Abstract
The expression of 21 novel genes located in the region from dnaA to abrB of the Bacillus subtilis chromosome was analyzed. One of the genes, yaaH, had a predicted promoter sequence conserved among SigE-dependent genes. Northern blot analysis revealed that yaaH mRNA was first detected from 2 h after the cessation of logarithmic growth (T2) of sporulation in wild-type cells and in spoIIIG (SigG−) and spoIVCB (SigK−) mutants but not in spoIIAC (SigF−) and spoIIGAB (SigE−) mutants. The transcription start point was determined by primer extension analysis; the −10 and −35 regions are very similar to the consensus sequences recognized by SigE-containing RNA polymerase. A YaaH-His tag fusion encoded by a plasmid with a predicted promoter for the yaaH gene was produced from T2 of sporulation in a B. subtilis transformant and extracted from mature spores, indicating that the yaaH gene product is a spore protein. Inactivation of the yaaH gene by insertion of an erythromycin resistance gene did not affect vegetative growth or spore resistance to heat, chloroform, and lysozyme. The germination of yaaH mutant spores in a mixture of l-asparagine, d-glucose, d-fructose, and potassium chloride was almost the same as that of wild-type spores, but the mutant spores were defective in l-alanine-stimulated germination. These results suggest that yaaH is a novel gene encoding a spore protein produced in the mother cell compartment from T2 of sporulation and that it is required for the l-alanine-stimulated germination pathway.
The gram-positive soil microorganism Bacillus subtilis initiates sporulation by dividing asymmetrically when nutrients are exhausted. Sporulation is a relatively simple model for cell differentiation, and its progress is marked by sequential and drastic changes in the physiological state of the cell. After asymmetric septation, the resultant larger and smaller cells are the mother cell and the forespore, respectively. As development proceeds, the mother cell engulfs the forespore and eventually lyses, releasing the mature spore. Mature spores are resistant to long periods of starvation, heat, toxic chemicals, lytic enzymes, and other factors that could damage a cell (12). Spores germinate and start growing when surrounding nutrients become available. Genes involved in this developmental system have been identified, and their biological functions have been analyzed (35, 37). These genes are mostly transcribed during sporulation by RNA polymerase containing developmentally specific sigma factors; these sigma factors, including SigF, SigE, SigG, and SigK, are temporally and spatially activated and regulate gene expression in a compartment-specific fashion (16, 37). The B. subtilis genome sequencing project revealed about 4,100 protein-coding genes, of which half have unknown functions (31). The identification of these genes will contribute useful information to the study of sporulation, germination, and spore dormancy of bacilli at the gene level. In the region from dnaA to abrB in the B. subtilis chromosome, 49 open reading frames (ORFs) were identified by the B. subtilis genome sequencing project (31), but 21 of them have not yet been analyzed. In order to discover novel genes involved in sporulation and/or germination, we systematically inactivated these genes and examined the resulting phenotypes and periods of expression. In this report, we describe the function of a gene, yaaH, which was revealed to be expressed only during sporulation.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and general techniques.
The B. subtilis and Escherichia coli strains used in this study are listed in Table 1. ASK202, ASK203, ASK204, and ASK205 are derivatives of 168 transformed with DNA from spoIIAC, spoIIGAB, spoIIIG, and spoIVCB mutants, respectively, obtained from P. Stragier. Oligonucleotide primers 8114F (5′-AGATCTTCGCTTCACAATACAGAA-3′) and 8114R (5′-AGATCTCTCGAGCTTAAATTCGTTAAAGGC-3′) were used to amplify a 424-bp segment internal to yaaH from the B. subtilis 168 chromosome. The PCR product was restricted at the BglII sites introduced by the primers and inserted into BamHI-restricted pMutin1 to create plasmid pMU114. Oligonucleotide primers 8114RTF (5′-AAGAAGCTTCCTAAGGACTGTATCGCG-3′) and 8141RTR (5′-GGAGGATCCGTGTCGCCTTGTTTTACCAC-3′) were used to amplify a 204-bp segment internal to yaaH from the B. subtilis 168 chromosome. The PCR product was restricted at the HindIII and BamHI sites introduced by the primers and inserted into BamHI- and HindIII-restricted pMutinT3 to create plasmid pMU114RT. pMutinT3 was pMutin1 into which the t1t2 terminator from the rrnB operon of E. coli had been introduced between the erythromycin resistance gene and the spac promoter (28, 44). pMU114 and pMU114RTR were introduced into strain 168 by transformation, a single crossover with selection for erythromycin resistance (0.5 μg/ml), yielding strains NIS8114 and NIS8114RT, respectively.
TABLE 1.
B. subtilis strains and plasmids
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| 168 | trpC2 | Laboratory stock |
| NIS8114 | trpC2 yaaH ΩpMU114 (erm) | This work |
| NIS8114RT | trpC2 yaaH ΩpMU114RT (erm) | This work |
| ASK202 | trpC2 pheA1 spoIIAC::kan | This work |
| ASK203 | trpC2 pheA1 spoIIGAB::kan | This work |
| ASK204 | trpC2 pheA1 spoIIIG::kan | This work |
| ASK205 | trpC2 pheA1 spoIVCB::erm | This work |
| ASK206 | trpC2 yaaH′-lacZ (cat) | This work |
| Plasmids | ||
| pMutin1 | bla erm t0 Pspac lacZ lacI | 44 (V. Vagner) |
| pMutinT3 | bla erm t1t2t0 Pspac lacZ lacI | 28 |
| pMU114 | bla erm t0yaaH′-lacZ lacI Pspac-′yaaH | This work |
| pMU114RT | bla erm t1t2t0yaaH′-lacZ lacI Pspac-′yaaH | This work |
| pTUBE1200H6 | tet | 38 |
| pYAAH8 | tet yaaI′ YaaH-His | This work |
A yaaH-lacZ translational fusion was constructed by fusion of the entire yaaH gene (from the promoter region to codon GTG), amplified with oligonucleotides HNF (5′-CCCCCCGGGGCTATAGCGGCGGAC-3′) and HNR (5′-CCCCCCGGGTCGAAACGTCTTTTTGACAAC-3′), to the initiation codon of a promoterless lacZ gene, which originated from a PstI fragment of pMC1871 (purchased from Pharmacia). The yaaH′-lacZ translational fusion was integrated by Campbell insertion at the yaaH locus, resulting in strain ASK206. Oligonucleotide primers YAAHM558 (5′-GATCTAGAGGAAACCCTCGCTAAA-3′) and YAAH1280R (5′-AAAGATCTAAACGTCTTTTTGACAACA-3′) were used to amplify a 723-bp segment including the yaaH gene and its 5′ upstream region from the B. subtilis 168 chromosome. The PCR product was restricted at the XbaI and BglII sites introduced by the primers and inserted into XbaI- and BamHI-restricted pTUBE1200H6 to create plasmid pYAAH8. pTUBE1200H6 is a multicopy vector having a tetracycline resistance gene, a multicloning site, and a replication origin, pAMα1, which is active in B. subtilis cells (38). pTUBE1200H6 and pYAAH8 were transformed into strain 168 with selection for tetracycline resistance (20 μg/ml) to produce the transformants pTUBE1200H6/168 and pYAAH8/168, respectively. B. subtilis strains were grown in Luria-Bertani (LB) and Difco sporulation (DS) media (34). E. coli was grown in Luria-Bertani medium. The conditions for the sporulation of B. subtilis and the method for the purification of mature spores have been described previously (2). Recombinant DNA methods were carried out as described by Sambrook et al. (33). Methods for preparing competent cells, for transformation, and for the preparation of chromosomal DNA of B. subtilis are described by Cutting and Vander Horn (9).
Northern analysis.
The cells were grown in DS medium at 37°C, and an aliquot was harvested by centrifugation. Total RNA was extracted from the cells as described previously (39). Aliquots containing 5 μg of total RNA were electrophoresed and blotted on a positively charged nylon membrane (Hybond-N+; Amersham). Hybridizations were performed with digoxigenin-labeled RNA probes (10 ng) according to the manufacturer’s instructions (Boehringer Mannheim Biochemicals). Hybridizations specific for yaaH mRNA were conducted with digoxigenin-labeled RNA probes synthesized in vitro with T7 RNA polymerase by use of PCR products amplified from pMU114. The primers used to introduce a promoter for T7 RNA polymerase for this amplification were 8114F and T7R (5′-TAATACGACTCACTATAGGGCGAAGTGTATCAACAAGCTGG-3′).
Primer extension analysis.
Total RNA was extracted from strain NIS8114RT growing in DS medium at 4 h after the onset of sporulation. In strain NIS8114RT, the yaaH promoter region is fused downstream of the BamHI cloning site to the promoterless lacZ gene of pMutinT3. The RNA sample was subjected to primer extension assays with digoxigenin-end-labeled primers specific for the sequences around the BamHI site and the lacZ gene of pMutinT3 (RT1 [5′-TGTATCAACAAGCTGGGGATC] and RT2 [5′-CCAGGGTTTTCCCGGTCGACC]). Therefore, these primers can detect yaaH-specific transcription. The RNA sample (20 μg) was incubated with the primers (1 pmol) for 60 min at 60°C and gradually cooled down to the ambient temperature over 90 min. After the addition of deoxynucleoside triphosphates (2.5 mM each) and reverse transcriptase (GIBCO BRL), the reaction mixtures were incubated for 60 min at 37°C. The cDNA products were electrophoresed through an 8% polyacrylamide–urea gel, blotted to a positively charged nylon membrane, and detected according to the manufacturer’s instructions (Boehringer). DNA ladders for the size marker were created with the same digoxigenin-end-labeled primers by use of a DIG Taq DNA sequencing kit (Boehringer).
Cellular localization of β-galactosidase activity.
An aliquot (2 μl) of a culture of strain ASK206 sporulated in DS medium was mixed with 12.5 mM fluorescein β-d-galactopyranoside (FDG; Sigma) solution (6 μl), and the mixture was incubated for 30 min at 37°C. The mixture was then transferred to a microscope slide coated with poly-l-lysine and stained with 4′,6-diamidino-2-phenylindole (DAPI). Fluorescence obtained from FDG and DAPI staining was observed under an Olympus AX70 fluorescence microscope with U-MNIBA and U-MWU mirror cube units, respectively. The images were captured with a cooled charge-coupled-device camera (PXL-1400; Photometrics) and obtained with IPLAB SPECTRUM image-processing software (Signal Analytics Corporation).
Preparation of spores.
Mature spores were prepared by culturing the bacteria in DS medium for 48 h at 37°C. The spores were harvested by centrifugation and purified by two washes in cold deionized water, lysozyme treatment (0.1 mg/ml) at 37°C for 10 min, and sonication (Nissei US-300 Ultrasonic Generator) six times at 4°C for 15 s each time. The resultant cells were washed with cold deionized water by repeated centrifugation until all cell debris and vegetative cells had been removed.
Spore resistance.
Cells were grown in DS medium at 37°C for 18 h after the end of exponential growth, and spore resistance was assayed as follows. The cultures were heated at 80°C for 30 min, treated with lysozyme (final concentration, 0.25 mg/ml) at 37°C for 10 min or treated with 10% (vol/vol) chloroform at room temperature for 10 min as described by Nicholson and Setlow (30), diluted in distilled water, plated on LB agar, and incubated overnight at 37°C. The numbers of survivors were determined by counting colonies.
Spore germination.
The purified spores were heat activated at 65°C for 15 min and then suspended in 50 mM Tris-HCl (pH 7.5) buffer to an optical density at 660 nm of 0.5. Either l-alanine (10 mM) or AGFK (3.3 mM l-asparagine, 5.6 mM d-glucose, 5.6 mM d-fructose, and 10 mM potassium chloride) was added. Germination was monitored by measurement of the decrease in the optical density at 660 nm of the spore suspension at 37°C for up to 90 min.
Solubilization of proteins from sporulating cells and mature spores.
For preparation of protein-containing extracts from sporulating cells, cultures (5 ml) were harvested every hour throughout sporulation and washed with 10 mM sodium phosphate buffer (pH 7.2). The pellets were suspended in 0.1 ml of lysozyme buffer (10 mM sodium phosphate [pH 7.2], 1% lysozyme), kept on ice for 5 min, solubilized in 0.1 ml of loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 10% glycerol, 0.05% bromophenol blue), and boiled for 5 min. For preparation of proteins from mature spores, spores were harvested 18 h after the cessation of logarithmic growth (T18) of sporulation and washed with 10 mM sodium phosphate buffer (pH 7.2). The pellets were suspended in 0.1 ml of lysozyme buffer, incubated at room temperature for 10 min, and washed with wash buffer (10 mM sodium phosphate [pH 7.2], 0.5 M NaCl). Spore proteins were solubilized in 0.1 ml of loading buffer and boiled for 5 min. The resulting samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (15% acrylamide) as described previously (1).
Immunoblotting.
For immunoblotting, proteins were transferred to a polyvinylidene difluoride membrane (Immobilon; 0.45-μm pore size; Millipore) and detected by use of rabbit immunoglobulin G (IgG) against the His tag (Qiagen) or SigK as the first antibody and donkey anti-rabbit IgG–horseradish peroxidase conjugate as the second antibody (Amersham). The antisera were diluted to 1/1,000 or 1/5,000 with 20 mM Tris-HCl (pH 7.6) buffer containing 0.8% NaCl and 0.5% Tween 80. Anti-SigK antiserum was provided by M. Fujita and Y. Sadaie (National Institute of Genetics, Mishima, Japan).
RESULTS
Identification of genes transcribed only at sporulation.
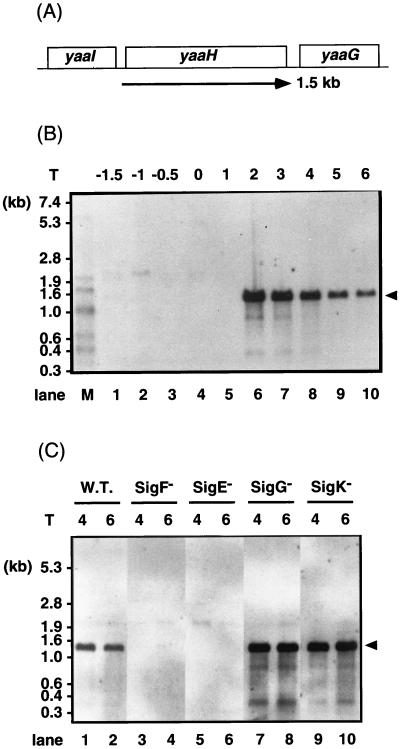
In the region from dnaA to abrB in the B. subtilis chromosome, 49 ORFs were identified by the B. subtilis genome sequencing project (31). The functions of 21 of these ORFs, yaaA, yaaB, yaaC, yaaD, yaaE, yaaF, yaaG, yaaH, yaaI, yaaJ, yaaK, yaaL, yaaN, yaaO, yaaQ, yaaR, yaaT, yabA, yabB, yabC, and yazA, have not yet been analyzed. Their transcription was analyzed by use of lacZ fusions constructed with integrational plasmid pMutin1, and a sporulation-specific gene, yaaH, was identified (3a). To confirm its expression pattern and transcription unit, total RNA was isolated from B. subtilis 168 and analyzed by Northern hybridization. The data in Fig. 1B showed that a single mRNA species of approximately 1.5 kb and which hybridized with a probe specific for yaaH was first detected from T2 of sporulation. From the nucleotide sequence, yaaH was predicted to be monocistronically transcribed (21, 31). The majority of genes induced during sporulation are transcribed by RNA polymerase containing sporulation-specific sigma factors. In order to determine which sigma factor was concerned with the transcription of yaaH, we performed Northern analysis with RNA prepared from sigma factor-deficient mutants (Fig. 1C). A probe specific for yaaH hybridized to a 1.5-kb mRNA in samples prepared from wild-type cells. This mRNA molecule was not detectable in spoIIAC and spoIIGAB mutants, which were deficient in SigF and SigE, respectively. On the other hand, the signal was still detectable in spoIIIG and spoIVCB mutants, which were deficient in SigG and SigK, respectively. SpoIIID was not essential for the transcription of yaaH (data not shown).
FIG. 1.
Analysis of yaaH mRNA by Northern hybridization. (A) Genome structure surrounding yaaH and the sizes of the ORFs. (B) Northern hybridization detecting the transcript of yaaH. Lanes: M, RNA molecular weight markers (digoxigenin labeled; 0.3 to 7.4 kb; Boehringer); 1 through 10, total RNA isolated from strain 168. T, harvesting times of cells, i.e., hours after the end of the exponential phase of growth. (C) Transcription of yaaH in strain 168 (spo+) (W.T.) (lanes 1 and 2) and in spoIIAC (SigF−) (lanes 3 and 4), spoIIGAB (SigE−) (lanes 5 and 6), spoIIIG (SigG−) (lanes 7 and 8), and spoIVCB (SigK−) (lanes 9 and 10) mutants analyzed by Northern hybridization. Total RNA was prepared from the cells at T4 (lanes 1, 3, 5, 7, and 9) and T6 (lanes 2, 4, 6, 8, and 10), respectively. The arrowheads indicate the position of mRNA hybridizing with the digoxigenin-labeled RNA probe.
Localization of the yaaH promoter.
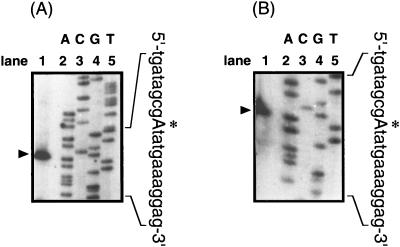
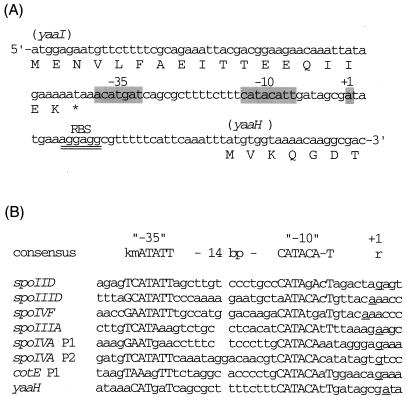
To localize precisely the yaaH promoter, primer extension analysis was carried out with RNA from sporulating cells of strain NIS8114RT. Two different primers were used for this analysis; both primers yielded the same transcription start site (Fig. 2). Transcription of yaaH starts 31 nucleotides (nt) upstream of the yaaH AUG codon, at an A residue (Fig. 3A). Sequences centered 10 and 35 nt upstream of the transcription start site are very similar to the −10 and −35 consensus sequences recognized by SigE, with appropriate spacing (14 nt) between these consensus sequences (Fig. 3B).
FIG. 2.
Determination of the transcription start site of yaaH by primer extension analysis. RNA prepared from sporulating cells of strain NIS8114 was hybridized with primers RT2 (A) and RT1 (B). Lanes labeled A, G, C, and T are DNA sequencing reactions with appropriate primers. Primer extension products are marked with arrowheads, and the transcription start site on the yaaH upstream sequence is marked with an asterisk and a capital letter.
FIG. 3.
Comparison of the 5′ upstream region of yaaH and a consensus sequence of the −10 and −35 regions recognized by SigE. (A) 5′ upstream sequence of the yaaH gene. The bases which match the consensus sequence are shaded. Double underlining indicates a ribosome binding site (RBS). (B) Comparison of the promoter sequence of yaaH and those of the genes dependent on SigE (13). k in the consensus sequence represents base T or G, m represents C or A, and r represents G or A. Consensus sequence is in uppercase letters; transcription start sites are underlined.
Determination of the compartment in which the YaaH-LacZ fusion protein is synthesized.
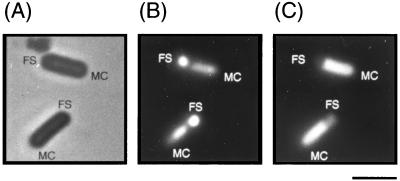
It has been shown that β-galactosidase activity can be detected in unfixed cells by use of fluorescence microscopy and the fluorogenic substrate FDG (22), and this technique allows the detection of compartment-specific transcription during sporulation. To determine the localization of yaaH gene expression during sporulation, we constructed a yaaH′-lacZ translational fusion and observed the fused gene product in transformants by fluorescence microscopy. The difference between the mother cell and the forespore could be recognized from the level of condensation of the nucleoid by staining with DAPI. As shown in Fig. 4B, the nucleoid in forespores appeared more compressed than that in mother cells. At the same time, fluorescence was detected in mother cells in strain ASK206 stained with FDG, as shown in Fig. 4C. On the other hand, no detectable fluorescence was observed in cells of strain 168 stained with FDG during sporulation (data not shown). From the results of Northern blotting, primer extension analysis, and microscopic analysis, we conclude that yaaH is specifically expressed by SigE RNA polymerase in mother cells during sporulation.
FIG. 4.
Detection of β-galactosidase activity of the YaaH-LacZ fusion in sporulating cells. Strain ASK206 carrying yaaH′-lacZ was cultured in DS medium and sampled 4 h after the onset of sporulation. The cells in the same view were observed by three different methods as described in Materials and Methods. (A) Phase-contrast image. (B and C) Fluorescence images of cells stained with DAPI (B) and FDG (C). FS, forespore; MC, mother cell; Bar, 2 μm.
Detection of the YaaH-His tag fusion in sporulating cells and mature spores.
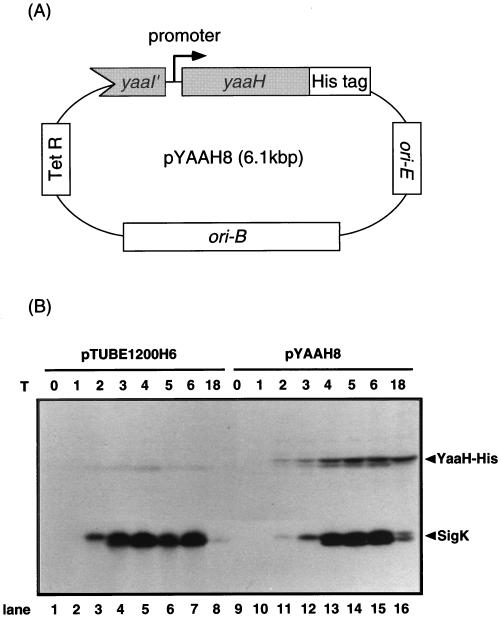
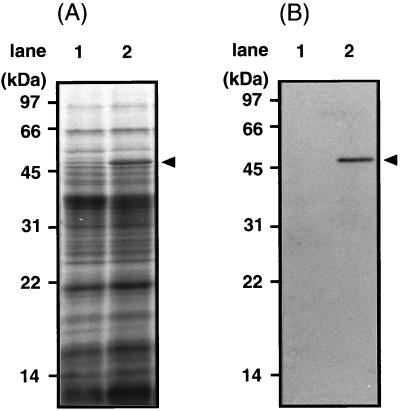
YaaH has many familial proteins (see Fig. 8). We used a YaaH-His tag fusion and His tag-specific antiserum in this study because antiserum against YaaH possibly binds not only to YaaH but also to its homologues. pTUBE1200H6 (38), a multicopy vector available in both E. coli and B. subtilis, was used as an expression vector. It has a tetracycline resistance gene and a multicloning site followed by a sequence encoding six consecutive histidine residues. To analyze the synthesis and location of YaaH protein in sporulating cells, plasmid pYAAH8, containing the yaaH gene and its promoter region, was constructed (Fig. 5A). The yaaH gene in this vector is fused to a sequence encoding six consecutive histidine residues (His tag) at its 3′ end, and the product, YaaH-His, is detectable with antiserum specific for the tag. pYAAH8 and a control vector, pTUBE1200H6, were introduced into B. subtilis 168 to generate transformants pYAAH8/168 and pTUBE1200H6/168, respectively. pYAAH8 is a multicopy plasmid which potentially produces more YaaH-His than a single-copy gene on the chromosome. The overproduction of YaaH-His was required for immunoblotting because the binding site for the anti-His tag antibody on this protein is limited. YaaH-His was produced from T2 of sporulation in pYAAH8/168 cells but not in those carrying the control vector (Fig. 5B). SigK, which was used as a control, was detectable from T2 of sporulation in both transformants (Fig. 5B). A band with a molecular mass of 49 kDa, corresponding to YaaH-His, was detectable in the protein extract from mature spores of pYAAH8/168 but not in that from pTUBE1200H6/168 mature spores (Fig. 6). It is unlikely that YaaH-His was sticking to the surface of spores due to its overproduction, because the spores used here were washed in the presence of 0.5 M sodium chloride as described in Materials and Methods. These results suggest that YaaH is a spore protein which is synthesized from T2 of sporulation.
FIG. 8.
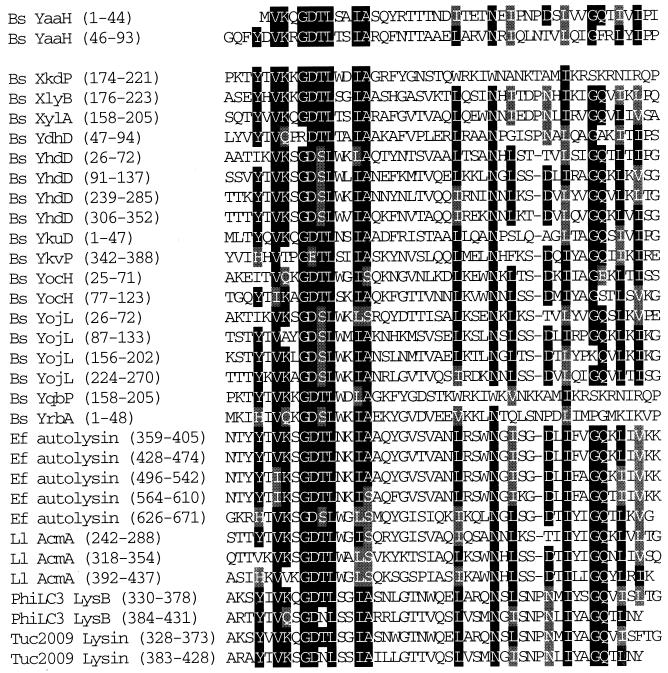
Comparison of proteins with amino acid sequences similar to the N-terminal portion of the YaaH protein. YaaH has two repeats of the consensus motif conserved among the bacterial lytic enzymes B. subtilis (Bs) XylA (23), Streptococcus faecalis (Ef) autolysin (4), Lactococcus lactis (Ll) AcmA (7), bacteriophage φLC3 (PhiLC3) LysB (5), and bacteriophage Tuc2009 lysin (3). B. subtilis XkdP, XlyB, YdhD, YhdD, YkuD, YkvP, YocH, YojL, YqbP, and YrbA, which have been identified by the B. subtilis genome sequencing project (21), also have the motif, but their characteristics are unknown. Identical and similar amino acid residues are indicated by black and gray boxes, respectively.
FIG. 5.
Detection of YaaH-His in sporulating cells. (A) pYAAH8 has replication origins available in both E. coli (ori-E) and B. subtilis (ori-B) cells, and it confers tetracycline resistance on these organisms. The yaaH gene in pYAAH8 is regulated by a promoter located upstream of the gene and fused to a sequence encoding six consecutive histidine residues (His tag). (B) B. subtilis 168 was transformed with control vector pTUBE1200H6 or pYAAH8 as described in Materials and Methods. Proteins prepared from the transformants were resolved by SDS-PAGE (15% acrylamide gel) and visualized by immunoblotting with antisera against the His tag and SigK. Arrowheads indicate the positions of the YaaH-His protein and SigK. T, harvesting times for cells (in hours).
FIG. 6.
Detection of YaaH-His in mature spores. Spore proteins were solubilized from B. subtilis 168 transformants carrying control vector pTUBE1200H6 (lane 1) or pYAAH8 (lane 2) as described in Materials and Methods. The proteins were resolved by SDS-PAGE (15% acrylamide gel) and visualized by Coomassie brilliant blue staining (A) or immunoblotting with anti-His tag antiserum (B). Arrowheads indicate the deduced molecular mass of the YaaH-His protein.
Properties of mutant spores.
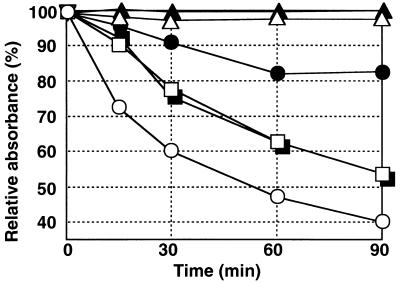
We characterized yaaH mutant cells; the vegetative growth of mutant cells in DS medium was the same as that of wild-type cells (data not shown). Mature spores prepared from the medium after 24 h of cultivation at 37°C showed resistance to heat, chloroform, and lysozyme, as did the wild-type spores (data not shown). The remarkable difference between yaaH mutant and wild-type cells was the spore germination response (Fig. 7). B. subtilis spores gradually lose the optical density of the suspension and heat resistance, as well as refractivity when observed under phase-contrast microscopy, when they are surrounded by appropriate germinants. Relative germination efficiency is evaluated by monitoring the optical density of the spore suspension and counting heat-resistant spores after incubation in the presence and absence of germinants. A reduction of the optical density of the spore suspension in Fig. 7 indicated that the wild-type spores germinated immediately during incubation with l-alanine and more slowly with AGFK. They lost heat resistance after incubation in the presence of l-alanine or AGFK (Table 2). The response of the mutant spores to germinant AGFK was the same as that of the wild-type spores when measured as the loss of optical density and heat resistance, while that to germinant l-alanine was lower than that of the wild-type spores (Fig. 7 and Table 2). About 25% of the mutant spores remained heat resistant after incubation in the presence of l-alanine for 90 min at 37°C. Using phase-contrast microscopy, we confirmed that almost all of the wild-type spores became phase bright to dark (fully germinated) after 90 min of incubation with l-alanine; in contrast, a few mutant spores became phase gray (partially germinated) under the same conditions (data not shown). These results suggested that yaaH is a gene required for the progress of the l-alanine-stimulated germination of spores.
FIG. 7.
Spore germination of B. subtilis 168 and a yaaH mutant. The germination of B. subtilis spores was monitored by measuring the optical density at 660 nm at the indicated times after the addition of l-alanine (circles), AGFK (squares), or control buffer (triangles). The efficiency of germination is expressed as relative absorbance. Open symbols, 168 (yaaH+); filled symbols, mutant NIS8114 (yaaH).
TABLE 2.
Heat resistance of spores after incubation in the presence and absence of germinantsa
| Genotype | No. of spores in the presence of:
|
||
|---|---|---|---|
| Tris-HCl buffer | AGFK | l-Alanine | |
| Wild type | 1.5 × 108 | 5.1 × 106 | 9.0 × 104 |
| yaaH mutant | 1.2 × 108 | 6.7 × 106 | 3.0 × 107 |
Spores were incubated in 10 mM Tris-HCl buffer (pH 7.0) without or with AGFK or l-alanine at 37°C for 90 min. The samples were heated at 80°C for 30 min and then spread on LB agar medium after appropriate dilution. The proportion of survivors was determined by counting colonies after 12 h of incubation at 37°C. The data represent the averages of three independent experiments.
DISCUSSION
Developmental gene expression during sporulation is unique to each of the two compartments (mother cell and forespore) and is regulated by compartment-specific sigma factors (35). SigE is the first of the alternative sigma factors to appear in the mother cell; SigF is its counterpart in the forespore (11). yaaH has a predicted SigE promoter (Fig. 3) and is transcribed from T2 of sporulation, when SigE is activated (Fig. 1). The mRNA of yaaH was detectable in the samples prepared from wild-type cells and SigG and SigK mutants but not in those prepared from SigE or SigF mutants (Fig. 1). SigF is essential for the activation of pro-SigE (35), and SigG is activated only in the forespore from stage III and directs the transcription of forespore-specific genes (32). SigK is activated exclusively in the mother cell from stage III to IV and directs the transcription of mother cell-specific genes (36). Both SigG and SigK require SigE for their activation directly or indirectly (32, 36). From these observations, we conclude that yaaH is expressed under the regulation of SigE RNA polymerase in the mother cell compartment.
A potential transcription terminator sequence is present in the downstream region of the yaaH gene (31). The molecular masses of mRNAs visualized by the probe for yaaH corresponded to that of the deduced transcript (Fig. 1). These results indicate that yaaH expression is independent from that of the downstream gene yaaG and that the phenotype of the yaaH mutant reflects only the function of yaaH.
Analysis with fusion proteins YaaH-LacZ and YaaH-His suggested that the YaaH protein is synthesized in the mother cell compartment and localizes in spores (Fig. 4, 5, and 6). The possibility of artificial incorporation of the YaaH protein into spores by overproduction was unlikely because of the following observations and results. Using the same strategy as in this work, we have previously shown that a spore coat protein, CotSA, cloned into a multicopy vector is selectively incorporated into spores (41). Its assembly is dependent on both CotE and CotS, and cotE or cotS mutant spores do not include CotSA (41). YaaH-His also remained in spores after washing with 0.5 M NaCl (Fig. 6). Moreover, analysis with a YaaH-green fluorescent protein (GFP) fusion (YaaH-GFP) and fluorescence microscopy showed that YaaH-GFP was detectable in mature spores (3a). These facts indicate that the assembly of YaaH-His into spores is not due to its overproduction. We have previously shown that not only coat proteins but also cortex proteins can be extracted from mature spores under our experimental conditions (40). A cortex protein of B. subtilis, YrbB, is extracted from mature spores and detected by immunoblotting after SDS-PAGE (40). Therefore, the results shown in Fig. 6 do not imply that the location of YaaH is limited to the spore coat.
No deduced signal sequence or hydrophobic regions could be found in the primary sequence of the YaaH protein, and the molecular mechanism for its assembly into spores is unknown. A database analysis showed that the YaaH protein has two repeats of the motif conserved among so-called cell wall binding proteins (Fig. 8). The motif is thought to be required for the cell wall binding ability of the proteins (20). B. subtilis proteins CwlF (PapQ), LytE, XkdP, XlyB, XylA, YdhD, YhdD, YkuD, YkvP, YocH, YojL, YqbP, and YrbA also have the same motif (15, 18, 21, 23, 24, 43). Except for the cell wall binding motif, the primary structure of the YaaH protein is not otherwise similar to almost all of these proteins. Over its entire length, YaaH shows slight similarity to B. subtilis YdhD, YkvQ, and YvbX, whose characteristics are also unknown (21). CwlF, LytE, and XylA were shown to be involved in cell wall hydrolysis (17, 23, 24). XlyB was predicted to encode an N-acetylmuramoyl-l-alanine amidase involved in defective prophage PBSX-mediated lysis (23). YrbA is produced from T2 of sporulation in the mother cell compartment and is involved in spore resistance to lysozyme and germination (42). The functions of XkdP, YdhD, YhdD, YkuD, YkvP, YocH, YojL, and YqbP are still unknown.
The inactivation of the yaaH gene did not impair vegetative growth or prevent the development of resistance to heat, chloroform, and lysozyme (data not shown). On the other hand, germination of the mutant spores induced by l-alanine was defective (Fig. 7). The process of spore germination in B. subtilis is dependent on the action of a germinant on a trigger site within the spore. Spores of B. subtilis 168 have two germination responses; one is activated by l-alanine alone, and the other is activated by AGFK. A hypothetical germination pathway and genes involved in germination stimulated by l-alanine or AGFK have been proposed (27). Genetic analysis indicates that germinant-specific germination mutants are classified into two groups. gerA mutants are defective specifically in response to l-alanine but germinate normally in AGFK, whereas in gerB, gerK, and fruB mutants, germination in response to l-alanine is normal but is not stimulated by AGFK (17, 26). These results suggest that the spore has two separate systems for detecting the alternative germination stimuli (27). The defect in yaaH mutant spores is thought to be limited to the l-alanine-stimulated germination pathway because the mutant spores showed a normal response to AGFK (Fig. 7).
About 20 genes are known to be required for the germination of B. subtilis spores (37). Among them, cotD, cotE, cotH, cotT, cotVWXYZ, cwlJ, and gerE are expressed only in the mother cell compartment (6, 10, 19, 25, 29, 45, 47). gerE encodes a DNA binding protein which regulates the expression of cot genes such as cotA, cotB, cotC, cotD, cotE, cotG, cotS, and cotX (8, 14, 37, 39, 46). We speculate that the function of YaaH is different from that of GerE, because YaaH does not have any known consensus motifs found among DNA binding proteins. Furthermore, YaaH lacks the motif found in the SleB protein family, including CwlJ and the specific cortex-hydrolyzing enzymes of Bacillus cereus and B. subtilis (19). The cotD, cotE, cotH, cotT, and cotVWXYZ genes encode spore coat proteins and/or regulators for coat protein assembly, and their mutant spores had morphological changes in coat structure and impaired germination (6, 10, 25, 29, 45, 47). yaaH showed no homology to those genes or to other reported spore coat genes. The mutant spores of yaaH appeared normal by phase-contrast microscopy (data not shown) and resistant to heat. We analyzed the protein extract from yaaH mutant spores by SDS-PAGE followed by Coomassie brilliant blue staining. Except for the absence of a band corresponding to the YaaH protein, no difference was found between the protein samples extracted from wild-type spores and yaaH mutant spores (data not shown). Electron microscopy also suggested that yaaH gene disruption did not alter the ultrastructure of spores (data not shown). Based on these results and on a similarity analysis of primary structure, we conclude that YaaH is a specific component of the system involved in the l-alanine-stimulated germination of B. subtilis spores.
ACKNOWLEDGMENTS
We thank Patrick Stragier for providing B. subtilis strains and Yoshito Sadaie and Masaya Fujita for providing antiserum against SigK. We thank Anne Moir for useful discussions and critical review and Michael G. Bramucci for critical reading of the manuscript. We also thank Kanae Fukuchi for technical assistance.
This work was supported by grant JPSP-RFTF96L00105 from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Abe A, Ogawa S, Koide K, Kohno T, Watabe K. Purification of Bacillus subtilis spore coat protein by electrophoretic elution procedure and determination of NH2-terminal amino acid sequence. Microbiol Immunol. 1993;53:809–812. doi: 10.1111/j.1348-0421.1993.tb01709.x. [DOI] [PubMed] [Google Scholar]
- 2.Abe A, Koide H, Kohno T, Watabe K. A Bacillus subtilis spore coat polypeptide gene, cotS. Microbiology. 1995;141:1433–1442. doi: 10.1099/13500872-141-6-1433. [DOI] [PubMed] [Google Scholar]
- 3.Arendt E K, Daly C, Fitzgerald G F, van de Guchte M. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of genes encoding lysin, a putative holin, and two structural proteins. Appl Environ Microbiol. 1994;60:1875–1883. doi: 10.1128/aem.60.6.1875-1883.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Asai, K., et al. Unpublished data.
- 4.Beliveau C, Potvin C, Trudel J, Asselin A, Bellemare G. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J Bacteriol. 1991;173:5619–5623. doi: 10.1128/jb.173.18.5619-5623.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birkeland N. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage phi-LC3: a dual lysis system of modular design. Can J Microbiol. 1994;40:658–665. doi: 10.1139/m94-104. [DOI] [PubMed] [Google Scholar]
- 6.Bourne N, Fitz-James P C, Aronson A I. Structural and germination defects of Bacillus subtilis spores with altered contents of a spore coat protein. J Bacteriol. 1991;173:6618–6625. doi: 10.1128/jb.173.20.6618-6625.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buist G, Kok J, Leenhouts K J, Dabrowska M, Venema G, Haandrikman A J. Molecular cloning and nucleotide sequencing of the gene encoding the major peptidoglycan hydrase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting S, Panzer S, Losick R. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J Mol Biol. 1989;207:393–404. doi: 10.1016/0022-2836(89)90262-3. [DOI] [PubMed] [Google Scholar]
- 9.Cutting S, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons, Ltd.; 1990. pp. 27–74. [Google Scholar]
- 10.Donovan W, Zheng L B, Sandman K, Losick R. Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol. 1987;196:1–10. doi: 10.1016/0022-2836(87)90506-7. [DOI] [PubMed] [Google Scholar]
- 11.Driks A, Losick R. Compartmentalized expression of a gene under the control of sporulation transcription factor sigma E in Bacillus subtilis. Proc Natl Acad Sci USA. 1991;88:9934–9938. doi: 10.1073/pnas.88.22.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gould G W. Mechanisms of resistance and dormancy. In: Hurst A, Gould G W, editors. The bacterial spore. Vol. 2. London, England: Academic Press, Ltd.; 1983. pp. 173–210. [Google Scholar]
- 13.Henriques A O, Bryan E M, Beall B W, Moran C P., Jr cse15, cse60, and csk22 are new members of mother-cell-specific sporulation regulons in Bacillus subtilis. J Bacteriol. 1997;179:389–398. doi: 10.1128/jb.179.2.389-398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland S K, Cutting S, Mandelstam J. The possible DNA-binding nature of the regulatory proteins, encoded by spoIID and gerE, involved in the sporulation of Bacillus subtilis. J Gen Microbiol. 1987;133:2381–2391. doi: 10.1099/00221287-133-9-2381. [DOI] [PubMed] [Google Scholar]
- 15.Holmberg C, Beijer L, Rutberg B, Rutberg L. Glycerol catabolism in Bacillus subtilis: nucleotide sequence of the genes encoding glycerol kinase (glpK) and glycerol-3-phosphate dehydrogenase (gipD) J Gen Microbiol. 1990;136:2367–2375. doi: 10.1099/00221287-136-12-2367. [DOI] [PubMed] [Google Scholar]
- 16.Igo M M, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- 17.Irie R, Okamoto T, Fujita Y. A germination mutant of Bacillus subtilis deficient in response to glucose. J Gen Appl Microbiol. 1982;28:345–354. [Google Scholar]
- 18.Ishikawa S, Hara Y, Ohnishi R, Sekiguchi J. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J Bacteriol. 1998;180:2549–2555. doi: 10.1128/jb.180.9.2549-2555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa S, Yamane K, Sekiguchi J. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J Bacteriol. 1998;180:1375–1380. doi: 10.1128/jb.180.6.1375-1380.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joris B, Englebert S, Chu C P, Kariyama R, Daneo-Moore L, Shockman G D, Ghuysen J M. Modular design of the Enterococcus hirae muramidase-2 and Streptococcus faecalis autolysin. FEBS Microbiol Lett. 1992;70:257–264. doi: 10.1016/0378-1097(92)90707-u. [DOI] [PubMed] [Google Scholar]
- 21.Kunst F, Ogasawara N, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 22.Lewis P J, Partridge S R, Errington J. Sigma factors, asymmetry, and the determination of cell fate in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:3849–3853. doi: 10.1073/pnas.91.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Longchamp P F, Mauel C, Karamata D. Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-l-alanine amidase gene of prophage PBSX. Microbiology. 1994;140:1855–1867. doi: 10.1099/13500872-140-8-1855. [DOI] [PubMed] [Google Scholar]
- 24.Margot P, Wahlen M, Gholamhuseinian A, Piggot P, Karamata D. The lytE gene of Bacillus subtilis 168 encodes a cell wall hydrolase. J Bacteriol. 1998;180:749–752. doi: 10.1128/jb.180.3.749-752.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moir A. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J Bacteriol. 1981;146:1106–1116. doi: 10.1128/jb.146.3.1106-1116.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moir A, Lafferty E, Smith D A. Genetic analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype with map location. J Gen Microbiol. 1979;111:165–180. doi: 10.1099/00221287-111-1-165. [DOI] [PubMed] [Google Scholar]
- 27.Moir A, Smith D A. The genetics of bacterial spore germination. Annu Rev Microbiol. 1990;44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- 28.Moriya S, Tsujikawa E, Hassan A K M, Asai K, Kodama T, Ogasawara N. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- 29.Naclerio G, Baccigalupi L, Zilhao R, De Felice M, Ricca E. Bacillus subtilis spore coat assembly requires cotH gene expression. J Bacteriol. 1996;178:4375–4380. doi: 10.1128/jb.178.15.4375-4380.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson W L, Setlow P. Sporulation, germination and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons, Ltd.; 1990. pp. 391–450. [Google Scholar]
- 31.Ogasawara N, Nakai S, Yoshikawa H. Systematic sequencing of the 180 kilobase region of the Bacillus subtilis chromosome containing the replication origin. DNA Res. 1994;1:1–14. doi: 10.1093/dnares/1.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Partridge S R, Errington J. The importance of morphological events and intercellular interactions in the regulation of prespore-specific gene expression during sporulation in Bacillus subtilis. Mol Microbiol. 1993;8:945–955. doi: 10.1111/j.1365-2958.1993.tb01639.x. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Schaeffer P, Millet J, Aubert J-P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stragier P, Bonamy C, Karamazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 36.Stragier P, Kunkel B, Kroos L, Losick R. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science. 1989;243:507–512. doi: 10.1126/science.2536191. [DOI] [PubMed] [Google Scholar]
- 37.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 38.Takamatsu H, Bunai K, Horinaka T, Oguro T, Nakamura K, Watabe K, Yamane K. Identification of a region required for binding to presecretory protein in Bacillus subtilis Ffh, a homologue of the 54-kDa subunit of mammalian signal recognition particle. Eur J Biochem. 1997;248:575–582. doi: 10.1111/j.1432-1033.1997.00575.x. [DOI] [PubMed] [Google Scholar]
- 39.Takamatsu H, Chikahiro Y, Kodama T, Koide H, Kozuka S, Tochikubo K, Watabe K. Spore coat protein, CotS, of Bacillus subtilis is synthesized under the regulation of SigK and GerE during development and is located in the inner coat layer of spores. J Bacteriol. 1998;180:2968–2974. doi: 10.1128/jb.180.11.2968-2974.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takamatsu H, Hiraoka T, Kodama T, Koide H, Kozuka S, Tochikubo K, Watabe K. Cloning of a novel gene yrbB, encoding a protein located in the spore integument of Bacillus subtilis. FEMS Lett. 1998;166:361–367. doi: 10.1111/j.1574-6968.1998.tb13913.x. [DOI] [PubMed] [Google Scholar]
- 41.Takamatsu H, Kodama T, Watabe K. Assembly of the CotSA coat protein into spores requires CotS in Bacillus subtilis. FEMS Microbiol Lett. 1999;174:201–206. doi: 10.1111/j.1574-6968.1999.tb13569.x. [DOI] [PubMed] [Google Scholar]
- 42.Takamatsu, H., T. Kodama, T. Nakayama, and K. Watabe. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 43.Takemaru K, Mizuno M, Sato T, Takeuchi M, Kobayashi Y. Complete nucleotide sequence of a skin element excised by DNA rearrangement during sporulation in Bacillus subtilis. Microbiology. 1995;141:323–327. doi: 10.1099/13500872-141-2-323. [DOI] [PubMed] [Google Scholar]
- 44.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Ichikawa H, Halberg R, Kroos L, Aronson A I. Regulation of the transcription of a cluster of Bacillus subtilis spore coat genes. J Mol Biol. 1994;240:405–415. doi: 10.1006/jmbi.1994.1456. [DOI] [PubMed] [Google Scholar]
- 46.Zheng L, Halberg R, Roels S, Ichikawa H, Kroos L, Losick R. Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes. J Mol Biol. 1992;226:1037–1050. doi: 10.1016/0022-2836(92)91051-p. [DOI] [PubMed] [Google Scholar]
- 47.Zheng L B, Donovan W P, Fitz-James P C, Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988;2:1047–1054. doi: 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]