Abstract
Endometrial carcinoma (EC) is the fourth most common cancer in women. Some long non-coding RNAs (lncRNAs) are regarded as potential prognostic biomarkers or targets for treatment of many types of cancers. We aim to screen prognostic-related lncRNAs and build a possible lncRNA signature which can effectively predict the survival of patients with EC. We obtained lncRNA expression profiling from the TCGA database. The patients were classified into training set and verification set. By performing Univariate Cox regression model, Robust likelihood-based survival analysis, and Cox proportional hazards model, we developed a risk score with the Cox co-efficient of individual lncRNAs in the training set. The optimum cut-off point was selected by ROC analysis. Patients were effectively divided into high-risk group and low-risk group according to the risk score. The OS of the low-risk patients was significantly prolonged compared with that of the high-risk group. At last, we validated this 11-lncRNA signature in the verification set and the complete set. We identified an 11-lncRNA expression signature with high stability and feasibility, which can predict the survival of patients with EC. These findings provide new potential biomarkers to improve the accuracy of prognosis prediction of EC.
Keywords: Noncoding RNA signature, endometrial carcinoma, prognostic, risk score, tumorigenesis
Introduction
Endometrial carcinoma (EC) is one of the most common gynecological malignancies and also the fourth most common cancer (about 4.8% of all cancers) in women. 1 Owing to the increase of obesity and an aging population, the incidence and mortality rates of endometrial cancer are increasing. EC is generally divided into two subtypes: estrogen-dependent subtype (type I) and estrogen-independent subtype (type II). Type I is the primary subtype, comprising about 80% of all endometrial cancer patients. Type II is more common in elder patients and comprises of more aggressive histologic subtypes. Although the overall prognosis of EC is good, there are still over 20% of women with EC dying of their disease.2–5 The FIGO staging system is an important benchmark which is conducive to both predicting prognosis and establishing therapy strategies. The ESMO-ESGO-ESTRO risk stratification has been used to assess the risk of poor prognosis of EC patients based on tumor characteristics. 6 However, recent studies have suggested that factors beyond classically established risk indicators might influence survival outcomes. 7 Patients may have quite different prognosis and treatment responses, although their morphologic features of the tumor are similar. A molecular-based classification reflecting the biological heterogeneity of cancer is necessary for precise prognostic evaluation of EC. Thus, it is essential to detect new biomarkers related to prognosis and treatment response.
Genomic studies have demonstrated that protein coding sequence are comprised only a small part (only 1.5%–2%) of the human genome, whereas a large part of the human genome are non-coding RNAs. 8 According to their transcript size, these non-coding RNAs can be divided into two major types: small non-coding RNAs and long non-coding RNAs (lncRNAs). Those under 200 nucleotides in length were called small ncRNAs and those consist of >200 nucleotides was called long noncoding RNA.9,10 LncRNAs have many biological functions such as regulation of gene expression, post-transcriptional processing, and transcription, as well as chromatin remodeling. 11 As revealed by recent studies, deregulation of lncRNA expression profile is associated with progression and survival outcomes of various cancers such as breast cancer, gastric cancer, prostate cancer, cervical carcinoma.12–16 Some lncRNAs like lnc-XLEC1 and TDRG1, were also considered to be involved in the development of EC.17,18 However, there is still limited evidence to indicate if lncRNAs signature derived from several lncRNAs can effectively evaluate the prognosis of patients with EC.
In this study, RNA sequencing (RNA-seq) data of endometrial carcinoma was downloaded from the Cancer Genome Atlas (TCGA) project to figure out prognosis related lncRNAs by using Cox regression analysis. Finally, 11-lncRNA signature based risk score was developed and validated with high stability. Patients can be effectively divided into low- and high-risk group of different long-term outcomes using this model. It might provide new avenues for prognostic prediction of EC and may also facilitate identification of potential therapeutic targets of EC.
Materials and methods
Data obtaining and preprocessing
Endometrial cancer RNA-seq data set and corresponding clinical follow-up data set were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). In total, we extracted information of 587 patients which recorded before 09-14-2017. We excluded some patients owing to the following reasons:
samples with lncRNA sequence data but without clinical data;
missing important clinical or biological data;
duplicate data.
We used a sample splitting method to randomly divide the patients into a training set to identify key lncRNAs (n = 257) and a verification set to verify the lncRNA signature (n = 256). Both mRNA profiles data and the corresponding clinical follow-up data of EC are publicly available and open-access.
According to the GENCODE project of version 22 (http://www.Gencodegenes.org), we extracted the lncRNA expression values from RNA-seq data. Altogether, 14,435 lncRNA profiles were obtained.
Screening of abundantly expressed lncRNA
We used R software (version 3.5.1) 19 and Bioconductor 20 for all statistical analyses in our whole study. Different Patients with the same cancer type had different prognosis and outcomes due to the different levels of lncRNA expression. Therefore, we screened abundantly expressed lncRNA by the following process. First, those lncRNAs expressed in >50% of all samples and expression level above 0 were selected and normalized by “calcNormFactors” function of “edgeR” package. 21 Then we filtered out low expressed genes with count-per-million (CPM), and lncRNAs with CPM higher than one at least in two samples were regarded as abundantly expressed lncRNA.
Identification and selection of lncRNAs relating to prognosis
Univariable Cox regression analysis was conducted by using “Survival” package in R to analyze the relationship between overall survival time and abundantly expressed lncRNA in the training set. 22 LncRNAs were considered statistically significant and selected as seed lncRNAs if their expressing significance p values is <0.05.
To increase the stability and reliability of the model, we performed robust likelihood-based survival analysis by using “Rbsurv” package in R.23–26 The process was as follows:
Randomly divide all 257 samples into the training set with N (1−p) samples and the verification set with N × p samples (p = 1/3). Perform Univariable Cox regression model in each identified prognostic relate lncRNA gene in the training set, and obtain the parameter estimate. Then evaluate log-likelihood with the parameter estimate in the validation set. This evaluation was performed for each gene.
Repeat the above procedure above 1000 times independently to get 1000 log-likelihoods for each gene. Select Gene with the largest mean log-likelihoods as the best gene, named gene (1). Then evaluate every two-gene model and select an optimum two-gene model, g (1) + g (2) to search the next best gene.
Continue the forward gene selection procedure until the fitting is impossible. Select an optimum model with the smallest AIC by computing Akaike information criterions (AICs) for all the candidate predictive models. Select the prognosis related lncRNAs meanwhile.
Development and validation of risk score formula
To understand the status of each lncRNA in survival prediction, we then performed multivariate Cox regression analysis by using “survival” package in R. Based on the regression analysis, regression coefficients of each of these lncRNAs were calculated by multivariable Cox regression analysis, a risk score formula was established in the training set. We calculated the risk score of each patient with this formula in the training set. Then by using “survivalROC” package in R, a receiver operating characteristic (ROC) curve and the area under the curve (AUC) values were also generated. 27 Consider the most lag sensitivity and specificity; we chose an optimum cut-off point based on the ROC curve. According to the optimum cut-off point, patients were divided into a high-risk group and low-risk group. With the Kaplan Meier estimate for multivariable analysis, the survival difference was assessed between the low and high-risk group, and the result was compared by using the log-rank test. We also used this risk score formula to validate on the verification set and the complete set.
Functional enrichment analysis of the prognosis associated lncRNA signature
The expression correlation between the prognosis associated lncRNA signature and each protein coding gene (PCGs) was examined by using “limma” R package 28 to calculate the Pearson correlation coefficient. The PCGs were considered as lncRNAs-related PCGs with |Pearson correlation coefficient| >0.5 and p-value <0.01.
Gene ontology (GO), including biological processes (BP), molecular functions (MF), and cellular components (CC) and genomes (Kyoto Encyclopedia of Genes and Genomes, KEGG) enrichment analyses of lncRNAs-related PCGs were performed to predict the biological function of the prognosis associated lncRNA signature by using “clusterProfiler” R package 29 and “org.Hs.eg.db” R package 30 with p-value cutoff = 0.05.
Results
Data source and preprocessing
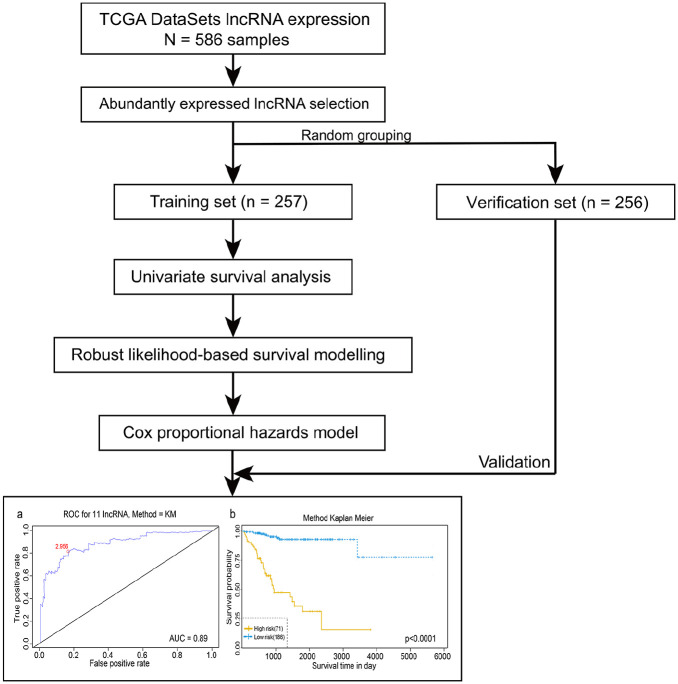
The procedure of our study was showed in Figure 1. In total, we obtained 15,427 lncRNA expression profiles from 586 patients’ samples with endometrial carcinoma from TCGA-UCEC database. We used a sample splitting method to randomly divide the patients into a training set to identify key lncRNAs (n = 257) and a verification set to verify the lncRNA signature (n = 256). The clinical features of these two group were showed in Table 1.
Figure 1.
Flow-process diagram of the whole study.
ROC: receiver operating characteristics; TCGA: the Cancer Genome Atlas.
Table 1.
Clinical features of training set (n = 257) and verification set (n = 256).
| Verification set | Training set | p | SMD | |
|---|---|---|---|---|
| Number | 256 | 257 | ||
| Age = >45 (%) | 246 (96.0) | 240 (93.4) | 0.256 | 0.114 |
| Clinical stage (%) | ||||
| Stage I | 1 (0.4) | 2 (0.7) | 0.98 | 0.211 |
| Stage IA | 80 (30.9) | 81 (31.1) | ||
| Stage IB | 72 (26.8) | 70 (25.6) | ||
| Stage IC | 14 (5.5) | 9 (3.3) | ||
| Stage II | 16 (6.2) | 14 (5.9) | ||
| Stage IIA | 2 (1.1) | 2 (1.1) | ||
| Stage IIB | 6 (2.6) | 5 (2.2) | ||
| Stage III | 1 (0.4) | 1 (0.4) | ||
| Stage IIIA | 19 (7.7) | 15 (7.0) | ||
| Stage IIIB | 3 (1.1) | 3 (1.1) | ||
| Stage IIIC | 14 (5.5) | 16 (6.2) | ||
| Stage IIIC1 | 9 (3.7) | 11 (4.4) | ||
| Stage IIIC2 | 9 (3.7) | 11 (4.4) | ||
| Stage IV | 2 (0.7) | 2 (0.7) | ||
| Stage IVA | 0 (0.0) | 2 (1.1) | ||
| Stage IVB | 9 (3.7) | 12 (4.8) | ||
| Histological type (%) | ||||
| Endometrioid endometrial adenocarcinoma | 199 (76.8) | 189 (72.9) | 0.523 | 0.098 |
| Mixed serous and endometrioid | 11 (4.0) | 11 (4.0) | ||
| Serous endometrial adenocarcinoma | 47 (19.1) | 57 (23.1) | ||
| Histological grade (%) | ||||
| G1 | 51 (19.9) | 42 (16.5) | 0.408 | 0.146 |
| G2 | 53 (20.2) | 63 (24.5) | ||
| G3 | 149 (58.5) | 144 (56.4) | ||
| High grade | 4 (1.5) | 7 (2.6) | ||
| Menopause status (%) | ||||
| Indeterminate | 9 (3.3) | 8 (2.9) | 0.368 | 0.2 |
| Peri | 11 (4.0) | 6 (2.2) | ||
| Post | 221 (81.2) | 221 (82.1) | ||
| Pre | 15 (5.5) | 20 (7.3) | ||
| OS = DEAD (%) | 45 (16.5) | 46 (16.8) | 1 | 0.008 |
Menopause status: Indeterminate: neither pre nor postmenopausal; Peri: 6 to 12 months since last menstrual period; Post: prior bilateral ovariectomy OR > 12 or since LMP with no prior hysterectomy; Pre: <6 months since LMP AND no prior bilateral ovariectomy AND not on estrogen replacement.
Identification and selection of lncRNAs relating to prognosis
In total, 6755 abundantly expressed lncRNA were screened out of 15,427 lncRNAs basing on our selection standard. Then we further randomized this 6755 lncRNA into a training set and a verification set. By using univariable Cox regression analysis, we identified 1146 differentially expressed lncRNA in the training set for further analysis. Table 1 showed the 20 lncRNAs with the lowest p-value.
Table 2 shows Top 20 lncRNAs significantly associated to the prognosis of patients in the training set
Table 2.
Top 20 lncRNAs significantly associated to the prognosis of patients in the training set (N = 257).
| LncRNA | HR | CI95 | Cox p value |
|---|---|---|---|
| LINC01759 | 0.58 | 0.47–0.71 | 0.000001 |
| LINC00475 | 0.41 | 0.29–0.59 | 0.000001 |
| AC128689.1 | 1.44 | 1.24–1.66 | 0.000001 |
| UBXN10_AS1 | 0.64 | 0.54–0.77 | 0.000001 |
| BX322234.1 | 1.86 | 1.44–2.4 | 0.000002 |
| LINC00501 | 1.68 | 1.36–2.08 | 0.000002 |
| AC025154.2 | 0.75 | 0.67–0.85 | 0.000003 |
| AC009005.1 | 0.72 | 0.62–0.83 | 0.000005 |
| UNQ6494 | 0.4 | 0.27–0.6 | 0.000005 |
| AC007991.2 | 1.57 | 1.29–1.9 | 0.000005 |
| AC084866.1 | 0.78 | 0.7–0.87 | 0.000009 |
| LINC00954 | 1.62 | 1.31–2 | 0.000009 |
| AP003306.1 | 0.45 | 0.31–0.64 | 0.000010 |
| PAX8_AS1 | 1.59 | 1.29–1.95 | 0.000012 |
| AC104662.3 | 0.56 | 0.43–0.72 | 0.000013 |
| AL353593.3 | 1.76 | 1.36–2.28 | 0.000018 |
| AC011294.1 | 1.78 | 1.37–2.32 | 0.000020 |
| AC007032.1 | 2.9 | 1.78–4.75 | 0.000021 |
| AC068987.4 | 1.39 | 1.2–1.62 | 0.000022 |
| AC015849.5 | 1.65 | 1.31–2.08 | 0.000023 |
By using robust likelihood-based survival analysis, the random data analysis was performed for 1000 times. Finally, 30 lncRNAs were picked out based on the AIC value for further analysis (Supplemental Table S1).
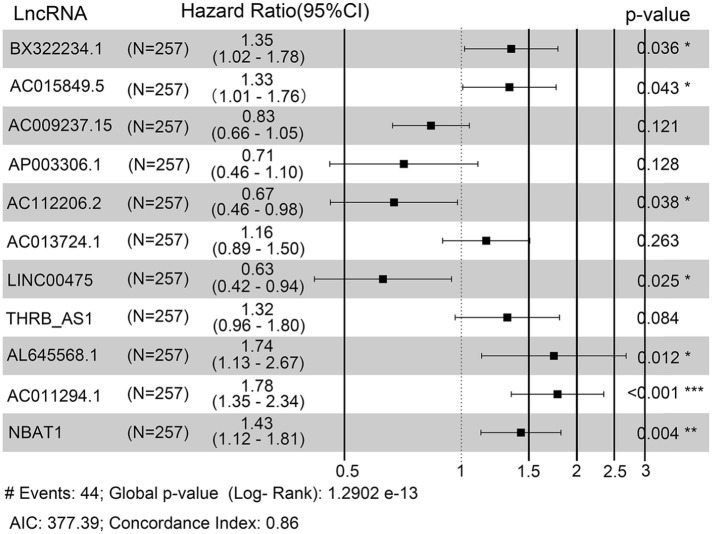
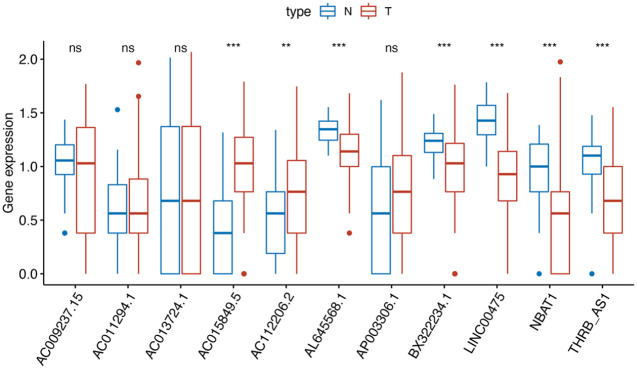
Then, we performed multivariate survival analysis to finger out the association between these 30 lncRNA and survival of endometrial carcinoma patients. Finally, the risk score based on an 11-lncRNA signature was established according to their Cox coefficients with p < 0.1 (Figure 2). Figure 3 showed that the expression levels of AC015849.5 and AC112206.2 were significantly higher in tumor samples, and the expression levels of AL645568.1, BX322234.1, LINC00475, NBAT1(poly A tail) and THRB-AS1 were higher in normal samples.
Figure 2.
Forest plot of 11-lncRNAs signature model in training set.
Figure 3.
Comparison of expression level of 11-lncRNAs between normal and cancer tissues.
Prognosis, impact, and stability of the 11-lncRNA signature model
We then performed multivariate survival analysis again on the 11-lncRNA signature based on their Cox coefficients to find out the relationship between this 11-lncRNA signature and the prognosis of endometrial carcinoma. By using “cox.zph” function of “survival” R package, we developed PH model of these 11 lncRNAs, and the global p-value calculated was 0.15117 (Supplemental Table S2).
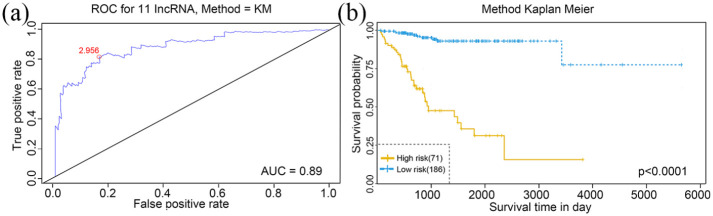
According to the ROC analysis, the AUC value of 11-lncRNA is 0.945(Figure 4(a)). Considering the maximal specificity and sensitivity, the optimum cut-off point was selected as 2.956.
Figure 4.
Receiver operating characteristic (ROC) analysis (a) and Kaplan–Meier survival analysis (b) of the 11 lncRNAs in training set. The red dot in part A represents the optimum cut-off point.
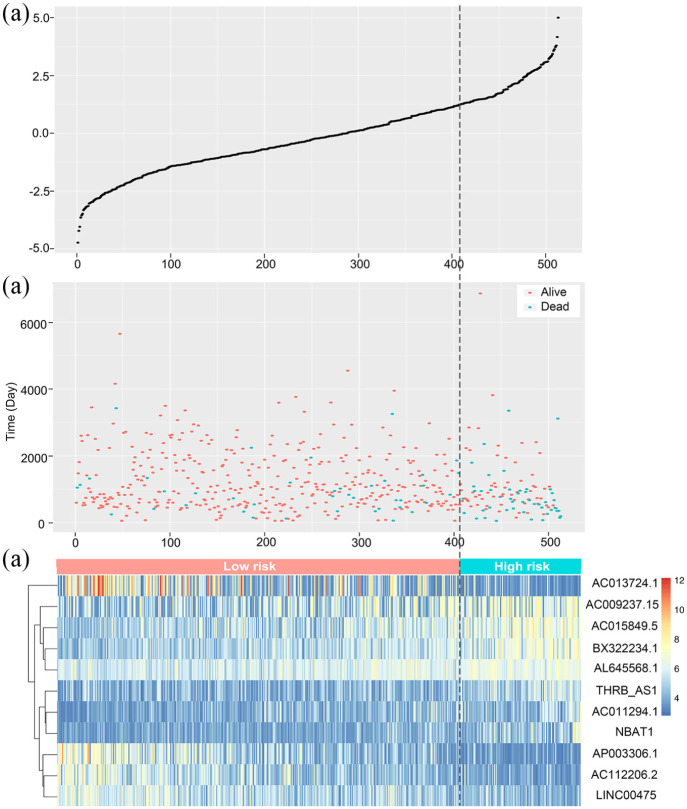
According to the optimum cut-off point, patients were classified into high and low risk groups. The result of the Kaplan-Meier curve and log-rank test showed that patients in the low-risk group had better long-term outcomes compared to those in the high-risk group (p < 0.0001) (Figure 4(b)). Figure 5 demonstrated the risk score distribution plot of lncRNA signature, patients’ survival plot and heat-map of the LncRNAs expression profiles which were plotted with the risk score of 11-lncRNA signature in the training set.
Figure 5.
The relationship between the expression levels of the 11 lncRNAs signature and risk scores: (a) risk score distribution plot of lncRNA signature, (b) patients’ survival in training set. The orange dot represents the alive samples and the blue dot represents the dead samples, and (c) heat-map of the lncRNA expression profiles.
Validity verification of 11-lncRNA signature model
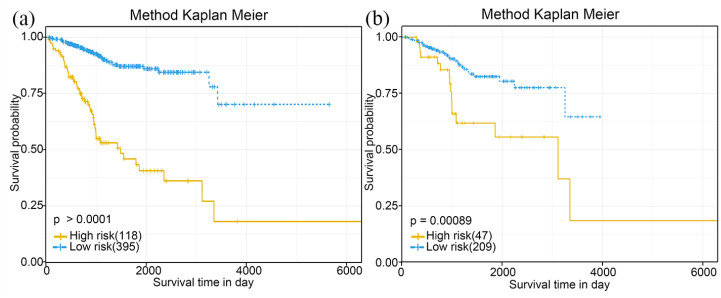
To confirm the prognosis impact of our model, the 11-lncRNA signature was validated with the same formula and the optimum cut-off point in the verification set. In consistent with our previous study, the Kaplan-Meier curve and log-rank test were performed again, and the result was shown in Figure 4. Both Kaplan-Meier curves showed that patients in the low-risk group had a prolonged survival time compared to those in the high-risk group (p = 0.0009 in the verification set, p < 0.0001 in the complete set) (Figure 6(a) and (b)).
Figure 6.
Verification of the prognostic 11-lncRNAs signature model in the verification set (N = 256) and the complete set (N = 513). Kaplan-Meier curve of patients from the complete set(a) and the verification set(b) by using the risk score based on the 11-lncRNA signature.
Functional enrichment analysis of the 11-lncRNA signature
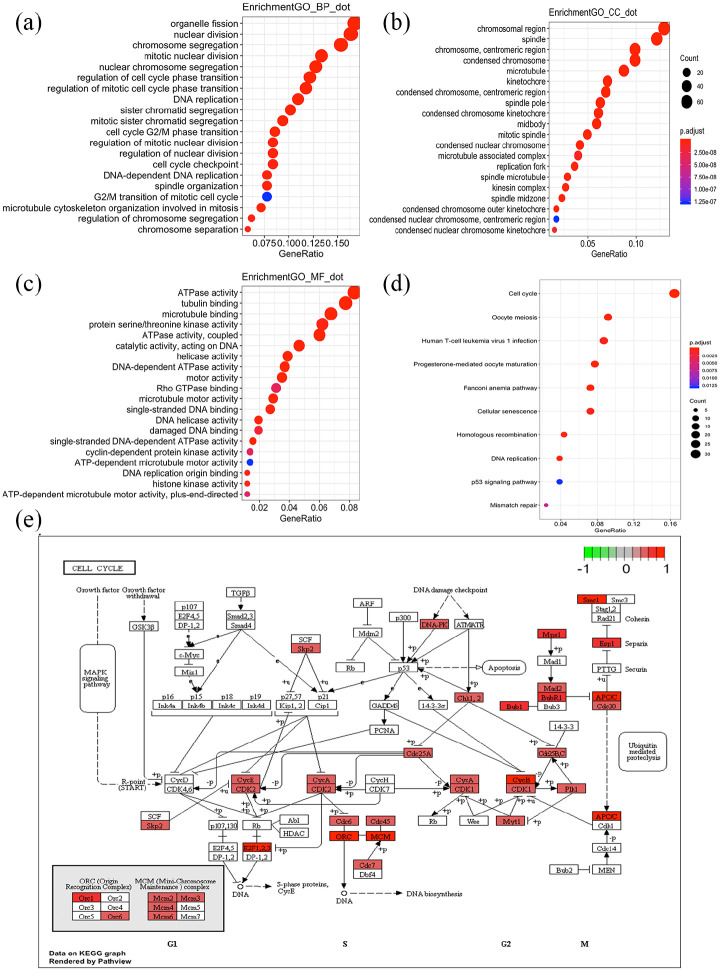
A total of 566 PCGs were selected as lncRNA-related PCGs (Supplemental Table S3). Totally, 422 GO terms (include 322 biological process (BP) terms, 72 cellular component (CC) terms, and 28 molecular function (MF) terms) (Supplemental Table S4) and 10 KEGG pathway terms (Supplemental Table S5) were enriched for these 566 PCGs. The most enriched BP terms were organelle fission (GO:0048285, 84 genes), nuclear division (GO:0000280, 82 genes), and chromosome segregation (GO:0007059 77 genes) (Figure 7(a)). For CC, the chromosomal region (GO:0098687, 68 genes) and spindle (GO:0005819, 64 genes) were the most enriched (Figure 7(b)). For MF, ATPase activity (GO:0016887, 43 genes) and tubulin binding (GO:0015631, 40 genes) were the most enrich (Figure 7(c)). As Figure 7(d), E shown, cell cycle pathway (hsa04110, 34 genes) was the most enriched KEGG pathway term. It might suggest that the 11-lncRNA signature was associated with mitotic cell cycle progression.
Figure 7.
Gene ontology enrichment analyses (a–c) of the 11-lncRNAs signature. BP, biological process; CC, cellular component; MF, molecular function. KEGG pathway enrichment analyses (d) of the 11-lncRNAs signature. Cell cycle pathway was the most enriched KEGG pathway term (e).
Discussion
Endometrial carcinoma, the fourth most common cancer in women, has been hot spots and difficulties in the field of cancer research. LncRNAs have been considered to play an important role in many human cancers including EC. 31 To provide a potential prognostic multi-lncRNA signature in endometrial carcinoma, we used recently available RNA-seq data from TCGA. Finally, a total of 11 lncRNAs related to the survival of patients were selected, included BX322234.1, AC015849.5, AC009237.15, AP003306.1, AC112206.2, AC013724.1, LINC00475, THRB_AS1, AL645568.1, AC011294.1, NBAT1 in the training set. Based on their Cox coefficients, an 11-lncRNA signature-based risk score was established. To improve the sensitivity and specificity of this model, we selected the optimum cut-off point by using the ROC analysis to divide patients into high and low-risk group. To validate the prognosis, impacts, and stability of the 11-lncRNA signature model, we used both the same risk score formulate and the optimum cut-off score in the verification set and the complete set. All of the studies above showed that the patients in low-risk group had a significantly prolonged survival time compared to those in the high-risk group and suggested that these lncRNAs may be involved in in the molecular pathogenesis and progression of endometrial carcinoma.
In our study, 11 prognosis-related-lncRNAs were identified in the training set. Among these 11 lncRNAs, except for two lncRNAs, AC112206.2 and LINC00475, all other lncRNA was significantly related to the short survival time of patients and high risk of EC. To the best of our knowledge, except for NBAT1 and LINC00475, other, nine lncRNAs were the first time to be identified as prognosis related lncRNA in EC. In previous studies, NBAT1(neuroblastoma associated transcript 1) was reported as a key lncRNA enriched in neuroblastomas, 32 breast cancer, 33 non-small cell lung cancer (NSCLC), 34 bladder cancer, 35 and gastric cancer. 36 The study in neuroblastomas suggested that by interacting with PRC2 (polycomb repressive complex 2) member EZH2 (enhancer of zeste2), NBAT1 play a crucial role in cell proliferation and invasion.34,36 They also suggested that reduced expression of NBAT1 is related to poor clinical outcome in neuroblastomas.32,37 Another study about breast cancer suggested that the expression level of NBAT1 was significantly associated with tumor metastasis in breast cancer. 33 However, the role of NBAT1 in endometrial carcinoma is still unknown, and it was likely to play a crucial role in the prognosis and treatment of EC. LINC00475 was regarded as metabolic pathway-related lncRNAs. A recent study revealed it was upregulated more than six fold in gastric cancer tissues compared with normal tissues, and it might represent novel biomarkers of gastric cancer. 38 In contrast, several lncRNAs, such as LINC01480, LINC00645, LINC00891, and LINC00702, which were confirmed specificity for EC significantly, 39 were not identified in this study. This may be caused by the different patients’ materials which were used, or the diverse statistics analytic methods chosen.
The most essential finding of our study was the setup of the 11-lncRNA signature and confirmation of its association with OS of EC patients. At the same time, the 11-lncRNA signature risk score was developed, and we found that patients with a low risk score had a significantly prolonged survival time compared to those with a high-risk score in the training set. This finding further emphasized the role of lncRNA-based risk score played in cancer prognosis studies.40,41 Furthermore, according to the maximal sensitivity and specificity of this 11-lncRNA signature, we selected the optimum cut-off point to divide the patient into high-risk and low-risk group that resulted in a more significant differential survival outcome.
According to the GO term enrichment analysis, the 11-lncRNA signature related PCGs were significantly enriched in functions about cell cycle, like organelle fission (GO:0048285), nuclear division (GO:0000280) and chromosome segregation (GO:0007059). KEGG pathway found that these PCGs were significantly enriched in cell cycle and fanconi anemia pathway.
However, this study also had limitations. The data we used in our research was solely obtained from TCGA dataset. In the future, large-scale, multicenter studies are needed to further validate the prognosis, impacts, and stability of the 11-lncRNAs signature model. Also, an integrative analysis combining the tumor staging system and other clinicopathologic parameters are necessary for Further studies. The biological function of these 11 lncRNAs in EC is required to be further investigated with more experiment and these further studies may provide potential targets for personalized treatment.
Conclusion
In conclusion, we constructed a prognosis associated model of endometrial cancer 11-lncRNA signature, to predict the OS time of EC patients. This signature can be a potential method for prognostic prediction and personalized therapy of endometrial cancer patients in future. At the same time, these 11 lncRNAs which was identified in our study represent high priority genes to be further investigated for functional significance in the development of EC.
Supplemental Material
Supplemental material, sj-xlsx-1-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress
Supplemental material, sj-xlsx-2-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress
Supplemental material, sj-xlsx-3-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress
Supplemental material, sj-xlsx-4-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress
Supplemental material, sj-xlsx-5-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress
Acknowledgments
We thank Dr. Jianming Zeng (University of Macau), and all the members of his bioinformatics team, biotrainee, for generously sharing their experience and codes.
Author biographies
Jing Wan, is a Gynae-oncologist, and her main research field is the basic and clinical study of endometrial cancer.
Peigen Chen is a Doctor of Medicine, his research field is the preservation of fertility for patients with gynecological tumors.
Yu Zhang is an Expert in the field of gynecological tumors, especially endometrial cancer, with many achievements.
Jie Ding is a Gynecological Expert and has published many results in the field of gynecological oncology.
Yuebo Yang is a Professor of Gynecology and has made outstanding contributions in the field of gynecological oncology.
Xiaomao Li is a Professor in Sun yat-sen university and has published more than 100 papers. His research interest includes the basic and clinical aspect of gy
Footnotes
Author contributions: Jing Wan and Peigen Chen carried out the study. Jing Wan also reviewed and edited the manuscript. Peigen Chen also analyzed and interpreted the data and drafted the manuscript. Yu Zhang and Jie Ding collected and analyzed the data. Yuebo Yang participated in the design and original draft writing. Xiaomao Li coordinated the study, participated in the design, and reviewed the manuscript. All authors read and approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Ethics approval: The data of this study are from TCGA database, and do not involve animal experiments and human specimens, and no ethics-related issues.
ORCID iDs: Yu Zhang  https://orcid.org/0000-0002-5632-9791
https://orcid.org/0000-0002-5632-9791
Xiaomao Li  https://orcid.org/0000-0001-6374-1488
https://orcid.org/0000-0001-6374-1488
Supplemental material: Supplemental material for this article is available online.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136(5): E359–E386. [DOI] [PubMed] [Google Scholar]
- 2.Jia N, Li Q, Tao X, et al. Enhancer of zeste homolog 2 is involved in the proliferation of endometrial carcinoma. Oncol Lett 2014; 8(5): 2049–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matias-Guiu X, Davidson B. Prognostic biomarkers in endometrial and ovarian carcinoma. Virchows Arch 2014; 464(3): 315–331. [DOI] [PubMed] [Google Scholar]
- 4.Zheng MJ, Gou R, Zhang WC, et al. Screening of prognostic biomarkers for endometrial carcinoma based on a ceRNA network. PeerJ 2018; 6: e6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol 1983; 15(1): 10–17. [DOI] [PubMed] [Google Scholar]
- 6.Ballester M, Bendifallah S, Darai E. European guidelines (ESMO-ESGO-ESTRO consensus conference) for the management of endometrial cancer. Bull Cancer2017; 104(12): 1032–1038. [DOI] [PubMed] [Google Scholar]
- 7.Bendifallah S, Canlorbe G, Collinet P, et al. Just how accurate are the major risk stratification systems for early-stage endometrial cancer? Br J Cancer 2015; 112(5): 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein LD. Human genome: end of the beginning. Nature 2004; 431(7011): 915–916. [DOI] [PubMed] [Google Scholar]
- 9.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011; 10: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol 2009; 21(3): 416–425. [DOI] [PubMed] [Google Scholar]
- 11.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet 2012; 3: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao X, Qin X, Li L, et al. A 15-long non-coding RNA signature to improve prognosis prediction of cervical squamous cell carcinoma. Gynecol Oncol 2018; 149(1): 181–187. [DOI] [PubMed] [Google Scholar]
- 13.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464(7291): 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu X, Tian X, Yu C, et al. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer 2016; 15(1): 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol 2013; 26(2): 155–165. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Rice K, Wang Y, et al. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology 2010; 151(3): 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Wang LL, Sun KX, et al. LncRNA TDRG1 enhances tumorigenicity in endometrial carcinoma by binding and targeting VEGF-A protein. Biochim Biophys Acta Mol Basis Dis 2018; 1864(9 Pt B): 3013–3021. [DOI] [PubMed] [Google Scholar]
- 18.Li F, Li H, Zhang L, et al. X chromosome-linked long noncoding RNA lnc-XLEC1 regulates c-Myc-dependent cell growth by collaborating with MBP-1 in endometrial cancer. Int J Cancer 2019; 145(4): 927–940. [DOI] [PubMed] [Google Scholar]
- 19.Team RC. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing, 2018. https://www.R-project.org/ [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5(10): R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26(1): 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Quigley J, Moreau T. Cox’s regression model: computing a goodness of fit statistic. Comput Methods Programs Biomed 1986; 22(3): 253–256. [DOI] [PubMed] [Google Scholar]
- 23.Renaud G, Stenzel U, Maricic T, et al. deML: robust demultiplexing of illumina sequences using a likelihood-based approach. Bioinformatics 2015; 31(5): 770–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Z, Zhang D, Duan Y, et al. A five-gene signature predicts overall survival of patients with papillary renal cell carcinoma. PLoS One 2019; 14(3): e0211491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kendall WL, Pollock KH, Brownie C. A likelihood-based approach to capture-recapture estimation of demographic parameters under the robust design. Biometrics 1995; 51(1): 293–308. [PubMed] [Google Scholar]
- 26.Wang JY, Tai JJ. Robust quantitative trait association tests in the parent-offspring triad design: conditional likelihood-based approaches. Ann Hum Genet 2009; 73(2): 231–244. [DOI] [PubMed] [Google Scholar]
- 27.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics 2005; 61(1): 92–105. [DOI] [PubMed] [Google Scholar]
- 28.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43(7): e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16(5): 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlson M. org.Hs.eg.db: genome wide annotation for Human. 2018. [Google Scholar]
- 31.Takenaka K, Chen BJ, Modesitt SC, et al. The emerging role of long non-coding RNAs in endometrial cancer. Cancer Genet 2016; 209(10): 445–455. [DOI] [PubMed] [Google Scholar]
- 32.Pandey GK, Kanduri C. Fighting neuroblastomas with NBAT1. Oncoscience 2015; 2(2): 79–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu P, Chu J, Wu Y, et al. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget 2015; 6(32): 32410–32425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng T, Li D, He Z, et al. Long noncoding RNA NBAT1 inhibits autophagy via suppression of ATG7 in non-small cell lung cancer. Am J Cancer Res 2018; 8(9): 1801–1811. [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Xie D, Zhang H. Long noncoding RNA neuroblastoma-associated transcript 1 gene inhibits malignant cellular phenotypes of bladder cancer through miR-21/SOCS6 axis. Cell Death Dis 2018; 9(10): 1042. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Yan J, Huang W, Huang X, et al. A negative feedback loop between long noncoding RNA NBAT1 and Sox9 inhibits the malignant progression of gastric cancer cells. Biosci Rep 2018; 38(6): BSR20180882. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Pandey GK, Mitra S, Subhash S, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 2014; 26(5): 722–737. [DOI] [PubMed] [Google Scholar]
- 38.Mo X, Wu Y, Chen L, et al. Global expression profiling of metabolic pathway-related lncRNAs in human gastric cancer and the identification of RP11-555H23.1 as a new diagnostic biomarker. J Clin Lab Anal 2019; 33(2): e22692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen BJ, Byrne FL, Takenaka K, et al. Transcriptome landscape of long intergenic non-coding RNAs in endometrial cancer. Gynecol Oncol 2017; 147(3): 654–662. [DOI] [PubMed] [Google Scholar]
- 40.Sana J, Radova L, Lakomy R, et al. Risk Score based on microRNA expression signature is independent prognostic classifier of glioblastoma patients. Carcinogenesis 2014; 35(12): 2756–2762. [DOI] [PubMed] [Google Scholar]
- 41.Steyerberg EW, Neville BA, Koppert LB, et al. Surgical mortality in patients with esophageal cancer: development and validation of a simple risk score. J Clin Oncol 2006; 24(26): 4277–4284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress
Supplemental material, sj-xlsx-2-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress
Supplemental material, sj-xlsx-3-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress
Supplemental material, sj-xlsx-4-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress
Supplemental material, sj-xlsx-5-sci-10.1177_00368504211006593 for Identification of the 11-lncRNA signatures associated with the prognosis of endometrial carcinoma by Jing Wan, Peigen Chen, Yu Zhang, Jie Ding, Yuebo Yang and Xiaomao Li in Science Progress