Abstract
Somatic and germline mutations in the dual zinc-finger transcription factor GATA3 are associated with breast cancers expressing the estrogen receptor (ER) and the autosomal dominant hypoparathyroidism–deafness–renal dysplasia syndrome, respectively. To elucidate the role of GATA3 in breast tumorigenesis, we investigated 40 breast cancers that expressed ER, for GATA3 mutations. Six different heterozygous GATA3 somatic mutations were identified in eight tumors, and these consisted of: a frameshifting deletion/insertion (944_945delGGinsAGC), an in-frame deletion of a key arginine residue (991_993delAGG), a seven-nucleotide frameshifting insertion (991_992insTGGAGGA), a frameshifting deletion (1196_1197delGA), and two frameshifting single nucleotide insertions (1224_1225insG found in three tumors and 1224_1225insA). Five of the eight mutations occurred in tumors that retained GATA3 immunostaining, indicating that absence of GATA3 immunostaining is an unreliable predictor of the presence of GATA3 mutations. Luciferase reporter assays, electrophoretic mobility shift assays, immunofluorescence, invasion and proliferation assays demonstrated that the GATA3 mutations resulted in loss (or reduction) of DNA binding, decrease in transactivational activity, and alterations in invasiveness but not proliferation. The 991_992insTGGAGGA (Arg330 frameshift) mutation led to a loss of nuclear localization, yet the 991_993delAGG (Arg330deletion) retained nuclear localization. Investigation of the putative nuclear localization signal (NLS) sites showed that the NLS of GATA3 does not conform to either a classical mono- or bi-partite signal, but contains multiple cooperative NLS elements residing around the N-terminal zinc-finger which comprises residues 264–288. Thus, approximately 20 % ER-positive breast cancers have somatic GATA3 mutations that lead to a loss of GATA3 transactivation activity and altered cell invasiveness.
Keywords: Nuclear Localization Signal, T47D Cell, Transactivation Activity, GATA3 Mutation, GATA3 Protein
Introduction
GATA3 belongs to a family of six mammalian GATA dual zinc-finger transcription factors (GATA1-6; Fig. 1a) that bind to the consensus 5′-(A/T)GATA(A/G)-3′ motif [1]. The C-terminal finger (ZnF2) is essential for DNA binding, whereas the N-terminal finger (ZnF1) helps stabilize this binding and physically interacts with other proteins such as the multi-type zinc-finger Friends of GATA (FOGs) [2]. GATA3 germline mutations are associated with the congenital hypoparathyroidism–deafness–renal dysplasia (HDR) syndrome in man [3–6], and somatic GATA3 mutations have been reported in breast cancer [7–13]. Thus, GATA3 has dual roles in development and oncogenesis. Indeed GATA3, in common with other GATA family members, plays important roles in vertebrate embryo organogenesis that includes the sympathetic nervous system, the mammary gland, parathyroid, kidney, inner ear, skin, and T cell lineages [14–16]. In oncogenesis, GATA3 overexpression has been reported in esophageal carcinoma, Hodgkin’s lymphoma, and pancreatic cancer [17–19], and underexpression is associated with cervical cancer and renal clear cell carcinoma [20–22]. However, in breast cancers GATA3 underexpression and overexpression have both been observed [23], and it has been reported that GATA3 is highly co-expressed with the estrogen receptor (ER) [20, 24]. Moreover, 70 GATA3 mutations (Fig. 1a) have been reported in breast tumors, and it has been observed that the incidence of GATA3 mutations is ∼5–20 % in breast cancers that immunostain for the ER [7, 9–13]. To further determine the role of GATA3 mutations and their altered function in breast cancers, we pursued combined mutational analysis and cellular studies of this transcription factor.
Fig. 1.
GATA3 mutations identified in breast cancer and HDR Syndrome. a Schematic representation of the genomic structure of the GATA3 gene. The human GATA3 gene consists of six exons that span 20 kb of genomic DNA and encode a 444-amino acid transcription factor which contains two transactivating domains (TA1 and TA2) and two zinc-fingers (ZnF1 and ZnF2). The sizes of exons 1, 2, 3, 4, 5, and 6 are 188, 610, 537, 146, 126, and 806 bp, respectively. The ATG (translation start) site is in exon 2 and the TAG (stop) site is in exon 6. The locations of 78 mutations (70, which have been identified or validated by Sanger DNA sequence analysis, from previous studies [7–13] and 8 in this report (asterisked)) found in breast cancers are shown above the genomic structure. The locations of 45 reported HDR mutations, and 6 reported whole deletions, are shown below. The arrow denotes the mutation confirmed in Fig. 2. b Alignment of amino acid residues surrounding zinc-finger regions, ZnF1 and ZnF2, between GATA family members. Basic residues C-terminal to ZnF1 are highly conserved in the GATA family members, whereas the basic residues N-terminal to ZnF1 are conserved in GATA 1–3 but not in GATA 4–6. Classical NLSs are typically small stretches of positively charged (basic) amino acids (arginine, R, and lysine, K), arranged as either monopartite (a single cluster) or bipartite (two clusters separated by a 10–12-amino acid spacer) sequences [58, 59], although there is no strict consensus sequence. The GATA3 NLS has been reported to involve ZnF1 residues 249 to 311 [40], whereas the GATA4 NLS has been more precisely defined and shown to involve the conserved residues R282, R283, R317, and R319, which are located in ZnF2 and its C-terminal region [39]. Four clusters of positively charged amino acids (K and R, shown in bold italics), similar to a classical NLS, are indicated (dashed underline), and four arginines (R) reported to be critical for nuclear targeting of GATA4 [39] are shown in bold. Residues 314 and 330 affected by mutations identified in breast tumors are boxed. The residues that form part of the zinc-fingers are shaded in gray. c Eighteen mutations, associated with HDR and 27 mutations [7–13] associated with breast cancer occurring in the two zinc-fingers and the adjacent C-terminal region, are detailed with the altered amino acids highlighted in black, and with every tenth amino acid numbered. fs frameshift, in inframe. Nonsense mutations E228X, R277X, and a deletion mutation at codon 201 lead to aberrant nuclear localization
Methods
Patients
Tumor samples were obtained from 40 patients diagnosed with breast cancer at St. Bartholomew’s and the Royal London National Health Service (NHS) Trust between 2005 and 2009. Informed consent was obtained from patients, and the study was granted NHS Research Ethics Committee approval (Central Office for Research Ethics Committee No. 06/Q0403/182). Blood samples for leukocyte DNA extraction were obtained from 55 unrelated Northern European individuals, using protocols approved by a Multicentre Research Ethics Committee (MREC/00/2/93).
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were retrieved for each patient and used to construct a tissue microarray (Beecher MTA1 machine; Alphelys TMA designer). Appropriate areas of invasive carcinomas were identified and three × 1-mm cores taken from each case. Cores of normal breast tissue were also included in the array as controls. Immunostaining was performed using the following antibodies and dilutions: ER (NCL-L-ER-6F11; Novocastra; 1:40), GATA3 (GATA3 HG3-31; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA; 1:200), and human epidermal growth factor 2 (Her2; c-erB-2 NCL-CBE-356; Novocastra, Newcastle-upon-Tyne, UK; 1:50) using reported methods [25]. Expression was scored, by two independent researchers (PG and JLJ), using the Allred Quick Score [26].
GATA3 Mutational Analysis
DNA was extracted from FFPE tumor tissue sections, matched non-involved lymph nodes, leukocytes and MCF-7 and T47D cells, as described [4, 27]. DNA was utilized with 13 GATA3 exon primer pairs for PCR amplification [4] and the PCR products examined for variants using high-resolution melt curve analysis (LightScanner® System, Idaho Technology Inc., Utah, USA) [28]. GATA3 DNA sequence analysis was undertaken on those samples that had variant melt curves, and abnormalities were confirmed by restriction endonuclease digestion or competitive allele-specific PCR incorporating a FRET quencher cassette (KBiosciences Competitive Allele–Specific PCR genotyping system (KASP), KBioscience, Herts, UK) [29] using independently obtained PCR products, as described previously [4].
Cell Lines and Tissue Culture
African green monkey kidney COS7 cells, which do not endogenously express GATA3, were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA), immediately expanded and frozen such that they could be revived for use; human ER-positive breast cancer ductal carcinoma (T47D) and adenocarcinoma (MCF-7) cells were obtained from the ATCC and independently authenticated by LGC Standards (Tracking No 710081047, May 2011) and stocks frozen for subsequent use. Cells were routinely maintained in DMEM or RPMI plus 2 mm l-glutamine and 10 % fetal bovine serum.
Plasmids
Full length (FL) wild-type and ZnF1 GATA3 constructs were sub-cloned into pcDNA3.1 (GATA3-pcDNA) (Invitrogen, Carlsbad, CA, USA) and the mammalian enhanced-green-fluorescent-protein (EGFP) expression vector (pEGFP-C1, BD Biosciences Clontech, Palo Alto, CA) to yield untagged and N-terminus EGFP tagged wild-type and mutant GATA3 proteins, respectively [4]. All mutations were generated using the QuikChange™ XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA) and the DNA sequences of the constructs verified, as previously reported [3, 4]. For luciferase assays, the pGL4 firefly luciferase reporter plasmid containing a GATA3 binding site in its promoter [15] and pRL-null Vector (Promega, Madison, WI, USA) were used as described [6].
Western Blot Analysis
COS7 cells were transiently transfected in six-well plates with 200 ng plasmid DNA (GATA3-pcDNA wild-type and mutant constructs) using FuGENE®6 transfection reagent (Roche Applied Science, Indianapolis, IN, USA), as previously described [4]. Forty-eight hours after transfection, cells were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 1 % NP-40, 0.1 % SDS, 0.5 % deoxycholate, 1 mM phenylmethylsulfonyl fluoride) and supplemented with protease inhibitors (Complete Mini, Roche). Western blot analysis utilizing the monoclonal antibody, HG3-31 anti-GATA3 (Santa Cruz Biotechnology Inc.), was used to detect the presence of GATA3 protein in the cell fractions [4]. An antibody against α-tubulin (Santa Cruz Biotechnology Inc.) was used to assess the quality of the subcellular fraction preparations as described [6].
Luciferase Reporter Assays
COS7, T47D, and MCF-7 cells were transiently transfected in 24-well plates with a total of 400 ng of plasmid DNA per well using FuGENE® 6 transfection reagent (Roche Applied Science, Indianapolis, IN, USA), as previously described [6]. The 400 ng of plasmid DNA consisted of: 200 ng/well of pGL4-GATA_CS, 100 ng/well of pRL-null to allow normalization of the data; and 100 ng of plasmid encoding wild-type and/or mutant GATA3-pcDNA and, when appropriate, empty vector pcDNA3.1 to keep the amount of transfected plasmid DNA constant [6]. Cells were harvested 48 h after transfection, lysed and luciferase activity was measured using the Dual Luciferase Reporter Assay (Promega, Madison, WI, USA) and the Turner Biosystems Veritas Microplate Luminometer [6]. Three experiments were carried out in triplicate, and data are presented as mean fold change compared to GATA3 WT ± standard error of the mean (SEM) of all experiments [6].
Proliferation and Invasion Assays
The T47D and MCF-7 cells were transiently transfected with 300 ng plasmid DNA (150 ng GATA3 wild-type and 150 ng mutant construct) using Genejuice Transfection Reagent (Novagen, Darmstadt, Germany) [30], and cell proliferation assays (Celltitre 96 Aqueous One Kit G5421, Promega, Southampton, UK) and cell invasion assays performed using transwell invasion assays, as described [25, 31].
Electrophoretic Mobility Shift Assay
COS7 cells were transfected with wild-type GATA3-pcDNA or a construct harboring one of the mutations. Nuclear protein extracts were prepared and used in shift assays as described [4–6].
Nuclear Localization Studies
COS7 cells were transfected with GATA3 wild-type and mutant constructs and immunocytochemistry performed as described [4, 6].
Statistical Analysis
Mean values ± SEM were calculated and analysis performed using unpaired Student’s t test for independent samples in which the Bonferroni correction for multiple testing was applied [32]. Distributions of GATA3 mutations were analyzed by the Chi-square test using the GraphPad QuickCalcs website: http://www.graphpad.com/quickcalcs/chisquared1.cfm (accessed July 2012).
Bioinformatics and Three-Dimensional Modeling
DNA sequence changes were compared to data from the National Heart Lung and Blood Institute Exome Sequencing Project (NHBLI-ESP), which provides the exome sequences from ∼5,400 samples [33]. The evolutionary conservation of the homology of GATA factors was examined using the multiple sequence alignment program ClustalW2 [34] with amino acid sequence data obtained from the NCBI database [35]. The programs PROSITE [36], PredictProtein (http://www.predictprotein.org/), and PSORT II (http://psort.hgc.jp) were used to identify putative nuclear localization signal sites (NLSs). Pymol [37] was used to visualize the three-dimensional model of human GATA3 ZnF2.
Results
Identification of GATA3 Mutations
Nine GATA3 DNA sequence abnormalities were identified and confirmed in 40 breast cancers, all of which showed nuclear staining for ER (Table 1, Fig. 2). These DNA sequence abnormalities consisted of eight heterozygous GATA3 mutations (Table 1, Fig. 1a) and one synonymous variant (Pro191Pro, c. 573 C>T). These DNA sequence abnormalities likely represent significant mutations as they: alter evolutionary conserved residues, e.g., Arg330 (Fig. 1b); were absent in our analysis of 110 alleles from the leukocyte DNA of 55 unrelated normal individuals; and were not reported in the NHBLI-ESP exome sequence database [33], thereby indicating that they were not functionally neutral polymorphisms which would be expected to occur in >1 % of the population (Fig. 2). Moreover, GATA3 mutational analysis using matching normal lymph node DNA which was available from three patients revealed that the GATA3 mutations 991_993delAGG, 1224_1225insG, and 1224_1225insA were absent in the normal lymph node DNA (Table 1, Fig. 2), thereby demonstrating that these GATA3 mutations were somatic. In addition, the GATA3 mutations identified in the remaining five tumors are also likely to be somatic mutations as the patients were not known to have the HDR syndrome, and these mutations were not found to be present in the leukocyte DNA of 55 unrelated individuals or in the NHBLI-ESP exome sequence database [33].
Table 1.
Details of GATA3 mutations identified in breast cancers and phenotypic details
| Tumora | Tumor size (mm)b | Grade (invasion)c | Her2 statusd | Recurrencee | Mutation numberf | DNA changeg | Exon | RE/ASh | Somatic origini | Amino acid changej | Predicted effectj | GATA3 mutant localizationk | GATA3 in tumorl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 9 | 1 | NK | No | 1 | c.944_945 del GGinsAGC | 5 | RE, BmgI | – | G314 fs353X | Fs, missense 39mer peptide, premature stop at codon 353; truncated protein with loss of ZnF2 | N, C | N |
| B | 85 | 3 | − | D | 2 | c.991_993 del AGG | 5 | RE, BseRI | Yes | R330del | Loss of Arg residue within ZnF2 | N | N |
| C | 40 | 3 | + | No | 3 | c.991_992 ins TGGAGGA | 5 | RE, FokI | – | R330 fs353X | Fs, missense 23mer peptide, premature stop at codon 353; truncated protein with loss of ZnF2 | N, C | – |
| D | 14 | 2 | − | No | 4 | c.1196_1197 delGA | 6 | RE, Hpy188 III | – | R399 fs353X | Fs, missense 106mer peptide; ZnF1/ZnF2 intact but C-terminal elongated to 505 amino acids | N, C | – |
| E | 16 | 3 | + | No | 5 | c.1224_1225 insG | 6 | RE, BslI | Yes | P408 fs506X | Fs, missense 98mer peptide; ZnF1/ZnF2 intact but C-terminal elongated to 506 amino acids | N, C | N |
| F | 17 | 3 | − | No | 5 | c.1224_1225 insG | 6 | RE, BslI | – | P408 fs506X | Fs, missense 98mer peptide; ZnF1/ZnF2 intact but C-terminal elongated to 506 amino acids | N, C | N |
| G | 19 | 3 | + | D | 5 | c.1224_1225 insG | 6 | RE, BslI | – | P408 fs506X | Fs, missense 98mer peptide; ZnF1/ZnF2 intact but C-terminal elongated to 506 amino acids | N, C | – |
| H | 24 | 3 | − | L,D | 6 | c.1224_1225 insA | 6 | AS | Yes | P408 fs506X | Fs, missense 98mer peptide; ZnF1/ZnF2 intact but C-terminal elongated to 506 amino acids | N, C | N |
aTumors harboring mutations are referred to by a letter, which is used for identification (e.g., in Fig. 2)
bAll tumors were invasive ductal carcinomas, except for tumor H which was an invasive lobular carcinoma
cGrade refers to the grading system [60]; tumors A, D, E, F, G had a negative lymph node status (i.e., no lymph node involvement), whilst B, C, and H had a positive lymph node status
dHuman epidermal growth factor receptor 2 (Her2) status described as +, positive; −, negative; or NK, not known
eRecurrence described as: L, local recurrence; D, distal recurrence; no, for no recurrence
fMutations are referred to by a number, which is used for identification (e.g., in Fig. 3 and in text)
gDel, deletion; ins, insertion; all mutations occurred as heterozygous mutations (Fig. 2)
hConfirmation of mutation by restriction enzyme digest (RE) or competitive allele-specific PCR (AS)
iMutation proven to be somatic (yes); −, normal tissue (e.g., lymph node) not available
jFs, frameshift; X, Stop codon
kNuclear localization data from mutant protein overexpressed in COS7 cells: N, nuclear; C, cytoplasmic
lImmunostaining performed on tumor sections: N, nuclear; −, absent
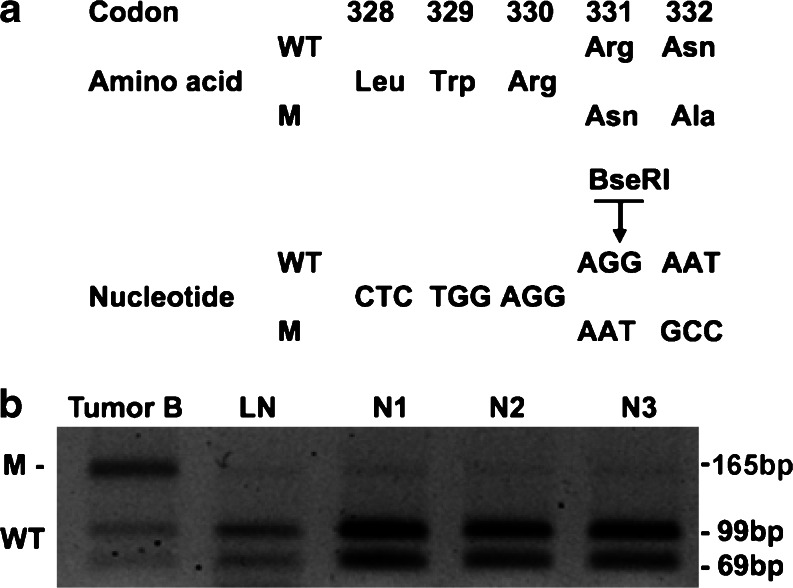
Fig. 2.
Identification of 991_993delAGG heterozygous mutation in breast tumor B (Table 1). a DNA sequence analysis revealed a heterozygous loss of AGG at codon 330, which was not present in normal (lymph node, LN) wild-type (WT) DNA from the patient. The WT DNA sequence involving codons 329 to 332 consisted of a short stretch (nucleotides 986–994) of a repeated sequence (GGA)3, which may explain the occurrence of two different mutations in tumors B and C (Table 1) at this site. The 991_993delAGG mutation resulted in the loss of a BseRI restriction endonuclease site. b PCR amplification and BseRI digestion result in two products of 69 and 99 bp from the wild-type allele, but only one product of 165 bp from the mutant allele. The tumor was heterozygous in having wild-type and mutant alleles, while the lymph node DNA from the patient was homozygous for the wild-type alleles. The absence of this 991_993delAGG mutation in 110 alleles from 55 unrelated normal European individuals (N1, N2, and N3 shown) indicated it is not a common DNA polymorphism
Four of the GATA3 mutations occurred in 5 of the 28 breast cancers that showed positive nuclear immunostaining for GATA3, whilst the remaining 3 GATA3 mutations were found in 3 of the 12 tumors that were negative for GATA3 (Table 1). These data suggest that absence of GATA3 immunostaining is not a reliable predictor for the presence of a GATA3 mutation. No relationship with standard clinicopathological factors or outcome was identified in this small cohort, with all eight patients alive at a median follow-up of 62 months (Table 1). However, GATA3 mutations were significantly associated with positive Her2 status, when compared to the entire series (p < 0.05), placing these tumors in the Luminal B category (Table 1).
All of the eight somatic GATA3 mutations associated with breast cancer are located within exons 5 and 6, consistent with the locations of the previously reported GATA3 mutations in breast cancer as 61 of the total 70 (i.e., 87 %) were also located within this region (Fig. 1a) [7–13]. This difference in the distribution of the somatic GATA3 mutations associated with breast cancer and the germline GATA3 mutations associated with the HDR syndrome, which was found to be statistically significant (p < 0.0001), may have functional consequences as the breast cancer GATA3 mutations are clustered around ZnF2 and the C-terminal domain, whereas the HDR GATA3 mutations are widespread (Fig. 1a). Furthermore, two of the GATA3 mutations (mutations 2 and 3 in tumors B and C) involve the residue Arg330, and four of the GATA3 mutations (mutations 5 and 6 in tumors E-H) involve the residue Ser408, which has also been recently reported in other ER-positive breast cancers [10–12], thereby suggesting that the DNA sequence encoding these residues may be more prone to mutations.
The six different GATA3 somatic mutations, associated with breast cancer (Fig. 1a), predict structurally significant changes (Table 1). Thus, the frameshifting insertion (991_992insTGGAGGA) and deletion–insertion (944_945delGGinsAGC) are predicted, if translated, to have truncated GATA3 proteins that lack part or all of ZnF2, respectively; these mutations are likely to result in a loss of DNA binding, as has been demonstrated for other such GATA3 mutations associated with the HDR syndrome [4]. However, the effects of the inframe deletion (991_993delAGG), which results in the loss of an evolutionary conserved arginine residue in ZnF2, and the frameshifting deletion (1196_1197delGA) and insertions (1224_1225insG and 1224_1225insA), which likely result in elongated missense proteins, are more difficult to predict, and these together with the two truncating mutations of ZnF2 were investigated further.
Transactivation, Cell Proliferation, and Cell Invasion Studies
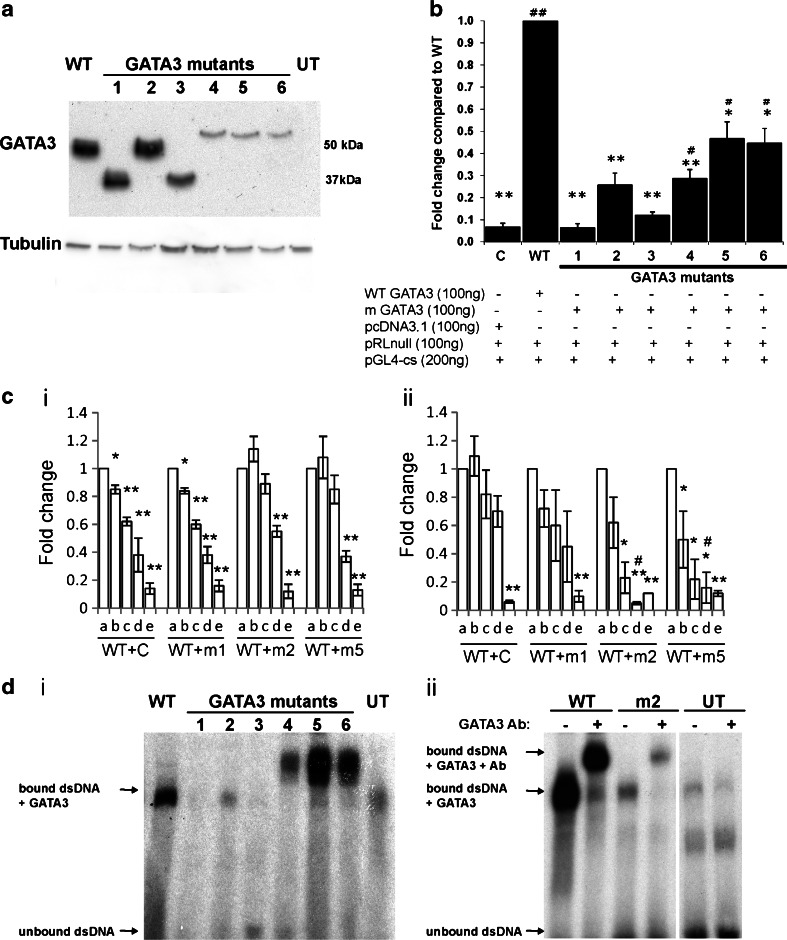
The effects of the GATA3 mutations on expression of GATA3 protein, within COS7 cells, were assessed using Western blot analysis (Fig. 3a). This demonstrated that the GATA3 wild-type and mutant 330delAGG were both 49 kDa in size and equally expressed; the 314delGGinsAGC and 330insTGGAGGA GATA3 mutants were smaller than the wild type (∼40 kDa) with either equal or reduced expression, respectively; and the 399delGA, 408insG, and 408insA GATA3 mutants, which were all larger (∼55 kDa) than the wild-type protein, had markedly reduced expressions. This suggests that mutant proteins 399delGA, 408insG, and 4080insA may all be less stable, and thus less abundant than wild-type proteins in cells. The consequences of the GATA3 mutants were further assessed by their effects on gene transactivation using luciferase reporter assays in COS7, T47D, and MCF-7 cells (Fig. 3b, c).These cells were chosen as they have differences in GATA3 expression, thereby facilitating investigation of the GATA3 mutants in different cellular environments. Thus, COS7 cells do not endogenously express GATA3, MCF-7 cells harbor a confirmed heterozygous GATA3 mutation, D336fs [7], and T47D cells do not harbor GATA3 mutations (data not shown). Furthermore, MCF-7 and T47D are ER-positive breast cancer cells and are therefore representative of cells in which the GATA3 mutations were detected. Expression of wild-type GATA3, in COS7 cells (Fig. 3b), resulted in a significant increase by 14-fold in relative luciferase activity, when compared with that of the empty pcDNA3.1 expression vector. The relative activities of the GATA3 mutants were compared to that of the wild-type GATA3 and found to be significantly reduced (p < 0.05). Indeed, the reduction in transactivation by the GATA3 mutants 944_945delGGinsAGC, 991_993delAGG, and 991_992insTGGAGGA was not significantly different to the empty vector negative control (p > 0.05), whereas the GATA3 mutants 1196_1197delGA, 1224_1225insG, and 1224_1225insA did have increased transactivation activity when compared to the empty pcDNA3.1 expression vector negative control (p < 0.05). These combined results indicate that the GATA3 mutants could be broadly divided into three classes: (1) 944_945delGGinsAGC and 991_992insTGGAGGA which resulted in truncated proteins with a complete loss of transactivation activity; (2) 991_993delAGG which resulted in an in-frame deletion and a loss of transactivation activity; and (3) 1196_1197delGA, 1224_1225insG, and 1224_1225insA which resulted in elongated proteins with a partial loss of transactivation activity (Fig. 3b). One GATA3 mutant, representative of each class, and comprising mutants 1, 2, and 5, was therefore selected for further study in the ER-positive breast cancer cell lines (Fig. 3c). This revealed that expression of wild-type GATA3, in the presence of endogenous GATA3, in the T47D and MCF-7 cells resulted in a significant increase by 7- and 16-fold, respectively, in relative luciferase activity when compared to the empty pcDNA3.1 expression vector. However, the relative activity of the GATA3 mutants when transfected alone was found to be significantly reduced when compared to that of the wild-type GATA3 in both types of cells (Fig. 3c). This reduction of transactivation activity by each of the GATA3 mutants was dose and cell dependent. Thus, co-transfection of wild-type and mutant GATA3 in T47D cells resulted in reduction of transactivation activity with low doses of 944_945delGGinsAGC, whereas 991_993delAGG and 1224_1225insG required higher doses to suppress transactivation activity (Fig. 3ci). In contrast, in MCF-7 cells that were co-transfected with wild-type and mutant GATA3, reduction of transactivation activity was observed with low doses of 991_993delAGG and 1224_1225insG, and only with high doses of 944_945delGGinsAGC (Fig. 3cii). These results indicated that the GATA3 mutants have different effects, which also vary in different cell types, and that the mutant GATA3 exerts a dominant negative action on wild-type GATA3, consistent with the observed heterozygous mutations in breast cancers (Fig. 2, Table 1).
Fig. 3.
Characterization of GATA3 mutants. a Western blot analysis of cell lysates from COS7 cells transiently transfected with wild-type (WT) and mutant GATA3 constructs to detect expression of GATA3 proteins using the HG3-31 antibody. Untransfected (UT) cells were used as controls, and Western blots with anti-α-tubulin were used to demonstrate equal loading of protein. The number of the GATA3 mutants, in panels a–d, refers to the mutations detailed in Table 1. GATA3 transactivation of reporter gene analyzed by luciferase reporter assays in (b) COS7 cells and (c). Breast cancer cells (i) T47D and (ii) MCF-7. WT and/or mutant GATA3 constructs and a pGL4-cs reporter vector containing a known GATA3 binding site were co-transfected into cells. For c, a–e refer to the relative dosage of wild-type (WT) and mutant (m1, m2, or m5) GATA3 construct, or pcDNA3.1 (C), to make a total of 100 ng plasmid: a 100 ng WT + 0 ng m (or 0 ng C); b 75 ng WT + 25 ng m (or 25 ng C); c 50 ng WT + 50 ng m (or 50 ng C); d 25 ng WT + 75 ng m (or 75 ng C); and e 0 ng WT + 100 ng m (or 100 ng C). Each experiment was carried out three times in triplicate. Data are shown as fold change relative to GATA3 wild-type ± standard error of the mean (n = 9), and p values calculated using the Student’s t test are for the fold change relative to WT GATA3 and pcDNA3.1 negative control (C). *p < 0.05 compared to WT GATA3, **p < 0.01 compared to WT GATA3, # p < 0.05 compared to pcDNA3.1, and ## p < 0.001 compared to pcDNA3.1. d Analysis of DNA-binding properties of GATA3 mutant proteins. (i) DNA binding of wild-type and mutant GATA3 proteins was assessed using EMSAs, where nuclear extracts were incubated with a radiolabelled (32P) double-stranded oligonucleotide containing the GATA3 consensus sequence. Equal amounts of nuclear lysate were loaded. Control binding reaction used untransfected cells. (ii) Supershift EMSA with wild-type (WT) and mutant GATA3 991_993delAGG (m2), and untransfected (UT) cell lysates where nuclear extracts were incubated with a radiolabelled (32P) double-stranded oligonucleotide containing the GATA3 consensus sequence, and with (+) or without (−) GATA3 antibody to test the specificity of the binding
The effects of these reductions in transactivation activity resulting from the GATA3 mutants on cellular proliferation and invasion were also investigated. Co-transfection of each of the three GATA3 mutants (944_945delGGinsAGC, 991_993delAGG, and 1224_1225insG) with wild-type GATA3 into T47D and MCF-7 cells did not reveal significant effects on cellular proliferation, when compared to transfection with wild-type GATA3 alone (Table 2). This is consistent with the study in the ER-negative, GATA3-negative breast cancer cell line MDA-MB-231 that reports that there is no significant difference in cell proliferation between control and GATA3-expressing cells [38]. However, co-transfection in T47D cells of wild-type GATA3 and GATA3 mutants 944_945delGGinsAGC and 1224_1225insG, but not 991_993delAGG, resulted in significant increases in invasion when compared to transfection with control vector only, although co-transfection of all three GATA3 mutants had significantly lower invasion when compared to transfection with wild-type GATA3 alone (Table 2). In contrast, transfection in MCF-7 cells revealed that only 991_993delAGG had a significantly higher invasion when compared to transfection with either wild-type GATA3 or control vector only; the invasion following co-transfection with wild-type and 944_945delGGinsAGC or 1224_1225insG did not differ significantly from that of wild-type GATA3 or control vector only (Table 2). These results of cell invasion are consistent with those of transactivation activity (Fig. 3c), in showing that the GATA3 mutants have different effects, which also vary in different cell types, and that the mutant GATA3 exerts a dominant negative action on wild-type GATA3.
Table 2.
Proliferation and cell invasion assays in T47D and MCF-7 cells
| Proliferationa | Invasiona | |||
|---|---|---|---|---|
| T47Db | MCF-7b | T47D | MCF-7 | |
| Control c | 1.00 ± 0.00 (n = 15) | 1.00 ± 0.00 (n = 17) | 1.00 ± 0.00 (n = 15) | 1.00 ± 0.00 (n = 9) |
| WT | 0.99 ± 0.00 (n = 15) | 0.98 ± 0.03 (n = 17) | 2.43 ± 0.06** (n = 15) | 1.05 ± 0.09 (n = 9) |
| WT + m1 | 1.03 ± 0.01 (n = 15) | 0.99 ± 0.01 (n = 17) | 1.60 ± 0.07*, ** (n = 15) | 0.88 ± 0.14 (n = 9) |
| WT + m2 | 1.02 ± 0.01 (n = 15) | 1.01 ± 0.02 (n = 17) | 0.85 ± 0.04* (n = 15) | 2.25 ± 0.10*, ** (n = 9) |
| WT + m5 | 1.03 ± 0.02 (n = 15) | 1.03 ± 0.04 (n = 17) | 2.14 ± 0.04*, ** (n = 15) | 1.07 ± 0.14 (n = 9) |
*p < 0.01 compared to wild-type GATA3; **p < 0.005 compared to control
aProliferation and invasion for WT and mutant GATA3 constructs were expressed as fold change with respect to control transfection with pcDNA3.1 alone. Mean ± standard error of the mean (n = number of experiments) are shown
bThe T47D and MCF-7 are ER-positive breast cancer cell lines; the MCF-7 cells have a GATA3 mutation, D336fs, whereas the T47D cells do not have any GATA3 abnormalities
cControl DNA (pcDNA3.1), WT – wild-type GATA3, and mutant GATA3 ( m1, m2, and m5 mutations—Table 1) constructs
DNA Binding and Subcellular Localization Studies
To establish the cause of the loss of transactivational activity (Fig. 3b, c), the six different GATA3 mutations were initially assessed for altered DNA binding by electrophoretic mobility shift assay (EMSAs; Fig. 3d). This revealed that the GATA3 mutants 944_945delGGinsAGC and 991_992insTGGAGGA had a loss of DNA binding, but 991_993delAGG, 1196_1197delGA, 1224_1225insG, and 1224_1225insA had retained DNA binding (Fig. 3di). However, the DNA binding by the mutant 991_993delAGG appeared to be reduced, and the specificity of this was therefore confirmed by a supershift EMSA (Fig. 3dii) which confirmed the presence of GATA3 in the protein–DNA complex. The partial or full loss of DNA binding by the GATA3 mutants 944_945delGGinsAGC, 991_993delAGG, and 991_992insTGGAGGA provides an explanation for the decrease of transactivational activity. However, 1196_1197delGA, 1224_1225insG, and 1224_1225insA had retained DNA binding, so we explored, by immunofluorescence studies, the possibility that these mutants may not be fully localized to the nucleus as an explanation for the observed reduced transactivation.
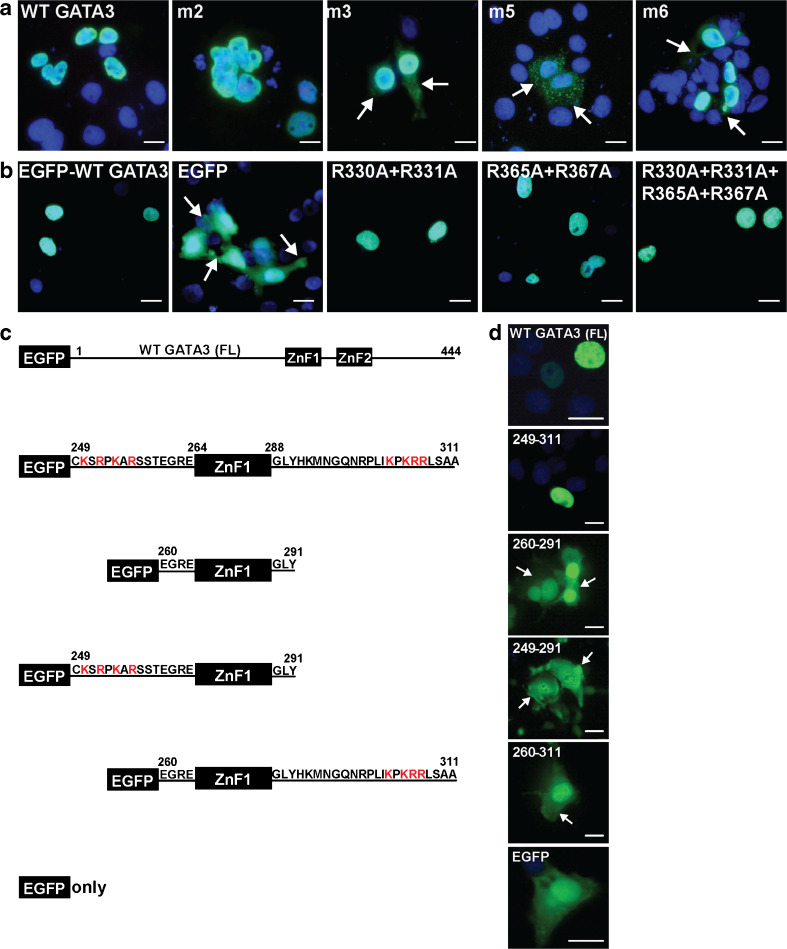
Immunofluorescence studies demonstrated that the GATA3 wild-type and the mutant 944_945delGGinsAGC and 991_993delAGG proteins localized to the nucleus (Fig. 4a), whereas the GATA3 mutant proteins 991_992insTGGAGGA, 1196_1197delGA, 1224_1225insG, and 1224_1225insA did not localize fully to the nucleus but instead located to the cytoplasm with a punctate expression pattern. These results indicate that GATA3 mutations involving residues 331–408, which encompass part of ZnF2 and the C-terminal domain (Fig 1b), disrupt nuclear localization, and this suggests that these residues may encode for a NLS. We therefore pursued studies to define the NLS of GATA3.
Fig. 4.
Immunofluorescence studies of GATA3 breast cancer mutants and engineered mutants of the putative NLSs. a Subcellular localization of wild-type (WT) and mutant GATA3 proteins identified in breast cancer (number refers to mutation detailed in Table 1) transfected in COS7 cells. The WT and GATA3 mutant 2 localized to the nucleus, whereas the GATA3 mutants 3, 4, and 5 localized to the nucleus and cytoplasm. Scale bar is 5 μm. b Subcellular localization of WT and engineered mutants using GATA3-EGFP constructs transfected into COS7 cells. Wild-type (WT), EGFP alone, and mutant GATA3 proteins: R330A + R331A, R356A + R367A, and R330A + R331A + R365A + R367A were investigated. Use of the GATA3-EGFP constructs revealed that EGFP was evenly distributed in the nucleus and cytoplasm (arrows), whereas full-length WT-GATA3 co-localized to the nucleus. In addition, all mutant GATA3-EGFP proteins co-localized to the nucleus, indicating that the ZnF2 residues R330, R331, R365, and R367 do not form an NLS. Scale bar is 5 μm. c Schematic representation of wild-type (WT) full-length GATA3 and partial GATA3 constructs of residues 249–311 encompassing ZnF1. The WT GATA3 (FL) and partial constructs were cloned downstream of the EGFP gene. Basic residues, arginine (R) and lysine (K) which represent critical residues in potential NLSs, are highlighted in red. d Subcellular localization of WT GATA3 (FL) and partial constructs in transfected COS7 cells, visualized by fluorescence microscopy. WT GATA3 (FL) localized to the nucleus, while EGFP-alone was evenly distributed in the nucleus and cytoplasm. GATA3 residues 249 to 311 which contained ZnF1 and the flanking basic residues co-localized to the nucleus. However, GATA3-ZnF1 alone (residues 260–291) and engineered proteins lacking the ZnF1 flanking N-terminal (residues 260–311) or C-terminal (residues 249–291) resulted in a loss of nuclear accumulation of EGFP (arrows) thereby confirming that the ZnF1 upstream and downstream sequences contain critical residues for nuclear localization. Scale bar is 5 μm
Defining the GATA3 Nuclear Localization Signal
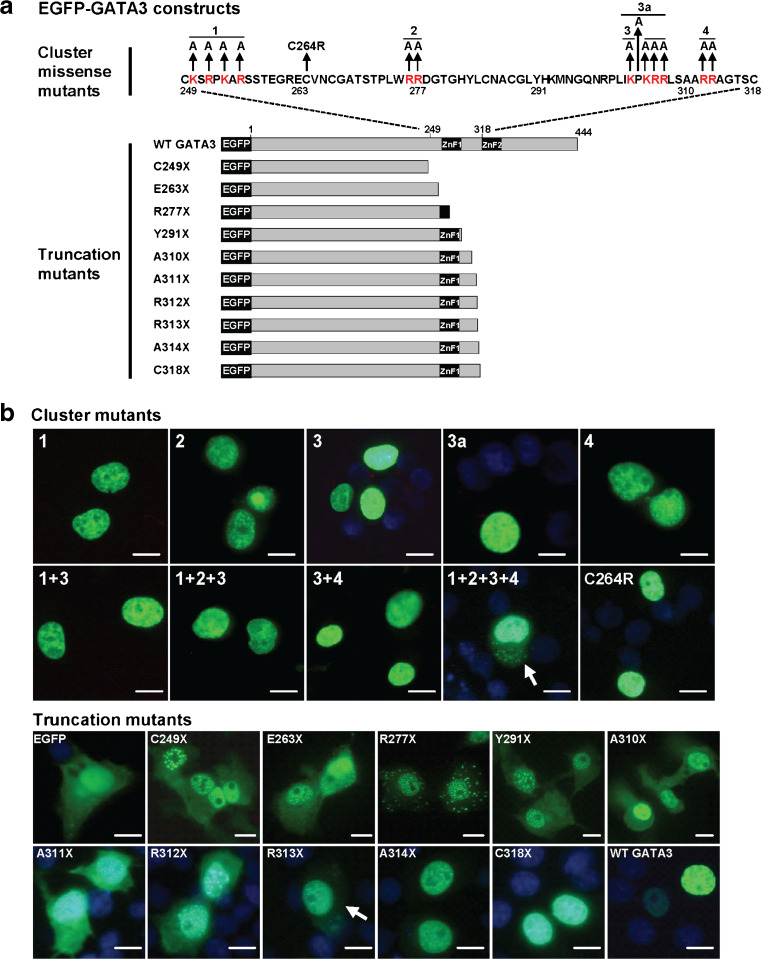
The NLS of GATA4 has been established, using EGFP-tagged constructs, to comprise the four arginine residues Arg282, Arg283, Arg317, and Arg319, which are equivalent to GATA3 residues Arg330, Arg331, Arg365, and Arg367 in ZnF2 and its C-terminal region, a region that is conserved in all GATA family members (Fig. 1b) [39]. Moreover, as the GATA3 991_992insTGGAGGA (Arg330fs) mutation resulted in a loss of nuclear localization, but the 991_993delAGG (Arg330del) mutation retained nuclear localization, we decided to first investigate these ZnF2 residues and the possibility that GATA3 has a similar NLS to the NLS in GATA4 [39]. For consistency, we used EGFP-tagged GATA3 protein, so that our studies were comparable. However, simultaneous mutation of these four equivalent ZnF2 residues, involving Arg330, Arg331, Arg365, and Arg367 in GATA3 (Arg330Ala + Arg331Ala + Arg365Ala + Arg367Ala), did not disrupt the nuclear localization of EGFP-GATA3 fusion protein which was similar to that of the wild-type (WT) EGFP-GATA3 (Fig. 4b). Thus, these results indicate that the GATA3 NLS does not involve the equivalent ZnF2 residues that form the GATA4 NLS site. These findings are also in agreement with the observation that the breast tumor mutation 991_993delAGG (Arg330del) did not disrupt nuclear localization (Fig. 4a). However, the GATA3 residues 249 to 311, which form ZnF1 and its N-terminal and C-terminal flanking sequences (Fig. 1b), were shown to be required for nuclear localization (Fig. 4b), thereby confirming findings of a previous study [40]. In addition, an HDR-associated mutation in the zinc-chelating cysteine, Cys264Arg, failed to disrupt the nuclear localization of the mutant EGFP-GATA3 (Fig. 5a, b), thereby suggesting that that GATA3 NLS is intrinsic to the amino acid sequence rather than the tertiary structure of ZnF1 or its ability to bind DNA. Analysis of the amino acid sequence of the GATA3 region 249 to 311 by protein domain prediction programs identified putative monopartite NLSs on either side of ZnF1. Thus, four clusters of positively charged amino acids (similar to the classical monopartite NLS) were identified (Fig. 5a), and these comprised: cluster 1 at position 250–256, N-terminal to the ZnF1, consisting of Lys-Ser-Arg-Pro-Lys-Ala-Arg residues; cluster 2 consisting of two adjacent residues Arg276 and Arg277 within ZnF1; cluster 3 at position 303–307, C-terminal to ZnF1, consisting of Lys-Pro-Lys-Arg-Arg residues; and cluster 4 consisting of two adjacent residues Arg312 and Arg313. The basic residues C-terminal to ZnF1 showed >95 % alignment between all GATA family members, while the basic residues N-terminal to ZnF1 are present in GATA 1–3 but not in GATA 4–6 (Fig. 1b). The importance of these clusters of basic amino acid sequences for nuclear import of GATA3 was assessed by introducing different combinations of point mutations, whereby each of the 12 positively charged arginine (Arg) and lysine (Lys) residues were altered to a neutral alanine (Ala) residue. Mutation of each of these four individual clusters did not disrupt nuclear targeting of EGFP-GATA3 (Fig. 5b). Cluster 3 has a Pro304, and as proline residues may be important for nuclear targeting [41], we mutated Pro304 together with basic residues within this cluster; these mutations also did not disrupt the nuclear localization of EGFP-GATA3 (Fig. 5b). Likewise, simultaneous mutations of clusters 1 and 3 (1 + 3, Fig. 5b); clusters 1, 2, and 3 (1 + 2 + 3, Fig. 5b); or clusters 3 and 4 (3 + 4, Fig. 5b) did not disrupt EGFP-GATA3 nuclear localization. However, combined mutations of all four clusters disrupted nuclear localization of the mutant EGFP-GATA3, whose cytoplasmic and nuclear distribution was similar to that of EGFP alone (1 + 2 + 3 + 4, Fig. 5b). These results indicate that the N-terminal and C-terminal sequences flanking ZnF1 are required for nuclear localization.
Fig. 5.
Nuclear localization of wild-type, missense, and truncation mutants of GATA3. a Schematic representation of wild-type (WT) and mutant missense and truncation GATA3 constructs. Four clusters (1–4) of basic positively charged amino acids which have similarities to the classical monopartite NLS were identified, and the arginine (R) and lysine (K) residues were altered to the neutral alanine (A). The HDR-associated missense mutation C264R was also studied. Truncation mutants were designed to facilitate analysis of ZnF1 and its N-terminal and C-terminal flanking sequence. b Subcellular localization of WT GATA3, missense and truncation GATA3 mutants visualized by fluorescence microscopy in transfected COS7 cells. Missense mutations of the four clusters either individually or in combinations (1 + 3, 1 + 2 + 3, or 3 + 4) and the C264R GATA3 mutation did not disrupt nuclear localization, whereas combined mutations of the four clusters (1 + 2 + 3 + 4), shown by arrow, disrupted nuclear localization. The truncation GATA3 mutants A314X and C318X localized to the nucleus, whereas all of the others were found to have nuclear and cytoplasmic localizations and similar to that of the EGFP alone. Scale bar is 5 μm
The requirement of an intact GATA3 ZnF1 and its flanking sequences was further explored by a series of truncation mutations (Fig. 5b). All truncation GATA3 mutations that lacked ZnF1 (Cys249Stop, Glu263Stop, and Arg277Stop) or the C-terminal flanking sequence up to residue Ala311 (Tyr291Stop, Ala310Stop, Ala311Stop) resulted in a loss of nuclear localization (Fig. 5b), whereas Ala314Stop did not disrupt nuclear localization (Fig. 5b), consistent with the results from the mutant 314delGA. Thus, the inclusion of Arg312 and Arg313 of cluster 4 (Fig. 5a), which conform to the classical NLS requirement for short stretches of basic residues, restored nuclear localization. However, missense mutations of these cluster 4 arginine residues (Arg312Ala + Arg313Ala) did not disrupt nuclear localization (Fig. 5b), thereby indicating that the other NLSs (e.g., basic amino acids in clusters 1, 2, and 3) contribute to targeting GATA3 protein to the nucleus and can compensate for the loss of Arg312 and Arg313.
Discussion
Our study has identified six different heterozygous GATA3 somatic mutations in 8 of 40 (20 %) of ER-positive breast cancers (Table 1). These consisted of an in-frame deletion of a key arginine residue (991_993delAGG) in ZnF2 (Fig. 2); a deletion/insertion (944_945delGGinsAGC); a seven-nucleotide insertion (991_992insTGGAGGA), leading to loss of ZnF2 and the C-terminal domain; a deletion (1196_1197delGA); and two single nucleotide insertions (1224_1225insG in three tumors, and 1224_1225insA) that have intact ZnF1 and ZnF2 domains, but elongated missense peptides. It is important to note that the presence (or absence) of GATA3 mutations in the breast cancer could not be predicted by the results of GATA3 immunostaining (Table 1). Thus, five of the eight (i.e., >60 %) breast cancers with GATA3 mutations showed immunostaining for GATA3 (Table 1). GATA3 immunostaining in these tumors may be due to expression of normal GATA3 protein from the wild-type allele, and/or the detection of mutant GATA3 protein by the GATA3 antibody. Four of these six different mutations are novel, with the remaining two mutations (1224_1225insA and 1224_1225insG) having recently been reported by exome capture sequencing studies [10–12]. In total, 53 novel GATA3 mutations have been previously reported in 70 ER-positive breast tumors comprising: 2 nonsense, 2 inframe insertion, 11 deletions leading to frameshifts, 27 insertions leading to frameshifts, 5 acceptor splice site mutations, 1 donor site mutation, and 5 missense mutations (Fig. 1a, [7–13]). The majority (>95 %) of these GATA3 mutations have been reported in luminal A or B ER+ breast cancers [8, 10, 11, 13] (Fig. 6), and 58 % of these are found in luminal A tumors and 39 % in luminal B tumors [8, 10, 11, 13], consistent with our findings which found 57 % of GATA3 mutations in luminal A tumors and 43 % in luminal B tumors. Luminal A tumors are associated with a very favorable and better prognosis than luminal B tumors (Fig. 6), and the contributions made by the presence or absence of GATA3 mutations to the prognosis remain to be elucidated.
Fig. 6.
Proportion of breast cancers defined by expression of the estrogen receptor (ER), human epidermal growth factor 2 (Her2) and GATA3, together with reported occurrence of GATA3 mutations and prognosis [8, 10, 11, 13]. Approximately 60 % of breast cancers express ER (ER+), while the remaining 40 % do not express ER (ER−), and 30 % express Her2 (Her2+), while the remaining 70 % do not express Her2 (Her2−) [23]. Breast cancers that are ER+/HER2−, ER+/Her2+, ER−/Her2+ and ER−/Her2− are referred to as luminal A, luminal B, Her2-enriched and basal subtypes, respectively [24]; and these tumor subtypes comprise 40, 20, 10, and 30 % of breast cancers, respectively [23]. GATA3 expression, as determined by immunostaining, is highest in luminal A tumors and lowest in Her2-enriched and basal tumors [23], and luminal A tumors have been reported to be associated with a very favorable prognosis, whereas the Her2-enriched and basal tumors are associated with increasingly unfavorable prognosis [23]. The occurrence of GATA3 mutations from 31 tumors (7 tumors from this study and 24 (indicated in parenthesis) from other studies [8, 10, 11, 13]) that report the breast cancer subtypes are shown. Thus, 58 % of the GATA3 mutations are found in luminal A tumors, 39 % in luminal B, 0 % in Her2-enriched tumors, and 3 % in basal tumors
The functional consequences of these 53 GATA3 mutations have not been investigated, and thus our study represents the first to investigate the effects of breast cancer GATA3 mutations on transcriptional activity, proliferation, and invasion. Of these reported 53 different GATA3 mutations, 48 (>90 %) fall into one of our three functionally defined classes (Fig. 3b) with 23 mutants (>45 %) belonging to class 1, 3 mutants (∼5 %) belonging to class 2, and 20 mutants (∼40 %) belonging to class 3, thereby illustrating the utility and generalizability of this classification. The functional effects of the remaining five mutations (<10 %), which consist of 1 in the 5′ untranslated region, three missense mutations in the C-terminal domain, and one in-frame insertion in the C-terminal domain, are more difficult to predict and require functional characterization using similar approaches to ones utilized by our study.
Our investigation of the functional consequences of the GATA3 mutations demonstrated that the GATA3 mutant proteins may result in deleterious effects by at least three mechanisms which include: an absence of nuclear localization, a lack of DNA binding, and a reduction in transactivation. Thus, the truncated GATA3 proteins 944_945delGGinsAGC and 991_992insTGGAGGA, which resulted in a loss of ZnF2 (Fig. 1c), were associated with an absence of DNA binding that led to a marked reduction (i.e., >85 %) of GATA3 transcriptional activity (Fig. 3b). The GATA3 mutant 991_993delAGG (Arg330del in ZnF2) did not affect nuclear localization (Fig. 4a) but instead significantly reduced DNA binding (Fig. 3d) and transactivational activity by ∼75 % (Fig. 3b, c). These findings are consistent with the results of three-dimensional modeling (Fig. 7) which revealed that in the GATA3 protein-bound DNA complex, the 330Arg residue is directly involved in DNA binding because of its location in the groove of the DNA. Finally, the GATA3 mutants 1196_1197delGA, 1224_1225insG, and 1224_1225insA, which result in mutant GATA3 proteins with intact ZnF2 domains and C-terminal basic domains but with an elongated missense C-terminal peptide (Fig. 3a), were able to bind to DNA (Fig. 3d), although this did not restore transactivational activity, which remained reduced by 55–70 % (Fig. 3b), and is likely due to decreased protein expression (Fig. 3a) and incomplete nuclear localization of the mutant GATA3 protein (Fig. 4a). This suggests that the mutant elongated missense tail may contain motifs that increase the sensitivity of the translated protein to endogenous proteases, thereby leading to retention in the proteasome, or that it may interfere with nuclear localization by disrupting interactions between GATA3 and importin, without affecting the residues that form part of the NLS. This mechanism, which would result in the observed decrease in transactivation (Fig. 3b), has similarities to that previously described for the HDR-associated GATA3 mutation, 407insC [5], and the engineered GATA1 mutants [42]. Finally, our studies reveal that GATA3 mutations may exert dominant effects on the ability of wild-type GATA3 to alter transactivation activity (Fig. 3b, c) and cellular invasiveness (Table 2), consistent with the report that GATA3 forms dimers [43].
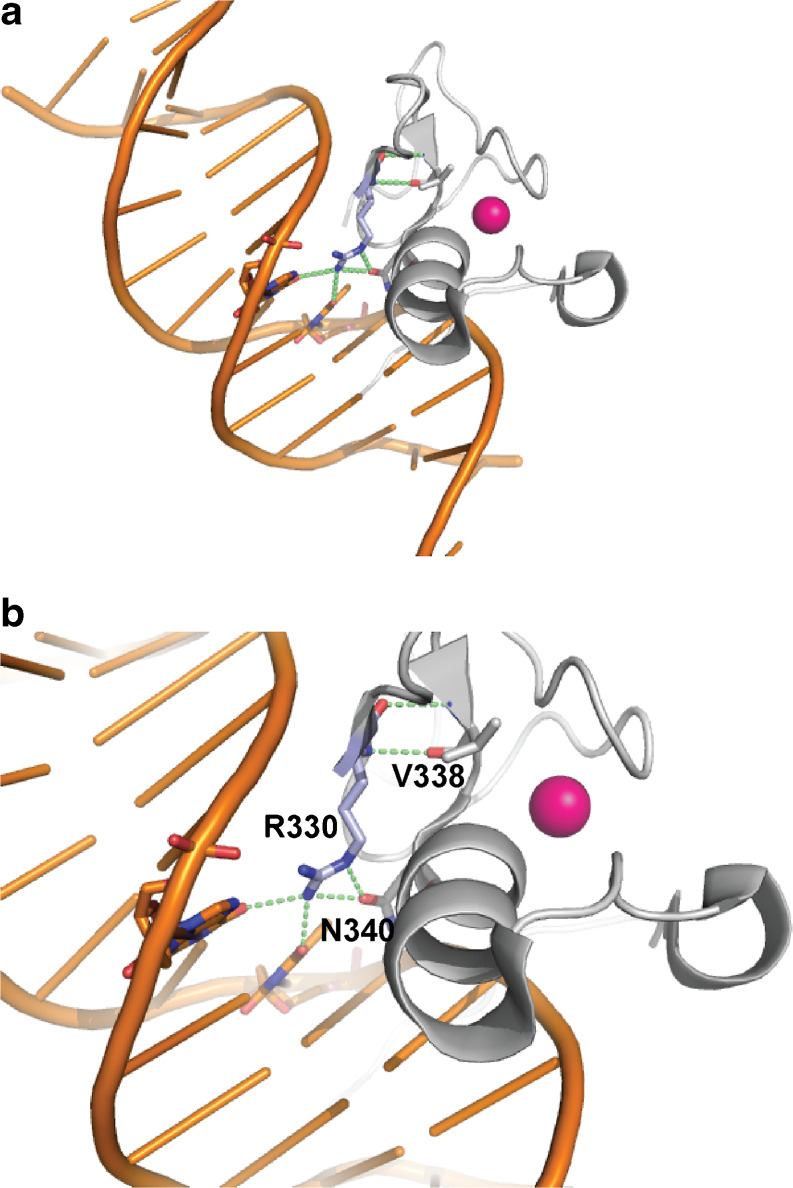
Fig. 7.
Three-dimensional structure of the human GATA3 ZnF2 based on the crystal structure of murine GATA3. The three-dimensional structure of murine Gata3 ZnF2 (residues 308–370) has been characterized, and this has 100 % identity to the human GATA3 ZnF2, thereby enabling its use to construct a three-dimensional model of human GATA3 ZnF2 (residues 309–371). a Cartoon model of GATA3 ZnF2 in complex with DNA, showing the protein (light grey), DNA double-helix (orange), and the coordinated Zn2+ atom (pink sphere). The residue Arg330 is shown in stick presentation colored by element (carbon, pale blue; oxygen, red; nitrogen, blue) and polar side-chain contacts (broken green lines) made by Arg330. b Enlarged view of the polar side-chain contacts made by Arg330 (R330). Arg330 is predicted to interact directly with DNA, forming hydrogen bonds with two DNA bases. In addition, Arg330 is also predicted to form hydrogen bonds to Val338 (V338) and Asn340 (N340), suggesting an additional role in the stabilization of the ZnF structure
The observed differences in cell invasion in the T47D and MCF-7 cell lines are consistent with those of a previous study reporting that GATA3 targets expression of the putative tumor suppressor caspase-14 in MCF-7 cells, but not T47D cells, and thereby negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells [44]. Such differences between these two breast cancer cell lines may be due to: the presence of a heterozygous GATA3 mutation (D336fs) in the MCF-7 cell line [7], and absence of GATA3 mutations in the T47D cell line; and variations in expression of other proteins such as E-cadherin, transforming growth factor beta (TGF-β) receptor II, and matrix-metalloproteinases that impact on cell invasion [45]. Thus, the T47D cells, which express GATA3 and have a reported increased expression of E-cadherin [46], showed increased invasion when transfected with wild-type GATA3 (Table 2), whereas the MCF-7 cells, which have reduced levels of E-cadherin compared to T47D [46], did not have increased cell invasion when transfected with wild-type GATA3. In addition, MCF-7 cells, but not T47D cells, express the TGF-β receptor II [47], thereby enabling the MCF-7 cells to respond to TGF-β, which induces the epithelial-to-mesenchymal transition of MCF-7 cells. Finally, the correlation in MCF-7 cells, but not T47D cells, between GATA3 expression and activated caspase-14 expression, which is a potent inducer of differentiation in mouse mammary epithelial cells and keratinocytes [44], further emphasizes the observed differences between these two breast cancer cells. Thus, these findings illustrate the complexity of utilizing these cell lines because of their differing expression of critical oncogenic proteins, putative tumor suppressors and receptors, in defining the specific effects of wild-type and mutant proteins in breast cancer.
The four GATA3 mutations 991_992insTGGAGGA, 1196_1197delGA, 1224_1225insG, and 1224_1225insA lead to abnormalities of nuclear localization of the mutant GATA3 proteins, and we therefore sought to define the NLS of GATA3. Our results show that the NLS of GATA3, which encompasses the ZnF1 region, differs from that of the NLS of GATA4 which encompasses ZnF2 [39], as mutations of the four critical arginine residues in ZnF2 and its C-terminal region (Fig. 4c, d) and truncating the GATA3 protein at Ala314, i.e., before ZnF2, did not disrupt nuclear accumulation (Fig. 5b). Such differences between GATA3 and GATA4 would be consistent with the classification of GATA family members into two sub-families, in which GATA1-3 form one sub-family, and GATA4-6 form another subfamily [48, 49]. Furthermore, our results have revealed that the NLS of GATA3 is spread throughout the amino acids of ZnF1 and its N-terminal and C-terminal flanking sequence (Fig. 1b), and is therefore unlike many other nuclear proteins which contain a classical NLS consisting of a short stretch of positive amino acids (Arg and Lys) [50]. Although ZnF1 and its flanking regions contain four clusters of positively charged amino acids (Fig. 1b and Fig. 5a ), which have features consistent with NLSs, it is important to note that none of these are able to function as an NLS on its own, and that it required combined mutations in all four clusters to prevent nuclear targeting (Fig. 5b). Moreover, while residues 249–311 may be sufficient for partial nuclear localization, the presence of Arg312 and Arg313 in the GATA3 protein is nevertheless required for complete nuclear accumulation to occur (Fig. 5b). Thus, multiple NLSs are required to direct nuclear accumulation of GATA3, and this is analogous to the situation for p53, which contains three NLSs which act independently and in an additive manner, such that deletion of all three is required to abolish nuclear localization, whereas deletion of any one of them results in a partial loss of nuclear accumulation [51].
Our finding of two GATA3 mutations involving the residue Arg330 and 4 GATA3 mutations involving the residue Ser408 in breast cancers from unrelated patients (Table 1) suggests that the DNA sequence encoding these residues may be more prone to mutations. Furthermore, an analysis of other GATA3 mutations reported in 70 breast cancers [7–13] also reveals that mutation may frequently involve these residues; for example, mutation of Ser408 has been reported in six tumors [10–12], and mutation of other residues such as Met294 has been reported in four tumors [10, 12]; Asp336 has been reported in two tumors [7, 10]; and mutations of the exon 4, 5, and 6 acceptor splice sites have been reported in four, six, and three tumors, respectively [7, 8, 10, 12]. Some of these recurrent mutations (Met294, Arg330, and Asp336) involve repetitive DNA sequences and a likely replication slippage model. Indeed, the DNA sequence in the vicinity of Arg330 (codons 229–332) contains a repeated sequence (GGA)3 (Fig. 2) all of which may lead to mispairing and DNA polymerase slippage [52] and thereby explain the occurrence of multiple independent mutations, as well as a frameshift deletion at Arg331 [12] and two reported synonymous changes at Arg229 and Arg330 [12, 33]. A similar replication slippage may account for the Met294 and Asp336 mutations as the 5′ DNA sequence contains repetitive sequences consisting of (A)4 and (G)4, respectively. Furthermore, examination of the 20 bp in the vicinity of the acceptor splice sites of exons 4, 5, and 6 reveals that they contain (T)4, (C)4, and (A)18, respectively, which would be prone to mutations by replication slippage. The occurrence of the ten GATA3 mutations [four from this study (Table 1)], which involve insertions of a nucleotide around residue Ser408 [10–12], is more difficult to explain as the DNA sequence does not contain any repeated sequence, tandem repeats, or CpG islands [5]. However, it has also been demonstrated that in regions where selection favors polymorphism, heterozygote instability increases the local polymorphism rate [53]. This may help to provide an explanation for the occurrence of the four mutations we identified at this residue, in addition to six previously reported mutations [10–12], as within 20 bp of Ser408 (residues 388–428), there are seven reported single nucleotide polymorphisms (SNPs): rs150229374, rs138679257, rs149351039, rs144824106, rs11567941, and two without dbSNP identifiers [33].
Approximately 88 % of the 78 GATA3 mutations identified in breast cancer (Fig. 1a) clustered in exons 5 and 6 which encode ZnF2 and the C-terminal domain, and this contrasts to the locations of the 51 germline HDR causing mutations which are widely distributed throughout the coding region and also involve ZnF1 [4–6], that binds to the cofactor Friend of GATA-2 (FOG2) [4] (Fig. 1a). The absence of GATA3-ZnF1 mutations in breast cancer (Fig. 1a, [7–9]) is difficult to explain as the GATA3-ZnF1 interaction with FOG2 has been shown, by studies of mice deleted for Fog2 or Gata3, to be of importance in regulating ER expression in the mouse mammary gland [54, 55]. However, the phenotypic differences between patients with HDR and breast cancers may instead be attributable to their respective germline and somatic origins, and the situation may be analogous to that reported for the alpha thalassemia/mental retardation syndrome X-linked (ATRX) gene [56]. Thus, germline ATRX mutations result in the X-linked alpha thalassemia mental retardation (ATR-X) syndrome [56], whereas somatic ATRX mutations have been identified in pancreatic neuroendocrine tumors (PNETs) [57]. Moreover, the germline ATRX mutations in ATR-X patients cluster in the ATRX-DNMT3-DNMT3L and helicase domains which are involved in protein–protein interactions, and the enzymatic function of the protein, respectively, whereas the somatic ATRX mutations identified in PNETS all occur within the helicase domain [57]. The basis of these different phenotypic effects related to the location of the mutation and its occurrence in germline or somatic cells remains to be elucidated, as well as the dual roles of GATA3 and ATRX in development and oncogenesis.
In summary, we have demonstrated the presence of GATA3 mutations in 20 % of ER-positive breast cancers, and defined their functional consequences on DNA binding, nuclear localization and transactivation activity, and cell invasiveness. In addition, we show that unlike many other nuclear proteins which contain a classical mono- or bi-partite signal, the NLS of GATA3 is complex and spread throughout the amino acids of ZnF1 and its adjacent flanking regions which contain four clusters of positive amino acids that all contribute to efficient nuclear targeting.
Acknowledgments
This work was supported by the Medical Research Council (MRC), UK (grant number G9825289/2004 and G1000467/2010). K.G. was an MRC-funded student.
Footnotes
Katherine U. Gaynor and Irina V. Grigorieva contributed equally to this work.
References
- 1.Simon MC. Gotta have GATA. Nat Genet. 1995;11:9–11. doi: 10.1038/ng0995-9. [DOI] [PubMed] [Google Scholar]
- 2.Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Esch H, Groenen P, Nesbit MA, Schuffenhauer S, Lichtner P, Vanderlinden G, Harding B, et al. GATA3 haplo-insufficiency causes human HDR syndrome. Nature. 2000;406:419–422. doi: 10.1038/35019088. [DOI] [PubMed] [Google Scholar]
- 4.Nesbit MA, Bowl MR, Harding B, Ali A, Ayala A, Crowe C, Dobbie A, et al. Characterization of GATA3 mutations in the hypoparathyroidism, deafness, and renal dysplasia (HDR) syndrome. J Biol Chem. 2004;279:22624–22634. doi: 10.1074/jbc.M401797200. [DOI] [PubMed] [Google Scholar]
- 5.Ali A, Christie PT, Grigorieva IV, Harding B, Van Esch H, Ahmed SF, Bitner-Glindzicz M, et al. Functional characterization of GATA3 mutations causing the hypoparathyroidism–deafness–renal (HDR) dysplasia syndrome: insight into mechanisms of DNA binding by the GATA3 transcription factor. Hum Mol Genet. 2007;16:265–275. doi: 10.1093/hmg/ddl454. [DOI] [PubMed] [Google Scholar]
- 6.Gaynor KU, Grigorieva IV, Nesbit MA, Cranston T, Gomes T, Gortner L, Thakker RV. A missense GATA3 mutation, Thr272Ile, causes the hypoparathyroidism, deafness, and renal dysplasia syndrome. J Clin Endocrinol Metab. 2009;94:3897–3904. doi: 10.1210/jc.2009-0717. [DOI] [PubMed] [Google Scholar]
- 7.Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerod A, et al. Mutation of GATA3 in human breast tumors. Oncogene. 2004;23:7669–7678. doi: 10.1038/sj.onc.1207966. [DOI] [PubMed] [Google Scholar]
- 8.Arnold JM, Choong DY, Thompson ER, Waddell N, Lindeman GJ, Visvader JE, Campbell IG, Chenevix-Trench G. Frequent somatic mutations of GATA3 in non-BRCA1/BRCA2 familial breast tumors, but not in BRCA1-, BRCA2- or sporadic breast tumors. Breast Cancer Res Treat. 2010;119:491–496. doi: 10.1007/s10549-008-0269-x. [DOI] [PubMed] [Google Scholar]
- 9.Chanock SJ, Burdett L, Yeager M, Llaca V, Langerod A, Presswalla S, Kaaresen R, et al. Somatic sequence alterations in twenty-one genes selected by expression profile analysis of breast carcinomas. Breast Cancer Res. 2007;9:R5. doi: 10.1186/bcr1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tominaga N, Naoi Y, Shimazu K, Nakayama T, Maruyama N, Shimomura A, Kim S J, Tamaki Y, Noguchi S. Clinicopathological analysis of GATA3-positive breast cancers with special reference to response to neoadjuvant chemotherapy. Ann Oncol. 2012;23(12):3051–7. doi: 10.1093/annonc/mds120. [DOI] [PubMed] [Google Scholar]
- 14.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 15.Grigorieva IV, Mirczuk S, Gaynor KU, Nesbit MA, Grigorieva EF, Wei Q, Ali A, et al. Gata3-deficient mice develop parathyroid abnormalities due to dysregulation of the parathyroid-specific transcription factor Gcm2. J Clin Invest. 2010;120:2144–2155. doi: 10.1172/JCI42021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 17.Shiga K, Shiga C, Sasano H, Miyazaki S, Yamamoto T, Yamamoto M, Hayashi N, Nishihira T, Mori S. Expression of c-erbB-2 in human esophageal carcinoma cells: overexpression correlated with gene amplification or with GATA-3 transcription factor expression. Anticancer Res. 1993;13:1293–1301. [PubMed] [Google Scholar]
- 18.Atayar C, Poppema S, Blokzijl T, Harms G, Boot M, van den Berg A. Expression of the T-cell transcription factors, GATA-3 and T-bet, in the neoplastic cells of Hodgkin lymphomas. Am J Pathol. 2005;166:127–134. doi: 10.1016/S0002-9440(10)62238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulbinas A, Berberat PO, Dambrauskas Z, Giese T, Giese N, Autschbach F, Kleeff J, Meuer S, Buchler MW, Friess H. Aberrant gata-3 expression in human pancreatic cancer. J Histochem Cytochem. 2006;54:161–169. doi: 10.1369/jhc.5A6626.2005. [DOI] [PubMed] [Google Scholar]
- 20.Hoch RV, Thompson DA, Baker RJ, Weigel RJ. GATA-3 is expressed in association with estrogen receptor in breast cancer. Int J Cancer. 1999;84:122–128. doi: 10.1002/(SICI)1097-0215(19990420)84:2<122::AID-IJC5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 21.Steenbergen RD, OudeEngberink VE, Kramer D, Schrijnemakers HF, Verheijen RH, Meijer CJ, Snijders PJ. Down-regulation of GATA-3 expression during human papillomavirus-mediated immortalization and cervical carcinogenesis. Am J Pathol. 2002;160:1945–1951. doi: 10.1016/S0002-9440(10)61143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ, Copland JA. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5:e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 25.Allen MD, Vaziri R, Green M, Chelala C, Brentnall AR, Dreger S, Vallath S, et al. Clinical and functional significance of alpha9beta1 integrin expression in breast cancer: a novel cell-surface marker of the basal phenotype that promotes tumour cell invasion. J Pathol. 2011;223:646–658. doi: 10.1002/path.2833. [DOI] [PubMed] [Google Scholar]
- 26.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 27.Krynska B, Del Valle L, Croul S, Gordon J, Katsetos CD, Carbone M, Giordano A, Khalili K. Detection of human neurotropic JC virus DNA sequence and expression of the viral oncogenic protein in pediatric medulloblastomas. Proc Natl Acad Sci U S A. 1999;96:11519–11524. doi: 10.1073/pnas.96.20.11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem. 2006;52:494–503. doi: 10.1373/clinchem.2005.063438. [DOI] [PubMed] [Google Scholar]
- 29.Jorde R, Schirmer H, Wilsgaard T, Joakimsen RM, Mathiesen EB, Njolstad I, Lochen ML, et al. Polymorphisms related to the serum 25-hydroxyvitamin d level and risk of myocardial infarction, diabetes, cancer and mortality. The Tromso Study. PLoS One. 2012;7:e37295. doi: 10.1371/journal.pone.0037295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hancox RA, Allen MD, Holliday DL, Edwards DR, Pennington CJ, Guttery DS, Shaw JA, Walker RA, Pringle JH, Jones JL. Tumour-associated tenascin-C isoforms promote breast cancer cell invasion and growth by matrix metalloproteinase-dependent and independent mechanisms. Breast Cancer Res. 2009;11:R24. doi: 10.1186/bcr2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas GJ, Lewis MP, Whawell SA, Russell A, Sheppard D, Hart IR, Speight PM, Marshall JF. Expression of the alphavbeta6 integrin promotes migration and invasion in squamous carcinoma cells. J Invest Dermatol. 2001;117:67–73. doi: 10.1046/j.0022-202x.2001.01379.x. [DOI] [PubMed] [Google Scholar]
- 32.Esapa CT, Head RA, Jeyabalan J, Evans H, Hough TA, Cheeseman MT, McNally EG, et al. A mouse with an N-Ethyl-N-Nitrosourea (ENU) induced Trp589Arg Galnt3 mutation represents a model for hyperphosphataemic familial tumoural calcinosis. PLoS One. 2012;7:e43205. doi: 10.1371/journal.pone.0043205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Exome Variant Server, NHLBI Exome Sequencing Project (ESP) (2011) Seattle, WA (URL: http://evs.gs.washington.edu/EVS/). Accessed Sept 2012
- 34.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLano WL. The PyMOL molecular graphics system. New York: Schrödinger, LLC; 2002. [Google Scholar]
- 38.Yan W, Cao QJ, Arenas RB, Bentley B, Shao R. GATA3 inhibits breast cancer metastasis through the reversal of epithelial–mesenchymal transition. J Biol Chem. 2010;285:14042–14051. doi: 10.1074/jbc.M110.105262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Philips AS, Kwok JC, Chong BH. Analysis of the signals and mechanisms mediating nuclear trafficking of GATA-4. Loss of DNA binding is associated with localization in intranuclear speckles. J Biol Chem. 2007;282:24915–24927. doi: 10.1074/jbc.M701789200. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z, Gu L, Romeo PH, Bories D, Motohashi H, Yamamoto M, Engel JD. Human GATA-3 trans-activation, DNA-binding, and nuclear localization activities are organized into distinct structural domains. Mol Cell Biol. 1994;14:2201–2212. doi: 10.1128/mcb.14.3.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoya Y, Kobayashi T, Koda T, Ikuta K, Kakinuma M, Kishi M. Two proline-rich nuclear localization signals in the amino- and carboxyl-terminal regions of the Borna disease virus phosphoprotein. J Virol. 1998;72:9755–9762. doi: 10.1128/jvi.72.12.9755-9762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang HY, Evans T. Distinct roles for the two cGATA-1 finger domains. Mol Cell Biol. 1992;12:4562–4570. doi: 10.1128/mcb.12.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bates DL, Chen Y, Kim G, Guo L, Chen L. Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J Mol Biol. 2008;381:1292–1306. doi: 10.1016/j.jmb.2008.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asselin-Labat ML, Sutherland KD, Vaillant F, Gyorki DE, Wu D, Holroyd S, Breslin K, et al. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol Cell Biol. 2011;31:4609–4622. doi: 10.1128/MCB.05766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 46.Holen I, Whitworth J, Nutter F, Evans A, Brown HK, Lefley DV, Barbaric I, Jones M, Ottewell PD. Loss of plakoglobin promotes decreased cell-cell contact, increased invasion, and breast cancer cell dissemination in vivo. Breast Cancer Res. 2012;14:R86. doi: 10.1186/bcr3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalkhoven E, Roelen BA, de Winter JP, Mummery CL, van den Eijnden-van Raaij AJ, van der Saag PT, van der Burg B. Resistance to transforming growth factor beta and activin due to reduced receptor expression in human breast tumor cell lines. Cell Growth Differ. 1995;6:1151–1161. [PubMed] [Google Scholar]
- 48.Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Exp Hematol. 1995;23:99–107. [PubMed] [Google Scholar]
- 49.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 50.Palmeri D, Malim MH. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaulsky G, Goldfinger N, Ben-Ze’ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990;10:6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogozin IB, Pavlov YI. Theoretical analysis of mutation hotspots and their DNA sequence context specificity. Mutat Res. 2003;544:65–85. doi: 10.1016/S1383-5742(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 53.Amos W. Heterozygosity and mutation rate: evidence for an interaction and its implications: the potential for meiotic gene conversions to influence both mutation rate and distribution. Bioessays. 2010;32:82–90. doi: 10.1002/bies.200900108. [DOI] [PubMed] [Google Scholar]
- 54.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manuylov NL, Smagulova FO, Tevosian SG. Fog2 excision in mice leads to premature mammary gland involution and reduced Esr1 gene expression. Oncogene. 2007;26:5204–5213. doi: 10.1038/sj.onc.1210333. [DOI] [PubMed] [Google Scholar]
- 56.Gibbons RJ, Wada T, Fisher CA, Malik N, Mitson MJ, Steensma DP, Fryer A, Goudie DR, Krantz ID, Traeger-Synodinos J. Mutations in the chromatin-associated protein ATRX. Hum Mutat. 2008;29:796–802. doi: 10.1002/humu.20734. [DOI] [PubMed] [Google Scholar]
- 57.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 59.Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-T. [DOI] [PubMed] [Google Scholar]
- 60.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]