Abstract
Breast cancer is primarily a hormone-dependent tumor that can be regulated by the status of steroid hormones, including estrogen and progesterone. Forkhead box P1 (FOXP1) is a member of the forkhead box transcription factor family and has been reported to be associated with various types of tumors. In the present study, we investigated the expression of FOXP1 in 133 human invasive breast cancers, obtained by core biopsy, by immunohistochemical analysis. Nuclear immunoreactivity of FOXP1 was detected in 89 cases (67%) and correlated positively with tumor grade and hormone receptor status, including estrogen receptor alpha (ERα) and progesterone receptor, and negatively with pathological tumor size. In ERα-positive MCF-7 breast cancer cells, we demonstrated that FOXP1 mRNA was upregulated by estrogen and increased ERα recruitment to ER binding sites identified by ChIP-on-chip analysis within the FOXP1 gene region. We also demonstrated that proliferation of MCF-7 cells was increased by exogenously transfected FOXP1 and decreased by FOXP1-specific siRNA. Furthermore, FOXP1 enhanced estrogen response element-driven transcription in MCF-7 cells. Finally, FOXP1 immunoreactivity was significantly elevated in relapse-free breast cancer patients treated with tamoxifen. These results suggest that FOXP1 plays an important role in proliferation of breast cancer cells by modulating estrogen signaling and that FOXP1 immunoreactivity could be associated with the estrogen dependency of clinical breast cancers, which may predict favorable prognosis in the patients treated with tamoxifen.
Keywords: FOXP1, Estrogen, ERα, Breast cancer, Tamoxifen, Recurrence
Introduction
Estrogen signaling pathways are involved in the growth and development of breast tumors through the activation of estrogen receptor alpha (ERα) [1]. Most breast cancers express high levels of ERα and exhibit estrogen-dependent proliferation. Estrogen receptors (ERs) are members of the nuclear receptor superfamily and regulate various cellular events, including cell growth and apoptosis, by acting as transcription factors activating the expression of target genes. Generally, ER-positive breast cancer has a better prognosis than ER-negative breast cancer, partly because of differing sensitivities to hormone therapy. Therefore, a comprehensive understanding of estrogen signaling pathways in breast cancer is required for both treatment and diagnosis of the disease.
The transcriptional activity of ERα is regulated by various coactivators and corepressors [2], as well as by interactions with other transcription factors, including the forkhead box (FOX) family. FOX family transcription factors influence ERα-regulated transcription by interaction with the ERα protein, as exemplified by FOXA1 [3]. Recent genome-wide studies aimed at identifying ERα and androgen receptor (AR) binding sites have shown that FOXA1 plays a role in regulating both of these nuclear receptor networks [4, 5]. FOXA1 is recognized as a pioneer transcription factor because chromatin binding by this protein can enable subsequent binding by the estrogen and androgen receptors [5, 6]. Of other FOX family members, FKHR (also known as FOXO1A) also binds directly to ERα and increases its transactivation through an estrogen response element [7].
FOXP1 is a FOX family member consisting of a winged-helix DNA-binding domain and an N-terminal transcriptional repression domain and represses its target genes by forming homodimers or heterodimers with FOXP2 and FOXP4 [8, 9]. Alteration of FOXP1 expression is associated with various types of tumors, including breast and prostate cancers [10–14]. FOXP1 may also be a critical transcription factor contributing to AR and ER signaling in a similar manner to FOXA1, although the functional roles of FOXP1 in estrogen signaling pathways, cancer cell proliferation, and its clinicopathological significance, remain to be elucidated.
In the present study, we evaluate the expression of FOXP1 in human breast cancers using immunohistochemistry and investigate the correlations between FOXP1 expression levels and clinicopathophysiological findings. Furthermore, we show that FOXP1 mRNA expression is induced by estrogen and ERα recruitment to ER binding sites in the FOXP1 gene region and is elevated in ERα-positive human breast cancer cells (MCF-7). Functional analyses also reveal that FOXP1 stimulates proliferation of MCF-7 cells and elevates ER-mediated transcription. Finally, negative FOXP1 immunoreactivity is associated with recurrence of tamoxifen-treated breast cancer. Our study provides a new insight into the association of FOXP1 in the estrogen signaling and therapeutic effect of tamoxifen in breast cancer.
Materials and Methods
Tissue Selection and Patient Characteristics
Between January 2005 and March 2006, 133 consecutive patients diagnosed with invasive breast cancer using a vacuum-assisted biopsy device (Mammotome®; Ethicon Endo-surgery Inc., Cincinnati, OH) at Saitama Medical University Hospital were included in a cohort study.
For nested-control study of the therapeutic effect of tamoxifen, 162 patients, diagnosed with primary breast cancer between 1989 and 1998, with or without distant recurrence, during or after adjuvant tamoxifen therapy, were identified from 3 institutions (National Hospital Organization Shikoku cancer center, Matsuyama, Japan; National Cancer Center Hospital, Tokyo, Japan; Tokyo Metropolitan Cancer and Infectious diseases Center Komagome Hospital, Tokyo, Japan). Relapse patients were defined as those with distant metastases within 5 years after surgery followed by tamoxifen treatment, and relapse-free as those without distant metastases. Eventually, 113 patients satisfied the criteria of our protocol and were included in this study.
Formalin-fixed paraffin-embedded sections obtained by biopsy or surgery were used in these studies. These studies were approved by the institutional review board at Saitama Medical University, and informed consent was obtained from all patients. The clinicopathological characteristics of the series are presented in Tables 1 and 2.
Table 1.
Relationship between immunoreactivity of FOXP1 and clinicopathological findings in invasive breast cancer (n = 133)
| Clinical findings | Immunoreactive score of FOXP1a | |||
|---|---|---|---|---|
| Positive (n = 89) | Negative (n = 44) | P value | ||
| Ageb (mean ± SD) | 57 ± 15 | 61 ± 13 | ||
| Age | ≤50 | 26 (19.5%) | 13 (9.8%) | 0.97 |
| 50< | 63 (47.4%) | 31 (23.3%) | ||
| Menopause | Pre | 27 (20.3%) | 11 (8.3%) | 0.52 |
| Post | 62 (46.6%) | 33 (24.8%) | ||
| Lymph node | Positive | 23 (21.3%) | 18 (16.7%) | 0.40 |
| Negative | 43 (39.8%) | 24 (22.2%) | ||
| pT | ≤20 mm | 37 (33.0%) | 33 (29.5%) | 0.014 |
| 20 mm< | 32 (28.6%) | 10 (8.9%) | ||
| Stage | I and II | 80 (60.1%) | 43 (32.3%) | 0.21 |
| III and IV | 9 (6.8%) | 1 (0.8%) | ||
| Grade | I | 33 (30.8%) | 12 (11.2%) | 0.015 |
| II and III | 31 (29.0%) | 31 (29.0%) | ||
| ER | Positive (PS ≥ 3) | 68 (51.1%) | 23 (17.3%) | 0.0048 |
| Negative (PS ≤ 2) | 21 (15.8%) | 21 (15.8%) | ||
| PgR | Positive (PS ≥ 3) | 44 (33.1%) | 13 (9.8%) | 0.029 |
| Negative (PS ≤ 2) | 45 (33.8%) | 31 (23.3%) | ||
| HER2 | Positive | 30 (25.6%) | 18 (15.4%) | 0.64 |
| Negative | 46 (39.3%) | 23 (19.7%) | ||
All other values represent the number and proportion of cases
ER estrogen receptor, PgR progesterone receptor, PS proportion score
aFOXP1 immunoreactive scores of 0, 2 and 3–8 were defined as negative and positive immunoreactivity, respectively
bData are presented as mean ± SD
Table 2.
Clinicopathological findings in adjuvant tamoxifen-treated invasive breast cancer patients followed up for 5 years after surgery (n = 113)
| Clinical findings | Relapse (n = 43) | Relapse free (n = 70) | P value | |
|---|---|---|---|---|
| Agea (mean ± SD) | 53.2 ± 9.9 | 55.2 ± 12.4 | 0.391 | |
| Age | ≤50 | 20 (17.7%) | 28 (24.8%) | 0.497 |
| 50< | 23 (20.3%) | 42 (37.2%) | ||
| pT | ≤30 mm | 22 (19.5%) | 52 (46.0%) | 0.012 |
| 30 mm< | 21 (18.6%) | 18 (15.9%) | ||
| Lymph node | Positive (n ≥ 1) | 30 (26.6%) | 30 (26.6%) | 0.005 |
| Negative (n = 0) | 13 (11.5%) | 40 (35.3%) | ||
| ERα | Positive | 39 (34.5%) | 64 (56.6%) | 0.835 |
| Negative | 4 (3.6%) | 6 (5.3%) | ||
| PgR | Positive | 36 (31.9%) | 60 (53.1%) | 0.987 |
| Negative | 7 (6.2%) | 10 (8.8%) | ||
| FOXP1 | Positive | 22 (19.5%) | 57 (50.4%) | <0.001 |
| Negative | 21 (18.6%) | 13 (11.5%) | ||
All other values represent the number and proportion of cases
pT pathological T stage, ERα estrogen receptor α, PgR progesterone receptor
aData are presented as mean ± SD
Antibodies
Affinity-purified rabbit polyclonal anti-FOXP1 antibody (anti-GX5050) was generated by the Genome Network Project at the Ministry of Education, Culture, Sports, Science and Technology, Japan (http://genomenetwork.nig.ac.jp) from serum derived from rabbits immunized with a peptide epitope consisting of amino acids 519–677 of the human FOXP1 protein. Anti-Myc antibody was purchased from Cell Signaling Technology (Beverly, MA).
Immunohistochemistry
Immunohistochemical analysis for FOXP1 was performed using an EnVision + visualization kit (Dako, Carpinteria, CA), as previously described [15]. Tissue sections (6 μm) were deparaffinized, rehydrated through graded ethanol, and rinsed in Tris-buffered saline with 0.05% Tween 20 (TBST). To retrieve antigens, sections were heated in an autoclave at 121°C for 15 min in 10 mM sodium citrate buffer (pH 6.0). Endogenous peroxidase activity was blocked using 0.3% hydrogen peroxide and the sections were incubated in 10% fetal bovine serum for 30 min. The primary antibody, a polyclonal antibody for FOXP1 (1:1,000 dilution), was applied and samples incubated at 4°C overnight. The sections were rinsed in TBST and incubated with EnVision + HRP-labeled polymer (anti-rabbit) for 1 h at room temperature. The antigen-antibody complex was visualized using the 3,3′-diaminobenzidine substrate kit for peroxidase (Vector laboratories, Burlingame, CA). Rabbit IgG was used in place of the primary antibody as a negative control.
Immunohistochemical Assessment
Slides were evaluated for the proportion (proportion score (PS): 0, none; 1, <1/100; 2, 1/100–1/10; 3, 1/10–1/3; 4, 1/3–2/3; and 5, >2/3) and staining intensity (intensity score (IS): 0, none; 1, weak; 2, moderate; and 3, strong) of positively stained cells. The total immunoreactivity score (TS: 0, 2–8) was determined as the sum of the proportion and intensity scores [16]. Two investigators (H. T. and A. O.) evaluated the tissue sections independently. If the immunoreactivity score differed between the two investigators, a third investigator (T. S.) evaluated the tissue sections, and the average immunoreactivity score was used. When the two investigators found it difficult to evaluate the TS of heterogeneous cancerous lesions, the third investigator made a deciding estimate. To identify potential correlation between FOXP1 expression in the malignant epithelium and clinicopathological characteristics, FOXP1 immunoreactive scores of 0, 2, and 3−8 were defined as negative and positive immunoreactivity, respectively.
Plasmid Construction and siRNA
Human FOXP1 (amino acids, 1–677) was C-terminally tagged with Myc, and human FOXA1 (amino acids, 2–472) was N-terminally tagged with Flag; both were subcloned into the pcDNA3 vector (pcDNA3-FOXP1-Myc and pcDNA3-Flag-FOXA1, respectively). Synthetic small interfering RNA (siRNA) duplexes targeting the human FOXP1 gene (Silencer® Select Pre-designed siRNA) and the luciferase reporter plasmid pGL2 (Luciferase GL2 Duplex) were purchased from Applied Biosystems (Carlsbad, CA) and Dharmacon (Lafayette, CO), respectively.
Cell culture and Transfection
MCF-7 cells were purchased from American Type Culture Collection (Rockville, MD) and maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37°C in 5% CO2. 17β-Estradiol (E2) was purchased from Sigma-Aldrich (St. Louis, MO). Transfection of FOXP1 was performed using 2 μg of pcDNA3-FOXP1-Myc and Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Whole cell extracts were analyzed by western blot analysis.
Quantitative Real-time Reverse Transcription Polymerase Chain Reaction
Total RNA extraction, first-strand cDNA synthesis, and quantitative PCR have been described elsewhere [15]. Primers are as follows: FOXP1, 5′-ACCGCTTCCATGGGAAATC-3′ and 5′-CCGTTCAGCTCTTCCCGTATT-3′; small heterodimer partner (SHP), 5′-TGGACTTCCTTGGTTTGGAC-3′ and 5′-TTCTGGTCCAATAAGCAGCC-3′; liver receptor homolog-1 (LRH-1), 5′-TGCAGGCAGTATCCCTCATC-3′ and 5′-AAATCCAACAATGCCAAAGC-3′; GAPDH, 5′-GGTGGTCTCCTCTGACTTCAACA-3′ and 5′-GTGGTCGTTGAGGGCAATG-3′. Fold induction of mRNA expression levels was determined by comparing estrogen-treated samples with those of the vehicle-treated control.
Western Blotting
Whole cell lysates were resolved by 10% denaturing SDS–PAGE and the blotted Immobilon-P Transfer Membrane (Millipore, Billerica, MA) was incubated with anti-FOXP1 antibody (anti-GX5050) or anti-β-actin antibody (Sigma). The band intensity in the captured images was quantified using the Scion Image program (Scion Corporation, Frederick, MD). The results were indicated as the mean ± SD of the relative intensity in three independent images.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay and quantitative real-time reversed transcription polymerase chain reaction (qRT-PCR) were performed as previously described [17, 18]. After 72-h hormone depletion, MCF-7 cells were treated with 100 nM E2 or vehicle (0.1% ethanol) for 45 min. Cells were fixed in 1% formaldehyde for 5 min at room temperature. Chromatin was sheared to an average size of 500 bp by sonication using a Bioruptor ultrasonicator (Cosmo-Bio, Tokyo, Japan). Lysates were rotated at 4°C overnight with a specific antibody against ERα. Salmon sperm DNA/protein A-agarose (Upstate Biotechnology, Lake Placid, NY) was added and incubated for 2 h. Precipitated DNA was used as a template for qPCR using an Applied Biosystems 7000 sequence detector (Foster City, CA) based on SYBR Green I fluorescence. A genomic fragment containing ERE in the enhancer region of TFF1 (405/393 bp from the transcription start site) was used as a positive control for ER binding [19]. Sequences of PCR primers are as follows: TFF1_ERE, 5′-TGAGATTCAGAAAGTCCCTCTTTC-3′ and 5′-TGGGCTTCATGAGCTCCTT-3′; FOXP1_ER_790, 5′-AACATCTGACAAATTATTGGGTGGTT-3′ and 5′-TGGCTTACCAGTTTAATGTCCCATA-3′; FOXP1_ER_791, 5′-AGGGTGAACCACAGCCTGTT-3′ and 5′-AAAGTGACAGTTTCCCAAGTACATGT-3′; FOXP1_ER_792, 5′-TGCAAGGTCTGTTTAACAGACACA-3′ and 5′-CCCCTTCATCCAAGCAAAAG-3′.
Luciferase Assay
MCF-7 cells were plated in 24-well culture plates at a density of 10,000 cells/well in phenol red-free medium containing 10% charcoal-stripped serum and transfected with 0.1 μg of ERE-tk-luc [20], together with 0.02 μg of pRL-cytomegalovirus (CMV; Promega, Madison, WI) using Lipofectamine 2000 reagent (Invitrogen). Twelve hours after transfection, cells were treated with 100 nM E2 or vehicle (ethanol) for 24 h, and luciferase activities were determined using a MicroLumatPlus microplate luminometer (Berthold Technologies, Bad Wildbad, Germany) and a Dual-Luciferase Assay System (Promega). Data are expressed as the mean (standard deviation (SD)) of 3 independent experiments performed in triplicate.
Cell Proliferation Assay
MCF-7 cells were seeded in 96-well plates at a density of 5000 cells/well in phenol red-free medium containing 5% charcoal-stripped serum for 24 h. Then, 0.2 μg of plasmid or 20 pmol of siRNA described above was transfected for 12 h and incubated with 100 nM E2 or vehicle for 120 h. Cell proliferation was examined using (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) (WST-8) assay kit (Nacalai Tesque, Kyoto, Japan) according to the manufacturer’s protocol.
Statistical Analyses
Correlation between the immunoreactivity score and clinicopathological characteristics was evaluated using the chi-square test. Differences between the 2 groups in luciferase and cell proliferation assays were analyzed using a 2-sample 2-tailed Student’s t test. P values < 0.05 were considered statistically significant. All data are presented in the text and figures as the mean (SD).
Results
Correlation of FOXP1 Immunoreactivity with Clinicopathological Features of Invasive Breast Cancer
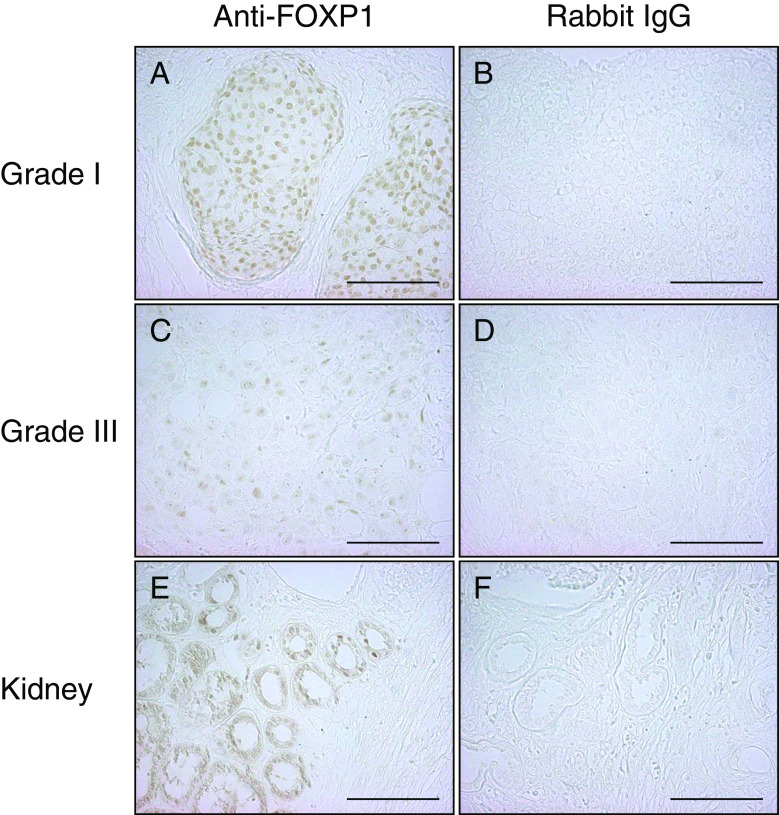
To investigate expression levels of FOXP1 protein in breast cancer, immunohistochemical analysis was performed using samples from 133 invasive breast cancers (Fig. 1). Positive nuclear immunoreactivity of FOXP1 was detected in 67% of breast cancer specimens (Fig. 1a, c). No signal was observed in the same specimens when rabbit IgG was used in immunostaining (Fig. 1b, d). We used kidney tissue as a control in this study (Fig. 1e, f), because FOXP1 was demonstrated to be expressed in normal kidney tissue by immunohistochemical analysis [10]. FOXP1 immunoreactivities in Grade III were significantly lower than those in Grade I in breast cancer. Namely, averaged Allred score of FOXP1 immunoreactivity in Grade I was 4.71, whereas that in Grade III was 2.73 (P = 0.01). In the luminal epithelium and myoepithelium, FOXP1 was strongly positive in almost all constituent cells. In addition, in stromal cells, FOXP1 was also strongly positive in almost all constituent cells. The nuclear immunoreactivity of FOXP1 was significantly correlated with ERα immunoreactivity (P = 0.0048, Table 1). The nuclear immunoreactivity of FOXP1 was also significantly correlated with tumor size (pT), histological grade, and progesterone receptor (PgR) (P = 0.014, 0.015, and 0.029, respectively), while no significant association was found with other clinicopathological characteristics (Table 1).
Fig. 1.
Immunohistochemistry of FOXP1 in breast cancer. Representative immunohistochemical staining of breast cancer tissues (a–d) and kidney (e, f) with anti-FOXP1 antibody (a, c, and e) or rabbit IgG (b, d, and f). Positive staining for FOXP1 was observed in the nuclei of breast cancer cells and kidney tubule cells. Immunoreactivity for FOXP1 tended to be strong in grade I breast cancer (a; total score, 8) and weaker in grade III breast cancer (c; total score, 3). Bar, 100 μm
Upregulation of FOXP1 mRNA by Estrogen Through the Putative ER Binding Sites in MCF-7 cells
We examined transcriptional regulation of the FOXP1 gene by estrogen in ERα-positive MCF-7 cells (Fig. 2a). FOXP1 mRNA level was significantly upregulated by 3-fold at 3 h after estrogen stimulation, suggesting that mRNA expression of FOXP1 is regulated by estrogen in breast cancer cells. FOXP1 protein was also upregulated by 1.73 ± 0.13 and 1.63 ± 0.18 fold at 12 and 24 h after estrogen stimulation, respectively, as confirmed by western blotting (Fig. 2b). In support of these results, genome-wide chromatin immunoprecipitation (ChIP)-on-chip analysis using MCF-7 cells demonstrated the presence of 3 estrogen receptor-binding sites (ERBSs; ER_790, 791, and 792) within the human FOXP1 gene region (Fig. 2c) [21]. To determine whether these ERBSs are bona fide ERBSs in MCF-7 cells, we performed a ChIP assay using the antibody specific for ERα. More than 2-fold ERα enrichment was observed on each of the 3 ERBSs after treatment with 100 nM estrogen for 45 min compared with controls treated with vehicle only in MCF-7 cells (Fig. 2d). ERα was enriched about sixfold on the ERBS within the TFF1 gene region, used as a positive control [19].
Fig. 2.
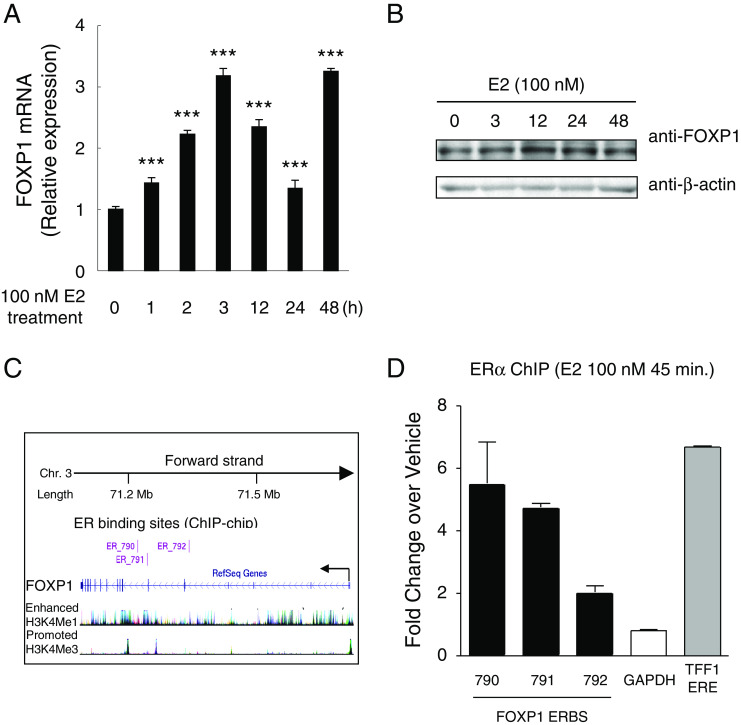
Transcriptional regulation of FOXP1 by estrogen in MCF-7 cells. a, b MCF-7 cells were treated with 100 nM 17β-estradiol (E2) for 48 h. Expression levels of FOXP1 mRNA (a) and protein (b) were examined at indicated time points by qRT-PCR and western blotting, respectively. The mRNA expression levels are shown as fold change over the expression level at 0 h. ***P < 0.001 compared with 0 h (by Student’s t test). c Schematic representation of the FOXP1 gene region in UCSC genome browser. Three ER binding sites (ERBSs) within the FOXP1 gene region were found by genome-wide ChIP-on-chip analysis [3]. d Estrogen-dependent recruitment of ERα to the FOXP1 ERBSs. MCF-7 cells were treated with 100 nM of E2 or vehicle for 45 min and then subjected to ChIP analysis with ERα-specific antibody. Immunoprecipitated DNA was examined by qPCR, and fold enrichments relative to vehicle control were plotted. The estrogen response element of the TFF1 gene (TFF1 ERE) was used as a positive control [19]
FOXP1 Contributes to Estrogen-Dependent Proliferation in Breast Cancer Cells
To further assess the role of FOXP1 in breast cancer, we performed both gain- and loss-of-function studies for FOXP1. Overexpression of FOXP1 was achieved by transient transfection of MCF-7 cells with pcDNA3-FOXP1-Myc for 1, 2, and 4 days and confirmed by immunoblotting (Fig. 3a). In the presence of estrogen, FOXP1-overexpressing MCF-7 cells exhibited a significantly higher growth rate compared with control cells expressing empty vector throughout the examined time points (Fig. 3b). In contrast, transient transfection of siRNA specific for FOXP1 in MCF-7 cells resulted in downregulation of FOXP1 expression as confirmed by qRT-PCR and western blotting after transfection (Fig. 3c). Growth of MCF-7 cells was suppressed by the FOXP1-specific siRNA treatment (Fig. 3d). These results indicate that FOXP1 promotes estrogen-dependent proliferation of breast cancer cells.
Fig. 3.
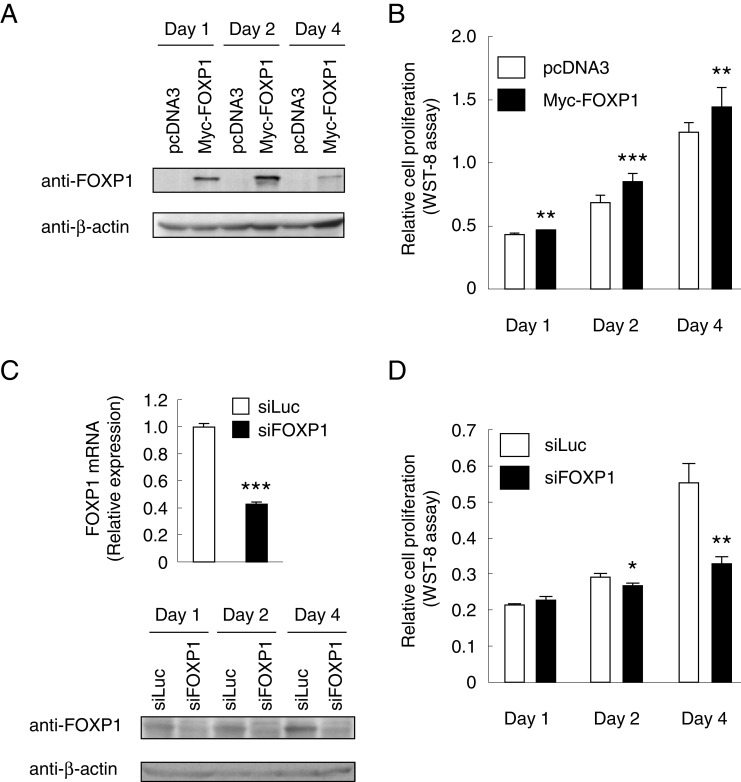
FOXP1 promotes estrogen-dependent proliferation of MCF-7 cells. a Total cell lysates from MCF-7 cells transfected with pcDNA3 or pcDNA3-FOXP1-Myc (Myc-FOXP1) for 1, 2, and 4 days were immunoblotted with anti-FOXP1 or anti-β-actin antibodies. b MCF-7 cells were transfected with pcDNA3-FOXP1-Myc for 24 h and then treated with 100 nM 17β-estradiol (E2) for 4 days. Relative cell proliferation at indicated time points was examined using a WST-8 assay kit. **P < 0.01 compared with control vector; ***P < 0.001 compared with control vector (Student’s t test). c Total RNA from MCF-7 cells transfected with siRNA specific for FOXP1 (siFOXP1) or luciferase (siLuc) for 48 h was examined by qRT-PCR (upper). *P < 0.05 compared with control siRNA (siLuc; Student’s t test). Total cell lysates from MCF-7 cells transfected with siFOXP1 or siLuc for 1, 2, and 4 days were immunoblotted with anti-FOXP1 or anti-β-actin antibodies (lower). d MCF-7 cells were transfected with siRNA specific for FOXP1 or luciferase and then treated with 100 nM 17β-estradiol (E2) for 4 days. Relative cell proliferation at indicated time points was examined as b. *P < 0.05 compared with control siRNA (siLuc; Student’s t test)
FOXP1 Promotes ER-Mediated Transcription
To examine whether FOXP1 influences ER-ERE-mediated transcription, a luciferase reporter vector containing an ERE (ERE-tk-luc) was introduced into MCF-7 cells with or without the FOXP1 expression vector (Fig. 4). The results indicate that FOXP1 significantly stimulated ER-ERE-mediated transactivation in MCF-7 cells when they were treated with estrogen. We also observed that the estrogen-dependent transactivation was elevated in response to increasing amounts of FOXA1 in MCF-7 cells (Fig. 4a). Consistent with these observations, we also evaluated the transcriptional regulations of previously reported estrogen-responsive genes, including SHP and LRH-1 [22, 23]. Upregulations of these genes were observed in FOXP1-overexpressing MCF-7 cells treated with 100 nM of estrogen (Fig. 4b, c). These results indicate that FOXP1 stimulates ER transcription activity in response to estrogen.
Fig. 4.

Effect of FOXP1 on ERE-mediated transcription and ER-target gene expressions in MCF-7 cells. a MCF-7 cells were transfected with a DNA mixture of 0.1 μg of estrogen response element (ERE)-tk-Luc, 0.02 μg of pRL-CMV, and increasing amounts of pcDNA3-FOXP1-Myc (Myc-FOXP1) or pcDNA3-Flag-FOXA1 (FOXA1). After a 12-h incubation, cells were treated with 17β-estradiol (E2; 100 nM) or vehicle (EtOH) for 24 h and then cell lysates were examined by luciferase assay. *P < 0.05; **P < 0.01; and ***P < 0.001 compared with control vector (pcDNA3) transfection (Student’s t test). b, c MCF-7 cells were transfected with pcDNA3 or pcDNA3-FOXP1-Myc (Myc-FOXP1) for 12 h and treated with E2 (100 nM) or EtOH for 24 h and then expression levels of SHP (b) and LRH-1 (c) were examined by qRT-PCR. *P < 0.05; ***P < 0.001 compared with control pcDNA3 (Student’s t test)
Positive FOXP1 Immunoreactivity Was Correlated with Favorable Prognosis for Distant Disease-Free Survival in Patients with Tamoxifen-Treated Breast Cancer
To examine the correlation of FOXP1 with the recurrence of breast cancer after endocrine therapy, immunohistochemical analysis was performed using samples from 113 patients with tamoxifen-treated invasive breast cancers, whose 5-year disease-free survival could be followed. Within 5 years after surgery, a relatively large fraction of relapse-free patients showed positive FOXP1 immunoreactivity (Table 2). These results suggest that the immunoreactivity for FOXP1 is positively associated with a favorable 5-year disease-free survival in patients with tamoxifen-treated breast cancer.
Discussion
Recent gene expression profiling studies have classified breast cancer into five intrinsic subtypes with unique molecular characteristics and prognostic significance [24, 25]. These include luminal subtypes A and B, HER2+/ER−, basal-like, and normal-like subtypes. Luminal subtypes A and B are ERα-positive breast cancers with subtype A expressing higher levels of ERα and having a better prognosis than subtype B [25]. FOXA1 expression correlates with luminal subtype A breast cancer and is a significant predictor of cancer-specific survival in patients with ER-positive tumors. The prognostic ability of FOXA1 in these low-risk breast cancers may prove to be useful in decisions regarding clinical treatment [26, 27].
FOXP1 is targeted by recurrent chromosome translocations, and its overexpression confers a poor prognosis in numerous types of lymphomas [28–32], suggesting that it functions as an oncogene. However, FOXP1 localizes to a tumor suppressor locus at 3p14.1 [10], and loss of FOXP1 expression in breast cancer is associated with a worse outcome [12]; this suggests that FOXP1 functions as a tumor suppressor in other tissue types. In the present study, we demonstrated that immunoreactivity for FOXP1 was significantly correlated with ERα immunoreactivity in breast cancer. FOXP1 immunoreactivity in breast cancers was also positively correlated with immunoreactivity for PgR, which is recognized as an estrogen-inducible gene [33]. These results suggest that FOXP1 is profoundly involved in estrogen signaling pathways in breast cancer. FOXP1 is associated with improved survival in primary and familial breast carcinoma [12, 34]. In addition, we showed that immunoreactivity for FOXP1 is positively associated with a favorable 5-year disease-free survival in patients with tamoxifen-treated breast cancer. Overall, FOXP1 expression is associated with a better clinical outcome and will be useful in deciding the clinical treatment for breast cancer, alongside FOXA1 [35, 36]. In the luminal epithelium and myoepithelium, FOXP1 was strongly positive in almost all constituent cells. In addition, in stromal cells, FOXP1 was also strongly positive in almost all constituent cells. These expression patterns of FOXP1 resembled those of ERβ [37, 38]. One explanation for this phenomenon is that FOXP1 might regulate ERβ expression levels, however, silencing of FOXP1 protein expression using siRNA did not reveal any effect on the expression of ERβ in the MCF-7 cell line [38]. Another possibility is that ERβ-dependent transcriptional activity is involved in FOXP1 gene expression. ERα and ERβ are assumed to have opposite effects on cellular function [39], postulating that FOXP1 may involved in antagonistic role of ERβ to ERα in these cells.
Consistent with the positive correlation between FOXP1 and ERα immunoreactivities, we demonstrated that FOXP1 mRNA expression was upregulated by estrogen in ERα-positive breast cancer MCF-7 cells. Supporting the role of estrogen in regulation of FOXP1, three ERBSs within the human FOXP1 chromosomal region, identified by genome-wide ChIP-on-chip analysis using MCF-7 cells [21], were significantly enriched by estrogen treatment in ChIP assays using the ERα antibody. Thus, it is possible that FOXP1 expression is regulated by ER through these ERBSs. FOXP1 mRNA level was significantly upregulated by threefold at 3 h after estrogen treatment, whereas irregularly regulated thereafter. Estrogen-mediated transcription of individual estrogen-responsive gene is distinctly and time-dependently regulated by promoter-specific associations of various factors, including ER, transcription factors other than ER, co-factors, chromatin remodeling factors, and general transcription factors. In addition, cyclic, proteasome-mediated turnover of ERα was assumed to permit continuous responses to changes in the concentration of estradiol [40], resulting in the temporal mRNA expression pattern of each gene. Indeed, a similar time-dependent mRNA expression pattern after estrogen stimulation is found in other estrogen-responsive genes including NRF-1 [41], postulating a common mechanism underlying the transcriptional regulation of these genes by estrogen.
The present study showed that FOXP1, as well as FOXA1, enhanced ERα-ERE-mediated transcription in breast cancer cells. One possible mechanism is that FOXP1 acts as a coregulator of ERα, because ligand-dependent activation of gene transcription by nuclear receptors such as ER and AR is dependent on the recruitment of coactivators. The α-helical LXXLL motif found in some coactivators is sufficient for ligand-dependent interaction with nuclear receptors, and this is present in the NH2 terminus of the FOXP1 protein [12]. Therefore, FOXP1 might physically associate with the ER through this motif. Interestingly, we previously reported that FOXP1 could interact with AR and act as a corepressor of AR-mediated transcription in an androgen-dependent manner [14]. These findings would also suggest that the coregulatory function of FOXP1 occurs in a nuclear receptor-specific manner. It would be worthwhile to identify the domains of the FOXP1 protein responsible for physical interaction with nuclear receptors and coregulatory effects on nuclear receptor-mediated gene expression.
FOXP1, 2, and 4 are highly related forkhead transcription factors expressed in various tissues, including the heart, lung, brain, and hematopoietic lineages. FOXP factors regulate cell proliferation in a cell context-dependent fashion [42]. Although regulation of cancer cell proliferation by FOXP1 has not previously been the subject of intensive study, FOXP1 overexpression was detected in gastric diffuse large B cell lymphoma, and FOXP1 knockdown efficiently blocked DLBCL proliferation [43]. In this study, FOXP1 was found to promote estrogen-dependent proliferation of MCF-7 cells, consistent with findings of a positive correlation between FOXP1 immunoreactivity and pT in breast cancer. In the endocardium, FOXP1 represses SOX17 expression, which, through regulation of β-catenin activity, controls FGF16/20 expression [44]. Furthermore, continued expression of FGF-20 is necessary for maintenance of the anchorage-independent growth state in RK3E cells, a rat epithelial cell line, transformed by β-catenin, implying that FGF-20 may be a critical element in oncogenesis induced by the Wnt signaling pathway [45]. Whether these regulation mechanisms are also involved in FOXP1-regulated breast cancer cell proliferation remains to be elucidated.
In summary, our results show that FOXP1 expression is induced by estrogen in breast cancer cells and that FOXP1 promotes cancer cell proliferation by enhancing ERE-mediated transcription. We further demonstrate that immunoreactivity for FOXP1 is positively correlated with favorable distant disease-free survival in the patients treated with adjuvant tamoxifen. These results suggest that pharmacological modulation of FOXP1 activity may be clinically useful to prevent and/or treat breast cancer and that evaluation of FOXP1 immunoreactivity may predict the therapeutic effect of tamoxifen on breast cancer.
Acknowledgments
We thank Saori Miyoshi, Kayoko Kanegae, and Wataru Sato, for their technical assistance. We also thank Tetsuya Fujimura, The University of Tokyo, for kindly providing human kidney tissue samples, and Shin-ichiro Horiguchi, Tokyo Metropolitan Cancer and Infectious diseases Center Komagome Hospital, for preparation of specimens. This work was supported in part by Grants-in-Aid for Cancer Research (21–4) from the Ministry of Health, Labor and Welfare; the Program for Promotion of Fundamental Studies in Health Science of the NIBIO; by grants of the Genome Network Project, the Cell Innovation Program and the Support Project of Strategic Research Center in Private Universities from the MEXT.
Conflict of Interest
The authors declare that they have no conflict of interest.
Abbreviations
- FOXP1
Forkhead box P1
- FOXA1
Forkhead box A1
- ERα
Estrogen receptor alpha
- PgR
Progesterone receptor
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- ERE
Estrogen response element
- siRNA
Small interfering RNA
- qRT-PCR
Quantitative real-time reverse transcriptase-polymerase chain reaction
References
- 1.Platet N, Cathiard AM, Gleizes M, Garcia M. Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit Rev Oncol Hematol. 2004;51:55–67. doi: 10.1016/j.critrevonc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Hall JM, McDonnell DP. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv. 2005;5:343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- 3.Carroll JS, Brown M. Estrogen receptor target gene: an evolving concept. Mol Endocrinol. 2006;20:1707–1714. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- 4.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistelinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grange T, Roux J, Rigaud G, Pictet R. Cell-type specific activity of two glucocorticoid responsive units of rat tyrosine aminotransferase gene is associated with multiple binding sites for C/EBP and a novel liver-specific nuclear factor. Nucleic Acids Res. 1991;19:131–139. doi: 10.1093/nar/19.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuur ER, Loktev AV, Sharma M, Sun Z, Roth RA, Weigel RJ. Ligand-dependent interaction of estrogen receptor-alpha with members of the forkhead transcription factor family. J Biol Chem. 2001;276:33554–33560. doi: 10.1074/jbc.M105555200. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang B, Lin D, Li C, Tucker P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem. 2003;278:24259–24268. doi: 10.1074/jbc.M207174200. [DOI] [PubMed] [Google Scholar]
- 10.Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P, Wood K, Cordell JL. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 2001;61:8820–8829. [PubMed] [Google Scholar]
- 11.Prown PJ, Ashe SL, Leich E, Burek C, Barrans S, Fenton JA, Jack AS, Pulford K, Rosenwald A, Banham AH. Potentially oncogenic B-cell activation induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL. Blood. 2008;111:2816–2824. doi: 10.1182/blood-2007-09-115113. [DOI] [PubMed] [Google Scholar]
- 12.Fox SB, Brown P, Han C, Ashe S, Leek RD, Harris AL, Banham AH. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor α and improved survival in primary human breast carcinomas. Clin Cancer Res. 2004;10:3521–3527. doi: 10.1158/1078-0432.CCR-03-0461. [DOI] [PubMed] [Google Scholar]
- 13.Banham AH, Boddy J, Launchbury R, Han C, Turley H, Malone PR, Harris AL, Fox SB. Expression of the forkhead transcriptional factor FOXP1 is associated both with hypoxia inducible factors (HIFs) and the androgen receptor in prostate cancer but is not directly regulated by androgens or hypoxia. Prostate. 2007;67:1091–1098. doi: 10.1002/pros.20583. [DOI] [PubMed] [Google Scholar]
- 14.Takayama K, Horie-Inoue K, Ikeda K, Urano T, Murakami K, Hayashizaki Y, Ouchi Y, Inoue S. FOXP1 is an androgen-responsive transcription factor that negatively regulates androgen receptor signaling in prostate cancer cells. Biochem Biophys Res Commun. 2008;374:388–393. doi: 10.1016/j.bbrc.2008.07.056. [DOI] [PubMed] [Google Scholar]
- 15.Ijichi N, Shigekawa T, Ikeda K, Horie-Inoue K, Fujimura T, Tsuda H, Osaki A, Saeki T, Inoue S. Estrogen-related receptor γ modulates cell proliferation and estrogen signaling in breast cancer. J Steroid Biochem Mol Biol. 2011;123:1–7. doi: 10.1016/j.jsbmb.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Allred DC, Clark GM, Elledge R, Fuqua SA, Brown RW, Chamness GC, Osborne CK, McGuire WL. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst. 1993;85:200–206. doi: 10.1093/jnci/85.3.200. [DOI] [PubMed] [Google Scholar]
- 17.Horie-Inoue K, Bono H, Okazaki Y, Inoue S. Identification and functional analysis of consensus androgen response elements in human prostate cancer cells. Biochem Biophys Res Commun. 2004;325:1312–1317. doi: 10.1016/j.bbrc.2004.10.174. [DOI] [PubMed] [Google Scholar]
- 18.Horie-Inoue K, Takayama K, Bono HU, Ouchi Y, Okazaki Y, Inoue S. Identification of novel steroid target genes through the combination of bioinformatics and functional analysis of hormone response elements. Biochem Biophys Res Commun. 2006;339:99–106. doi: 10.1016/j.bbrc.2005.10.188. [DOI] [PubMed] [Google Scholar]
- 19.Giamarchi C, Solanas M, Chailleux C, Augereau P, Vignon F, Rochefort H, Richard-Foy H. Chromatin structure of the regulatory regions of pS2 and cathepsin D genes in hormone-dependent and -independent breast cancer cell lines. Oncogene. 1999;18:533–541. doi: 10.1038/sj.onc.1202317. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda K, Ogawa S, Tsukui T, Horie-Inoue K, Ouchi Y, Kato S, Muramatsu M, Inoue S. Protein phosphatase 5 is a negative regulator of estrogen receptor-mediated transcription. Mol Endocrinol. 2004;18:1131–1143. doi: 10.1210/me.2003-0308. [DOI] [PubMed] [Google Scholar]
- 21.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 22.Lai K, Harnish DC, Evans MJ. Estrogen receptor α regulates expression of the orphan receptor small heterodimer partner. J Biol Chem. 2003;278:36418–36429. doi: 10.1074/jbc.M303913200. [DOI] [PubMed] [Google Scholar]
- 23.Annicotte JS, Chavey C, Servant N, Teyssier J, Bardin A, Licznar A, Badia E, Pujol P, Vignon F, Maudelonde T, Lazennec G, Cavailles V, Fajas L. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene. 2005;24:8167–8175. doi: 10.1038/sj.onc.1208950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 25.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lønning P, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, Dunn S, Huntsman DG, Nakshatri H. FOXA1 expression in breast cancer–correlation with luminal subtype A and survival. Clin Cancer Res. 2007;13:4415–4421. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
- 27.Thorat MA, Marchio C, Morimiya A, Savage K, Nakshatri H, Reis-Filho JS, Badve S. Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J Clin Pathol. 2008;61:327–332. doi: 10.1136/jcp.2007.052431. [DOI] [PubMed] [Google Scholar]
- 28.Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood. 2004;104:2933–2935. doi: 10.1182/blood-2004-03-1209. [DOI] [PubMed] [Google Scholar]
- 29.Banham AH, Connors JM, Brown PJ, Cordell JL, Ott G, Sreenivasan G, Farinha P, Horsman DE, Gascoyne RD. Expression of the FOXP1 transcription factor is strongly associated with inferior survival in patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2005;11:1065–1072. [PubMed] [Google Scholar]
- 30.Sagaert X, De Paepe P, Libbrecht L, Vanhentenrijk V, Verhoef G, Thomas J, Wlodarska I, De Wolf-Peeters C. Forkhead box protein P1 expression in mucosa-associated lymphoid tissue lymphomas predicts poor prognosis and transformation to diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:2490–2497. doi: 10.1200/JCO.2006.05.6150. [DOI] [PubMed] [Google Scholar]
- 31.Goatly A, Bacon CM, Nakamura S, Ye H, Kim I, Brown PJ, Ruskoné-Fourmestraux A, Cervera P, Streubel B, Banham AH, Du MQ. FOXP1 abnormalities in lymphoma: translocation breakpoint mapping reveals insights into deregulated transcriptional control. Mod Pathol. 2008;21:902–911. doi: 10.1038/modpathol.2008.74. [DOI] [PubMed] [Google Scholar]
- 32.Hoeller S, Schneider A, Haralambieva E, Dirnhofer S, Tzankov A. FOXP1 protein overexpression is associated with inferior outcome in nodal diffuse large B-cell lymphomas with non-germinal centre phenotype, independent of gains and structural aberrations at 3p14.1. Histopathology. 2010;57:73–80. doi: 10.1111/j.1365-2559.2010.03600.x. [DOI] [PubMed] [Google Scholar]
- 33.Kastner P, Krust A, Turcotte B, Stropp U, Tora L, Gronemeyer H, Chambon P. Two distinct estrogen-regulated promoters generate transcripts encoding the two functionally different human progesterone receptor forms A and B. EMBO J. 1990;9:1603–1614. doi: 10.1002/j.1460-2075.1990.tb08280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rayoo M, Yan M, Takano EA, Bates GJ, Brown PJ, Banham AH, Fox SB. Expression of the forkhead box transcription factor FOXP1 is associated with oestrogen receptor alpha, oestrogen receptor beta and improved survival in familial breast cancers. J Clin Pathol. 2009;62:896–902. doi: 10.1136/jcp.2009.065169. [DOI] [PubMed] [Google Scholar]
- 35.Wolf I, Bose S, Williamson EA, Miller CW, Karlan BY, Koeffler HP. FOXA1: growth inhibitor and a favorable prognostic factor in human breast cancer. Int J Cancer. 2007;120:1013–1022. doi: 10.1002/ijc.22389. [DOI] [PubMed] [Google Scholar]
- 36.Habashy HO, Powe DG, Rakha EA, Ball G, Paish C, Gee J, Nicholson RI, Ellis IO. Forkhead-box A1 (FOXA1) expression in breast cancer and its prognostic significance. Eur J Cancer. 2008;44:1541–1551. doi: 10.1016/j.ejca.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 37.Skliris GP, Carder PJ, Lansdown MR, Speirs V. Immunohistochemical detection of ERβ in breast cancer: towards more detailed receptor profiling? Br J Cancer. 2001;84:1095–1098. doi: 10.1054/bjoc.2001.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bates GJ, Fox SB, Han C, Launchbury R, Leek RD, Harris AL, Banham AH. Expression of the forkhead transcription factor FOXP1 is associated with that of estrogen receptorβ in primary invasive breast carcinomas. Breast Cancer Res Treat. 2008;111:453–459. doi: 10.1007/s10549-007-9812-4. [DOI] [PubMed] [Google Scholar]
- 39.Sotoca AM, van den Berg H, Vervoort J, van der Saag P, Ström A, Gustafsson JA, Rietjens I, Murk AJ. Influence of cellular ERα/ERβ ratio on the ERα-agonist induced proliferation of human T47D breast cancer cells. Toxicol Sci. 2008;105:303–311. doi: 10.1093/toxsci/kfn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Métivier R, Penot G, Hübner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/S0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 41.Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of nuclear respiratory factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. 2008;22:609–622. doi: 10.1210/me.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang B, Weidenfeld J, Lu MM, Maika S, Kuziel WA, Morrisey EE, Tucker PW. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004;131:4477–4487. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- 43.Craig VJ, Cogliatti SB, Imig J, Renner C, Neuenschwander S, Rehrauer H, Schlapbach R, Dirnhofer S, Tzankov A, Müller A. Myc-mediated repression of microRNA-34a promotes high grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117(23):6227–6236. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Li S, Yuan L, Tian Y, Weidenfeld J, Yang J, Liu F, Chokas AL, Morrisey EE. Foxp1 coordinates cardiomyocyte proliferation through both cell-autonomous and nonautonomous mechanisms. Genes Dev. 2010;24:1746–1757. doi: 10.1101/gad.1929210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE. FGF-20 and DKK1 are transcriptional targets of β-catenin and FGF-20 is implicated in cancer and development. EMBO J. 2005;24:73–84. doi: 10.1038/sj.emboj.7600460. [DOI] [PMC free article] [PubMed] [Google Scholar]