Abstract
The Cel5 cellulase (formerly known as endoglucanase Z) from Erwinia chrysanthemi is a multidomain enzyme consisting of a catalytic domain, a linker region, and a cellulose binding domain (CBD). A three-dimensional structure of the CBDCel5 has previously been obtained by nuclear magnetic resonance. In order to define the role of individual residues in cellulose binding, site-directed mutagenesis was performed. The role of three aromatic residues (Trp18, Trp43, and Tyr44) in cellulose binding was demonstrated. The exposed potential hydrogen bond donors, residues Gln22 and Glu27, appeared not to play a role in cellulose binding, whereas residue Asp17 was found to be important for the stability of Cel5. A deletion mutant lacking the residues Asp17 to Pro23 bound only weakly to cellulose. The sequence of CBDCel5 exhibits homology to a series of five repeating domains of a putative large protein, referred to as Yheb, from Escherichia coli. One of the repeating domains (Yheb1), consisting of 67 amino acids, was cloned from the E. coli chromosome and purified by metal chelating chromatography. While CBDCel5 bound to both cellulose and chitin, Yheb1 bound well to chitin, but only very poorly to cellulose. The Yheb protein contains a region that exhibits sequence homology with the catalytic domain of a chitinase, which is consistent with the hypothesis that the Yheb protein is a chitinase.
Cellulose and chitin are composed of 1,4-β-linked pyranose units (in chitin, the C-2 hydroxyl groups have been replaced by N-acetyl residues) whose glycosidic bonds are hydrolyzed by cellulases and chitinases, respectively. Generally, these enzymes exhibit multidomain structures. The gram-negative bacterium Erwinia chrysanthemi is a plant pathogen that produces an endoglucanase, referred to as Cel5 (formerly known as endoglucanase Z). Cel5 consists of an N-proximal 287-amino-acid catalytic domain, a 35-amino-acid linker region, and a 62-amino-acid cellulose-binding domain (CBD). Catalytic domains and CBDs have been grouped into families on the basis of sequence homology (13, 16, 33). The catalytic domain of E. chrysanthemi Cel5 is in glycoside hydrolase family 5, and CBDCel5 is in CBD family V.
The three-dimensional structure of CBDCel5 has been solved by nuclear magnetic resonance (NMR) spectroscopy (8). Other CBD structures determined by NMR spectroscopy are those of (i) family I CBDCBH1 from Trichoderma reesei cellobiohydrolase I (20, 24), (ii) family I CBDEG1 from T. reesei endoglucanase I (25), (iii) family II CBDCex from Cellulomonas fimi xylanase-glucanase (38), and (iv) family IV CBDN1 from C. fimi β-1,4-glucanase (18). The structures determined by X-ray crystallography are (i) family IIIa CBDCip from the scaffoldin subunit of Clostridium thermocellum (35) and (ii) family IIIc CBDE4 from Thermomonospora fusca endo/exocellulase E4 (31).
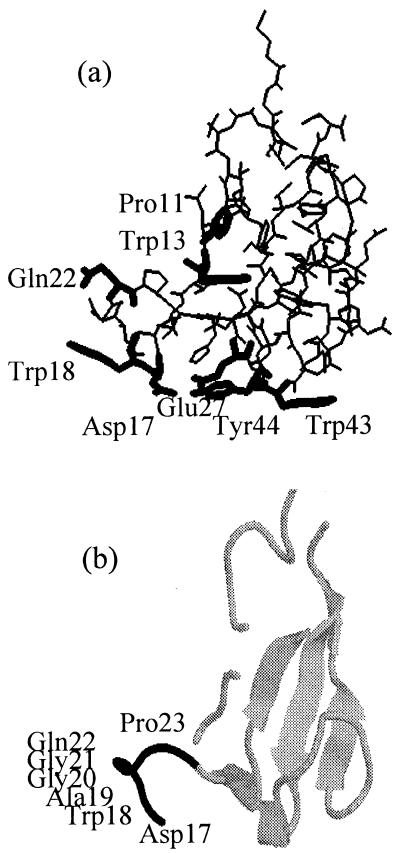
CBDCel5 exhibits a boot-shaped structure based mainly on a triple antiparallel β-sheet perpendicular to a less-ordered summital loop (see Fig. 1). There are five amino acids (Asp17, Trp18, Glu27, Trp43, and Tyr44) that potentially interact with the glucose chain of cellulose and contribute directly to the binding. In the present work, we have constructed a series of CBDCel5 mutants in which these amino acids were independently replaced by a functionally neutral alanine residue.
FIG. 1.
(a) Three-dimensional structure of CBDCel5. The residues that were mutated have been highlighted. (b) Backbone structure of CBDCel5. The highlighted residues represent the region that was deleted in the mutant Cel5ΔD17-P23.
Additionally, we investigated the structural role of residues present in the turn of the boot-shaped model (cisPro11, Trp13). Also, the less-ordered loop region was investigated (Gln22Ala, Cel5ΔAsp17-Pro23).
The region covering residues 29 to 61 corresponds to the β-sheet core of CBDCel5 and exhibits homology with sequences Yheb1 to -5 of an Escherichia coli hypothetical protein (8). Yheb1 to -5 are repeating domains of a 94-kDa protein in the Bfr-TufA intergenic region (1). In this paper, we describe the cellulose- and chitin-binding properties of the E. coli Yheb1 protein.
MATERIALS AND METHODS
Media.
The media used were Luria-Bertani (LB) and M9CAS (7). A cellulase phenotype was tested by using minimal medium plates supplemented with 0.2% glycerol, 0.2 g of yeast extract per liter, and 10 g of carboxymethyl cellulose per liter.
Strains.
The following E. coli strains were used in this study: (i) DH5α: F′ endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA (NaIr) relA1 Δ(lacIZYA-argF) U169 deoR [φ80 dlac Δ(lacZ) M15]; (ii) BL21(DE3): F− ompT hsdSB(rB− mB−) gal dcm, DE3 (a λ prophage carrying the T7 RNA polymerase gene); and (iii) AD494(DE3) (Novagen): Δara-leu7697 ΔlacX74 ΔphoAPvuII phoR ΔmalF3 F′ [lac+ (lacIq) pro] trxB::kan, DE3.
Plasmids.
Plasmid pSZ is a pBluescript SK− plasmid (Stratagene) carrying the E. chrysanthemi cel5 gene on a SalI-EcoRI restriction fragment. CBDCel5 was subcloned into plasmid pET22b(+) to construct pECZ. pET22b(+) (Novagen) has a T7 promoter, a C-terminal tag of six histidine residues, and a pelB leader sequence designed for the export of target proteins into the periplasm. The Yheb1 cognate DNA fragment was cloned into pET22b(+) to construct pECYB1. pCPP2006 is a plasmid carrying the E. chrysanthemi out genes that enables E. coli strains to secrete Cel5 into the surrounding medium (15).
Genetic techniques and DNA manipulations.
Plasmid DNA preparation and bacterial transformation were performed by routine procedures (9, 32). Site-directed mutagenesis of plasmid pSZ to generate point mutations in cel5 was performed by the method of Kunkel et al. (21), as presented in the Boehringer kit. Alternatively, a recombinant PCR method (2) was used. The double mutant (W43AY44A) was produced by PCR amplification of the entire pSZ plasmid by mutagenic primers (mutated residues in boldface) divergently orientated but overlapping (underlined) at their 5′ ends (mutagenic oligonucleotide, 5′-TCgCTgCCCggAACggATgCggTggCCgCgTTTgCggTATACAggTTg-3′; anchor oligonucleotide, 5′-gCATCCgTTCCgggCAgCgATTCCTCCTggACgCAggTTg-3′). The triple mutant was then produced by a second recombinant PCR mutagenesis with the double mutant as a template (mutagenic oligonucleotide, 5′-gTTATgAgTAggCTgCCCgCCCgCggCgTCTTTgCTAACCCAgTTg-3′; anchor oligonucleotide, 5′-gCgggCAgCCTACTCATAACgAAgCAggCCAATCgATCg-3′). PCR was performed with Expand High Fidelity DNA polymerase (Boehringer Mannheim) in the presence of 5% dimethyl sulfoxide. Treatment with DpnI destroyed the parental Dam-methylated DNA, whereas the PCR-synthesized molecules remained intact. The resulting linear DNA molecules were transformed into E. coli DH5α (a recA strain).
The internal deletion of residues Asp17 to Pro23 was performed with two separate PCRs to amplify the DNA at each side of the required deletion. The oligonucleotide primers were designed to produce two fragments in which the residues D17 to P23 were substituted for a leucine residue and a HindIII site was introduced. The following oligonucleotide primers were used: Del1, 5′-CgAATTgggTACCgggCCCCCCCTC-3′; Del2, 5′-AgTAAgCTTgCTAACCCAgTTggggTAAAC-3′; Del5, 5′-ggTTAgCAAgCTTACTCATAACgAAgCAggCCA-3′; and NOBA, 5′-gAACTAgTggCTCCCCCgggg-3′. After PCR, the HindIII-digested fragments were ligated and cloned into the pBluescript SK− vector.
The Yheb1 cognate DNA fragment was isolated from the E. coli chromosome and cloned into pET22b(+) to construct pECYB1. The oligonucleotide primers were designed to delete the pelB leader sequence and contained NdeI and XhoI restriction sites, respectively: NDEYB1, 5′-gATggCATATgATggAAgCATggAATAAC-3′; and XYHEB1, 5′-ggATTCTCgAgTTCgCAggAAAgTgTATT-3′. pECYB1 was transformed into E. coli AD494 competent cells, which are thioredoxin reductase mutants that enable disulfide bond formation in the cytoplasm.
The presence of the desired mutation and the absence of any secondary mutations were verified by sequencing.
Purification of CBDCel5(His)6 and Yheb1(His)6.
To express the wild-type CBDCel5 under a T7 promoter, E. coli BL21(DE3) cells carrying pECZ were grown at 37°C in 2 liters of M9CAS medium with 100 μg of ampicillin per ml. The expression of CBDCel5(His)6 was induced by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at mid-log phase. After 4 h of incubation, cells were harvested and the culture supernatant was filtered before being loaded onto a metal chelating column.
E. coli AD494 cells transformed with pECYB1 were grown at 37°C in 2 liters of M9CAS medium and induced with IPTG, as described above. The cells were disrupted by three passages through a French pressure cell at 5,000 lb/in2. Cell debris was pelleted by centrifugation, and the resulting supernatant was loaded onto a metal chelating column.
Chromatography was performed with a HiTrap metal chelating column (Pharmacia) charged with Ni2+. The bound protein was eluted with a pH step gradient from pH 7.2 to 4. Fractions, typically 1 ml, were collected and assayed for CBDCel5(His)6 or Yheb1(His)6 by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting. Peak fractions were desalted and concentrated by using an Ultrafree-15 centrifugal filter unit with a 5,000-NMWL (molecular weight of 5,000 cutoff) polysulfone membrane (Millipore).
Periplasmic preparations.
Strains were grown in LB or M9CAS medium at 37°C (with IPTG induction when appropriate) up to an optical density at 600 nm of 2. After centrifugation, the cell pellet was suspended in a solution containing 20 mM Tris-HCl (pH 8), 20% sucrose, 1 mM EDTA, 1 mg of lysozyme per ml, 1 mM phenylmethylsulfonyl fluoride, and 0.02 mg of DNase I per ml (50 U). Osmotic shock was performed by freezing the cell suspensions at −20°C followed by incubation at 37°C for 15 min. After centrifugation, the supernatant corresponded to the periplasmic preparation.
Enzyme activity and protein assays.
Assays were performed in HEPES buffer (100 mM [pH 7.4]) with p-nitrophenyl β-d-cellobioside at a final concentration of 20 mM (3). The assays were performed in 96-well enzyme-linked immunosorbent assay plates incubated at 37°C with agitation (160 rpm) over a course of 40 h. Release of p-nitrophenol was monitored in a Titertek Multiskan apparatus with a 405-nm filter. A molar extinction coefficient of 16,000 was determined for p-nitrophenol at 405 nm. Enzyme activity was expressed as μmol of pNP liberated per h. Protein assays were performed by the method of Bradford (6).
Binding tests.
Avicel PH-101 and chitin (purified from crab shells) were obtained from Fluka and Sigma, respectively. Bacterial crystalline cellulose (BC) from Nata de coco is a food-grade commercial cellulose (Fujicco Co., Kobe, Japan). The cubes of cellulose were extensively washed and then homogenized in a Waring blender to form a uniform suspension of cellulose (5). Samples (200 μl, typically 0 to 50 μg of protein) were mixed with BC, Avicel, and chitin at final concentrations of 1, 15, and 10 mg/ml, respectively. Assay tubes were mixed by slow vertical rotation at 4°C for 2 h. The cellulose was pelleted by centrifugation, and the resulting supernatant represented the unbound Cel5. The pellet was washed with 400 μl of 20 mM Tris-HCl (pH 7.5), followed by 400 μl of 20 mM Tris-HCl (pH 7.5)–0.5 M NaCl. The washed pellet was resuspended in 40 μl of loading buffer, heated to 95°C for 15 min, and then subjected to SDS-PAGE and Western blotting. It is noteworthy that the unbound fractions (volume, 400 μl) were 10 times less concentrated than the bound fractions (40 μl).
SDS-PAGE.
SDS-PAGE with homogeneous 12.5% or 20% polyacrylamide gels was carried out by using a Pharmacia Phast system. Low-molecular-weight (6,500 to 45,000) color markers were from Sigma.
Western blotting.
Proteins were Western blotted onto a Hybond nitrocellulose membrane (Amersham). The membrane was incubated with antibodies raised against Cel5 and then with goat anti-rabbit immunoglobulin G (IgG) conjugated to alkaline phosphatase. For His-tagged proteins, the primary antibody was anti-His (Invitrogen), and the secondary antibody was goat anti-mouse IgG. Alkaline phosphatase activity was detected by adding 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium.
RESULTS
Stability of Cel5 mutants.
Ten point mutants were constructed in which the wild-type residue was substituted for by an alanine residue. The mutations obtained in cel5 in the pSZ vector were Pro11Ala, Trp13Ala, Asp17Ala, Trp18Ala, Gln22Ala, Glu27Ala, Trp43Ala, Tyr44Ala, Trp43AlaTyr44Ala, and Trp18AlaTrp43AlaTyr44Ala (Fig. 1a). Also, a mutant referred to as Cel5ΔAsp17-Pro23 was constructed in which the region encompassing residues Asp17 to Pro23 was deleted (Fig. 1b).
The effect of point mutations on the stability of the protein was observed by Western blot analysis of overnight cultures (data not shown). The mutations cis-Pro11Ala and Trp13Ala were both destabilizing. The stability of the point mutants Trp18Ala, Gln22Ala, Glu27Ala, Trp43Ala, and Tyr44Ala was not significantly affected. In contrast, substitution of Asp17 by alanine significantly affected the stability. Also, the mutant Cel5ΔAsp17-Pro23 was less stable than the wild-type Cel5 structure.
BC binding isotherms of Cel5 mutants.
The binding affinities of the various mutant forms of CBDCel5 were investigated with mutations in the complete Cel5 (i.e., in the presence of the catalytic domain and linker region). An advantage of this approach was that high levels of purity were not necessary, because the binding affinity could be quantified by measuring the catalytic activity with p-nitrophenyl β-d-cellobioside as the substrate. Routinely, the mutants produced in the E. coli periplasm were used for the binding tests. Alternatively, culture supernatants of E. coli cells containing the pCPP2006 plasmid, which carries the E. chrysanthemi out genes necessary for secretion from the cell, were used as the starting material.
Two different cellulose substrates were used for the binding tests. Avicel has approximately 50 to 60% crystallinity, whereas BC is a highly crystalline (approximately 70%) form of cellulose I. The advantage of using BC is that it is more homogeneous than Avicel and has a higher surface/volume ratio. These are not substrates for Cel5 hydrolyzing activity, and so there would have been no hydrolysis during the binding test assays (29).
The cellulose binding affinities of wild-type and mutant forms of Cel5 were analyzed by binding-isotherm measurements. Since the exact number of possible binding sites was unknown, the saturation curves were compared semiquantitatively, and no apparent affinity constants were calculated. The free Cel5 was measured directly (by pNPCase assay), and the bound Cel5 was calculated by subtracting the free Cel5 from the total Cel5 activity in the sample used for the binding test. Values representing the amount of unbound activity were reproducible to about 5 to 10% for duplicate experiments. The binding tests were performed for 2 h at 4°C.
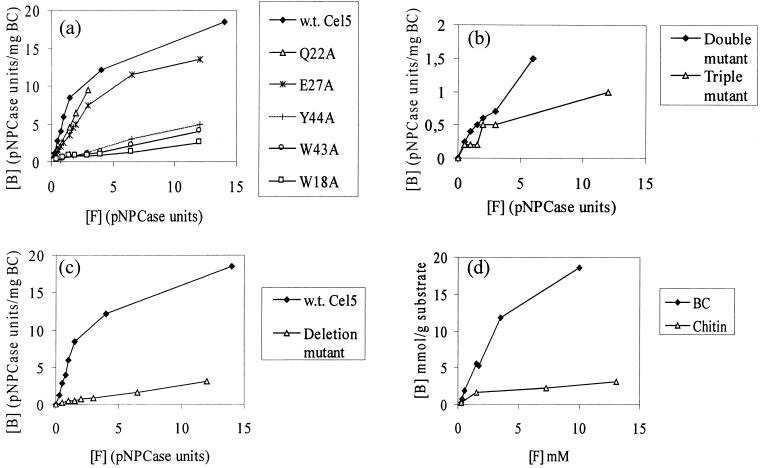
The BC binding isotherms of wild-type Cel5, Trp18Ala, Gln22Ala, Glu27Ala, Trp43Ala, and Tyr44Ala were determined (Fig. 2a). The single mutations Gln22Ala and Glu27Ala had no significant effect on the binding affinity. The binding affinity of the mutant Asp17Ala could not be determined, because the mutation affected the stability, and only low levels of activity could be detected. On substitution for the respective aromatic residues (Trp18, Trp43, and Tyr44) with an alanine residue, the binding was significantly affected in each case. From the results of the effect of the single mutations, three aromatic residues appeared to be responsible for the binding affinity of the CBDCel5. Binding isotherms were determined for the double mutant Trp43AlaTyr44Ala and the triple mutant Trp18AlaTrp43AlaTyr44Ala (Fig. 2b). Binding levels of the triple mutant were particularly low and could simply represent the level of nonspecific binding. The binding data were supported by Western blot analysis, which was used to confirm that Cel5 had not undergone proteolysis to separate the two domains for any of the mutants.
FIG. 2.
Binding isotherms on BC at 4°C in 20 mM Tris buffer at pH 7.5 for wild-type (w.t.) Cel5 and single mutants of Cel5 (a), double (W43AY44A) and triple (W18AW43AY44A) mutants of Cel5 (b), the deletion mutant Cel5ΔD17-P23 (c), and pure CBDCel5(His)6 (BC and chitin) (d). [B] and [F] stand for bound and free, respectively.
Cellulose and chitin binding of wild-type Cel5 and Cel5ΔAsp17-Pro23.
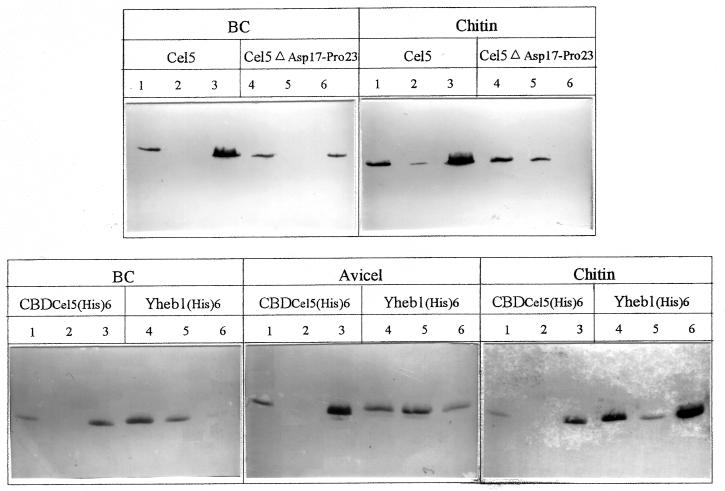
The residue Asp17, which seemed to play an important role in the structure of Cel5, is found in the less-ordered loop region of the protein. Furthermore, on alignment of the sequence of CBDCel5 with various chitin-binding domains, the sequence corresponding to the loop region of CBDCel5 was absent in the chitin-binding domains (8). To investigate the contribution of this region to the overall function of CBDCel5, a cel5 mutation was constructed such that the encoded product lacked residues Asp17 to Pro23 of the CBD domain. The binding isotherms for wild-type Cel5 and Cel5ΔAsp17-Pro23 were determined by using 1 mg of BC per ml as a substrate (Fig. 2c). The binding affinity of Cel5ΔAsp17-Pro23 to BC was significantly lower than that with wild-type Cel5. Additionally, the binding of wild-type Cel5 and Cel5ΔAsp17-Pro23 to BC and chitin was investigated by using Western blot analyses (Fig. 3). The weaker binding affinity to BC of the deletion mutant Cel5ΔAsp17-Pro23 compared to that of the wild-type Cel5 was confirmed. Furthermore, the mutant Cel5ΔAsp17-Pro23 bound only poorly to chitin, the majority being detected in the unbound fraction (lane 5).
FIG. 3.
The upper blots show the binding of wild-type Cel5 and Cel5ΔAsp17-Pro23 to BC (1 mg/ml) and chitin (10 mg/ml). The lower blots show the binding of CBDCel5(His)6 and Yheb(His)6 to BC (1 mg/ml), Avicel (15 mg/ml), and chitin (10 mg/ml). Binding at 4°C for 2 h was analyzed on gels and by immunoblotting as described in Materials and Methods. Lanes: 1 and 4, starting sample; 2 and 5, unbound; 3 and 6, bound.
Purification of wild-type CBDCel5(His)6 and Yheb1(His)6.
An unexpected relatedness was observed previously between CBDCel5 and a series of repeating domains in a hypothetical E. coli Yheb protein (8). Therefore, we decided to carry out a functional comparison of CBDCel5 and one of the Yheb domains, Yheb1. The pECZ construct allowed for the production of processed and disulfide-bonded CBDCel5(His)6 in the periplasm of E. coli recombinant cells. When the cells were grown in M9CAS medium, a significant proportion of the CBDCel5 was found in the culture supernatant, which was used for the purification. Typical yields were 0.5 to 1 mg of pure CBDCel5(His)6 per liter of culture. The one-dimensional NMR spectrum of CBDCel5(His)6 was compared to that of CBDCel5, as prepared by the method of Brun et al. (7), and it was observed that the His tag had not affected the structure (data not shown).
The Yheb1 cognate DNA fragment was cloned from the E. coli chromosome, and the 67-amino-acid protein was purified. Yheb1(His)6 was produced in the cytoplasm of E. coli AD494 cells, and the cell extracts were used to purify the protein. Typical yields were 1 to 2 mg of pure Yheb1(His)6 per liter of culture.
Binding tests of pure CBDCel5(His)6 and pure Yheb1(His)6.
BC and chitin binding isotherms of pure CBDCel5(His)6 were determined (Fig. 2d). A saturating level of CBDCel5(His)6 on cellulose was not quite achieved, but the isotherm seems to plateau at a value in excess of 25 μmol/g of cellulose. Approximately, 10 times more CBDCel5 is bound by BC than by the same quantity of chitin.
Western blots were used to analyze BC (1 mg/ml), Avicel (15 mg/ml), and chitin (10 mg/ml) binding of pure CBDCel5(His)6 and Yheb1(His)6 (Fig. 3). Under the experimental conditions stated, the pure CBDCel5(His)6 bound well to BC, chitin, and Avicel, and no unbound CBDCel5(His)6 was detected (lane 2). Pure Yheb1(His)6 bound with high affinity to chitin (lane 6). On comparing the Western blots, binding of Yheb1(His)6 to Avicel resulted in approximately equal portions of bound (lane 6) and unbound (lane 5) protein, but the binding to BC was poor, with the majority of Yheb1(His)6 detected in the unbound fraction (lane 5).
Experiments with CBDCel5, which did not contain a His tag, revealed that the His tag did not affect the binding affinity of CBDCel5 (data not shown).
DISCUSSION
In this work, we studied the CBD of the Cel5 cellulase from E. chrysanthemi. Isolated CBDCel5 appears to be functionally and structurally analogous to CBDCel5 in the full-length Cel5. The one-dimensional NMR spectra of Cel5, CBDCel5, and CBDCel5(His)6 indicated that the tryptophan side chains are in similar environments, suggesting that the same folding pattern occurs (7). Therefore, we decided to investigate the role of individual amino acids in the CBDCel5 in the presence of the catalytic and linker region, which is the natural form of the enzyme.
There are essentially three effects that could contribute to the binding of a CBD to the cellulose surface. One possibility is that aromatic residues in the CBD interact with the pyranosyl rings of one of the cellulose chains. The interaction between sugar and aromatic moieties is common in carbohydrate binding proteins (34). A second possible interaction is the hydrogen bonding of polar surface atoms on the CBD. The polar residues may play a role in either stabilizing other cellulose-binding residues by hydrogen bonding or by hydrogen bonding directly with cellulose. For example, Asn and Gln residues on the flat face of fungal CBDs are highly conserved, and these residues participate in hydrogen-bonding interactions with cellulose (17, 25). A third possibility is that hydrophobic effects could contribute to the binding. It has been shown that the binding of T. reesei CBDCBH1 to cellulose was significantly affected by high ionic strength (30), which suggested that the interaction with cellulose included a hydrophobic effect.
The NMR structure of CBDCel5 led us to predict five amino acids (Asp17, Trp18, Glu27, Trp43, and Tyr44) that potentially interact with the glucose chain of cellulose and contribute directly to the binding (Fig. 1a). Here we have demonstrated the contribution of the three aromatic residues (Trp18, Trp43, and Tyr44) to binding affinity. The mutant proteins exhibited much lower affinity for BC than the wild-type Cel5. The very low binding affinity observed with the triple mutant could simply reflect nonspecific binding. All three surface tryptophans of the CBD from Pseudomonas fluorescens subsp. cellulosa xylanase A were found to play a role in binding both soluble and insoluble ligands (27). Furthermore, all of the CBDs of which the three-dimensional structures have been solved and that bind crystalline cellulose have been found to exhibit a planar surface comprising three aromatic residues (8, 22, 24, 25, 32, 38).
Two residues on the flat face of Cel5 (Asp17 and Glu27) were predicted to have a possible role in hydrogen bonding interactions. The mutation Glu27Ala did not affect the binding affinity, despite being in a position in which there is the possibility of hydrogen bonding either directly with cellulose or with residue Tyr44 (Fig. 1a). The role of Asp17 in cellulose binding could not be evaluated, since the mutation affected the stability of Cel5.
The residues cis-Pro11 and Trp13 were both predicted to be important for the stability of CBDCel5, due to their position in the hydrophobic core of the structure (Fig. 1a), and this was found to be true. Pro11 is the only Pro residue of CBDCel5 that exhibits a cis conformation and thus would be expected to have an important biological function.
The CBDCel5 structure is essentially a triple antiparallel β-sheet perpendicular to a summital loop, which is one of the most disordered parts of the CBDCel5 structure (8). A type I turn has been defined between Trp18 and Gly21, and deletion of this region would be expected to affect the structure (Fig. 1b). The deletion of residues Asp17 to Pro23 resulted in a lower binding affinity for BC, compared to that of wild-type Cel5. In addition to Trp18, there are two other potential binding residues in this loop region (Asp17 and Gln22). Asp17 was found to have a structural role (see above), while the mutation Gln22Ala produced no effect on binding affinity. This is consistent with the NMR model, since Gln22 is not directly on the cellulose-binding surface (Fig. 1a). Therefore, the reduction in binding affinity of Cel5ΔAsp17-Pro23 can be attributed to the loss of one of the three tryptophan residues (Trp18), which plays an important role in cellulose binding.
The structural information about CBDCel5 coupled with sequence alignments has shown that CBDCel5 exhibits sequence similarity to the E. coli Yheb1 to -5 sequences and to modules thought to be chitin-binding domains (8). A search of the Swiss Protein Data Bank identified several chitinases and endoglucanases with significant similarity to regions of the translated open reading frame of the E. coli hypothetical Yheb protein. A region of the E. coli Yheb protein corresponding to the putative catalytic domain exhibited sequence homology with a Saccharopolyspora erythraea chitinase, which belongs to family 18 of glycosyl hydrolases (19). We purified the 67-amino-acid Yheb1 protein. Under the assay conditions used, Yheb1 bound to chitin, but bound only poorly to Avicel and BC. Therefore, it is likely that the E. coli Yheb protein is a multidomain chitinase consisting of a family 18 catalytic domain, a linker region, and a series of five chitin-binding domains. Yheb1 to -5 are probably all chitin-binding domains, since the chitin binding of Yheb1 has been demonstrated and there is very high identity (∼60%) with the other Yheb repeats. The presence of more than one CBD in a cellulase has been reported for endoglucanase C from Cellulomonas fimi (10) and the cellulase B from Cellvibrio mixtus (11). Also, it has been reported that two CBDs, contained within a single polypeptide, can interact synergistically (23). It is noteworthy that multidomain chitinases often have, in addition to catalytic domains and chitin-binding domains, other reiterated domains. Fibronectin type III-like domains and cadherin-like domains have been found in several chitinases (4, 12, 26, 36, 37).
Here we report that the family V CBDCel5 binds chitin. Chitin and cellulose have similar structures, and the finding that the CBDCel5 can bind to chitin demonstrates that the N-acetyl substitutions on the glucose residues of chitin do not block the interaction with the CBDCel5. There have been previous reports of CBDs that bind to chitin. The family III CBDCbpA from Clostridium cellulovorans has a high affinity for chitin (14), as has the family II CBDCex of an endoglucanase from C. fimi (28). Also, the chitin-binding domain from a Clostridium paraputrificum chitinase has been shown to bind to cellulose (26). The fact that chitin binding has been demonstrated with CBDs from different families suggests that the mechanisms of binding may be common for chitin-binding domains and CBDs and that they may have the same evolutionary origin. A striking feature of the alignment of CBDCel5 with the E. coli Yheb1 to -5 sequences and several other chitin-binding domains (8) was that the loop of residues Asp17 to Pro23 was only present in CBDCel5. Yheb1, which binds only poorly to BC, binds chitin at approximately the same efficiency as CBDCel5. Thus, a simple hypothesis was that the Asp17-Pro23 loop was necessary for cellulose binding but not chitin binding. However, the mutant Cel5ΔD17-P23 was affected in both cellulose and chitin binding affinity, compared to the wild-type CBDCel5. This illustrates that the situation is not as simple as had been predicted, and it seems likely that the loop region participates in both cellulose and chitin binding via the residue Trp18.
ACKNOWLEDGMENTS
This work was supported by a short-term FEBS fellowship and a Marie Curie fellowship awarded to H. Simpson as part of the European Community Fourth Framework Programme, as well as grants from the CNRS, the University Aix-Marseille II, and the EEC Biotech Programme BIO4-CT97-2303.
We thank A. Marteau for constructing the pECZ plasmid.
REFERENCES
- 1.Andrews S C, Harrison P M, Guest J R. Cloning, sequencing, and mapping of the bacterioferritin gene (bfr) of Escherichia coli K-12. J Bacteriol. 1989;171:3940–3947. doi: 10.1128/jb.171.7.3940-3947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansaldi M, Lepelletier M, Mejean V. Site-specific mutagenesis by using an accurate recombinant polymerase chain reaction method. Anal Biochem. 1996;234:110–111. doi: 10.1006/abio.1996.0060. [DOI] [PubMed] [Google Scholar]
- 3.Barras F, Bortoli-German I, Bauzan M, Rouvier J, Gey C, Heyraud A, Henrissat B. Stereochemistry of the hydrolysis reaction catalysed by endoglucanase Z from Erwinia chrysanthemi. FEBS Lett. 1992;300:145–148. doi: 10.1016/0014-5793(92)80183-h. [DOI] [PubMed] [Google Scholar]
- 4.Blaak H, Schnellmann J, Walter S, Henrissat B, Schrempf H. Characteristics of an exochitinase from Streptomyces olivaceoviridis, its corresponding gene, putative protein domains and relationship to other chitinases. Eur J Biochem. 1993;214:659–669. doi: 10.1111/j.1432-1033.1993.tb17966.x. [DOI] [PubMed] [Google Scholar]
- 5.Boisset, C. Personal communication.
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brun E, Gans P, Marion D, Barras F. Overproduction, purification and characterisation of the cellulose-binding domain of the Erwinia chrysanthemi secreted endoglucanase EGZ. Eur J Biochem. 1995;231:142–148. [PubMed] [Google Scholar]
- 8.Brun E, Moriaud F, Gans P, Blackledge M J, Barras F, Marion D. Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry. 1997;36:16074–16086. doi: 10.1021/bi9718494. [DOI] [PubMed] [Google Scholar]
- 9.Chung C T, Miller R H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988;16:3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coutinho J B, Gilkes N R, Warren R A, Kilburn D G, Miller R C. The binding of Cellulomonas fimi endoglucanase C (CenC) to cellulose and Sephadex is mediated by the N-terminal repeats. Mol Microbiol. 1992;6:1243–1253. doi: 10.1111/j.1365-2958.1992.tb01563.x. [DOI] [PubMed] [Google Scholar]
- 11.Fontes C M, Clarke J H, Hazlewood G P, Fernandes T H, Gilbert T H, Ferreira L M. Identification of tandemly repeated type VI cellulose-binding domains in an endoglucanase from the aerobic soil bacterium Cellvibrio mixtus. Appl Microbiol Biotechnol. 1998;49:552–559. doi: 10.1007/s002530051212. [DOI] [PubMed] [Google Scholar]
- 12.Fujii T, Miyashita K. Multiple domain structure in a chitinase gene (chiC) of Streptomyces lividans. J Gen Microbiol. 1993;139:677–686. doi: 10.1099/00221287-139-4-677. [DOI] [PubMed] [Google Scholar]
- 13.Gilkes N R, Henrissat B, Kilburn D G, Miller R C, Jr, Warren R A J. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein M A, Takagi M, Hashida S, Shoseyov O, Doi R H, Segel I H. Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A. J Bacteriol. 1993;175:5762–5768. doi: 10.1128/jb.175.18.5762-5768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He S Y, Lindeberg M, Chatterjee A K, Collmer A. Cloned Erwinia chrysanthemi out genes enable Escherichia coli to selectively secrete a diverse family of heterologous proteins to its milieu. Proc Natl Acad Sci USA. 1991;88:1079–1083. doi: 10.1073/pnas.88.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffren A-M, Teeri T T, Teleman O. Molecular dynamics of fungal cellulose-binding domains: differences in molecular rigidity but a preserved cellulose binding surface. Protein Eng. 1995;8:443–450. doi: 10.1093/protein/8.5.443. [DOI] [PubMed] [Google Scholar]
- 18.Johnson P, Joshi M, Tomme P, Kilburn D, McIntosh L. Structure of the N-terminal cellulose-binding domain of Cellulomonas fimi Cen C. 2. NMR and ultraviolet spectroscopy. Biochemistry. 1996;35:13895–13906. doi: 10.1021/bi961186a. [DOI] [PubMed] [Google Scholar]
- 19.Kamei K, Yamamura Y, Hara S, Ikenaka T. Amino acid sequence of chitinase from Streptomyces erythraeus. J Biochem. 1989;105:979–985. doi: 10.1093/oxfordjournals.jbchem.a122791. [DOI] [PubMed] [Google Scholar]
- 20.Kraulis P, Clore M, Nilges M, Jones A, Pettersson G, Knowles J, Gronenborn A. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. Biochemistry. 1989;28:7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel T A, Bebenek K, McClay J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–138. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 22.Linder M, Mattinen M-L, Kontteli M, Stahlberg G, Drakenberg T, Reinikainen T, Pettersson G, Annila A. Identification of functionally important amino acids in the cellulose binding domain of Trichoderma reesei cellobiohydrolase I. Protein Sci. 1995;4:1056–1064. doi: 10.1002/pro.5560040604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linder M, Salovuori I, Ruohonen L, Teeri T T. Characterization of a double cellulose-binding domain. J Biol Chem. 1996;271:21268–21272. doi: 10.1074/jbc.271.35.21268. [DOI] [PubMed] [Google Scholar]
- 24.Mattinen M L, Kontteli M, Kerovuo J, Linder M, Annila A, Lindeberg G, Reinikainen T, Drakenberg T. Three-dimensional structures of three engineered cellulose-binding domains of cellobiohydrolase I from Trichoderma reesei. Protein Sci. 1997;6:294–303. doi: 10.1002/pro.5560060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattinen M L, Linder M, Drakenberg T, Annila A. Solution structure of the cellulose-binding domain of endoglucanase I from Trichoderma reesei and its interaction with cello-oligosaccharides. Eur J Biochem. 1998;256:279–286. doi: 10.1046/j.1432-1327.1998.2560279.x. [DOI] [PubMed] [Google Scholar]
- 26.Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J Bacteriol. 1997;179:7306–7314. doi: 10.1128/jb.179.23.7306-7314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy T, Simpson P, Williamson P, Hazlewood G P, Gilbert H J, Orosz L. All three surface tryptophans in type IIa cellulose binding domains play a pivotal role in binding both soluble and insoluble ligands. FEBS Lett. 1998;429:312–316. doi: 10.1016/s0014-5793(98)00625-5. [DOI] [PubMed] [Google Scholar]
- 28.Ong E, Gilkes N R, Miller R C, Warren R A J, Kilburn D G. The cellulose-binding domain (CBDCex) of an exoglucanase from Cellulomonas fimi: production in Escherichia coli and characterisation of the polypeptide. Biotechnol Bioeng. 1993;42:401–409. doi: 10.1002/bit.260420402. [DOI] [PubMed] [Google Scholar]
- 29.Py B, Bortoli-German I, Haiech J, Chippaux M, Barras F. Cellulase EGZ of Erwinia chrysanthemi: structural organisation and importance of His98 and Glu133 residues for catalysis. Protein Eng. 1991;4:325–333. doi: 10.1093/protein/4.3.325. [DOI] [PubMed] [Google Scholar]
- 30.Reinikainen T, Teleman O, Teeri T T. Effects of pH and high ionic strength on the adsorption and activity of native and mutated cellobiohydrolase I from Trichoderma reesei. Proteins. 1995;22:392–403. doi: 10.1002/prot.340220409. [DOI] [PubMed] [Google Scholar]
- 31.Sakon J, Irwin D, Wilson D B, Karplus P A. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nature Struct Biol. 1997;4:810–818. doi: 10.1038/nsb1097-810. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Tomme P, Warren R A J, Gilkes N R. Cellulose hydrolysis of bacteria and fungi. Adv Microb Physiol. 1995;37:1–81. doi: 10.1016/s0065-2911(08)60143-5. [DOI] [PubMed] [Google Scholar]
- 34.Toone E J. Structure and energetics of protein-carbohydrate complexes. Curr Opin Struct Biol. 1994;4:719–728. [Google Scholar]
- 35.Tormo J, Lamed R, Chirino A, Morag E, Bayer E, Shoham Y, Steitz T. Crystal structure of a bacterial family III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 36.Tsujibo H, Orikoshi H, Shiotani K, Hayashi M, Umeda J, Miyamoto K, Imada C, Okami Y, Inamori Y. Characterization of chitinase C from a marine bacterium, Alteromonas sp. strain O-7, and its corresponding gene and domain structure. Appl Environ Microbiol. 1998;64:472–478. doi: 10.1128/aem.64.2.472-478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe T, Ito Y, Yamada T, Hashimoto M, Sekine S, Tanaka H. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin. J Bacteriol. 1994;176:4465–4472. doi: 10.1128/jb.176.15.4465-4472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu G-Y, Ong E, Gilkes N, Kilburn D, Muhandiram D, Harris-Brandts M, Carver J, Kay L, Harvey T. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:6993–7009. [PubMed] [Google Scholar]