Abstract
The dramatically increased prevalence of breast cancer after menopause is of great concern and is correlated with elevated local levels of estrogens. This is mainly due to an increase in aromatase expression driven by its proximal promoter II (PII). We have previously demonstrated that the CREB co-activator CRTC2 binds directly to PII and stimulates its activity via mechanisms involving LKB1-AMPK in response to prostaglandin E2 (PGE2). There are three members of the CRTC family (CRTC1-3) and this study aimed to characterize the role of other CRTCs in the activation of aromatase PII. The expression and subcellular localization of CRTCs were examined in preadipocytes using qPCR and immunofluorescence. Under basal conditions, CRTC1 expression was the lowest, whereas CRTC3 transcripts were present at higher levels. Basally, CRTC2 and CRTC3 were mainly cytoplasmic and PGE2 caused their nuclear translocation. Reporter assays and chromatin immunoprecipitation (ChIP) were performed to assess the effect of CRTCs on PII activity and binding. Basal PII activity was significantly increased with all CRTCs. Forskolin (FSK)/phorbol 12-myristate 13-acetate (PMA), to mimic PGE2, resulted in a further significant increase in PII activity with all CRTCs, with CRTC2 and CRTC3 having greater effects. This was consistent with ChIP data showing an increased binding of CRTCs to PII with FSK/PMA. Moreover, gene silencing of CRTC2 and CRTC3 significantly reduced the FSK/PMA-mediated stimulation of aromatase activity. Interestingly, CRTCs acted cooperatively with CREB1 to increase PII activity, and both CREs were found to be essential for the maximal induction of PII activity by CRTCs. Phosphorylation of CRTC2 at its AMPK target site, Ser 171, dictated its subcellular localization, and the activation of aromatase PII in preadipocytes. In conclusion, this study demonstrates that aromatase regulation in primary human breast preadipocytes involves more than one CRTC.
Keywords: Postmenopausal Breast Cancer, Aromatase Expression, CREB Target Gene, Primary Human Preadipocytes, Regulate Aromatase Expression
Introduction
The incidence of breast cancer cases increases dramatically with advancing age, and 70 % of postmenopausal breast cancers are estrogen receptor positive. This is largely due to an increase in locally produced estrogens within the breast adipose via the enhanced expression of aromatase. This occurs as a result of the promoter switching from the distal promoter I.4 to the alternative proximal promoters II (PII) and I.3 on the CYP19A1 gene in response to tumor-derived factors such as prostaglandin E2 (PGE2) [reviewed in 1]. The activity of aromatase PII is largely regulated by the transcription factor CREB1 via cAMP-dependent mechanisms [2]. CREB1 has been shown to bind to two distinct CRE-like sequences, namely CRE1 and CRE2 (approximately 80 bp upstream of CRE1), within the PII region of the CYP19A1 gene using the mouse 3T3-L1 preadipocyte cell line and primary human preadipocytes, and this interaction is increased in the presence of cAMP in 3T3-L1 preadipocytes [2]. The transactivation potential of CREB1 is largely mediated by phosphorylation at Ser133, located in the kinase-inducible domain, by protein kinase A (PKA) upon stimulation with cAMP [3]. Obesity is a well-defined risk factor for developing breast cancer [4, 5] and a body mass index greater than 30 is found to be associated with a twofold increased risk of breast cancer. Higher estrogen production in postmenopausal women has been shown to account for this obesity-associated breast cancer risk [6]. Interestingly, a recent study has also demonstrated that the high levels of PGE2 produced from inflamed breast tissues from overweight/obese women correlate with both aromatase expression and activity [7].
Nevertheless, the PKA-mediated phosphorylation of CREB1 at Ser133 is not sufficient to stimulate the activation of all CREB target genes [8]. A family of CREB co-activators, termed CRTCs (CREB-regulated transcription co-activators) and previously known as TORCs (transducers of regulated CREB activity) has recently been identified using high-throughput screens for modulators of CRE luciferase reporters [9–12]. There are three members of the CRTC family, namely CRTC1, CRTC2, and CRTC3. All three members possess an N-terminal CREB binding domain, a central regulatory region, a splicing domain, and a C-terminal transactivation domain [reviewed in 13]. The highly conserved N-terminal coiled coil domain of the CRTCs is known to interact with the bZip domain of CREB [10]. Furthermore, CRTCs have shown to act as co-activators of CREB independent of its phosphorylation status at Ser-133 [10]. CRTCs also demonstrate their ability to increase the interaction with the TAF(II)130 component of TFIID, which directs an increase expression of CRE target genes [10].
The nuclear translocation of CRTC proteins through their lack of phosphorylation or dephosphorylation is a critical and conserved step in the upregulation of CREB target genes [11]. Moreover, this is induced by increasing intracellular cAMP or calcium levels [11]. Both the nuclear localization sequence and nuclear export sequence motifs are found to be conserved within the three CRTC family members and they tend to shuttle in and out of the nucleus under resting conditions [12]. CRTC2 has been previously shown to be mainly sequestered in the cytoplasm under basal conditions through phosphorylation-dependent interactions with 14-3-3 proteins [12]. In that case, dephosphorylation of CRTC2 at Serine 171 (Ser171) in response to cAMP agonists and calcium drives its nuclear translocation by disrupting CRTC2:14-3-3 complexes [12]. AMP-activated protein kinase (AMPK), which acts as a master regulator of energy homeostasis, and salt-inducible kinase (SIK) have been shown to directly phosphorylate CRTC family members [12, 14–16]. Studies undertaken in kidney cells have demonstrated that active AMPK can phosphorylate CRTC2 at Ser171, resulting in its cytoplasmic accumulation [15]. CRTC1 in Caenorhabditis elegans, with conserved phosphorylation sites at Ser 76 and Ser 179, has also been shown to be a direct target of AMPK [16]. SIK1 has the ability to phosphorylate all CRTCs [14, 15], while other studies have shown that SIK2 can phosphorylate CRTC2 at Ser171 and promote its association with 14-3-3 proteins [12].
We have previously demonstrated the existence of a relationship between CRTC2 and PII-driven aromatase expression in primary human breast preadipocytes in the context of postmenopausal breast cancer [17]. In that case, FSK/PMA-mediated repression of LKB1/AMPK led to the nuclear translocation of CRTC2, as well as the increased binding and activation of aromatase promoter PII. Importantly, mutation of the proximal CRE abolished the CRTC2-mediated induction of aromatase promoter PII activity, confirming that CRTC2 acts primarily via CREB-dependent mechanisms. However, it remains to be clarified as to whether other CRTCs play a role in regulating PII-driven aromatase expression. This study aimed to characterize the role of CRTCs in regulating the activation of aromatase PII in response to tumor-derived and obesity-related factor, PGE2.
Materials and methods
Plasmids
The CYP19A1 PII-516 luciferase reporter plasmid, which contains 502 bp (−516 to −14) of the proximal promoter PII was generated as previously described [18]. The pCMV.CREB1 plasmid was purchased from Promega (USA). The flag-tagged CRTC-pcDNA vectors were obtained from Mark Montminy from Salk Institute, generated as described previously [10].
Mutagenic primers for CRE mutants of PII luciferase reporter plasmid and phosphorylation site mutants of CRTC2 were designed using Agilent Technologies’s web-based QuickChange Primer Design Application available at website https://www.genomics.agilent.com/CollectionSubpage.aspx?PageType=Tool&SubPageType=ToolQCPD&PageID=15. Designed primer sequences were proxy CRE mut PII F: 5′-TTTGGCTTTCAATTGGGAATGGAATTCACTCTACCCACTCAAGGGC-3′, proxy CRE mut PII R: 5′-GCCCTTGAGTGGGTAGAGTGAATTCCATTCCCAATTGAAAGCCAAA-3′, distal CRE mut PII F: 5′-CCTAAACAAAACCTGCTGATGGATTCACAAAATGACTCCACCTCT-3′, distal CRE mut PII R: 5′-AGAGGTGGAGTCATTTTGTGAATCCATCAGCAGGTTTTGTTTAGG-3′, CRTC2 S171A F: 5′-CACTTAACAGGACAAGCGCTGACTCTGCTCTTCAC-3′, CRTC2 S171A R: 5′-GTGAAGAGCAGAGTCAGCGCTTGTCCTGTTAAGTG-3′, CRTC2 S171D F: 5′-TGCACTTAACAGGACAAGCGATGACTCTGCTCTTCACACA-3′ and CRTC2 S171D R: 5′-TGTGTGAAGAGCAGAGTCATCGCTTGTCCTGTTAAGTGCA-3′. Site-directed mutagenesis was performed using QuickChange® II Site-Directed Mutagenesis Kit (Agilent Technologies, USA) according to the manufacturer’s instructions. Briefly, thermal cycling was used to generate a mutant plasmid containing staggered nicks using two synthetic primers both containing desired mutations. The product was then treated with DpnI endonuclease to digest parental methylated and hemimethylated DNA. Following digestion, the nicked vector containing the desired mutations was transformed into XL1-blue supercompetent cells.
Cell culture
Primary human breast preadipocytes were isolated after collagenase treatment of whole subcutaneous adipose tissue obtained from women undergoing reduction mammoplasty and cultured in Waymouth’s medium (Invitrogen, USA), as previously described [19]. The studies have been approved by Southern Health Human Research Ethics Committee B. COS-7 (African green monkey kidney preadipocyte-like cell line) and 3T3-L1 (mouse preadipocyte cell line) cells were obtained from the ATCC, USA and grown in Dulbecco’s modified eagle medium (DMEM, Invitrogen, USA) supplemented with 10 % fetal calf serum, 50 U/ml penicillin, and 50 μg/ml streptomycin at 37 °C in 5 % CO2, as recommended. Before treatments, cells were serum starved for 24 h in DMEM containing 0.1 % bovine serum albumin, 50 U/ml penicillin, and 50 μg/ml streptomycin. Treatments included PGE2 (1 μM), FSK (25 μM)/PMA (4 nM), and 5-aminoimidazole-4-carboxamide 1-β-d-ribofuranoside (AICAR, 0.5 mM; an AMPK activator), purchased from Sigma-Aldrich (USA).
Reverse transcription and quantitative PCR
Total RNA was extracted from cultured primary human breast preadipocytes in six-well plates using the RNeasy Mini Kit (QIAGEN, Germany), treated with DNaseI (Ambion, USA) and quantified using the NanoDrop 1000 Spectrophotometer (Thermo Scientific, USA). Of the RNA, 0.3–1 μg was reverse transcribed using AMV RT Kit using oligo-dT primer (Promega, USA) as directed by the manufacturer. cDNAs were amplified on the LightCycler using LightCycler FastStart DNA Master SYBR Green l kit (Roche, Germany). Quantification of human CRTC1, CRTC2, CRTC3, aromatase and housekeeping genes, β-actin or 18s, transcript expression was carried out using primers hCRTC1 F: 5′-CAGTCCCAGGAATGGAAGAG-3′, hCRTC1 R: 5′-GCAGACGGGAAGATGTTGAT-3′, hCRTC2 F: 5′-TGACTTCAACCTGGGGAATC-3′, hCRTC2 R: 5′-GTGGGTCAAGTTCTGGTGGT-3′, hCRTC3 F: 5′-ACAACTGTGGGAGACCAAGG-3′, hCRTC3 R: 5′-GTGTTCAAGGTCCCCAAGAA-3′, hArom F: 5′-TTGGAAATGCTGAACCCGAT-3′, hArom R: 5′-CAGGAATCTGCCGTGGGGAT-3′, β-actin F: 5′-TGCGTGACATTAAGGAGAAG-3′, β-actin R: 5′-GCTCGTAGCTCTTCTCCA-3′, 18S-F: 5′-CGGCTACCACATCCAAGGAA-3′ and 18S-R: 5′GCTGGAATTACCGCGGCT-3′. Cycling conditions were one cycle at 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, 59 °C for 5 s and 72 °C for 6 s for CRTC1 and CRTC3, 40 cycles of 95 °C for 10 s, 59 °C for 6 s and 72 °C for 6 s for CRTC2, 40 cycles of 95 °C for 10 s, 60 °C for 5 s and 72 °C for 10 s for aromatase and 30 cycles of 95 °C for 10 s, 59 °C for 5 s and 72 °C for 10 s for β-actin or 18s. All the samples were quantified using standards of known concentrations and corrected for abundance with the housekeeping gene β-actin or 18s.
Immunofluorescence and confocal imaging
The subcellular localization of endogenously expressed CRTC proteins in primary human breast preadipocytes was visualized using immunofluorescence and confocal microscopy, as previously described [20]. The cells were grown on coverslips and treated with PGE2 for 24 h. Immunofluorescence was performed using CRTC1 (2501, 1/200 dilution), CRTC2 (3826, 1/200 dilution) and CRTC3 (2768, 1/200 dilution) antibodies from Cell Signaling Technology, USA and lamin B1 + B2 antibody (1/1000 dilution, Abcam, USA) for nuclear stain and visualized using secondary antibodies, alexa fluor-546 (red) and -488 (green) from Invitrogen (USA), respectively, using confocal microscopy (Olympus Optical Co Ltd, Japan). In order to visualize the subcellular localization of phosphorylation site mutants of CRTC2, 3T3-L1 cells were transfected with 2 μg of GFP-tagged CRTC2 mutants using Amaxa Cell line Nucleofector kit V, program T-030 (Lonza Cologne GmbH, Germany) as directed by manufacturer.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed on primary human breast preadipocytes treated with FSK/PMA for 45 min, to examine the endogenous binding of CRTCs to aromatase PII using the ChIP-IT express kit (Active Motif, USA) according to manufacturer’s guide with minor modifications [17]. Briefly, cells were fixed in PBS containing 1 % formaldehyde, followed by termination of reaction by adding glycine. Isolated chromatin was then sheared using sonication at 20 % amplitude, seven times for 30 s pulses. Immunoprecipitation performed overnight with CRTC1 (sc-46270×, Santa Cruz Biotechnology, USA), CRTC2 (sc-46272×, Santa Cruz Biotechnology), CRTC3 (2768, Cell Signaling Technology, USA) antibodies and IgG as a control. Immunoprecipitated and reverse cross-linked chromatin was analyzed by quantitative PCR and the reaction was stopped within the linear range of amplification. The products were then run on agarose gel and visualized by ethidium bromide staining.
Reporter gene assays
COS-7 cells, grown in six-well plates were transfected with either 470 ng of the wild-type CYP19A1 PII-516 luciferase reporter construct, or proximal, distal, or double CRE-mutated CYP19A1 PII-516 luciferase reporter constructs, together with 35 ng of CRTCs-pcDNA or CREB1-pcDNA. 3T3-L1 cells were transfected using Cell line Nucleofector kit V, program T-030 (Lonza Cologne GmbH, Germany), with 1 μg of CYP19A1 PII-516 luciferase reporter construct and 1 μg of wild type (wt) or phosphorylation site mutant of CRTC2, as directed by the manufacturer. β-galactosidase control vector was co-transfected into COS-7 cells. Transfected cells were serum starved for 24 h, followed by 24 h treatment with FSK/PMA. Luciferase reporter assays were carried out using the Dual-Glo Luciferase Assay System (Promega, USA) according to the manufacturer’s guide and data was normalized to β-galactosidase activity in COS-7 cells, and total protein using Pierce BCA protein assay kit (Thermo Scientific, USA) in 3T3-L1 cells.
Aromatase activity assay (tritiated water release assay)
Primary human breast preadipocytes in six-well plates were transfected with 2 μg of CRTCs-pcDNA using Cell line Nucleofector kit V, program T-030 (Lonza Cologne GmbH, Germany) or 3 μl of CRTC siRNA (CRTC1 siRNA sc-45600, CRTC2 siRNA sc-45832, CRTC3 siRNA sc-90206, control siRNA-A sc-37007 from Santa Cruz Biotechnology) using lipofectamine transfection reagent (Invitrogen, USA), as directed by the manufacturer. Preadipocytes were serum-starved for 24 h and treated with FSK/PMA. Aromatase activity in preadipocytes was measured using the tritiated water-release assay using androst-4-ene-3, 17-dione (NET926001MC, PerkinElmer, USA) as a substrate, as previously described [21]. Specific activity was normalized to total protein amount.
Statistical analysis
All data are reported as mean ± standard error and analyzed by student’s t test of group means using GraphPad Prism version 5.00 (GraphPad, La Jolla, CA, USA). Statistical significance was defined as *, P < 0.05; **, P ≤ 0.005; ***, P ≤ 0.0005; and ****, P ≤ 0.0001.
Results
CRTCs bind to and activate aromatase PII
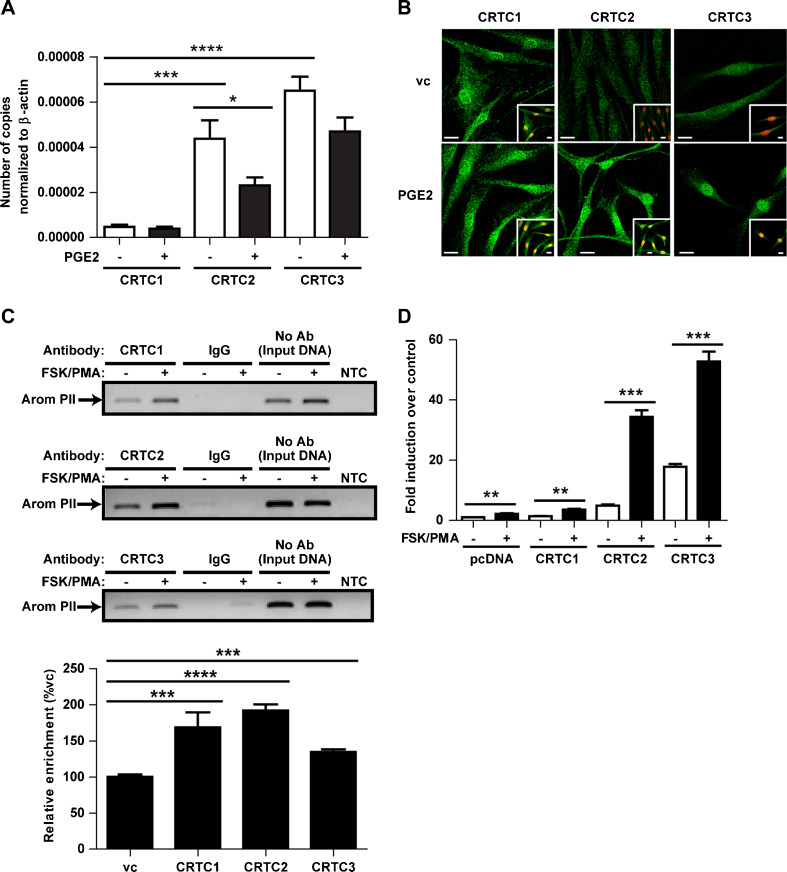
Relative levels of CRTC transcripts and the effect of PGE2 on transcript expression of CRTCs were examined in primary breast preadipocytes. Under basal conditions, CRTC3 transcript was expressed at higher levels than CRTC2 followed by CRTC1 (Fig. 1a). CRTC2 mRNA was significantly decreased in response to PGE2, and no change observed for CRTC1 and CRTC3 with treatment (Fig. 1a). The subcellular localization of endogenously expressed CRTCs was examined using immunofluorescence and confocal microscopy in human breast preadipocytes after treatment with PGE2. Under resting conditions, CRTC2 and CRTC3 were mainly found within the cytoplasm and CRTC1 appeared to be predominantly perinuclear. Interestingly, with PGE2 treatment, a marked increase in staining for CRTC2 and CRTC3 was observed in the nucleus, whereas CRTC1 appeared to remain perinuclear (Fig. 1b). To demonstrate the interaction between CRTCs and aromatase PII, chromatin immunoprecipitation was utilized (Fig. 1c). Treatment with FSK/PMA, a mimetic of PGE2, resulted in an increased binding of the CRTCs to aromatase PII compared to vehicle control, consistent with the nuclear import of CRTCs in the presence of PGE2. In order to assess the effect of the CRTCs on aromatase PII activity, reporter assays were carried out in COS-7 cells (Fig. 1d). Cells transfected with each CRTC resulted in an increase in PII activity compared to pcDNA control vector under resting conditions. Moreover, treatment with FSK/PMA caused a further significant increase in PII activity with all three CRTCs and CRTC2 had the highest fold induction of PII activity with treatment compared to CRTC1 and CRTC3, although CRTC3 showed the highest induction.
Fig. 1.
Role of CRTCs in aromatase PII activation. a Relative abundance of CRTC transcripts in primary breast preadipocytes with and without PGE2 treatment. b Confocal images of immunofluorescence on endogenous CRTC proteins (green) in primary breast preadipocytes with and without PGE2 treatment, lamin B1 + B2 nuclear stain in red. c Chromatin immunoprecipitation (ChIP) showing endogenous binding of CRTCs to aromatase PII in human breast preadipocytes with and without FSK/PMA treatment. d Reporter assays demonstrating the effect of CRTCs on aromatase PII activity with and without FSK/PMA treatment. vc Vehicle control; mean ± SEM, n = 3 for qPCR and reporter assays; confocal images are representative of the majority of cells examined; scale bar 20.0 μm; all experiments repeated twice
CRTCs stimulate aromatase activity in preadipocytes
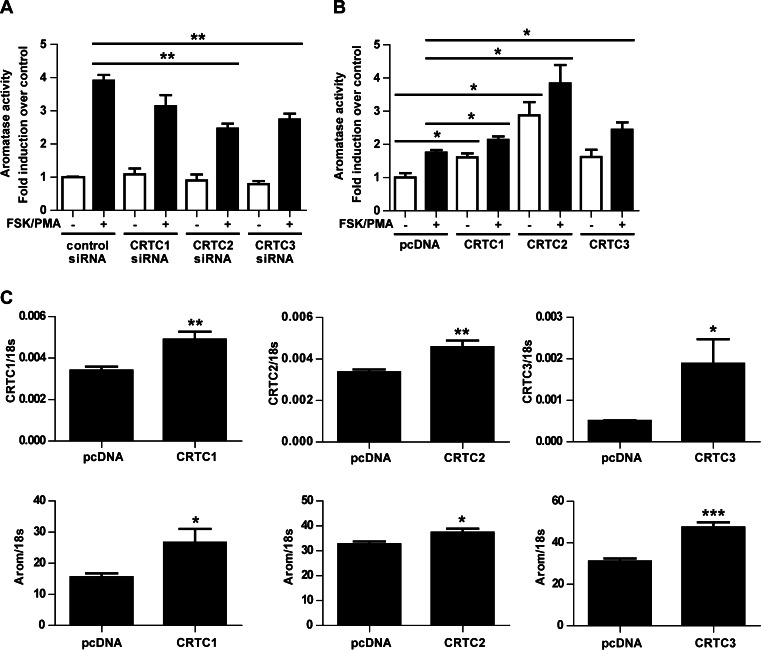
To determine the effect of CRTCs on aromatase activity, aromatase activity assays were performed in preadipocytes whereby CRTCs were either silenced or overexpressed. Interestingly, the FSK/PMA-mediated stimulation of aromatase activity was significantly reduced when CRTCs were knocked down (Fig. 2a) and increased when CRTCs were overexpressed (Fig. 2b), with CRTC2 appearing to have the greatest effect. Furthermore, aromatase transcript expression was significantly increased when CRTCs were overexpressed (Fig. 2c).
Fig. 2.
Role of CRTCs in regulating aromatase expression and activity in primary human breast preadipocytes. Aromatase activity assays demonstrating the effect of silencing (a) and overexpressing (b) CRTCs on the FSK/PMA-mediated stimulation of aromatase activity. c Effect of CRTC overexpression on aromatase transcript expression. Mean ± SEM, n = 3, repeated twice
CREB1 shows an additive effect with CRTCs to increase aromatase PII activity
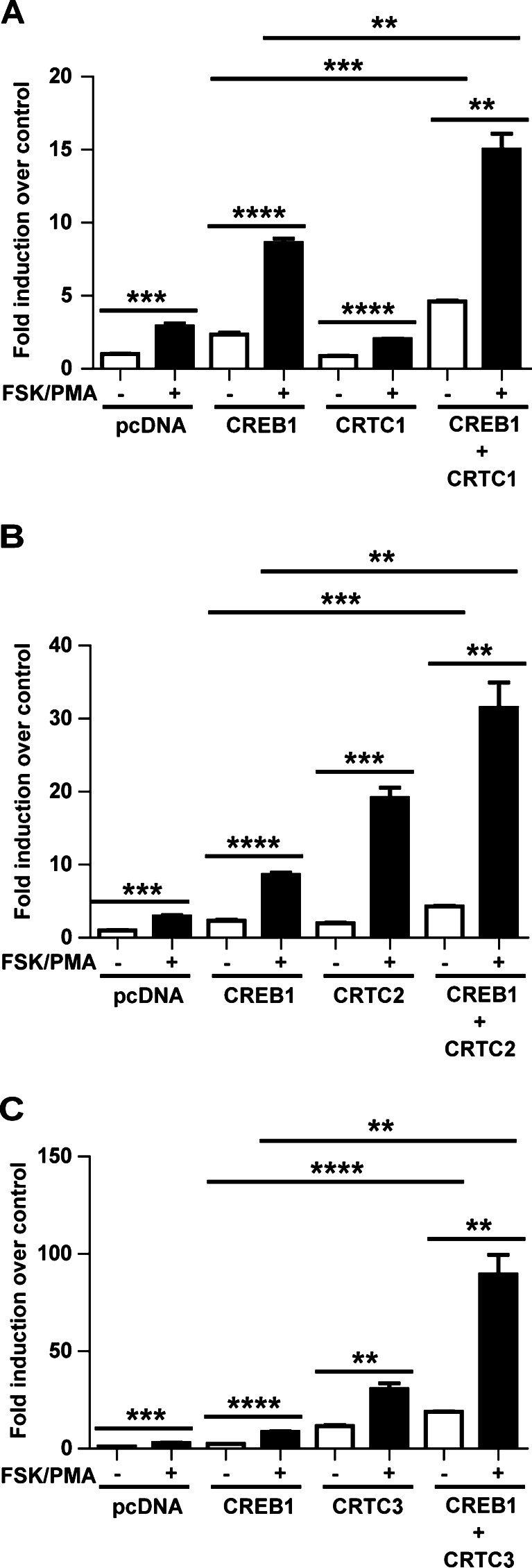
In order to determine the combined effect of CREB1 and CRTCs on aromatase PII activity, reporter assays were performed in COS-7 cells (Fig. 3). Results demonstrated that the cells transfected with CREB1 led to a significant increase in PII activity, and the effect of each CRTC family member on the CREB1-mediated aromatase PII activity was additive.
Fig. 3.
Role of CRTCs and CREB1 in the regulation aromatase PII activity. Reporter assays demonstrating the effect of CREB1 and a CRTC1, b CRTC2, and c CRTC3 on aromatase PII activity with or without FSK/PMA treatment. Mean ± SEM, n = 3, repeated twice
Both CREs on aromatase PII contribute to the maximal induction by CRTCs
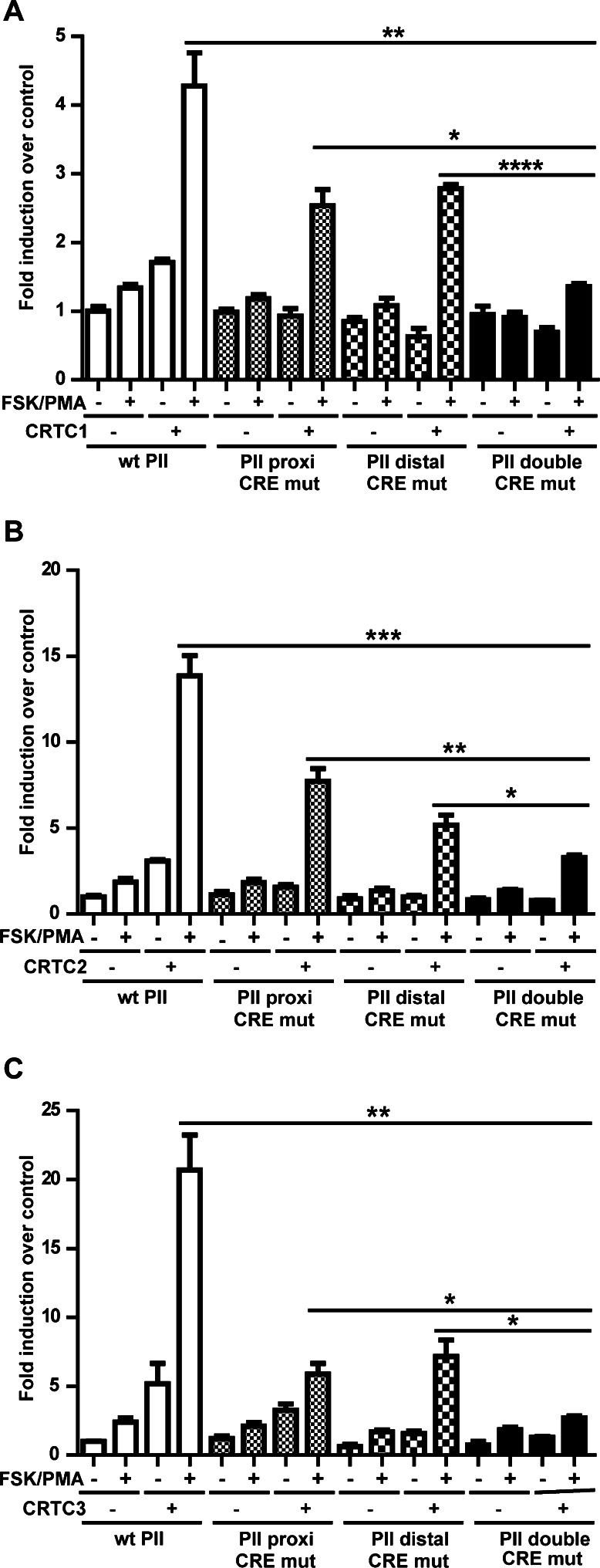
To determine the contribution of each CRE to the CRTC-mediated activity of aromatase PII, reporter assays were performed using CRE mutant reporter constructs in COS-7 cells (Fig. 4). There was a significant reduction in promoter activity in cells transfected with CRTC constructs and proximal CRE-mutated or distal-CRE mutated PII reporter constructs compared to wt PII reporter construct in response FSK/PMA. Interestingly, the effect on PII activity was further reduced significantly in cells transfected with double CRE-mutated PII reporter construct compared to cells transfected with CRTC constructs and proximal or distal CRE-mutated PII reporter constructs with FSK/PMA treatment, suggesting that both CREs are essential for the maximal induction of PII activity by CRTCs.
Fig. 4.
Role of CREs in the activation of aromatase PII. Reporter assays demonstrating the effect of proximal and distal CREs with a CRTC1, b CRTC2, and c CRTC3 on aromatase PII activity with or without FSK/PMA treatment. Mean ± SEM, n = 3, repeated twice
Phosphorylation of CRTC2 at Serine 171 dictates its subcellular localization and the activation of aromatase promoter II in preadipocytes
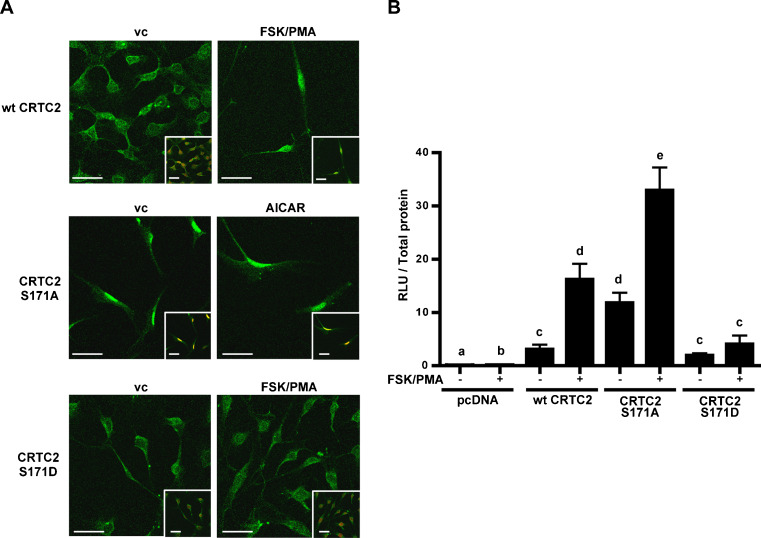
The effect of mutating CRTC2 at Ser171 (AMPK phosphorylation site) on its subcellular localization (Fig. 5a) and on the activation of PII (Fig. 5b) was examined in 3T3-L1 preadipocytes using immunofluorescence/confocal imaging and reporter assays, respectively. Results demonstrated that the CRTC2-GFP fusion protein was mainly localized in the cytoplasm under resting conditions, however FSK/PMA treatment, resulted in its nuclear translocation. The S171A mutant, that mimics a dephosphorylated state, was nuclear under both resting conditions and after AICAR treatment, which stimulates AMPK activity. Conversely, the S171D mutant, that mimics a phosphorylated state, was cytoplasmic under both resting conditions and after FSK/PMA treatment. PII luciferase assays revealed that treatment of wt CRTC2-transfected cells with FSK/PMA led to a significant increase in PII activity. Interestingly, transfection with the S171A mutant in the absence of FSK/PMA resulted in PII activity similar to that observed in cells transfected with wt CRTC2 treated with FSK/PMA. Conversely, the S171D mutant was unable to cause an increase in PII activity, even in the presence of FSK/PMA.
Fig. 5.
Effect of phosphorylating CRTC2 at Ser171 on its subcellular localization and the activation of aromatase PII in preadipocytes. a Confocal images on overexpressed GFP-tagged wt, S171A and S171D of CRTC2 (green) and immunofluorescence on lamin B1 + B2 nuclear stain in red, with or without FSK/PMA or AICAR treatment. b Reporter assays demonstrating the effect of wt, S171A and S171D of CRTC2 on aromatase PII activity with or without FSK/PMA treatment. vc Vehicle control, RLU relative luciferase units, β-gal β-galactosidase activity. Mean ± SEM, n = 3 for reporter assays, confocal images are representative of the majority of cells examined, scale bar represents 50.0 μm, all experiments repeated twice
Discussion
This study provides evidence that all three CRTC family members can activate aromatase PII in a CRE-dependent manner, together with CREB1, in the context of postmenopausal breast cancer. Recent advances in the treatment of breast cancer using aromatase inhibitors (AIs) have led to the decline in breast cancer-associated mortality; however, inhibition of aromatase throughout the body leads to unwanted side effects. Understanding the cellular and molecular mechanisms behind the PII-specific expression of aromatase will allow estrogen production to be targeted specifically within the breast, thereby circumventing any currently observed side-effects associated with AIs. Therefore, this study identifies the CRTC–CREB interaction as a potential therapeutic target for the treatment of obesity-related, estrogen-dependent, postmenopausal breast cancer.
The expression and regulation of CRTCs in preadipocytes has previously not been characterized. In this study, we demonstrate that CRTC3 transcript is highly abundant and CRTC1 is relatively low. In response to PGE2, known to be elevated within breast tissue in obesity and cancer, CRTC2 transcripts were significantly reduced, but no significant effect on CRTC1 and CRTC3 transcript expression was detected. Further studies will be required to elucidate the mechanisms behind this observation. Moreover, FSK/PMA, to mimic PGE2, resulted in a significant increase in binding of each CRTC to aromatase PII as well as an increase in PII activity in CRTC transfected preadipocytes. The effect of CRTC2 and CRTC3, however, were more pronounced than that of CRTC1, consistent with changes in subcellular localization of the CRTCs observed in response to PGE2. These results are also consistent with previous findings by Wang et al., who demonstrated that CRTC2 is recruited to the CREs on the PEPCK promoter in response to FSK treatment [22].
CREB1 is found to be overexpressed in breast cancer, prostate cancer, non-small-cell lung cancer and acute leukemia [23]. CREB1 transcript expression has been shown to be almost five times higher in adipose tissue of breasts containing a tumor as compared to normal breast adipose [2]. A study by Chhabra et al. reported that CREB1 is expressed in both tumor and normal breast adipose tissues, with higher levels in tumor tissues [24]. CREB1 transcript expression is also found to be higher in ductal carcinoma as compared to lobular and other breast carcinoma, node-positive tumors compared to node-negative tumors, and patients with a poor prognosis and with metastasis compared to cancer-free patients [24]. Moreover, breast cancer patients with higher CREB1 levels are shown to have significantly shorter disease-free survival [24]. In the current study, we have demonstrated for the first time that CREB1 and each CRTC act additively to induce aromatase PII. Given the importance of overexpression of CREB1 in cancer development and progression, this additive effect of CREB1 and CRTCs helps to further clarify the mechanism of action and the effects behind it.
Proximal and distal CREs within aromatase PII have been previously reported to play a pivotal role in CREB-mediated PII activation [2]. Interestingly, the current report shows that both proximal and distal CREs are essential for the maximal induction of aromatase PII by CRTCs using reporter assays with mutated proximal and distal CRE sites. This result differs from our previous findings demonstrating that mutation of the proximal CRE was sufficient to abolish the CRTC2-mediated induction of PII [17]. This may be due to differences in the level of stimulation of PII by CRTC2 or to the different cell types used in each experiment. Nevertheless, it is clear that CRTCs act via CREs on PII.
Site-directed mutagenesis and reporter assays also revealed that the phosphorylation of CRTC2 at Ser171 dictates its subcellular localization and directly affects its ability to activate aromatase PII. Taken with previous results demonstrating that PGE2 leads to a decrease in AMPK activity and as a consequence the increased nuclear localization of CRTC2 [17], this study identifies the LKB1/AMPK/CRTC2 pathway as a potential target for breast cancer treatment. Moreover, the overexpression of LKB1 has been shown to be associated with cytoplasmic sequestration of CRTC2, and knockdown of LKB1 resulted in the nuclear translocation of CRTC2 [17].
In summary, we have identified that all three members of the CRTC family contribute to an increase in PII activity, in conjunction with CREB1, in a CRE-dependent manner, suggesting a critical role for CRTCs in regulating aromatase expression in human breast preadipocytes. However, additional studies are required to test the possibility of targeting CRTC–CREB interactions to treat and possibly to prevent obesity-related, postmenopausal breast cancer.
Acknowledgments
This research was supported by NHMRC (Australia) Project Grant #GNT1005735 to KAB and ERS, the Victorian Government, through the Victorian Cancer Agency funding of the Victorian Breast Cancer Research Consortium to ERS and KAB, and by the Victorian Government Operational Infrastructure Support Program. NUS is supported by a Faculty Postgraduate Research Scholarship (FPRS), Monash University (2009–2012). KAB is supported by an NHMRC (Australia) Career Development Award GNT1007714. ERS is supported by an NHMRC (Australia) Senior Principal Research Fellowship GNT0550900. We thank Prof. Mark Montminy for providing CRTC–pcDNA plasmids. PHI Data Audit 12–16.
References
- 1.Simpson ER, Brown KA. Obesity, aromatase and breast cancer. Expert Rev Endocrinol Metab. 2011;6(3):383–395. doi: 10.1586/eem.11.35. [DOI] [PubMed] [Google Scholar]
- 2.Sofi M, et al. Role of CRE-binding protein (CREB) in aromatase expression in breast adipose. Breast Canc Res Treat. 2003;79(3):399–407. doi: 10.1023/A:1024038632570. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 4.Roberts DL, Dive C, Renehan AG. Biological mechanisms linking obesity and cancer risk: new perspectives. Annu Rev Med. 2010;61:301–316. doi: 10.1146/annurev.med.080708.082713. [DOI] [PubMed] [Google Scholar]
- 5.Harvie M, Hooper L, Howell AH. Central obesity and breast cancer risk: a systematic review. Obes Rev. 2003;4(3):157–173. doi: 10.1046/j.1467-789X.2003.00108.x. [DOI] [PubMed] [Google Scholar]
- 6.Stoll BA. Upper abdominal obesity, insulin resistance and breast cancer risk. Int J Obes Relat Metab Disord. 2002;26(6):747–753. doi: 10.1038/sj.ijo.0801998. [DOI] [PubMed] [Google Scholar]
- 7.Subbaramaiah K, et al. Increased levels of COX-2 and prostaglandin E2 contribute to elevated aromatase expression in inflamed breast tissue of obese women. Cancer Discov. 2012;2(4):356–365. doi: 10.1158/2159-8290.CD-11-0241. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iourgenko V, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci U S A. 2003;100(21):12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conkright MD, et al. TORCs: transducers of regulated CREB activity. Molecular Cell. 2003;12(2):413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Bittinger MA, et al. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr Biol. 2004;14(23):2156–2161. doi: 10.1016/j.cub.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119(1):61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12(3):141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh Y, et al. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS J. 2006;273(12):2730–2748. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- 15.Koo S-H, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437(7062):1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 16.Mair W, et al. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470(7334):404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown KA, et al. Subcellular localization of cyclic AMP-responsive element binding protein-regulated transcription coactivator 2 provides a link between obesity and breast cancer in postmenopausal women. Cancer Res. 2009;69(13):5392–5399. doi: 10.1158/0008-5472.CAN-09-0108. [DOI] [PubMed] [Google Scholar]
- 18.Michael MD, et al. Ad4BP/SF-1 regulates cyclic AMP-induced transcription from the proximal promoter (PII) of the human aromatase P450 (CYP19) gene in the ovary. J Biol Chem. 1995;270(22):13561–13566. doi: 10.1074/jbc.270.22.13561. [DOI] [PubMed] [Google Scholar]
- 19.Ackerman GE, et al. Aromatization of androstenedione by human adipose tissue stromal cells in monolayer culture. J Clin Endocrinol Metab. 1981;53(2):412–417. doi: 10.1210/jcem-53-2-412. [DOI] [PubMed] [Google Scholar]
- 20.Brown KA, et al. Metformin inhibits aromatase expression in human breast adipose stromal cells via stimulation of AMP-activated protein kinase. Breast Cancer Res Treat. 2010;123(2):591–596. doi: 10.1007/s10549-010-0834-y. [DOI] [PubMed] [Google Scholar]
- 21.Lephart ED, Simpson ER. Assay of aromatase activity. Methods Enzymol. 1991;206:477–483. doi: 10.1016/0076-6879(91)06116-K. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Targeted disruption of the CREB coactivator Crtc2 increases insulin sensitivity. Proc Natl Acad Sci U S A. 2010;107(7):3087–3092. doi: 10.1073/pnas.0914897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao X, et al. Targeting CREB for cancer therapy: friend or foe. Curr Cancer Drug Targets. 2010;10(4):384–391. doi: 10.2174/156800910791208535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chhabra A, et al. Expression of transcription factor CREB1 in human breast cancer and its correlation with prognosis. Oncol Rep. 2007;18(4):953–958. [PubMed] [Google Scholar]