Abstract
HIV-1 capsid (CA) is an attractive target for its indispensable roles in the viral life cycle. We report the design, synthesis, and mechanistic study of a novel series of 2-piperazineone peptidomimetics as HIV capsid modulators by mimicking the structure of host factors binding to CA. F-Id-3o was the most potent compound from the synthesized series, with an anti-HIV-1 EC50 value of 6.0 μM. However, this series of compounds showed a preference for HIV-2 inhibitory activity, in which Id-3o revealed an EC50 value of 2.5 μM (anti-HIV-2 potency), an improvement over PF74. Interestingly, F-Id-3o did bind HIV-1 CA monomers and hexamers with comparable affinity, unlike PF74, consequently showing antiviral activity in the early and late stages of the HIV-1 lifecycle. Molecular dynamics simulations shed light on F-Id-3o and Id-3o binding modes within the HIV-1/2 CA protein and provide a possible explanation for the increased anti-HIV-2 potency. Metabolic stability assays in human plasma and human liver microsomes indicated that although F-Id-3o has enhanced metabolic stability over PF74, further optimization is necessary. Moreover, we utilized computational prediction of drug-like properties and metabolic stability of F-Id-3o and PF74, which correlated well with experimentally derived metabolic stability, providing an efficient computational pipeline for future preselection based on metabolic stability prediction. Overall, the 2-piperazineone-bearing peptidomimetics are a promising new chemotype in the CA modulators class with considerable optimization potential.
We report the design, synthesis, and mechanistic study of a novel series of 2-piperazineone peptidomimetics as HIV capsid modulators by mimicking the structure of host factors binding to CA.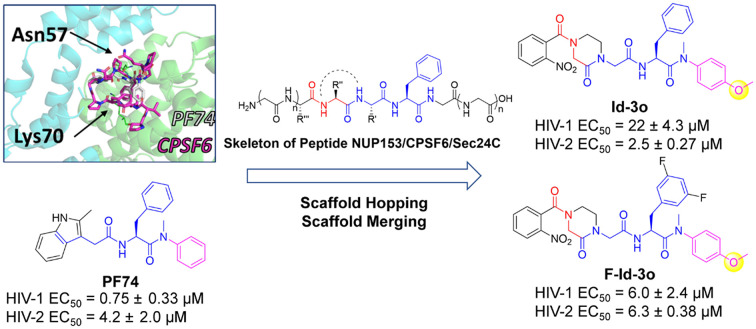
1. Introduction
Acquired immunodeficiency syndrome (AIDS), mainly caused by the pathogen of human immunodeficiency virus type 1 (HIV-1), remains a significant cause of death globally.1 HIV-2 infection, which contributes to the global AIDS burden, is found predominantly in West African nations. HIV-2 originates from simian immunodeficiency virus (SIV) transmission by sooty mangabey (SIVsmm).2,3 HIV-2 infection is less pathogenic than HIV-1 in infected individuals.4,5 However, HIV/AIDS is still one of the most severe epidemics caused by viruses. According to WHO, in 2021, 38.4 million people live with HIV worldwide, while 650 000 people died of HIV-related illnesses.6 Though combined antiretroviral therapy (cART) can reduce the viral load below the detection limit, the rapid emergence of drug resistance7–10 and the toxicity11 of existing drugs are urging us to search for novel anti-HIV drugs. Ideally, new drugs for HIV/AIDS should act via novel mechanisms to meet the increasing clinical needs.12,13
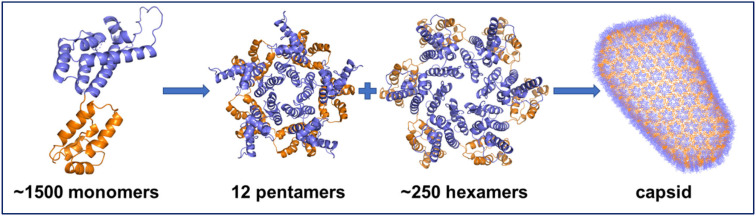
HIV-1 capsid (CA) is a research hotspot for its distinctive structure and unique roles in the HIV life cycle.14–16 HIV-1 CA is composed of the N-terminal domain (NTD), C-terminal domain (CTD), which are connected by a flexible linker (Fig. 1).12,17 Protein–protein interactions within different domains are indispensable to the assembly of the complete CA. NTD–NTD interaction and NTD–CTD interaction facilitate the formation of pentamers and hexamers, while CTD–CTD interaction drives the assembly of the whole capsid.18,19 Ultimately, about 1500 copies of monomeric CA form about 12 pentamers and 250 hexamers, which combine into a complete CA cone. The mature, assembled capsid protects the viral enzymes and genome during the early life steps of the viral life cycle until the uncoating event.20,21
Fig. 1. Structures of CA monomer (PDB ID: 4XFX), pentamer (PDB ID: 5MCY), hexamer (PDB ID: 4XFX), and the assembled cone (PDB ID: 3J3Q). The NTD is shown in purple, and the CTD is shown in orange.
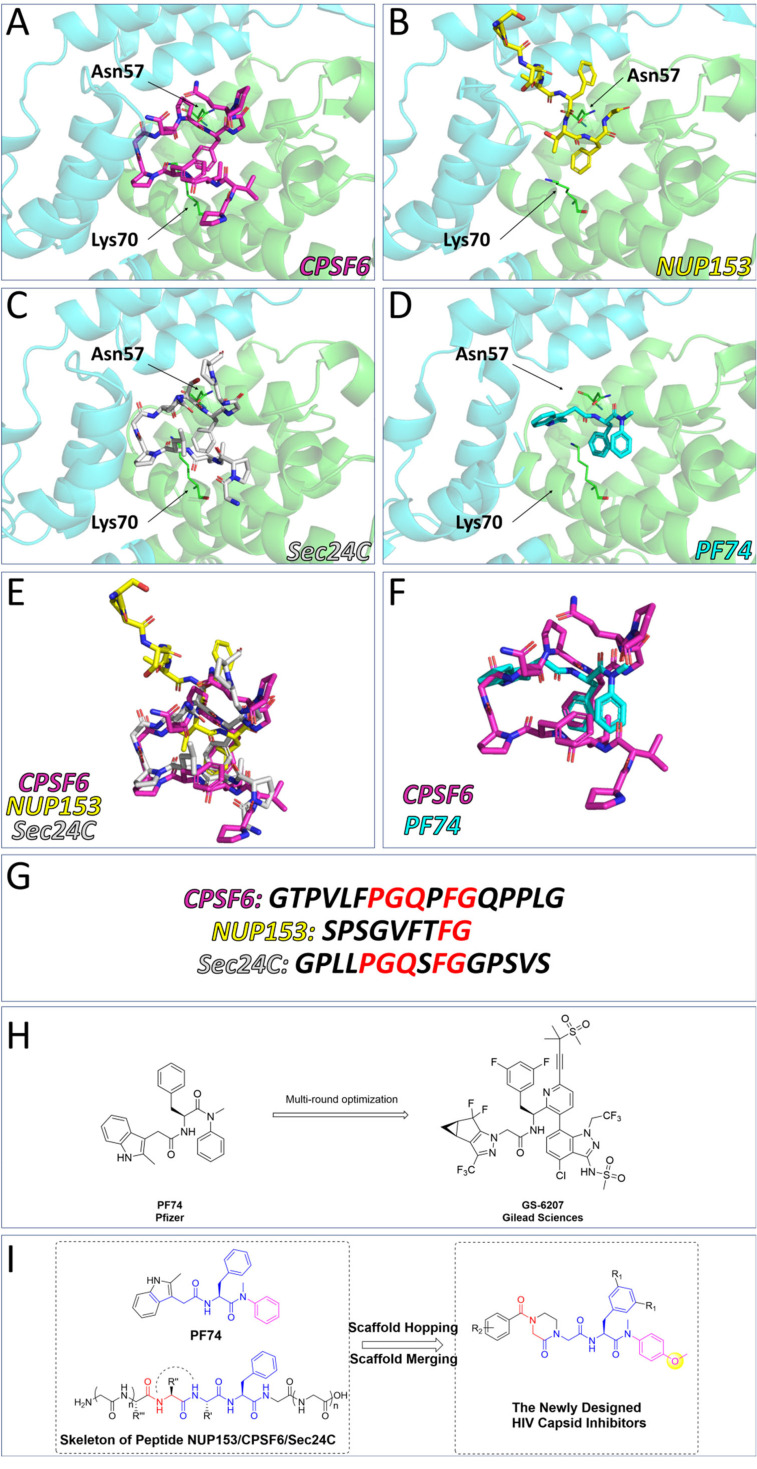
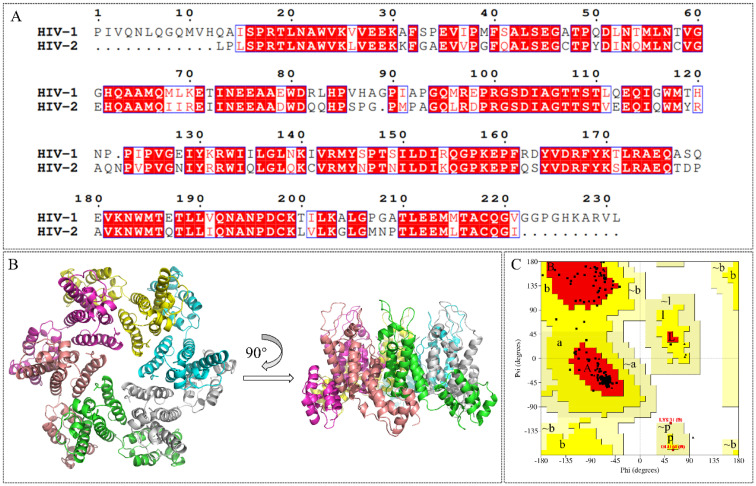
Various host factors interact with capsid in the intracellular transport and nuclear entry processes.22–26 These include NUP153, which mediates the entry of the capsid into the nucleus, and CPSF6 and Sec24C, which help stabilize the capsid to prevent premature uncoating. Coincidentally, the above three host factors interact with CA at the same site within the NTD–CTD interface, the binding site for numerous CA modulators (Fig. 2).27–29PF74 is a widely studied capsid modulators reported by Pfizer Inc., which binds to the same site with the above three host factors, abandoned due to low anti-HIV activity and poor stability.30–32 Due to its novel antiviral mechanism, PF74 has been used as a scaffold for optimization in addition to a probe to decipher HIV-1 biology.33–36PF74 has a characteristic phenylalanine-glycine (FG) motif that mimics motifs in CPSF6/NUP153 and Sec24C. Notably, the phenylalanine at this position is vital for the orientation of the small molecule within the interprotomer pocket. GS-6207 (Lenacapavir) is a compound derived from PF74 reported by Gilead Science, showing robust antiviral activity in MT-4 cells (EC50 = 105 pM) and binding to the same site with PF74.37 Similar to PF74, GS-6207 stabilizes the capsid, resulting in a buildup of intact core in the cytoplasm, and the binding of NUP153 and CPSF6 to the CA was interfered by GS-6207.38 In addition to its strong antiviral activity, the stable metabolic stability of GS-6207 is another great breakthrough, allowing for once-6-month injection therapy.12 This drug has been approved by the European Community and becomes the first drug targeting CA. However, the complex and laborious synthesis scheme limits its industrialization. At the same time, excessive molecular weight is also a point that needs to be decreased. Therefore, we hypothesized that further mimicry of the host cell peptides using PF74 as a base scaffold would result in novel capsid modulators worthy of additional optimization.
Fig. 2. Binding modes within the HIV-1 CA interprotomer pocket of (A) CPSF6 peptide (in purple, PDB ID: 6AY9), (B) NUP153 peptide (in yellow, PBD ID: 6AYA), (C) Sec24C peptide (in grey, PDB ID: 6PU1), (D) PF74 (in cyan, PDB ID: 5HGL). (E) Structural alignment of binding modes for CPSF6 peptide (purple), NUP153 peptide (yellow), and Sec24C peptide (grey). (F) Structural alignment of binding modes of CPSF6 peptide (purple) and PF74 (cyan). (G) The amino acid sequences of CPSF6 peptide, NUP153 peptide, and Sec24C peptide. (H) The structures of PF74 and GS-6207. (I) The design strategy of the peptidomimetic.
Several features of PF74 are considered crucial for the correct orientation of the molecule within the interprotomer pocket. The methylaniline of PF74 forms an important cation–pi interaction with Lys70 and is considered to be one such vital feature, and we retained the phenylalanine–methylaniline skeleton of PF74 during the design of chemotypes. For the side chain extending towards the CTD, we introduced the protein skeleton to compete with host factors for binding sites. Simultaneously, we limited the structural flexibility through a cyclization and scaffold hopping strategy to reduce the peptide-like property of the compounds and improve metabolic stability. Referring to the structure of GS-6207 and several reported compounds,33,39–43 we introduced difluoro substitution at the benzene meta-position of phenylalanine to explore the structure–activity relationships of these compounds. Finally, we introduced hydrogen bond donors/receptors into the compounds to enhance affinity and antiviral activity. We designed and synthesized a series of 2-piperazineone-containing peptidomimetics as novel HIV capsid modulators based on the above design strategy. We tested the antiviral activities of the compounds and studied the mechanism of antiviral action of the compounds through surface plasmon resonance (SPR) interaction analysis, the single-round infection assay, and molecular dynamics simulations. Moreover, the stabilities in HLM and plasma were experimentally obtained, and the physicochemical properties of lead compound F-Id-3o and Id-3o were computationally predicted.
2. Chemistry
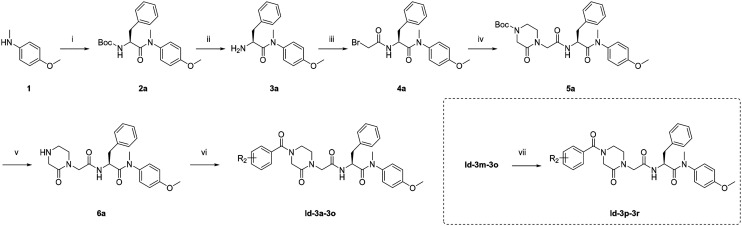
As shown in Scheme 1, starting from commercially available 4-methoxy-N-methylaniline (1), the target compounds were prepared via a concise and well-established synthetic route as outlined below. Treating of 1 with Boc-l-phenylalanine and 2-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU) in N,N-diisopropylethylamine (DIEA) and dichloromethane (DCM) afforded 2a, followed by removal of tert-butyloxycarbonyl (Boc) protection resulted in the formation of free amine 3a. The intermediate 4a was obtained by acylation of 3a with bromoacetic acid in DCM solution. The nucleophilic substitution of 2-piperazineone with 4a in N,N-dimethylformamide(DMF) resulted in 5a. Then remove the Boc and obtain 6a. Finally, triethylamine (TEA), 6a, and benzoyl chloride substituted by different groups were added to DCM to afford compounds Id-3a-3o. Other target compounds, Id-3p-3r, were prepared by a hydrogenation reduction of the nitro group of Id-3m-3o.
Scheme 1. Preparation of Id-3a-3r. Reagents and conditions: (i) Boc-l-phenylalanine, HATU, DIEA, DCM, 0 °C to r.t.; (ii) trifluoroacetic acid, DCM, r.t.; (iii) bromoacetic acid, HATU, DIEA, DCM, 0 °C to r.t.; (iv) 2,5-piperazinedione, K2CO3, DMF, 55 °C. (v) Trifluoroacetic acid, DCM, r.t.; (vi) substituted benzoyl chloride, TEA, DCM, 0 °C to r.t.: (vii) H2, Pd, DCM, r.t.
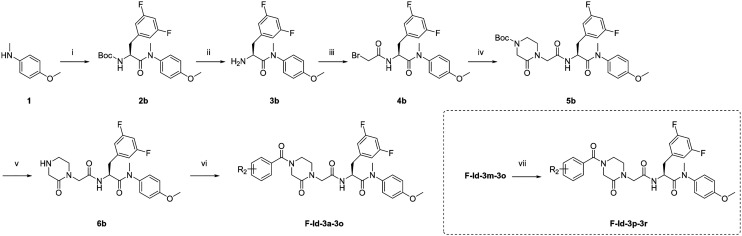
As shown in Scheme 2, starting from commercially available 4-methoxy-N-methylaniline (1), the target compounds were prepared via a concise and well-established synthetic route as outlined below. Treating of 1 with Boc-3,5-difluoro-l-phenylalanine and HATU in DIEA and DCM afforded 2b, followed by removal of Boc protection resulted in the formation of free amine 3b. The intermediate 4 was obtained by acylation of 3b with bromoacetic acid in DCM solution. The nucleophilic substitution of 2-piperazineone with 4b in DMF resulted in 5b. Then remove the Boc and obtain 6. Finally, TEA, 6b and benzoyl chloride substituted by different groups were added to DCM to afford compounds F-Id-3a-3o. The other target compounds, F-Id-3p-3r, were prepared by a hydrogenation reduction of the nitro group of F-Id-3m-3o.
Scheme 2. Preparation of F-Id-3a-3r. Reagents and conditions: (i) Boc-3,5-difluoro-l-phenylalanine, HATU, DIEA, DCM, 0 °C to r.t.; (ii) trifluoroacetic acid, DCM, r.t.; (iii) bromoacetic acid, HATU, DIEA, DCM, 0 °C to r.t.; (iv) 2,5-piperazinedione, K2CO3, DMF, 55 °C. (v) Trifluoroacetic acid, DCM, r.t.; (vi) substituted benzoyl chloride, TEA, DCM, 0 °C to r.t.: (vii) H2, Pd, DCM, r.t.
3. Results and discussion
3.1. In vitro anti-HIV assays and SARs analysis
All the target compounds were tested for antiviral activities and cytotoxicities using MT-4 cells infected by HIV-1 IIIB or HIV-2 ROD. EC50 and CC50 values for each compound are shown respectively in Table 1. PF74 was utilized as the control drug in this assay.
Anti-HIV activity and cytotoxicity in MT-4 cells infected with HIV-1 IIIB and HIV-2 ROD.
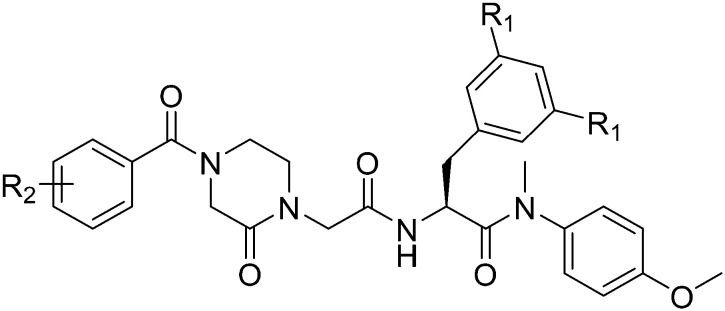
| |||||||
|---|---|---|---|---|---|---|---|
| Compounds | R1 | R2 | EC50a (μM) | CC50b (μM) | SIc | ||
| HIV-1 IIIB | HIV-2 ROD | HIV-1 IIIB | HIV-2 ROD | ||||
| Id-3a | H | H | 28 ± 7.8 | 3.6 ± 0.30 | >2.4 × 102 | >8.6 | >67 |
| Id-3b | H | 4-F | >77 | 11 ± 1.2 | 77 ± 28 | <1 | 7 |
| Id-3c | H | 3-F | 23 ± 4.8 | 4.6 ± 1.5 | >2.3 × 102 | >10 | >50 |
| Id-3d | H | 2-F | 24 ± 3.1 | 7.8 ± 2.0 | 1.4 × 102 ± 64 | 5.8 | 18 |
| Id-3e | H | 4-Cl | >8.9 | >9.0 | 9.0 ± 4.2 | <1 | <1 |
| Id-3f | H | 4-Br | >11 | >11 | 11 ± 2.9 | <1 | <1 |
| Id-3g | H | 4-CH3 | 33 ± 8.6 | 9.2 ± 1.1 | 64 ± 22 | 1.9 | 7.0 |
| Id-3h | H | 4-CH3O | 26 ± 4.1 | 21 ± 8.2 | 1.1 × 102 ± 34 | 4.2 | 5.2 |
| Id-3i | H | 4-CF3 | ≥37 | ≥41 | 42 ± 19 | ≤1.0 | ≤1.0 |
| Id-3j | H | 3-CF3 | >6.7 | >6.7 | 6.7 ± 3.3 | <1 | <1 |
| Id-3k | H | 2-CF3 | 21 ± 3.6 | 2.9 ± 1.8 | 1.3 × 102 ± 41 | 6.2 | 45 |
| Id-3l | H | 4-COOCH3 | >1.9 × 102 | 24 ± 6.2 | 2.0 × 102 ± 6.1 | <1 | 8.3 |
| Id-3m | H | 4-NO2 | >16 | >16 | 16 ± 6.3 | <1 | <1 |
| Id-3n | H | 3-NO2 | >24 | >24 | 24 ± 3.2 | <1 | <1 |
| Id-3o | H | 2-NO2 | 22 ± 4.3 | 2.5 ± 0.27 | 1.8 × 102 ± 14 | 8.2 | 72 |
| Id-3p | H | 4-NH2 | 21 ± 6.0 | 28 ± 4.1 | >2.3 × 102 | >11 | >8.2 |
| Id-3q | H | 3-NH2 | ≥2.0 × 102 | 27 ± 6.8 | >2.3 × 102 | ≤1 | >8.5 |
| Id-3r | H | 2-NH2 | 23 ± 5.7 | 5.4 ± 1.0 | 1.6 × 102± 38 | 7.0 | 30 |
| F-Id-3a | F | H | 25 ± 8.7 | 12 ± 4.0 | 1.1 × 102 ± 36 | 4.4 | 4.4 |
| F-Id-3b | F | 4-F | 20 ± 6.9 | 21 ± 13 | 77 ± 3.0 | 3.8 | 3.7 |
| F-Id-3c | F | 3-F | 25 ± 9.3 | 13 ± 6.6 | 99 ± 28 | 4.0 | 7.6 |
| F-Id-3d | F | 2-F | 22 ± 5.6 | 5.3 ± 1.4 | 1.4 × 102 ± 56 | 6.4 | 26 |
| F-Id-3e | F | 4-Cl | >19 | >19 | 19 ± 4.1 | <1 | <1 |
| F-Id-3f | F | 4-Br | >19 | >19 | 19 ± 6.0 | <1 | <1 |
| F-Id-3g | F | 4-CH3 | 32 ± 7.8 | 35 ± 3.0 | 74 ± 29 | 2.3 | 2.1 |
| F-Id-3h | F | 4-CH3O | 24 ± 3.4 | 31 ± 6.0 | 96 ± 32 | 4.0 | 3.1 |
| F-Id-3i | F | 4-CF3 | >18 | >18 | 18 ± 5.6 | <1 | <1 |
| F-Id-3j | F | 3-CF3 | >3.7 | >3.7 | 3.7 ± 1.4 | <1 | <1 |
| F-Id-3k | F | 2-CF3 | 12 ± 5.8 | 5.0 ± 0.81 | 1.1 × 102 ± 3.6 | 9.2 | 22 |
| F-Id-3l | F | 4-COOCH3 | 18 ± 3.5 | 26 ± 3.0 | 1.2 × 102 ± 55 | 6.7 | 4.6 |
| F-Id-3m | F | 4-NO2 | >18 | >18 | 18 ± 5.6 | <1 | <1 |
| F-Id-3n | F | 3-NO2 | 34 ± 5.0 | >47 | 47 ± 15 | 1.4 | <1 |
| F-Id-3o | F | 2-NO2 | 6.0 ± 2.4 | 6.3 ± 0.38 | 1.6 × 102 ± 45 | 27 | 25 |
| F-Id-3p | F | 4-NH2 | 18 ± 3.6 | 32 ± 5.6 | >2.2 × 102 | >12 | >6.9 |
| F-Id-3q | F | 3-NH2 | 23 ± 10 | 16 ± 2.1 | >2.2 × 102 | >10 | >14 |
| F-Id-3r | F | 2-NH2 | 18 ± 4.1 | 8.6 ± 0.77 | 1.6 × 102 ± 37 | 8.9 | 19 |
| PF74 | — | — | 0.75 ± 0.33 | 4.2 ± 2.0 | 32 ± 3.0 | 43 | 7.6 |
EC50: the concentration of the compound required to achieve 50% protection of MT-4 cells against HIV-induced cytotoxicity effect, determined in at least triplicate against HIV in MT-4 cells.
CC50: the concentration of the compound required to reduce the viability of uninfected cells by 50%, determined in at least triplicate against HIV in MT-4 cells; values were averaged from at least three independent experiments.
SI: selectivity index, the ratio of CC50/EC50.
3.1.1. In vitro anti-HIV activity
Generally, most of the newly synthesized compounds showed anti-HIV activity. Most compounds showed increased activity in inhibiting HIV-2 than HIV-1. 11 compounds (Id-3a, Id-3c, Id-3d, Id-3g, Id-3k, Id-3o, Id-3r, F-Id-3d, F-Id-3k, F-Id-3o, F-Id-3r) exhibited superior or comparable anti-HIV-2 activity to PF74. The anti-HIV-2 activity of Id-3o (EC50 = 2.5 ± 0.27 μM) was 2-fold higher as compared to PF74 (EC50 = 4.2 ± 2.0 μM). Interestingly, the anti-HIV-1 activity of all compounds did not exceed PF74 (EC50 = 0.75 ± 0.33 μM). F-Id-3o is the most potent compound from our series regarding anti-HIV-1 activity, with an EC50 value of 6.0 ± 2.4 μM, approximately 8-fold lower than PF74. However, F-Id-3o showed a superior toxicity profile compared to PF74, which has a higher selectivity index.
3.1.2. SAR of R1 group
When R1 was substituted by fluorine, the anti-HIV-1 activities of most compounds were increased. For example, the activity of F-Id-3o (EC50 = 6.0 ± 2.4 μM) is greatly improved relative to that of Id-3o (EC50 = 22 ± 4.3 μM). Nonetheless, certain degree of improvement of the cytotoxicity of some compounds are observed simultaneously (e.g., F-Id-3a (CC50 = 1.1 × 102 ± 36 μM) < Id-3a (CC50 > 2.4 × 102 μM), F-Id-3c (CC50 = 99 ± 28 μM) < Id-3c (CC50 > 2.3 × 102 μM) and F-Id-3l (CC50 = 1.2 × 102 ± 55 μM) < Id-3l (CC50 = 1.9 × 102 ± 6.3 μM)). In contrast, when R1 was not substituted, the compounds were more efficient to HIV-2 (e.g., F-Id-3a (EC50 = 12 ± 4.0 μM) > Id-3a (EC50 = 3.6 ± 0.30 μM), F-Id-3o (EC50 = 6.3 ± 0.38 μM) > Id-3o (EC50 = 2.5 ± 0.27 μM)).
3.1.3. SAR of R2 when R1 was not substituted by fluorine
When R2 was para-substituted, all compounds, except Id-3g, Id-3h, and Id-3p, lost their inhibitory activities against HIV-1, which compelled the electron-withdrawing groups substituting at the para-position was unfavorable to enhance the anti-HIV-1 activity. By comparing the EC50 of the four above compounds exhibiting anti-HIV-1 activity, we found that the enhancement of anti-HIV-1 activity (Id-3q > Id-3h > Id-3g) is parallel to that of electron-donating ability (–NH2 > –OCH3 > –CH3). The substitution position of R2 was also crucial for anti-HIV-1 activity. For the electron-withdrawing groups (–F, –CF3, and –NO2), the ortho-substitution was more sensitive to HIV-1 inhibition than the meta- and para-substitution, considering the SD and the toxicity. Conversely, the electron-donating group (–NH2) substituting at the para or ortho position has a more significant effect than the meta position.
Unlike anti-HIV-1 activity, any para- or meta-substitution of R2 was detrimental to anti-HIV-2 activity. The substitution of the electron-withdrawing groups at the ortho position was detected to have stronger anti-HIV-2 activity, and the stronger the electron-withdrawing ability (–NO2 > –CF3 > –H) was, the stronger the antiviral activity (Id-3o > Id-3k > Id-3a).
3.1.4. SAR of R2 when R1 was substituted by fluorine
When R1 was substituted by fluorine and R2 was substituted at the para-position with electron-withdrawing groups, most of the compounds lost their inhibitory activity against HIV-1. However, other active compounds with para-substitution did not show noticeable activity improvement or any SAR rule. In addition, meta-substitution did not result in any increase in anti-HIV-1 activity relative to the unsubstituted compound. When the ortho-position was subsisted by a strong electron-withdrawing group, the anti-HIV-1 activity of the compound was improved to a certain extent (F-Id-3o > F-Id-3k > F-Id-3a).
Any substitution at the para- and meta-position was also considered negative for enhancing anti-HIV-2 activity. In contrast, the ortho-position substitution was particularly significant for the improvement of anti-HIV-2 activity, especially when it was substituted by electron-withdrawing groups (F-Id-3k > F-Id-3d > F-Id-3o > F-Id-3r > F-Id-3a).
3.2. Compounds interact with HIV-1 CA as determined by surface plasmon resonance (SPR)
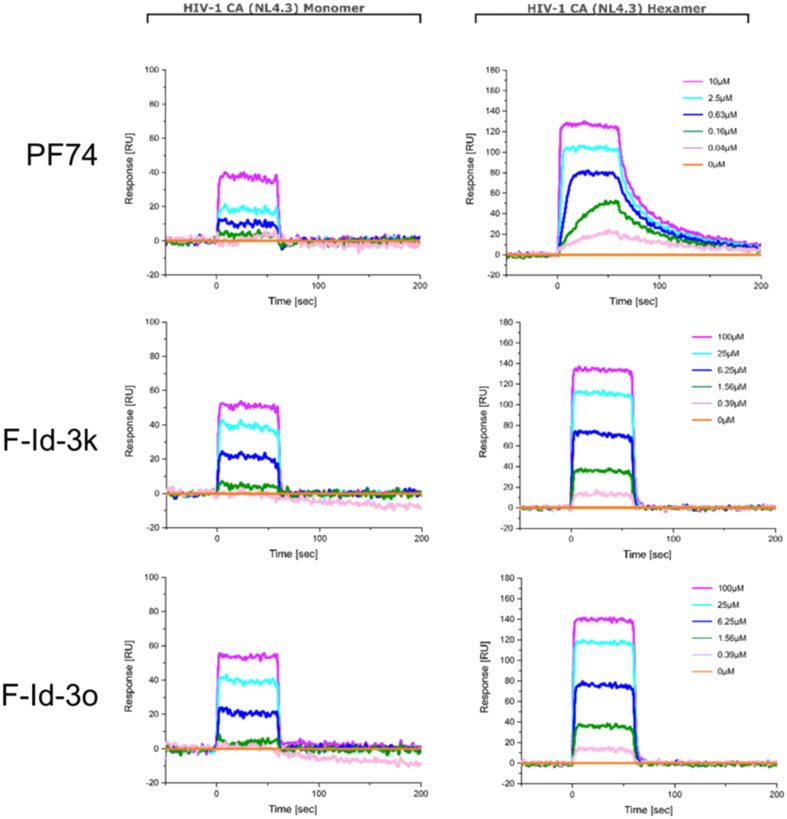
Since most of the compounds that exhibited anti-HIV activity had a similar skeleton, the most potent compounds (F-Id-3k and F-Id-3o) were chosen for affinity determination via SPR utilizing monomeric and hexameric HIV-1 CA protein with PF74 as an in-line control.
As shown in Table 2, the equilibrium dissociation constant (KD) values revealed that all three compounds tested preferentially bind to HIV-1 CA hexamers rather than CA monomers. However, F-Id-3k and F-Id-3o showed decreased affinities for CA hexamers and monomers compared to PF74, explaining the decrease in anti-HIV-1 activities of the two novel modulators compared to PF74. Meanwhile, for CA hexamers, the koff values of F-Id-3k and F-Id-3o were about 14-time higher than that of PF74, suggesting faster dissociation. As found in our previous study, the decrease in koff value was positively correlated with the increase in antiviral activity,44 illustrating the low anti-HIV-1 activity of F-Id-3k and F-Id-3o from a kinetic point of view (Fig. 3).
SPR results of F-Id-3k, F-Id-3o, and PF74 binding to monomeric and hexameric CA.
| Compounds | K D a (μM) | Ratiob | k off c (1/s) | |
|---|---|---|---|---|
| Monomer | Hexamer | |||
| F-Id-3k | 12 ± 1.3 | 5.3 ± 0.55 | 2.2 | 0.25 ± 0.013 |
| F-Id-3o | 13 ± 2.3 | 5.2 ± 0.52 | 2.5 | 0.25 ± 0.045 |
| PF74 | 3.4 ± 1.3 | 0.16 ± 0.041 | 21.4 | 0.018 ± 0.0039 |
All values represent the average response from three replicates. Errors represent standard deviation (SD).
Ratio = KMonomerD/KHexamerD highlights the preference for HIV-1 CA monomer or hexamer binding.
k off = off-rate measured for HIV-1 CA hexamer.
Fig. 3. SPR sensorgrams of F-Id-3k and F-Id-3o binding to two variants of the CA protein (monomer and disulfide-stabilized hexamer), respectively, with PF74 as the reference.
3.3. The novel modulators exhibit both early and late-stage inhibition activity
Modulators binding at the hexameric NTD–CTD interface typically exert a dual-stage inhibition effect, simultaneously inhibiting both the early and late stages of the HIV-1 life cycle. In order to characterize the antiviral effect of F-Id-3o on both stages, we performed a single-round infection assay (SRI assay).
Table 3 shows the results of the SRI assay. F-Id-3o exhibited strong dual-stage inhibiting activity at high concentrations, related to its ability to bind to monomers and hexamers, as shown earlier by SPR. In the early stage, F-Id-3o binds to the hexamer of the capsid, preventing the binding of host factors that compete for this binding site within the interprotomer pocket, thereby interfering with the normal uncoating process. However, in the later stage, the predominant form of CA is monomeric in the context of the Gag polyprotein. Compounds bound to the monomeric CA form can interfere with the stability of the NTD–CTD interface that has been or will be formed—ultimately affecting Gag assembly at the plasma membrane for virus budding. As previously mentioned, PF74 was shown to have both early and late inhibitory properties, but in this assay, it only exhibited early-stage inhibiting activity. This is not contradictory, as we have previously shown that PF74 interacts with the monomeric form of CA with a KD of approximately 3.4 μM. Therefore, the utilization of 1 μM in our assay set-up may not be sufficient to see the late-stage effects of PF74.
Results of single-round infection assay.
| Compounds | Concentration (μM) | % infectiona | |
|---|---|---|---|
| Early stage | Late stage | ||
| F-Id-3o | 60 | 8.1 ± 4.7 | 10 ± 7.7 |
| PF74 | 1 | −0.12 ± 0.17 | 1.5 × 102 ± 10 |
| DMSO | — | 1.0 × 102 ± 8.4 | 1.0 × 102 ± 11 |
Infections are an average of 3 replicates with error bars indicating standard error of the mean (SEM).
3.4. Molecular dynamics (MD) simulations study
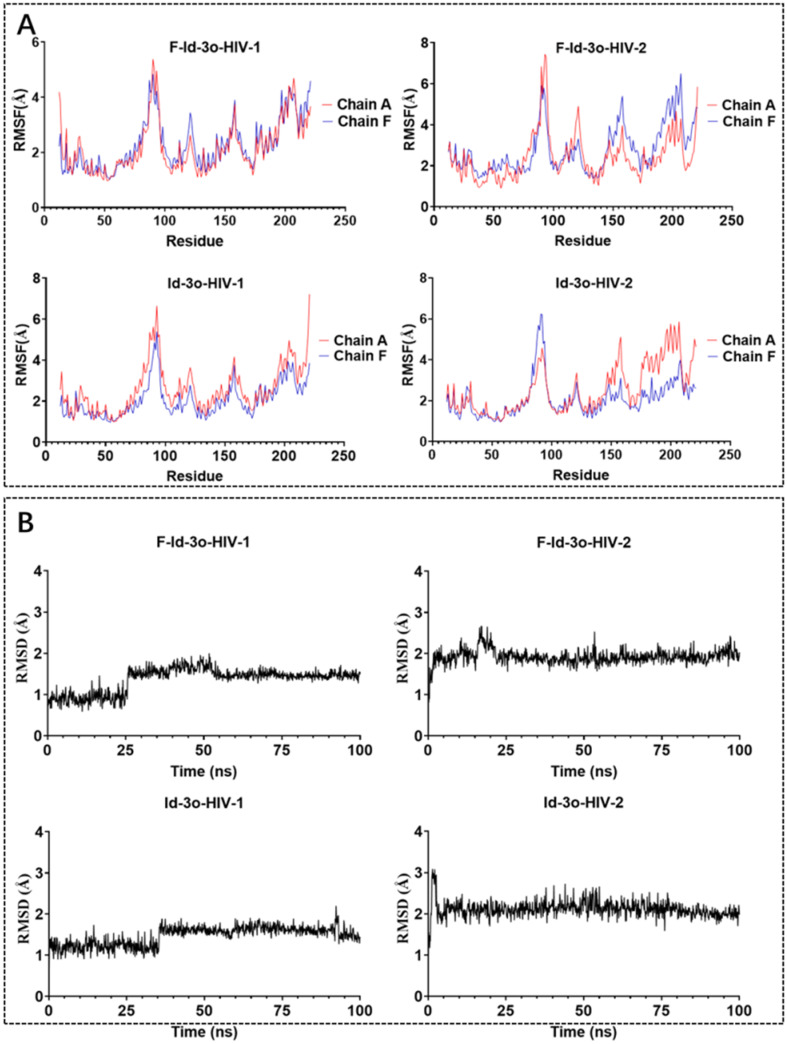
In order to study the binding modes of our lead compounds with proteins and to explain the selectivity of these compounds for HIV-1/2, we performed molecular dynamics simulations. Since the complete crystal structure of the HIV-2 capsid is not yet reported, we performed homology modeling. Subsequently, we performed 100 ns molecular dynamics simulations for Id-3o and F-Id-3o bound to HIV-1/2 capsid hexamers.
As shown in Fig. 4A, the sequence of CA of HIV-2 ROD is highly homologous to HIV-1 CA; therefore, we chose HIV-1 CA hexamer (PDB ID: 4XFZ) as a template for homology modeling to obtain HIV-2 CA hexamer (Fig. 4B). In Fig. 4C, the vast majority of residues modeled were in the favored (A, B and L regions) or allowed regions (a, b, l and p regions), revealing that the HIV-2 capsid hexamer model obtained by homology modeling is acceptable.
Fig. 4. (A) Sequence alignment of HIV-1 CA and HIV-2 CA. (B) The HIV-2 capsid hexamer model obtained by homology modeling. (C) Ramachandran plot of the HIV-2 capsid hexamer model. The HIV-1 CA with PDB ID: 4XFZ was used as a template structure for homology modeling.
F-Id-3o and Id-3o were selected for MD simulations since they were the most active against HIV-1 or HIV-2. Since every monomer of the CA hexamer and the ligands between them are the same, we selected the first group of dimers (chain A and chain F) and their ligands. Root mean square fluctuation (RMSF) of amino acids was calculated and represented in Fig. 5A to investigate the deviated amino acids. Most amino acids in the four MD simulations have deviated from the original structure. The deviation of amino acids shows that HIV CA has different conformations induced by the deviated ligands. This also indicates that the ligands could have different binding modes to HIV CA. To investigate the conformations of the two ligands bound to HIV-1 and -2 CA, their root mean square deviation (RMSD) were calculated and shown in Fig. 5B. The figures show that the two ligands have deviated from the docked conformer and have predominant stable conformation after 100 ns. However, the RMSD of Id-3o bound to HIV-2 CA is the most stable among the four, and there is not much fluctuation, which indicates that the binding of Id-3o to HIV-2 CA may be relatively stable, especially compared with F-Id-3o binding to HIV-2 CA. This may explain the better anti-HIV-2 activity of Id-3p relative to the analogues with fluorine atom substitution in R1.
Fig. 5. (A) RMSF (heavy atoms) of amino acids of HIV CA monomers in reference to the MD simulation. (B) RMSD (heavy atoms) of the bound Id-3o and F-Id-3o in reference to the docked conformer.
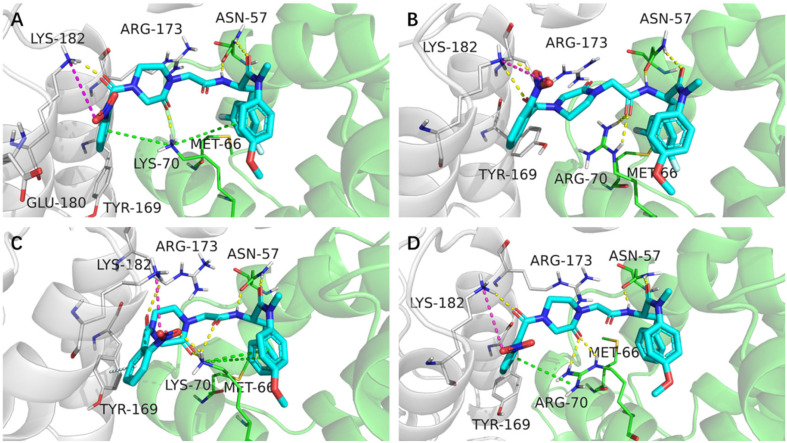
The whole trajectory was clustered to find the protein and the inhibitor conformations. The trajectory was clustered according to the two ligands to investigate their binding to the binding sites. Fig. 6 shows representative structures of the most dominant cluster. According to Fig. 5, these combination conformations were stable for a long time during the MD simulation.
Fig. 6. Binding of F-Id-3o and Id-3o in the most populated clusters. (A) F-Id-3o binds to HIV-1 CA. (B) F-Id-3o binds to HIV-2 CA. (C) Id-3o binds to HIV-1 CA. (D) Id-3o binds to HIV-2 CA. Yellow dashes: hydrogen bonds; green dashes: pi–cation; cyan dashes: pi–pi stacking; magenta dashes: salt bridges. The CTD is shown in gray and NTD in green.
The bonding forces between F-Id-3o/Id-3o and HIV-1/2 CA were investigated in the representative structure of the predominant cluster (Fig. 6). The two ligands have similar binding modes, engaging hydrogen bonds with Asn57, which is crucial in both HIV-1 and HIV-2 CA. Simultaneously, salt bridges introduced by the nitro group with Lys182 and the hydrogen bonds formed by carbonyl with the same residue are beneficial for compound stabilization by the CTD. Note that Lys70 in the HIV-1 capsid corresponds to Arg70 in the HIV-2 capsid. Although both are basic amino acids, their effect on binding capacity may be dramatic. Arginine is more basic and has more polar hydrogens than lysine, resulting in a stronger hydrogen bond formation and a weaker cation–pi formation profile.45 Taking together these observations could explain why our novel series of compounds prefer suppressing HIV-2 over HIV-1.
3.5. Computational assessment of drug-like properties, metabolic stability, and toxity
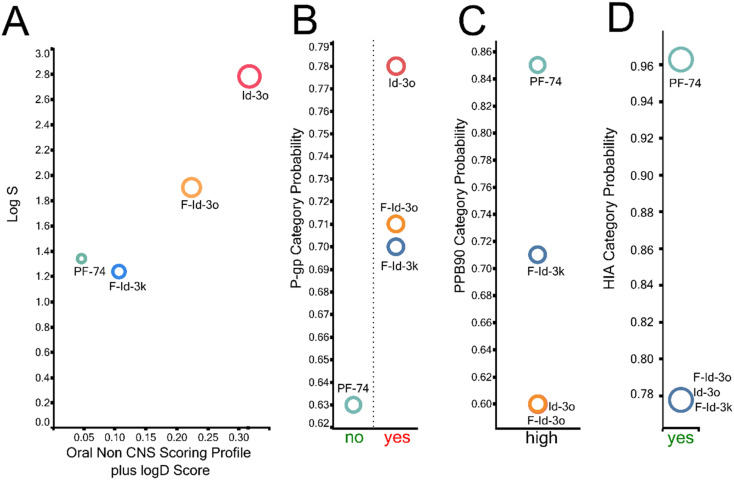
PF74 suffers from a number of problems that limit its clinical use, and these primarily relate to its drug-like properties. Therefore, we sought to analyze the new compounds' predicted ADME properties (absorption, distribution, metabolism, and excretion) and compare them with PF74 (Fig. 7). To accomplish this comparison, we used in silico prediction of drug-like metrics of the results as implemented in the oral non-central nervous system (CNS) drug profile in StarDrop 7 (Optibrium, Ltd., Cambridge, UK).46 This profile consists of several models, and for simplicity, a probabilistic scoring algorithm combines the model predictions in the oral non-CNS drug profile into an overall score. For reference, scores range from 0 to 1, with 0 suggesting extremely non-druglike and 1 suggesting the perfect drug.
Fig. 7. (A) Plot showing the StarDrop (Optibrium, Ltd., Cambridge, UK)-derived log S versus a multimetric oral non-CNS profile score. Importance: log S = 0.9, HIA = 0.85, log P = 0.6, log D = 0.6, hERG pIC50 = 0.4, 2D6 affinity category = 0.3, 2C9 pKi = 0.3, P-gp category = 0.3, PPB90 category = 0.2, BBB category = 0.11, BBB log(brain : blood) = 0.11 (B) P-gp category, (C) PPB90 = plasma protein binding >90%, (D) HIA = human intestinal absorption category, and probability of prediction of F-Id-3k, F-Id-3o, I3o and PF74.
Optibrium's oral non-CNS drug profile is composed of the following metrics: log S (intrinsic aqueous solubility); classification for human intestinal absorption; log P (octanol/water); hERG (human ether-à-go-go-related gene) pIC50 (mammalian cells); cytochrome P450 CYP2D6 classification; cytochrome P450 CYP2C9 pKi values; classification of P-glycoprotein transport; classification of blood–brain barrier (BBB) penetration; and predicted BBB penetration value. The models and their respective importance to the profile are shown in Fig. 7. As can be seen, F-Id-3o and Id-3o display improved aqueous solubility compared to PF74 as judged by the log S, which improves overall bioavailability. Overall, F-Id-3o and Id-3o show improved oral non-CNS drug profile scores, primarily due to improved solubility, low plasma protein binding, and favorable hERG pIC50 compared to PF74.
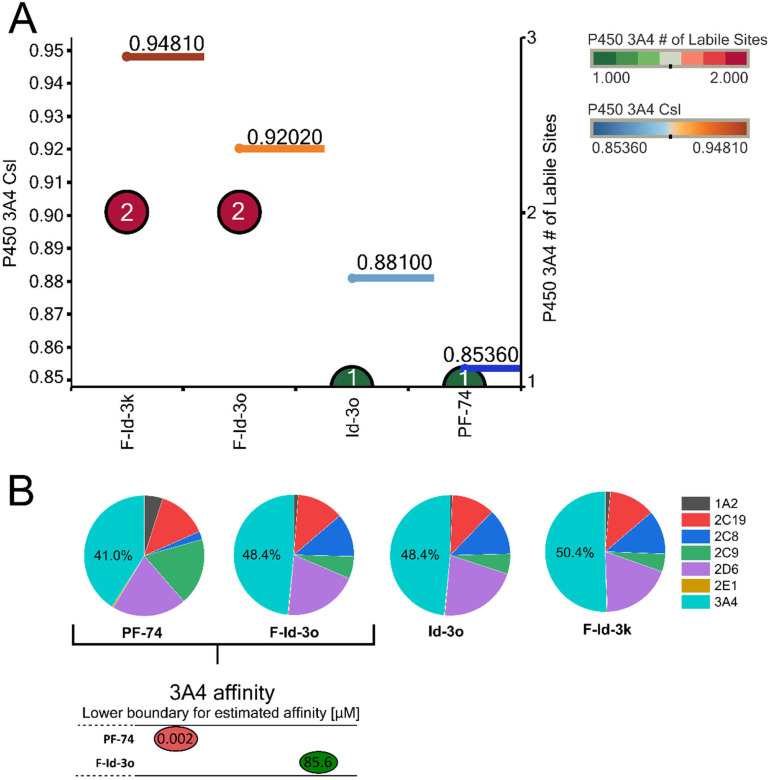
A major hurdle for PF74's development is its poor metabolic stability.28 Orally administered drugs must first pass the intestinal wall, followed by the portal circulation to the liver before reaching the bloodstream. Both sides are locations for first-pass metabolism and can adversely metabolize drugs before adequate plasma concentrations are reached. Therefore, we next sought to computationally investigate whether or not our compounds had improved predicted metabolic stability. We employed a computational analysis first demonstrated to be an accurate indicator of metabolic stability by the Cocklin group.47,48 This analysis uses the P450 module in StarDrop 7 (Optibrium, Ltd., Cambridge, UK)48,49 to predict each compound's major metabolizing Cytochrome P450 isoforms using the WhichP450™ model,50 followed by affinity prediction to that isoform using the HYDE function in SeeSAR (BioSolveIT Gmbh, Germany). The results of this analysis are shown in Fig. 8.
Fig. 8. Computational prediction of metabolic stability. (A) Overall composite site lability (CSL) score and number of labile sites (for metabolism). A lower CSL score indicates a more stable molecule. The prediction was achieved using the StarDrop (version 7) P450 module. (B) Prediction of the major metabolizing CYP isoforms. All compounds are predicted to be metabolized by the 3A4 isoform, including PF74. A lower boundary for predicted 3A4 affinity of F-Id-3o and PF74 using the hydrogen bond and dehydration scoring function (HYDE) implemented in SeeSAR11.2.
All compounds, including PF74, are predicted to be primarily metabolized by the CYP3A4 isoform (Fig. 8B). We, therefore, analyzed the predicted metabolic lability of our compounds and PF74 by the CY3A4 isoform by comparing the overall composite site lability (CSL) score and the number of labile sites. The CSL score reflects the overall efficiency of metabolism of the molecule by combining the labilities of individual sites within the compound. The number of labile sites between our compounds and PF74 is not significantly different; however, the CSL score indicated increased metabolic stability in the following order F-Id-3k, F-Id-3o, Id-3o, and PF74 (Fig. 8A), with PF74 displaying the lowest CSL score indicating higher metabolic stability.
In addition to the CSL score and number of labile sites, which assume that all compounds bind with similar affinity to the CYP3A4 isoform, other factors such as compound reduction rate and actual binding affinity to the CYP3A4 isoform can infer metabolic stability. Moreover, binding affinity can also be influenced by intrinsic compound properties such as size and lipophilicity. Therefore, we performed predictive binding affinity calculations using the HYdrogen bond and DEhydration (HYDE) energy scoring function in SeeSAR 11.2 (BioSolveIT Gmbh, Germany).51 For this analysis, we used the structure of the human CYPA4 bound to an inhibitor (PDB ID: 4D78).52 The HYDE scoring function in SeeSAR provides a range of affinities, spanning an upper and lower limit. Therefore, we used the lower limit as the affinity parameter to compare F-Id-3o and PF74 (Fig. 8B), which resulted in an affinity of 85.9 μM for F-Id-3o and 2 nM for PF74. Combining the results from these predictions (CSL scores, labile sites, and predicted CYP3A4 affinity), this analysis indicates that compound F-Id-3o should have slightly greater metabolic stability than PF74, primarily due to the significantly lower CYP3A4 affinity. Going forward, this approach provides a useful multi-parameter optimization filtering mechanism for designing new compounds and reduces the time and cost of synthesis.
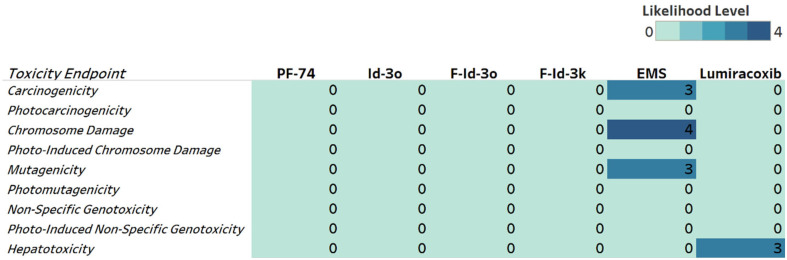
Next, we evaluated the potential toxicity associated with nitroaromatic compounds (NACs). NACs are known to have genotoxic activity and can seriously threaten humans and the environment.53–57 NACs can directly interact with the human genome via DNA interactions. Fortunately, those interactions can be computationally described with physicochemical properties within the molecule. To include genotoxicity and hepatotoxicity endpoints in our multiparameter optimization, we evaluated the toxicity endpoints of F-Id-3k, F-Id-3o, I3o, and PF74 using the Derek Nexus module within Stardrop V7. Derek Nexus is a knowledge- and rule-based expert system for semi-quantitatively estimating DNA reactive moieties within molecules. Based on this prediction, none of our compounds, including PF74, show any concerning likelihood of genotoxicity or hepatotoxicity (Fig. 9). In contrast, and used as positive controls in our prediction, ethyl methanesulfonate (EMS) and lumiracoxib are known to have in vivo genotoxic and hepatotoxic effects.
Fig. 9. Genotoxicity and hepatotoxicity endpoints for selected compounds. The highlighted table shows the StarDrop V7 (Optibrium, Ltd., Cambridge, UK)-derived toxicity endpoints using the Derek Nexus module based on a knowledge-based prediction. Structural alerts within the molecule with a level of likelihood for concerns are based on precedence from experimental data. Likelihood level 0 = no report; 1 = inactive; 2 = equivocal; 3 = plausible; 4 = probable. Ethyl methanesulfonate (EMS) was used as a positive control for genotoxicity,58 and lumiracoxib as a positive control for hepatotoxicity endpoints.59.
3.6. Human plasma and liver microsome (HLM) stability
Equipped with computational predictions for drug-like properties and metabolic stability, we next performed metabolic stability assays using human liver microsomes.
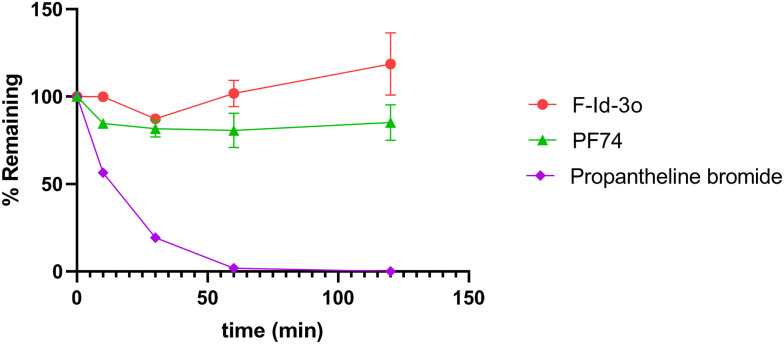
Poor metabolic stability prevents the further clinical application of PF74. Therefore, we tested the human plasma and liver microsome stability of F-Id-3o with PF74 and propantheline bromide as in-line control. In the two assays, F-Id-3o shows more robust metabolic stability than PF74. As shown in Fig. 10, the remaining rate of F-Id-3o in human plasma was always higher than that of PF74. Simultaneously, according to Table 4, the t1/2 of F-Id-3o (t1/2 = 3.8 min) was 7.6 times longer than that of PF74 (t1/2 = 0.5 min) as well as the clearance rate of F-Id-3o (CLint(mic) = 3.7 × 102 μL min−1 mg−1) was significantly lower than that of PF74 (CLint(mic) = 2.9 × 103 μL min−1 mg−1), probably due to the introduction of piperazinone to prevent the metabolism of the peptidomimetics to a certain extent. Overall, our experimentally derived evaluation of metabolic stability is in good agreement with the previously described computational prediction approach. Therefore, our computational pipeline provides an excellent approach for prescreening potential lead compounds in future studies.
Fig. 10. Human plasma stability assay. Experiments were performed in triplicate. % remaining = 100 × (PAR at appointed incubation time/PAR at time T0). PAR is the peak area ratio of a test compound to the internal standard, and accuracy should be within 80–120% of the indicated value.
Metabolic stability assay in human liver microsomes.
| Sample | HLM (final concentration of 0.5 mg protein per mL) | |||||
|---|---|---|---|---|---|---|
| R 2 a | t 1/2 b (min) | CLint(mic)c (μL min−1 mg−1) | CLint(liver)d (mL min−1 kg−1) | Remaining (T = 60 min) | Remaining (NCFe = 60 min) | |
| F-Id-3o | 0.9520 | 3.8 | 3.7 × 102 | 3.3 × 102 | 0.23% | 96% |
| PF74 | 1.0000 | 0.48 | 2.9 × 103 | 2.6 × 103 | 0.0% | 113% |
| Testosterone | 0.9982 | 17 | 83 | 74 | 7.9% | 91% |
| Diclofenac | 0.9947 | 3.7 | 3.7 × 102 | 3.3 × 102 | 0.0% | 97% |
| Propafenone | 0.9350 | 5.0 | 2.8 × 102 | 2.5 × 102 | 0.0% | 94% |
R 2 is the correlation coefficient of the linear regression to determine the kinetic constant (see raw data worksheet in the ESI†).
t 1/2 is half-life.
CLint(mic) = 0.693/t1/2/mg microsome protein per ml.
CLint(liver) = CLint(mic) × mg microsomal protein/g liver weight × g liver weight/kg body weight.
NCF: abbreviation of no co-factor. No NADPH is added to NCF samples (replaced by buffer) during the 60-minute incubation. If the NCF remaining is less than 60%, then possibly non-NADPH-dependent metabolism occurs.
4. Conclusion
This study has designed, synthesized, and evaluated a novel series of peptidomimetics as HIV capsid modulators by mimicking the structure of host factors binding to CA. Most of the synthesized compounds displayed anti-HIV activity. F-Id-3o is the most potent HIV-1 inhibitor among the compounds, with an EC50 value of 6.0 μM. Unexpectedly, this series of compounds showed a preference for HIV-2 inhibitory than HIV-1, in which Id-3o revealed an anti-HIV-2 EC50 value of 2.5 μM, ∼10-times its anti-HIV-1 activity. The SPR assay based on HIV-1 CA indicated that F-Id-3o has a nearly comparable affinity for CA monomer and hexamer compared to PF74. Its lower affinity and faster off-rate for HIV-1 CA may be the reason for its reduced anti-HIV-1 activity, which needs attention for future studies for peptidomimetics. Combining the SPR and single-round infection assay, we conclude that F-Id-3o shares a dual-stage inhibitor profile. Our work identifies F-Id-3o as a novel HIV-1 CA modulators with a similar mechanistic profile for the dual-stage inhibition to other reported CA modulators. In addition, the binding modes of Id-3o and F-Id-3o to HIV-1/2 CA were predicted by the molecular dynamic simulation, partly explaining the HIV-2 preference.
We then utilized a previously published computational workflow to predict drug-like properties and metabolic stability by comparing F-Id-3o and PF74 and validated our prediction using stability assays in human plasma and human liver microsomes. Our prediction was in formidable agreement with the experimental results for metabolic stability and again highlighted the potential of utilizing this computational pipeline to preselect promising lead compounds prior to exhaustive and expensive experimental testing. Computational prediction and experimental validation of metabolic stability indicated that although F-Id-3o has enhanced metabolic stability over PF74, further optimization is necessary for improvements.
To improve antiviral activity, we plan to introduce substituents at appropriate molecule positions to form a more efficient interaction network within the interprotomer pocket of the CA protein. In addition, due to the peptidomimetic nature, groups such as amides may be responsible for the poor metabolic stability, and the replacement of amides with more stable groups using medicinal chemistry strategies such as bioisosteres may play a role in improving metabolic stability and enhancing antiviral activity. Overall, F-Id-3o and Id-3o are attractive chemotypes that provide opportunities for further optimization of potency and metabolic stability as novel lead compounds for HIV-1/2 capsid modulators.
5. Experimental section
5.1. Chemistry
1H NMR and 13C NMR spectra were recorded on Bruker AV-400 spectrometer or Bruker AV-600 spectrometer using solvents as indicated (DMSO-d6). Chemical shifts were reported in δ values (ppm) with tetramethylsilane (TMS) as the internal reference, and J values were reported in hertz (Hz). Melting points (mp) were determined on a micromelting point apparatus and were uncorrected. TLC was performed on silica gel GF254 for TLC (Merck), and spots were visualized by iodine vapor or irradiation with UV light (λ = 254 nm). Flash column chromatography was performed on a column packed with silica gel60 (200–300 mesh). Thin-layer chromatography was performed on pre-coated HUANGHAI_HSGF254, 0.15–0.2 mm TLC-plates. Solvents were of reagent grade and were purified and dried by standard methods when necessary. The concentration of the reaction solutions involved the use of a rotary evaporator at reduced pressure. The solvents of DCM, TEA and methanol etc., were obtained from Sinopharm Chemical Reagent Co., Ltd (SCRC), which were of AR grade. The key reactants, including 4-methoxy-N-methylaniline, N-(tert-butoxycarbonyl)-l-phenylalanine etc. were purchased from Bide Pharmatech Co., Ltd or Shanghai Haohong Scientific Co., Ltd. The purity of final representative compounds was checked by HPLC and was >95%.
5.1.1. General procedure for the synthesis of 2a and 2b
To a solution of (tert-butoxycarbonyl)-l-phenylalanine or (tert-butoxycarbonyl)-3,5-difluoro-l-phenylalanine (1.5 eq.) in 20 mL DCM was added HATU (1.5 eq.) at 0 °C, and the mixture was stirred for 0.5 h. Subsequently, DIEA (3 eq.) and 4-methoxy-N-methylaniline (1 eq.) were added to the mixture and then stirred at room temperature for another 2 h (monitored by TLC). The reaction solution was initially washed with saturated sodium bicarbonate and extracted with DCM (3 × 20 mL), and dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to afford a corresponding crude product, purified by flash column chromatography to afford intermediates 2a and 2b.
5.1.1.1. tert-Butyl (S)-(1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)carbamate (2a)
Yellow oil, yield: 81%. ESI-MS: m/z 384.85 (M + 1)+. C22H28N2O4 [384.48].
5.1.1.2. tert-Butyl (S)-(3-(3,5-difluorophenyl)-1-((4-methoxyphenyl) (methyl)amino)-1-oxopropan-2-yl)carbamate (2b)
Yellow oil, yield: 85%. 1H NMR (400 MHz, DMSO-d6): δ 7.31 (d, J = 8.4 Hz, 2H, Ph–H), 7.13–7.04 (m, 3H, Ph–H), 7.01 (d, J = 9.5 Hz, 1H, Ph–H), 6.44 (d, J = 8.3 Hz, 2H, Ph–H), 4.20–4.10 (m, 1H, CH), 3.81 (s, 3H, OCH3), 3.14 (s, 3H, NCH3), 2.82–2.61 (m, 2H, PhCH2), 1.29 (s, 9H, C(CH3)). 13C NMR (150 MHz, DMSO-d6) δ 171.56 (C O), 162.47 (dd, 1JCF = 245.8, 3JCF = 13.7 Hz), 159.18, 155.59 (C O), 143.13, 136.06, 129.26, 115.37, 112.31 (dd, 2JCF = 19.5, 4JCF = 3.3 Hz), 102.23 (t, 2JCF = 25.4 Hz), 78.52, 55.96, 53.10, 37.81, 36.85, 28.58. ESI-MS: m/z 421.07 (M + 1)+, 443.17 (M + 23)+. C22H26F2N2O4 [420.46].
5.1.2. General procedure for the synthesis of 3a and 3b
Trifluoroacetic acid (5.0 eq.) was added dropwise to the corresponding substituted intermediate 2a or 2b (1.0 eq.) in 30 mL DCM and stirred at room temperature for 1 h (monitored by TLC). Then, the resulting mixture solution was alkalized to pH ∼7 with saturated sodium bicarbonate solution and then extracted with DCM (3 × 30 mL), dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to afford corresponding crude products 3a and 3b.
5.1.2.1. (S)-2-Amino-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (3a)
Yellow oil, yield: 82%. ESI-MS: m/z 285.16 (M + 1)+. C17H20N2O2 [284.36].
5.1.2.2. (S)-2-Amino-3-(3,5-difluorophenyl)-N-(4-methoxyphenyl)-N-methylpropanamide (3b)
Yellow oil, yield: 83%. 1H NMR (400 MHz, DMSO-d6): δ 7.10–6.93 (m, 5H, Ph–H), 6.57 (h, J = 4.1 Hz, 2H, Ph–H), 3.78 (s, 3H, OCH3), 3.35 (dd, J = 7.6, 5.9 Hz, 1H, CH), 3.09 (s, 3H, NCH3), 2.74 (dd, J = 13.1, 5.8 Hz, 1H, PhCH), 2.54–2.45 (m, 1H, PhCH), 1.82 (s, 2H, NH2). 13C NMR (150 MHz, DMSO-d6) δ 174.57 (C O), 162.50 (dd, 1JCF = 245.5, 3JCF = 13.3 Hz), 158.96, 143.82 (t, 3JCF = 9.3 Hz), 136.31, 128.95, 115.21, 112.50 (dd, 2JCF = 19.7, 4JCF = 4.6 Hz), 101.96 (t, 2JCF = 25.4 25.8 Hz), 55.91, 52.99, 41.27, 37.48. ESI-MS: m/z 321.11 (M + 1)+, m/z 343.25 (M + 23)+. C17H18F2N2O2 [320.34].
5.1.3. General procedure for the synthesis of 4a and 4b
Bromoacetic acid (1.2 eq.) and HATU (1.5 eq.) were mixed in 15 mL DCM and stirred in an ice bath for 0.5 h. Then, the corresponding substituted intermediate 3a or 3b (1 eq.) and DIEA (2 eq.) were slowly added to the above solution at 0 °C. The reaction system was then stirred at room temperature for an additional 0.5 h (monitored by TLC). The resulting mixture was initially washed with saturated sodium bicarbonate and extracted with DCM (3 × 20 mL), and dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to afford a corresponding crude product, purified by flash column chromatography to afford intermediate 4a and 4b.
5.1.3.1. (S)-2-(2-Bromoacetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (4a)
White oil, yield: 75%. 1H NMR (600 MHz, DMSO-d6): δ 8.62 (d, J = 7.9 Hz, 1H, NH), 7.22–7.16 (m, 3H, Ph–H), 7.05 (d, J = 8.3 Hz, 2H, Ph–H), 6.96 (d, J = 9.2 Hz, 2H, Ph–H), 6.88 (d, J = 6.2 Hz, 2H, Ph–H), 4.44 (td, J = 8.4, 5.6 Hz, 1H, CH), 3.82 (d, J = 2.4 Hz, 2H, CH2), 3.79 (s, 3H, OCH3), 3.10 (s, 3H, NCH3), 2.87 (dd, J = 13.5, 5.5 Hz, 1H, PhCH), 2.65 (dd, J = 13.5, 8.7 Hz, 1H, PhCH). 13C NMR (150 MHz, DMSO-d6): d 171.05 (C O), 165.90 (C O), 159.07, 137.62, 135.89, 129.36, 129.06, 128.61, 126.95, 115.18, 55.92, 52.10, 37.94, 37.77, 29.51. ESI-MS: m/z 405.4 (M + 1)+. C19H21BrN2O3 [405.29].
5.1.3.2. (S)-2-(2-Bromoacetamido)-3-(3,5-difluorophenyl)-N-(4-methoxyphenyl)-N-methylpropanamide (4b)
White solid, yield: 80%. 1H NMR (400 MHz, DMSO-d6): δ 8.70 (d, J = 8.0 Hz, 1H, NH), 7.22 (d, J = 8.8 Hz, 2H, Ph–H), 7.04 (d, J = 8.6 Hz, 3H, Ph–H), 6.52 (d, J = 6.3 Hz, 2H, Ph–H), 4.44 (qd, J = 8.6, 4.6 Hz, 1H, CH), 3.81 (s, 2H, BrCH2), 3.80 (s, 3H, OCH3), 3.13 (s, 3H, NCH3), 2.89 (dd, J = 13.7, 4.6 Hz, 1H, PhCH), 2.69 (dd, J = 13.7, 9.3 Hz, 1H, PhCH). 13C NMR (150 MHz, DMSO-d6): d 170.57 (C O), 166.05 (C O), 162.55 (dd, 1JCF = 245.9, 3JCF = 13.3 Hz), 159.24, 142.20 (t, 3JCF = 9.5 Hz), 135.79, 129.10, 115.31, 112.42 (dd, 2JCF = 19.8, 4JCF = 5.0 Hz), 102.50 (t, 2JCF = 25.6 Hz), 55.95, 51.60, 37.76, 37.31, 29.32. ESI-MS: m/z 443.15 (M + 2)+, m/z 463.16 (M – 1 + 23)+. C17H18F2N2O2 [441.27].
5.1.4. General procedure for the synthesis of 5a and 5c
Under ice bath, the corresponding substituted key intermediate 4a or 4b (1 eq.), 1-Boc-3-oxopiperazine (1.2 eq.), K2CO3 (2 eq.) were dissolved in the solution of DMF (6 mL). The resulting mixture was then stirred at 40 °C (monitored by TLC). Then the reaction mixture was diluted with water (20 mL), and the combined phase was washed with ethyl acetate (3 × 20 mL), dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to give the corresponding crude product which was purified by flash column chromatography to afford products 5a and 5b.
5.1.4.1. tert-Butyl (S)-4-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-3-oxopiperazine-1-carboxylate (5a)
White solid, yield: 81%. ESI-MS: m/z 523.09 (M − 1)−. C28H36N4O6 [524.62].
5.1.4.2. tert-Butyl (S)-4-(2-((3-(3,5-difluorophenyl)-1-((4-methoxyphenyl) (methyl) amino)-1-oxopropan-2-yl)amino)-2-oxoethyl)-3-oxopiperazine-1-carboxylate (5b)
White solid, yield: 88%. 1H NMR (400 MHz, DMSO-d6): δ 8.42 (d, J = 8.0 Hz, 1H, NH), 7.24 (d, J = 8.3 Hz, 2H, Ph–H), 7.02 (d, J = 7.1 Hz, 3H, Ph–H), 6.49 (d, J = 6.3 Hz, 2H, Ph–H), 4.46 (td, J = 8.8, 4.2 Hz, 1H, CH), 4.00–3.85 (m, 4H, CH2 × 2), 3.79 (s, 3H, OCH3), 3.50 (s, 2H, CH2), 3.16 (t, J = 5.4 Hz, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.6, 4.4 Hz, 1H, PhCH), 2.69 (dd, J = 13.6, 9.5 Hz, 1H, PhCH), 1.41 (s, 9H, C(CH3)3). 13C NMR (150 MHz, DMSO-d6) δ 170.85 (C O), 167.70 (C O), 167.62 (C O), 162.53 (dd, 1JCF = 245.9, 3JCF = 13.5 Hz), 159.19, 153.63 (C O), 142.55 (t, 3JCF = 9.6 Hz), 135.86, 129.13, 115.30, 112.35 (dd, 2JCF = 20.0, 4JCF = 5.0 Hz), 102.44 (t, 2JCF = 26.0 Hz), 80.08, 55.93, 51.42, 51.33, 48.77, 47.11, 40.63, 37.76, 37.25, 28.44. ESI-MS: m/z 561.00 (M + 1)+, 583.19 (M + 23)+, 599.16 (M + 39)+, 559.22 (M − 1)−. C28H34F2N4O6 [560.60].
5.1.5. General procedure for the synthesis of 6a and 6b
Trifluoroacetic acid (5.0 eq.) was added dropwise to the corresponding substituted intermediate 5a or 5b (1.0 eq.) in 30 mL DCM and stirred at room temperature for 1 h (monitored by TLC). Then, the resulting mixture solution was alkalized to pH ∼7 with saturated sodium bicarbonate solution and then extracted with DCM (3 × 30 mL), dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to afford corresponding crude products 6a and 6b.
5.1.5.1. (S)-N-(4-methoxyphenyl)-N-methyl-2-(2-(2-oxopiperazin-1-yl)acetamido)-3-phenylpropanamide (6a)
Yellow oil, yield: 71%. 1H NMR (600 MHz, DMSO-d6) δ 8.23 (d, J = 8.0 Hz, 1H, NH), 7.21–7.07 (m, 5H, Ph–H), 6.97 (d, J = 9.0 Hz, 2H, Ph–H), 6.86 (d, J = 6.3 Hz, 2H, Ph–H), 4.48 (td, J = 8.6, 5.1 Hz, 1H, CH), 3.87 (q, J = 16.1 Hz, 2H, CH2), 3.79 (s, 3H, OCH3), 3.24 (s, 2H, CH2), 3.10 (s, 3H, NCH3), 3.04 (t, J = 5.7 Hz, 2H, CH2), 2.85 (dd, J = 13.6, 5.1 Hz, 2H, CH2), 2.82 (d, J = 5.7 Hz, 1H, PhCH), 2.66 (dd, J = 13.6, 9.0 Hz, 1H, PhCH), 1.24 (s, 1H, NH). 13C NMR (100 MHz, DMSO-d6) δ 170.94 (C O), 168.23 (C O), 168.03 (C O), 162.51 (dd, 1JC–F = 245.8, 3JC–F = 13.3 Hz), 159.18 (C O), 142.60 (t, 3JC–F = 9.4 Hz), 135.89, 129.17, 115.28, 112.58–112.08 (m), 102.44 (t, 2JC–F = 25.7 Hz), 55.93, 51.43, 50.24, 48.80, 48.66, 42.88, 37.75, 37.16. HRMS: m/z 425.2162 (M + 1)+. C23H28N4O4 [424.2111].
5.1.5.2. (S)-3-(3,5-difluorophenyl)-N-(4-methoxyphenyl)-N-methyl-2-(2-(2-oxopiperazin-1-yl)acetamido)propanamide (6b)
Yellow oil, yield: 67%. 1H NMR (400 MHz, DMSO-d6) δ 8.31 (d, J = 8.0 Hz, 1H, NH), 7.26 (d, J = 8.5 Hz, 2H, Ph–H), 7.03 (d, J = 8.3 Hz, 3H, Ph–H), 6.50 (h, J = 4.6 Hz, 2H, Ph–H), 4.46 (td, J = 8.8, 4.2 Hz, 1H, CH), 3.87 (d, J = 1.9 Hz, 2H, CH2), 3.80 (s, 3H, OCH3), 3.21 (s, 2H, CH2), 3.13 (s, 3H, NCH3), 3.04 (t, J = 5.4 Hz, 2H, CH2), 2.86 (dd, J = 13.6, 4.2 Hz, 1H, PhCH), 2.81 (q, J = 4.9 Hz, 2H, CH2), 2.70 (dd, J = 13.9, 9.6 Hz, 1H, PhCH), 1.24 (s, 1H, NH). 13C NMR (100 MHz, DMSO-d6) δ 170.94 (C O), 168.23 (C O), 168.03 (C O), 162.51 (dd, 1JCF = 245.8, 3JCF = 13.3 Hz), 159.18, 142.60 (t, 3JCF = 9.4 Hz), 135.89, 129.17, 115.28, 112.56–112.15 (m), 102.44 (t, 2JCF = 25.7 Hz), 55.93, 51.43, 50.24, 48.80, 48.66, 42.88, 37.75, 37.16. HRMS: m/z 461.1993 (M + 1)+, 921.3886 (2M + 1)+. C23H26F2N4O4 [460.1922].
5.1.6. General procedure for the synthesis of Id-3a-3r and F-Id-3a-3r
Under ice bath, the key intermediate 6a or 6b (1 eq.), corresponding substituted benzoyl chloride (1.5 eq.), TEA (2 eq.) were dissolved in the solution of DCM (10 mL). The resulting mixture was then stirred at room temperature (monitored by TLC). Then the reaction mixture was extracted with DCM (20 mL), and the combined organic phase was washed with saturated NaCl solution (3 × 20 mL), dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to give the corresponding crude product, which was purified by recrystallization or preparation thin layer chromatography to afford product Id-3a-3o and F-Id-3a-3o. Id-3m-3o or F-Id-3m-3o and 10% Pd·C (10% w/w) were dissolved in methanol (5 mL) and DCM (5 mL) and the solution degassed and stirred at room temperature for 2 h under H2. The mixture was filtered and concentrated, and the resulting residue were purified by recrystallization or preparation thin layer chromatography to provide the target compounds Id-3p-3r and F-Id-3p-3r.
5.1.6.1. (S)-2-(2-(4-Benzoyl-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3a)
White solid, yield: 67%. m.p.: 89–90 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.43 (d, J = 8.0 Hz, 1H, NH), 7.62–7.40 (m, 5H, Ph–H), 7.26–7.08 (m, 5H, Ph–H), 6.98 (d, J = 8.5 Hz, 2H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 4.47 (q, J = 7.7, 7.1 Hz, 1H, CH), 4.13 (d, J = 15.0 Hz, 1H, CH), 4.10–3.83 (m, 4H, CH2 × 2), 3.79 (s, 3H, OCH3), 3.53 (s, 1H, CH), 3.15 (s, 2H, CH2), 3.11 (s, 3H, NCH3), 2.86 (dd, J = 13.5, 4.9 Hz, 1H, PhCH), 2.65 (dd, J = 13.5, 9.4 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.42 (C O), 167.57 (C O), 159.03 (C O), 137.97, 135.94, 130.49, 129.33, 129.15, 128.98, 128.60, 127.55, 126.87, 115.17, 55.90, 51.89, 48.64, 37.79. HRMS: m/z 529.2446 (M + 1)+, 551.2269 (M + 23)+. C30H32N4O5 [528.2373].
5.1.6.2. (S)-2-(2-(4-(4-Fluorobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3b)
White solid, yield: 70%. m.p.: 87–89 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.42 (d, J = 8.0 Hz, 1H, NH), 7.52 (t, J = 6.9 Hz, 2H, Ph–H), 7.31 (t, J = 8.8 Hz, 2H, Ph–H), 7.16 (dt, J = 12.5, 7.6 Hz, 5H, Ph–H), 6.98 (d, J = 8.4 Hz, 2H, Ph–H), 6.84 (d, J = 7.2 Hz, 2H, Ph–H), 4.46 (td, J = 8.7, 5.0 Hz, 1H, CH), 4.24–4.00 (m, 2H, CH2), 3.93 (s, 2H, CH2), 3.79 (s, 3H, OCH3), 3.55 (s, 2H, CH2), 3.22–3.13 (m, 2H, CH2), 3.11 (s, 3H, NCH3), 2.85 (dd, J = 13.5, 4.9 Hz, 1H, PhCH), 2.65 (dd, J = 13.5, 9.4 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.42(C O), 167.58(C O), 159.02(C O), 137.97, 135.94, 130.27, 129.32, 129.15, 128.60, 126.87, 116.07, 115.86, 115.16, 55.90, 51.90, 48.63, 37.79. HRMS: m/z 547.2354 (M + 1)+, 569.2167 (M + 23)+. C30H31FN4O5 [546.2278].
5.1.6.3. (S)-2-(2-(4-(3-Fluorobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3c)
White solid, yield: 68%. m.p.: 92–93 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.41 (d, J = 8.1 Hz, 1H, NH), 7.55 (q, J = 6.8, 6.1 Hz, 1H, Ph–H), 7.49–7.40 (m, 1H, Ph–H), 7.33 (q, J = 7.9, 7.4 Hz, 2H, Ph–H), 7.15 (dt, J = 20.9, 7.2 Hz, 5H, Ph–H), 6.97 (d, J = 8.1 Hz, 2H, Ph–H), 6.84 (d, J = 7.2 Hz, 2H, Ph–H), 4.47 (dq, J = 8.8, 4.8 Hz, 1H, CH), 4.20 (s, 1H, CH), 4.05–3.81 (m, 4H, CH2 × 2), 3.79 (s, 3H, OCH3), 3.50–3.40 (m, 1H, CH), 3.17 (dd, J = 27.2, 5.7 Hz, 2H, CH2), 3.11 (s, 3H, NCH3), 2.86 (dd, J = 13.5, 4.9 Hz, 1H, PhCH), 2.65 (dt, J = 13.8, 7.5 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.39(C O), 167.52(C O), 164.94(C O), 161.93 (d, 1JCF = 463.7 Hz), 159.03(C O), 137.93, 135.94, 129.42, 129.32, 129.14, 128.58, 126.84, 125.49, 116.42 (d, 2JCF = 21.1 Hz), 115.17, 55.90, 51.79, 48.68, 47.35, 46.09, 43.78, 37.79. HRMS: m/z 547.2352 (M + 1)+, 569.2181 (M + 23)+. C30H31FN4O5 [546.2278].
5.1.6.4. (S)-2-(2-(4-(2-Fluorobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3d)
Yellow oil, yield: 75%. 1H NMR (400 MHz, DMSO-d6) δ 8.41 (d, J = 8.0 Hz, 1H, NH), 7.53 (q, J = 7.3 Hz, 1H, Ph–H), 7.33 (dd, J = 20.0, 10.3 Hz, 3H, Ph–H), 7.15 (dd, J = 13.6, 7.7 Hz, 5H, Ph–H), 6.98 (d, J = 8.4 Hz, 2H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 4.47 (q, J = 9.7, 8.9 Hz, 1H, CH), 4.15 (s, 1H, CH), 4.10–3.83 (m, 4H, CH2 × 2), 3.79 (s, 3H, OCH3), 3.51 (s, 1H, CH), 3.16 (s, 2H, CH2), 3.11 (s, 3H, NCH3), 2.86 (dd, J = 13.3, 4.6 Hz, 1H, PhCH), 2.65 (dd, J = 13.6, 9.4 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.42 (C O), 167.56 (C O), 162.28 (d, 1JCF = 245.0 Hz), 159.03 (C O), 137.97, 135.95, 131.34, 131.26, 129.32, 129.15, 128.60, 126.87, 115.16, 55.90, 48.65, 37.79. HRMS: m/z 547.2353 (M + 1)+, 569.2178 (M + 23)+. C30H31FN4O5 [546.2278].
5.1.6.5. (S)-2-(2-(4-(4-Chlorobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3e)
White solid, yield: 71%. m.p.: 60–61 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.42 (d, J = 8.0 Hz, 1H, NH), 7.55 (d, J = 6.6 Hz, 2H, Ph–H), 7.49 (d, J = 8.2 Hz, 2H, Ph–H), 7.24–7.09 (m, 5H, Ph–H), 6.99 (d, J = 8.4 Hz, 2H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 4.49 (d, J = 7.9 Hz, 1H, CH), 4.14 (s, 1H, CH), 3.98 (m, 4H, CH2 × 2), 3.80 (s, 3H, OCH3), 3.54 (s, 1H, CH), 3.17 (s, 2H, CH2), 3.12 (s, 3H, NCH3), 2.86 (dd, J = 13.8, 4.9 Hz, 1H, PhCH), 2.66 (dd, J = 13.5, 9.7 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.41 (C O), 168.21 (C O), 167.57 (C O), 159.03 (C O), 137.97, 135.95, 134.30, 132.00, 131.61, 129.62, 129.32, 129.15, 129.07, 128.60, 126.87, 115.17, 65.50, 55.90, 51.89, 48.65, 37.79, 30.48, 19.12, 14.02. HRMS: m/z 563.2051 (M + 1)+. C30H31ClN4O5 [562.1983].
5.1.6.6. (S)-2-(2-(4-(4-Bromobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3f)
White solid, yield: 62%. m.p.: 167–168 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.42 (d, J = 8.0 Hz, 1H, NH), 7.69 (d, J = 6.4 Hz, 2H, Ph–H), 7.41 (d, J = 8.0 Hz, 2H, Ph–H), 7.23–7.10 (m, 5H, Ph–H), 6.99 (d, J = 8.3 Hz, 2H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 4.52–4.42 (m, 1H, CH), 4.15 (s, 1H, CH), 4.11–3.83 (m, 4H, CH2 × 2), 3.80 (s, 3H, OCH3), 3.53 (s, 1H, CH), 3.17 (s, 2H, CH2), 3.12 (s, 3H, NCH3), 2.86 (dd, J = 13.7, 5.0 Hz, 1H, PhCH), 2.71–2.61 (m, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.41 (C O), 167.57 (C O), 159.02 (C O), 137.96, 135.94, 131.99, 129.82, 129.31, 129.14, 128.60, 126.87, 115.16, 65.50, 55.90, 55.38, 51.90, 48.64, 37.79, 32.67, 19.12. HRMS: m/z 609.1539 (M + 1)+. C30H31BrN4O5 [606.1478].
5.1.6.7. (S)-N-(4-Methoxyphenyl)-N-methyl-2-(2-(4-(4-methylbenzoyl)-2-oxopiperazin-1-yl)acetamido)-3-phenylpropanamide (Id-3g)
White solid, yield: 65%. m.p.: 91–92 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.41 (d, J = 8.0 Hz, 1H, NH), 7.34 (d, J = 7.7 Hz, 2H, Ph–H), 7.28 (d, J = 7.8 Hz, 2H, Ph–H), 7.15 (dd, J = 16.3, 7.4 Hz, 5H, Ph–H), 6.98 (d, J = 8.4 Hz, 2H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 4.47 (td, J = 8.7, 4.9 Hz, 1H, CH), 4.09 (s, 2H, CH2), 4.01–3.87 (m, 2H, CH2), 3.79 (s, 3H, OCH3), 3.57 (d, J = 58.4 Hz, 2H, CH2), 3.21–3.13 (m, 2H, CH2), 3.11 (s, 3H, NCH3), 2.86 (dd, J = 13.5, 4.9 Hz, 1H, PhCH), 2.65 (dd, J = 13.6, 9.3 Hz, 1H, PhCH), 2.35 (s, 3H, PhCH3). 13C NMR (100 MHz, DMSO-d6) δ 171.41 (C O), 169.34, 167.57 (C O), 165.18, 159.03 (C O), 140.28, 137.97, 135.95, 132.56, 129.43, 129.33, 129.14, 128.60, 127.71, 126.87, 115.17, 55.90, 51.87, 48.64, 37.78, 21.41, 19.13. HRMS: m/z 543.2600 (M + 1)+, 565.2407 (M + 23)+. C31H34N4O5 [542.2529].
5.1.6.8. (S)-2-(2-(4-(4-Methoxybenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3h)
White solid, yield: 66%. m.p.: 83–85 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.41 (d, J = 8.0 Hz, 1H, NH), 7.42 (d, J = 8.1 Hz, 2H, Ph–H), 7.23–7.09 (m, 5H, Ph–H), 6.99 (dd, J = 10.7, 8.5 Hz, 4H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 4.47 (td, J = 8.6, 5.0 Hz, 1H, CH), 4.10 (s, 2H, CH2), 3.94 (d, J = 5.1 Hz, 2H, CH2), 3.80 (s, 3H, OCH3), 3.79 (s, 3H, OCH3), 3.68 (d, J = 20.3 Hz, 2H, CH2), 3.17 (d, J = 6.2 Hz, 2H, CH2), 3.11 (s, 3H, NCH3), 2.86 (dd, J = 13.6, 5.0 Hz, 1H, PhCH), 2.65 (dd, J = 13.5, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.41 (C O), 169.13, 167.58 (C O), 165.22, 161.05 (C O), 159.03 (C O), 137.97, 135.95, 129.74, 129.33, 129.14, 128.60, 127.38, 126.87, 115.17, 114.20, 55.90, 55.78, 51.88, 48.64, 37.79. HRMS: m/z 559.2548 (M + 1)+, 581.2372 (M + 23)+. C31H34N4O6 [558.2478].
5.1.6.9. (S)-N-(4-Methoxyphenyl)-N-methyl-2-(2-(2-oxo-4-(4-(trifluoromethyl)benzoyl)piperazin-1-yl)acetamido)-3-phenylpropanamide (Id-3i)
White solid, yield: 77%. m.p.: 63–64 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.42 (d, J = 8.0 Hz, 1H, NH), 7.85 (d, J = 8.2 Hz, 2H, Ph–H), 7.67 (d, J = 5.7 Hz, 2H, Ph–H), 7.27–7.06 (m, 5H, Ph–H), 6.98 (d, J = 8.5 Hz, 2H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 4.47 (q, J = 9.1, 8.3 Hz, 1H, CH), 4.22 (dd, J = 13.9, 7.4 Hz, 2H, CH2), 4.08–3.93 (m, 2H, CH2), 3.90 (d, J = 14.1 Hz, 1H, CH), 3.79 (s, 3H, OCH3), 3.50 (s, 1H, CH), 3.17 (s, 2H, CH2), 3.11 (s, 3H, NCH3), 2.86 (dd, J = 13.5, 4.7 Hz, 1H, PhCH), 2.66 (dd, J = 13.3, 9.7 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.42 (C O), 167.58 (C O), 159.02 (C O), 137.96, 135.93, 132.00, 130.59, 129.31, 129.14, 128.60, 128.35, 126.87, 126.03, 115.15, 65.50, 55.89, 51.91, 48.65, 46.35, 44.31, 37.78, 30.47, 19.12, 14.01. HRMS: m/z 597.2321 (M + 1)+, 619.2132 (M + 23)+. C31H31F3N4O5 [596.2247].
5.1.6.10. (S)-N-(4-Methoxyphenyl)-N-methyl-2-(2-(2-oxo-4-(3-(trifluoromethyl)benzoyl)piperazin-1-yl)acetamido)-3-phenylpropanamide (Id-3j)
White solid, yield: 74%. m.p.: 67–68 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.42 (d, J = 8.0 Hz, 1H, NH), 7.88 (d, J = 7.6 Hz, 1H, Ph–H), 7.84–7.69 (m, 3H, Ph–H), 7.28–7.08 (m, 5H, Ph–H), 6.99 (d, J = 8.4 Hz, 2H, Ph–H), 6.85 (d, J = 7.3 Hz, 2H, Ph–H), 4.48 (q, J = 7.7, 6.9 Hz, 1H, CH), 4.18 (s, 1H, CH), 4.13–3.84 (m, 4H, CH2 × 2), 3.80 (s, 3H, OCH3), 3.52 (s, 1H, CH), 3.18 (s, 2H, CH2), 3.12 (s, 3H, NCH3), 2.86 (dd, J = 13.6, 4.9 Hz, 1H, PhCH), 2.66 (dd, J = 13.5, 9.5 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.41 (C O), 167.57 (C O), 159.03 (C O), 137.97, 135.95, 130.23, 129.31, 129.15, 128.59, 126.85, 125.68, 115.16, 55.90, 48.65, 37.78. HRMS: m/z 597.2319 (M + 1)+, 619.2135 (M + 23)+. C31H31F3N4O5 [596.2247].
5.1.6.11. (S)-N-(4-Methoxyphenyl)-N-methyl-2-(2-(2-oxo-4-(2-(trifluoromethyl)benzoyl)piperazin-1-yl)acetamido)-3-phenylpropanamide (Id-3k)
White solid, yield: 72%. m.p.: 93–94 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.40 (d, J = 7.8 Hz, 1H, NH), 7.85 (d, J = 7.8 Hz, 1H, Ph–H), 7.78 (t, J = 7.6 Hz, 1H, Ph–H), 7.69 (t, J = 7.8 Hz, 1H, Ph–H), 7.58–7.47 (m, 1H, Ph–H), 7.23–7.05 (m, 5H, Ph–H), 6.97 (d, J = 8.4 Hz, 2H, Ph–H), 6.85 (d, J = 7.5 Hz, 2H, Ph–H), 4.48 (t, J = 7.6 Hz, 1H, CH), 4.39–4.21 (m, 1H, CH), 4.21–3.98 (m, 2H, CH2), 3.97–3.82 (m, 2H, CH2), 3.78 (s, 3H, OCH3), 3.76–3.48 (m, 1H, CH), 3.31–3.16 (m, 2H, CH2), 3.10 (s, 3H, NCH3), 2.86 (dd, J = 13.4, 4.6 Hz, 1H, PhCH), 2.65 (dd, J = 13.8, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.38 (C O), 167.50 (C O), 164.88 (C O), 159.02 (C O), 137.93, 135.94, 133.47, 130.44, 130.35, 129.34, 129.32, 129.13, 128.62, 128.57, 127.99, 127.09, 126.91, 126.84, 125.51, 115.14, 55.89, 51.79, 50.36, 48.70, 46.97, 46.37, 45.93, 43.82, 40.64, 40.43, 40.22, 40.01, 39.81, 39.60, 39.39, 37.79. HRMS: m/z 597.2321 (M + 1)+, 619.2138 (M + 23)+. C31H31F3N4O5 [596.2247].
5.1.6.12. Methyl (S)-4-(4-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-3-oxopiperazine-1-carbonyl)benzoate (Id-3l)
White solid, yield: 69%. m.p.: 90–92 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.42 (d, J = 8.0 Hz, 1H, NH), 8.04 (d, J = 7.8 Hz, 2H, Ph–H), 7.58 (d, J = 7.9 Hz, 2H, Ph–H), 7.26–7.06 (m, 5H, Ph–H), 6.98 (d, J = 8.4 Hz, 2H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 4.47 (q, J = 6.9, 5.7 Hz, 1H, CH), 4.20 (d, J = 16.7 Hz, 1H, CH), 4.17–3.90 (m, 4H, CH2 × 2), 3.88 (s, 3H, COOCH3), 3.79 (s, 3H, OCH3), 3.49 (s, 1H, CH), 3.16 (s, 2H, CH2), 3.11 (s, 3H, NCH3), 2.86 (dd, J = 13.4, 4.7 Hz, 1H, PhCH), 2.65 (dd, J = 13.5, 9.6 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.87 (C O), 167.72 (C O), 159.18 (C O), 148.19, 135.83, 130.79, 129.32, 129.17, 128.60, 125.12, 115.28, 112.47, 112.22, 102.48, 55.93, 51.51, 48.73, 40.64, 40.43, 40.22, 40.02, 39.81, 39.60, 39.39, 37.76, 37.14. HRMS: m/z 587.2500 (M + 1)+, 609.2326 (M + 23)+. C32H34N4O7 [586.2427].
5.1.6.13. (S)-N-(4-Methoxyphenyl)-N-methyl-2-(2-(4-(4-nitrobenzoyl)-2-oxopiperazin-1-yl)acetamido)-3-phenylpropanamide (Id-3m)
White solid, yield: 80%. m.p.: 104–105 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.40 (d, J = 8.0 Hz, 1H, NH), 8.31 (d, J = 6.5 Hz, 2H, Ph–H), 7.72 (d, J = 8.2 Hz, 2H, Ph–H), 7.24–7.06 (m, 5H, Ph–H), 6.98 (d, J = 8.4 Hz, 2H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 4.47 (q, J = 7.0, 6.4 Hz, 1H, CH), 4.23 (s, 1H, CH), 4.10–3.81 (m, 4H, CH2 × 2), 3.79 (s, 3H, OCH3), 3.49 (s, 1H, CH), 3.18 (s, 2H, CH2), 3.11 (s, 3H, NCH3), 2.86 (dd, J = 13.8, 4.9 Hz, 1H, PhCH), 2.66 (t, J = 11.4 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.41 (C O), 167.56 (C O), 159.04 (C O), 148.57, 141.81, 137.96, 135.96, 129.32, 129.14, 128.92, 128.60, 126.87, 124.27, 115.17, 55.91, 51.87, 48.68, 47.28, 37.79. HRMS: m/z 574.2300 (M + 1)+, 596.2117 (M + 23)+. C30H31N5O7 [573.2223].
5.1.6.14. (S)-N-(4-Methoxyphenyl)-N-methyl-2-(2-(4-(3-nitrobenzoyl)-2-oxopiperazin-1-yl)acetamido)-3-phenylpropanamide (Id-3n)
White solid, yield: 62%. m.p.: 86–87 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.41 (d, J = 8.0 Hz, 1H, NH), 8.34 (d, J = 8.1 Hz, 1H, Ph–H), 8.28 (s, 1H, Ph–H), 7.91 (d, J = 7.4 Hz, 1H, Ph–H), 7.78 (t, J = 7.9 Hz, 1H, Ph–H), 7.26–7.08 (m, 5H, Ph–H), 6.99 (d, J = 8.4 Hz, 2H, Ph–H), 6.86 (d, J = 7.1 Hz, 2H, Ph–H), 4.54–4.44 (m, 1H, CH), 4.27–4.14 (m, 1H, CH), 4.12–3.84 (m, 4H, CH2 × 2), 3.80 (s, 3H, OCH3), 3.54 (s, 1H, CH), 3.20 (s, 2H, CH2), 3.12 (s, 3H, NCH3), 2.87 (dd, J = 13.3, 4.8 Hz, 1H, PhCH), 2.66 (dd, J = 13.6, 9.4 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.42 (C O), 167.59 (C O), 159.04 (C O), 148.19, 137.97, 135.95, 131.99, 130.79, 129.32, 129.14, 128.60, 126.87, 115.17, 55.90, 51.90, 48.66, 37.79, 19.12. HRMS: m/z 574.2297 (M + 1)+, m/z 596.2119 (M + 23)+. C30H31N5O7 [573.2223].
5.1.6.15. (S)-N-(4-Methoxyphenyl)-N-methyl-2-(2-(4-(2-nitrobenzoyl)-2-oxopiperazin-1-yl)acetamido)-3-phenylpropanamide (Id-3o)
White solid, yield: 65%. m.p.: 69–70 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.41 (d, J = 8.3 Hz, 1H, NH), 8.24 (t, J = 9.2 Hz, 1H, Ph–H), 7.89 (t, J = 7.7 Hz, 1H, Ph–H), 7.75 (t, J = 8.0 Hz, 1H, Ph–H), 7.59 (t, J = 7.9 Hz, 1H, Ph–H), 7.25–7.09 (m, 5H, Ph–H), 6.99 (t, J = 6.8 Hz, 2H, Ph–H), 6.86 (t, J = 6.8 Hz, 2H, Ph–H), 4.49 (h, J = 5.5, 4.8 Hz, 1H, CH), 4.20 (s, 1H, CH), 4.06–3.81 (m, 4H, CH2 × 2), 3.80 (d, J = 4.1 Hz, 3H, OCH3), 3.41 (d, J = 5.0 Hz, 1H, CH), 3.30–3.14 (m, 2H, CH2), 3.11 (s, 3H, NCH3), 2.87 (dt, J = 13.4, 3.6 Hz, 1H, PhCH), 2.67 (dq, J = 14.1, 8.6, 7.6 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.40 (C O), 167.55 (C O), 164.89 (C O), 159.02 (C O), 145.70, 145.65, 137.93, 135.93, 135.54, 132.18, 132.03, 132.00, 131.92, 131.19, 131.13, 129.44, 129.34, 129.32, 129.14, 128.63, 128.59, 128.48, 126.92, 126.85, 125.35, 115.16, 65.50, 55.89, 51.87, 51.82, 50.10, 48.74, 48.59, 47.08, 46.27, 46.04, 43.70, 37.79, 30.48, 19.12, 14.02. HRMS: m/z 574.2292 (M + 1)+, m/z 596.2115 (M + 23)+. C30H31N5O7 [573.2223].
5.1.6.16. (S)-2-(2-(4-(4-Aminobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3p)
White solid, yield: 52%. m.p.: 150–151 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.38 (d, J = 8.0 Hz, 1H, NH), 7.17 (d, J = 8.1 Hz, 5H, Ph–H), 7.12 (d, J = 8.0 Hz, 2H, Ph–H), 6.98 (d, J = 8.4 Hz, 2H, Ph–H), 6.85 (d, J = 7.0 Hz, 2H, Ph–H), 6.58 (s, 2H, Ph–H), 5.59 (s, 2H, PhNH2), 4.48 (t, J = 6.8 Hz, 1H, CH), 4.08 (s, 2H, CH2), 4.00–3.85 (m, 2H, CH2), 3.79 (s, 3H, OCH3), 3.66 (s, 2H, CH2), 3.15 (s, 2H, CH2), 3.11 (s, 3H, NCH3), 2.85 (dd, J = 13.5, 4.9 Hz, 1H, PhCH), 2.65 (dd, J = 13.5, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.41 (C O), 167.58 (C O), 165.47 (C O), 159.04 (C O), 151.47, 137.97, 135.96, 129.90, 129.34, 129.14, 128.59, 126.85, 121.28, 115.18, 113.13, 65.50, 55.90, 51.84, 48.64, 47.11, 37.80, 30.49, 19.12. HRMS: m/z 544.2555 (M + 1)+, m/z 566.2380 (M + 23)+. C30H33N5O5 [543.2482].
5.1.6.17. (S)-2-(2-(4-(3-Aminobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3q)
White solid, yield: 55%. m.p.: 84–85 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.38 (d, J = 7.9 Hz, 1H, NH), 7.23–7.10 (m, 5H, Ph–H), 7.07 (d, J = 7.8 Hz, 1H, Ph–H), 6.97 (d, J = 8.3 Hz, 2H, Ph–H), 6.85 (d, J = 7.2 Hz, 2H, Ph–H), 6.64 (d, J = 8.2 Hz, 1H, Ph–H), 6.58 (s, 1H, Ph–H), 6.50 (d, J = 7.5 Hz, 1H, Ph–H), 5.27 (s, 2H, PhNH2), 4.48 (d, J = 6.9 Hz, 1H, CH), 4.07 (s, 2H, CH2), 4.01–3.84 (m, 2H, CH2), 3.78 (s, 3H, OCH3), 3.72–3.44 (m, 2H, CH2), 3.14 (s, 2H, CH2), 3.11 (s, 3H, NCH3), 2.85 (dd, J = 13.9, 4.8 Hz, 1H, PhCH), 2.65 (dd, J = 13.4, 9.5 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.40 (C O), 167.55 (C O), 159.04 (C O), 149.30, 137.95, 136.13, 135.97, 129.39, 129.33, 129.14, 128.60, 126.87, 115.68, 115.19, 114.47, 55.91, 51.84, 48.67, 45.77, 40.67, 40.46, 40.26, 40.05, 39.84, 39.63, 39.42, 37.80, 30.71, 19.12, 14.01. HRMS: m/z 544.2559 (M + 1)+, m/z 566.2385 (M + 23)+. C30H33N5O5 [543.2482].
5.1.6.18. (S)-2-(2-(4-(2-Aminobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (Id-3r)
White solid, yield: 61%. m.p.: 85–86 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.40 (d, J = 8.1 Hz, 1H, NH), 7.20 (s, 1H, Ph–H), 7.19–7.10 (m, 5H, Ph–H), 7.02 (d, J = 7.5 Hz, 1H, Ph–H), 6.98 (d, J = 8.5 Hz, 2H, Ph–H), 6.86 (d, J = 7.2 Hz, 2H, Ph–H), 6.72 (d, J = 8.0 Hz, 1H, Ph–H), 6.58 (t, J = 7.4 Hz, 1H, Ph–H), 5.28 (s, 2H, PhNH2), 4.48 (q, J = 8.5 Hz, 1H, CH), 4.08 (s, 2H, CH2), 3.94 (s, 2H, CH2), 3.80 (s, 3H, OCH3), 3.60 (s, 2H, CH2), 3.21–3.14 (m, 2H, CH2), 3.11 (s, 3H, NCH3), 2.86 (dd, J = 13.5, 4.9 Hz, 1H, PhCH), 2.66 (dd, J = 13.4, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.41 (C O), 167.59 (C O), 165.42 (C O), 159.02 (C O), 146.37, 137.95, 135.95, 131.99, 130.91, 129.33, 129.14, 128.59, 128.36, 126.87, 118.76, 116.02, 115.94, 115.16, 55.90, 51.84, 48.64, 46.52, 40.65, 40.44, 40.23, 40.02, 39.81, 39.60, 39.39, 37.79, 30.48, 19.12, 14.02. HRMS: m/z 544.2552 (M + 1)+, m/z 566.2372 (M + 23)+. C30H33N5O5 [543.2482].
5.1.6.19. (S)-2-(2-(4-Benzoyl-2-oxopiperazin-1-yl)acetamido)-3-(3,5-difluorophenyl)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3a)
White solid, yield: 73%. m.p.: 69–71 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.45 (d, J = 8.0 Hz, 1H, NH), 7.55–7.42 (m, 5H, Ph–H), 7.25 (d, J = 8.3 Hz, 2H, Ph–H), 7.03 (d, J = 8.4 Hz, 3H, Ph–H), 6.50 (d, J = 7.7 Hz, 2H, Ph–H), 4.49 (dd, J = 8.9, 4.1 Hz, 1H, CH), 4.16 (s, 2H, CH2), 3.95 (s, 2H, CH2), 3.80 (s, 3H, OCH3), 3.57 (s, 2H, CH2), 3.23 (s, 2H, CH2), 3.14 (s, 3H, NCH3), 2.88 (dd, J = 13.7, 4.2 Hz, 1H, PhCH), 2.70 (dd, J = 13.7, 9.5 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 169.26 (C O), 167.68 (C O), 162.51 (dd, 1JCF = 245.9, 3JCF = 13.3 Hz), 159.18 (C O), 142.59 (d, 3JCF = 9.1 Hz), 135.83, 135.50, 130.49, 129.16, 128.97, 127.53, 115.29, 112.35 (d, 2JCF = 24.7 Hz), 102.48 (t, 2JCF = 25.6 Hz), 55.93, 51.47, 48.71, 37.76, 37.19. HRMS: m/z 565.2253 (M + 1)+, m/z 587.2077 (M + 23)+. C30H30F2N4O5 [564.2184].
5.1.6.20. (S)-3-(3,5-Difluorophenyl)-2-(2-(4-(4-fluorobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3b)
White solid, yield: 68%. m.p.: 83–85 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.44 (d, J = 8.0 Hz, 1H, NH), 7.52 (t, J = 6.8 Hz, 2H, Ph–H), 7.35–7.27 (m, 2H, Ph–H), 7.25 (d, J = 8.2 Hz, 2H, Ph–H), 7.02 (d, J = 8.5 Hz, 3H, Ph–H), 6.49 (d, J = 7.7 Hz, 2H, Ph–H), 4.47 (td, J = 8.9, 4.2 Hz, 1H, CH), 4.11 (s, 2H, CH2), 3.94 (s, 2H, CH2), 3.79 (s, 3H, OCH3), 3.74–3.39 (m, 2H, CH2), 3.23 (d, J = 5.4 Hz, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.7, 4.2 Hz, 1H, PhCH), 2.69 (dd, J = 13.7, 9.6 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 168.33 (C O), 167.70 (C O), 163.34 (d, 1JCF = 235.7 Hz), 162.51 (dd, 1JCF = 246.1, 3JCF = 13.2 Hz), 159.17 (C O), 142.60 (d, 3JCF = 10.0 Hz), 135.83, 131.88, 130.25, 129.17, 115.97 (d, 2JCF = 21.6 Hz), 115.28, 112.35 (d, 2JCF = 24.9 Hz), 102.50 (t, 2JCF = 26.0 Hz), 55.92, 51.49, 48.69, 37.76, 37.15. HRMS: m/z 583.2167 (M + 1)+, m/z 605.1990 (M + 23)+. C30H29F3N4O5 [582.2090].
5.1.6.21. (S)-3-(3,5-Difluorophenyl)-2-(2-(4-(3-fluorobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3c)
White solid, yield: 66%. m.p.: 79–80 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.45 (s, 1H, NH), 7.53 (q, J = 7.3 Hz, 1H, Ph–H), 7.32 (dt, J = 17.9, 7.9 Hz, 3H, Ph–H), 7.25 (d, J = 8.5 Hz, 2H, Ph–H), 7.02 (d, J = 8.5 Hz, 3H, Ph–H), 6.50 (d, J = 7.7 Hz, 2H, Ph–H), 4.47 (td, J = 8.8, 4.2 Hz, 1H, CH), 4.16 (s, 2H, CH2), 3.94 (s, 2H, CH2), 3.79 (s, 3H, OCH3), 3.54 (s, 2H, CH2), 3.23 (s, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.8, 4.2 Hz, 1H, PhCH), 2.70 (dd, J = 13.7, 9.6 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 167.69 (C O), 163.79 (d, 1JCF = 245.8 Hz), 161.07 (dd, 1JCF = 245.5, 3JCF = 14.6 Hz), 159.17 (C O), 142.55, 135.83, 131.29 (d, 3JCF = 8.2 Hz), 129.17, 115.28, 112.35 (d, 2JCF = 24.6 Hz), 102.48 (t, 2JCF = 28.5 Hz), 55.92, 51.50, 48.70, 37.75, 37.14. HRMS: m/z 583.2167 (M + 1)+, m/z 605.1989 (M + 23)+. C30H29F3N4O5 [582.2090].
5.1.6.22. (S)-3-(3,5-Difluorophenyl)-2-(2-(4-(2-fluorobenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3d)
White solid, yield: 64%. m.p.: 80–82 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.43 (d, J = 8.0 Hz, 1H, NH), 7.54 (q, J = 7.3 Hz, 1H, Ph–H), 7.44 (t, J = 7.3 Hz, 1H, Ph–H), 7.32 (q, J = 7.7, 7.0 Hz, 2H, Ph–H), 7.24 (d, J = 8.1 Hz, 2H, Ph–H), 7.12–6.94 (m, 3H, Ph–H), 6.49 (d, J = 7.7 Hz, 2H, Ph–H), 4.47 (td, J = 8.7, 4.1 Hz, 1H, CH), 4.20 (d, J = 5.2 Hz, 1H, CH), 3.95 (d, J = 9.6 Hz, 2H, CH2), 3.85 (s, 2H, CH2), 3.79 (s, 3H, OCH3), 3.46 (q, J = 5.0 Hz, 1H, CH), 3.23 (dt, J = 36.8, 5.5 Hz, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.6, 4.0 Hz, 1H, PhCH), 2.69 (dd, J = 13.6, 9.6 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.85 (C O), 167.65 (C O), 164.98 (C O), 162.93 (d, 1JCF = 245.5 Hz), 162.07 (d, 1JCF = 246.6 Hz), 159.17 (C O), 142.53 (t, 3JCF = 9.3 Hz), 135.82, 132.41, 129.44, 129.16, 125.49 (d, 3JCF = 2.7 Hz), 116.41 (d, 2JCF = 21.4 Hz), 115.28, 112.35 (dd, 2JCF = 17.9, 4JCF = 6.6 Hz), 102.48 (t, 2JCF = 24.6 Hz), 55.92, 51.47, 48.75, 47.47, 46.10, 43.78, 37.76, 37.18. HRMS: m/z 583.2162 (M + 1)+, m/z 605.1981 (M + 23)+. C30H29F3N4O5 [582.2090].
5.1.6.23. (S)-2-(2-(4-(4-Chlorobenzoyl)-2-oxopiperazin-1-yl)acetamido)-3-(3,5-difluorophenyl)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3e)
White solid, yield: 62%. m.p.: 99–100 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.45 (d, J = 8.0 Hz, 1H, NH), 7.54 (d, J = 8.9 Hz, 2H, Ph–H), 7.48 (d, J = 7.9 Hz, 2H, Ph–H), 7.25 (d, J = 8.2 Hz, 2H, Ph–H), 7.03 (d, J = 8.4 Hz, 3H, Ph–H), 6.49 (d, J = 7.8 Hz, 2H, Ph–H), 4.46 (td, J = 8.8, 4.2 Hz, 1H, CH), 4.15 (s, 2H, CH2), 3.94 (s, 2H, CH2), 3.79 (s, 3H, OCH3), 3.56 (s, 2H, CH2), 3.22 (s, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.7, 4.3 Hz, 1H, PhCH), 2.70 (dd, J = 13.7, 9.5 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.87 (C O), 168.23 (C O), 167.71 (C O), 162.49 (dd, 1JCF = 246.1, 3JCF = 13.2 Hz), 159.16 (C O), 142.53 (t, 3JCF = 9.6 Hz), 135.81, 135.21, 134.25, 129.58, 129.17, 129.08, 115.27, 112.34 (dd, 2JCF = 18.3, 4JCF = 6.4 Hz), 102.50 (t, 2JCF = 25.5 Hz), 55.91, 51.51, 48.69, 37.75, 37.12. HRMS: m/z 599.1870 (M + 1)+, m/z 621.1689 (M + 23)+. C30H29ClF2N4O5 [598.1795].
5.1.6.24. (S)-2-(2-(4-(4-Bromobenzoyl)-2-oxopiperazin-1-yl)acetamido)-3-(3,5-difluorophenyl)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3f)
White solid, yield: 79%. m.p.: 94–95 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.45 (d, J = 8.0 Hz, 1H, NH), 7.68 (d, J = 8.0 Hz, 2H, Ph–H), 7.41 (d, J = 8.0 Hz, 2H, Ph–H), 7.25 (d, J = 8.2 Hz, 2H, Ph–H), 7.03 (d, J = 8.3 Hz, 3H, Ph–H), 6.50 (d, J = 7.7 Hz, 2H, Ph–H), 4.47 (dt, J = 9.1, 4.3 Hz, 1H, CH), 4.20–3.99 (m, 2H, CH2), 3.94 (s, 2H, CH2), 3.79 (s, 3H, OCH3), 3.77–3.46 (m, 2H, CH2), 3.22 (s, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.7, 4.1 Hz, 1H, PhCH), 2.70 (dd, J = 13.7, 9.7 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.87 (C O), 167.71 (C O), 159.16 (C O), 135.81, 132.00, 129.17, 115.27, 112.44, 112.12, 55.92, 51.51, 48.73, 40.59, 40.39, 40.18, 39.97, 39.76, 39.55, 39.34, 37.75, 37.34. HRMS: m/z 645.1347 (M + 1)+, m/z 667.1160 (M + 23)+. C30H29BrF2N4O5 [642.1289].
5.1.6.25. (S)-3-(3,5-Difluorophenyl)-N-(4-methoxyphenyl)-N-methyl-2-(2-(4-(4-methylbenzoyl)-2-oxopiperazin-1-yl)acetamido)propanamide (F-Id-3g)
White solid, yield: 77%. m.p.: 82–83 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.45 (d, J = 8.0 Hz, 1H, NH), 7.34 (d, J = 7.7 Hz, 2H, Ph–H), 7.30–7.21 (m, 4H, Ph–H), 7.03 (d, J = 8.3 Hz, 3H, Ph–H), 6.50 (d, J = 7.8 Hz, 2H, Ph–H), 4.47 (q, J = 8.9 Hz, 1H, CH), 4.11 (s, 2H, CH2), 3.94 (s, 2H, CH2), 3.80 (s, 3H, OCH3), 3.58 (s, 2H, CH2), 3.23 (d, J = 5.4 Hz, 2H, CH2), 3.13 (s, 3H, NCH3), 2.92–2.83 (m, 1H, PhCH), 2.74–2.64 (m, 1H, PhCH), 2.36 (s, 3H, PhCH3). 13C NMR (100 MHz, DMSO-d6) δ 170.87 (C O), 167.79 (C O), 167.71 (C O), 162.49 (dd, 1JCF = 245.9, 3JCF = 13.6 Hz), 159.16 (C O), 142.54, 135.80, 132.49, 129.80, 129.60, 129.44, 129.17, 127.71, 115.27, 112.34 (dd, 2JCF = 18.5, 4JCF = 5.9 Hz), 102.50 (t, 2JCF = 25.4 Hz), 55.91, 55.37, 51.50, 48.67, 37.75, 37.13, 32.68, 21.59, 21.40. HRMS: m/z 579.2410 (M + 1)+, m/z 601.2228 (M + 23)+. C31H32F2N4O5 [578.2341].
5.1.6.26. (S)-3-(3,5-Difluorophenyl)-2-(2-(4-(4-methoxybenzoyl)-2-oxopiperazin-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3h)
White solid, yield: 71%. m.p.: 74–75 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.42 (d, J = 8.1 Hz, 1H, NH), 7.42 (d, J = 7.4 Hz, 2H, Ph–H), 7.24 (d, J = 8.3 Hz, 2H, Ph–H), 7.01 (t, J = 7.4 Hz, 5H, Ph–H), 6.50 (d, J = 7.8 Hz, 2H, Ph–H), 4.47 (td, J = 8.9, 3.9 Hz, 1H, CH), 4.10 (s, 2H, CH2), 3.94 (d, J = 4.4 Hz, 2H, CH2), 3.80 (s, 3H, OCH3), 3.79 (s, 3H, PhOCH3), 3.68 (s, 2H, CH2), 3.23 (d, J = 5.5 Hz, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.6, 4.3 Hz, 1H, PhCH), 2.69 (dd, J = 13.7, 9.6 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 167.70 (C O), 162.50 (dd, 1JCF = 246.1, 3JCF = 13.3 Hz), 161.06 (C O), 159.17 (C O), 142.55 (t, 3JCF = 9.6 Hz), 135.83, 129.72, 129.17, 127.36, 115.28, 114.21, 112.35 (dd, 2JCF = 18.5, 4JCF = 6.1 Hz), 102.50 (t, 2JCF = 25.4 Hz), 65.49, 55.92, 55.77, 51.48, 48.68, 37.76, 37.17, 30.48, 19.12. HRMS: m/z 595.2364 (M + 1)+, m/z 617.2175 (M + 23)+. C31H32F2N4O6 [594.2290].
5.1.6.27. (S)-3-(3,5-Difluorophenyl)-N-(4-methoxyphenyl)-N-methyl-2-(2-(2-oxo-4-(4-(trifluoromethyl)benzoyl)piperazin-1-yl)acetamido)propanamide (F-Id-3i)
White solid, yield: 68%. m.p.: 81–82 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.44 (d, J = 8.0 Hz, 1H, NH), 7.85 (d, J = 8.0 Hz, 2H, Ph–H), 7.67 (d, J = 7.9 Hz, 2H, Ph–H), 7.25 (d, J = 8.3 Hz, 2H, Ph–H), 7.02 (d, J = 8.6 Hz, 3H, Ph–H), 6.50 (d, J = 7.7 Hz, 2H, Ph–H), 4.48 (dd, J = 8.9, 4.1 Hz, 1H, CH), 4.21 (d, J = 13.3 Hz, 1H, CH), 3.95 (s, 4H, CH2 × 2), 3.79 (s, 3H, OCH3), 3.52 (s, 1H, CH), 3.30–3.18 (m, 2H, CH2), 3.13 (s, 3H, NCH3), 2.88 (dd, J = 13.8, 4.2 Hz, 1H, PhCH), 2.70 (dd, J = 13.7, 9.6 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.87 (C O), 167.70 (C O), 162.50 (dd, 1JCF = 245.6, 3JCF = 13.1 Hz), 159.17 (C O), 142.54 (t, 3JCF = 9.7 Hz), 139.71, 135.83, 128.38, 126.04, 125.72, 115.28, 112.34 (dd, 2JCF = 18.3, 4JCF = 6.4 Hz), 102.50 (t, 2JCF = 25.4 Hz), 55.92, 51.50, 48.72, 47.40, 46.34, 44.37, 37.75, 37.14. HRMS: m/z 633.2135 (M + 1)+, m/z 655.1953 (M + 23)+. C31H29F5N4O5 [632.2058].
5.1.6.28. (S)-3-(3,5-Difluorophenyl)-N-(4-methoxyphenyl)-N-methyl-2-(2-(2-oxo-4-(3-(trifluoromethyl)benzoyl)piperazin-1-yl)acetamido)propanamide (F-Id-3j)
White solid, yield: 65%. m.p.: 84–85 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.43 (d, J = 8.0 Hz, 1H, NH), 7.87 (d, J = 7.7 Hz, 1H, Ph–H), 7.81 (s, 1H, Ph–H), 7.74 (dt, J = 15.3, 7.7 Hz, 2H, Ph–H), 7.25 (d, J = 8.3 Hz, 2H, Ph–H), 7.02 (d, J = 8.2 Hz, 3H, Ph–H), 6.49 (d, J = 7.8 Hz, 2H, Ph–H), 4.47 (td, J = 8.9, 4.2 Hz, 1H, CH), 4.18 (s, 1H, CH), 4.11–3.82 (m, 4H, CH2 × 2), 3.79 (s, 3H, OCH3), 3.65–3.43 (m, 1H, CH), 3.24 (s, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 10.3, 3.1 Hz, 1H, PhCH), 2.70 (dd, J = 13.6, 9.6 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 167.70 (C O), 162.50 (dd, 1JCF = 246.1, 3JCF = 13.5 Hz), 159.17 (C O), 142.54 (t, 3JCF = 9.6 Hz), 135.83, 131.54, 130.22, 129.96, 129.64, 129.32, 129.16, 127.08, 125.67, 124.45, 122.96, 115.27, 112.34 (dd, 2JCF = 17.9, 4JCF = 6.1 Hz), 102.47 (t, 2JCF = 25.6 Hz), 55.91, 51.50, 48.71, 37.75, 37.14. HRMS: m/z 633.2136 (M + 1)+, m/z 655.1953 (M + 23)+. C31H29F5N4O5 [632.2058].
5.1.6.29. (S)-3-(3,5-Difluorophenyl)-N-(4-methoxyphenyl)-N-methyl-2-(2-(2-oxo-4-(2-(trifluoromethyl)benzoyl)piperazin-1-yl)acetamido)propanamide (F-Id-3k)
White solid, yield: 62%. m.p.: 87–88 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.41 (d, J = 8.1 Hz, 1H, NH), 7.84 (d, J = 7.9 Hz, 1H, Ph–H), 7.78 (t, J = 7.6 Hz, 1H, Ph–H), 7.69 (t, J = 7.7 Hz, 1H, Ph–H), 7.57–7.48 (m, 1H, Ph–H), 7.24 (d, J = 8.2 Hz, 2H, Ph–H), 7.10–6.94 (m, 3H, Ph–H), 6.50 (t, J = 7.1 Hz, 2H, Ph–H), 4.47 (t, J = 11.2 Hz, 1H, CH), 4.34–4.05 (m, 2H, CH2), 4.04–3.83 (m, 3H, CH2, CH), 3.79 (s, 3H, OCH3), 3.76–3.49 (m, 1H, CH2), 3.29–3.17 (m, 2H, CH), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 12.9, 3.9 Hz, 1H, PhCH), 2.69 (dd, J = 13.6, 9.4 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.85 (C O), 167.65 (C O), 164.91 (C O), 162.49 (dd, 1JCF = 246.9, 3JCF = 12.8 Hz), 159.17 (C O), 142.54, 135.83, 133.52, 133.46, 130.44, 130.35, 129.16, 127.97, 127.08, 125.72, 115.27, 112.35 (d, 2JCF = 24.8 Hz), 102.45, 55.92, 51.47, 50.35, 46.51, 45.94, 43.82, 37.77, 37.18. HRMS: m/z 633.2136 (M + 1)+, m/z 655.1953 (M + 23)+. C31H29F5N4O5 [632.2058].
5.1.6.30. Methyl (S)-4-(4-(2-((3-(3,5-difluorophenyl)-1-((4-methoxyphenyl)(methyl)amino)-1-oxopropan-2-yl)amino)-2-oxoethyl)-3-oxopiperazine-1-carbonyl)benzoate (F-Id-3l)
White solid, yield: 66%. m.p.: 82–83 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.44 (d, J = 8.0 Hz, 1H, NH), 8.04 (d, J = 7.9 Hz, 2H, Ph–H), 7.58 (d, J = 7.9 Hz, 2H, Ph–H), 7.25 (d, J = 8.3 Hz, 2H, Ph–H), 7.02 (d, J = 8.6 Hz, 3H, Ph–H), 6.49 (d, J = 7.7 Hz, 2H, Ph–H), 4.47 (dt, J = 9.4, 4.7 Hz, 1H, CH), 4.19 (s, 2H, CH2), 3.95 (s, 3H, CH2, CH), 3.88 (s, 3H, COOCH3), 3.79 (s, 3H, OCH3), 3.52 (s, 1H, CH), 3.24 (d, J = 15.3 Hz, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.7, 4.2 Hz, 1H, PhCH), 2.70 (dd, J = 13.6, 9.5 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 167.70 (C O), 166.13 (C O), 162.50 (dd, 1JCF = 246.2, 3JCF = 13.7 Hz), 159.17 (C O), 142.54 (t, 3JCF = 9.3 Hz), 135.83, 131.04, 129.84, 129.17, 127.84, 115.28, 112.35 (dd, 2JCF = 18.0, 4JCF = 6.3 Hz), 102.49 (t, 2JCF = 26.0 Hz), 55.93, 52.85, 51.50, 48.71, 37.76, 37.14. HRMS: m/z 623.2313 (M + 1)+, m/z 645.2133 (M + 23)+. C32H32F2N4O7 [622.2239].
5.1.6.31. (S)-3-(3,5-Difluorophenyl)-N-(4-methoxyphenyl)-N-methyl-2-(2-(4-(4-nitrobenzoyl)-2-oxopiperazin-1-yl)acetamido)propanamide (F-Id-3m)
White solid, yield: 76%. m.p.: 86–87 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.44 (d, J = 8.1 Hz, 1H, NH), 8.31 (d, J = 8.2 Hz, 2H, Ph–H), 7.73 (d, J = 8.2 Hz, 2H, Ph–H), 7.25 (d, J = 8.3 Hz, 2H, Ph–H), 7.03 (d, J = 8.6 Hz, 3H, Ph–H), 6.49 (d, J = 7.7 Hz, 2H, Ph–H), 4.48 (dd, J = 8.8, 4.2 Hz, 1H, CH), 4.30–4.06 (m, 2H, CH2), 3.95 (s, 3H, CH2, CH), 3.80 (s, 3H, OCH3), 3.51 (s, 1H, CH), 3.25 (d, J = 15.8 Hz, 2H, CH2), 3.13 (s, 3H, NCH3), 2.88 (dd, J = 13.6, 4.1 Hz, 1H, PhCH), 2.70 (dd, J = 13.7, 9.6 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 167.69 (C O), 167.43 (C O), 162.51 (dd, 1JCF = 246.2, 3JCF = 13.7 Hz), 159.18 (C O), 142.54 (t, 3JCF = 9.5 Hz), 141.82, 135.84, 132.19, 131.98, 129.16, 128.90, 124.27, 115.29, 112.34 (dd, 2JCF = 18.5 Hz, 4JCF = 6.1 Hz), 102.49 (t, 2JCF = 25.5 Hz), 65.49, 55.93, 51.49, 48.73, 37.76, 37.16, 30.48, 19.12. HRMS: m/z 610.2106 (M + 1)+, m/z 632.1932 (M + 23)+. C30H29F2N5O7 [609.2035].
5.1.6.32. (S)-3-(3,5-Difluorophenyl)-N-(4-methoxyphenyl)-N-methyl-2-(2-(4-(3-nitrobenzoyl)-2-oxopiperazin-1-yl)acetamido)propanamide (F-Id-3n)
White solid, yield: 81%. m.p.: 92–93 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.36 (d, J = 8.0 Hz, 1H), 8.25 (d, J = 8.4 Hz, 1H, NH), 8.19 (s, 1H, Ph–H), 7.82 (d, J = 7.7 Hz, 1H, Ph–H), 7.69 (t, J = 7.8 Hz, 1H, Ph–H), 7.17 (d, J = 8.2 Hz, 2H, Ph–H), 6.94 (d, J = 8.2 Hz, 3H, Ph–H), 6.41 (d, J = 7.7 Hz, 2H, Ph–H), 4.39 (d, J = 10.4 Hz, 1H, CH), 4.11 (s, 2H, CH2), 3.87 (s, 2H, CH2), 3.76 (s, 1H, CH), 3.71 (s, 3H, OCH3), 3.47 (s, 1H, CH), 3.17 (d, J = 10.3 Hz, 2H, CH), 3.05 (s, 3H, NCH3), 2.84–2.73 (m, 1H, PhCH), 2.61 (dd, J = 13.8, 9.5 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.87 (C O), 167.73 (C O), 166.97 (C O), 162.49 (dd, 1JCF = 245.9, 3JCF = 13.5 Hz), 159.16 (C O), 148.16, 142.53 (t, 3JCF = 9.4 Hz), 137.08, 135.81, 133.89, 130.80, 129.18, 125.13, 122.67, 115.27, 112.34 (dd, 2JCF = 18.2, 4JCF = 6.1 Hz), 102.50 (t, 2JCF = 25.8 Hz), 55.92, 51.53, 48.71, 46.46, 37.75, 37.09, 32.67, 14.55. HRMS: m/z 610.2108 (M + 1)+, m/z 632.1928 (M + 23)+. C30H29F2N5O7 [609.2035].
5.1.6.33. (S)-3-(3,5-Difluorophenyl)-N-(4-methoxyphenyl)-N-methyl-2-(2-(4-(2-nitrobenzoyl)-2-oxopiperazin-1-yl)acetamido)propanamide (F-Id-3o)
White solid, yield: 73%. m.p.: 98–99 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.43 (d, J = 7.3 Hz, 1H, NH), 8.23 (t, J = 8.9 Hz, 1H, Ph–H), 7.88 (t, J = 7.6 Hz, 1H, Ph–H), 7.74 (t, J = 7.9 Hz, 1H, Ph–H), 7.58 (t, J = 8.5 Hz, 1H, Ph–H), 7.25 (dd, J = 8.4, 4.4 Hz, 2H, Ph–H), 7.02 (d, J = 8.5 Hz, 3H, Ph–H), 6.50 (t, J = 6.9 Hz, 2H, Ph–H), 4.48 (dq, J = 9.1, 4.7 Hz, 1H, CH), 4.19 (s, 1H, CH), 3.96 (s, 2H, CH2), 3.85 (s, 2H, CH2), 3.79 (d, J = 4.4 Hz, 3H, OCH3), 3.42 (q, J = 5.4 Hz, 2H, CH2), 3.17 (s, 1H, CH), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.4, 3.9 Hz, 1H, PhCH), 2.70 (t, J = 11.5 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 167.68 (C O), 167.64 (C O), 162.50 (dd, 1JCF = 256.7, 3JCF = 13.7 Hz), 159.18 (C O), 145.72, 142.53 (t, 3JCF = 9.2 Hz), 135.84, 135.52, 132.02, 131.18, 129.16, 128.62, 128.48, 125.36, 125.32, 115.29, 112.35 (dd, 2JCF = 17.3, 4JCF = 6.2 Hz), 102.47, 55.93, 51.48, 50.09, 47.19, 46.05, 37.77, 37.19. HRMS: m/z 610.2105 (M + 1)+, m/z 632.1930 (M + 23)+. C30H29F2N5O7 [609.2035].
5.1.6.34. (S)-2-(2-(4-(4-Aminobenzoyl)-2-oxopiperazin-1-yl)acetamido)-3-(3,5-difluorophenyl)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3p)
White solid, yield: 62%. m.p.: 90–91 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.45 (d, J = 8.1 Hz, 1H, NH), 7.25 (d, J = 8.3 Hz, 2H, Ph–H), 7.17 (d, J = 8.1 Hz, 2H, Ph–H), 7.03 (d, J = 8.4 Hz, 3H, Ph–H), 6.56 (d, J = 8.1 Hz, 2H, Ph–H), 6.50 (d, J = 7.7 Hz, 2H, Ph–H), 5.62 (s, 2H, NH2), 4.46 (td, J = 8.7, 4.0 Hz, 1H, CH), 4.09 (s, 2H, CH2), 3.93 (d, J = 7.3 Hz, 2H, CH2), 3.79 (s, 3H, OCH3), 3.68 (t, J = 5.5 Hz, 2H, CH2), 3.21 (d, J = 5.3 Hz, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.6, 4.3 Hz, 1H, PhCH), 2.69 (dd, J = 13.6, 9.8 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 169.97 (C O), 167.72 (C O), 162.49 (dd, 1JCF = 245.8, 3JCF = 13.1 Hz), 159.16 (C O), 151.07, 150.23, 142.55, 135.81, 129.90, 129.17, 121.50, 115.28, 112.36 (dd, 2JCF = 17.6, 4JCF = 6.6 Hz), 102.49 (t, 2JCF = 25.5 Hz), 55.91, 51.48, 48.64, 47.23, 37.75, 37.15. HRMS: m/z 580.2371 (M + 1)+, m/z 602.2186 (M + 23)+. C30H31F2N5O5 [579.2293].
5.1.6.35. (S)-2-(2-(4-(3-Aminobenzoyl)-2-oxopiperazin-1-yl)acetamido)-3-(3,5-difluorophenyl)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3q)
White solid, yield: 53%. m.p.: 95–96 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.43 (d, J = 8.0 Hz, 1H, NH), 7.24 (d, J = 8.2 Hz, 2H, Ph–H), 7.08 (t, J = 7.7 Hz, 1H, Ph–H), 7.02 (d, J = 8.5 Hz, 3H, Ph–H), 6.64 (d, J = 8.1 Hz, 1H, Ph–H), 6.58 (s, 1H, Ph–H), 6.50 (d, J = 7.6 Hz, 3H, Ph–H), 5.28 (s, 2H, NH2), 4.53–4.41 (m, 1H, CH), 4.06 (s, 2H, CH2), 3.94 (d, J = 8.6 Hz, 2H, CH2), 3.79 (s, 3H, OCH3), 3.60 (s, 2H, CH2), 3.21 (t, J = 5.3 Hz, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.5, 4.2 Hz, 1H, PhCH), 2.69 (dd, J = 13.5, 9.5 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 167.69 (C O), 162.51 (dd, 1JCF = 245.9, 3JCF = 13.1 Hz), 159.17 (C O), 149.30, 142.54 (t, 3JCF = 9.6 Hz), 136.10, 135.83, 129.39, 129.16, 115.69, 115.29, 112.36 (dd, 2JCF = 19.3, 4JCF = 5.4 Hz), 102.49 (t, 2JCF = 25.0 Hz), 55.93, 51.47, 48.70, 37.76, 37.21. HRMS: m/z 580.2371 (M + 1)+. C30H31F2N5O5 [579.2293].
5.1.6.36. (S)-2-(2-(4-(2-aminobenzoyl)-2-oxopiperazin-1-yl)acetamido)-3-(3,5-difluorophenyl)-N-(4-methoxyphenyl)-N-methylpropanamide (F-Id-3r)
White solid, yield: 55%. m.p.: 79–80 °C. 1H NMR (400 MHz, DMSO-d6) δ 8.42 (d, J = 8.1 Hz, 1H, NH), 7.24 (d, J = 8.3 Hz, 2H, Ph–H), 7.12 (t, J = 7.8 Hz, 1H, Ph–H), 7.02 (d, J = 8.7 Hz, 4H, Ph–H), 6.71 (d, J = 8.2 Hz, 1H, Ph–H), 6.57 (t, J = 7.4 Hz, 1H, Ph–H), 6.49 (d, J = 7.7 Hz, 2H, Ph–H), 5.28 (s, 2H, NH2), 4.46 (td, J = 8.7, 4.4 Hz, 1H, CH), 4.07 (s, 2H, CH2), 3.93 (s, 2H, CH2), 3.79 (s, 3H, OCH3), 3.66 (s, 2H, CH2), 3.21 (s, 2H, CH2), 3.13 (s, 3H, NCH3), 2.87 (dd, J = 13.5, 4.1 Hz, 1H, PhCH), 2.69 (dd, J = 13.5, 9.7 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 170.86 (C O), 168.90 (C O), 167.71 (C O), 162.50 (dd, 1JCF = 245.7, 3JCF = 13.7 Hz), 159.16 (C O), 146.37, 142.54 (t, 3JCF = 9.6 Hz), 135.83, 130.92, 129.17, 128.33, 118.74, 115.28, 112.36 (dd, 2JCF = 18.1, 4JCF = 6.7 Hz), 102.49 (t, 2JCF = 25.4 Hz), 55.92, 51.47, 48.67, 47.24, 37.77, 37.18. HRMS: m/z 580.2368 (M + 1)+, m/z 602.2190 (M + 23)+. C30H31F2N5O5 [579.2293].
5.2. In vitro anti-HIV assay in MT-4 cells
Evaluation of the antiviral activity of the compounds against HIV in MT-4 cells was performed using the MTT assay as described below. Stock solutions (10 × final concentration) of test compounds were added in 25 μL volumes to two series of triplicate wells to allow simultaneous evaluation of their effects on mock- and HIV-infected cells at the beginning of each experiment. Serial 5-fold dilutions of test compounds were made directly in flat-bottomed 96-well microtiter trays using a Biomek 3000 robot (Beckman Instruments, Fullerton, CA). Untreated HIV- and mock-infected cell samples were included as controls. HIV stock (50 μL) at 100–300 CCID50 (50% cell culture infectious doses) or culture medium was added to either the infected or mock-infected wells of the microtiter tray. Mock-infected cells were used to evaluate the effects of the test compound on uninfected cells to assess the test compounds' cytotoxicity. Exponentially growing MT-4 cells were centrifuged for 5 minutes at 220 g, and the supernatant was discarded. The MT-4 cells were resuspended at 6 × 105 cells per mL, and 50 μL volumes were transferred to the microtiter tray wells. Five days after infection, the viability of mock-and HIV-infected cells was examined spectrophotometrically using the MTT assay. The MTT assay is based on the reduction of yellow colored 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Acros Organics) by mitochondrial dehydrogenase activity in metabolically active cells to a blue-purple formazan that can be measured spectrophotometrically. The absorbances were read in an eight-channel computer-controlled photometer (Infinite M1000, Tecan), at two wavelengths (540 and 690 nm). All data were calculated using the median absorbance value of three wells. The 50% cytotoxic concentration (CC50) was defined as the concentration of the test compound that reduced the absorbance (OD540) of the mock-infected control sample by 50%. The concentration achieving 50% protection against the cytopathic effect of the virus in infected cells was defined as the 50% effective concentration (EC50).
5.3. Determination of the mechanism of representative compounds
5.3.1. Cells
Human embryonic kidney 293 T (a gift from Dr. Irwin Chaiken, Drexel University, Philadelphia, PA) were cultured in Dulbecco's modified Eagle's medium (DMEM), 10% FBS, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin and 2 mM l-glutamine. Human astroglioma U87 cells stably expressing CD4/CXCR4 (obtained from Prof. Hongkui Deng, Peking University, and Prof. Dan Littman, New York University, USA, through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH)60,61 were cultured in DMEM supplemented with 10% FBS, 100 U ml−1 penicillin, 100 μg mL−1 streptomycin and 2 mM l-glutamine, 300 μg mL−1 G418 (Thermo Scientific, Waltham, MA) and 1 μg mL−1 puromycin (Thermo Scientific). Cells were incubated continuously, unless otherwise stated, at 37 °C in a humidified 5% CO2/95% air environment.
5.3.2. Proteins
IgG b12 anti-HIV-1 gp120 was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti-HIV-1 gp120 monoclonal (IgG1 b12) from Dr. Dennis Burton and Carlos Barbas; p24 was produced in-house as previously described.62 Briefly, a vector containing C-terminally His-tagged HIV-1NL4-3CA (a gift from Dr. Eric Barklis, Oregon Health and Science University, Portland, OR) was transformed into BL21-Codon Plus (DE3)-RIL competent cells (Agilent Technologies, Wilmington, DE) and grown up in autoinduction ZYP-5052 medium overnight with shaking (225 rpm) at 30 °C.63 Bacterial cultures were spun down at 7000 rpm, and the supernatant was discarded. Cell pellets were resuspended in PBS and lysed via sonication. The resultant supernatant was clarified and immediately applied to a Talon cobalt resin affinity column (Clonetech Laboratories, Mountain View, CA). Protein was eluted using 1X PBS with 250 mM imidazole. Purified CA–H6 monomers were dialyzed overnight into 20 mM Tris-HCl pH 8.0 at 4 °C, concentrated to 120 μM, flash-frozen in liquid nitrogen, aliquoted, and stored at −80 °C. The CA hexamer was generated by introducing mutations at the following sites: A14C, E45C, W184A, and M185A through site-directed mutagenesis (Stratagene). The CA hexamer construct was expressed and purified following the same protocol as described above. After purification, the CA–H6 hexamers were dialyzed into 200 mM β-ME followed by sequential dialyzes to remove the β-ME to allow for hexamer assembly slowly.
5.3.3. Production of pseudotyped viruses
Single-round infectious envelope-pseudotyped luciferase-reporter viruses were produced by dual transfection of two vectors (3 μg of vector 1 and 4 μg of vector 2) in 6-well plated 293 T cells (1 × 106 cells per well).61 Vector 1 is an envelope-deficient HIV-1 pNL4-3-Luc + R-E plasmid that carries the luciferase-reporter gene.64 Vector 2 is a plasmid expressing the HIV-1BG505 gp160 Env.65 Transfections of these vectors were carried out via calcium phosphate (ProFection Mammalian Transfection System, Promega, Madison, WI) for 5 h. Following the 5 h transfection incubation, DNA-containing medium was removed, cells were washed with DMEM and replenished with fresh culture media. Supernatants containing pseudovirus were collected 72 h post-transfection, clarified, filtered, aliquoted and stored at −80 °C.
5.3.4. SPR direct interaction analysis
All binding assays were performed on a ProteOn XPR36 SPR Protein Interaction Array System (Bio-Rad Laboratories, Hercules, CA). The instrument temperature was set at 25 °C for all kinetic analyses. ProteOn GLH sensor chips were preconditioned with two short pulses each (10 s) of 50 mM NaOH, 100 mM HCl, and 0.5% sodium dodecyl sulfide. Then the system was equilibrated with PBS-T buffer (20 mM sodium phosphate, 150 mM NaCl, and 0.005% polysorbate 20, pH 7.4). The surface of a GLH sensor chip was activated with a 1 : 100 dilution of a 1 : 1 mixture of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (0.2 M) and sulfo-N-hydroxysuccinimide (0.05 M). Immediately after chip activation, the HIV-1 NL4-3 capsid protein constructs, purified as described above, were prepared at a concentration of 100 μg mL−1 in 10 mM sodium acetate, pH 5.0 and injected across ligand flow channels for 5 min at a flow rate of 30 μL min−1. Then, after unreacted protein had been washed out, excess active ester groups on the sensor surface were capped by a 5 min injection of 1 M ethanolamine HCl (pH 8.0) at a flow rate of 5 μL min−1. A reference surface was similarly created by immobilizing a non-specific protein (IgG b12 anti-HIV-1 gp120; was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti-HIV-1 gp120 monoclonal (IgG1 b12) from Dr. Dennis Burton and Carlos Barbas) and was used as a background to correct non-specific binding. To prepare a compound for direct binding analysis, compound stock solutions, along with 100% DMSO, and totalling 30 μL was made to a final volume of 1 mL by addition of sample preparation buffer (PBS, pH 7.4). Preparation of analyte in this manner ensured that the concentration of DMSO was matched with that of running buffer with 3% DMSO. Serial dilutions were then prepared in the running buffer (PBS, 3% DMSO, 0.005% polysorbate 20, pH 7.4) and injected at a flow rate of 100 μl min−1, for a 1 min association phase, followed by up to a 5 min dissociation phase using the “one-shot kinetics” capability of the Proteon instrument. Data were analyzed using the ProteOn Manager software version 3.0 (Bio-Rad). The responses from the reference flow cell were subtracted to account for the nonspecific binding and injection artefacts. Experimental data were fitted to a simple 1 : 1 binding model (where applied). The average kinetic (dissociation [kd] rates) and equilibrium parameters generated from 3 replicates were used to define the off-rates and equilibrium dissociation constant (KD).
5.3.5. Single-round infection (SRI) assay
The single-round HIV-1 infection assay details have been published previously.64,66,67 Briefly, U87.CD4.CXCR4 (1.2 × 104 cells per well) target cells were seeded in 96-well luminometer-compatible tissue culture plates (Greiner Bio-one). After 24 hours compound, DMSO (vehicle control for compounds, Sigma) was mixed with pseudotyped viruses (normalized to p24 content). The mixture was added to the target cells and incubated for 48 hours at 37 °C. Following this, the media was removed from each well, and the cells were lysed by the addition of 50 μL per well of luciferase lysis buffer (Promega) and one freeze–thaw cycle. A GloMax 96 microplate luminometer (Promega) was used to measure the luciferase activity of each well after the addition of 50 μL per well of luciferase assay substrate (Promega).
5.3.6. Viral late-stage infection assay
Single-round infectious specific envelope-pseudotyped luciferase-reporter viruses were produced from 293 T cells61 in the presence of 100 μM compound or DMSO (a vehicle control for compounds, Sigma). After 48 hours of incubation at 37 °C, the resulting culture supernatants containing pseudotype virus stocks were diluted twenty-fold and then used to infect U87.CD4.CXCR4 target cells. Target cells with or without pseudotyped viruses were incubated for 48 hours at 37 °C. Subsequently, the media was removed from each well, and the cells were lysed by the addition of 50 μl of luciferase lysis buffer (Promega) and one freeze–thaw cycle. A GloMax 96 microplate luminometer (Promega) was used to measure the luciferase activity of each well after the addition of 50 μl of luciferase assay substrate (Promega). Compound induced effects are manifested as a decrease in infectivity in the target cells (measured as luciferase activity), normalized against the infectivity of virus produced from DMSO (vehicle control) treated cells.
5.4. Molecular dynamics simulation
5.4.1. Homology-modeling
Due to the lack of HIV-2 CA hexamer crystal structure, we have employed SWISS-MODEL to homology-modeling HIV-2 CA hexamer and used all default settings. The FASTA documents of HIV-2 ROD CA came from UniProt (https://www.uniprot.org).
5.4.2. Protein preparation
The HIV-1 CA hexamer (PDB ID: 4XFZ) and HIV-2 CA homology-modeled were uploaded and prepared in the Protein Preparation (Maestro, Schrödinger, LLC, New York, NY, 2021), in which the redundant water molecules were removed and whole protein was modified according to the default settings.
5.4.3. System preparation and docking
All molecules were prepared using LigPrep (Maestro, Schrödinger, LLC, New York, NY, 2021). In our whole docking study, default parameters were used, and the best-ranked compounds were retained for subsequent analysis.
5.4.4. Dynamics simulation
Using the result with the best-ranked docking results for each compound as the initial conformation. Water addition, salt addition operations and molecular dynamics simulations were performed using System Builder and Molecular Dynamics (Maestro, Schrödinger, LLC, New York, NY, 2021) using the default parameters.
5.5. Metabolic stability in human liver microsomes and human plasma
Details of the analytical method and raw data are given in the ESI.†
Author contributions
Xujie Zhang and Lin Sun contributed equally to this work. Xujie Zhang, Lin Sun, Shujing Xu, Tianguang Huang, Dang Ding, Chuanfeng Liu and Xiangyi Jiang synthesized the compounds. Simon Cocklin and Alexej Dick performed the SPR and SRI assays. Erik De Clercq and Christophe Pannecouque evaluated the anti-HIV activity. Xujie Zhang, Fabao Zhao and Alexej Dick did the computation work. Peng Zhan and Xinyong Liu contributed to the conceptualization and methodology. The manuscript was written through contributions of all authors. All of the authors approved the final version of the manuscript.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support from National Natural Science Foundation of China (NSFC No. 82173677, 82204196), the Key Project of NSFC for International Cooperation (No. 81420108027), Science Foundation for Outstanding Young Scholars of Shandong Province (ZR2020JQ31), Qilu Young Scholars Program of Shandong University, the Taishan Scholar Program at Shandong Province, Shandong Provincial Natural Science Foundation (ZR2022QH015) and NIH/NIAID grant R01AI150491 (Cocklin, PI, Salvino, Co-I).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3md00134b
References
- Wang Z. Cherukupalli S. Xie M. Wang W. Jiang X. Jia R. Pannecouque C. De Clercq E. Kang D. Zhan P. Liu X. J. Med. Chem. 2022;65:3729–3757. doi: 10.1021/acs.jmedchem.1c01758. [DOI] [PubMed] [Google Scholar]
- Gao F. Bailes E. Robertson D. L. Chen Y. Rodenburg C. M. Michael S. F. Cummins L. B. Arthur L. O. Peeters M. Shaw G. M. Sharp P. M. Hahn B. H. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- Chen Z. Telfier P. Gettie A. Reed P. Zhang L. Ho D. D. Marx P. A. J. Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil A. Sarr A. D. Sankalé J. L. Meloni S. T. Mboup S. Kanki P. J. Virol. 2007;81:5325–5330. doi: 10.1128/JVI.02625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlink R. Kanki P. Thior I. Travers K. Eisen G. Siby T. Traore I. Hsieh C. C. Dia M. C. Gueye E. H. et al. . Science. 1994;265:1587–1590. doi: 10.1126/science.7915856. [DOI] [PubMed] [Google Scholar]
- WHO, Data and statistics, https://www.unaids.org/en/resources/fact-sheet
- Governa P. Manetti F. Eur. J. Med. Chem. 2022;229:114078. doi: 10.1016/j.ejmech.2021.114078. [DOI] [PubMed] [Google Scholar]
- Smith S. J. Ferris A. Zhao X. Pauly G. Schneider J. P. Burke, Jr. T. R. Hughes S. H. Viruses. 2021;13:2501. doi: 10.3390/v13122501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J. Guo J. Kang D. Li Z. Wang G. Wu J. Zhang Z. Fang H. Hou X. Huang Z. Li G. Lu X. Liu X. Ouyang L. Rao L. Zhan P. Zhang X. Zhang Y. Chin. Chem. Lett. 2020;31:1695–1708. doi: 10.1016/j.cclet.2020.03.028. [DOI] [Google Scholar]
- Li G. Wang Y. De Clercq E. Acta Pharm. Sin. B. 2022;12:1567–1590. doi: 10.1016/j.apsb.2021.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana L. N. Anand A. R. Sethuraman S. Krishnan U. M. J. Controlled Release. 2014;192:271–283. doi: 10.1016/j.jconrel.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Xu S. Sun L. Ding D. Tao Y. Kang D. Liu X. Zhan P. Future Med. Chem. 2022;14:605–607. doi: 10.4155/fmc-2022-0008. [DOI] [PubMed] [Google Scholar]
- Ma Y. Frutos-Beltrán E. Kang D. Pannecouque C. De Clercq E. Menéndez-Arias L. Liu X. Zhan P. Chem. Soc. Rev. 2021;50:4514–4540. doi: 10.1039/D0CS01084G. [DOI] [PubMed] [Google Scholar]
- Zhang M. J. Stear J. H. Jacques D. A. Böcking T. Biophys. Rev. 2022;14:23–32. doi: 10.1007/s12551-021-00922-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner G. Weaver N. Anokhin B. Spearman P. Viruses. 2022;14:478. doi: 10.3390/v14030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedán A. Caroe E. R. Barr G. C. R. Bishop K. N. Viruses. 2021;13:1425. doi: 10.3390/v13081425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. Zhang X. Xu S. Huang T. Song S. Cherukupalli S. Zhan P. Liu X. Eur. J. Med. Chem. 2021;217:113380. doi: 10.1016/j.ejmech.2021.113380. [DOI] [PubMed] [Google Scholar]
- Chen N. Y. Zhou L. Gane P. J. Opp S. Ball N. J. Nicastro G. Zufferey M. Buffone C. Luban J. Selwood D. Diaz-Griffero F. Taylor I. Fassati A. Retrovirology. 2016;13:28. doi: 10.1186/s12977-016-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. Biochemistry. 2016;55:2539–2552. doi: 10.1021/acs.biochem.6b00159. [DOI] [PubMed] [Google Scholar]
- Pornillos O. Ganser-Pornillos B. K. Yeager M. Nature. 2011;469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A. Yamashita M. Retrovirology. 2021;18:32. doi: 10.1186/s12977-021-00577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek K. A. Yücel S. S. Li X. Engelman A. PLoS Pathog. 2013;9:e1003693. doi: 10.1371/journal.ppat.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedán A. Donaldson C. D. Caroe E. R. Cosnefroy O. Taylor I. A. Bishop K. N. PLoS Pathog. 2021;17:e1009484. doi: 10.1371/journal.ppat.1009484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano D. A. Peng K. Laketa V. Börner K. Jost K. L. Lucic B. Glass B. Lusic M. Müller B. Kräusslich H. G. eLife. 2019;8:e41800. doi: 10.7554/eLife.41800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A. Ode H. Nohata K. Ohmori H. Nakayama E. E. Iwatani Y. Shioda T. Virology. 2019;532:118–126. doi: 10.1016/j.virol.2019.04.012. [DOI] [PubMed] [Google Scholar]
- Rebensburg S. V. Wei G. Larue R. C. Lindenberger J. Francis A. C. Annamalai A. S. Morrison J. Shkriabai N. Huang S. W. KewalRamani V. Poeschla E. M. Melikyan G. B. Kvaratskhelia M. Nat. Microbiol. 2021;6:435–444. doi: 10.1038/s41564-021-00868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques D. A. McEwan W. A. Hilditch L. Price A. J. Towers G. J. James L. C. Nature. 2016;536:349–353. doi: 10.1038/nature19098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. P. Francis A. C. Meuser M. E. Mankowski M. Ptak R. G. Rashad A. A. Melikyan G. B. Cocklin S. J. Drug Des. Res. 2018;5:1070. [PMC free article] [PubMed] [Google Scholar]
- Xu J. P. Branson J. D. Lawrence R. Cocklin S. Bioorg. Med. Chem. Lett. 2016;26:824–828. doi: 10.1016/j.bmcl.2015.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair W. S. Pickford C. Irving S. L. Brown D. G. Anderson M. Bazin R. Cao J. Ciaramella G. Isaacson J. Jackson L. Hunt R. Kjerrstrom A. Nieman J. A. Patick A. K. Perros M. Scott A. D. Whitby K. Wu H. Butler S. L. PLoS Pathog. 2010;6:e1001220. doi: 10.1371/journal.ppat.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahani R. L. Diana-Rivero R. Vernekar S. K. V. Wang L. Du H. Zhang H. Castaner A. E. Casey M. C. Kirby K. A. Tedbury P. R. Xie J. Sarafianos S. G. Wang Z. Viruses. 2021;13:479. doi: 10.3390/v13030479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Casey M. C. Vernekar S. K. V. Sahani R. L. Kirby K. A. Du H. Zhang H. Tedbury P. R. Xie J. Sarafianos S. G. Wang Z. Acta Pharm. Sin. B. 2021;11:810–822. doi: 10.1016/j.apsb.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. Wu G. Zalloum W. A. Meuser M. E. Dick A. Sun L. Chen C. H. Kang D. Jing L. Jia R. Cocklin S. Lee K. H. Liu X. Zhan P. RSC Adv. 2019;9:28961–28986. doi: 10.1039/C9RA05869A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. Zalloum W. A. Meuser M. E. Jing L. Kang D. Chen C. H. Tian Y. Zhang F. Cocklin S. Lee K. H. Liu X. Zhan P. Eur. J. Med. Chem. 2018;158:478–492. doi: 10.1016/j.ejmech.2018.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuser M. E. Reddy P. A. N. Dick A. Maurancy J. M. Salvino J. M. Cocklin S. J. Med. Chem. 2021;64:3747–3766. doi: 10.1021/acs.jmedchem.0c01810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Sun L. Meuser M. E. Zalloum W. A. Xu S. Huang T. Cherukupalli S. Jiang X. Ding X. Tao Y. Kang D. De Clercq E. Pannecouque C. Dick A. Cocklin S. Liu X. Zhan P. Eur. J. Med. Chem. 2021;226:113848. doi: 10.1016/j.ejmech.2021.113848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link J. O. Rhee M. S. Tse W. C. Zheng J. Somoza J. R. Rowe W. Begley R. Chiu A. Mulato A. Hansen D. Singer E. Tsai L. K. Bam R. A. Chou C. H. Canales E. Brizgys G. Zhang J. R. Li J. Graupe M. Morganelli P. Liu Q. Wu Q. Halcomb R. L. Saito R. D. Schroeder S. D. Lazerwith S. E. Bondy S. Jin D. Hung M. Novikov N. Liu X. Villaseñor A. G. Cannizzaro C. E. Hu E. Y. Anderson R. L. Appleby T. C. Lu B. Mwangi J. Liclican A. Niedziela-Majka A. Papalia G. A. Wong M. H. Leavitt S. A. Xu Y. Koditek D. Stepan G. J. Yu H. Pagratis N. Clancy S. Ahmadyar S. Cai T. Z. Sellers S. Wolckenhauer S. A. Ling J. Callebaut C. Margot N. Ram R. R. Liu Y. P. Hyland R. Sinclair G. I. Ruane P. J. Crofoot G. E. McDonald C. K. Brainard D. M. Lad L. Swaminathan S. Sundquist W. I. Sakowicz R. Chester A. E. Lee W. E. Daar E. S. Yant S. R. Cihlar T. Nature. 2020;584:614–618. doi: 10.1038/s41586-020-2443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester S. M. Wei G. Zhao H. Adu-Ampratwum D. Iqbal N. Courouble V. V. Francis A. C. Annamalai A. S. Singh P. K. Shkriabai N. Van Blerkom P. Morrison J. Poeschla E. M. Engelman A. N. Melikyan G. B. Griffin P. R. Fuchs J. R. Asturias F. J. Kvaratskhelia M. Science. 2020;370:360–364. doi: 10.1126/science.abb4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy S. S., Chou C.-H., Link J. O. and Tse W. C., WO2015130964A1, Gilead Sciences, Inc., USA, 2015 [Google Scholar]
- Bondy S. S., Cannizzaro C. E., Chou C.-H., Halcomb R. L., Hu Y. E., Link J. O., Liu Q., Schroeder S. D., Tse W. C. and Zhang J. R., WO2013006738A1, Gilead Sciences, Inc., USA, 2013 [Google Scholar]
- Bekerman E., Blair W. S., Chiu A., Cihlar T., Levine D. J., Tse W. C., Yant S. R. and Zheng J. X., WO2021108544A1, Gilead Sciences, Inc., USA, 2021 [Google Scholar]
- Bauer L. E., Gorman E. M., Mulato A. S., Rhee M. S., Rowe C. W., Sellers S. P., Stefanidis D., Tse W. C., Yant S. R. and Chiu A., WO2020018459A1, Gilead Sciences, Inc., USA, 2020 [Google Scholar]
- Xu S. Sun L. Zalloum W. A. Zhang X. Huang T. Ding D. Tao Y. Zhao F. Gao S. Kang D. De Clercq E. Pannecouque C. Dick A. Cocklin S. Liu X. Zhan P. Chin. Chem. Lett. 2023;34(3):107611. doi: 10.1016/j.cclet.2022.06.034. [DOI] [Google Scholar]
- Meuser M. E. Murphy M. B. Rashad A. A. Cocklin S. Molecules. 2018;23:1940. doi: 10.3390/molecules23081940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K. Woo S. M. Siu T. Cortopassi W. A. Duarte F. Paton R. S. Chem. Sci. 2018;9:2655–2665. doi: 10.1039/C7SC04905F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall M. Champness E. Obrezanova O. Leeding C. Chem. Biodiversity. 2009;6:2144–2151. doi: 10.1002/cbdv.200900148. [DOI] [PubMed] [Google Scholar]
- Tuyishime M. Danish M. Princiotto A. Mankowski M. K. Lawrence R. Lombart H. G. Esikov K. Berniac J. Liang K. Ji J. Ptak R. G. Madani N. Cocklin S. Bioorg. Med. Chem. Lett. 2014;24:5439–5445. doi: 10.1016/j.bmcl.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadsheh R. Meuser M. E. Cocklin S. Molecules. 2020;25:1430. doi: 10.3390/molecules25061430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyzack J. D. Hunt P. A. Segall M. D. J. Chem. Inf. Model. 2016;56:2180–2193. doi: 10.1021/acs.jcim.6b00233. [DOI] [PubMed] [Google Scholar]
- Hunt P. A. Segall M. D. Tyzack J. D. J. Comput.-Aided Mol. Des. 2018;32:537–546. doi: 10.1007/s10822-018-0107-0. [DOI] [PubMed] [Google Scholar]
- Reulecke I. Lange G. Albrecht J. Klein R. Rarey M. ChemMedChem. 2008;3:885–897. doi: 10.1002/cmdc.200700319. [DOI] [PubMed] [Google Scholar]
- Kaur P. Chamberlin A. R. Poulos T. L. Sevrioukova I. F. J. Med. Chem. 2016;59:4210–4220. doi: 10.1021/acs.jmedchem.5b01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic P. Somanathan R. J. Appl. Toxicol. 2014;34:810–824. doi: 10.1002/jat.2980. [DOI] [PubMed] [Google Scholar]
- Koske D. Goldenstein N. I. Kammann U. Aquat. Toxicol. 2019;217:105345. doi: 10.1016/j.aquatox.2019.105345. [DOI] [PubMed] [Google Scholar]
- Knasmüller S. Kerklaan P. Mohn G. R. Mutat. Res. 1986;164:9–17. doi: 10.1016/0165-1161(86)90037-3. [DOI] [PubMed] [Google Scholar]
- Hasspieler B. M. Haffner G. D. Adeli K. J. Toxicol. Environ. Health. 1997;52:137–148. doi: 10.1080/00984109708984057. [DOI] [PubMed] [Google Scholar]
- Malejka-Giganti D. Parkin D. R. Decker R. W. Niehans G. A. Bliss R. L. Churchwell M. I. Beland F. A. Int. J. Cancer. 2008;122:1958–1965. doi: 10.1002/ijc.23352. [DOI] [PubMed] [Google Scholar]
- Sega G. A. Mutat. Res. 1984;134:113–142. doi: 10.1016/0165-1110(84)90007-1. [DOI] [PubMed] [Google Scholar]
- Pillans P. I. Ghiculescu R. A. Lampe G. Wilson R. Wong R. Macdonald G. A. J. Gastroenterol. Hepatol. 2012;27:1102–1105. doi: 10.1111/j.1440-1746.2011.07036.x. [DOI] [PubMed] [Google Scholar]
- Björndal A. Deng H. Jansson M. Fiore J. R. Colognesi C. Karlsson A. Albert J. Scarlatti G. Littman D. R. Fenyö E. M. J. Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner I. Sierra L. J. Fraser A. K. Maciunas L. Mankowski M. K. Vinnik A. Fedichev P. Ptak R. G. Martín-García J. Cocklin S. ChemMedChem. 2013;8:426–432. doi: 10.1002/cmdc.201200577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortagere S. Madani N. Mankowski M. K. Schön A. Zentner I. Swaminathan G. Princiotto A. Anthony K. Oza A. Sierra L. J. Passic S. R. Wang X. Jones D. M. Stavale E. Krebs F. C. Martín-García J. Freire E. Ptak R. G. Sodroski J. Cocklin S. Smith, 3rd A. B. J. Virol. 2012;86:8472–8481. doi: 10.1128/JVI.05006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Protein Expression Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Connor R. I. Chen B. K. Choe S. Landau N. R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Hoffenberg S. Powell R. Carpov A. Wagner D. Wilson A. Kosakovsky Pond S. Lindsay R. Arendt H. Destefano J. Phogat S. Poignard P. Fling S. P. Simek M. Labranche C. Montefiori D. Wrin T. Phung P. Burton D. Koff W. King C. R. Parks C. L. Caulfield M. J. J. Virol. 2013;87:5372–5383. doi: 10.1128/JVI.02827-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocklin S. Gopi H. Querido B. Nimmagadda M. Kuriakose S. Cicala C. Ajith S. Baxter S. Arthos J. Martín-García J. Chaiken I. M. J. Virol. 2007;81:3645–3648. doi: 10.1128/JVI.01778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. Choe S. Walker R. Di Marzio P. Morgan D. O. Landau N. R. J. Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.