ABSTRACT
Escherichia coli sequence type 131 (ST131) is a globally dominant multidrug-resistant clone, although its clinical impact on patients with bloodstream infection (BSI) is incompletely understood. This study aims to further define the risk factors, clinical outcomes, and bacterial genetics associated with ST131 BSI. A prospectively enrolled cohort study of adult inpatients with E. coli BSI was conducted from 2002 to 2015. Whole-genome sequencing was performed with the E. coli isolates. Of the 227 patients with E. coli BSI in this study, 88 (39%) were infected with ST131. Patients with E. coli ST131 BSI and those with non-ST131 BSI did not differ with respect to in-hospital mortality (17/82 [20%] versus 26/145 [18%]; P = 0.73). However, in patients with BSI from a urinary tract source, ST131 was associated with a numerically higher in-hospital mortality than patients with non-ST131 BSI (8/42 [19%] versus 4/63 [6%]; P = 0.06) and increased mortality in an adjusted analysis (odds ratio of 5.85; 95% confidence interval of 1.44 to 29.49; P = 0.02). Genomic analyses showed that ST131 isolates primarily had an H4:O25 serotype, had a higher number of prophages, and were associated with 11 flexible genomic islands as well as virulence genes involved in adhesion (papA, kpsM, yfcV, and iha), iron acquisition (iucC and iutA), and toxin production (usp and sat). In patients with E. coli BSI from a urinary tract source, ST131 was associated with increased mortality in an adjusted analysis and contained a distinct repertoire of genes influencing pathogenesis. These genes could contribute to the higher mortality observed in patients with ST131 BSI.
KEYWORDS: ST131, bacteremia, bloodstream infections, Escherichia coli, urinary tract infection
INTRODUCTION
Escherichia coli bloodstream infections (BSI) are associated with increased health care costs, infection recurrence, and increased mortality that is often due to antibacterial resistance (1–3). Much of the increasing E. coli antibacterial resistance has been driven by the global spread of the genetic clone sequence type 131 (ST131) (4, 5). E. coli ST131 has been the subject of significant research; yet, important questions remained unanswered.
First, the risk factors for E. coli ST131 infection are incompletely understood. Previous studies have shown that patients with E. coli ST131 and those with non-ST131 BSI have generally similar baseline characteristics, although risk factors for ST131 acquisition have included recent surgery (6, 7), infection source (8–10), and prior antibiotic use (7). One area that has not received significant attention, however, is how geospatial factors influence acquisition of E. coli ST131 BSI. For example, it is unclear how a patient’s local community (e.g., where the patient lives) influences the bacterium with which they are infected. Second, the influence of the ST131 genotype on patient outcomes in E. coli BSI is unclear. Prior work has demonstrated an association between ST131 and persistent or recurrent infection (11–13), but no firm association between ST131 and increased mortality has yet been established. One study showed an association between ST131 and mortality in an adjusted analysis (14), while multiple others did not (6, 9–11, 15). Third, we have an incomplete understanding of the virulence genes and flexible genomic islands that distinguish ST131 strains from non-ST131 strains. A better understanding of the genetic factors that have led to the global success of this clone may help to combat its further spread.
In this work, we generated a large cohort of prospectively enrolled patients with E. coli BSI at Duke University Medical Center from 2002 to 2015. Detailed clinical data and outcomes were collected from each patient, and the bacterial isolates were sequenced by the J. Craig Venter Institute. This cohort was used to address three hypotheses. First, ST131 isolates would be associated with specific geospatial locations. Second, ST131 would be associated with increased mortality relative to non-ST131 isolates in E. coli BSI after adjustment for patient factors. Third, ST131 would be associated with a specific set of virulence genes and flexible genomic islands (fgi) that are differentially present in ST131 isolates. Given that E. coli ST131 has been demonstrated to have an association with treatment failure in urinary tract infections (13), we paid particular attention to the subgroup of patients with BSI from a urinary tract source.
MATERIALS AND METHODS
Clinical data.
The patient clinical data and bacterial isolates were obtained from the Duke Blood Stream Infection Biorepository (BSIB). The BSIB contains prospectively collected clinical data on greater than 3,500 unique inpatients at Duke University Hospital with monomicrobial Gram-negative bacterial BSI since 2002. Consent was obtained from all study participants, and the study was approved by the Duke University Institutional Review Board. Definitions are outlined in Text S1 in the supplemental material.
Whole-genome sequencing and bioinformatics analyses.
The whole-genome sequencing and bioinformatics analyses are detailed in Text S1.
Statistical analysis.
In the unadjusted analyses, continuous variables were reported as means with standard deviations (SDs) and compared with t tests. Dichotomous variables were reported as counts and percentages and compared with Fisher’s exact or chi-square tests as appropriate. In the adjusted analyses, logistic regression models were generated to determine associations. Outcomes of interest in the adjusted models included the presence of ST131 (as opposed to non-ST131 E. coli BSI) and in-hospital mortality. Model covariates included age, gender, race, route of infection (e.g., hospital-acquired infection, etc.), source of infection, hematopoietic or solid organ transplant, diabetes mellitus, recent corticosteroid use (within 30 days before BSI), HIV, recent surgery (within 30 days before BSI), days to effective antibiotics, ST131 genotype, and chronic health Acute Physiology and Chronic Health Evaluation II (APACHE-II) score. The chronic health portion of the APACHE-II describes whether a patient has severe organ system insufficiency on hospital admission. These covariates were selected to broadly encompass the clinical factors known or thought to influence BSI outcome. Model covariates with near-significant P values (P ≤ 0.15) in a univariable analysis were included in the final multivariable logistic regression models. In the analysis of virulence genes associated with ST131 compared with non-ST131, we performed a Bonferroni correction to account for multiple comparisons. P values less than 0.05 were considered significant. The geospatial analysis is described in Text S1.
Data availability.
All of the genomes determined in this study are available at NCBI under BioProject number PRJNA290784.
RESULTS
Patient characteristics.
In total, 227 patients with E. coli BSI were included in this study. Of these, 88 (39%) were infected with ST131. Patients with ST131 BSI, compared with those with non-ST131 infections, were more likely to have a health care-associated route of infection (either hospital-acquired or healthcare-associated, community-acquired; 80/88 [91%] versus 100/139 [72%]; P = 0.003) (Table 1). Otherwise, patients with ST131 and non-ST131 E. coli BSI had similar characteristics (Table 1). As expected, ST131 isolates exhibited higher antibiotic resistance to fluoroquinolones (77/88 [88%] versus 31/139 [22%]; P < 0.0001) and were more commonly multidrug resistant (77/88 [88%] versus 73/139 [53%]; P < 0.0001) (Table 1). Interestingly, however, ST131 and non-ST131 isolates did not differ with respect to ceftriaxone resistance (17/88 [19%] versus 24/139 [19%]; P = 0.73). In total, 43 patients (19%) did not survive to discharge. Patients with E. coli ST131 BSI, compared with those with non-ST131 BSI, did not differ with respect to in-hospital mortality (17/82 [20%] versus 26/145 [18%]; P = 0.73) or in any of the listed complications of BSI (Table 1). The only two cases of recurrent BSI were caused by ST131, although this did not meet statistical significance given the low sample size (2/88 [2%] versus 0/139 [0%]; P = 0.15). The first case of E. coli ST131 BSI was in 2004, and the number of such infections increased over time (Fig. S1 in the supplemental material).
TABLE 1.
Characteristics of patients with E. coli bloodstream infections and bacteria
| Patient/bacterial characteristicsa | ST131 N = 88 n (%) | Non-ST131 N = 139 n (%) | P valued |
|---|---|---|---|
| Age (mean [SD]) | 62 (14) | 63 (15) | 0.34 |
| Male gender | 50 (57) | 73 (53) | 0.59 |
| Race | 0.29 | ||
| White | 52 (59) | 96 (69) | |
| Black | 28 (32) | 32 (23) | |
| Other | 8 (9) | 11 (8) | |
| Hemodialysis dependent | 10 (11) | 10 (7) | 0.34 |
| Diabetes mellitus | 36 (41) | 41 (29) | 0.09 |
| Corticosteroid use within 30 days | 21 (24) | 29 (21) | 0.62 |
| Hematopoietic or solid organ transplant | 9 (10) | 17 (12) | 0.83 |
| HIV | 2 (2) | 1 (1) | 0.56 |
| Surgery within 30 days | 24 (27) | 30 (22) | 0.34 |
| Source | 0.24 | ||
| Urine/pyelonephritis | 42 (48) | 63 (45) | |
| Abscess | 2 (2) | 5 (4) | |
| Pneumonia | 5 (6) | 3 (2) | |
| Skin/soft tissue infection | 3 (3) | 8 (6) | |
| Biliary tract | 10 (11) | 12 (8) | |
| Line | 1 (1) | 6 (4) | |
| Otherb | 13 (15) | 13 (9) | |
| Unidentified source | 12 (14) | 31 (22) | |
| Route | 0.003 | ||
| Hospital acquired | 19 (22) | 22 (16) | |
| Community acquired, healthcare associated | 61 (69) | 78 (56) | |
| Community acquired, non-health care associated | 8 (9) | 39 (28) | |
| APACHE-II acute physiology score (mean [SD]) | 8 (6) | 8 (6) | 0.86 |
| APACHE-II chronic health score (mean [SD]) | 4 (2) | 4 (2) | 0.08 |
| Days to effective antibiotic therapy | 0.11 | ||
| 0 | 51 (58) | 88 (63) | |
| 1 | 20 (23) | 31 (22) | |
| 2 | 5 (6) | 13 (9) | |
| ≥3 | 12 (14) | 7 (5) | |
| Antibiotic resistance | |||
| Ceftriaxone-resistant | 17 (19) | 24 (19) | 0.73 |
| Fluoroquinolone-resistant | 77 (88) | 31 (22) | <0.0001 |
| Trimethoprim-sulfamethoxazole resistant | 52 (59) | 48 (35) | 0.004 |
| Carbapenem-resistant | 0 | 0 | 1.00 |
| Multidrug resistant | 77 (88) | 73 (53) | <0.0001 |
| Extensively drug resistant | 0 | 0 | 1.00 |
| Patient outcomes | ST131 N = 88 n (%) | Non-ST131 N = 139 n (%) | P value |
| In-hospital mortality | 18 (20) | 25 (18) | 0.73 |
| Recurrent bloodstream infection | 2 (2) | 0 (0) | 0.15 |
| Complicationsc | 44 (50) | 66 (47) | 0.78 |
| Septic shock | 20 (22) | 33 (24) | 1.00 |
| Acute kidney injury | 32 (36) | 39 (28) | 0.24 |
| ALI/ARDS | 5 (6) | 11 (7.9) | 0.60 |
| Disseminated intravascular coagulation | 2 (2) | 4 (3) | 1.00 |
Patients were stratified by whether they had an infection with E. coli sequence type 131 (ST131) or another E. coli ST; ARDS, acute respiratory distress syndrome; ALI, acute lung injury; APACHE, Acute Physiology and Chronic Health Evaluation; SD, standard deviation.
The “Other” category includes bloodstream infections from sources such as septic arthritis, sinusitis, etc. that do not fit into the predefined categories.
Defined as present if ≥1 of the below complications were suffered.
P values less than 0.05 are in bold.
Risk factors for E. coli ST131 BSI.
A multivariable logistic regression model revealed that the only risk factor for ST131 BSI, relative to non-ST131 BSI, was health care exposure. Compared to patients with community-acquired, non-health care-associated infections, both hospital-acquired (odds ratio [OR] of 4.03; 95% confidence interval [CI] of 1.52 to 11.49; P = 0.006) and community-acquired, health care-associated infections (OR of 3.58; 95% CI of 1.60 to 8.90; P = 0.003) were associated with E. coli ST131 BSI relative to non-ST131 BSI (Table 2). We did not find a significant association between geography and the odds of ST131 infection (P = 0.75 for significance of smoothed coordinate terms), and models incorporating patient geospatial coordinates were not significantly different than aspatial models.
TABLE 2.
Adjusted analysis of factors influencing acquisition of E. coli ST131 bloodstream infection
| Patient characteristics | Odds ratio | 95% CI | P valuec |
|---|---|---|---|
| Racea | |||
| Black | 1.54 | 0.80 to 2.97 | 0.190 |
| Other | 1.36 | 0.47 to 3.81 | 0.550 |
| Diabetes mellitus | 1.50 | 0.83 to 2.72 | 0.180 |
| Route of acquisitionb | |||
| Hospital acquired | 4.03 | 1.52 to 11.49 | 0.006 |
| Community acquired, health care associated | 3.58 | 1.60 to 8.90 | 0.003 |
| Chronic APACHE-II score | 1.08 | 0.94 to 1.24 | 0.280 |
Reference is white race.
Reference is community acquired, non-health care associated.
P values less than 0.05 are in bold.
Outcomes in patients with E. coli BSI.
We generated a multivariable logistic regression model of in-hospital mortality in patients with E. coli BSI (Table S1). The covariate of ST131 BSI (relative to patients with non-ST131 BSI) was not included in the final model as it was associated with a P value greater than 0.15 in a univariable analysis (OR of 1.17; 95% CI of 0.59 to 2.29; P = 0.64). Age (OR of 1.04; 95% CI of 1.01 to 1.07; P = 0.02) and unidentified source of BSI (OR of 2.88; 95% CI of 1.09 to 8.20; P = 0.02) were independently associated with increased mortality.
Subgroup analysis of patients with BSI from a urinary tract source.
Given that E. coli ST131 has been demonstrated to have an association with treatment failure in urinary tract infections (13), we performed a subgroup analysis on patients with BSI from a urinary tract source. There were 105 (46%) such patients. Of these 105 patients, 42 (40%) were infected with E. coli ST131. Patients with ST131 BSI, compared with those with non-ST131 BSI, more often had either hospital-acquired or community-associated, health care-associated infections (36/42 [86%] versus 37/63 [59%]; P = 0.001) and had recent surgery (14/42 [33%] versus 5/63 [8%]; P = 0.002) (Table S2). BSI with ST131 was associated with a numerically higher in-hospital mortality than non-ST131 BSI, although this did not meet statistical significance (8/42 [19%] versus 4/63 [6%]; P = 0.06). In the adjusted analysis, however, E. coli ST131 BSI was associated with increased mortality (OR of 5.85; 95% CI of 1.44 to 29.49; P = 0.02) (Table 3). Other factors associated with increased mortality included age (OR of 1.08; 95% CI of 1.03 to 1.16; P = 0.0007) and APACHE-II chronic health score (OR of 1.59; 95% CI of 1.09 to 2.90; P = 0.04). The final model did not contain the variable for days to effective antibiotic therapy, as this covariate was associated with a P value of 0.38 in a univariable analysis. However, the association of ST131 with increased in-hospital mortality remained significant when days to effective antibiotic therapy was included as an additional covariate in the model.
TABLE 3.
Adjusted analysis of factors influencing in-hospital mortality in patients with E. coli bloodstream infections with a urinary tract source
| Variable | Odds ratio | 95% CI | P valuea |
|---|---|---|---|
| Age | 1.08 | 1.03 to 1.16 | 0.007 |
| Chronic health APACHE-II score | 1.59 | 1.09 to 2.90 | 0.040 |
| ST131 | 5.85 | 1.44 to 29.49 | 0.020 |
P values less than 0.05 are in bold.
Comparative genomic analysis of ST131 versus non-ST131 E. coli BSI isolates.
To further investigate why the ST131 genotype was associated with increased mortality in patients with BSI from a urinary tract source, we used whole-genome sequencing to identify the serotypes, flexible genomic islands (fgis), and virulence genes associated with ST131 relative to non-ST131 strains. This analysis was limited to 193 E. coli strains for which we had high quality sequences with adequate coverage (193/227 [85%]). ST131 isolates primarily had an O25:H4 serotype (62/68 [91%]). This serotype was rare among non-ST131 isolates (2/125 [2%]). ST131 primarily contained allele 30 of the type 1 fimbriae adhesin gene fimH (i.e., fimH30; 56/68 [82%]). ST131 was associated with 11 fgis that contained 255 genes (Table S3). The fgis ranged in size from 1 to 59 genes. The primary putative function of the fgis associated with ST131 was adhesion. One of the fgis (n = 44 genes; fgi 33 in Table S3) encodes a second flagellar system (i.e., the FLAG-2 locus), a second fgi (n = 8 genes; fgi 335 in Table S3) encodes putative fimbriae-associated genes, and the third adhesion locus encodes a flagellin fliC gene (n = 1 gene; fgi 5626 in Table S3).
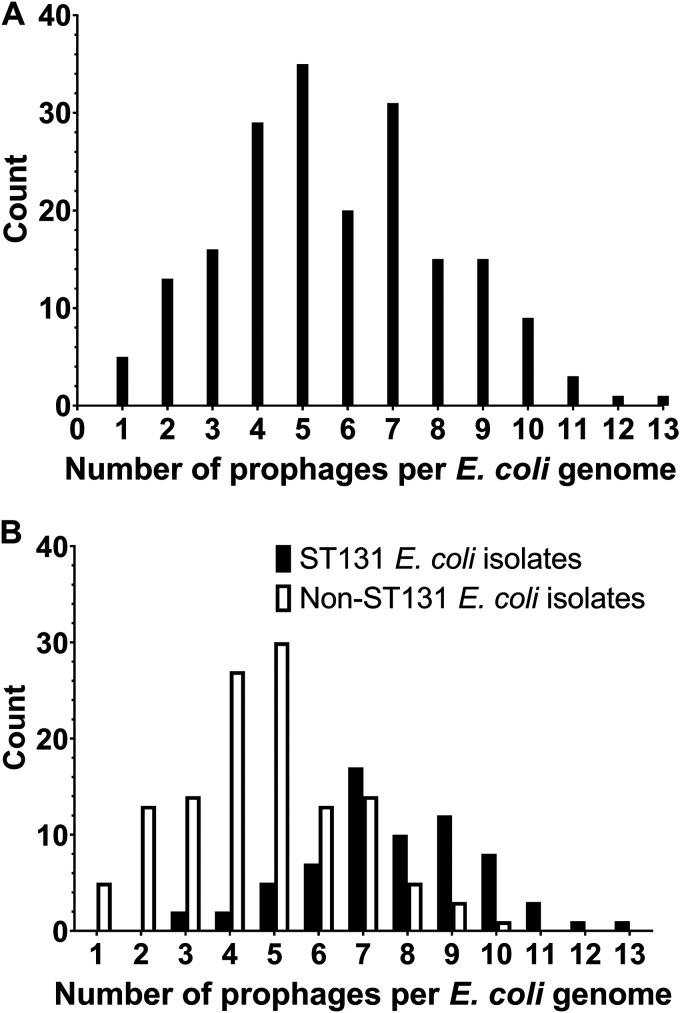
Of the 11 fgis associated with ST131, 4 (36%) were homologous to putative prophages. Genes within these four fgis accounted for 71% (161/227) of all genes in ST131-associated fgis (fgi 22, 27, 59, and 65 in Table S3). Given the high proportion of ST131-associated fgis that contained prophages, we performed a full genomic analysis of prophages in this set of E. coli BSI isolates using Phage_Finder. In total, 1,110 prophages greater than 10 kilobases (kb) were identified, and they grouped into 46 clusters by average nucleotide identity. The number of prophages per E. coli BSI isolate ranged from 1 to 13 (mean of 5.8 and standard deviation of 2.4; Fig. 1A). ST131 isolates contained a significantly higher number of prophages than non-ST131 isolates (mean of 7.7 [standard deviation of 2.0] versus mean of 4.7 [standard deviation of 1.9], respectively; P < 0.0001; Fig. 1B). The numbers of putative prophages identified for each E. coli BSI isolate are shown in Fig. S2. We determined if the presence of any specific prophage clusters was associated with patient mortality. We found that presence of prophage cluster 36 was associated with increased patient in-hospital mortality (cluster 36 in-hospital mortality 43% [10/23]; all other in-hospital mortality 18% [30/170]; P = 0.01). This association remained significant after adjusting for patient and treatment variables (OR of 4.14; 95% CI of 1.72 to 13.56; P = 0.009). However, prophage cluster 36 was not identified in any ST131 E. coli isolates. VirulenceFinder (16) was used to identify putative virulence genes within this prophage cluster. Genes iss (increased serum survival gene) and putative heat-labile enterotoxin eltIIAB-c2 were associated with prophage cluster 36. Gene iss has been extensively studied and is associated with increased serum survival and pathogenicity in chick sepsis models (17–19). Deletion of iss reduces production of the capsule, which in turn increases susceptibility to complement-mediated serum killing (20). The putative heat-labile enterotoxin eltIIAB-c2, or LTII-c2, has not been well studied, although it has been associated with increased cytotoxicity (21).
FIG 1.
Distribution of number of prophages per E. coli BSI isolate. (A) Histograms of all E. coli included in this study (n = 193). (B) E. coli BSI isolates stratified by sequence type 131 (ST131) or not. E. coli ST131 was associated with a higher number of prophage clusters per genome (P < 0.0001).
We further examined putative virulence genes in the entire cohort using VirulenceFinder. In total, 6,444 virulence genes were identified in the 193 E. coli BSI isolates. The BSI isolates had a mean of 33.4 virulence genes (standard deviation [SD] of 6.6). ST131 isolates, on average, had fewer virulence genes than non-ST131 isolates (mean of 31.3 [SD of 2.9] versus mean of 34.6 [SD of 7.6]; P < 0.0001). However, ST131 had a distinct repertoire of virulence genes relative to non-ST131. Ten putative virulence genes were associated with ST131 relative to non-ST131. These 10 genes have diverse roles in pathogenesis, such as adhesion, iron acquisition, and toxin production (Table 4). The adhesion genes included a polysaccharide capsule synthesis gene kpsM, an adhesin gene iha, and fimbrial genes papA and yfcV. The iron acquisition genes iucC and iutT are the synthase and receptor, respectively, for the siderophore aerobactin. The toxin genes usp and sat have been shown to exhibit activity against mammalian cells (22, 23).
TABLE 4.
Virulence genes associated with E. coli ST131 bloodstream infection isolates compared with non-ST131 bloodstream infection isolates
| Gene | No. (%) of ST131 strains that contain gene N = 68 | No. (%) of non-ST131 strains that contain gene N = 125 | Annotation | Adjusted P valuea |
|---|---|---|---|---|
| hha | 67 (99) | 75 (60) | Hemolysin expression modulator | 1.41E−08 |
| iha | 65 (96) | 45 (36) | Adherence protein | 2.02E−15 |
| iucC | 64 (94) | 66 (53) | Aerobactin synthetase | 8.43E−08 |
| iutA | 64 (94) | 65 (52) | Ferric aerobactin receptor | 3.63E−08 |
| kpsMII K5 | 43 (68) | 16 (13) | Polysialic acid transport protein; group 2 capsule | 1.52E−10 |
| ompT | 68 (100) | 101 (81) | Outer membrane protease | 1.80E−03 |
| papA F43 | 57 (84) | 29 (23) | Major pilin subunit F43 | 2.00E−14 |
| sat | 64 (94) | 45 (36) | Serine protease autotransporters of Enterobacteriaceae (SPATE) | 1.71E−14 |
| usp | 68 (100) | 71 (57) | Uropathogenic specific protein | 3.85E−11 |
| yfcV | 67 (99) | 74 (59) | Fimbrial protein | 1.27E−08 |
Adjusted P value refers to P value after applying a Bonferroni correction to account for multiple comparisons.
DISCUSSION
In this study, we performed a genomic analysis on a large set of clinical E. coli BSI isolates that were procured from prospectively enrolled and well-characterized patients at a large US institution from 2002 to 2015. Our aims were to better understand the risk factors for and outcomes associated with E. coli ST131 BSI. This study revealed several key findings.
First, we found that ST131 was associated with increased mortality in patients with E. coli BSI from a urinary tract source. This is an area of controversy as one study showed an association between ST131 and mortality (14), while multiple others did not (6, 9–11, 15). When E. coli BSI from all sources were considered together, no association between ST131 and increased mortality was identified, underscoring the importance of stratification by infection source. We performed a subgroup analysis on patients with E. coli BSI from a urinary tract source because portal of entry is a well-described factor in BSI outcomes, logistic regression models may not adequately adjust for all covariates such as source of BSI, and ST131 has been demonstrated to be associated with treatment failure in urinary tract infections (13). Interestingly, the association between ST131 and increased mortality remained even after “days to appropriate antibiotic therapy” was added to the model. This suggests that ST131 may be associated with increased virulence or there is a confounding clinical factor that is not represented in the adjusted analysis.
Second, we performed a genomic analysis of E. coli BSI isolates to identify genetic factors that may be contributing to the observed increased mortality in patients with E. coli ST131. To our knowledge, this is the first study to specifically examine prophage content associated with ST131 isolates, and we identified increased prophage content within ST131 isolates. We also examined the fgis and virulence genes associated with ST131. ST131, compared with non-ST131, contained fgis encoding putative virulence factors, such as a second flagellar system and genes involved in adhesion, iron uptake, and toxin production. While there has been speculation that the second putative flagellar system (i.e., FLAG-2) plays a role in pathogenesis (24), functional studies have not clearly shown such a role (25, 26). However, some of the genes associated with ST131 could impact virulence and mortality in patients with E. coli BSI. For example, capsular genes such as kpsM could influence capsule production and hence serum survival, iron uptake genes such as iucC and iutA could influence survival in the blood, and the toxins usp and sat have been shown to exhibit activity against mammalian cells (22, 23). The virulence genes associated with ST131 in this study have all been identified in at least one prior study (4, 27–29).
This study had several limitations. First, all patients were enrolled from a single institution. Differences in patient population and treatment practices at other institutions may influence the mortality associated with ST131 BSI, and so these results require validation at additional sites. Second, information on detailed management decisions (e.g., timing/volume of intravenous fluids) that could have influenced patient outcomes is not available for each patient. Some such management decisions, such as the use of carbapenems to treat ceftriaxone-resistant E. coli BSI (30), have likely changed since this historical cohort was enrolled. However, the number of days to appropriate antibiotic therapy was measured and was similar between groups. Third, the sample size may have limited our ability to identify a relationship between ST131 BSI and a patient’s home address. However, the lack of a spatial relationship suggests that factors explaining outcomes are not geographically distributed in this study population.
In conclusion, we show that after adjustment for patient demographics, medical comorbidities, and treatment factors, the mortality associated with E. coli ST131 BSI from a urinary tract source is higher than that of non-ST131 BSI from a urinary tract source. This result provides some evidence for increased virulence of the ST131 genotype in E. coli urinary tract infections complicated by BSI.
ACKNOWLEDGMENTS
V.G.F. reports personal fees from Novartis, Debiopharm, Genentech, Achaogen, Affinium, Medicines Co., MedImmune, Bayer, Basilea, Affinergy, Janssen, Contrafect, Regeneron, Destiny, Amphliphi Biosciences, Integrated Biotherapeutics, C3J, Armata, Valanbio, Akagera, Aridis, and Roche, grants from National Institutes of Health (NIH), MedImmune, Allergan, Pfizer, Advanced Liquid Logics, Theravance, Novartis, Merck, Medical Biosurfaces, Locus, Affinergy, Contrafect, Karius, Genentech, Regeneron, Deep Blue, Basilea, and Janssen, royalties from UpToDate, stock options from Valanbio and ArcBio, honoraria from Infectious Diseases of America for his service as Associate Editor of Clinical Infectious Diseases, and a patent on sepsis diagnostics pending. J.T.T. reports being a Scientific Advisor for Resonantia Diagnostics, Inc. All other authors report no conflicts of interest.
This work was supported in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services under Award Number U19AI110819 and the CDC Prevention Epicenter Program U54CK000603. V.G.F. was supported by 1R01-AI165671-01. J.T.T. was supported by K08-AI171183-01.
Author contributions included conception and design (A.B., M.D.A., V.G.F., and J.T.T.), acquisition of data (G.S., P.M.L., K.H., F.R., L.B., T.H.C., M.D.A., D.E.F., and J.T.T.), analysis and interpretation of data (A.B., G.S., P.M.L., K.H., L.B., T.H.C., M.D.A., V.G.F., D.E.F., and J.T.T.), and manuscript preparation (A.B., G.S., P.M.L., K.H., F.R., L.B., T.H.C., M.D.A., V.G.F., D.E.F., and J.T.T.).
Footnotes
Supplemental material is available online only.
Contributor Information
Vance G. Fowler, Jr., Email: vance.fowler@duke.edu.
Patricia J. Simner, Johns Hopkins University
REFERENCES
- 1.Melzer M, Petersen I. 2007. Mortality following bacteraemic infection caused by extended spectrum beta-lactamase (ESBL) producing E. coli compared to non-ESBL producing E. coli. J Infect 55:254–259. doi: 10.1016/j.jinf.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Thaden JT, Li Y, Ruffin F, Maskarinec SA, Hill-Rorie JM, Wanda LC, Reed SD, Fowler VG. 2017. Increased costs associated with bloodstream infections caused by multidrug-resistant Gram-negative bacteria are due primarily to patients with hospital-acquired infections. Antimicrob Agents Chemother 61:e01709-16. doi: 10.1128/AAC.01709-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antimicrobial Resistance Collaborators. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 5.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho SY, Kang C-I, Cha MK, Wi YM, Ha YE, Chung DR, Lee NY, Peck KR, Song J-H, Korean Network for Study on Infectious Diseases . 2015. Clinical features and treatment outcomes of bloodstream infections caused by extended-spectrum beta-lactamase-producing Escherichia coli sequence type 131. Microb Drug Resist 21:463–469. doi: 10.1089/mdr.2014.0261. [DOI] [PubMed] [Google Scholar]
- 7.Ha YE, Kang C-I, Cha MK, Park SY, Wi YM, Chung DR, Peck KR, Lee NY, Song J-H. 2013. Epidemiology and clinical outcomes of bloodstream infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in patients with cancer. Int J Antimicrob Agents 42:403–409. doi: 10.1016/j.ijantimicag.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 8.Peirano G, Pitout JD. 2014. Fluoroquinolone-resistant Escherichia coli sequence type 131 isolates causing bloodstream infections in a Canadian region with a centralized laboratory system: rapid emergence of the H30-Rx sublineage. Antimicrob Agents Chemother 58:2699–2703. doi: 10.1128/AAC.00119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung H-C, Lai C-H, Lin J-N, Huang C-K, Liang S-H, Chen W-F, Shih Y-C, Lin H-H, Wang J-L. 2012. Bacteremia caused by extended-spectrum-beta-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob Agents Chemother 56:618–622. doi: 10.1128/AAC.05753-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu YH, Cheng MF, Lai CH, Lin HH, Hung CH, Wang JL. 2014. The role of sequence type (ST) 131 in adult community-onset non-ESBL-producing Escherichia coli bacteraemia. BMC Infect Dis 14:579. doi: 10.1186/s12879-014-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee R, Johnston B, Lohse C, Porter SB, Clabots C, Johnson JR. 2013. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect Control Hosp Epidemiol 34:361–369. doi: 10.1086/669865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JR, Thuras P, Johnston BD, Weissman SJ, Limaye AP, Riddell K, Scholes D, Tchesnokova V, Sokurenko E. 2016. The pandemic H30 subclone of Escherichia coli sequence type 131 is associated with persistent infections and adverse outcomes independent from its multidrug resistance and associations with compromised hosts. Clin Infect Dis 62:1529–1536. doi: 10.1093/cid/ciw193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Can F, Azap OK, Seref C, Ispir P, Arslan H, Ergonul O. 2015. Emerging Escherichia coli O25b/ST131 clone predicts treatment failure in urinary tract infections. Clin Infect Dis 60:523–527. doi: 10.1093/cid/ciu864. [DOI] [PubMed] [Google Scholar]
- 14.Wang J-L, Lee C-C, Lee C-H, Lee N-Y, Hsieh C-C, Hung Y-P, Tang H-J, Ko W-C. 2018. Clinical impact of sequence type 131 in adults with community-onset monomicrobial Escherichia coli bacteremia. J Clin Med 7:508. doi: 10.3390/jcm7120508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morales-Barroso I, López-Cerero L, Molina J, Bellido M, Navarro MD, Serrano L, González-Galán V, Praena J, Pascual A, Rodríguez-Baño J. 2017. Bacteraemia due to non-ESBL-producing Escherichia coli O25b:H4 sequence type 131: insights into risk factors, clinical features and outcomes. Int J Antimicrob Agents 49:498–502. doi: 10.1016/j.ijantimicag.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binns MM, Davies DL, Hardy KG. 1979. Cloned fragments of the plasmid ColV,I-K94 specifying virulence and serum resistance. Nature 279:778–781. doi: 10.1038/279778a0. [DOI] [PubMed] [Google Scholar]
- 18.Nolan LK, Horne SM, Giddings CW, Foley SL, Johnson TJ, Lynne AM, Skyberg J. 2003. Resistance to serum complement, iss, and virulence of avian Escherichia coli. Vet Res Commun 27:101–110. doi: 10.1023/a:1022854902700. [DOI] [PubMed] [Google Scholar]
- 19.Gibbs PS, Maurer JJ, Nolan LK, Wooley RE. 2003. Prediction of chicken embryo lethality with the avian Escherichia coli traits complement resistance, colicin V production, and presence of the increased serum survival gene cluster (iss). Avian Dis 47:370–379. doi: 10.1637/0005-2086(2003)047[0370:POCELW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 20.Biran D, Sura T, Otto A, Yair Y, Becher D, Ron EZ. 2021. Surviving serum: the Escherichia coli iss gene of extraintestinal pathogenic E. coli is required for the synthesis of group 4 capsule. Infect Immun 89:e0031621. doi: 10.1128/IAI.00316-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nawar HF, King-Lyons ND, Hu JC, Pasek RC, Connell TD. 2010. LT-IIc, a new member of the type II heat-labile enterotoxin family encoded by an Escherichia coli strain obtained from a nonmammalian host. Infect Immun 78:4705–4713. doi: 10.1128/IAI.00730-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nipič D, Podlesek Z, Budič M, Črnigoj M, Žgur-Bertok D. 2013. Escherichia coli uropathogenic-specific protein, Usp, is a bacteriocin-like genotoxin. J Infect Dis 208:1545–1552. doi: 10.1093/infdis/jit480. [DOI] [PubMed] [Google Scholar]
- 23.Guyer DM, Radulovic S, Jones FE, Mobley HL. 2002. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect Immun 70:4539–4546. doi: 10.1128/IAI.70.8.4539-4546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren CP, Beatson SA, Parkhill J, Pallen MJ. 2005. The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J Bacteriol 187:1430–1440. doi: 10.1128/JB.187.4.1430-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakkanat A, Totsika M, Schaale K, Duell BL, Lo AW, Phan M-D, Moriel DG, Beatson SA, Sweet MJ, Ulett GC, Schembri MA. 2015. The role of H4 flagella in Escherichia coli ST131 virulence. Sci Rep 5:16149. doi: 10.1038/srep16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Fu J, Liu C, Chen J, Sun M, Chen H, Tan C, Wang X. 2017. Characterization and distinction of two flagellar systems in extraintestinal pathogenic Escherichia coli PCN033. Microbiol Res 196:69–79. doi: 10.1016/j.micres.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Nicolas-Chanoine M-H, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park Y-J, Lavigne J-P, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 28.Lavigne J-P, Vergunst AC, Goret L, Sotto A, Combescure C, Blanco J, O'Callaghan D, Nicolas-Chanoine M-H. 2012. Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One 7:e34294. doi: 10.1371/journal.pone.0034294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boll EJ, Overballe-Petersen S, Hasman H, Roer L, Ng K, Scheutz F, Hammerum AM, Dungu A, Hansen F, Johannesen TB, Johnson A, Nair DT, Lilje B, Hansen DS, Krogfelt KA, Johnson TJ, Price LB, Johnson JR, Struve C, Olesen B, Stegger M. 2020. Emergence of enteroaggregative Escherichia coli within the ST131 lineage as a cause of extraintestinal infections. mBio 11:e00353-20. doi: 10.1128/mBio.00353-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris PNA, Tambyah PA, Lye DC, Mo Y, Lee TH, Yilmaz M, Alenazi TH, Arabi Y, Falcone M, Bassetti M, Righi E, Rogers BA, Kanj S, Bhally H, Iredell J, Mendelson M, Boyles TH, Looke D, Miyakis S, Walls G, Al Khamis M, Zikri A, Crowe A, Ingram P, Daneman N, Griffin P, Athan E, Lorenc P, Baker P, Roberts L, Beatson SA, Peleg AY, Harris-Brown T, Paterson DL, MERINO Trial Investigators and the Australasian Society for Infectious Disease Clinical Research Network (ASID-CRN) . 2018. Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E. coli or Klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: a randomized clinical trial. JAMA 320:984–994. doi: 10.1001/jama.2018.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download jcm.00199-23-s0001.docx, DOCX file, 0.9 MB (891.4KB, docx)
Data Availability Statement
All of the genomes determined in this study are available at NCBI under BioProject number PRJNA290784.