Abstract
Arginine-rich dipeptide repeat proteins (R-DPRs), abnormal translational products of a GGGGCC hexanucleotide repeat expansion in C9ORF72, play a critical role in C9ORF72-related amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), the most common genetic form of the disorders (c9ALS/FTD). R-DPRs form liquid condensates in vitro, induce stress granule formation in cultured cells, aggregate, and sometimes coaggregate with TDP-43 in postmortem tissue from patients with c9ALS/FTD. However, how these processes are regulated is unclear. Here, we show that loss of poly(ADP-ribose) (PAR) suppresses neurodegeneration in c9ALS/FTD fly models and neurons differentiated from patient-derived induced pluripotent stem cells. Mechanistically, PAR induces R-DPR condensation and promotes R-DPR–induced stress granule formation and TDP-43 aggregation. Moreover, PAR associates with insoluble R-DPR and TDP-43 in postmortem tissue from patients. These findings identified PAR as a promoter of R-DPR toxicity and thus a potential target for treating c9ALS/FTD.
Introduction
A GGGGCC (G4C2) hexanucleotide repeat expansion in the C9ORF72 gene is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (1, 2). Two unique pathological hallmarks of C9ORF72-mediated ALS and frontotemporal lobar degeneration (FTLD), the neuropathological diagnosis of FTD, are foci of repeat-containing transcripts and aggregation of abnormal translation products of these transcripts, dipeptide repeat proteins (DPRs) (3–8). Among the five DPR species—namely, poly(glycine- arginine) (GR), poly(glycine-alanine) (GA), poly(glycine-proline) (GP), poly(proline-alanine) (PA), and poly(proline-arginine) (PR)— the arginine-rich DPRs (R-DPRs) poly(GR) and poly(PR) are particularly toxic when overexpressed in cell or animal models (9–16). However, what regulates R-DPR aggregation is unclear.
Stress granules (SGs) are cytoplasmic RNA/protein condensates assembled in cells upon diverse cellular stressors (17). Upon stress, ribosomes disassemble, and mRNAs are embedded into SGs enriched in RNA binding proteins, whose liquid-liquid phase separation (LLPS) mediates SG assembly (18–20). Normally, SGs are dynamic and can disassemble when stress is removed (17, 21); however, SGs with aberrant dynamics are related to the aggregation of SG proteins TDP-43, Fused in sarcoma (FUS), and other heterogeneous nuclear ribonucleoproteins (hnRNPs), which is a pathological hallmark of ALS and FTD, including c9ALS/FTD (22–25).
Previous studies identified a critical role of SGs in R-DPR–mediated cytotoxicity. R-DPRs interact with many SG proteins, and their overexpression causes the formation of aberrant, poorly dynamic SGs in cells without additional stress (11, 14, 26). Furthermore, chemically synthesized R-DPRs can undergo LLPS, recruit SG proteins, and cause SG protein precipitation when added into cell lysates (26). In agreement with these data, poly(GR) localizes to SGs in cells, promotes the aggregation of recombinant TDP-43 in vitro, and coaggregates with TDP-43 and the SG protein eIF3η in postmortem tissue from patients with c9ALS/FTD (27). We have previously found that inhibiting SG assembly by genetic or pharmacological approaches suppresses R-DPR–induced cytotoxicity or neurodegeneration in cellular or animal models (14). Together, these findings suggest that R-DPRs promote aberrant SG formation and protein aggregation, which contributes to neurodegeneration. However, how these processes are regulated is unclear.
Poly(ADP-ribose) (PAR) is a highly dynamic polymer that posttranslationally modifies proteins, a process known as PARylation. It is tightly regulated by PAR polymerases (PARPs) and PAR glycohydrolase (PARG), which synthesize and degrade PAR, respectively (28). As a posttranslational modification, PAR plays an essential role in cellular physiology, including SG assembly. Many SG proteins are PARylated or bind to PAR (29, 30). However, elevated PAR can be toxic. For example, PARP1 hyperactivation can induce a special type of programmed cell death called parthanatos, whereas loss of PARG, respectively, causes neurodegeneration or embryonic lethality in Drosophila or mice (31–33). In c9ALS/FTD, elevated nuclear PAR was reported in neurons from patients (34), but whether and how PAR relates to c9ALS/FTD pathogenesis is unclear.
Here, we show that loss of PARP1 (fly homolog: Parp) function suppresses R-DPR aggregation and neurodegeneration in c9ALS/FTD fly models and/or neurons differentiated from patient-derived induced pluripotent stem cells (iPSCs). Mechanistically, PAR interacts with R-DPRs, induces their condensation, and enhances their ability to induce aberrant SG formation and TDP-43 aggregation. Furthermore, PAR associates with detergent-insoluble poly(GR) and pathologic TDP-43 in frontal cortex tissue from patients with c9ALS/FTD. Together, our data support a role of PAR in promoting R-DPR toxicity and suggest that it might be a therapeutic target.
Results
Loss of Parp/PARP1 function suppresses neurodegeneration in c9ALS/FTD models
In a previously published screen, we identified 102 genes whose RNA interference (RNAi) strongly or moderately suppress eye degeneration in a Drosophila model of c9ALS/FTD, in which flies express 30 repeats of G4C2 [(G4C2)30] using the upstream activated sequence (UAS)/ Galectin 4 (GAL4) system (35) under the control of glass multiple reporter (GMR)–GAL4 (36). Comparing these results to RNAi screen results from independent studies using other fly models of c9ALS/FTD (11, 37), we found that parp, the fly homolog of human PARP1, is one of the only four genes identified from more than one screen (fig. S1A).
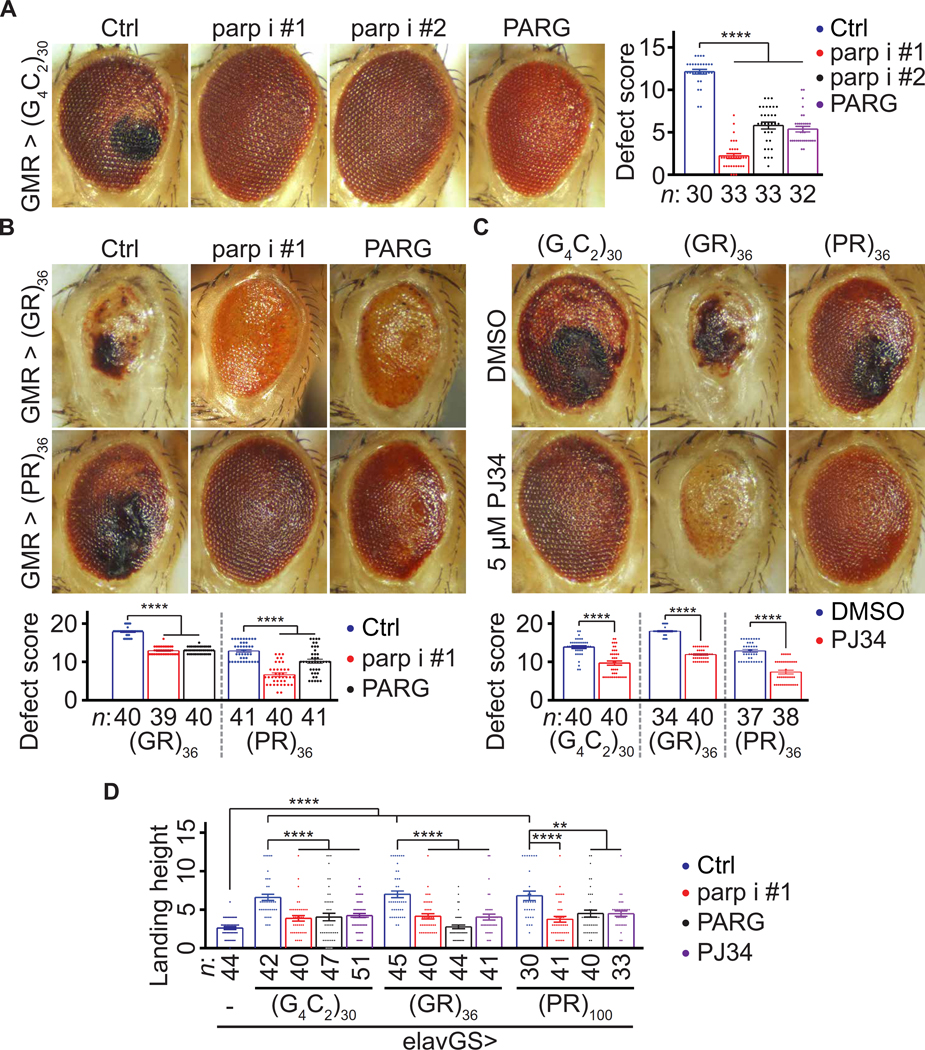
To verify that parp RNAi suppresses neurodegeneration in fly models of c9ALS/FTD, we tested two independent parp RNAi lines and showed that they both suppressed eye degeneration in the (G4C2)30 model (Fig. 1A). Furthermore, overexpressing PARG, which decreased PAR (31, 32), also suppressed eye degeneration in the (G4C2)30 flies (Fig. 1A). Next, we tested whether parp RNAi suppresses eye degeneration in other c9ALS/FTD fly models. Among all five DPR species, only the R-DPRs, poly(GR) and poly(PR), cause eye degeneration in flies (9, 11). Parp RNAi suppressed eye degeneration in flies expressing 36 repeats of GR or PR [(GR)36 or (PR)36] using alternative non-G4C2 repeat codons under the control of GMR-GAL4 (Fig. 1B) without decreasing R-DPR (fig. S1B). Furthermore, PARG overexpression (Fig. 1B) or feeding (G4C2)30, (GR)36, or (PR)36 flies with PJ34, a fly Parp and human PARP1/2 inhibitor (38), also suppressed eye degeneration (Fig. 1C). Together, our data suggest that decreasing PAR suppresses eye degeneration in multiple fly models of c9ALS/FTD. Next, we tested whether decreasing PAR suppresses locomotion defects in fly models of c9ALS/FTD. Previously, we showed that, using a gene-switch system (elavGS), pan-neuronal expression of (G4C2)30 impairs flight of aged flies (36). Here, we show that using alternative codons, pan-neuronal expression of (GR)36 and 100 repeats of PR [(PR)100], also caused flight defects in aged flies (Fig. 1D).
Fig. 1. Loss of Parp/PARP1 function suppresses neurodegeneration in fly models of c9ALS/FTD.
(A) Fly eyes expressing (G4C2)30, under the control of GMRGAL4, with or without parp RNAi (parp i) or PARG cDNA. Right: Fly eye defects were scored using a published method (79). Briefly, points were added if there was a complete loss of interommatidial bristles, necrotic patches, retinal collapse, loss of ommatidial structure, and/or depigmentation of the eye. (B) Fly eyes expressing (GR)36 or (PR)36 with or without parp RNAi or PARG cDNA. Quantification of the eye scores at the bottom. (C) Eyes of flies expressing (G4C2)30, (GR)36, or (PR)36 and fed with DMSO or PJ34. Quantified at the bottom. (D) Flight assays (see Materials and Methods). Oneway analysis of variance (ANOVA) with Dunnett’s tests (A, B, and D) and Student’s t tests (C). **P < 0.01 and ****P < 0.0001. Means ± SEM.
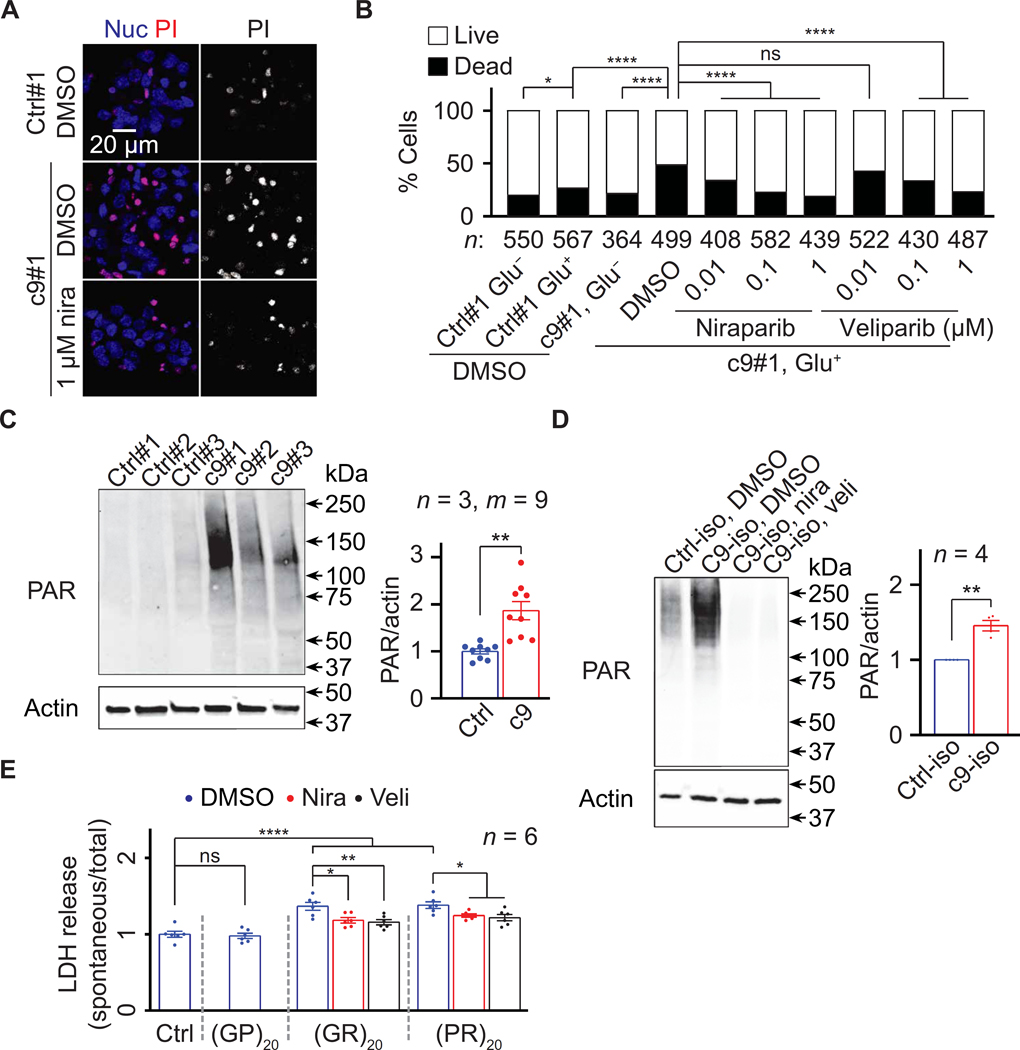
Furthermore, parp RNAi, PARG overexpression, or PJ34 feeding suppressed these defects, suggesting that down-regulating PAR mitigates locomotion defects in multiple fly models of c9ALS/FTD. To validate our findings above in a patient-derived model, we used four c9ALS/FTD iPSC-derived neuron (iPSN) lines along with three age- and sex-matched control lines and an isogenic control line (fig. S2A). We used a previously established protocol to generate iPSNs expressing several neuronal markers, including ISL1, NK6 Homeobox 1 (NKX6.1), SMI32, and Tubulin Beta 3 Class III (TUBB3), 32 days after the onset of differentiation (39–41). A common neuronal defect implicated in ALS/FTD, including c9ALS/FTD, is hyper- sensitivity to glutamate-induced excitotoxicity (2, 7, 42, 43). Using propidium iodide (PI) staining to label dead neurons, we previously showed that a 4-hour treatment of c9ALS/FTD iPSNs with 10 μM glutamate caused more cell death compared to control iPSNs (39).
Here, we verified these findings (Fig. 2, A and B, and fig. S2C) and further showed that a 5-day pretreatment with the PARP1/2 inhibitors niraparib or veliparib reduced PAR (fig. S2B) and glutamate-induced neuronal death (Fig. 2, A and B, and fig. S2C). Furthermore, PARP1 RNAi or PARG overexpression also suppressed glutamate-induced neuronal death in c9ALS/FTD iPSN line #1 (fig. S2D), suggesting that decreasing PAR could reduce neurotoxicity in c9ALS/FTD iPSNs. Consistent with these data, c9ALS/FTD iPSNs exhibited elevated PAR compared to the control iPSNs (Fig. 2, C and D).
Fig. 2. Loss of Parp/PARP1 function suppresses neurodegeneration in c9ALS/FTD iPSNs.
(A) Control line #1 (Ctrl#1) or c9ALS/FTD line #1 (c9#1) iPSNs pretreated with DMSO or niraparib (nira), treated with glutamate, and stained with propidium iodide (PI; red) and NucBlue (Nuc; blue). (B) Quantification of glutamate toxicity assays on Ctrl#1 and c9#1 iPSNs pretreated with DMSO, nira, or veliparib (veli). For each condition, three visual frames were imaged, and total numbers of cells were counted. The experiments were biologically replicated twice. (C and D) Immunoblots of lysates from three age and sexmatched pairs (C) and an isogenic pair (D) of Ctrl and c9 iPSNs. In (C), three replicates of three iPSN pairs (n = 3, total m = 9 data points) were used for statistical analyses. (E) LDH assays on Ctrl#1 iPSNs pretreated with DMSO or 5 μM nira or veli and treated with 10 μM DPRs. χ2 tests (B), Student’s t tests (C and D), and oneway ANOVA with Dunnett’s tests (E). ns, not significant. *P < 0.05, **P < 0.01, and ****P < 0.0001. Means ± SEM.
Previous studies showed that treating U-2 osteosarcoma (OS) cells with chemically synthesized peptides of 20 repeats of GR or PR [(GR)20 or (PR)20] caused cytotoxicity, as measured by the enzymatic MTT assay (10). To directly assess R-DPR toxicity in iPSNs, we treated a control iPSN line (Ctrl#1) with (GR)20 or (PR)20 and evaluated cytotoxicity using an enzymatic assay that measures the release of lactate dehydrogenase (LDH) from neurons due to damaged plasma membranes (44). As shown in Fig. 2E, a 2-day treatment with 5 μM (GR)20 or (PR)20, but not 20 repeats of GP, [(GP)20], a nontoxic DPR (9, 45), enhanced LDH release in these iPSNs. Furthermore, a 1-day pretreatment with 5 μM niraparib or veliparib reduced (GR)20- or (PR)20-induced PAR up regulation and LDH release without affecting intracellular poly(GR) or poly(PR) (Fig. 2E and fig. S2, E to G). In addition, adding (GR)20 reversed niraparib- and veliparib-mediated suppression of glutamate-induced neuronal death in c9ALS/FTD iPSN line #1 (fig. S2H). In summary, our data suggest that down-regulating PAR suppresses neuronal defects in both fly and iPSN models of c9ALS/FTD.
PAR binds to R-DPRs and induces their condensation in vitro
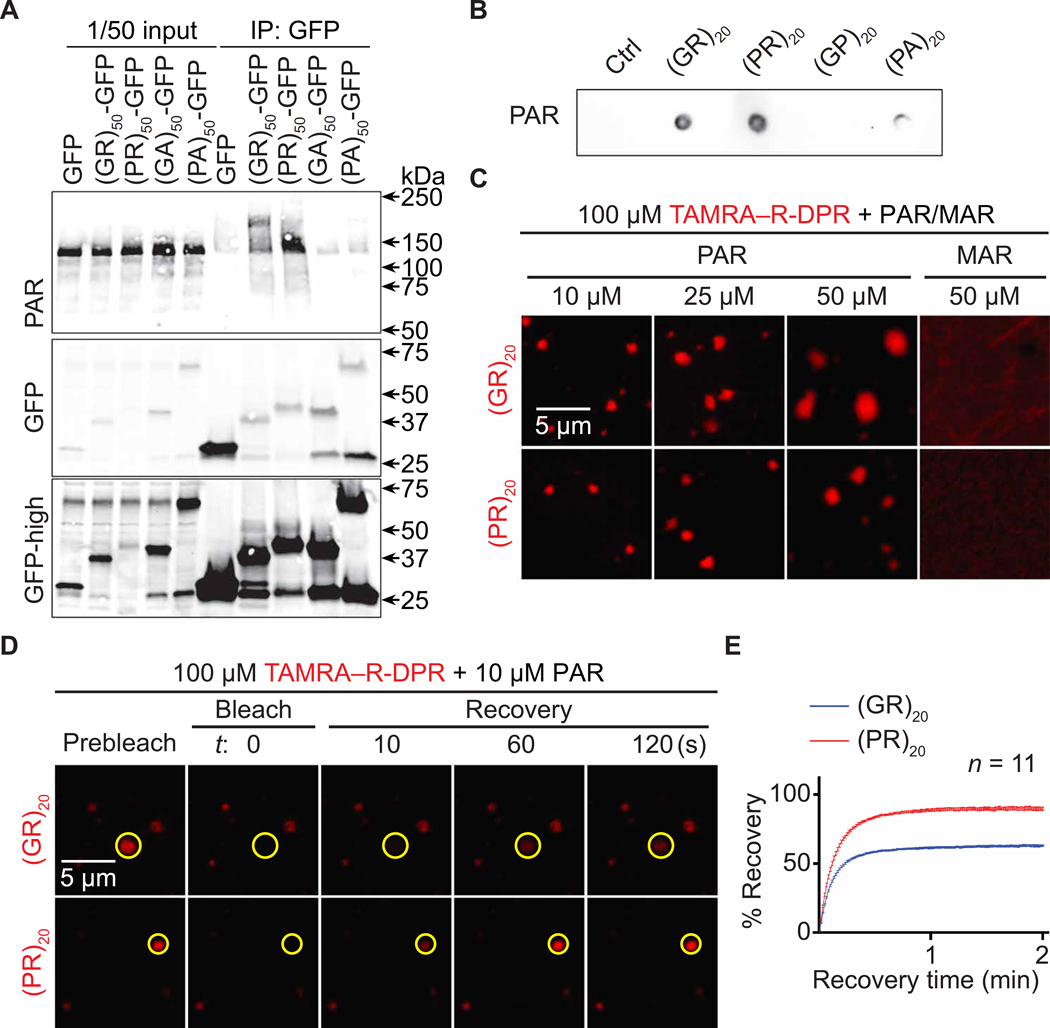
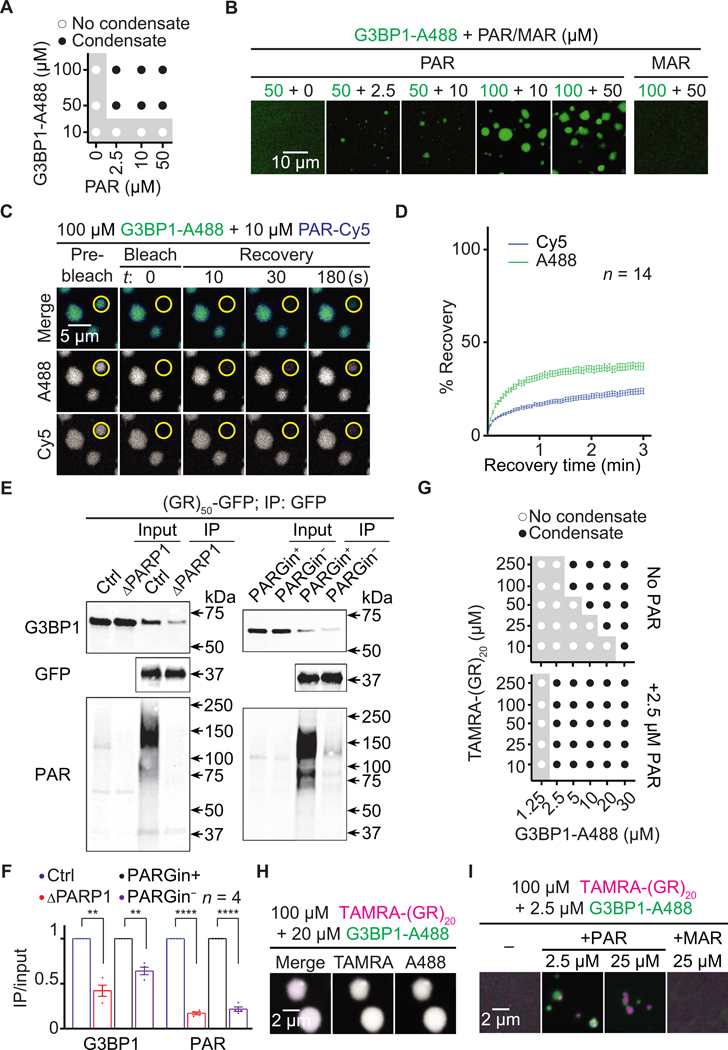
PAR was shown to bind to TDP-43, FUS, hnRNP A1, α-synuclein, and several other proteins to promote their condensation via either LLPS or aggregation and to contribute to TDP-43 and α-synuclein toxicity (30, 46–55). As R-DPR aggregation is a pathological hallmark of c9ALS/FTD (3–6, 8), we hypothesized that PAR could bind R-DPRs, thus promoting their condensation. PAR is negatively charged, and many PAR-binding protein domains are enriched in positively charged amino acids such as arginine (50, 56). To test whether PAR binds to R-DPRs, we first performed coimmunoprecipitation (co-IP) assays on human embryonic kidney (HEK) 293T cells transiently overexpressing green fluorescent protein (GFP)–tagged 50 repeats of DPRs [(DPR)50-GFP] using alternative codons. To inhibit ectopic PARP and PARG activation caused by cell lysis, which can skew co-IP results (56, 57), we added PJ34 and the PARG inhibitor PDD00017273 (PARGin) to our cell lysis and IP buffers. As shown in Fig. 3A, (GR)50- and (PR)50-GFP coimmunoprecipitated with PAR, suggesting that PAR can associate with R-DPRs in cells. Ribonuclease treatment did not reduce this association (fig. S3A), suggesting that the effect was not due to RNA-mediated mechanisms. Compared to the control and other DPRs, (GR)20 and (PR)20 showed stronger binding to chemically synthesized PAR in dot blot binding assays (Fig. 3B and fig. S3B), suggesting that PAR directly interacts with R-DPRs in vitro. RNA, which is negatively charged and structurally similar to PAR, can promote R-DPR LLPS in a phosphate buffer (26). Thus, we tested whether synthetic PAR could promote R-DPR LLPS in the same buffer. As shown by differential interference contrast (DIC) images in fig. S3 (C and D), PAR, but not mono(ADP-ribose) (MAR), caused (GR)20 or (PR)20 to phase separate into condensates without crowding agents. These condensates were about 1 to 2 μm in diameter. To better study these condensates, we labeled (GR)20 or (PR)20 with the 5-carboxytetramethylrhodamine (TAMRA) fluorophore. Similar to unlabeled R-DPRs, PAR, but not MAR, caused TAMRA–R-DPRs to phase separate into condensates without crowding agents (Fig. 3C).
Fig. 3. PAR induces R-DPR condensation in vitro.
(A) Co-IP of PAR and DPRs in HEK293T cells. (B) Dot blot binding assays of PAR and DPRs. (C) TAMRA–R-DPR (red) mixed with PAR (10 to 50 μM MAR equivalent) or MAR. (D and E) FRAP assays on PAR and TAMRA–R-DPR (red) condensates. Yellow circles indicate bleached and analyzed areas. Total R-DPRs (1 μM) were labeled. Buffer for (C to E): 61.5 mM K2HPO4 and 38.5 mM KH2PO4
Using fluorescent recovery after photobleaching (FRAP) assays, we photobleached individual TAMRA-R-DPR condensates and found that the fluorescence was partially recovered, suggesting that a fraction of TAMRA–R-DPR was mobile (Fig. 3, D and E). To study the relationship of R-DPRs and PAR in these condensates, we labeled PAR with the Cy5 fluorophore using a previously published method (58). Of interest, we observed Cy5 emission excited by lasers with a TAMRA excitation wavelength (fig. S3, E and F), suggesting Förster resonant energy transfer (FRET) from TAMRA-(GR)20 or TAMRA- (PR)20 to PAR-Cy5. We verified the FRET with the acceptor photo- bleaching method (59), showing that photobleaching the Cy5 signal restored TAMRA intensity (fig. S3, G and H). Because FRET typically occurs between molecules within a 10-nm distance range to allow bimolecular interactions (59), these data confirmed that PAR directly interacted with R-DPRs. Together, our data suggest that PAR binds to R-DPRs and promotes their condensation in vitro.
Loss of PARP1 activity suppresses R-DPR–induced SG formation
SGs play a critical role in R-DPR toxicity (11, 13, 14, 26). Overexpression of R-DPRs induces spontaneous formation of aberrant SGs in cells (11, 14, 26), whereas inhibiting SG assembly by genetic or pharmacological approaches suppresses R-DPR–mediated cellular defects (14). Since PARP1 inhibitors suppress or delay SG assembly induced by DNA damage or arsenite stress (29, 30), we hypothesized that loss of PARP1 activity could mitigate R-DPR– induced SG formation.
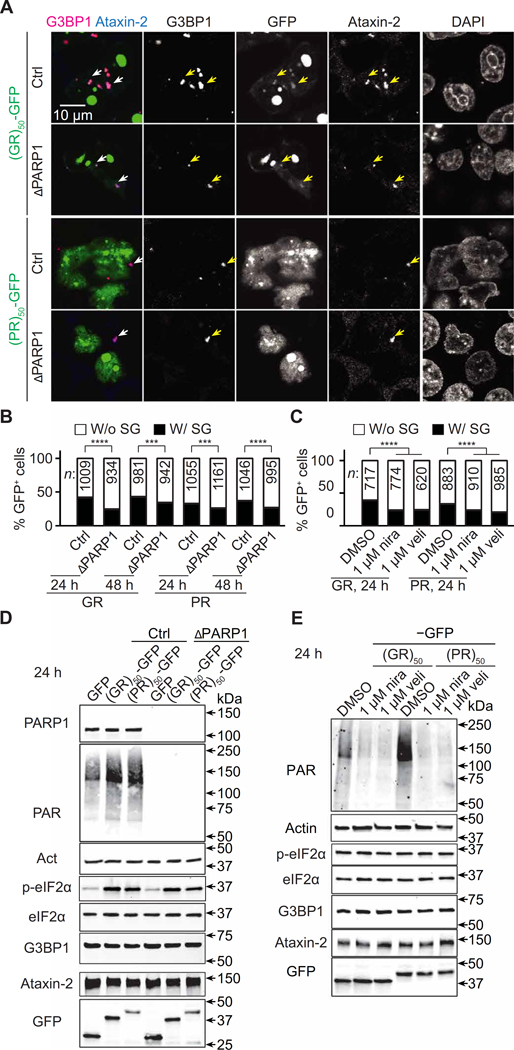
To study R-DPR–induced SG formation, we stained HEK293T cells overexpressing (GR)50- or (PR)50-GFP for 8, 24, or 48 hours with the SG markers Ras GTPase-activating protein-binding protein 1 (G3BP1) and Ataxin-2. We did not observe SGs after 8 hours of over- expression (~150 cells counted); how- ever, about 30 to 40% of GFP-expressing cells exhibited SGs after 24 or 48 hours (Fig. 4, A and B), suggesting that R-DPRs induced SG formation in a relatively slow manner compared to arsenite, which induces SG formation within an hour. Consistent with previous studies (11), (GR)50-GFP, but not (PR)50 GFP, localized to SGs (Fig. 4A and fig. S4A).
Fig. 4. Loss of PARP1 activity suppresses R-DPR–induced SG formation.
(A) Control (Ctrl) or PARP1 KO (ΔPARP1) HEK293T cells transfected with R-DPR–GFP (green) were immunofluorescently stained for G3BP1 (magenta), Ataxin-2 (blue), and DAPI 24 hours after transfection. Arrowheads indicate SGs. Note that (GR)50, but not (PR)50, localizes to SGs. (B and C) Quantification of R-DPR–GFP–expressing HEK293T cells that exhibit SGs. (B) Control (Ctrl) or PARP1 KO (ΔPARP1) cells. (C) Ctrl cells treated with DMSO, nira, or veli. DMSO or the PARP1 inhibitors were added to the cells 2 hours before transfection and remained in the culture media until fixation or cell lysis. For each condition, 20 to 30 visual fields from three biological replicates were randomly selected and imaged. The total numbers of GFP-positive cells with or without SGs were separately counted. χ2 tests. ***P < 0.001 and ****P < 0.0001. (D and E) Western blots on HEK293T cells overexpressing R-DPR–GFP for 24 hours. (D) Ctrl or ΔPARP1 cells. (E) Ctrl cells treated with DMSO, nira, or veli.
Next, we tested whether PARP1 knockout (KO) could mitigate R-DPR– induced SG formation. As shown in Fig. 4 (A and B) and fig. S4B, PARP1 KO decreased the percentage of cells with SGs after 24 or 48 hours of (GR)50- or (PR)50-GFP overexpression. We chose the 24-hour time point for our subsequent analyses. Similar to PARP1 KO, a 2-hour pretreatment of 1 μM niraparib or veliparib decreased the percent of cells with SGs after 24 hours of (GR)50- or (PR)50-GFP overexpression (Fig. 4C). PARP1 KO or PARP inhibitors did not decrease GFP, G3BP1, or Ataxin-2 amounts (Fig. 4, D and E, and fig. S4C). Together, our data suggest that loss of PARP1 activity reduces R-DPR–induced SG formation.
PAR promotes G3BP1 condensation and interaction with poly(GR)
Next, we studied the mechanism responsible for PARP1-mediated SG formation. Previous studies suggested an essential role of eukaryotic Initiation Factor 2 α (eIF2α) phosphorylation, as an eIF2α kinase inhibitor or phospho-dead mutation prevents SG formation induced by R-DPR overexpression (14, 26). However, PARP1 KO or inhibitors did not decrease phospho-eIF2α in cells expressing (GR)50- or (PR)50-GFP (Fig. 4, D and E, and fig. S4C), suggesting that PARP1 does not promote eIF2α phosphorylation.
SG assembly is mediated by the condensation of SG proteins, among which G3BP1 plays a pivotal role (18–20). Upon RNA binding, G3BP1 undergoes LLPS, which drives SG assembly. Double KO of G3BP1 and its homolog G3BP2 completely suppresses arsenite- or R-DPR–induced SGs, whereas overexpressing G3BP1 alone induces SGs without additional stress (11, 14, 26, 60). Given that PAR is structurally similar to RNA, we hypothesized that it could also promote G3BP1 LLPS, thereby driving SG formation. Consistent with this hypothesis, G3BP1 can be PARylated and bind to PAR (29, 49, 56, 61), and immunofluorescent staining has detected PAR in arsenite-induced SGs (30).
To test this hypothesis, we mixed synthetic PAR with Alexa-488– labeled recombinant G3BP1 (G3BP1-A488). As shown in Fig. 5 (A and B) and fig. S5 (A and B), PAR, but not MAR, caused G3BP1-A488 to phase separate without crowding agents. Using PAR-Cy5, together with FRAP assays, we showed that a fraction of both PAR and G3BP1 in the condensates is mobile (Fig. 5, C and D), suggesting that PAR promotes G3BP1 LLPS.
Fig. 5. PAR promotes G3BP1 condensation and interaction with poly(GR).
(A) Phase diagram and (B) images of Alexa 488–labeled recombinant G3BP1 (G3BP1-A488; green) mixed with PAR (2.5 to 50 μM MAR equivalents) or 50 μM MAR. (C and D) FRAP assays on G3BP1-A488 (green) and PARCy5 (blue) condensates. Yellow circles indicate bleached and analyzed areas. (E) Co-IP of (GR)50-GFP and endogenous G3BP1 in control (Ctrl) or PARP1 KO (ΔPARP1) HEK293T cells or in Ctrl HEK293T cells with or without the PARG inhibitor PDD00017273 (PARGin+ or PARGin−, respectively) in the lysis and IP buffer. Quantified in (F). Student’s t tests. **P < 0.01; ****P < 0.0001. Means ± SEM. (G) Phase diagram and (H and I) images of TAMRA-(GR)20 (magenta) mixed with G3BP1-A488 (green) and PAR (2.5 to 25 μM MAR equivalents) or 25 μM MAR. Total molecules (1 μM) were fluorescently labeled. Buffer for condensation assays: PBS.
Compared to chemical stressors, R-DPRs promote SG assembly via an additional mechanism—they directly interact with some SG proteins. Thus, their LLPS can trigger the condensation of these SG proteins (11, 26). Using co-IP followed by mass spectrometry analyses, a prior study identified 196 proteins interacting with (GR)50- or (PR)50-GFP in HEK293T cells, most of which are SG proteins (11). For some of these proteins, including TIA-1, hnRNP A1, and FUS, (GR)20 and/or (PR)20 have been shown to promote their condensation (11, 26). These mass spectrometry analyses also identified G3BP1 as an R-DPR interactor. Given the importance of G3BP1 in SG assembly, we hypothesized that R-DPRs might also promote its condensation. Furthermore, because PAR binds to R-DPRs, and G3BP1 is PARylated, we hypothesized that PAR could modulate R-DPR/ G3BP1 interactions and enhance the ability of R-DPRs to promote G3BP1 condensation.
First, we used Western blot to verify the interaction between R-DPRs and G3BP1. As shown in fig. S5C, (GR)50-GFP, but not (PR)50-GFP, strongly interacted with G3BP1, consistent with the localization of (GR)50, but not (PR)50, to SGs (11, 14) (Fig. 4A). As such, also given that poly(GR), but not other DPR, inclusions correlate with neurodegeneration and clinicopathological subtypes (16, 62) and that poly(GR), but not poly(PR), coaggregates with SG proteins in vivo (15, 27), we focused on poly(GR) for our subsequent studies. To test whether PAR modulated the interaction of G3BP1 with (GR)50-GFP, we performed co-IP assays either in PARP1 KO cells or with PARGin removed from buffers. As shown in Fig. 5 (E and F), both methods reduced PAR and inhibited this interaction, suggesting that PAR enhances G3BP1 interaction with poly(GR).
To test whether poly(GR) also promotes G3BP1 condensation, we mixed TAMRA-(GR)20 with G3BP1-A488 and found that they formed condensates without a crowding agent (Fig. 5, G and H, and fig. S5D). Moreover, PAR promoted this process by inducing G3BP1/ poly(GR) condensate formation at conditions in which the proteins do not form condensates without PAR (Fig. 5, G and I, and fig. S5, E and F). These data suggest that PAR promotes G3BP1 condensation and interaction with poly(GR).
PAR promotes poly(GR)-induced TDP-43 aggregation
SG assembly can trigger TDP-43 aggregation (22). We and others have shown that poly(GR) promotes the aggregation of recombinant TDP-43, colocalizes with TDP-43 in SGs in cultured cells, and coaggregates with TDP-43 in postmortem tissue from patients with c9ALS/FTD (11, 14, 27). Together, these findings suggest that poly(GR) contributes to c9ALS/FTD pathogenesis by promoting TDP-43 aggregation.
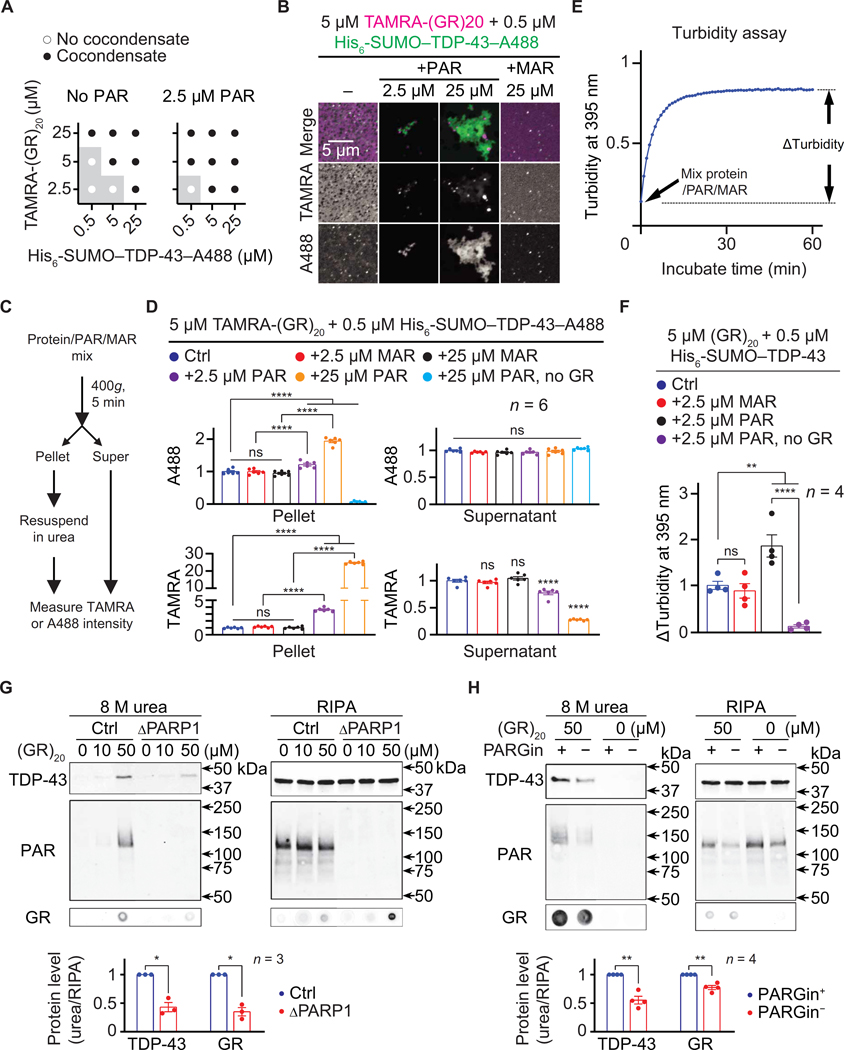
To investigate whether PAR modulates poly(GR)-induced TDP-43 aggregation, we mixed TAMRA-(GR)20 and A488-labeled recombinant TDP-43 (TDP-43–A488), with or without PAR. As shown in Fig. 6 (A and B), TAMRA-(GR)20 and TDP-43–A488 formed irregularly shaped, aggregate-like condensates. Furthermore, PAR, but not MAR, promoted this process by inducing condensate formation at conditions in which the proteins do not form condensates without PAR. To quantify the amount of proteins in the condensates, we pelleted the condensates by centrifugation, dissolved them in urea buffer, and measured A488 and TAMRA fluorescent intensities (Fig. 6C). As shown in Fig. 6D, PAR, but not MAR, strongly increased the A488 signal in the pellet compared to the control, suggesting that PAR can promote poly(GR)-induced TDP-43 condensation. Of importance, little A488 signal was detected in the pellet if TAMRA- (GR)20 was not included in the initial mixture (Fig. 6D), suggesting that PAR alone was unable to cause TDP-43 condensation in this condition.
Fig. 6. PAR promotes poly(GR)-induced TDP-43 aggregation.
(A) Phase diagram and (B) images of TAMRA-(GR)20 (magenta) mixed with His6-SUMO–TDP-43–A488 (green) and PAR (2.5 to 25 μM MAR equivalents) or 25 μM MAR. Total proteins were fluorescently labeled. Buffer: PBS. (C) The TDP-43 precipitation assay. (D) Quantification of A488 and TAMRA intensities. (E) The turbidity assay. (F) Quantification of increases in 395-nm absorbance. (G and H) Immunoblot of RIPA and urea fractions of control (Ctrl) or PARP1 KO (ΔPARP1) HEK293T cell lysate (G) or Ctrl HEK293T cell lysate with or without the PARG inhibitor PDD00017273 (PARGin+ or PARGin−, respectively; H), treated with (GR)20. Quantification reported below. One-way ANOVA with Tukey’s tests (D and F) and Student’s t tests (G and H). *P < 0.05, **P < 0.01, and ****P < 0.0001. Means ± SEM.
To further test whether PAR promotes poly(GR)-induced TDP-43 condensation, we used a turbidity assay, which measures the increase in solution turbidity due to condensate formation (63). We mixed non- fluorescently labeled (GR)20 and TDP-43 with or without PAR or MAR in solution and measured the increase in turbidity after a 60-min incubation (Fig. 6E). As shown in Fig. 6F, PAR, but not MAR, caused higher turbidity increase compared to controls. Also, little increase was observed if (GR)20 was not included in the mix (Fig. 6F), consistent with our data in Fig. 6D. Together, these data strongly suggest that PAR promotes poly(GR)-induced TDP-43 condensation in vitro.
Next, we tested whether PAR promotes poly(GR)-induced TDP-43 aggregation in cultured cells. To induce endogenous TDP-43 aggregation, we used a previously published method in which chemically synthesized poly(GR) peptides were added to radioimmunoprecipitation assay (RIPA) buffer lysates of cells (64). As shown in Fig. 6 (G and H) and fig. S6, 50 μM (GR)20, but not (GP)20, caused endogenous TDP-43 and PAR, as well as (GR)20 itself, to become RIPA insoluble but urea soluble. Furthermore, either PARP1 KO or removing PARG in from the cell lysis buffer reduced TDP-43 and (GR)20 abundance in the urea versus RIPA fractions (Fig. 6, G and H), suggesting that PAR promotes poly(GR) and TDP-43 aggregation in cell lysates. In summary, our data suggest that PAR promotes poly(GR)-induced TDP-43 condensation and aggregation in vitro and in cell lysates, respectively.
Loss of PARP1/Parp suppresses R-DPR aggregation in C9ALS/FTD flies
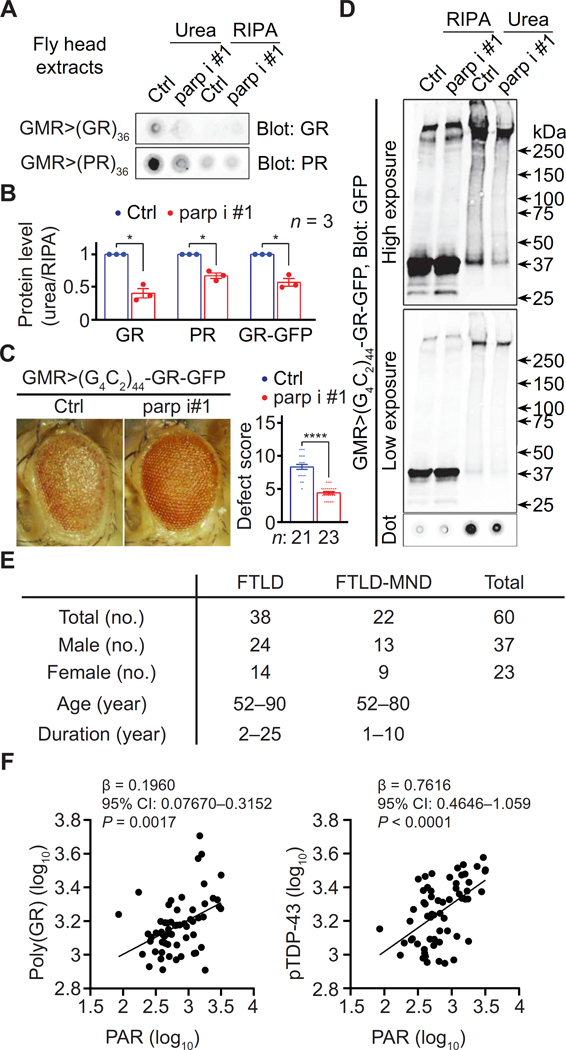
To study whether PAR contributed to R-DPR aggregation in vivo, we performed dot blot on RIPA and urea extracts from aged fly heads expressing (GR)36 or (PR)36, with or without parp RNAi, under the control of GMR-GAL4. As shown in Fig. 7 (A and B) and S1B, parp RNAi reduced R-DPR in the urea versus RIPA fractions but not total R-DPR. Another fly model of c9ALS/FTD expresses 44 G4C2 repeats with a C-terminal GFP tag in the poly(GR) reading frame (65), which allows easy detection of poly(GR). We found that parp RNAi also suppressed eye degeneration (Fig. 7C) and poly(GR)-GFP in the urea versus RIPA fractions in this fly model (Fig. 7D), but not total GFP (fig. S1B). Together, our data suggest that parp loss suppresses R-DPR aggregation in fly models of c9ALS/FTD.
Fig. 7. PAR and protein aggregation in vivo.
(A) Dot blots of RIPA and urea fractions of head extract from flies expressing R-DPRs without (Ctrl) or with coexpressing parp RNAi (parp i) #1. (B) Quantification of (A) and (D). (C) Fly eyes expressing 44 G4C2 repeats with a C-terminal GFP tag in the poly(GR) reading frame [(G4C2)44- GR-GFP] with or without coexpressing parp RNAi. (D) Immunoblot of RIPA and urea fractions of head extract from flies expressing (G4C2)44-GR-GFP with or without coexpressing parp i #1. Student’s t tests. *P < 0.05 and ****P < 0.0001. Means ± SEM.(E) c9ALS/FTD patient information. (F) Association of insoluble PAR with insoluble poly(GR) (left) or insoluble TDP-43 phosphorylated at S409/410 (pTDP-43) (right) in frontal cortex lysates from C9ORF72-repeat expansion carriers with frontotemporal lobar degeneration (FTLD) with or without motor neuron disease (MND). CI, confidence interval. Multiple linear regression.
PAR associates with poly(GR) and TDP-43 aggregation in patient postmortem frontal cortices
To better probe the human relevance of our findings above, we investigated the relationships among insoluble PAR, poly(GR), and TDP-43 in postmortem frontal cortex tissue from 60 C9ORF72 repeat expansion carriers neuropathologically diagnosed with FTLD or FTLD with motor neuron disease (MND) (Fig. 7E and table S1).
We measured PAR and poly(GR) in RIPA-insoluble, urea-soluble frontal cortex fractions by dot blot followed by densitometry analyses. In addition, we used an established immunoassay (66) to measure TDP-43 with phosphoserines at 409 or 410 (pTDP-43), a pathologic form of TDP-43 (67, 68). As shown in Fig. 7F, insoluble PAR was positively associated with both poly(GR) and pTDP-43 in analyses adjusting for age, sex, and disease subtype (FTLD versus FTLD- MND), further supporting a relationship of PAR with poly(GR) and TDP-43 aggregation.
Discussion
R-DPRs play a critical role in c9ALS/FTD pathogenesis. Both in vitro and in vivo evidence suggest that they exert their toxicity, at least in part, by causing aberrant SG formation and inducing the aggregation of SG proteins such as TDP-43 (11, 13, 26, 27). However, how these processes are regulated is unclear. Here, we found that loss of PARP1 activity suppresses R-DPR–induced SG formation. Furthermore, PAR promotes R-DPR condensation and poly(GR) coaggregation with TDP-43, suggesting that it might contribute to R-DPR toxicity. Remaining questions include whether loss of PARylation suppresses R-DPR–induced SG formation and whether this can be rescued by restoring PAR or PARylation.
PARP1 acts as a first responder that detects DNA damage (28). Poly(GR) can cause DNA damage by increasing mitochondrial re- active oxygen species, whereas poly(GA) and G4C2 repeat RNAs can respectively disrupt the DNA repair machinery and generate R-loops, DNA/RNA complexes that can cause double-strand breaks (69, 70). Thus, further studies can test whether DNA damage causes increased PAR in c9ALS/FTD.
Protein aggregation is a common feature in neurodegenerative diseases. Previous studies showed that PAR promotes a-synuclein aggregation and toxicity in a Parkinson’s disease model (48). Here, we show that PAR promotes R-DPR–induced TDP-43 aggregation and that PARP1 RNAi or PARP1 inhibitors suppress neurode- generation in fly and iPSN models of c9ALS/FTD. When combined, these findings suggest an important role of PAR in promoting protein aggregation and neurodegeneration. It ought also be mentioned that, in addition to promoting protein aggregation, excessive PAR causes parthanatos and inhibits axonal growth (32, 71). In contrast, loss of PARP1 activity increases nicotinamide adenine dinucleotide (NAD), which can be beneficial to neurons (72). Hence, inhibiting PARP1 may suppress c9ALS/FTD neurodegenerative via multiple mechanisms.
Previous studies suggested that PAR can play either deleterious or beneficial roles in TDP-43 toxicity, depending on the research systems used. Knockdown of PARPs, PARG overexpression, or PARP1/2 inhibition suppresses neurodegeneration caused by TDP-43 overexpression in Drosophila or cultured neurons (30, 34, 47), suggesting that PAR contributes to TDP-43 toxicity. However, PAR also prevents pathological phase separation and aggregation and pro- motes the physiological LLPS of TDP-43 in vitro (46, 47), suggesting a beneficial role. We show here that PAR by itself does not trigger TDP-43 aggregation. Also, although poly(GP) causes PAR up-regulation, it does not result in cytotoxicity or TDP-43 aggregation. Conversely, PAR promotes poly(GR)-induced TDP-43 aggregation and con- tributes to poly(GR) toxicity. Hence, how PAR affects TDP-43 toxicity likely depends on additional TDP-43 regulators. TDP-43 aggregates in patient tissue contain other proteins (13, 27), and future studies can focus on how these proteins—PAR, poly(GR), and TDP-43—affect their coaggregation.
SG assembly is mediated by a network of inter and intramolecular interactions among SG proteins (18). Because many SG proteins are PARylated or bind to PAR (56, 61), PAR may foster SG assembly by providing additional valences to the network, thereby promoting condensation. Previous studies have shown that PAR induces LLPS of SG proteins TDP-43, and hnRNP A1 and PARP1 inhibitors respectively delay or suppress arsenite or DNA damage–induced SG assembly (29, 30). Consistent with this notion, we show that PAR induces G3BP1 condensation in vitro, and loss of PARP1 activity reduces R-DPR–induced SG assembly, suggesting a general role of PAR in protein condensation and SG formation. A remaining question is how PARP1, a mainly nuclear protein, causes PARylation of SG proteins in the cytoplasm. One possibility is that these proteins are PARylated in the nucleus before translocating the SGs; alternatively, they can be PARylated by cytoplasmic PARP1 or PARPs activated by PARP1.
SGs can trigger the aggregation of TDP-43 and other SG proteins and thus are believed to contribute to ALS/FTD pathogenesis. Although c9ALS/FTD iPSNs do not constitutively form SGs, they are constitutively under a certain degree of stress, as indicated by a mild increase in phospho-eIF2a (14). Furthermore, loss of Ataxin-2 or SG inhibitors suppresses toxicity in yeast, animal, and iPSN models of c9- and TDP-43–ALS/FTD (14, 73–76). Here, we show that loss of PARP1 activity reduces SG formation and suppresses neurodegeneration in fly and cellular models of c9ALS/FTD, supporting a role of SG formation in pathogenesis. As several PARP1 inhibitors are Food and Drug Administration–approved or in advanced clinical trials for cancer treatment, and veliparib is neuro- protective in a mouse model of Parkinson’s disease without detrimental side effects (48), these findings suggest potential clinical translations. As SGs were also implicated in neurodegeneration observed in mouse models of other diseases, including tauopathies, a prion disease, and a vanishing white matter disease (74, 77, 78), PAR likely also contributes to their pathogenesis. Hence, targeting PAR may be a therapeutic approach for these diseases.
To further test the potential of PAR as a therapeutic target for ALS and FTD, it is important to validate our findings in mammalian models in vivo. Several mouse models of c9- and TDP-43–ALS/FTD are available (13, 27, 73, 79), and future studies can test whether PARP1 KO or PARP inhibitors ameliorate neurodegeneration and behavioral defects in these models. Moreover, although nuclear PAR is up-regulated in brain neurons of patients with ALS/FTD (34), it is unclear about the amount of cytoplasmic PAR, which is more relevant to SG assembly and the aggregation of DPRs and TDP-43. In addition, it is unclear whether PAR in the cerebrospinal fluid is up-regulated in patients with c9ALS/FTD. Last, potential deleterious effects of PARP1 inhibition needs to be considered given the essential roles of PAR in cellular physiology. For instance, PARP1 inhibitors can suppress DNA damage repair and kill cells (28). In addition, PAR is essential to nucleolar structure and function (80). Thus, it is important to identify safe doses of PARP inhibitors when treating neurons.
Materials and Methods
Study design
The goals of this study were to: (i) test the role of PARP1/PAR in fly and iPSN models of c9ALS/FTD, (ii) determine how PARP1/PAR affects R-DPR condensation in vitro and SG assembly in cultured cells, and (iii) investigate how PAR affects poly(GR) and TDP-43 aggregation in postmortem tissue of patients with c9ALS/FTD. As such, we performed fly eye degeneration and flight assays, toxicity assays in iPSNs, protein/PAR condensation assays, FRAP analyses, immunofluorescent staining, and Western blots. We also measured insoluble PAR, poly(GR), and pTDP-43 amounts in patient post- mortem tissue.
Sample size for postmortem studies was determined on the basis of availability, whereas that for other studies was determined on the basis of previously published papers and current accepted standards according to journal policies. No statistical analysis was used to pre- determine sample size. All experiments were done with a minimum number of replicates based on previous expertise in statistical analyses of similar experimental datasets, and the statistical analysis demonstrates that our sample sizes revealed differences between groups. All data were unbiasedly collected, and no data were excluded from the study. The findings in this study were collected from multiple independent experiments and were reliably reproduced. The n numbers of each samples are indicated in the figure legend. Investigators were blinded to genotypes for the iPSN and cell culture experiments but not the fly experiments. Human tissues were collected at Mayo Clinic with the approval of the Institutional Review Board. All patient information is Health Insurance Portability and Accountability Act compliant.
Statistical analysis
To quantify fluorescent or Western blot intensities, certain areas/ bands were circled, and the intensities were measured using ImageJ/ Fiji. Statistical analyses were performed using the GraphPad Prism software. Tests used and levels of significance for each experiment are explained in each figure legend.
Supplementary Material
Fig. S1. Parp RNAi suppress neurodegeneration in c9ALS/FTD fly models.
Fig. S2. PARP1 inhibitors suppress toxicity in iPSN models of c9ALS/FTD.
Fig. S3. PAR interacts with R-DPRs.
Fig. S4. PARP1 loss of function suppresses R-DPR-induced SG formation.
Fig. S5. PAR promotes G3BP1 condensation.
Fig. S6. Poly(GR) induces TDP-43 aggregation in cell lysates.
Fig. S7. Verification of the isogenic control and c9ALS/FTD iPSN pair.
Table S1. Patient Demographic Information
Table S2. Materials and Methods
Data file S1. Raw data
Acknowledgments:
We are grateful to all patients who agreed to donate postmortem tissue. We thank the following individuals or entities for reagents or services: J. Paul Taylor (G3BP1 bacterial expression construct), X. Wen and D. Trotti (DPR-GFP constructs), Bloomington Drosophila Stock Center (NIH P40OD018537, fly stocks), and R. Cole and the Johns Hopkins mass spectrometry core.
Funding:
This work was funded by the NIH-NINDS/NIA R01NS117461 (to K.Z., Y.-J.Z., and T.F.G.), DoD W81XWH-21-1-0082 (K.Z.), Target ALS (to K.Z., T.E.L., J.D.R., L.G., and T.F.G.), The Frick Foundation for ALS Research (to K.Z. and L.G.), Center for Biomedical Discovery at Mayo Clinic (to K.Z.), NIH R01NS085207 (to J.D.R.), NIH R01NS099320 (to J.D.R.), NIH P01NS099114 (to J.D.R. and T.F.G.), NIH R01NS094239 (to J.D.R.), NIH U24NS078736 (to J.D.R.), Muscular Dystrophy Association (to J.D.R.), ALS Association (to J.D.R. and T.E.L.), Answer ALS (to J.D.R.), The Robert Packard Center for ALS Research (to J.D.R. and T.E.L.), NIH R01NS082563 (to T.E.L.), NIH R01NS094239 (to T.E.L.), NIH P30NS050274 (to T.E.L.), ALS Association (to T.E.L.), RF1NS121143 (to L.G.), Ralph and Marian Falk Medical Research Trust (to L.G.), NIH R01GM104135 (to A.K.L.L.), NIH R37NS067525 (to V.L.D. and T.M.D.), NIH P01NS084974 (to Y.-J.Z. and T.F.G.), NIH R01NS121125 (to T.F.G.), and NIH K99NS123242 (to A.N.C.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
All data are available in the main text or the Supplementary Materials.
References and Notes:
- 1.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson M, Finch NCA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P,Hsiung G-YR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten J, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M; Consortium ITALSGEN, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy, Singleton J, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor J, A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mori K, Arzberger T, Grässer FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, van Broeckhoven C, Kremmer E, Kretzschmar HA, Haass C, Edbauer D, Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, van Broeckhoven C, Haass C, Edbauer D, The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook E, Subramony SH, Ostrow LW, Rothstein JD, Troncoso JC, Ranum LPW, RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A. 110, E4968–E4977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul III JW, Rademakers R, Boylan KB, Dickson DW, Petrucelli L, Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD, RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PEA, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, Cosio DM, van Blitterswijk M, Lee WC, Rademakers R, Boylan KB, Dickson DW, Petrucelli L, Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizielinska S, Grönke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, Cleverley K, Nicoll AJ, Pickering-Brown S, Dols J, Cabecinha M, Hendrich O, Fratta P, Fisher EMC, Partridge L, Isaacs AM, C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL, Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, Maxwell BA, Kim NC, Temirov J, Moore J, Kolaitis RM, Shaw TI, Bai B, Peng J, Kriwacki RW, Taylor JP, C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, McKnight SL, Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y-L, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, Zhang X, Prudencio M, Chew J, Cook CN, Daughrity LM, Tong J, Song Y, Pickles SR, Castanedes-Casey M, Kurti A, Rademakers R, Oskarsson B, Dickson DW, Hu W, Gitler AD, Fryer JD, Petrucelli L, Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 24, 1136–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, Taylor JP, Gitler AD, Rothstein JD, Lloyd TE, Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang YJ, Guo L, Gonzales PK, Gendron TF, Wu Y, Jansen-West K, O’Raw AD, Pickles SR, Prudencio M, Carlomagno Y, Gachechiladze MA, Ludwig C, Tian R, Chew J, DeTure M, Lin WL, Tong J, Daughrity LM, Yue M, Song Y, Andersen JW, Castanedes-Casey M, Kurti A, Datta A, Antognetti G, McCampbell A, Rademakers R, Oskarsson B, Dickson DW, Kampmann M, Ward ME, Fryer JD, Link CD, Shorter J, Petrucelli L, Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 363, eaav2606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakae N, Bieniek KF, Zhang YJ, Ross K, Gendron TF, Murray ME, Rademakers R, Petrucelli L, Dickson DW, Poly-GR dipeptide repeat polymers correlate with neurodegeneration and clinicopathological subtypes in C9ORF72-related brain disease. Acta Neuropathol. Commun. 6, 63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Protter DSW, Parker R, Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang P, Mathieu C, Kolaitis R-M, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, Yu J, Martin EW, Mittag T, Kim HJ, Taylor JP, G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, Wei M-T, Whitney G, Lyons SM, Anderson P, Jacobs WM, Ivanov P, Brangwynne CP, Competing protein-RNA interaction networks control multiphase intracellular organization. Cell 181, 306–324.e28 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlüßler R, Kim K, Trussina IREA, Wang J, Mateju D, Poser I, Maharana S, Ruer-Gruß M, Richter D, Zhang X, Chang Y-T, Guck J, Honigmann A, Mahamid J, Hyman AA, Pappu RV, Alberti S, Franzmann TM, RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y, Protter DSW, Rosen MK, Parker R, Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li YR, King OD, Shorter J, Gitler AD, Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandes N, Nero L, Lyons S, Ivanov P, Mittelmeier T, Bolger T, Buchan J, Stress granule assembly can facilitate but is not required for TDP-43 cytoplasmic aggregation. Biomolecules 10, 1367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp M-D, Simons M, Niessing D, Madl T, Dormann D, Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi A-S, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP, Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, de Decker M, Jaspers T, Ryan VH, Janke AM, Baatsen P, Vercruysse T, Kolaitis R-M, Daelemans D, Taylor JP, Kedersha N, Anderson P, Impens F, Sobott F, Schymkowitz J, Rousseau F, Fawzi NL, Robberecht W, van Damme P, Tompa P, van den Bosch L, Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook CN, Wu Y, Odeh HM, Gendron TF, Jansen-West K, del Rosso G, Yue M, Jiang P, Gomes E, Tong J, Daughrity LM, Avendano NM, Castanedes-Casey M, Shao W, Oskarsson B, Tomassy GS, McCampbell A, Rigo F, Dickson DW, Shorter J, Zhang Y-J, Petrucelli L, C9orf72 poly(GR) aggregation induces TDP-43 proteinopathy. Sci. Transl. Med. 12, eabb3774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray Chaudhuri A, Nussenzweig A, The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610–621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isabelle M, Gagné JP, Gallouzi IE, Poirier GG, Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)- dependent following exposure to MNNG-induced DNA alkylation. J. Cell Sci. 125(Pt 19), 4555–4566 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Duan Y, du A, Gu J, Duan G, Wang C, Gui X, Ma Z, Qian B, Deng X, Zhang K, Sun L, Tian K, Zhang Y, Jiang H, Liu C, Fang Y, PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 29, 233–247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, Miwa M, Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 101, 82–86 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM, Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. U.S.A. 103, 18308–18313 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stöger T, Poirier GG, Dawson VL, Dawson TM, Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 101, 17699–17704 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGurk L, Mojsilovic-Petrovic J, van Deerlin VM, Shorter J, Kalb RG, Lee VM, Trojanowski JQ, Lee EB, Bonini NM, Nuclear poly(ADP-ribose) activity is a therapeutic target in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 6, 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brand AH, Perrimon N, Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). [DOI] [PubMed] [Google Scholar]
- 36.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu SL, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, Rothstein JD, The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman LD, Prudencio M, Kramer NJ, Martinez-Ramirez LF, Srinivasan AR, Lan M, Parisi MJ, Zhu Y, Chew J, Cook CN, Berson A, Gitler AD, Petrucelli L, Bonini NM, Toxic expanded GGGGCC repeat transcription is mediated by the PAF1 complex in C9orf72-associated FTD. Nat. Neurosci. 22, 863–874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murawska M, Hassler M, Renkawitz-Pohl R, Ladurner A, Brehm A, Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLOS Genet. 7, e1002206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyne AN, Zaepfel BL, Hayes L, Fitchman B, Salzberg Y, Luo E-C, Bowen K, Trost H, Aigner S, Rigo F, Yeo GW, Harel A, Svendsen CN, Sareen D, Rothstein JD, G4C2 repeat RNA initiates a POM121-mediated reduction in specific nucleoporins in C9orf72 ALS/FTD. Neuron 107, 1124–1140.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baxi EG, Thompson T, Li J, Kaye JA, Lim RG, Wu J, Ramamoorthy D, Lima L, Vaibhav V, Matlock A, Frank A, Coyne AN, Landin B, Ornelas L, Mosmiller E, Thrower S, Farr SM, Panther L, Gomez E, Galvez E, Perez D, Meepe I, Lei S, Mandefro B, Trost H, Pinedo L, Banuelos MG, Liu C, Moran R, Garcia V, Workman M, Ho R, Wyman S, Roggenbuck J, Harms MB, Stocksdale J, Miramontes R, Wang K, Venkatraman V, Holewenski R, Sundararaman N, Pandey R, Manalo D-M, Donde A, Huynh N, Adam M, Wassie BT, Vertudes E, Amirani N, Raja K, Thomas R, Hayes L, Lenail A, Cerezo A, Luppino S, Farrar A, Pothier L, Prina C, Morgan T, Jamil A, Heintzman S, Jockel-Balsarotti J, Karanja E, Markway J, McCallum M, Joslin B, Alibazoglu D, Kolb S, Ajroud-Driss S, Baloh R, Heitzman D, Miller T, Glass JD, Patel-Murray NL, Yu H, Sinani E, Vigneswaran P, Sherman AV, Ahmad O, Roy P, Beavers JC, Zeiler S, Krakauer JW, Agurto C, Cecchi G, Bellard M, Raghav Y, Sachs K, Ehrenberger T, Bruce E, Cudkowicz ME, Maragakis N, Norel R, van Eyk JE, Finkbeiner S, Berry J, Sareen D, Thompson LM, Fraenkel E, Svendsen CN, Rothstein JD, Answer ALS, a large-scale resource for sporadic and familial ALS combining clinical and multi-omics data from induced pluripotent cell lines. Nat. Neurosci. 25, 226–237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coyne AN, Baskerville V, Zaepfel BL, Dickson DW, Rigo F, Bennett F, Lusk CP, Rothstein JD, Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci. Transl. Med. 13, eabe1923 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y, Lin S, Staats KA, Li Y, Chang W-H, Hung S-T, Hendricks E, Linares GR, Wang Y, Son EY, Wen X, Kisler K, Wilkinson B, Menendez L, Sugawara T, Woolwine P, Huang M, Cowan MJ, Ge B, Koutsodendris N, Sandor KP, Komberg J, Vangoor VR, Senthilkumar K, Hennes V, Seah C, Nelson AR, Cheng T-Y, Lee S-JJ, August PR, Chen JA, Wisniewski N, Hanson-Smith V, Belgard TG, Zhang A, Coba M, Grunseich C, Ward ME, van den Berg LH, Pasterkamp RJ, Trotti D, Zlokovic BV, Ichida JK, Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat. Med. 24, 313–325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothstein JD, Martin LJ, Kuncl RW, Decreased glutamate transport by the brain and spinal cord in amyotrophic lateral sclerosis. N. Engl. J. Med. 326, 1464–1468 (1992). [DOI] [PubMed] [Google Scholar]
- 44.Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu Y-F, Katzman RB, Gass J, Murray ME, Shinohara M, Lin W-L, Garrett A, Stankowski JN, Daughrity L, Tong J, Perkerson EA, Yue M, Chew J, Castanedes-Casey M, Kurti A, Wang ZS, Liesinger AM, Baker JD, Jiang J, Lagier-Tourenne C, Edbauer D, Cleveland DW, Rademakers R, Boylan KB, Bu G, Link CD, Dickey CA, Rothstein JD, Dickson DW, Fryer JD, Petrucelli L, C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci. 19, 668–677 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, Pasinelli P, Ichida JK, Trotti D, Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGurk L, Gomes E, Guo L, Shorter J, Bonini NM, Poly(ADP-ribose) engages the TDP-43 nuclear-localization sequence to regulate granulo-filamentous aggregation. Biochemistry 57, 6923–6926 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGurk L, Gomes E, Guo L, Mojsilovic-Petrovic J, Tran V, Kalb RG, Shorter J, Bonini NM, Poly(ADP-Ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 71, 703–717.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kam TI, Mao X, Park H, Chou SC, Karuppagounder SS, Umanah GE, Yun SP, Brahmachari S, Panicker N, Chen R, Andrabi SA, Qi C, Poirier GG, Pletnikova O, Troncoso JC, Bekris LM, Leverenz JB, Pantelyat A, Ko HS, Rosenthal LS, Dawson TM, Dawson VL, Poly(ADP-ribose) drives pathologic a-synuclein neurodegeneration in Parkinson’s disease. Science 362, eaat8407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vivelo CA, Wat R, Agrawal C, Tee HY, Leung AKL, ADPriboDB: The database of ADP-ribosylated proteins. Nucleic Acids Res. 45, D204–D209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teloni F, Altmeyer M, Readers of poly(ADP-ribose): Designed to be fit for purpose. Nucleic Acids Res. 44, 993–1006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask M-BD, Streicher W, Jungmichel S, Nielsen ML, Lukas J, Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S, A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Kawashima K, Izawa M, Poly(ADP-ribose) synthesis in nucleoli and ADP-ribosylation of nucleolar proteins in mouse ascites tumor cells in vitro. J. Biochem. 89, 1889–1901 (1981). [DOI] [PubMed] [Google Scholar]
- 54.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P, Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499 (2011).\ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guetg C, Santoro R, Noncoding RNAs link PARP1 to heterochromatin. Cell Cycle 11, 2217–2218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dasovich M, Beckett MQ, Bailey S, Ong S-E, Greenberg MM, Leung AKL, Identifying poly(ADP-ribose)-binding proteins with photoaffinity-based proteomics. J. Am. Chem. Soc. 143, 3037–3042 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D-S, Camacho CV, Nagari A, Malladi VS, Challa S, Kraus WL, Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol. Cell 75, 1270–1285.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ando Y, Elkayam E, McPherson RL, Dasovich M, Cheng S-J, Voorneveld J, Filippov DV, Ong S-E, Joshua-Tor L, Leung AKL, ELTA: Enzymatic labeling of terminal ADP-ribose. Mol. Cell 73, 845–856.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Munster EB, Kremers GJ, Adjobo-Hermans MJW, Gadella TWJ Jr., Fluorescence resonance energy transfer (FRET) measurement by gradual acceptor photobleaching. J. Microsc. 218(Pt 3), 253–262 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney G, Ivanov P, Anderson P, G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ayyappan V, Wat R, Barber C, Vivelo CA, Gauch K, Visanpattanasin P, Cook G, Sazeides C, Leung AKL, ADPriboDB 2.0: An updated database of ADP-ribosylated proteins. Nucleic Acids Res. 49, D261–D265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saberi S, Stauffer JE, Jiang J, Garcia SD, Taylor AE, Schulte D, Ohkubo T, Schloffman CL, Maldonado M, Baughn M, Rodriguez MJ, Pizzo D, Cleveland D, Ravits J, Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. 135, 459–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O’Donovan K, Fare CM, Diaz Z, Singh N, Zhang ZC, Coughlin M, Sweeny EA, De Santis ME, Jackrel ME, Rodell CB, Burdick JA, King OD, Gitler AD, Lagier-Tourenne C, Pandey UB, Chook YM, Taylor JP, Shorter J, Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hutten S, Usluer S, Bourgeois B, Simonetti F, Odeh HM, Fare CM, Czuppa M, Hruska-Plochan M, Hofweber M, Polymenidou M, Shorter J, Edbauer D, Madl T, Dormann D, Nuclear import receptors directly bind to arginine-rich dipeptide repeat proteins and suppress their pathological interactions. Cell Rep. 33, 108538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goodman LD, Prudencio M, Srinivasan AR, Rifai OM, Lee VMY, Petrucelli L, Bonini NM, eIF4B and eIF4H mediate GR production from expanded G4C2 in a Drosophila model for C9orf72-associated ALS. Acta Neuropathol. Commun. 7, 62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prudencio M, Gonzales PK, Cook CN, Gendron TF, Daughrity LM, Song Y, Ebbert MTW, van Blitterswijk M, Zhang Y-J, Jansen-West K, Baker MC, De Ture M, Rademakers R, Boylan KB, Dickson DW, Petrucelli L, Link CD, Repetitive element transcripts are elevated in the brain of C9orf72 ALS/FTLD patients. Hum. Mol. Genet. 26, 3421–3431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hasegawa M, Arai T, Nonaka T, Kametani F, Yoshida M, Hashizume Y, Beach TG, Buratti E, Baralle F, Morita M, Nakano I, Oda T, Tsuchiya K, Akiyama H, Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 64, 60–70 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neumann M, Kwong LK, Lee EB, Kremmer E, Flatley A, Xu Y, Forman MS, Troost D, Kretzschmar HA, Trojanowski JQ, Lee VM-Y, Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 117, 137–149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walker C, Herranz-Martin S, Karyka E, Liao C, Lewis K, Elsayed W, Lukashchuk V, Chiang S-C, Ray S, Mulcahy PJ, Jurga M, Tsagakis I, Iannitti T, Chandran J, Coldicott I, de Vos KJ, Hassan MK, Higginbottom A, Shaw PJ, Hautbergue GM, Azzouz M, el-Khamisy SF, C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat. Neurosci. 20, 1225–1235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, Petrucelli L, Miller BL, Almeida S, Gao F-B, Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron 92, 383–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brochier C, Jones JI, Willis DE, Langley B, Poly(ADP-ribose) polymerase 1 is a novel target to promote axonal regeneration. Proc. Natl. Acad. Sci. U.S.A. 112, 15220–15225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorrentino V, Romani M, Mouchiroud L, Beck JS, Zhang H, D’Amico D, Moullan N, Potenza F, Schmid AW, Rietsch S, Counts SE, Auwerx J, Enhancing mitochondrial proteostasis reduces amyloid- proteotoxicity. Nature 552, 187–193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano G. Auburger, Pulst SM, Taylor JP Rigo F, Gitler AD, Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Apicco DJ, Ash PEA, Maziuk B, Blang CL, Medalla M, Abdullatif AA, Ferragud A, Botelho E, Ballance HI, Dhawan U, Boudeau S, Cruz AL, Kashy D, Wong A, Goldberg LR, Yazdani N, Zhang C, Ung CY, Tripodis Y, Kanaan NM, Ikezu T, Cottone P, Leszyk J, Li H, Luebke J, Bryant CD, Wolozin B, Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat. Neurosci. 21, 72–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elden AC, Kim H-J, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rüb U, Auburger G, Trojanowski JQ, Lee VM-Y, van Deerlin VM, Bonini NM, Gitler AD, Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, Lee VM-Y, Finkbeiner S, Gitler AD, Bonini NM, Therapeutic modulation of eIF2a phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat. Genet. 46, 152–160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Halliday M, Radford H, Sekine Y, Moreno J, Verity N, le Quesne J, Ortori CA, Barrett DA, Fromont C, Fischer PM, Harding HP, Ron D, Mallucci GR, Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis. 6, e1672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wong YL, Bon LL, Basso AM, Kohlhaas KL, Nikkel AL, Robb HM, Donnelly-Roberts DL, Prakash J, Swensen AM, Rubinstein ND, Krishnan S, McAllister FE, Haste NV, O’Brien JJ, Roy M, Ireland A, Frost JM, Shi L, Riedmaier S, Martin K, Dart MJ, Sidrauski C, eIF2B activator prevents neurological defects caused by a chronic integrated stress response. eLife 8, e42940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang Y-J, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, Bieniek KF, Bauer PO, Whitelaw EC, Rousseau L, Stankowski JN, Stetler C, Daughrity LM, Perkerson EA, Desaro P, Johnston A, Overstreet K, Edbauer D, Rademakers R, Boylan KB, Dickson DW, Fryer JD, Petrucelli L, C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348, 1151–1154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boamah EK, Kotova E, Garabedian M, Jarnik M, Tulin AV, Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLOS Genet. 8, e1002442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, Lee VMY, Forman MS, Taylor JP, TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 30, 7729–7739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ababneh NA, Scaber J, Flynn R, Douglas A, Barbagallo P, Candalija A, Turner MR, Sims D, Dafinca R, Cowley SA, Talbot K, Correction of amyotrophic lateral sclerosis related phenotypes in induced pluripotent stem cell-derived motor neurons carrying a hexanucleotide expansion mutation in C9orf72 by CRISPR/Cas9 genome editing using homology-directed repair. Hum. Mol. Genet. 29, 2200–2217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ame JC, Hakmé A, Quenet D, Fouquerel E, Dantzer F, Schreiber V, Detection of the nuclear poly(ADP-ribose)-metabolizing enzymes and activities in response to DNA damage. Methods Mol. Biol. 464, 267–283 (2009). [DOI] [PubMed] [Google Scholar]
- 83.Bischof J, Maeda RK, Hediger M, Karch F, Basler K, An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gendron TF, van Blitterswijk M, Bieniek KF, Daughrity LM, Jiang J, Rush BK, Pedraza O, Lucas JA, Murray ME, Desaro P, Robertson A, Overstreet K, Thomas CS, Crook JE, Castanedes-Casey M, Rousseau L, Josephs KA, Parisi JE, Knopman DS, Petersen RC, Boeve BF, Graff-Radford NR, Rademakers R, Lagier-Tourenne C, Edbauer D, Cleveland DW, Dickson DW, Petrucelli L, Boylan KB, Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol. 130, 559–573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Parp RNAi suppress neurodegeneration in c9ALS/FTD fly models.
Fig. S2. PARP1 inhibitors suppress toxicity in iPSN models of c9ALS/FTD.
Fig. S3. PAR interacts with R-DPRs.
Fig. S4. PARP1 loss of function suppresses R-DPR-induced SG formation.
Fig. S5. PAR promotes G3BP1 condensation.
Fig. S6. Poly(GR) induces TDP-43 aggregation in cell lysates.
Fig. S7. Verification of the isogenic control and c9ALS/FTD iPSN pair.
Table S1. Patient Demographic Information
Table S2. Materials and Methods
Data file S1. Raw data
Data Availability Statement
All data are available in the main text or the Supplementary Materials.