Abstract
The signal recognition particle (SRP) targeting pathway is required for the efficient insertion of many polytopic inner membrane proteins (IMPs) into the Escherichia coli inner membrane, but in the absence of SRP protein export proceeds normally. To define the properties of IMPs that impose SRP dependence, we analyzed the targeting requirements of bitopic IMPs that are structurally intermediate between exported proteins and polytopic IMPs. We found that disruption of the SRP pathway inhibited the insertion of only a subset of bitopic IMPs. Studies on a model bitopic AcrB-alkaline phosphatase fusion protein (AcrB 265-AP) showed that the SRP requirement for efficient insertion correlated with the presence of a large periplasmic domain (P1). As previously reported, perturbation of the SRP pathway also affected the insertion of a polytopic AcrB-AP fusion. Even exhaustive SRP depletion, however, failed to block the insertion of any AcrB derivative by more than 50%. Taken together, these data suggest that many proteins that are normally targeted by SRP can utilize alternative targeting pathways and that the structure of both hydrophilic and membrane-spanning domains determines the degree to which the biogenesis of a protein is SRP dependent.
The signal recognition particle (SRP) was first identified in mammalian cells as a soluble factor that is required for the entry of virtually all proteins into the secretory pathway. Mammalian SRP, a ribonucleoprotein complex composed of six polypeptides and a 300-nucleotide RNA, guides or “targets” nascent polypeptides to transport sites in the endoplasmic reticulum (ER) (reviewed in reference 52). The SRP 54-kDa subunit (SRP54) specifically binds to the N-terminal signal sequences of secreted proteins (23, 25) and the transmembrane (TM) domains of integral membrane proteins (18) as they emerge from translating ribosomes and then releases them after interaction with a heterodimeric receptor in the ER membrane (11, 46). Release of nascent chains appears to be coupled to their insertion into a transport channel or “translocon” (7, 12, 17), the core of which is a heterotrimer called the Sec61p complex (13).
Although homologs of SRP and its receptor have been identified in every organism that has been examined and have been shown to target proteins to both the yeast ER (14, 34) and the bacterial inner membrane (8, 45, 48), it is clear that unicellular organisms also possess alternative targeting pathways (15, 24, 43, 53). In Escherichia coli, SRP is comprised solely of an SRP54 homolog (Ffh) and a small RNA (4.5S RNA) (37, 41), and the SRP receptor consists of a single subunit (FtsY) (1, 42). To date, these factors have been shown to be required only for the efficient insertion of polytopic inner membrane proteins (IMPs) into the inner membrane (8, 45, 48). Most or all exported proteins can be targeted to transport sites by chaperones such as SecB that simply keep their passengers in a loosely folded, transport-competent conformation. Chaperones differ from SRP in that they bind in a relatively promiscuous fashion to sequences within the mature region of presecretory proteins (6, 10) and in that they appear to function at least in part in a posttranslational fashion (26). Despite the multiplicity of targeting pathways in yeasts and bacteria, essentially all proteins are thought to be transported across the membrane by a single translocon (reviewed in reference 40). Together with the peripheral membrane protein SecA, the SecY complex, which is closely related to the Sec61p complex (12, 16), forms the core of the translocon in prokaryotes.
The features of an IMP that require utilization of the SRP targeting pathway for efficient insertion have not yet been identified. Several studies have suggested that bacterial SRP, like eukaryotic SRP (51), recognizes proteins on the basis of hydrophobicity. Experiments in which the interaction of E. coli SRP with model nascent chains has been assessed by UV-cross-linking and preprotein translocation assays (38, 49, 50) have demonstrated a correlation between signal sequence hydrophobicity and SRP binding in vitro. These studies, together with recent in vivo experiments on the targeting of an M13 procoat derivative (H1-procoat) that has an unusually hydrophobic signal sequence (9), imply that the presence of a highly hydrophobic segment is sufficient to route a protein into the SRP targeting pathway. These experiments do not examine, however, whether the hydrophobicity of IMPs is the distinguishing feature that necessitates targeting by SRP for efficient insertion. Indeed, the observation that the insertion of some IMPs is not detectably affected by disruption of the SRP pathway (48) suggests that certain proteins that are targeted by SRP under normal growth conditions can also utilize alternative targeting pathways effectively. All of the IMPs that have been shown to require SRP for efficient membrane insertion are complex proteins that contain multiple transmembrane (TM) domains (8, 48) or, in the case of H1-procoat, one TM domain and an unusually long and hydrophobic signal sequence (9). Thus, it is unclear whether the SRP requirement is due to the presence of multiple TM domains, key individual TM domains, or hydrophilic domains that lie outside the membrane.
In this report we describe experiments designed to identify the features of an E. coli IMP that obligate utilization of the SRP targeting pathway. To simplify interpretation of the experiments, we examined the targeting requirements of model bitopic proteins that contain only a single TM domain. These proteins differ from exported proteins in two important respects. First, their TM domain, which serves as a targeting signal, is longer and much more hydrophobic than most signal peptides. Second, they contain cytoplasmic and periplasmic segments that may be structurally distinct from the mature domains of exported proteins. We found that the biogenesis of only some bitopic proteins was significantly affected by disruption of the SRP targeting pathway. Genetic engineering of one of the affected proteins (an AcrB derivative) revealed that the SRP requirement for efficient insertion is dictated by the periplasmic domain and not by the TM domain. Furthermore, we found that different methods of disrupting the SRP pathway, including exhaustive SRP depletion, only partially blocked the insertion of both bitopic and polytopic IMPs. Considered together, these results show that a variety of factors, in addition to hydrophobicity, can contribute to SRP dependence and suggest that the SRP pathway facilitates, but is not absolutely required for, the insertion of many IMPs.
MATERIALS AND METHODS
Reagents, media, and bacterial manipulations.
E. coli MC4100 [F− araD139 Δ(argF-lac)U169 rpsL150 relA1 thi fib5301 deoC1 ptsF25 rbsR] and WAM113 [MC4100 ara+ ffh::kan-1 λ(Para-ffh Apr)] have been described previously (4, 36). Medium preparation and basic bacterial manipulations were performed by standard methods (31). Selective media contained 100 μg of ampicillin or 40 μg of chloramphenicol per ml. Indicator plates for alkaline phosphatase (AP) activity contained 40 μg of 5-bromo-4-chloro-3-indolylphosphate (BCIP; Boehringer Mannheim) per ml. Taq polymerase (AmpliTaq; Perkin-Elmer) was used to amplify DNA fragments by PCR. The antiserum to AP was obtained from 5 Prime→3 Prime.
Plasmid construction.
All AP fusions used in this study were subcloned into pACYC184 or its derivative pNU74 (48). The construction of AP fusions to AcrB at amino acid residue 576 (AcrB 576-AP) and to Tsr at amino acid residue 164 (Tsr 164-AP) has been described (29, 48). The bitopic AcrB 265-AP fusion was constructed by first digesting plasmid S215 (48) with NruI and SalI to delete most of AcrB. The resulting plasmid was then cut with SalI and ligated to a BsiHKAI-XhoI fragment of plasmid pHI-1 (20) containing the AP gene by using the oligonucleotide adapters 5′-TCGAAGGCGATATTACTGCACCCGGCGGTGCT-3′ and 5′-CCGCCGGGTGCAGTAATATCGCCT-3′ to create plasmid pJN4. A fragment containing the fusion was isolated by digesting pJN4 with SmaI and HindIII and was ligated to pACYC184 digested with NruI and HindIII to create pJN6. The YfgA 139-AP fusion was constructed by using plasmid S356 (48), which contains the YfgA gene embedded in a fragment of genomic DNA (GenBank accession number ECAE000338; bp 1301 to 2939). S356 was digested with BlpI and SalI and ligated to the AP-containing fragment of pHI-1 described above by using the oligonucleotide adapters 5′-TCAGCAGGGCGATATTACTGCACCCGGCGGTGCT-3′ and 5′-CCGCCGGGTGCAGTAATATCGCCCTGC-3′. The MdoH 173-AP fusion was constructed from plasmid E140 (48) by random TnphoA insertion (28).
To facilitate replacement of the TM domain of AcrB 265-AP, the PCR-based megaprimer method (44) was used to introduce EagI sites flanking the AcrB membrane anchor in a version of pJN4 in which a BstEII-HindIII fragment had been deleted (pJN4.1). The resulting plasmid was designated pJN7. The 5′ EagI site produces a silent CGC-to-CGG mutation at codon 8. The 3′ EagI site creates a leucine-to-arginine mutation at codon 30 (CTG to CGG). A SmaI to HindIII fragment of pJN7 was subcloned into pNU74 digested with EcoRV and HindIII to create pLD1. The AcrB TM domain was removed from pLD1 by digestion with EagI, and complementary oligonucleotides 5′-GGCCGATTGTGACCAGCT TACTGCTGGT T T TGGCCG T T T T TGGCCT T T TACAACTGACATCAGGCGGTC-3′ and 5′-GGCCGACCGCCTGATGTCAGTTGTAAAAGGCCAAAAACGGCCAAAACCAGCAGTAAGCTGGTCACAATC-3′ encoding the first TM domain of Tsr were inserted to create pJN8. The synthetic portion of pJN8 was sequenced to ensure that no spurious mutations had been introduced.
To generate the deletions of AcrB 265-AP used here, pLD1 and pJN6 were digested with appropriate restriction enzymes and religated by using oligonucleotide adapters to maintain the proper reading frame. pLD1 was digested with EagI to delete amino acid residues 9 to 30 (AcrB 265Δ1-AP; pJN17). pJN6 was digested with EcoRV to delete residues 102 to 153 (AcrB 265Δ2-AP; pJN11), XmnI and EcoRV to delete residues 102 to 188 (AcrB 265Δ3-AP; pJN15), AatII and XmnI to delete residues 197 to 262 (AcrB 265Δ4-AP, pJN16), AatII and BsrGI to delete residues 82 to 264 (AcrB 265Δ5-AP; pJN9), AatII and KpnI to delete residues 88 to 260 (AcrB 265Δ6-AP; pJN13), and AatII and EcoRV to delete residues 105 to 263 (AcrB 265Δ7-AP; pJN12).
Pulse-chase labeling, immunoprecipitation, and immunoblots.
For experiments in which a dominant lethal ftsY allele was overexpressed, MC4100 transformed with pTRC-ftsY(G385A) (48) and a plasmid encoding an AP fusion were grown to an optical density at 550 nm (OD550) of 0.4 in M9 medium containing 0.4% glucose and all l-amino acids except methionine and cysteine. For experiments in which Ffh was depleted, WAM113 cells transformed with a plasmid encoding AcrB 576-AP were diluted from an overnight culture to an OD550 of 0.005 in M9 medium supplemented with 0.2% fructose, amino acids, and either 0.2% arabinose or 0.2% glucose and then grown to log phase. At appropriate times, aliquots of cells were removed and labeled with Tran35S-label (ICN), spheroplasted, treated with proteinase K, and trichloroacetic acid (TCA) precipitated as described previously (48).
Immunoprecipitations, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and phosphorimaging were performed as described previously (32, 48). To measure Ffh levels, cells were removed at various times from WAM113 cultures, proteins were precipitated with TCA, and immunoblotting with an anti-Ffh antiserum was performed as previously described (48).
RESULTS
SRP is required for the efficient insertion of a subset of bitopic IMPs.
To determine whether the presence of multiple TM domains dictates SRP dependence, we analyzed the membrane insertion of a variety of single-TM-domain-containing proteins (bitopic proteins) after disruption of the SRP pathway. All of the bitopic proteins examined have a type II (N terminus inside) orientation. To conduct these experiments, we truncated several polytopic proteins and one naturally occurring bitopic protein (YfgA, a protein of unknown function) and constructed in-frame periplasmic fusions to AP. We then analyzed the insertion of the fusion proteins by using a simple protease protection assay in which exogenously added protease releases the AP domains of properly inserted proteins but is unable to digest fusion proteins remaining in the cytoplasm (47). Plasmids encoding the AP fusion proteins were introduced into MC4100, and the cells were grown to mid-log phase. The function of the SRP pathway was then disrupted by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to induce the expression of a dominant lethal ftsY allele (G385A). Previous studies have shown that synthesis of the mutant FtsY protein blocks the activity of the SRP pathway more rapidly than Ffh depletion and is equally effective (48). After expression of the ftsY(G385A) allele was induced, cells were labeled with radioactive amino acids. The insertion of newly synthesized AP fusion proteins was then analyzed by treating spheroplasted cells with protease and immunoprecipitating AP-containing polypeptides.
We found that the insertion of only two of four different bitopic IMP-AP fusion proteins was affected by disruption of the SRP pathway. Although AcrB 265-AP (AP fusion to multidrug efflux pump) and Tsr 164-AP (AP fusion to methyl-accepting chemotaxis protein I) are superficially similar proteins that both contain a very short cytoplasmic tail and a large periplasmic segment between the membrane and the AP moiety, the insertion of only AcrB 265-AP was affected by overexpression of the ftsY(G385A) allele (Fig. 1A and B). A significant amount of protease-resistant AcrB 265-AP fusion protein was observed in pulse-labeled cells treated with IPTG and was still visible after a 5-min chase (Fig. 1A). As reported previously (48), protease-protected Tsr 164-AP could not be detected in cells that were induced to express the mutant ftsY allele (Fig. 1B). YfgA 139-AP and MdoH 173-AP are also structurally related in that they both contain long cytoplasmic tails and a very short periplasmic linker between the membrane and the AP moiety, but only the insertion of YfgA 139-AP was affected by disruption of the SRP pathway (Fig. 1C). Overexpression of the ftsY(G385A) allele had no detectable effect on the insertion of MdoH 173-AP (Fig. 1D). The biogenesis defects that we observed were not merely due to an increase in ftsY expression because the overexpression of wild-type ftsY had no effect on IMP insertion (data not shown). Taken together, these results demonstrate that the efficient insertion of some bitopic IMPs requires SRP but also that the presence of a TM domain is not sufficient to confer SRP dependence. Furthermore, because the insertion of only one member of each structural pair required SRP for efficient insertion, these results indicate that SRP dependence is not determined by a simple structural feature such as a long periplasmic domain.
FIG. 1.
Insertion of a subset of bitopic IMPs is inhibited by overexpression of ftsY(G385A). MC4100 cells transformed with pTRC-ftsY(G385A) and a second plasmid encoding an AP fusion were grown to mid-log phase. The cultures were then divided in half; one portion was left untreated (lanes 1 and 2), and IPTG was added to the other portion (lanes 3 and 4) to induce ftsY(G385A) overexpression. The insertion of AcrB 265-AP (A), Tsr 164-AP (B), YfgA 139-AP (C), and MdoH 173-AP (D) was analyzed 40 min after IPTG addition by pulse-chase labeling and immunoprecipitation of AP-containing polypeptides from protease-treated spheroplasts as described in Materials and Methods. The length of the chase is indicated. The diagrams illustrate the structure of each IMP. In all cases, the N terminus is located in the cytoplasm, and the C terminus is in the periplasm.
SRP dependence is conferred by the periplasmic domain of AcrB 265-AP.
In comparing the sequences of the bitopic fusion proteins in the experiments described above, we noticed a correlation between the hydrophobicity of the TM domains and the dependence on SRP for efficient insertion. As measured on the GES scale by the TopPred II computer application set at the default parameters (5), the hydrophobicity scores of the predicted TM domains of AcrB 265-AP and YfgA 139-AP were 2.27 and 2.35, respectively, whereas the TM domains of Tsr 164-AP and MdoH 173-AP scored only 1.88 and 1.27, respectively. To test whether the SRP dependence of AcrB 265-AP biogenesis is due to the extreme hydrophobicity of its TM domain, we replaced the native membrane anchor with the TM domain of Tsr 164-AP. We first engineered restriction sites into the plasmid encoding AcrB 265-AP to facilitate removal of the TM segment. In order to create these sites, it was necessary to introduce a leucine-to-arginine mutation at amino acid residue 30 immediately following the TM domain. Subsequently, the segment encoding the TM domain of the resulting construct (AcrB* 265-AP) was excised and replaced with oligonucleotides encoding the TM domain of Tsr 164-AP to create AcrB*Σ 265-AP (Fig. 2).
FIG. 2.
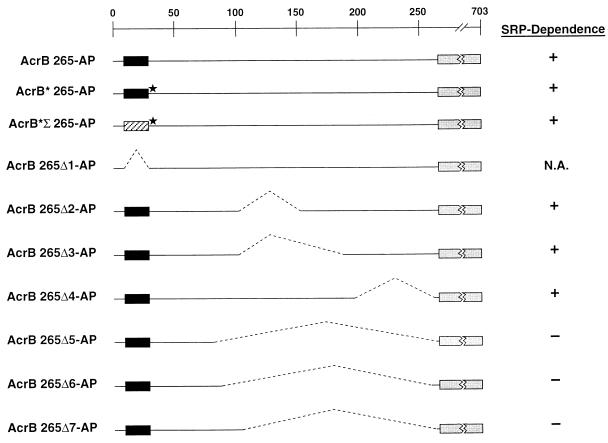
Summary of the AcrB 265-AP derivatives used in this study. The AcrB 265-AP protein (top line) contains a TM domain (amino acid residues 9 to 29 [solid rectangle]) and an AP moiety fused at residue 265 (shaded rectangle). The construction of all derivatives is described in Materials and Methods. AcrB* 265-AP (second line) contains a leucine-to-arginine point mutation (★) at residue 30, and AcrB*Σ 265-AP (third line) contains both the L30R mutation and the first TM domain of Tsr (hatched rectangle) in place of the native AcrB TM domain. The location of deletions in seven additional derivatives (Δ1–Δ7) is indicated by dashed lines. The effect of the disruption of the SRP pathway on the insertion of each derivative is indicated. +, Defect observed; −, defect not observed; N.A., not testable.
We found that disruption of the SRP pathway produced a similar effect on the insertion of AcrB 265-AP, AcrB* 265-AP, and AcrB*Σ 265-AP (Fig. 3A to C). As in the experiments described above, the insertion of these proteins was examined in MC4100 after the induction of overexpression of the ftsY(G385A) allele. In each case, a significant amount of protease-protected fusion protein was observed in pulse-labeled cells after a 2-min chase. Less of the full-length protein was observed after a 10-min chase, presumably because it was slowly degraded in the cytoplasm and/or inserted into the membrane. To confirm that the predicted TM domain of AcrB 265-AP serves as the only targeting signal in the protein, we deleted amino acids 9 to 30 to create AcrB 265Δ1-AP (Fig. 2). As expected, the AcrB 265Δ1-AP protein was completely resistant to protease digestion both in cells that expressed the ftsY(G385A) allele and in control cells (Fig. 3D), indicating that the protein was localized exclusively in the cytoplasm. Taken together, these results suggest that neither the degree of hydrophobicity nor the sequence composition of a TM domain is sufficient to dictate the targeting requirements of a bitopic IMP.
FIG. 3.
Insertion of an AcrB 265-AP derivative containing a Tsr TM domain is inhibited by overexpression of ftsY(G385A). The insertion of AcrB 265-AP (A), AcrB* 265-AP (B), AcrB*Σ 265-AP (C), and AcrB 265Δ1-AP (D) in MC4100 was analyzed as described in the legend to Fig. 1. IPTG was added to the cells in lanes 3 and 4 to induce ftsY(G385A) overexpression. The length of the chase is indicated. The position of the point mutation in AcrB* 265-AP and AcrB*Σ 265-AP is indicated (★), and each distinct TM domain is represented by different shading.
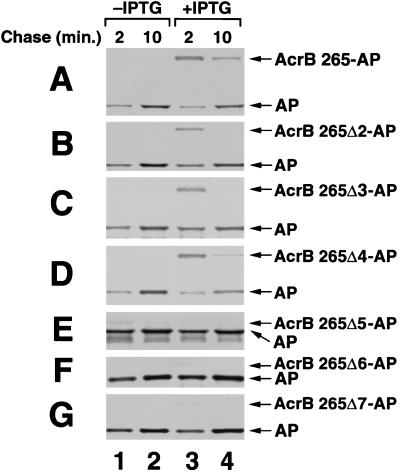
Further experiments revealed that the SRP dependence of AcrB 265-AP insertion is conferred by the 235-amino-acid residue periplasmic (P1) domain situated between the membrane and the AP moiety. To determine whether the presence of the P1 domain influences the targeting requirements of the protein, we examined the biogenesis of a series of deletion mutants (Fig. 2) after inhibiting the SRP pathway. Small deletions of approximately 50 to 90 residues in the middle or in the C-terminal end of the P1 domain had little or no effect on SRP dependence. A similar proportion of radiolabeled AcrB 265-AP, AcrB 265Δ2-AP, AcrB 265Δ3-AP, and AcrB 265Δ4-AP remained in the cytoplasm of cells that overproduced the ftsY(G385A) allele (Fig. 4A to D). The effect of deleting amino acids 35 to 76 from the N terminus of the P1 domain could not be evaluated because the mutant protein was partially protease protected even in wild-type MC4100, presumably because a minority of the protein was inserted with a topology in which the AP moiety remained in the cytoplasm (data not shown). By contrast, the insertion of AcrB 265-AP mutants with P1 domain deletions of more than 150 residues was completely SRP independent. The insertion of three different mutant fusion proteins of this type was as efficient in cells that expressed the ftsY(G385A) allele as in control cells (Fig. 4E to G). Although these results do not pinpoint a specific sequence in the AcrB 265-AP P1 domain that confers SRP dependence, they do provide evidence that the amino acid composition or overall structure of this domain prevents the protein from utilizing alternative targeting mechanisms effectively.
FIG. 4.
Large deletions in the AcrB 265-AP periplasmic domain abolish SRP dependence. The insertion of AcrB 265-AP (A), AcrB 265Δ2-AP (B), AcrB 265Δ3-AP (C), AcrB 265Δ4-AP (D), AcrB 265Δ5-AP (E), AcrB 265Δ6-AP (F), and AcrB 265Δ7-AP (G) in MC4100 was analyzed as described in the legend to Fig. 1. The deletions in each plasmid are summarized in Fig. 2. IPTG was added to the cells in lanes 3 and 4 to induce ftsY(G385A) overexpression. The length of the chase is indicated.
Exhaustive SRP depletion does not completely abolish the insertion of a polytopic IMP.
In all experiments that we performed with bitopic AcrB-AP and YfgA-AP fusions, overexpression of the ftsY(G385A) allele produced only partial insertion defects. In all previous studies, however, disruption of the SRP targeting pathway only partially blocked the insertion of polytopic IMPs as well (8, 9, 48). Although different methods have been used to eliminate SRP activity, each method has produced similar effects on IMP insertion. These results raise the possibility that the biogenesis of even extremely hydrophobic polytopic IMPs is not entirely SRP dependent. To ensure that these intermediate effects were not due to an incomplete disruption of the SRP pathway, we examined the insertion of the polytopic AcrB 576-AP fusion under extreme conditions in which virtually no SRP remained in the cells.
We found that even after severe depletion of SRP, a large amount of AcrB 576-AP was still inserted into the inner membrane. In these experiments, a strain that contains the ffh gene under control of the tight araBAD promoter (WAM113) was first transformed with a plasmid encoding the fusion protein. Cells were then grown in minimal medium as previously described (48) either in the presence or in the absence of 0.2% arabinose, which drives the expression of ffh. Glucose was used as a carbon source for cultures that did not contain arabinose to further reduce background ffh expression via catabolite repression. Cultures containing glucose grew slightly better than those containing arabinose for about five generations, or 5 h under the conditions used here (Fig. 5A). After that time, a strong growth defect was observed in cultures that contained glucose. The level of Ffh present in the cultures at various times was monitored by Western blot (Fig. 5B). After 5 h of growth in the absence of inducer, cells contained less than 1% as much Ffh as control cells. Because WAM113 grown in the presence of 0.2% arabinose contain approximately twice as much Ffh as MC4100 (data not shown); this result implies that cells grown in glucose medium contain about 2% as much Ffh as wild-type cells. It has previously been shown that the insertion of AcrB 576-AP and other IMPs is only partially blocked at this time point (48). After 7 h, WAM113 cells grown in the absence of arabinose contained less than 0.5% as much Ffh as MC4100. Despite the presence of very little detectable Ffh and the profound growth defect (Fig. 5A, arrow), the insertion of AcrB 576-AP was still only partially blocked. After pulse labeling and a 15-min chase, cells grown in glucose contained about half as much properly assembled fusion protein in the inner membrane as did control cells (Fig. 5C, lanes 3 and 6). These results strongly suggest that alternative targeting pathways can partially substitute for the SRP pathway to facilitate biogenesis of the AcrB protein.
FIG. 5.
Exhaustive depletion of SRP does not completely block the insertion of a polytopic IMP-AP fusion. (A) Growth of WAM113 transformed with a plasmid expressing AcrB 576-AP in the presence of arabinose (▴) or glucose (○). (B) Cells grown in the presence of arabinose or glucose in the experiment depicted in panel A were removed from cultures at the indicated time point, and the Ffh levels were measured by Western blot. The amount of Ffh in cells grown in glucose is expressed as a percentage of the amount of Ffh in cells grown in arabinose at the indicated time point. (C) The insertion of AcrB 576-AP was examined in the experiment depicted in panel A after 7 h of Ffh depletion (arrow) by pulse-chase labeling and immunoprecipitation of AP-containing polypeptides from protease-treated spheroplasts as described in Materials and Methods. Lanes: 1 to 3, cells grown in arabinose; 4 to 6, cells grown in glucose. The length of the chase is indicated.
DISCUSSION
In this report we provide evidence that a combination of structural features, rather than the mere presence of one or more TM domains, determines whether the efficient biogenesis of an E. coli IMP is dependent on the SRP targeting pathway. This conclusion emerged from experiments in which the fate of model IMP-AP fusions was examined after severe inhibition of the SRP pathway. We found that the efficient membrane insertion of some, but not all, bitopic IMPs required SRP activity. Whereas these results demonstrate that the presence of multiple TM domains is not a prerequisite for SRP dependence, they also show that the presence of a single highly hydrophobic targeting signal is not sufficient to require utilization of the SRP pathway. Detailed analysis of a model bitopic protein revealed that SRP dependence was due to the presence of a periplasmic domain. Furthermore, we observed that the insertion of a highly hydrophobic protein that contains seven TM domains was only partially blocked even after extensive SRP depletion. Taken together, these results suggest the existence of an alternative targeting pathway and imply that hydrophobicity is only one of a number of factors that determine the degree to which a protein is required to utilize the SRP targeting pathway for efficient insertion into the membrane. Although all of our experiments were performed with IMP-AP fusions, the observation that the insertion of an IMP that resembles a native protein is also only partially SRP dependent (9) suggests that access to SRP-independent targeting pathways was not due to the presence of an AP domain. Nevertheless, it will be important to confirm our results with IMPs that lack a large globular domain derived from an exported protein.
The observation that major determinants of SRP dependence reside in hydrophilic segments of IMPs leads to the unexpected conclusion that the biogenesis of at least some SRP substrates can be rescued by alternative targeting pathways in the absence of SRP. Although the experiments presented here only address the consequences of eliminating the SRP pathway, numerous studies on mammalian (51), yeast (30, 33), chloroplast (19), and bacterial (9, 38, 49, 50) SRPs strongly suggest that SRP binds to nascent polypeptide chains that exceed a threshold hydrophobicity. These studies predict that the hydrophobicity of TM domains should route most, if not all, E. coli IMPs into the SRP pathway. Whereas under normal physiological conditions it is possible that SRP provides the primary targeting pathway for IMPs by binding to TM domains as they emerge during translation, our results indicate that disruption of the SRP targeting pathway affects the biogenesis of only a subset of these proteins. By contrast, it has been shown in yeast cells that there is a correspondence between those proteins that are most likely to be targeted by SRP (namely, those that have particularly hydrophobic signal sequences) and those whose translocation is most SRP dependent (33). Thus, the SRP dependence of E. coli proteins, unlike eukaryotic proteins, cannot be predicted solely on the basis of the primary amino acid sequence.
Clues as to why SRP is required for the targeting of bitopic proteins such as YfgA 139-AP and AcrB 265-AP emerge from a consideration of the mechanism by which a periplasmic or cytoplasmic domain might affect membrane insertion. The simplest interpretation of the data is that, in the absence of SRP, the hydrophilic domains of these proteins either promote folding into an insertion-incompetent conformation or mask the TM segment so that it cannot interact with the translocon. An implication of the deletion analysis of AcrB 265-AP is that amino acids 188 to 263 play a pivotal role in determining the targeting requirements of the protein (Fig. 2 and Fig. 4C and G). These data suggest that, in the absence of SRP, a large polypeptide segment beyond the TM domain is synthesized in the cytoplasm and can ultimately contribute to nonproductive folding. E. coli SRP appears to bind to the targeting sequences of nascent chains as soon as they are exposed to the cytoplasm (9), however, and probably prevents AcrB 265-AP from following a nonproductive pathway by targeting it to the membrane before a critical length of polypeptide is synthesized. IMPs and exported proteins may differ in their targeting requirements in part because the hydrophilic domains of IMPs have evolved to fold as membrane-anchored units and misfold if they interact with TM segments in the cytoplasm. Unlike IMPs, exported proteins appear to be well adapted to using SRP-independent targeting pathways. The available evidence suggests that signal sequences prevent the formation of a tightly folded structure and thereby maintain translocation competence after a large portion or all of a preprotein is synthesized (35).
Although our results indicate that the presence of a large hydrophilic domain plays an important role in determining the targeting requirements of an IMP, it is likely that the presence of multiple TM domains also contributes to the SRP dependence of polytopic IMP biogenesis. Unlike AcrB, some polytopic IMPs that require SRP for efficient insertion (e.g., lactate permease and α-ketoglutarate permease) contain only relatively short hydrophilic tails and loops (48). Presumably SRP is required to target nascent polytopic IMPs to the membrane soon after synthesis of the first TM domain in order to prevent aggregation caused by the interaction of TM domains in the cytoplasm. Despite the prospect of misfolding or aggregation, however, the biogenesis of both bitopic and polytopic IMPs appears to be only partially SRP dependent. Even after Ffh was exhaustively depleted, the insertion of an IMP that contains both the P1 domain and seven TM domains (AcrB 576-AP) was reduced by only about 50%. Given that the concentration of Ffh has been estimated to be in the range of 200 to 1,000 copies per cell (21), it is likely that only a trace of Ffh remained after the 99.5% depletion that was achieved in our experiments. By contrast, a less-effective depletion of SecA in a closely related strain causes a quantitative block of AcrB 265-AP and AcrB 576-AP insertion, as well as protein export (39), indicating that much stronger effects are possible under certain experimental conditions. Partial SRP dependence might be due to a competition between the acquisition of an insertion-incompetent state, which is determined by the folding of a protein during its biosynthesis, and proper targeting of the protein by less-efficient mechanisms.
Considered together with the results of previous studies, the data presented here suggest an explanation for the essentiality of the SRP targeting pathway in E. coli (2, 27, 36). Although there appears to be a basal level of IMP insertion in the absence of SRP, the Ffh/4.5S RNA particle may play a critical role in cell physiology by simply expediting the delivery of nascent chains to the translocon. That is, cells may require SRP either because their growth is sensitive to the concentration of key IMPs that reside in the inner membrane or because the aggregation of IMPs in the cytoplasm that would result from the loss of SRP is highly toxic. Given that the chaperone-based targeting pathways may have evolved in rapidly growing organisms to increase the efficiency of protein translocation by facilitating the uncoupling of translation and translocation (22), it might have been expected that E. coli metabolism would be tuned to bypass the SRP pathway. Presumably, molecular chaperones cannot effectively duplicate the function of SRP although they, too, are thought to bind to exposed hydrophobic regions of passenger proteins (3). Indeed, the conservation of a specialized ribonucleoprotein-based targeting machinery suggests that chaperones cannot bind productively to IMPs or cannot deliver them to the translocon.
ACKNOWLEDGMENTS
We are grateful to Linda Diehl for expert technical assistance. We also thank George Poy for oligonucleotide synthesis and Brenda Peculis for helpful comments on the manuscript.
REFERENCES
- 1.Bernstein H D, Poritz M A, Strub K, Hoben P J, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown S, Fournier M J. The 4.5S RNA gene of Escherichia coli is essential for cell growth. J Mol Biol. 1984;178:533–550. doi: 10.1016/0022-2836(84)90237-7. [DOI] [PubMed] [Google Scholar]
- 3.Bukau B, Horwich A L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 4.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 5.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 6.Collier D N, Bankaitis V A, Weiss J B, Bassford P J., Jr The antifolding activity of SecB promotes the export of the E. coli maltose-binding protein. Cell. 1988;53:273–283. doi: 10.1016/0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 7.Connolly T, Rapiejko P J, Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- 8.de Gier J W, Mansournia P, Valent Q A, Phillips G J, Luirink J, von Heijne G. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 1996;399:307–309. doi: 10.1016/s0014-5793(96)01354-3. [DOI] [PubMed] [Google Scholar]
- 9.de Gier J W L, Scotti P A, Sääf A, Valent Q A, Kuhn A, Luirink J, von Heijne G. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:14646–14651. doi: 10.1073/pnas.95.25.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gannon P M, Li P, Kumamoto C A. The mature portion of Escherichia coli maltose-binding protein (MBP) determines the dependence of MBP on SecB for export. J Bacteriol. 1989;171:813–818. doi: 10.1128/jb.171.2.813-818.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorlich D, Prehn S, Hartmann E, Kalies K U, Rapoport T A. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- 13.Gorlich D, Rapoport T A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 14.Hann B C, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- 15.Hansen W, Garcia P D, Walter P. In vitro protein translocation across the yeast endoplasmic reticulum: ATP-dependent posttranslational translocation of the prepro-alpha-factor. Cell. 1986;45:397–406. doi: 10.1016/0092-8674(86)90325-9. [DOI] [PubMed] [Google Scholar]
- 16.Hartmann E, Sommer T, Prehn S, Gorlich D, Jentsch S, Rapoport T A. Evolutionary conservation of components of the protein translocation complex. Nature. 1994;367:654–657. doi: 10.1038/367654a0. [DOI] [PubMed] [Google Scholar]
- 17.High S, Andersen S S, Gorlich D, Hartmann E, Prehn S, Rapoport T A, Dobberstein B. Sec61p is adjacent to nascent type I and type II signal-anchor proteins during their membrane insertion. J Cell Biol. 1993;121:743–750. doi: 10.1083/jcb.121.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.High S, Flint N, Dobberstein B. Requirements for the membrane insertion of signal-anchor type proteins. J Cell Biol. 1991;113:25–34. doi: 10.1083/jcb.113.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.High S, Henry R, Mould R M, Valent Q, Meacock S, Cline K, Gray J C, Luirink J. Chloroplast SRP54 interacts with a specific subset of thylakoid precursor proteins. J Biol Chem. 1997;272:11622–11628. doi: 10.1074/jbc.272.17.11622. [DOI] [PubMed] [Google Scholar]
- 20.Inouye H, Michaelis S, Wright A, Beckwith J. Cloning and restriction mapping of the alkaline phosphatase structural gene (phoA) of Escherichia coli and generation of deletion mutants in vitro. J Bacteriol. 1981;146:668–675. doi: 10.1128/jb.146.2.668-675.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen C G, Pedersen S. Concentrations of 4.5S RNA and Ffh protein in Escherichia coli: the stability of Ffh protein is dependent on the concentration of 4.5S RNA. J Bacteriol. 1994;176:7148–7154. doi: 10.1128/jb.176.23.7148-7154.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnsson N, Varshavsky A. Ubiquitin-assisted dissection of protein transport across membranes. EMBO J. 1994;13:2686–2698. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krieg U C, Walter P, Johnson A E. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumamoto C A, Beckwith J. Evidence for specificity at an early step in protein export in Escherichia coli. J Bacteriol. 1985;163:267–274. doi: 10.1128/jb.163.1.267-274.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurzchalia T V, Wiedmann M, Girshovich A S, Bochkareva E S, Bielka H, Rapoport T A. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- 26.Lee C, Beckwith J. Cotranslational and posttranslational protein translocation in prokaryotic systems. Annu Rev Cell Biol. 1986;2:315–336. doi: 10.1146/annurev.cb.02.110186.001531. [DOI] [PubMed] [Google Scholar]
- 27.Luirink J, ten Hagen-Jongman C M, van der Weijden C C, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manoil C. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 29.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 30.Matoba S, Ogrydziak D M. Another factor besides hydrophobicity can affect signal peptide interaction with signal recognition particle. J Biol Chem. 1998;273:18841–18847. doi: 10.1074/jbc.273.30.18841. [DOI] [PubMed] [Google Scholar]
- 31.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 32.Newitt J A, Bernstein H D. A mutation in the Escherichia coli secY gene that produces distinct effects on inner membrane protein insertion and protein export. J Biol Chem. 1998;273:12451–12456. doi: 10.1074/jbc.273.20.12451. [DOI] [PubMed] [Google Scholar]
- 33.Ng D T, Brown J D, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogg S C, Poritz M A, Walter P. Signal recognition particle receptor is important for cell growth and protein secretion in Saccharomyces cerevisiae. Mol Biol Cell. 1992;3:895–911. doi: 10.1091/mbc.3.8.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park S, Liu G, Topping T B, Cover W H, Randall L L. Modulation of folding pathways of exported proteins by the leader sequence. Science. 1988;239:1033–1035. doi: 10.1126/science.3278378. [DOI] [PubMed] [Google Scholar]
- 36.Phillips G J, Silhavy T J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 37.Poritz M A, Bernstein H D, Strub K, Zopf D, Wilhelm H, Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- 38.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi H-Y, Bernstein H D. SecA is required for the insertion of inner membrane proteins targeted by the E. coli signal recognition particle. J Biol Chem. 1999;274:8993–8997. doi: 10.1074/jbc.274.13.8993. [DOI] [PubMed] [Google Scholar]
- 40.Rapoport T A, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- 41.Ribes V, Romisch K, Giner A, Dobberstein B, Tollervey D. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990;63:591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- 42.Romisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 43.Rothblatt J A, Meyer D I. Secretion in yeast: reconstitution of the translocation and glycosylation of alpha-factor and invertase in a homologous cell-free system. Cell. 1986;44:619–628. doi: 10.1016/0092-8674(86)90271-0. [DOI] [PubMed] [Google Scholar]
- 44.Sarkar G, Sommer S S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 45.Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 46.Tajima S, Lauffer L, Rath V L, Walter P. The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J Cell Biol. 1986;103:1167–1178. doi: 10.1083/jcb.103.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Traxler B, Lee C, Boyd D, Beckwith J. The dynamics of assembly of a cytoplasmic membrane protein in Escherichia coli. J Biol Chem. 1992;267:5339–5345. [PubMed] [Google Scholar]
- 48.Ulbrandt N D, Newitt J A, Bernstein H D. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 49.Valent Q A, de Gier J-W L, von Heijne G, Kendall D A, ten Hagen-Jongman C M, Oudega B, Luirink J. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol Microbiol. 1997;25:53–64. doi: 10.1046/j.1365-2958.1997.4431808.x. [DOI] [PubMed] [Google Scholar]
- 50.Valent Q A, Kendall D A, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walter P, Johnson A E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 53.Waters M G, Blobel G. Secretory protein translocation in a yeast cell-free system can occur posttranslationally and requires ATP hydrolysis. J Cell Biol. 1986;102:1543–1550. doi: 10.1083/jcb.102.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]