Abstract
Breast cancer is primarily a hormone-dependent tumor that can be regulated by the status of the steroid hormones estrogen and progesterone. Forkhead box A1 (FOXA1) is a member of the forkhead box transcription factor family and functions as a pioneer factor of the estrogen receptor (ER) in breast cancer. In the present study, we demonstrate that FOXA1 mRNA was upregulated by estrogen and that estrogen receptor-α (ERα) recruitment to ER-binding sites in the vicinity of the FOXA1 gene was increased by estrogen in ERα-positive MCF-7 breast cancer cells. The estrogen-induced FOXA1 upregulation was repressed by 4-hydroxytamoxifen treatment. We also demonstrated that the proliferation and the migration of MCF-7 cells were decreased by FOXA1-specific small interfering RNA (siRNA; siFOXA1). Furthermore, siFOXA1 decreased the estrogen response element-driven transcription and the estrogen-dependent upregulation of ERα target genes in MCF-7 cells. Next, the immunohistochemical analyses of FOXA1 were performed using two groups of breast cancer specimens. The nuclear immunoreactivity of FOXA1 was detected in 80 (74 %) of 108 human invasive breast cancers and was negatively correlated with tumor grade and positively correlated with hormone receptor status, including ERα and progesterone receptor, pathological tumor size, and immunoreactivity of FOXP1, another FOX family transcription factor. FOXA1 immunoreactivity was significantly elevated in the relapse-free breast cancer patients treated with tamoxifen. Notably, the double-positive immunoreactivities of FOXA1 and FOXP1 were significantly associated with a favorable prognosis for the relapse-free and overall survival of patients with tamoxifen-treated breast cancer, with lower P values compared with FOXA1 or FOXP1 immunoreactivity alone. These results suggest that FOXA1 plays an important role in the proliferation and migration of breast cancer cells by modulating estrogen signaling and that the double-positive immunoreactivities of FOXA1 and FOXP1 are associated with a favorable prognosis of tamoxifen-treated breast cancer.
Keywords: Breast Cancer, Breast Cancer Cell, Luminal Breast Cancer, FOXA1 Expression, Conventional ChIP Assay
Introduction
Estrogen signaling pathways regulate various cellular events, including cell growth and apoptosis, through the activation of estrogen receptor alpha (ERα) [1]. ERα functions as a transcription factor to activate the target gene expression. Clinically, ERα is noted as the defining feature of luminal breast cancers, which comprise most breast cancers. Luminal breast cancers are generally treated with endocrine therapies, including the antiestrogen tamoxifen which acts as an antagonist for the estrogen receptor (ER) in breast cancer cells. Due to the sensitivity to endocrine therapy, ER-positive luminal breast cancers are considered to have better prognosis than ER-negative breast cancers. However, resistance for antiestrogen therapies is acquired in a substantial fraction of recurrent breast cancers. Identification of the factors involved in drug resistance, recurrence, or poor prognosis of breast cancer would be useful for the treatment and the diagnosis of the disease.
The transcriptional activity of ERα is regulated by various co-activators and co-repressors [2], as well as by interactions with other transcription factors, including the forkhead box (FOX) family. FOX family transcription factors influence ERα-regulated transcription by interacting with the ERα protein, as exemplified by FOXA1 [3]. Recent global gene expression studies of breast cancer revealed that a high FOXA1 expression was positively correlated with ERα and PR, but negatively correlated with histological grade and proliferation markers [4–6]. In addition, FOXA1 expression was associated with better cancer-specific survival, indicating that it is a better predictor of survival in breast cancer [4–6]. Recent genome-wide studies aimed at identifying ERα and androgen receptor (AR)-binding sites have shown that FOXA1 plays a role in regulating both of these nuclear receptor networks [7,8]. FOXA1 is recognized as a pioneer transcription factor because chromatin binding by this protein can enable subsequent binding by the ER and AR [8,9]. More recently, genome-wide mapping of ERα-, AR-, and FOXA1-binding events in breast and prostate cancer cells by the use of high-throughput sequencing has uncovered additional details of transcriptional control mechanisms in the nuclear receptor-mediated transcription involved in several collaborative factors, including TLE1 and AP-2γ [10,11].
Gene expression profiling studies have classified breast cancer into five intrinsic subtypes with unique molecular characteristics and prognostic significance [12,13]: the luminal A and B, HER2+/ER−, basal-like, and normal-like subtypes. Luminal subtypes A and B are ERα-positive breast cancers, with subtype A expressing higher levels of ERα and having a better prognosis than subtype B [13]. FOXA1 expression correlates with luminal subtype A breast cancer, and the immunoreactivity of FOXA1 is shown as a significant predictor of cancer-specific survival in patients with ER-positive tumors [5]. The prognostic ability of FOXA1 in these low-risk breast cancers may prove to be useful in clinical treatment decisions [5,6].
The altered expression of FOXP1, another FOX family member, is associated with various types of tumors, including breast cancer [14–18]. We recently demonstrated that FOXP1 plays an important role in the proliferation of breast cancer cells by modulating estrogen signaling, and FOXP1 immunoreactivity could be associated with the immunoreactivity of ERα and PR in breast cancer, which may predict favorable prognosis in patients treated with tamoxifen [19]. However, the clinical significance of FOXA1 on tamoxifen resistance in breast cancer and the correlation between FOXA1 and FOXP1 remain to be elucidated.
In the present study, we show that FOXA1 mRNA expression is induced by estrogen and that ERα recruitment to the ER-binding sites in the FOXA1 gene region is elevated by estrogen in ERα-positive MCF-7 human breast cancer cells. Functional analyses reveal that the knockdown of FOXA1 impairs the proliferation and migration of MCF-7 cells as well as ER-mediated transcription and estrogen-dependent upregulation of ERα target genes in MCF-7 cells. Furthermore, we evaluated the FOXA1 expression in human breast cancers using immunohistochemistry and investigated the correlations between FOXA1 expression levels and clinicopathophysiological findings. Finally, FOXA1 immunoreactivity is negatively associated with the recurrence of tamoxifen-treated breast cancer. Together with our previous report on FOXP1 [19], the double-positive immunoreactivities of FOXA1 and FOXP1 are significantly associated with longer survival in patients with tamoxifen-treated breast cancer. Our study provides new insight into the association between FOXA1 and FOXP1 in estrogen signaling and the prognosis of tamoxifen-treated breast cancer.
Materials and Methods
Tissue Selection and Patient Characteristics
Between January 2005 and March 2006, 108 consecutive patients with invasive breast cancer diagnosed using a vacuum-assisted biopsy device (Mammotome, Ethicon Endo-surgery Inc., Cincinnati, OH) at the Saitama Medical University Hospital were included in a cohort study.
For a nested control study of the therapeutic effect of tamoxifen on recurrent breast cancer, 162 patients with breast cancer diagnosed between 1989 and 1998 with or without distant metastases during or after tamoxifen therapy were identified from three institutions (National Hospital Organization Shikoku Cancer Center, Matsuyama, Japan; National Cancer Center Hospital, Tokyo, Japan; and Tokyo Metropolitan Cancer and Infectious Diseases Center, Komagome Hospital, Tokyo, Japan). Patients experiencing relapse were defined as those with distant metastases within 5 years after surgery followed by tamoxifen treatment, whereas those who were considered relapse-free were those without distant metastases. A total of 113 patients met our protocol criteria and were included in this study.
Formalin-fixed paraffin-embedded tissue sections obtained by biopsy or surgery were used in these studies. These studies were approved by the institutional review board of Saitama Medical University, and informed consent was obtained from all the patients. The clinicopathological characteristics of the series are presented in Tables 1 and 2.
Table 1.
Relationship between the immunoreactivity of FOXA1 and clinicopathological findings in invasive breast cancer (n = 108)
| Clinical findings | Immunoreactivity score of FOXA1a | |||
|---|---|---|---|---|
| Positive (n = 80) | Negative (n = 28) | P value | ||
| Ageb (mean ± SD) | 59.9 ± 14.8 | 57.1 ± 14.2 | ||
| ≤50 | 23 (21.3 %) | 9 (8.3 %) | 0.92 | |
| >50 | 57 (52.8 %) | 19 (17.6 %) | ||
| pT | ≤20 mm | 44 (48.9 %) | 9 (10.0 %) | 0.047 |
| >20 mm | 23 (25.6 %) | 14 (15.5 %) | ||
| Stage | I, II | 73 (68.2 %) | 28(26.2 %) | 0.31 |
| III, IV | 6 (5.6 %) | 0 (0.0 %) | ||
| Grade | I, II | 58 (70.7 %) | 16 (19.5 %) | 0.024 |
| III | 3 (3.7 %) | 5 (6.1 %) | ||
| ER | Positive (PS ≥ 3) | 56 (51.9 %) | 13 (12.0 %) | 0.025 |
| Negative (PS ≤ 2) | 24 (22.2 %) | 15 (13.9 %) | ||
| PgR | Positive (PS ≥ 3) | 36 (33.3 %) | 5 (4.6 %) | 0.020 |
| Negative (PS ≤ 2) | 44 (40.8 %) | 23 (21.3 %) | ||
| HER2 | Positive | 17 (18.5 %) | 9 (9.8 %) | 0.37 |
| Negative | 51 (55.4 %) | 15 (16.3 %) | ||
| Lymph node | Positive | 28 (31.5 %) | 9 (10.1 %) | 0.92 |
| Negative | 41 (46.1 %) | 11 (12.3 %) | ||
| FOXP1a | Positive | 58 (55.2 %) | 13 (12.4 %) | 0.027 |
| Negative | 21 (20.0 %) | 13 (12.4 %) | ||
All other values represent the number and proportion of cases
ER estrogen receptor, PgR progesterone receptor, TS total score, PS proportion score
aFOXA1 and FOXP1 immunoreactivity scores of 0, 2, and 3–8 were defined as negative and positive immunoreactivity, respectively
bData are presented as the mean ± SD
Table 2.
Clinicopathological findings in adjuvant tamoxifen-treated invasive breast cancer patients followed up for 5 years after surgery (n = 113)
| Clinical findings | Relapse (n = 43) | Relapse-free (n = 70) | P value | |
|---|---|---|---|---|
| Agea (mean ± SD) | 53.2 ± 9.9 | 55.2 ± 12.4 | 0.39 | |
| Age | ≤50 | 20 (17.7 %) | 28 (24.8 %) | 0.50 |
| >50 | 23 (20.3 %) | 42 (37.2 %) | ||
| pT | ≤30 mm | 22 (19.5 %) | 52 (46.0 %) | 0.012 |
| >30 mm | 21 (18.6 %) | 18 (15.9 %) | ||
| Lymph node | Positive (n ≥ 1) | 30 (26.5 %) | 30 (26.5 %) | 0.005 |
| Negative (n = 0) | 13 (11.5 %) | 40 (35.4 %) | ||
| ERα | Positive | 39 (34.5 %) | 64 (56.6 %) | 0.84 |
| Negative | 4 (3.6 %) | 6 (5.3 %) | ||
| PgR | Positive | 36 (31.9 %) | 60 (53.1 %) | 0.99 |
| Negative | 7 (6.2 %) | 10 (8.8 %) | ||
| FOXA1 | Positive | 19 (16.8 %) | 51 (45.1 %) | 0.002 |
| Negative | 24 (21.2 %) | 19 (16.8 %) | ||
| FOXP1 | Positive | 22 (19.5 %) | 57 (50.4 %) | <0.001 |
| Negative | 21 (18.6 %) | 13 (11.5 %) | ||
All other values represent the number and proportion of cases
pT pathological T stage, ERα estrogen receptor-α, PgR progesterone receptor
aData are presented as the mean ± SD
Antibodies
Mouse monoclonal anti-FOXA1 antibody was purchased from Abcam (Cambridge, UK). Affinity-purified rabbit polyclonal anti-FOXP1 antibody (anti-GX5050) was generated by the Genome Network Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan (http://genomenetwork.nig.ac.jp), from serum derived from rabbits immunized with a peptide epitope consisting of amino acids 519–677 of the human FOXP1 protein.
Immunohistochemistry
The immunohistochemical analyses for FOXA1 and FOXP1 were performed using an EnVision+ visualization kit (Dako, Carpinteria, CA) as previously described [20]. The tissue sections (6 μm) were deparaffinized, rehydrated through graded ethanol, and rinsed in Tris-buffered saline with 0.05 % Tween-20 (TBST). For antigen retrieval, the sections were heated in an autoclave at 121 °C for 5 min in 10 mM sodium citrate buffer (pH 6.0). The sections were blocked with endogenous peroxidase (0.3 % H2O2) and incubated in 10 % fetal bovine serum for 30 min. The primary antibody, a monoclonal antibody for FOXA1 (1:2,000 dilution) or a polyclonal antibody for FOXP1 (1:1,000 dilution), was applied, and the samples were incubated overnight at 4 °C. The sections were rinsed in TBST and incubated with EnVision+ horseradish peroxidase-labeled polymer for 1 h at room temperature. The antigen–antibody complex was visualized using the 3,3′-diaminobenzidine substrate kit for peroxidase (Vector Laboratories, Burlingame, CA). Mouse or rabbit immunoglobulin G was used in place of the primary antibody as a negative control.
Immunohistochemical Assessment
The slides were evaluated for the proportion [proportion score (PS): 0, none; 1, <1/100; 2, 1/100–1/10; 3, 1/10–1/3; 4, 1/3–2/3; and 5, >2/3] and the staining intensity [intensity score (IS): 0, none; 1, weak; 2, moderate; and 3, strong] of positively stained cells. The total immunoreactivity score (TS = 0, 2–8) was determined as the sum of the proportion and intensity scores [21]. Two investigators (H.T. and A.O.) evaluated the tissue sections independently. If the immunoreactivity score differed between the two investigators, a third investigator (T.S.) evaluated the tissue sections and the mean immunoreactivity score was used. When the two investigators found it difficult to evaluate the TS of heterogeneous cancerous lesions, the third investigator made the deciding estimate. To identify potential correlations between the FOXA1 or FOXP1 expression in the malignant epithelium and clinicopathological characteristics, immunoreactivity scores of 0 or 2 and 3–8 were defined as negative and positive immunoreactivity, respectively.
Small Interfering RNA
Synthetic small interfering RNA (siRNA) duplexes targeting the human FOXA1 gene (Silencer Select Pre-designed siRNA) and the luciferase reporter plasmid pGL2 (Luciferase GL2 Duplex) used as a negative control were purchased from Applied Biosystems (Carlsbad, CA) and Dharmacon (Lafayette, CO), respectively.
Cell Culture and Transfection
MCF-7 cells were purchased from the American Type Culture Collection (Rockville, MD) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10 % fetal bovine serum (FBS) at 37 °C in 5 % CO2. 17β-Estradiol (E2) and 4-hydroxytamoxifen (4-OHT) were purchased from Sigma-Aldrich (St. Louis, MO). The transfection of siRNA was performed using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol.
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction
Total RNA extraction, first-strand cDNA synthesis, and quantitative PCR (qPCR) protocols have been described elsewhere [20]. The primers for FOXA1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), progesterone receptor (PgR), and growth regulation by estrogen in breast cancer 1 (GREB1) are as follows: FOXA1, 5′-AGGTGTGTATTCCAGACCCG-3′ and 5′-TTGACGGTTTGGTTTGTGTG-3′; GAPDH, 5′-GGTGGTCTCCTCTGACTTCAACA-3′ and 5′-GTGGTCGTTGAGGGCAATG-3′; PgR, 5′-CGTGCCTATCCTGCCTCTCA-3′ and 5′-CCGCCGTCGTAACTTTCG-3′; GREB1, 5′-CGACCATCGGCTTTAGGTATCTT-3′ and 5′-CCACCCTTTGTGGCGTTTT-3′. The fold induction of the mRNA expression levels was determined by comparing the estrogen-treated samples with the vehicle-treated controls.
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation (ChIP) assay and the qPCR were performed as previously described [22,23]. After 72-h hormone depletion, MCF-7 cells were treated with 100 nM E2 or vehicle (0.1 % ethanol) for 45 min. The cells were fixed in 1 % formaldehyde for 5 min at room temperature. Chromatin was sheared to a mean size of 500 bp by sonication using a Bioruptor ultrasonicator (Cosmo-Bio, Tokyo, Japan). The lysates were rotated overnight at 4 °C, with an ERα-specific antibody (sc-543, Santa Cruz Biotechnologies, Santa Cruz, CA). Protein A-agarose (Upstate Biotechnology, Lake Placid, NY) was added and incubated for 2 h. Precipitated DNA was used as a template for qPCR using an ABI PRISM 7000 sequence detector (Applied Biosystems, Foster City, CA) based on SYBR Green I fluorescence. A genomic fragment containing ERE in the enhancer region of trefoil factor 1 (TFF1; 405/393 bp from the transcription start site) was used as a positive control for ER binding [24]. The sequences of PCR primers are as follows: TFF1_ERE, 5′-TGAGATTCAGAAAGTCCCTCTTTC-3′ and 5′-TGGGCTTCATGAGCTCCTT-3′; FOXA1_ER_8127, 5′-TGGGTCAGGAAGAAAACATCTGT-3′ and 5′-TGTGGAAACACCCAGACGAA-3′; FOXA1_ER_8128, 5′-GAGGACGGCCCGACTTG-3′ and 5′-CACCGGTTTATATCTTTATGACATAACTTT-3′; GAPDH, 5′-GGTGGTCTCCTCTGACTTCAACA-3′ and 5′-GTGGTCGTTGAGGGCAATG-3′; Myoglobin, 5′-AAGTTTGACAAGTTCAAGCACCTG-3′ and 5′-TGGCACCATGCTTCTTTTAAGTC-3′.
Luciferase Assay
The MCF-7 cells were plated in 24-well culture plates at a density of 10,000 cells per well in phenol red-free medium containing 10 % charcoal-stripped FBS and transfected with 0.1 μg of a luciferase reporter vector containing ERE (ERE-TK-LUC) [25] and 0.02 μg of pRL-CMV (Promega, Madison, WI), together with siRNA for FOXA1 using the Lipofectamine 2000 reagent (Invitrogen). Twelve hours after the transfection, the cells were treated with 100 nM E2 or vehicle (ethanol) for 24 h, and the luciferase activities were then determined using the MicroLumatPlus microplate luminometer (Berthold Technologies, Bad Wildbad, Germany) and the Dual-Luciferase Assay System (Promega). The data are expressed as the mean ± SD of three independent experiments performed in triplicate.
Cell Proliferation Assay
The MCF-7 cells were seeded in 96-well plates at a density of 5,000 cells per well in DMEM containing 10 % FBS for 24 h. Then, 20 pmol of the siRNA described above was transfected for 12 h and incubated for 96 h. Cell proliferation was examined using a (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt; WST-8) assay kit (Nacalai Tesque, Kyoto, Japan), performed according to the manufacturer’s protocol.
Cell Migration Assay
To measure the cell migration activity, migration assays were performed using the Cell Culture Inserts for 24-well plates (8.0-μm pore; BD Falcon, BD Biosciences, Franklin Lakes, NJ). Before cell culturing, the inside of the insert membranes was coated with purified fibronectin (BD Falcon) in PBS at a concentration of 10 μg/mL for 30 min at room temperature. After the transfection of siRNA for 48 h, the MCF-7 cells were collected and resuspended in DMEM containing 10 % FBS. The MCF-7 cells were subsequently added to the upper chamber at 3 × 105 cells per well. After the 24-h incubation, the cells on the upper surface of the membrane were completely wiped away with a cotton swab. The cells on the lower surfaces of the membrane were fixed with 100 % methanol, the insert membranes were cut and stained with Giemsa stain solution (Wako, Tokyo, Japan), and the permeating cells within five randomly selected areas were counted under an inverted microscope. At least three independent experiments were performed for all the conditions. The data are shown as the mean ± SD.
Statistical Analyses
The differences between the two groups were analyzed using a two-sample, two-tailed Student’s t test. Value of P <0.05 was considered statistically significant. All data are presented in the text and the figures as the mean ± SD. The correlation between the immunoreactivity score and the clinicopathological characteristics was evaluated using the chi-square test. Relapse-free and overall survival curves were obtained using the Kaplan–Meier method and verified using the log-rank (Mantel–Cox) test. Univariate and multivariate analyses were performed using a logistic regression model with the JMP 9 software (SAS Institute, Cary, NC), and P < 0.05 was regarded as statistically significant.
Results
Estrogen Upregulates FOXA1 mRNA Expression and ERα Recruitment to the Putative ER-Binding Sites Within the FOXA1 Gene Region in MCF-7 Cells
We examined the transcriptional regulation of the FOXA1 gene by 17β-estradiol (E2) in the ERα-positive MCF-7 cells (Fig. 1a). The FOXA1 mRNA level was significantly upregulated twofold at 3 h and threefold at 48 h after E2 stimulation, while E2-mediated upregulation of FOXA1 was repressed by 4-OHT (Fig. 1b). These results suggest that the mRNA expression of FOXA1 is transcriptionally regulated by estrogen in breast cancer cells owing to the relatively early response (by 1 h). In support of these results, the genome-wide ChIP-on-chip analysis using the MCF-7 cells reported by Carroll et al. [26] demonstrated the presence of two estrogen receptor-binding sites (ERBSs; ER_8127 and 8128) within a 3-kb distance from the start or end point of the gene (Fig. 1c). To determine whether these ERBSs are bona fide ERBSs in the MCF-7 cells, we performed a conventional ChIP assay using ERα-specific antibody in comparison with a known estrogen response element of TFF1 as a positive control and the coding region of myoglobin (MB) as a negative control (normalized by the coding region of GAPDH) [24]. As shown in Fig. 1d, basal estrogen-independent ERα binding on ER_8128 in the FOXA1 promoter region is larger than that on ER_8127 in the downstream region, and substantial E2-induced ERα recruitment is observed in both ER_8127 and ER_8128. These data suggest that both ERBSs contribute to the estrogen-dependent transcription of FOXA1.
Fig. 1.
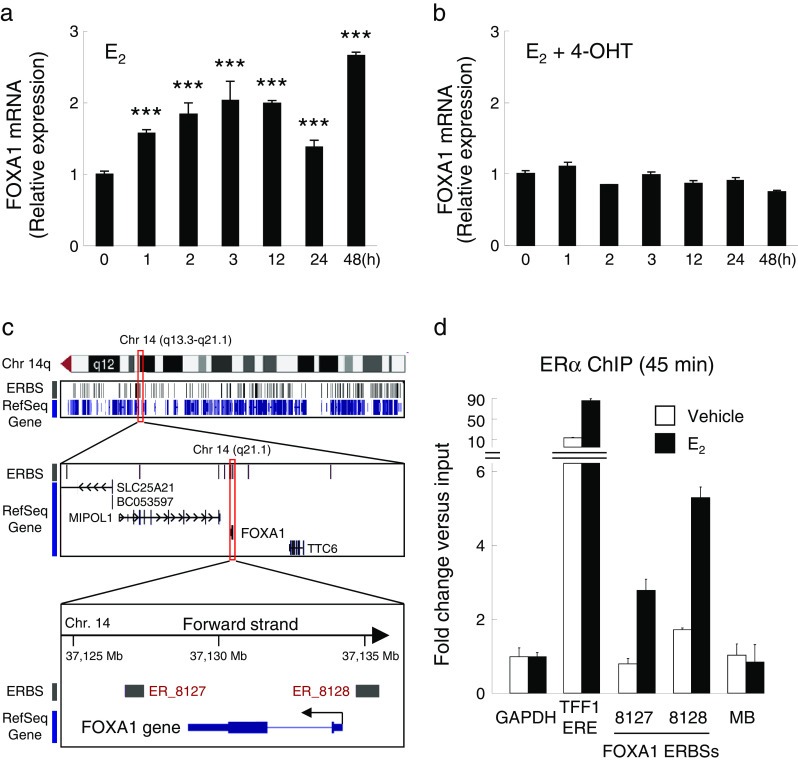
Estrogen-induced expression of FOXA1 in the MCF-7 cells. a, b MCF-7 cells were treated with 100 nM 17β-estradiol (E2) (a) or with 100 nM E2 and 1 nM 4-OHT (b) for 48 h. The FOXA1 mRNA level was examined at indicated time points by qRT-PCR; the results are shown as a fold change over the expression level at 0 h. ***P < 0.001 compared with 0 h (by Student’s t test). c Schematic representation of estrogen receptor-binding sites (ERBSs) in the vicinity of FOXA1 gene in chromosome 14. Cytogenetic location of the FOXA1 gene in chromosome 14 and UCSC Genome Browser view customized with estrogen-dependent ERα binding sites (ERBSs) determined by ChIP-on-chip at the 14q region [26] are shown. Two significant ERBSs (ER_8127 and 8128) at a cutoff value P < 1e−3 are identified in the vicinity of FOXA1 gene region. d Estrogen-dependent recruitment of ERα to the FOXA1 ERBSs. The MCF-7 cells were treated with 100 nM of E2 or vehicle for 45 min and then subjected to ChIP analysis with the ERα-specific antibody. Immunoprecipitated DNA was quantified by qPCR; the results were represented as the fold enrichment of ERα occupancy in the immunoprecipitated DNAs vs. input DNAs. The estrogen response element of the Trefoil factor 1 (TFF1) gene (TFF1 ERE) and non-ERBS within the myoglobin (MB) gene region were used as the positive and negative controls, respectively
FOXA1 Contributes to Breast Cancer Cell Proliferation and Migration
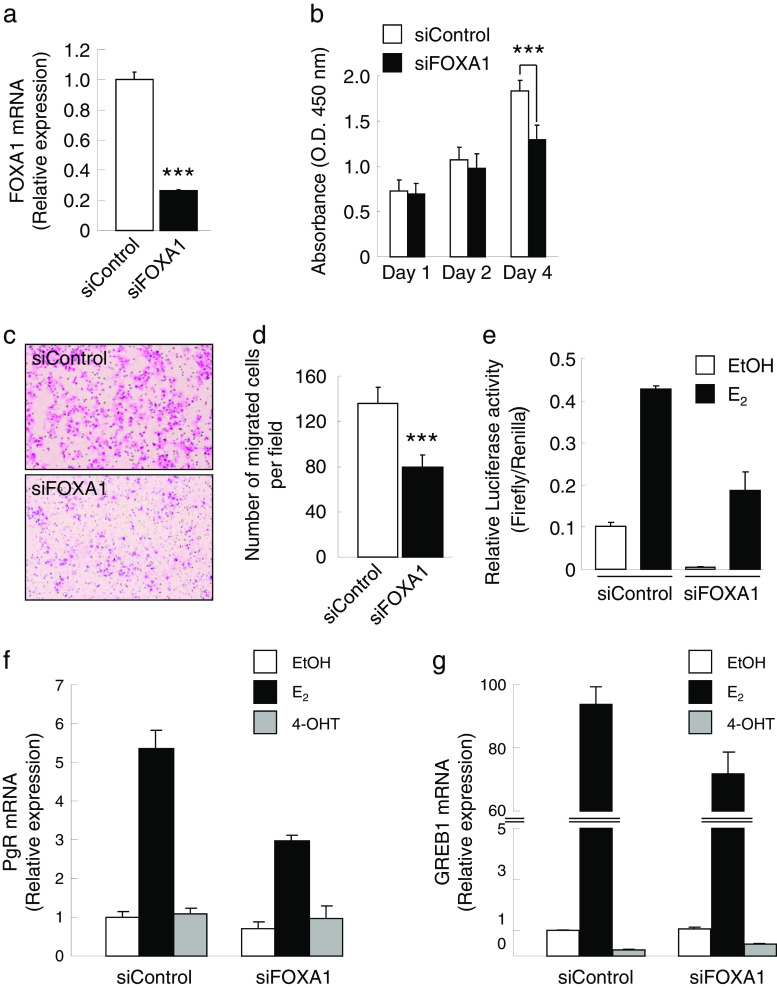
To assess the role of FOXA1 in breast cancer, we performed loss-of-function studies for it. Transient transfection of siFOXA1 in MCF-7 cells resulted in a downregulated FOXA1 expression compared with that of the control siRNA (siControl), as confirmed by qRT-PCR (Fig. 2a). The MCF-7 cell growth was suppressed by the siFOXA1 treatment (Fig. 2b). Furthermore, the siFOXA1 treatment resulted in a decreased MCF-7 cell migration (Fig. 2c, d). These results indicate that FOXA1 regulates breast cancer cell proliferation and migration.
Fig. 2.
Knockdown of FOXA1 suppresses cell proliferation and migration of the MCF-7 cells. a Total RNA was prepared from the MCF-7 cells transfected with 20 nM of siRNA specific for FOXA1 (siFOXA1) or control siRNA (siControl) for 48 h; the FOXA1 mRNA level was examined by quantitative reverse transcription polymerase chain reaction (qRT-PCR). ***P < 0.001 compared with the siControl (Student’s t test). b MCF-7 cells were transfected with 20 nM of siFOXA1 or siControl for 4 days in Dulbecco’s modified Eagle medium (DMEM). The relative cell proliferation at indicated time points was examined using a WST-8 assay kit and determined photometrically as the absorbance at OD = 450 nm. ***P < 0.001 compared with the siControl (Student’s t test). c, d The MCF-7 cells were transfected with 20 nM of siFOXA1 or siControl for 48 h in DMEM, and then the cells were transferred into transwells for an additional 48 h. The representative photographs (c) and numbers of migrated cells (d) are shown. e Effect of FOXA1 on estrogen response element (ERE)-mediated transcription in the MCF-7 cells. The MCF-7 cells were transfected with a DNA mixture of 0.1 μg of ERE-TK-LUC, 0.02 μg of pRL-CMV, and 20 nM of siFOXA1 or siControl. After 12 h, the cells were treated with 17β-estradiol (100 nM) or vehicle (EtOH) for 24 h, and then the cell lysates were prepared for and used in the luciferase assay. f, g Effect of siFOXA1 on the expression of ERα target genes. MCF-7 cells were transfected with 20 nM of siFOXA1 or siControl. After 12 h, cells were treated with E2, 4-OHT, or EtOH for 24 h. mRNA levels of progesterone receptor (PgR) (f) and growth regulation by estrogen in breast cancer 1 (GREB1) (g) were examined by qRT-PCR; the results are shown as a fold change over EtOH treatment
FOXA1 Regulates ER-Mediated Transcription
To examine whether FOXA1 influences ER-ERE-mediated transcription, ERE-TK-LUC was introduced into the MCF-7 cells together with siFOXA1 or siControl (Fig. 2e). The results indicate that siFOXA1 reduced ER-ERE-mediated transactivation in the presence or absence of estrogen in MCF-7 cells. To further confirm the contribution of FOXA1 to ER-mediated transcriptional regulation, we evaluated the expressions of endogenous ERα target genes including PgR and GREB1 in the presence or absence of FOXA1 siRNA (Fig. 2f, g). We showed that E2-induced expressions of PgR and GREB1 were reduced by FOXA1 siRNA treatment, suggesting that FOXA1 affects E2-mediated target gene activation.
Correlation Between FOXA1 Immunoreactivity and the Clinicopathological Features of Invasive Breast Cancer
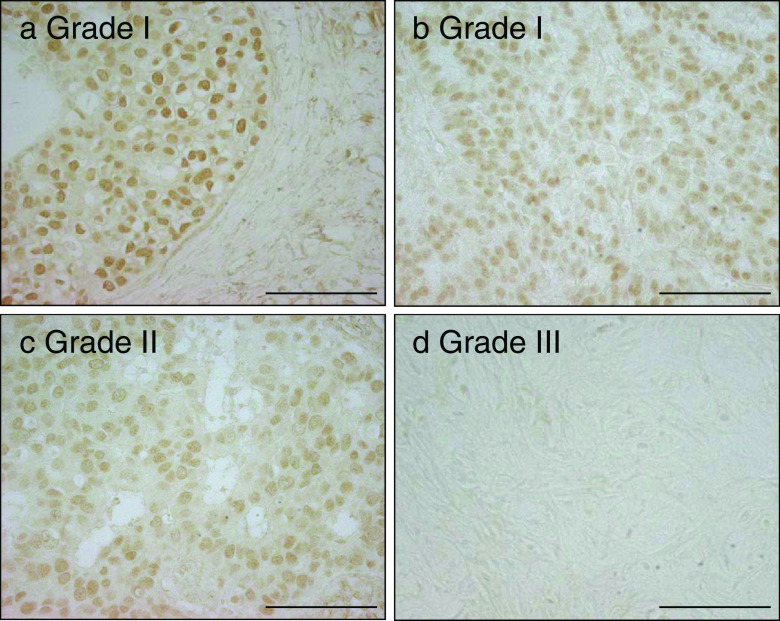
To investigate the expression levels of the FOXA1 protein in breast cancer, immunohistochemical analysis was performed using samples from 108 invasive breast cancers. Representative images of FOXA1 immunostaining are shown in Fig. 3. Positive nuclear immunoreactivity of FOXA1 was detected in 74 % of breast cancer specimens (Table 1). FOXA1 immunoreactivity in grade III breast cancer was significantly lower than that in grade I breast cancer. Namely, the mean Allred score of FOXA1 immunoreactivity in grade I was 5.83, whereas that in grade III was 2.38 (P = 0.002). The nuclear immunoreactivity of FOXA1 was significantly correlated with ERα immunoreactivity (P = 0.025, Table 1). The positive nuclear immunoreactivity of FOXA1 was also significantly correlated with pathological tumor size (pT), low histological grade, and positive immunoreactivity of PgR and FOXP1 (P = 0.047, 0.024, 0.020, and 0.027, respectively), whereas no significant association was found with other clinicopathological characteristics (Table 1).
Fig. 3.
Immunohistochemistry of FOXA1 in breast cancer. The representative images of the immunohistochemical staining of breast cancer tissues with the anti-FOXA1 antibody are shown. Positive staining for FOXA1 was observed in the nuclei of the breast cancer cells. The FOXA1 immunoreactivities in grade III breast cancer were significantly lower than those in grade I breast cancer (5.83 vs. 2.38, P = 0.002). Bar, 100 μm
Positive FOXA1 Immunoreactivity is Correlated with Good Prognosis for the Relapse-Free Survival in Patients with Tamoxifen-Treated Breast Cancer
To examine the correlation of FOXA1 with the recurrence of breast cancer after endocrine therapy, immunohistochemical analysis was performed using samples from 113 patients with tamoxifen-treated invasive breast cancers whose relapse-free survival could be followed. Within 5 years after the surgery, a relatively large fraction of the relapse-free patients showed positive FOXA1 immunoreactivity (Table 2). Furthermore, patients with positive immunoreactivity for FOXA1 showed good relapse-free and overall survival, with significant differences compared with FOXA1-negative patients (Fig. 4a, b, respectively).
Fig. 4.
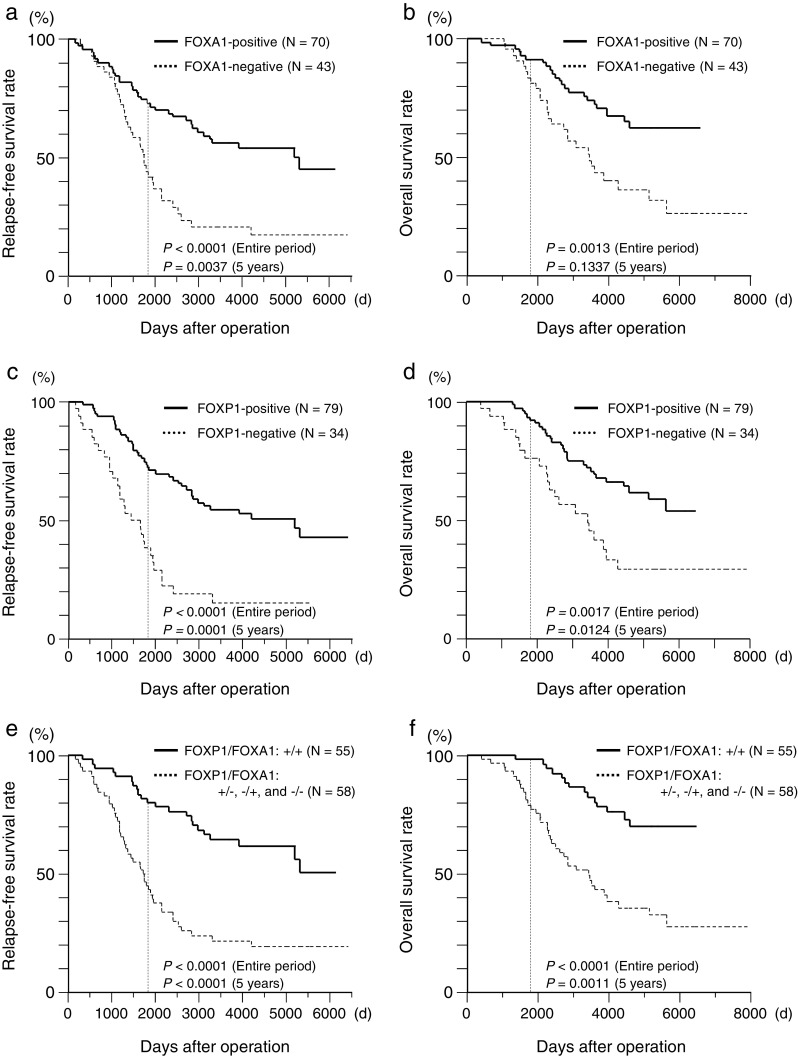
Relapse-free survival and overall survival according to the FOXA1 and/or FOXP1 immunoreactivities in the patients with tamoxifen-treated breast cancer with or without distant metastasis within 5 years after surgery or entire follow-up period (n = 113). The Kaplan–Meier survival curves were plotted using JMP 9 software, and the statistical significances were determined using the log-rank test. a, b Patients with the positive FOXA1 immunoreactivity showed good relapse-free survival and overall survival (a and b, respectively). c, d Relapse-free survival and overall survival according to the FOXP1 immunoreactivity in the patients with tamoxifen-treated breast cancer with or without distant metastasis within 5 years after surgery or entire follow-up period. Patients with positive FOXP1 immunoreactivity showed good relapse-free survival and overall survival (c and d, respectively). e, f Patients with double-positive FOXA1 and FOXP1 immunoreactivities showed better relapse-free survival and overall survival (e and f, respectively)
We recently reported that FOXP1, another FOX family member, was also correlated with the relapse-free survival of the patients with tamoxifen-treated breast cancer [19]. In this study, patients with positive immunoreactivity for FOXP1 also showed good relapse-free and overall survivals, with significant differences compared with the FOXP1-negative patients (Fig. 4c, d, respectively). Finally, we divided the patients into two groups: those with double-positive FOXA1 and FOXP1 immunoreactivities and those with the FOXA1-positive/FOXP1-negative, FOXA1-negative/FOXP1-positive, and FOXA1-negative/FOXP1-negative immunoreactivities. It is notable that patients with double-positive FOXA1 and FOXP1 immunoreactivities showed good relapse-free and overall survival with significant differences compared with the FOXA1-positive/FOXP1-negative, FOXA1-negative/FOXP1-positive, and FOXA1-negative/FOXP1-negative immunoreactivities (Fig. 4e, f, respectively).
Table 3 shows the results of the univariate and multivariate proportional analyses of the relapse-free and overall survival rates associated with FOXA1 and FOXP1 immunoreactivities and the patients’ clinicopathological characteristics. For the relapse-free survival, either FOXP1 or FOXA1 immunoreactivity was found to be a significant prognostic predictor through the univariate analysis (P = 0.001 and 0.002, respectively), whereas only FOXP1 immunoreactivity was a significantly better prognostic predictor based on multivariate analysis (P = 0.026, odds ratio = 0.35, 95 % confident index = 0.14–0.88; Table 3, relapse-free survival). On the other hand, neither FOXP1 nor FOXA1 was significantly associated with overall survival based on the multivariate analysis (Table 3, overall survival). It is an interesting finding that double-positive FOXP1 and FOXA1 immunoreactivities were significantly associated with favorable prognosis for the relapse-free and overall survival with lower P values compared with either FOXP1 or FOXA1 immunoreactivity based on the multivariate analyses (P = 0.002 and 0.002 in Table 4, respectively). These results indicate that the double-positive FOXA1 and FOXP1 immunoreactivities are favorable prognostic predictors of the relapse-free and overall survivals in patients with tamoxifen-treated breast cancer.
Table 3.
Univariate and multivariate analyses of relapse-free survival and overall survival in tamoxifen-treated patients with breast cancer (n = 113)
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| Odds ratio (95 % CI) | P value | Odds ratio (95 % CI) | P value | |
| Relapse-free survival | ||||
| FOXA1 (positive vs. negative) | 0.30 (0.13–0.65) | 0.002a | 0.44 (0.18–1.07) | 0.070 |
| FOXP1 (positive vs. negative) | 0.24 (0.10–0.55) | 0.001a | 0.35 (0.14–0.88) | 0.026 |
| pT (≤30 vs. >30 mm) | 0.36 (0.16–0.80) | 0.013a | 0.59 (0.24–1.45) | 0.25 |
| Lymph node (0 vs. ≥1) | 0.33 (0.14–0.72) | 0.005a | 0.51 (0.21–1.23) | 0.14 |
| ERα (positive vs. negative) | 0.91 (0.25–3.76) | 0.90 | ||
| PgR (positive vs. negative) | 0.86 (0.30–2.55) | 0.77 | ||
| Age (≤50 vs. >50 years) | 1.30 (0.60–2.82) | 0.50 | ||
| Overall survival | ||||
| FOXA1 (positive vs. negative) | 0.41 (0.13–1.27) | 0.12 | ||
| FOXP1 (positive vs. negative) | 0.27 (0.08–0.84) | 0.024a | 0.34 (0.99–1.11) | 0.072 |
| pT (≤30 vs. >30 mm) | 0.24 (0.07–0.76) | 0.015a | 0.30 (0.08–0.97) | 0.044 |
| Lymph node (0 vs. ≥1) | 0.41 (0.13–1.31) | 0.14 | ||
| ERα (positive vs. negative) | 0.53 (0.12–3.76) | 0.47 | ||
| PgR (positive vs. negative) | 2.51 (0.45–47.1) | 0.34 | ||
| Age (≤50 vs. >50 years) | 1.97 (0.64–6.39) | 0.24 | ||
CI confidence interval, ERα estrogen receptor-α, PgR progesterone receptor
aData were considered significant in the univariate analyses and examined in the multivariate analyses
Table 4.
Univariate and multivariate analyses including double-positive FOXA1 and FOXP1 immunoreactivities of relapse-free survival and overall survival in tamoxifen-treated patients with breast cancer (n = 113)
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| Odds ratio (95 % CI) | P value | Odds ratio (95 % CI) | P value | |
| Relapse-free survival | ||||
| FOXA1/FOXP1 (+/+ vs. +/−, −/+, and −/−) | 0.20 (0.09–0.46) | <0.001a | 0.26 (0.11–0.62) | 0.002 |
| pT (≤30 vs. >30 mm) | 0.36 (0.16–0.80) | 0.013a | 0.55 (0.23–1.34) | 0.19 |
| Lymph node (0 vs. ≥1) | 0.33 (0.14–0.72) | 0.005a | 0.46 (0.19–1.10) | 0.081 |
| ERα (positive vs. negative) | 0.91 (0.25–3.76) | 0.90 | ||
| PgR (positive vs. negative) | 0.86 (0.30–2.55) | 0.77 | ||
| Age (≤50 vs. >50 years) | 1.30 (0.60–2.82) | 0.50 | ||
| Overall survival | ||||
| FOXA1/FOXP1 (+/+ vs. +/−, −/+, and −/−) | 0.06 (0.003–0.34) | <0.001a | 0.08 (0.004–0.44) | 0.002 |
| pT (≤30 vs. >30 mm) | 0.24 (0.07–0.76) | 0.015a | 0.36 (0.10–1.20) | 0.096 |
| Lymph node (0 vs. ≥1) | 0.41 (0.13–1.31) | 0.14 | ||
| ERα (positive vs. negative) | 0.53 (0.12–3.76) | 0.47 | ||
| PgR (positive vs. negative) | 2.51 (0.45–47.1) | 0.34 | ||
| Age (≤50 vs. >50 years) | 1.97 (0.64–6.39) | 0.23 | ||
CI confidence interval, ERα estrogen receptor-α, PgR progesterone receptor
aData were considered significant in the univariate analyses and examined in the multivariate analyses
Discussion
In the present study, the FOXA1 mRNA expression was found to be upregulated by estrogen in ERα-positive MCF-7 breast cancer cells. In addition, ERα recruitment to ERBSs within the FOXA1 gene region was significantly enriched by estrogen treatment in the ChIP assays using the ERα antibody. In the reporter assay, knockdown of FOXA1 impaired ERα-ERE-mediated transcription in the MCF-7 cells. Together with these findings and our previous report on FOXP1 [19], it is possible that ERα receives positive feedback regulation from FOXA1 and FOXP1 in breast cancer cells.
FOXA1-binding sites were detected in 50 % of genes that are regulated by ER, and FOXA1 depletion partially attenuated the estrogen response in breast cancer cells [7,27,28] responsible for the transcriptional regulation of various ERα-target genes, including TFF1, GREB1, and cyclin D1. We showed previously that FOXP1 overexpression upregulates the transcription of ERα target genes including LRH-1 and SHP in breast cancer cells [19]. Notably, the binding site for ERα was identified to overlap with that for FOXA1 in the far upstream of the LRH-1 gene region [26]. These findings would suggest that FOXA1 and FOXP1 have a partially redundant function of modulating ERα-mediated transcription. It would be worthwhile to identify the genome-wide binding sites of FOXP1 to elucidate the functional correlation between FOXA1 and FOXP1 in ERα-mediated transcription in breast cancer cells.
In our previous data, FOXP1 immunoreactivity was significantly elevated in tamoxifen-treated breast cancer patients without relapse compared with those with relapse (P < 0.001) [19]. FOXP1 immunoreactivity is also an independent prognostic factor for relapse-free survival by both univariate and multivariate analyses in the present study (P = 0.001 and 0.026, respectively, Table 3). With regard to FOXA1 immunoreactivity, a significant association with relapse-free survival was shown by univariate analysis (P = 0.002, Table 3). Consistent with our observation in part, Mehta et al. [29] recently reported that FOXA1 is an independent prognostic marker for ER-positive breast cancer. Although significant correlations are not observed between overall survival and either FOXA1 or FOXP1 immunoreactivities, we found that double-positive staining of FOXA1 and FOXP1 is a powerful independent prognostic factor for both relapse-free and overall survival using univariate and multivariate analyses, independent of ERα status of the tumors in the tamoxifen-treated patients. These results suggest that the combined analyses of the FOXA1 and FOXP1 immunoreactivities provide powerful prognostic indicators for the tamoxifen-treated patients with ERα-positive breast cancers.
In the context of ER/FOXA1-driven signaling in breast cancer cells, it remains to be clarified whether the estrogen-mediated induction of FOXA1 contributes to the increased proliferation and whether the migration of cells or the estrogen-mediated proliferation leads to the enhancement of FOXA1 induction. In the present study, we showed that the estrogen-dependent upregulation of FOXA1 was repressed by 4-hydroxytamoxifen. Migration study also showed that the siRNA-mediated knockdown of FOXA1 leads to the reduction of cell migration, suggesting that ER-regulated FOXA1 plays a role in the pathophysiology of breast cancer cells. Our conventional ChIP assay data may also support our idea as the early response of ERα recruitment was observed in the ER-binding sites in the vicinity of FOXA1 gene region. In terms of FOXA1 protein expression, Laganière et al. [27] showed that the estrogen upregulates also FOXA1 protein level in MCF-7 cells 4–8 h after stimulation. Taken together, these data suggest that FOXA1 functions as an ER target in the MCF-7 cells, although it might also be possible that the estrogen-dependent proliferation at late phase will affect the expression profile of FOXA1.
Our clinical data revealed that positive FOXA1 immunoreactivity is correlated with good relapse-free and overall survival in patients with tamoxifen-treated breast cancer, which is consistent with the previous gene expression profiling study [13] as FOXA1 expression is correlated with luminal subtype A with higher levels of ERα expression and having a better prognosis than subtype B. Similar to FOXA1, another ER target cyclin D1 has been shown as a marker of good prognosis in previous studies [30,31], although this molecule is a positive cell cycle regulator and responsible for the estrogen-dependent proliferation in MCF-7 cells [32]. In contrast, Ross-Innes et al. [33] recently proposed that FOXA1 also plays a role in the differential ER-binding events in the tumors with poor outcome. Notably, it has been shown that ERα signals including ERα occupancy and estrogen-dependent cell proliferation are FOXA1-dependent in both tamoxifen-sensitive and tamoxifen-refractory MCF-7 cells by performing the siRNA-mediated knockdown study [28]. Further study will be required to answer the question whether ER/FOXA1-driven proliferation is associated with tumor recurrence in various disease stages.
Recent genome-wide study revealed that most of ER–chromatin interactions and gene expression changes depended on the presence of FOXA1 [28]. FOXA1 influenced genome-wide chromatin accessibility under different ligand conditions, including estrogen and tamoxifen in breast cancer cells [28]. Thus, it is suggested that FOXA1 is a major determinant of estrogen–ER activity and endocrine response in breast cancer cells. Taken together with our data showing that FOXA1 and FOXP1 have analogous functions on the ER-mediated transcription and clinicopathological significance in breast cancer, it can be speculated that FOXP1 may have an important role in the regulation of ER activity and tamoxifen response in conjunction with FOXA1 in breast cancer cells.
In summary, our results show that FOXA1 expression was induced by estrogen in breast cancer cells and that FOXA1 promoted cancer cell proliferation and migration by enhancing ERE-mediated transcription. We further demonstrated that immunoreactivities for FOXA1 and FOXP1 were positively correlated with the relapse-free survival for tamoxifen resistance in breast cancer. These results suggest that pharmacological modulation of FOXA1 and FOXP1 activities may be clinically useful in the prevention and/or treatment of breast cancer and that the combined evaluation of FOXA1 and FOXP1 immunoreactivities may predict the prognosis of tamoxifen-treated patients with breast cancer.
Acknowledgments
We thank Ms. Saori Miyoshi, Ms. Kayoko Kanegae, and Mr. Wataru Sato for their technical assistance. We also thank Dr. Shin-ichiro Horiguchi, Tokyo Metropolitan Cancer and Infectious diseases Center Komagome Hospital, for the preparation of specimens. This work was supported in part by Grants-in-Aid for Cancer Research (21-4) from the Ministry of Health, Labor and Welfare; the Program for Promotion of Fundamental Studies in Health Science of the NIBIO; grants from JSPS, grants of the Cell Innovation Program and the Support Project of Strategic Research Center in Private Universities from the MEXT; Institutional Grant from the Medical Research Center, Saitama Medical University (no. 23-1-1-09, NI).
Conflict of Interest
The authors declare that they have no conflict of interest.
Abbreviations
- FOXA1
Forkhead box A1
- FOXP1
Forkhead box P1
- ERα
Estrogen receptor alpha
- PgR
Progesterone receptor
- GREB1
Growth regulation by estrogen in breast cancer 1
- TFF1
Trefoil factor 1
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- ERE
Estrogen response element
- siRNA
Small interfering RNA
- qRT-PCR
Quantitative real-time reverse transcriptase polymerase chain reaction
References
- 1.Platet N, Cathiard AM, Gleizes M, Garcia M. Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit Rev Oncol Hematol. 2004;51:55–67. doi: 10.1016/j.critrevonc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Hall JM, McDonnell DP. Coregulators in nuclear estrogen receptor action: from concept to therapeutic targeting. Mol Interv. 2005;5:343–357. doi: 10.1124/mi.5.6.7. [DOI] [PubMed] [Google Scholar]
- 3.Carroll JS, Brown M. Estrogen receptor target gene: an evolving concept. Mol Endocrinol. 2006;20:1707–1714. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- 4.Habashy HO, Powe DG, Rakha EA, Ball G, Paish C, Gee J, Nicholson RI, Ellis IO. Forkhead-box A1 (FOXA1) expression in breast cancer and its prognostic significance. Eur J Cancer. 2008;44:1541–1551. doi: 10.1016/j.ejca.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 5.Badve S, Turbin D, Thorat MA, Morimiya A, Nielsen TO, Perou CM, Dunn S, Huntsman DG, Nakshatri H. FOXA1 expression in breast cancer—correlation with luminal subtype A and survival. Clin Cancer Res. 2007;13:4415–4421. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
- 6.Thorat MA, Marchio C, Morimiya A, Savage K, Nakshatri H, Reis-Filho JS, Badve S. Forkhead box A1 expression in breast cancer is associated with luminal subtype and good prognosis. J Clin Pathol. 2008;61:327–332. doi: 10.1136/jcp.2007.052431. [DOI] [PubMed] [Google Scholar]
- 7.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistelinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grange T, Roux J, Rigaud G, Pictet R. Cell-type specific activity of two glucocorticoid responsive units of rat tyrosine aminotransferase gene is associated with multiple binding sites for C/EBP and a novel liver-specific nuclear factor. Nucleic Acids Res. 1991;19:131–139. doi: 10.1093/nar/19.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes KA, Hurtado A, Brown GD, Launchbury R, Ross-Innes CS, Hadfield J, Odom DT, Carroll JS (2011) Transducin-like enhancer protein 1 mediates estrogen receptor binding and transcriptional activity in breast cancer cells. Proc Natl Acad Sci U S A 2011. doi:10.1073/pnas.1018863108 [DOI] [PMC free article] [PubMed]
- 11.Tan SK, Lin ZH, Chang CW, Varang V, Chng KR, Pan YF, Yong EL, Sung WK, Cheung E. AP-2γ regulates oestrogen receptor-mediated long-range chromatin interaction and gene transcription. EMBO J. 2011;30:2569–2581. doi: 10.1038/emboj.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 13.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lønning P, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P, Wood K, Cordell JL. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 2001;61:8820–8829. [PubMed] [Google Scholar]
- 15.Prown PJ, Ashe SL, Leich E, Burek C, Barrans S, Fenton JA, Jack AS, Pulford K, Rosenwald A, Banham AH. Potentially oncogenic B-cell activation induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL. Blood. 2008;111:2816–2824. doi: 10.1182/blood-2007-09-115113. [DOI] [PubMed] [Google Scholar]
- 16.Fox SB, Brown P, Han C, Ashe S, Leek RD, Harris AL, Banham AH. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor α and improved survival in primary human breast carcinomas. Clin Cancer Res. 2004;10:3521–3527. doi: 10.1158/1078-0432.CCR-03-0461. [DOI] [PubMed] [Google Scholar]
- 17.Banham AH, Boddy J, Launchbury R, Han C, Turley H, Malone PR, Harris AL, Fox SB. Expression of the forkhead transcriptional factor FOXP1 is associated both with hypoxia inducible factors (HIFs) and the androgen receptor in prostate cancer but is not directly regulated by androgens or hypoxia. Prostate. 2007;67:1091–1098. doi: 10.1002/pros.20583. [DOI] [PubMed] [Google Scholar]
- 18.Takayama K, Horie-Inoue K, Ikeda K, Urano T, Murakami K, Hayashizaki Y, Ouchi Y, Inoue S. FOXP1 is an androgen-responsive transcription factor that negatively regulates androgen receptor signaling in prostate cancer cells. Biochem Biophys Res Commun. 2008;374:388–393. doi: 10.1016/j.bbrc.2008.07.056. [DOI] [PubMed] [Google Scholar]
- 19.Shigekawa T, Ijichi N, Ikeda K, Horie-Inoue K, Shimizu C, Saji S, Aogi K, Tsuda H, Osaki A, Saeki T, Inoue S. FOXP1, an estrogen-inducible transcription factor, modulates cell proliferation in breast cancer cells and 5-year recurrence-free survival of patients with tamoxifen-treated breast cancer. Horm Cancer. 2011;2:286–297. doi: 10.1007/s12672-011-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ijichi N, Shigekawa T, Ikeda K, Horie-Inoue K, Fujimura T, Tsuda H, Osaki A, Saeki T, Inoue S. Estrogen-related receptor γ modulates cell proliferation and estrogen signaling in breast cancer. J Steroid Biochem Mol Biol. 2011;123:1–7. doi: 10.1016/j.jsbmb.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Allred DC, Clark GM, Elledge R, Fuqua SA, Brown RW, Chamness GC, Osborne CK, McGuire WL. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst. 1993;85:200–206. doi: 10.1093/jnci/85.3.200. [DOI] [PubMed] [Google Scholar]
- 22.Horie-Inoue K, Bono H, Okazaki Y, Inoue S. Identification and functional analysis of consensus androgen response elements in human prostate cancer cells. Biochem Biophys Res Commun. 2004;325:1312–1317. doi: 10.1016/j.bbrc.2004.10.174. [DOI] [PubMed] [Google Scholar]
- 23.Horie-Inoue K, Takayama K, Bono HU, Ouchi Y, Okazaki Y, Inoue S. Identification of novel steroid target genes through the combination of bioinformatics and functional analysis of hormone response elements. Biochem Biophys Res Commun. 2006;339:99–106. doi: 10.1016/j.bbrc.2005.10.188. [DOI] [PubMed] [Google Scholar]
- 24.Giamarchi C, Solanas M, Chailleux C, Augereau P, Vignon F, Rochefort H, Richard-Foy H. Chromatin structure of the regulatory regions of pS2 and cathepsin D genes in hormone-dependent and -independent breast cancer cell lines. Oncogene. 1999;18:533–541. doi: 10.1038/sj.onc.1202317. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda K, Ogawa S, Tsukui T, Horie-Inoue K, Ouchi Y, Kato S, Muramatsu M, Inoue S. Protein phosphatase 5 is a negative regulator of estrogen receptor-mediated transcription. Mol Endocrinol. 2004;18:1131–1143. doi: 10.1210/me.2003-0308. [DOI] [PubMed] [Google Scholar]
- 26.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 27.Laganière J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguère V. From the cover: location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta RJ, Jain RK, Leung S, Choo J, Nielsen T, Huntsman D, Nakshatri H, Badve S. FOXA1 is an independent prognostic marker for ER-positive breast cancer. Breast Canc Res Treat. 2012;131:881–890. doi: 10.1007/s10549-011-1482-6. [DOI] [PubMed] [Google Scholar]
- 30.Hwang TS, Han HS, Hong YC, Lee HJ, Paik NS. Prognostic value of combined analysis of cyclin D1 and estrogen receptor status in breast cancer patients. Pathol Int. 2003;53:74–80. doi: 10.1046/j.1440-1827.2003.01441.x. [DOI] [PubMed] [Google Scholar]
- 31.Boström P, Söderström M, Palokangas T, Vahlberg T, Collan Y, Carpen O, Hirsimäki P. Analysis of cyclins A, B1, D1 and E in breast cancer in relation to tumour grade and other prognostic factors. BMC Res Notes. 2009;2:140. doi: 10.1186/1756-0500-2-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prall OW, Rogan EM, Musgrove EA, Watts CK, Sutherland RL. c-Myc or cyclin D1 mimics estrogen effects on cyclin E-cdk2 activation and cell cycle reentry. Mol Cell Biol. 1998;18:4499–4508. doi: 10.1128/mcb.18.8.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, Ali S, Chin SF, Palmieri C, Caldas C, Carroll JS. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]