosine kinase phosphorylation site in the carboxy-terminal region of the protein (Fig. 1).
pagR is cotranscribed with pagA.
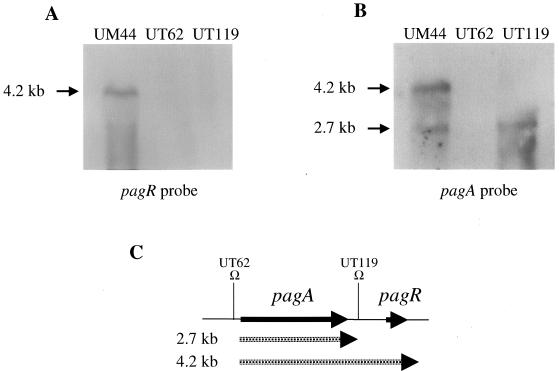
Northern hybridization experiments were performed to detect mRNA corresponding to the pagR gene. Biotinylated DNA probes corresponding to sequences internal to pagR and pagA were hybridized to RNA isolated from the parental strain, UM44, and mutants UT62 and UT119. Results are shown in Fig. 2. Hybridization of the pagR probe with UM44 RNA revealed a single band representing a 4.2-kb transcript, while hybridization of UM44 RNA with the pagA probe resulted in two bands corresponding to 4.2- and 2.7-kb transcripts. Mutations in UT62 and UT119 are shown schematically in Fig. 2C. UT62 carries an Ωkm-2 insertion that prevents transcription from the major transcription start site of pagA (12). RNA isolated from UT62 did not hybridize to the pagR or pagA probes. UT119 is a mutant containing an Ωkm-2 insertion in a BamHI restriction site (Fig. 1) between the pagA and pagR genes. No transcripts were detected when UT119 RNA was probed with pagR. When the pagA probe was hybridized to RNA from UT119, only the smaller, 2.7-kb transcript was detected. Taken together, these results indicate that transcription from the pagA promoter results in a monocistronic transcript corresponding to pagA and a bicistronic transcript corresponding to pagA and pagR (see Fig. 2C).
FIG. 2.
Northern blot detection of pagR and pagA mRNA. RNA samples (20 μg) from cells grown at 37°C in 5% CO2 were probed with biotinylated DNA probes corresponding to sequences internal to pagR (A) and pagA (B). Transcript sizes are indicated. (C) The relative locations of the pagA and pagR mRNA transcripts and the Ωkm-2 insertions in UT62 and UT119 are shown.
The size of the monocistronic transcript is indicative of termination between the pagA and pagR genes. Welkos et al. (33) proposed that a nucleotide sequence beginning 43 bp downstream of the pagA coding sequence represents a transcription terminator (Fig. 1 and 3A). The sequence could facilitate formation of a hairpin structure with a 19-bp stem and a calculated free energy of −22.2 kcal/mol. We performed RNase T2 protection assays to determine if the 3′ end of the short transcript mapped near this site. A riboprobe extending from the center of pagR to 114 bp upstream of the inverted repeat sequence (riboprobe 1, Fig. 3A) was hybridized to RNA from UM44 and subsequently digested with RNase T2. Two small, protected fragments were observed (Fig. 3B, lane 1). The sizes of the fragments, 162 and 167 nt, indicate that they are pagA transcripts with 3′ ends mapping 3 and 8 bp downstream of the inverted repeat sequence. As expected, the full-length probe was also protected from digestion with RNase T2 due to hybridization with the bicistronic (pagA-pagR) transcript (not shown in the figure). These results indicate that termination occurs at the predicted site but is incomplete. The inverted repeat sequence can therefore be considered an attenuator.
Transcription of pagA from the major start site, P1, is dependent upon the trans-acting regulatory gene, atxA (12). In addition, steady-state levels of pagA mRNA are increased when cells are grown in elevated levels of CO2-bicarbonate compared to growth in air (2, 12, 25). RNase T2 protection assays were performed to confirm that control of pagR transcription was similar to that of the pagA gene. A riboprobe extending from within pagR to a BamHI restriction site located between the pagA and pagR genes (riboprobe 2, Fig. 3A) was hybridized to RNA isolated from UM44, UT53, UT53 (pUTE34), and UT119 and subsequently digested with RNase T2. Results are shown in Fig. 3C. All RNase-protected hybridization products were of similar size, indicating complete protection of the probe, as expected. The comparison of the intensities of bands obtained by using RNA from UM44 cultured in 5% CO2 and RNA from UM44 cultured in air confirmed that the pagR transcript is CO2-regulated. No transcript was detected by using RNA isolated from the atxA-null mutant, UT53. pagR transcript was detected in RNA isolated from UT53 carrying atxA in trans on pUTE34, although the amount was lower than the wild-type level. It has been shown previously that the steady-state level of pagA transcripts in strains overexpressing atxA in trans is significantly lower than that detected in the UM44 parent strain (6). No pagR transcript was detected from UT119, confirming that there are no pagR transcripts that initiate between pagA and pagR. Thus, pagR is cotranscribed with pagA and is expressed in a manner similar to pagA.
pagR represses pagA expression.
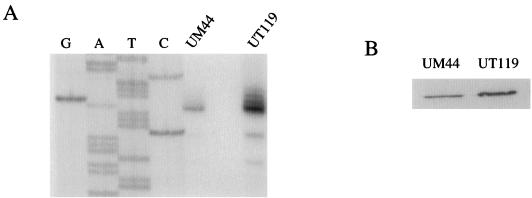
Considering that pagR is cotranscribed with pagA and that the predicted PagR protein has sequence similarity to regulators of transcription, we hypothesized that pagR affects expression of pagA in a feedback control loop. Primer extension experiments were performed to measure steady-state levels of pagA transcripts in UM44 and UT119 (pagR) grown at 37°C in 5% CO2. Results of a representative experiment are shown in Fig. 4A. In repeated experiments, the level of pagA transcript was 2.5- to 7.0-fold higher in UT119 than in UM44, indicating that pagR represses pagA expression. It is not clear why the level of repression varied in different experiments. Identical RNA samples used to measure expression of atxA (see below) did not exhibit such variation. Western blottings were performed to determine if the increase in pagA expression in the pagR mutant also led to an increase in PA protein level (Fig. 4B). Levels of PAs from UT119 lysates were 2.0-fold higher than those from UM44 lysates.
FIG. 4.
Regulation of pagA by pagR. Cultures for isolation of RNA and protein were incubated in 5% CO2. Strains are as indicated. (A) Primer extension experiments were performed with 20 μg of RNA. The end-labeled primer was complementary to a 33-bp sequence located 32 nt downstream of the first nucleotide of the pagA translational start site. Lanes G, A, T, and C correspond to the dideoxy sequencing reaction carried out with the same oligonucleotide primer. (B) Western blot showing PA detected in cell lysates.
pagR represses atxA expression.
The pagA gene is positively regulated in trans by atxA (12, 29). We tested for pagR-controlled expression of atxA. Results of primer extension experiments are shown in Fig. 5. The steady-state level of atxA mRNA was increased twofold in UT119 (pagR) compared to that in UM44, indicating that pagR represses atxA at a low level.
FIG. 5.
Regulation of atxA by pagR. Primer extensions were performed as described in the legend for Fig. 4. The end-labeled primer was complementary to a 27-bp sequence located 55 bp downstream of the first nucleotide of the translational start site of the atxA gene. Strains are as indicated.
UT119 contains an Ωkm-2 insertion between the pagA and pagR genes and does not express pagR. To confirm that the UT119 phenotype was due to the lack of pagR expression, we attempted to complement UT119 with a plasmid containing pagR under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter Pspac (36). After induction with IPTG, the pagR transcript in UT119 containing the recombinant plasmid was barely detectable, suggesting that the truncated transcript was unstable. Therefore, we tested for complementation of the UT119 phenotype using two different constructs. Plasmid pUTE334 contains a 5.6-kb pXO1 fragment containing pagA and pagR. Plasmid pUTE366 contains the same pXO1 fragment with a frameshift mutation within pagR. As shown in Fig. 5, pUTE334 complemented the mutation in UT119, restoring atxA expression to the level observed in the parent strain, UM44. UT119 was not complemented by pUTE366. These results show that the UT119 phenotype is due to disruption of pagR expression.
DISCUSSION
In this study, we determined that a previously unknown gene, pagR, is cotranscribed with the toxin gene pagA (previously known as pag) and negatively controls expression of the operon. Incomplete transcription termination near the 3′ end of an inverted repeat sequence downstream of pagA results in mono- and bicistronic transcripts containing pagA mRNA. There is no evidence that attenuation at this site is regulated. Expression of pagR appears to mimic pagA expression. In light of our results, we refer to the structural gene for PA as pagA. Thus, the pag operon is composed of pagA and pagR.
The predicted amino acid sequence of the putative PagR protein is similar to those of regulatory proteins NolR from R. meliloti (13) and CadC from L. monocytogenes (14). NolR and CadC contain helix-turn-helix motifs and are predicted to be DNA-binding proteins. If the PagR protein binds DNA specifically, the PagR target may be within the promoter region of pagA or within the promoter region of some other gene that affects pagA expression.
A pagR mutant shows elevated expression of pagA and atxA, a trans-acting positive regulator of pagA. Yet it is unlikely that increased pagA expression in a pagR mutant is attributed simply to increased AtxA levels. Results of previous studies indicate that AtxA levels are not limiting for expression of pagA. Strains carrying multiple copies of the pagA promoter synthesize normal levels of PA (24). Furthermore, a 10-fold increase in atxA expression has a negative effect on pagA expression, resulting in a 60 to 70% decrease in PA levels (6). Thus, PagR may control expression of the pag operon independent of AtxA or in conjunction with AtxA and some other regulatory protein(s).
Our data indicate that pagR functions to limit pagA expression under culture conditions which are optimal for synthesis of protective antigen. In experiments not presented here, we tested virulence of the pagR mutant in a mouse model for anthrax. Subcutaneous inoculation of mice with high doses of spores of the toxigenic noncapsulated Sterne strain results in a lethal disease (20). In this model, the 50% lethal dose for the pagR mutant was the same as that for the parent strain (11). This result indicates that increased PA synthesis by the pagR strain does not increase virulence in the mouse model. It is also possible that pagR function during infection differs from that observed during culture of B. anthracis in vitro.
The pagA gene is the only toxin gene that is known to be transcribed in an operon. The 5′ ends of transcripts corresponding to the cya and lef genes have been mapped (7); however, the sizes of the cya and lef transcripts have not been determined. Sequence analysis of regions downstream of cya and lef does not indicate that there are putative transcription regulators adjacent to these genes. A 332-bp ORF lies 286 bp downstream of cya and in the same orientation (accession no. AF003936). The predicted protein product of this ORF has sequence similarity to the purine salvage pathway enzyme adenine phosphoribosyltransferase of many organisms. A 303-bp ORF is located 1.4-kb downstream of lef and in the same orientation (accession no. AF031382). The predicted protein product of this ORF has no homology to any sequences in GenBank.
The pagA gene is coordinately expressed with the other toxin genes, cya and lef, in response to the same signals, i.e., CO2-bicarbonate and temperature, and the same activator, atxA. It is possible that pagR also regulates the cya and lef genes. Cataldi et al. (5) examined EF activity and LF protein level in the culture supernatant of a B. anthracis strain harboring an erythromycin resistance cassette in the pagA gene. EF activity was increased twofold, while LF protein level was unchanged, compared to that in the strain without the cassette (5). If the insertion in the pagA mutant had a polar effect on pagR expression, it is possible that the observed increase in EF activity was due to increased expression of cya in the absence of pagR expression. In future experiments we will address the mechanism for pagR-mediated repression and determine whether or not pagR also regulates the other toxin genes.
ACKNOWLEDGMENTS
We thank Jean-Claude Sirard and Michele Mock of the Institut Pasteur for performing virulence assays. We are grateful to Malcolm Winkler for helpful discussions and use of equipment and Heidi Kaplan for critical reading of the manuscript.
This work was supported by Public Health Service grant AI33537 from the National Institutes of Health.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1996. [Google Scholar]
- 2.Bartkus J M, Leppla S H. Transcriptional regulation of the protective antigen gene of Bacillus anthracis. Infect Immun. 1989;57:2295–2300. doi: 10.1128/iai.57.8.2295-2300.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camilli A, Portnoy D A, Youngman P. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol. 1990;172:3738–3744. doi: 10.1128/jb.172.7.3738-3744.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cataldi A, Fouet A, Mock M. Regulation of pag gene expression in Bacillus anthracis: use of a pag-lacZ transcriptional fusion. FEMS Microbiol Lett. 1992;98:89–94. doi: 10.1016/0378-1097(92)90137-d. [DOI] [PubMed] [Google Scholar]
- 5.Cataldi A, Labruyere E, Mock M. Construction and characterization of a protective antigen-deficient Bacillus anthracis strain. Mol Microbiol. 1990;4:1111–1117. doi: 10.1111/j.1365-2958.1990.tb00685.x. [DOI] [PubMed] [Google Scholar]
- 6.Dai Z, Koehler T M. Regulation of anthrax toxin activator gene (atxA) expression in Bacillus anthracis: temperature, not CO2/bicarbonate, affects AtxA synthesis. Infect Immun. 1997;65:2576–2582. doi: 10.1128/iai.65.7.2576-2582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai Z, Sirard J-C, Mock M, Koehler T M. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol Microbiol. 1995;16:1171–1181. doi: 10.1111/j.1365-2958.1995.tb02340.x. [DOI] [PubMed] [Google Scholar]
- 8.Green B D, Battisti L, Koehler T M, Thorne C B. Demonstration of a capsule plasmid in Bacillus anthracis. Infect Immun. 1985;49:291–297. doi: 10.1128/iai.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guignot J, Mock M, Fouet A. AtxA activates the transcription of genes harbored by both Bacillus anthracis virulence plasmids. FEMS Microbiol Lett. 1997;147:203–207. doi: 10.1111/j.1574-6968.1997.tb10242.x. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmaster A R, Koehler T M. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect Immun. 1997;65:3091–3099. doi: 10.1128/iai.65.8.3091-3099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koehler, T. M. Unpublished results.
- 12.Koehler T M, Dai Z, Kaufman-Yarbray M. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J Bacteriol. 1994;176:586–595. doi: 10.1128/jb.176.3.586-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondorosi E, Pierre M, Cren M, Haumann U, Buire M, Hoffmann B, Schell J, Kondorosi A. Identification of NolR, a negative transacting factor controlling the nod regulon in Rhizobium meliloti. J Mol Biol. 1991;222:885–896. doi: 10.1016/0022-2836(91)90583-r. [DOI] [PubMed] [Google Scholar]
- 14.Lebrun M, Audurier A, Cossart P. Plasmid-borne cadmium resistance genes in Listeria monocytogenes are similar to cadA and cadC of Staphylococcus aureus and are induced by cadmium. J Bacteriol. 1994;176:3040–3048. doi: 10.1128/jb.176.10.3040-3048.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leppla S H. Anthrax toxins. In: Moss J, Iglewski B, Vaughan M, Tu A T, editors. Bacterial toxins and virulence factors in disease. New York, N.Y: Marcel Dekker; 1995. pp. 543–572. [Google Scholar]
- 16.Leppla S H. Production and purification of anthrax toxin. Methods Enzymol. 1988;165:103–116. doi: 10.1016/s0076-6879(88)65019-1. [DOI] [PubMed] [Google Scholar]
- 17.Mock M, Labruyere E, Glaser P, Danchin A, Ullmann A. Cloning and expression of the calmodulin-sensitive Bacillus anthracis adenylate cyclase in Escherichia coli. Gene. 1988;64:277–284. doi: 10.1016/0378-1119(88)90342-3. [DOI] [PubMed] [Google Scholar]
- 18.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petosa C, Collier R J, Klimpel K R, Leppla S H, Liddington R C. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 20.Pezard C, Berche P, Mock M. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect Immun. 1991;59:3472–3477. doi: 10.1128/iai.59.10.3472-3477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ristroph J D, Ivins B E. Elaboration of Bacillus anthracis antigens in a new, defined culture medium. Infect Immun. 1983;39:483–486. doi: 10.1128/iai.39.1.483-486.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson D, Leppla S H. Molecular cloning and expression in Escherichia coli of the lethal factor gene of Bacillus anthracis. Gene. 1986;44:71–78. doi: 10.1016/0378-1119(86)90044-2. [DOI] [PubMed] [Google Scholar]
- 23.Robertson D L, Tippetts M T, Leppla S H. Nucleotide sequence of the Bacillus anthracis edema factor gene (cya): a calmodulin-dependent adenylate cyclase. Gene. 1988;73:363–371. doi: 10.1016/0378-1119(88)90501-x. [DOI] [PubMed] [Google Scholar]
- 24.Sirard J-C, Mock M, Fouet A. Molecular tools for the study of transcriptional regulation in Bacillus anthracis. Res Microbiol. 1995;146:729–737. doi: 10.1016/0923-2508(96)81069-2. [DOI] [PubMed] [Google Scholar]
- 25.Sirard J-C, Mock M, Fouet A. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J Bacteriol. 1994;176:5188–5192. doi: 10.1128/jb.176.16.5188-5192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thorne C B. Genetics of Bacillus anthracis. In: Leive L, editor. Microbiology. Washington, D.C: American Society for Microbiology; 1985. pp. 56–62. [Google Scholar]
- 27.Thorne C B, Belton F C. An agar-diffusion method for titrating Bacillus anthracis immunizing antigen and its application to a study of antigen production. J Gen Microbiol. 1957;17:505–516. doi: 10.1099/00221287-17-2-505. [DOI] [PubMed] [Google Scholar]
- 28.Tsui H-C T, Pease A J, Koehler T M, Winkler M E. Detection and quantitation of RNA transcribed from bacterial chromosomes and plasmids. In: Adolph K W, editor. Molecular microbiology techniques, Part A. San Diego, Calif: Academic Press Inc.; 1994. pp. 179–204. [Google Scholar]
- 29.Uchida I, Hornung J M, Thorne C B, Klimpel K R, Leppla S H. Cloning and characterization of a gene whose product is a trans-activator of anthracis toxin synthesis. J Bacteriol. 1993;175:5329–5338. doi: 10.1128/jb.175.17.5329-5338.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uchida I, Makino S-I, Sekizaki T, Terakado N. Cross-talk to the genes for Bacillus anthracis capsule synthesis by atxA, the gene encoding the trans-activator of anthrax toxin synthesis. Mol Microbiol. 1997;23:1229–1240. doi: 10.1046/j.1365-2958.1997.3041667.x. [DOI] [PubMed] [Google Scholar]
- 31.Vietri N J, Marrero R, Hoover T A, Welkos S L. Identification and characterization of a trans-activator involved in the regulation of encapsulation by Bacillus anthracis. Gene. 1995;152:1–9. doi: 10.1016/0378-1119(94)00662-c. [DOI] [PubMed] [Google Scholar]
- 32.Vodkin M H, Leppla S H. Cloning of the protective antigen gene of Bacillus anthracis. Cell. 1983;34:693–697. doi: 10.1016/0092-8674(83)90402-6. [DOI] [PubMed] [Google Scholar]
- 33.Welkos S L, Lowe J R, Eden-McCutchan F, Vodkin M, Leppla S H, Schmidt J J. Sequence and analysis of the DNA encoding protective antigen of Bacillus anthracis. Gene. 1988;69:287–300. doi: 10.1016/0378-1119(88)90439-8. [DOI] [PubMed] [Google Scholar]
- 34.Wesche J, Elliott J L, Falnes P O, Olsnes S, Collier R J. Characterization of membrane translocation by anthrax protective antigen. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]
- 35.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 36.Yansura D F, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]