Abstract
Background:
The role of viruses is well known in health and disease. The aim of this report was to describe the profile of viruses in the gut of healthy Saudi children.
Methods:
In 20 randomly selected school age children from Riyadh, stool samples were collected in cryovials and stored at −80° C. At the time of analysis, the samples were sent by express mail in a temperature-controlled container to the laboratory in the USA, Viral DNA was isolated and shotgun metagenomic sequencing was performed. The abundance of each organism was expressed as an average relative percentage across the viral phylogenetic tree from phyla to species.
Results:
The median age of the children was 11.3 (range 6.8–15.4) years, and 35% were males. Caudovirales were the most abundant bacteriophage order (77%) and Siphoviridae, Myoviridae, and Podoviridae families predominated, accounting for 41%, 25%, and 11%, respectively. Among the viral bacteriophage species, the most abundant were the Enterobacteria phages.
Conclusion:
The profile and abundance of the gut virome in healthy Saudi children reveal important differences from the literature. Further studies from different populations with larger sample sizes are needed to understand the role of gut viruses in the pathogenesis of disease in general and in the response to fecal microbiota therapy in particular.
Keywords: Children, microbiome, Saudi Arabia, virome
INTRODUCTION
The gut virome includes all the nucleic acids (DNA and RNA) of the virus-like particles. Quantitatively, the virome is at least equal to bacteria, and may outnumber bacterial cells in the gut.[1,2] The virome is dominated by bacteriophages which are viruses that infect bacteria. Bacteriophages can be lytic or lysogenic.[3] Lytic viruses penetrate bacteria and control the genetic replication to produce virions that are released and may infect new bacteria. Lysogenic viruses integrate into the genome of bacteria without lysing (killing) them. Thus, the ability of phages to transfer genes from one host to another can lead to increased diversity of viral species, increased antibiotic resistance, and/or induction of virulence factors in the host bacteria.[4] Other phages may alter the antigenicity of their hosts by modifying the O-antigen component. In the era of fecal microbiota therapy (FMT), studies on the role of the viral component of fecal samples of healthy donors and their effects on the response to FMT are needed.[5,6] Accordingly, the characterization of the virome profile in health is the first step. As with the bacterial component of the microbiome, the genetic makeup of an individual’s virome is influenced by diet, nutrition status, health, socioeconomic group, geographical location, age, lifestyle, season, and medication.[7,8,9] Studies on the role of dietary lifestyle in the gut virome profile suggested variations between populations with different dietary lifestyles, indicating the need for studies from different populations.[8,9,10,11] In this study, we used shotgun metagenomic DNA sequencing (untargeted sequencing) of purified viral samples from healthy children.[12] The objective was to characterize the profile of bacteriophages and DNA eukaryotic viruses in a cohort of healthy Saudi children, a Middle eastern population. RNA eukaryotic viruses were not analyzed.
SUBJECTS AND METHODS
The study population
The study was performed at King Khalid University Hospital, King Saud University Medical City, King Saud University; and King Fahad Medical City Children Hospital, Ministry of Health, Riyadh, the Kingdom of Saudi Arabia (KSA). Stool samples were collected from healthy schoolchildren taken from a larger random sample recruited for a mass screening study.[13] The children were on a normal family diet at the time of sample collection.
Sample collection and storage
Stool samples were collected in cryovials and stored at −80°C. At the time of analysis, the samples were sent by express mail in a temperature-controlled container filled with dry ice until delivery, to the laboratory where metagenomic, bioinformatics, and statistical analyses were performed (CosmosID, Rockville, MD, USA).
DNA isolation and sequencing
DNA was isolated using the DNeasy PowerSoil DNA kit (Qiagen, Hilden, Germany), with each process done according to the manufacturer’s instructions. Isolated viral DNA was quantified by Qubit (Thermo Fisher Scientific, Waltham, MA, USA).
DNA libraries were prepared using the Illumina Nextera XT library preparation kit, according to the manufacturer’s protocol. Library quantity and quality were assessed with Qubit and Tapestation (Agilent Technologies, Santa, Clara, CA, USA). Libraries were then sequenced on an HiSeq platform (2 × 150 bp; Illumina, San Diego, CA, USA).
Bioinformatic and abundance analysis
Unassembled sequencing reads were directly analyzed with the CosmosID bioinformatics platform (CosmosID Inc., Rockville, MD, USA) described elsewhere for microbiome analysis and quantification of each organism’s relative abundance.[14,15,16,17] Briefly, the system uses curated genome databases and a high-performance data-mining algorithm that rapidly disambiguates hundreds of millions of metagenomic sequence reads into the discrete microorganisms engendering the sequences.
The abundance of each organism was calculated and expressed as an average relative percentage across the viral phylogenetic tree from phyla to species.
The datasets generated during this study are available in the NCBI SRA. Access link: http://www.ncbi.nlm.nih.gov/bioproject/757365.
Ethical approval
This study was approved by the Institutional Review Board of the College of Medicine, King Saud University Riyadh, Kingdom of Saudi Arabia (no. 14/4464/IRB). All children and/or their parents gave informed consent and/or assent for participation in the study.
RESULTS
The study population
Twenty healthy Saudi children were enrolled. The median age was 11.3 (range 6.8–15.4) years, and 35% were males. The weight average and range were 46.9 (20-76) kg and the BMI average and range were 19.8 (12.5-28.0) kg/m2. The children were on a normal Saudi family diet dominated by the consumption of rice, bread, red meat, and chicken. In addition, the children frequently consumed fast food and sweetened gaseous drinks but rarely fruit or vegetables.
The abundance of viruses
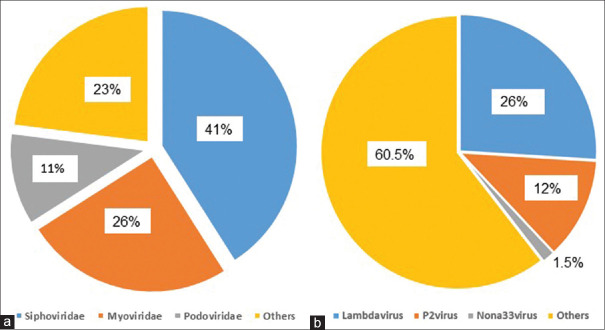
The profile and abundance in this study were determined by shotgun analysis of the DNA of viral particles only and did not include RNA viruses. Among 206 sequenced taxa, only 24 (11.7%) were not identified in the available database and therefore were designated unidentified. Caudovirales were the most abundant bacteriophage order (77%). The abundance of the top families and genera is illustrated in Figure 1. Among the list of viral families, Siphoviridae, Myoviridae, and Podoviridae families predominated, accounting for 41%, 25%, and 11%, respectively. Similarly, the most abundant genera included Lambdavirus, P2virus, and Nona33virus accounting for 26%, 12%, and 1.5%, respectively [Table 1]. The abundance of all the identified bacteriophage species is shown in Table 2. Among the Enterobacteria phages, the most abundant species were Enterobacteria phage BP-4795, Enterobacteria phage YYZ-2008, Enterobacteria phage mEp460, and Enterobacteria phage P88 accounting for 6.6%, 5.4%, 3.3%, and 3.3%, respectively. The most abundant Escherichia phages included Escherichia phage TL-2011b (2.5%), Escherichia virus P2 (2.4%), Escherichia virus HK022 (2.1%), and Escherichia virus If1 (1.8%), whereas Lactobacillus phage KC5a was the most abundant lactobacillus phage (2.9%). Among Lactococcus phages, Lactococcus phage ul36 was the most abundant (1.7%) and Salmonella phage RE-2010 was the most abundant among the Salmonella phages (1%). Shigella phage SfIV was the most abundant Shigella phage (1.4%) and Streptococcus phage 20617 was the most abundant Streptococcus phage (15%).
Figure 1.
Illustration of the abundance of the top families and genera. Panel (a) shows the predominance of the Siphoviridae family (41%) and the others (23%) cover all other family members with abundance less than 11% each. Panel (b) shows the predominance of the Lambdavirus genera (26%) and the others refer to all other genera with abundance less than 1.5% each
Table 1.
Viral abundance from order to genera level
| Level | Organism | Abundance | Level | Organism | Abundance |
|---|---|---|---|---|---|
| Order | Caudovirales | 0.77 | Genera | Lambdavirus | 0.26 |
| Family | Geminiviridae | 0.0008 | Genera | Muvirus | 0.001 |
| Family | Inoviridae | 0.018 | Genera | N15virus | 1.40−05 |
| Family | Myoviridae | 0.25 | Genera | Nona33virus | 0.015 |
| Family | Podoviridae | 0.11 | Genera | P1virus | 0.01 |
| Family | Retroviridae | 9.95−05 | Genera | P22virus | 0.014 |
| Family | Siphoviridae | 0.41 | Genera | P2virus | 0.12 |
| Genera | Begomovirus | 0.0008 | Genera | Pa6virus | 4.55−05 |
| Genera | C2virus | 0.0001 | Genera | Phietavirus | 8.90−05 |
| Genera | Cc31virus | 0.0012 | Genera | Phifelvirus | 0.0001 |
| Genera | Epsilon15virus | 0.033 | Genera | Rb69virus | 5.70−05 |
| Genera | Gammaretrovirus | 9.95−05 | Genera | Sfi11virus | 0.004 |
| Genera | Hp1virus | 0.0001 | Genera | Sfi21dt1virus | 0.005 |
| Genera | Jerseyvirus | 0.0002 | Genera | Tl2011virus | 0.006 |
Table 2.
Abundance of viral species
| No. | Organism | Abundance | No. | Organism | Abundance |
|---|---|---|---|---|---|
| 1 | Bacteroides phage B124-14 | 0.006 | 61 | Lactobacillus phage LF1 | 0.001 |
| 2 | Bacteroides phage B40-8 | 0.002 | 62 | Lactobacillus phage Lrm1 | 6.35-05 |
| 3 | Cronobacter phage ENT39118 | 1.75-05 | 63 | Lactobacillus phage phiadh | 0.0001 |
| 4 | Cronobacter phage ENT47670 | 9.00-06 | 64 | Lactobacillus prophage Lj771 | 0.0002 |
| 5 | Cronobacter phage phiES15 | 2.00-05 | 65 | Lactobacillus prophage Lj965 | 1.35-05 |
| 6 | Enterobacter virus CC31 | 0.0008 | 66 | Lactococcus phage 1706 | 9.50-06 |
| 7 | Enterobacter virus PG7 | 0.0004 | 67 | Lactococcus phage 340 | 1.35-05 |
| 8 | Enterobacteria phage 933W | 0.014 | 68 | Lactococcus phage bIBB29 | 2.95-05 |
| 9 | Enterobacteria phage BP-4795 | 0.066 | 69 | Lactococcus phage bIL170 | 2.45-05 |
| 10 | Enterobacteria phage cdtI | 0.030 | 70 | Lactococcus phage bIL285 | 0.001 |
| 11 | Enterobacteria phage fiAA91-ss | 0.024 | 71 | Lactococcus phage bIL286 | 0.002 |
| 12 | Enterobacteria phage HK106 | 0.003 | 72 | Lactococcus phage bIL309 | 0.001 |
| 13 | Enterobacteria phage HK140 | 0.003 | 73 | Lactococcus phage bIL310 | 0.01 |
| 14 | Enterobacteria phage HK225 | 0.001 | 74 | Lactococcus phage bIL311 | 0.011 |
| 15 | Enterobacteria phage HK446 | 0.002 | 75 | Lactococcus phage bIL312 | 0.006 |
| 16 | Enterobacteria phage HK542 | 0.002 | 76 | Lactococcus phage bIL67 | 9.40-05 |
| 17 | Enterobacteria phage HK544 | 0.006 | 77 | Lactococcus phage BK5-T | 0.001 |
| 18 | Enterobacteria phage HK629 | 0.002 | 78 | Lactococcus phage BM13 | 0.004 |
| 19 | Enterobacteria phage HK630 | 0.002 | 79 | Lactococcus phage c2 | 2.35-05 |
| 20 | Enterobacteria phage HK633 | 0.003 | 80 | Lactococcus phage jm2 | 5.50-06 |
| 21 | Enterobacteria phage IME10 | 0.007 | 81 | Lactococcus phage P008 | 5.00-06 |
| 22 | Enterobacteria phage mEp043 | 0.001 | 82 | Lactococcus phage P335 sensu lat | 0.001 |
| 23 | Enterobacteria phage mEp235 | 0.0004 | 83 | Lactococcus phage phiLC3 | 0.001 |
| 24 | Enterobacteria phage mEp237 | 0.003 | 84 | Lactococcus phage r1t | 0.002 |
| 25 | Enterobacteria phage mEp460 | 0.035 | 85 | Lactococcus phage TP901-1 | 0.006 |
| 26 | Enterobacteria phage mEpX1 | 9.35-05 | 86 | Lactococcus phage Tuc2009 | 0.007 |
| 27 | Enterobacteria phage mEpX2 | 0.0004 | 87 | Lactococcus phage ul36 | 0.017 |
| 28 | Enterobacteria phage P4 | 0.023 | 88 | Lambdavirus_u_s | 0.0004 |
| 29 | Enterobacteria phage P88 | 0.033 | 89 | Leuconostoc phage Lmd1 | 2.80-05 |
| 30 | Enterobacteria phage phiP27 | 0.003 | 90 | Leuconostoc phage P793 | 5.50-06 |
| 31 | Enterobacteria phage RB3 | 2.40-05 | 91 | Leuconostoc phage phiLN03 | 0.0008 |
| 32 | Enterobacteria phage SfV | 0.015 | 92 | Leuconostoc phage phiLN04 | 0.001 |
| 33 | Enterobacteria phage ST104 | 0.0002 | 93 | Leuconostoc phage phiLN12 | 4.75-05 |
| 34 | Enterobacteria phage YYZ-2008 | 0.054 | 94 | Murine leukemia virus | 3.50-06 |
| 35 | Enterobacterial phage mEp213 | 7.90-05 | 95 | Pectobacterium phage ZF40 | 2.15-05 |
| 36 | Enterobacterial phage mEp234 | 0.002 | 96 | Phage Gifsy-2 | 1.50-05 |
| 37 | Enterobacterial phage mEp390 | 0.001 | 97 | Propionibacterium virus P1001 | 4.55-05 |
| 38 | Escherichia phage D108 | 0.001 | 98 | Salmonella phage epsilon34 | 0.003 |
| 39 | Escherichia phage HK639 | 0.0001 | 99 | Salmonella phage Fels-1 | 4.50-05 |
| 40 | Escherichia phage HK75 | 0.001 | 100 | Salmonella phage Fels-2 | 0.001 |
| 41 | Escherichia phage P13374 | 0.002 | 101 | Salmonella phage FSL SP-004 | 0.002 |
| 42 | Escherichia phage TL-2011b | 0.025 | 102 | Salmonella phage g341c | 4.00-06 |
| 43 | Escherichia phage vB_EcoM | 1.50-05 | 103 | Salmonella phage HK620 | 0.004 |
| 44 | Escherichia virus 933W | 0.012 | 104 | Salmonella phage RE-2010 | 0.01 |
| 45 | Escherichia virus 9g | 3.50-06 | 105 | Salmonella phage SE1 | 0.0002 |
| 46 | Escherichia virus HK022 | 0.021 | 106 | Salmonella phage SE2 | 0.0002 |
| 47 | Escherichia virus HK97 | 0.003 | 107 | Salmonella phage SPN9CC | 0.0003 |
| 48 | Escherichia virus HX01 | 2.00-05 | 108 | Salmonella phage SSU5 | 0.008 |
| 49 | Escherichia virus If1 | 0.01 | 109 | Salmonella phage ST64B | 9.00-05 |
| 50 | Escherichia virus JS09 | 2.45-05 | 110 | Salmonella phage vB_SemP_Emek | 0.001 |
| 51 | Escherichia virus Min27 | 0.001 | 111 | Salmonella phage Vi II-E1 | 3.50-05 |
| 52 | Escherichia virus N15 | 1.40-05 | 112 | Salmonella virus Epsilon15 | 7.85-05 |
| 53 | Escherichia virus P1 | 0.01 | 113 | Salmonella virus P22 | 0.0002 |
| 54 | Escherichia virus P2 | 0.02 | 114 | Salmonella virus PsP3 | 0.002 |
| 55 | Escherichia virus phiV10 | 0.01 | 115 | Salmonella virus SPN1S | 3.50-06 |
| 56 | Escherichia virus RB69 | 1.25-05 | 116 | Shigella phage Sf6 | 0.006 |
| 57 | Escherichia virus TL2011 | 0.01 | 117 | Shigella phage SfII | 0.01 |
| 58 | Klebsiella phage phiKO2 | 0.0002 | 118 | Shigella phage SfIV | 0.013 |
| 59 | Lactobacillus phage KC5a | 0.03 | 119 | Spleen focus-forming virus | 9.60-05 |
| 60 | Lactobacillus phage Lc-Nu | 0.0003 | 120 | Staphylococcus phage Ipla5 | 8.90-05 |
| 121 | Streptococcus phage 20617 | 0.15 | 138 | Streptococcus phage O1205 | 0.0001 |
| 122 | Streptococcus phage 2972 | 6.30-05 | 139 | Streptococcus phage PH 10 | 0.001 |
| 123 | Streptococcus phage 315.1 | 5.25-05 | 140 | Streptococcus phage PH 15 | 6.70-05 |
| 124 | Streptococcus phage 315.2 | 0.0002 | 141 | Streptococcus phage phiBHN167 | 4.50-06 |
| 125 | Streptococcus phage 315.6 | 3.90-05 | 142 | Streptococcus phage Sfi11 | 0.0007 |
| 126 | Streptococcus phage 5093 | 0.0007 | 143 | Streptococcus phage Sfi19 | 0.0007 |
| 127 | Streptococcus phage 7201 | 0.001 | 144 | Streptococcus phage Sfi21 | 0.0004 |
| 128 | Streptococcus phage 858 | 0.001 | 145 | Streptococcus phage SM1 | 0.0003 |
| 129 | Streptococcus phage Abc2 | 0.001 | 146 | Streptococcus phage TP-778L | 0.001 |
| 130 | Streptococcus phage Alq132 | 0.002 | 147 | Streptococcus phage TP-J34 | 7.70-05 |
| 131 | Streptococcus phage DCC1738 | 0.0002 | 148 | St×2-converting phage 1717 | 0.083 |
| 132 | Streptococcus phage DT1 | 0.001 | 149 | St×2-converting phage 86 | 0.014 |
| 133 | Streptococcus phage EJ-1 | 0.0004 | 150 | Synechococcus phage S-CBS1 | 1.80-05 |
| 136 | Streptococcus phage M102 | 3.70-05 | 151 | Vibrio phage pYD38-A | 0.0002 |
| 137 | Streptococcus phage MM1 | 0.0001 | 152 | Yersinia phage L-413C | 0.046 |
| 153 | Watermelon chlorotic stunt virus | 0.001 |
DISCUSSION
Knowledge of the viral profile in healthy individuals is a prerequisite for the study of the role of viruses in disease pathogenesis and etiology. Bacteriophages are the most abundant viruses in humans and infection of bacteria by phages can alter microbiota structure by killing host cells or altering their phenotype, contributing either to the maintenance of intestinal homeostasis or causing microbial imbalance and development of chronic infectious and autoimmune diseases.
To our knowledge, this is the first report on gut viral profiles in healthy Saudi children, a Middle Eastern population who have different cultures and dietary lifestyles than their Western counterparts. Our findings that bacteriophages were the most abundant viruses and Caudovirales were the most abundant order (77%), are consistent with the results of several reviews.[18,19,20,21,22] Interestingly, crAssphages, (cross assembly phage; members of the Caudovirales) were not found in the fecal samples of our children, a finding contrary to reports of the abundance of more than 50% of the human gut samples.[23,24,25] The explanation of this important variation is not clear at present. It is possible that the lack of detection of this virus and others in our sample is related to age, ethnicity, culture, dietary lifestyle, or geographic differences.[26,27] The significance of these new viruses in health or disease is still not clear.[28] Nevertheless, our results are consistent with reports of the predominance of bacteriophages of the Siphoviridae, Podoviridae, and Myoviridae families. Microviridae are less abundant in infants but rise in abundance with age.[29,30] In addition, the profile of phage species in this report is consistent with some studies, reporting that phages of the early bacterial colonizers, including Escherichia, Klebsiella, Enterococcus, Staphylococcus, and Streptococcus species, were some of the most abundant early virome members in children.[31,32]
Similarities with previous reports include the predominance of the bacteriophages Caudovirales order; the Siphoviridae, Podoviridae, and Myoviridae families; the Escherichia, Klebsiella, Enterococcus, Staphylococcus, and Streptococcus species. The most important difference was the lack of cross assembly phage in our study.
Our study has a few limitations including the relatively small sample size which may be acceptable for this first report of the gut virome in Saudi children. In addition, the limitation to DNA viruses is recognized.
In conclusion, the profile and abundance of the intestinal virome in healthy Saudi children reveal similarities and distinctive features as illustrated in the literature. Further studies from different populations with larger sample sizes are needed to advance knowledge of the importance of gut viruses in the pathogenesis of disease in general and their role in the response to FMT in particular.
Financial support and sponsorship
The Deanship of Scientific Research, King Saud University.
Conflicts of interest
There are no conflicts of interest.
Acknowledgement
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through Research Group No (RGP-1441-007).
REFERENCES
- 1.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10:607–17. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA. 2013;110:10771–6. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–81. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol. 2010;59:145–55. [PubMed] [Google Scholar]
- 5.Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W, et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2017;67:634–43. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anonye BO. Commentary: Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Front Cell Infect Microbiol. 2018;8:104. doi: 10.3389/fcimb.2018.00104. doi:10.3389/fcimb. 2018.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim MS, Bae JW. Spatial disturbances in altered mucosal and luminal gut viromes of diet induced obese mice. Environ Microbiol. 2016;18:1498–510. doi: 10.1111/1462-2920.13182. [DOI] [PubMed] [Google Scholar]
- 8.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. The human gut virome: Inter-individual variation and dynamic response to diet. Genome Research. 2011;21:1616–25. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogilvie LA, Caplin J, Dedi C, Diston D, Cheek E, Bowler L, et al. Comparative (meta) genomic analysis and ecological profiling of human gut-specific bacteriophage fB124-14. PLoS One. 2012;7:e35053. doi: 10.1371/journal.pone.0035053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes A, Blanton LV, Cao S, Zhao G, Manary M, Trehan I, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci U S A. 2015;112:11941–6. doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart E. A roadmap to the human virome. PLoS Pathog. 2013;9:e1003146. doi: 10.1371/journal.ppat.1003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–3. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Hussaini A, Troncone R, Khormi M, AlTuraiki M, Alkhamis W, Alrajhi M, et al. Mass screening for celiac disease among school-aged children: Toward exploring celiac iceberg in Saudi Arabia. J Pediatr Gastroenterol Nutr. 2017;65:646–65. doi: 10.1097/MPG.0000000000001681. [DOI] [PubMed] [Google Scholar]
- 14.Ottesen A, Ramachandran P, Reed E, White JR, Hasan N, Subramanian P, et al. Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol. 2016;16:275. doi: 10.1186/s12866-016-0894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasan NA, Young BA, Minard-Smith AT, Saeed K, Li H, Heizer EM, et al. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS One. 2014;9:e97699. doi: 10.1371/journal.pone.0097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–52. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponnusamy D, Kozlova EV, Sha J, Erova TE, Azar SR, Fitts EC, et al. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc Natl Acad Sci U S A. 2016;113:722–7. doi: 10.1073/pnas.1523817113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aggarwala V, Liang G, Bushman FD. Viral communities of the human gut:metagenomic analysis of composition and dynamics. Mob DNA. 2017;8:12. doi: 10.1186/s13100-017-0095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shkoporov AN, Hill C. Bacteriophages of the human gut: The “known unknown”of the microbiome. Cell Host Microbe. 2019;25:195–209. doi: 10.1016/j.chom.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 20.Carding SR, Davis N, Hoyles L. Review article: The human intestinal virome in health and disease. Aliment Pharmacol Ther. 2017;46:800–15. doi: 10.1111/apt.14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim ES, Wang D, Holtz LR. The bacterial microbiome and virome milestones of infant development. Trends Microbiol. 2016;24:801–10. doi: 10.1016/j.tim.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–50. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutilh B, Cassman N, McNair K, Sanchez SE, Silva GGZ, Boling L, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:1–11. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerin E, Shkoporov A, Stockdale SR, Clooney AG, Ryan FJ, Sutton TDS, et al. Biology and taxonomy of crAss-like bacteriophages, the most abundant virus in the human gut. Cell Host Microbe. 2018;24:653–64. doi: 10.1016/j.chom.2018.10.002. e6. [DOI] [PubMed] [Google Scholar]
- 25.Edwards RA, Vega AA, Norman HM, Ohaeri M, Levi K, Dinsdale EA, et al. Global phylogeography and ancient evolution of the widespread human gut virus crAssphage. Nat Microbiol. 2019;4:1727–36. doi: 10.1038/s41564-019-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory AC, Zablocki O, Zayed AA, Howell A, Bolduc B, Sullivan MB. The Gut Virome Database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe. 2020;28:724–40. doi: 10.1016/j.chom.2020.08.003. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rampelli S, Turroni S, Schnorr SL, Soverini M, Quercia S, Barone M, et al. Characterization of the human DNA gut virome across populations with different subsistence strategies and geographical origin. Environ Microbiol. 2017;19:4728–35. doi: 10.1111/1462-2920.13938. [DOI] [PubMed] [Google Scholar]
- 28.Mukhopadhya I, Segal JP, Carding SR, Hart AL, Hold GL. The gut virome: the ‘missing link’ between gut bacteria and host immunity? Ther Adv Gastroenterol. 2019;12:1–17. doi: 10.1177/1756284819836620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang G, Zhao C, Zhang H, Mattei L, Sherrill-Mix S, Bittinger K, et al. The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature. 2020;581:470–4. doi: 10.1038/s41586-020-2192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 2015;21:1228–34. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Baumann-Dudenhoeffer AM, D’Souza AW, Tarr PI, Warner BB, Dantas G. Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat Med. 2018;24:1822–9. doi: 10.1038/s41591-018-0216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]