Abstract
Actinobacteria are ubiquitous bacteria undergoing complex developmental transitions coinciding with antibiotic production in response to stress or nutrient starvation. This transition is mainly controlled by the interaction between the second messenger c-di-GMP and the master repressor BldD. To date, the upstream factors and the global signal networks that regulate these intriguing cell biological processes remain unknown. In Saccharopolyspora erythraea, we found that acetyl phosphate (AcP) accumulation resulting from environmental nitrogen stress participated in the regulation of BldD activity through cooperation with c-di-GMP. AcP-induced acetylation of BldD at K11 caused the BldD dimer to fall apart and dissociate from the target DNA and disrupted the signal transduction of c-di-GMP, thus governing both developmental transition and antibiotic production. Additionally, practical mutation of BldDK11R bypassing acetylation regulation could enhance the positive effect of BldD on antibiotic production. The study of AcP-dependent acetylation is usually confined to the control of enzyme activity. Our finding represents an entirely different role of the covalent modification caused by AcP, which integrated with c-di-GMP signal in modulating the activity of BldD for development and antibiotic production, coping with environmental stress. This coherent regulatory network might be widespread across actinobacteria, thus has broad implications.
Graphical Abstract
Graphical Abstract.
INTRODUCTION
Soil-dwelling actinobacteria are Gram-positive bacteria producing numerous antibiotics or secondary metabolites that are commercially or clinically relevant, such as antibacterial, antiviral, antitumor, and antifungal properties (1). Members of this genus face with drastic changes in key nutrients including carbon and nitrogen sources, phosphate, oxygen, iron, sulfur, etc., and secondary metabolites are produced in response to environmental conditions, including nutrient starvation.
Like all living cells, actinobacteria have to be able to sense, respond to, and adapt to these stressful conditions. This requires appropriate sensors for external/internal stimuli and signal transduction processes driving transcriptomic and proteomic changes that allow them to repair or avoid cellular damage, which are mainly subject to transcriptional regulation mediated by multiple interactions of pleiotropic and cluster-situated transcription factors that work in networks and cascades (2). Environmental stress thus turns into metabolic cues within the bacterial cell and results in an unbalanced metabolism of primary and secondary metabolism, causing a series of consequences for growth, cell differentiation, reproduction, and secondary metabolite biosynthesis. Pleiotropic transcription factors including GlnR, PhoP and MtrA, can sense the corresponding nutrient status and control hundreds of reactions in actinobacterial cells directly or indirectly (2–8). Moreover, nutrient limitation or starvation also induces stringent regulation of the transcription factors themselves through covalent modification (such as acylation and phosphorylation) and noncovalent interaction with nucleotide second messengers (such as c-di-GMP). Increasing carbon flux or nitrogen starvation is verified to trigger global protein acetylation via acetyltransferase, which regulates the function of transcription factors, such as GlnR and PhoP, exhibiting strict transcriptional and posttranslational control of nutrient assimilation, which is fundamental for actinobacterial survival (2,9).
The stringent response to stress or nutrient starvation in actinobacteria is closely linked to complex morphological differentiation and the production of secondary metabolites, including antibiotics (10,11). Morphological differentiation follows a genetically programmed developmental pathway from spore formation of aerial mycelium through multiple septum formation to the final release of dormant spores. BldD is identified as a key developmental regulator that coordinates this pathway (12–14). BldD is an autoregulatory and master regulator that oversees the entire regulatory cascade through the repression of a global regulon of nearly 170 sporulation-related genes during vegetative growth (15,16). In particular, actinobacteria are renowned for the production of clinically important antibiotics and other bioactive compounds, which is genetically and temporally coordinated with development. Thus, BldD also pleiotropically influences antibiotic production in addition to causing loss of aerial mycelium formation (12,14,15). Additional interactions on the regulatory backbone are provided by the nucleotide second messenger c-di-GMP, which functions as a corepressor of BldD to control the decision to initiate the developmental program (17). A high level of c-di-GMP blocks the formation of aerial hyphae and spores until a drop in the level of c-di-GMP relieves BldD-mediated repression of the regulatory cascade (17–19). Thus, a governing regulation controlling the switch from vegetative growth to sporulation was formed (Figure 1A). In all domains of life, nucleotide-based second messengers allow a rapid integration of external and internal signals into regulatory pathways that control cellular responses to changing conditions. However, for BldD, at the top of the entire development process, it remains largely unknown whether other upstream factors or regulatory mechanisms could affect its function and c-di-GMP signal transduction during drastic changes in nutrients and the consequent variation in growth, differentiation and secondary metabolite biosynthesis.
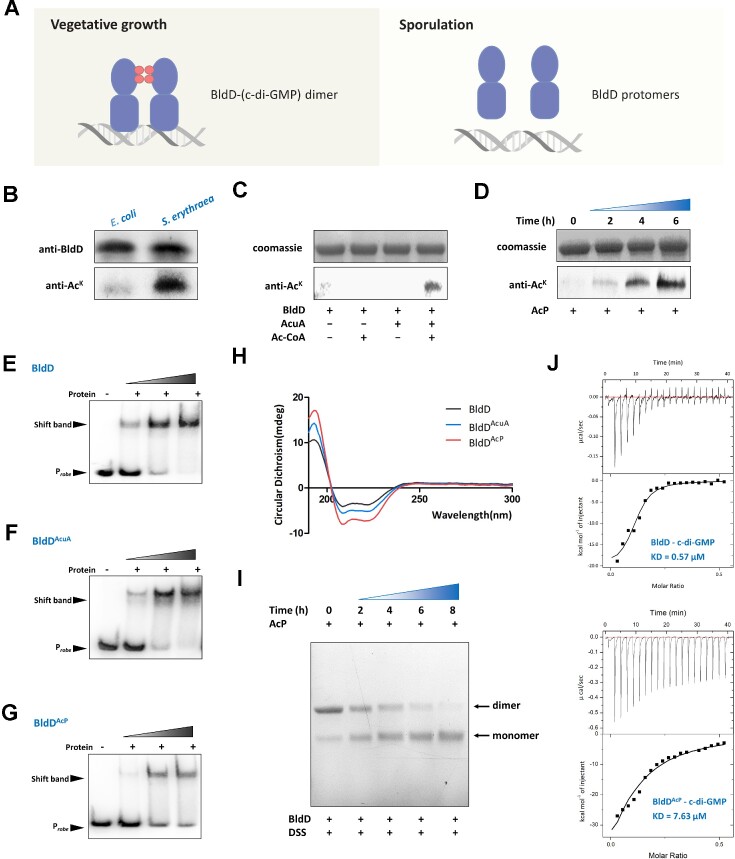
Figure 1.
AcP-induced acetylation of BldD impairs its DNA-binding activity and interaction with c-di-GMP. (A) c-di-GMP through the repressor BldD to control development. (B) Acetylation level of BldD in S. erythraea. His-tagged BldD overexpressed in E. coli was used as a control. (C) In vitro acetylation of His-tagged BldD protein incubated with or without AcuA and Ac-CoA at 37°C for 2 h. (D) In vitro acetylation of His-tagged BldD protein with 10 mM AcP for various lengths of time (0, 2, 4 and 6 h) at 37°C. The acetylation levels were determined by Western blotting using a specific anti-AcK antibody. EMSA of BldD (E), AcuA-dependent acetylated BldD (F) and AcP-dependent acetylated BldD (G) binding to the bldD gene promoter. (H) Circular dichroism spectra of BldD, AcuA-dependent acetylated BldD and AcP-dependent acetylated BldD. (I) Crosslinking after incubation with AcP for various lengths of time at 37°C. (J) Characterization of the interaction of c-di-GMP with BldD and AcP-acetylated BldD using ITC. Titration of c-di-GMP (100 μM) into BldD (40 μM).
Here, we show that in addition to c-di-GMP-induced multimerization, BldD is under the direct regulation of AcP-induced covalent modification resulting from nitrogen limitation, thus revealing a link between protein acetylation and c-di-GMP signaling in S. erythraea. Specifically, we found that AcP-dependent acetylation of BldD led to loss of function in dimerization as well as interaction with c-di-GMP, which caused weakened BldD–DNA binding and inhibited its transcriptional activity. AcP-dependent acetylation on K11 of BldD was then demonstrated to govern the inactivation of BldD, leading to derepression of the BldD regulon of sporulation genes and repression of the BldD regulon of secondary metabolite biosynthesis genes, thereby controlling morphological differentiation and antibiotic production. It was observed that this regulatory function of acetylated BldD on K11 is highly conserved across streptomycetes and some other genera of actinobacteria. Our results present an unidentified signal transduction mechanism through the interaction between AcP and c-di-GMP signaling that allows a global cellular response to environmental changes.
MATERIALS AND METHODS
Strains and culture conditions
All strains and plasmids used in this work are listed in Supplementary Table S2. S. erythraea was cultured in TSB medium at 30°C and 220 rpm for 48 h for seed stock preparation. Nearly 500 μl was then transferred to minimal Evans medium containing 25 mM TES (N‐(Tris(hydroxymethyl)methyl)‐2‐ aminoethanesulfonic acid sodium salt), 2 mM citric acid, 10 mM KCl, 0.25 mM CaCl2, 1.25 mM MgCl2, 2 mM Na2SO4, 1 mM Na2MoO4, 0.5% trace elements (0.02 mM MnSO4·4H2O, 6 μM ZnSO4·7H2O, 0.02 mM H3BO3, 1 μM KI, 2 μM Na2MoO4·2H2O, 0.05 mM CuSO4·5H2O, 0.05 mM CoCl2·6H2O), 2.5% (m/v) glucose, 2 mM NaH2PO4, pH 7.2 and supplemented with 1 mM, 5 mM, 15 mM (NH4)2 SO4 for the indicated biochemical tests.
Overproduction and purification of proteins in vitro
The bldD gene was amplified from the genomic DNA of S. erythraea by PCR using the primers listed in Supplementary Table S3. After restriction digestion with EcoRI and HindIII, the gene coding for BldD (SACE_2077) was cloned into pET28a. The protein was expressed by the E. coli BL21(DE3) strain and purified as previously described (20). His-tagged protein was purified by Ni-NTA Superflow columns (Qiagen), and eluted with 250 mM imidazole (in 50 mM NaH2PO4, 300 mM NaCl, pH 8.0). The protein concentration was determined using BCA Protein Assay Kit (TIANGEN) with BSA as the standard.
Electrophoretic mobility shift assay (EMSA)
DNA fragment spanning the 152 bp promoter region from upstream of bldD gene was amplified by PCR using the primers listed in Supplementary Table S3. The PCR products were modified with biotin with primer (5′biotinAGCCAGTGGCGATAAG-3′). The biotin-labeled PCR products were purified with the PCR purification kit (Shanghai TransGen Biotech, China). EMSA was carried out as previously described (7) with a chemiluminescent EMSA kit (Beyotime Biotechnology, China). The binding reaction contained 10 mM Tris–HCl (pH 8.0), 25 mM MgCl2, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.01% Nonidet P40, 50 μg ml−1 poly[d(I-C)], and 10% glycerol. After binding, the samples were separated on a nondenaturing PAGE gel in an ice bath of 0.5× Tris–borate–EDTA at 100 V.
In vitro protein acetylation assays
The in vitro enzymatic acetylation assays with AcuA and nonenzymatic acetylation with AcP were performed as previously described (20). The reaction was performed in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (pH 7.5) containing 200 μM tris(2-carboxyethyl) phosphine (TCEP) hydrochloride, and 10 μg purified BldD with 0.2 μM AcuA and 25 μM Ac-CoA for enzymatic acetylation or 10 mM AcP for nonenzymatic acetylation. After the reaction, the acetylated protein was isolated by SDS-PAGE and analyzed by Western blotting and LC−MS/MS.
Western blot analysis
Western blot analysis was performed as previously described (20). Protein lysates were separated by 10% SDS-PAGE and then transferred to the PVDF membranes for 60 min at 100 V. After blocking with 3% BSA in PBST buffer (phosphate buffered saline containing 0.1% Tween-80) at room temperature for 1 h, anti‐acetyl lysine (hereafter Anti‐AcK) antibody (PTM-102) purchased from PTM BioLab (HangZhou Jingjie) diluted 1:15000 in TBST/5% BSA was used. The blot was performed via ImageQuant LAS 4000 system (GE Healthcare, UK) after chemiluminescent HRP substrate treatment.
Identification of acetylated lysine residues by mass spectrometry
Protein samples were separated by SDS-PAGE. The bands containing BldD were destained and dehydrated for further digestion and LC−MS/MS as described in detail in previous studies (21). The gel bands were sliced and destained in 50% ethanol treatment. After being fully dehydrated in 100% ACN, samples were reduced by 10 mM DTT at 56°C for 40 min and then alkylated by 15 mM iodoacetamide (IAA) in darkness for another 40 min. After that, the gels were washed in washing buffer (50% ACN/50% mM NH4HCO3 (v/v)) and proteins were digested by trypsin at an enzyme to substrate ratio of 1:40 for 16 h. The tryptic peptides were extracted in 50% ACN/ 5% TFA, 75%/0.1% TFA and 100% TFA in sequence. The samples were then dissolved in solvent A (0.1% (v/v) formic acid and 2% acetonitrile in water) and analyzed by the orbitrap Fusion mass spectrometer in two technical replicates. The raw data were converted to mgf files and then analyzed by Mascot search engine (v 2.3.01, Matrix Science). The search parameters were enzymes, trypsin/P; missed cleavage, 2; fixed modification, Carbamidomethyl (C); variable modifications, acetylation (K), oxidation (M) and acetyl (protein N-terminal); peptide mass tolerance, 10 ppm; fragment mass tolerance, 0.5 Da. Areas under the curve (AUCs) of the precursor ion peak were used to evaluate the peptide intensity. Unmodified peptide (AVVVGSYER) was used for protein level normalization. The ratios of the acylated peptides were normalized to the ratios of their corresponding protein levels. Normalized ratios of the peptides were used for further analysis.
Site‐directed mutagenesis of BldD acetylated‐site mutants
With the primers listed in Supplementary Table S3 and the Fast mutagenesis system (Transgen Biotech, China), the mutants (K11Q, K11R) of bldD were introduced into the pET28a(+)::bldD plasmid. Purified proteins were analyzed by SDS-PAGE, and the concentration was determined with BCA Protein Assay Kit (TIANGEN) with BSA as the standard.
Circular dichroism spectroscopy
Through ultrafiltration, the proteins (0.2 mg/ml) were dissolved in a modified PBS buffer containing 1.4 M KF, 100 mM K2HPO4, and 18 mM KH2PO4. The far-UV CD spectra were recorded at 20°C using a Chirascan Plus instrument (Applied Photophysics) and were collected from 190 to 260 nm using a rectangular quartz cell with a 1 mm path length. The spectra were analyzed for secondary structure content using CDNN CD spectra deconvolution software (Applied Photophysics).
Immunoprecipitation (IP) and immunoblotting (IB)
Purification of BldD protein from S. erythraea through IP was performed as previously described (20) using the specific anti-BldD polyclonal antibody (Solarbio, China), the acetylation level was tested by immunoblotting (IB) using the anti‐AcK antibody (PTM-102) purchased from PTM BioLab (HangZhou Jingjie). Binding was visualized using an ECL western blotting method. After ECL detection, films were scanned by MF-ChemiBIS software, version 3.2 (DNR Bio-Imaging Systems, Israel), and quantified with ImageJ software.
Overexpression of BldD and its mutants in S. erythraea
The bldD gene and its mutants (BldDK11Q, BldDK11R) with His-tag were amplified by PCR using the primers listed in Supplementary Table S3. After restriction digestion with NdeI and XbaI, the gene coding for BldD (SACE_2077) was cloned into the integrative plasmid pIB139 (22), gaining the plasmids pIB139-bldD, pIB139-bldDK11Q, and pIB139-bldDK11R. The plasmid was introduced into wild-type S. erythraea through PEG-mediated protoplast transformation. The strains were verified by PCR and DNA sequencing.
ChIP-seq and data analysis
The ObldD and ObldDK11Q strains were grown for the appropriate length of time. Formaldehyde was added to cultures at a final concentration of 1% (vol/vol), and incubation was continued for 30 min. Glycine was then added to a final concentration of 125 mM to stop the cross-linking. The samples were left at room temperature for 10 min and washed twice in the precooled TBS buffer (pH 7.5) containing 20 mM Tris–HCl and 150 mM NaCl. The samples were then transferred to the ChIP-seq performed by E-GENE Tech Co., Ltd (Shenzhen) using an anti-BldD polyclonal antibody. For the data analysis, in brief, fastp software (v0.20.0) was used to trim adaptors and remove low-quality reads to get high-quality clean reads. Clean reads were aligned to the reference genome using bowtie2 software (v2.2.4). MACS2 software (v2.2.7.1) was used for peak calling. Bedtools software (v2.30.0) was used for peak annotation based on GTF annotation files. Homer (v4.11) software was used to identify motifs. MAnorm2 (v1.2.0) software was used to identify differentially enriched regions. The enriched peaks were visualized in IGV (v2.14.1) software.
RNA preparation and real-time RT-PCR
RNA preparation and real-time RT-PCR were performed as previously described (20) utilizing the primers listed in Supplementary Table S2. Total RNA was extracted and purified from the collected cell samples using the RNeasy Mini Kit (Qiagen, Valencia, CA). About 1 μg of total RNA was reverse transcribed using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Shiga, Japan). For real-time RT-PCR, the SYBR premix Ex Taq™ GC Kit (Perfect Real Time, Takara) was used, and about 100 ng cDNA was added to a 20 μl volume of PCR reaction. The transcription values of each gene were normalized relative to the value for the internal control gene 16S rRNA (SACE_8101) using the comparative Ct method. For each gene, the relative expression value at the first time point was defined as 1. Gene expression values were determined in triplicate.
Cross‐linking experiments
The BldD proteins were dialyzed into cross-linking buffer (100 mM NaH2PO4, 150 mM NaCl, pH 8). 60 μM BldD, 1 mM disuccinimidyl suberate (DSS) in dimethylsulfoxide (DMSO) and 120 μM c-di-GMP were incubated at room temperature for 60 min (17). Samples were fractionated on 15% SDS-PAGE gels and visualized by Coomassie staining. All experiments were performed at least twice.
Phenotypic analysis
S. erythraea was cultured on R2YE agar plates (23). Erythromycin yield was analyzed by bioassay with Bacillus subtilis as the indicator bacteria. Two hundred microliters of liquid was absorbed from the bacterial solution, and measured of OD600 by a microplate reader (BioTek, USA). Colonies were grown for 96 h on R2YE agar and prepared for transmission electron microscopy of thin sections. Each specimen was examined with a Hitachi HT7700 (Japan) transmission electron microscope.
Erythromycin determination
Erythromycin was extracted from the 50 ml TSB cultures for 120 h. Erythromycin concentration was measured using an Agilent 1100 HPLC System with a C18 column (5 μm, 250 × 4.6 mm), which was equilibrated with 45% solution A (K2HPO4, 30 mM, pH 8.0) and 55% solution B (acetonitrile). An isocratic program was carried out at a flow rate of 1 ml·min−1 using a UV detector at 215 nm.
AcP measurement
The AcP level was determined as described (24). The luminescence was determined with microplate reader (Biotek, USA). A 100 μl clarified sample was mixed with 1 μl MgCl2 (100 mM), 30 μl ADP (100 μM), and 1.5 μl acetate kinase (0.4 μg/μl). After incubation at 37°C for 90 min, 50 μl of reaction solution was mixed with 50 μl of CellTiter-Glo (Promega) reagent. ATP (0.1–10 μM) was used for the standard curve.
C-di-GMP quantification
Cell cultures of 5 mL were harvested by centrifugation at 4000 g and 4°C for 10 min and washed with 1 × PBS twice. Cells were then harvested by centrifugation and resuspended in 2 ml of PBS buffer. Sonication was used to disrupt the cells, and cell debris were removed by centrifugation at 7200 g for 10 min. The supernatants were used for c-di-GMP quantification with the ELISA kits (LAMI, Shanghai). The data were obtained by a microplate reader (BioTek, USA).
Biolayer interferometry (BLI) assay
The biosensor, Streptavidin (SA) purchased from ForteBio, was used in this work. The loading buffer (pH 8.0) contained 10 mM HEPES, 2 mM MgCl2, 0.1 mM EDTA and 200 mM KCl, and the running buffer contained an extra 10 μg/ml BSA and 0.02% Tween-20. The biotin-labeled DNA probe used was the same as the EMSAs. The DNA probe was stored in loading buffer, and His-tagged BldD was stored in running buffer during the BLI assay with SA sensors. Samples were then detected within the OptiPlate-96 Black Opaque (PerkinElmer).
ITC experiment
Purified BldD protein was titrated with c-di-GMP in ITC200 All samples were prepared in the PBS buffer (pH 8.0). Typically, the titrant concentration in the syringe was 100–300 μM, and the titrant concentration in the reaction cell was 10–50 μM. Titration was conducted at 25°C using a multiple injection method with 150 s intervals. The obtained data were integrated, corrected, and analyzed using the MicroCal PEAQ-ITC Analysis Software with a single-site binding model.
RESULTS
The master regulator BldD was acetylated both in vivo and in vitro
Acetylation dynamically changes in accordance with intracellular carbon and nitrogen availability, which affects signal transduction or transcription through its various substrates, including metabolic enzymes as well as transcriptional regulators (25). To test whether BldD is a substrate of acetylation, the acetylation status of BldD protein expressed by S. erythraea was immunoprecipitated with anti-BldD antibody and subjected to western blotting analysis. His-tagged BldD overexpressed in E. coli was used as a control. As shown in Figure 1B, BldD from S. erythraea was significantly acetylated, whereas no obvious acetylation was found in the BldD protein from E. coli. In vitro acetylation was also tested using the endogenous acyltransferase AcuA identified previously (20) and acetyl-phosphate (AcP). As shown in Figure 1C and D, BldD could be acetylated via enzymatic acetylation catalyzed directly by AcuA as well as nonenzymatic acetylation with AcP in vitro. These results demonstrated that the BldD protein was a new acetylation substrate both in vitro and in vivo.
AcP-induced acetylation of BldD strongly impairs its DNA-binding activity and interaction with c-di-GMP
BldD binds a tetrameric c-di-GMP to the C-terminal domain, which in turn drives or enhances BldD dimerization. The dimerized BldD binds to its target promoters to repress a broad regulon of sporulation- and secondary metabolism-related genes, including even bldD itself (26). To investigate the effect of acetylation on the DNA-binding activity of the BldD regulator, electrophoretic mobility shift assays (EMSAs) were performed using the BldD protein after acetylation with AcuA and AcP. As shown in Figure 1E, F and G, AcuA-acetylated BldD (BldDAcuA) performed similarly to unacetylated BldD, while AcP-acetylated BldD (BldDAcP) showed significantly weak DNA binding, as evidenced by the faint mobility shift. The results suggested that AcP-dependent acetylation might be the major resulting effect on the DNA-binding activity of BldD. To verify this, the BldD, BldDAcuA and BldDAcP proteins were examined by circular dichroism assay. The far-UV circular dichroism spectra showed decreases in ellipticity at 195–220 nm, indicating an increase in the α-helical content of BldDAcuA and BldDAcP and suggesting that acetylation altered the secondary structure of BldD (Figure 1H). Moreover, AcP-dependent acetylation caused a more remarkable influence on the BldD structure with significantly higher α-helicity and a concomitant decrement in antiparallel and random coil structures compared with BldDAcuA (Figure 1H, Supplementary Table S1). The BldD and BldDAcP proteins were then subjected to a chemical cross-linking assay, and less BldD became cross-linked in an acetylation time-dependent manner in the presence of AcP (Figure 1I). These results indicated that AcP-dependent acetylation dominated the impairment of BldD DNA-binding activity. Similarly, the interaction between BldDAcP and c-di-GMP detected by isothermal titration calorimetry (ITC) experiments (Figure 1J) also showed a ∼15-fold drop in the c-di-GMP-binding affinity compared to BldD (KD from 0.57 to 7.63 μM). This conclusion was further evaluated in that AcP-dependent acetylation functioned as the major mechanism inhibiting both the DNA-binding activity and the interaction with c-di-GMP of BldD.
BldD is inactive mainly upon acetylation of K11
To determine the acetylation sites of BldD proteins, the in vitro and in vivo acetylated BldD proteins were subjected to trypsin digestion, the resulting peptides were analyzed by tandem mass spectrometry. As listed in Table 1 and Supplementary Figures S2-14, five acetylated peptides containing K35, K53, K83, K98 and K119 were identified in AcuA-dependent acetylated BldD, and six acetylated peptides containing K6, K11, K35, K53, K83, K98 and K119 were identified in AcP-dependent acetylated BldD, while only one peptide, ALGGKLR containing K11, was identified in endogenous BldD. The main reason for the single K11 identified to be acetylated in vivo might be due to the low stoichiometric acetylation of other sites, as acetylation dynamic changes over time in response to the nutritional status of the bacterial environment. K11 is located in the N-terminal domain, which mediates DNA binding and dimerization. In addition, the α1 helix containing K11 participates in the formation of a hydrophobic core and forms a structural scaffold that anchors the HTH DNA-binding motif consisting of α2 and α3 helices (27).
Table 1.
The acetylated sites of BldD under the different conditions
| In vitro | |||
|---|---|---|---|
| Protein BldD | In vivo | Enzymatic (AcuA) | Non-enzymatic (AcP) |
| K11 | K35, K53, K83, K98, K119 | K6, K11, K35, K53, K83, K98, K119 | |
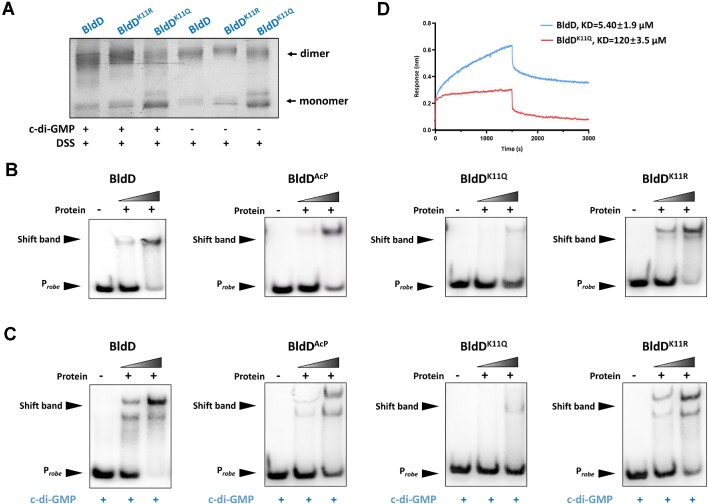
We next investigated whether K11 was involved in the DNA-binding activity or the interaction with c-di-GMP. By introducing substitutions at the acetylated site K11 to generate variants BldDK11Q and BldDK11R based on the principle that glutamine (Q) can serve as a structural mimic for acetyl-lysine and arginine (R) serves as the genetic mimic of unacetylated lysine (28). In the absence of c-di-GMP, we found that BldDK11R showed equivalent or similar intensities of cross-linking bands comparable to those obtained for BldD (Figure 2A). However, BldDK11Q mutants displayed a significantly reduced dimer form and an increased monomer form, a similar result was observed in the presence of c-di-GMP, such that BldDK11Q abolished the enhancement of c-di-GMP effects on the BldD dimer (Figure 2A). These results revealed that acetylation of the K11 residue was required for BldD dimerization and its interaction with c-di-GMP. The EMSA (Figure 2B and C) led to the same conclusion: BldDK11R performed similarly to unacetylated BldD, and c-di-GMP strongly induced BldD binding to DNA, consistent with a previous report that c-di-GMP enhances the DNA binding activity of BldD (17,29). While the BldDK11Q showed significantly weak DNA-binding as evidenced by the faint mobility shift. To confirm this, we measured the KD between BldD/BldDK11Q and DNA using biolayer interferometry assay. As Figure 2D showed, the BldD-DNA had a binding affinity of KD ∼5.4 μM, whereas the KD was ∼120 μM, a >22-fold increase for BldDK11Q-DNA, which was consistent with the data shown in Figure 2B. These results indicated that BldD is inactive mainly upon AcP-dependent acetylation at K11.
Figure 2.
BldD is inactive mainly upon acetylation of K11. (A) Crosslinking of BldD and its mutants with or without c-di-GMP. (B) EMSA of BldD and its mutants binding to the bldD gene promoter in the absence of c-di-GMP. (C) EMSA of BldD and its mutants binding to the bldD gene promoter with 1 μM c-di-GMP. (D) Biolayer interferometry assay of purified His-BldD and BldDK11Q with the bldD gene promoter.
Acetylation of K11 inhibits the global outcome of BldD binding to its targeted genes
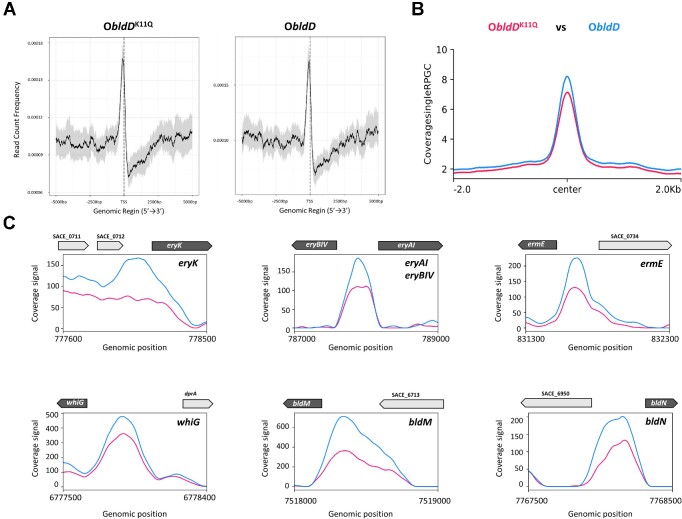
The observed association between acetylation of K11 and BldD function suggested a potential effect of K11 acetylation on the target genes and the related metabolic pathways in vivo. To address this, we constructed S. erythraea strains overproducing mutant BldD and native BldD using the E. coli–S. erythraea integrative shuttle vector pIB139 (22). The corresponding strains ObldDK11Q, ObldDK11R and ObldD were obtained. The ObldDK11Q and ObldD strain used as a control were selected and subjected to the ChIP-seq experiments using a specific anti-BldD antibody. The total (nonimmunoprecipitated) input DNA from each strain was also subjected to sequencing. Analysis of the ChIP-seq data revealed that both ObldDK11Q and ObldD signals were widely distributed at transcription start sites (TSSs) with a sharp single peak (see Figure 3A). The total detected peaks showed downregulated binding signals in the ObldDK11Q strain compared with the ObldD strain (Figure 3B). Collectively, representative results from the visualization and verification of the related genes during exposure to BldDK11Q overexpression illustrate the ChIP-seq peak changes at the individual gene level (Figure 3C). Among the genes that were most severely affected by K11 acetylation of BldD, several are involved in morphological differentiation including bldM, bldN and whiG, and antibiotic production, such as ermE, eryK, eryAI and eryBIV (Figure 3C). To verify that the effects were caused by BldD acetylation, the growth behavior and bldD transcription levels of the WT, ObldDK11Q and ObldD strains were tested. There were no differences observed in bacterial growth, the bldD transcription levels in the ObldDK11Q and ObldD strains were comparable and significantly higher than the transcription of background bldD (Supplementary Figure S1A and B). Thus, we concluded that acetylation of K11 inhibits the global outcome of BldD binding to its targets, which might therefore contribute to the changes in morphological differentiation and erythromycin production.
Figure 3.
ChIP-seq analysis of BldD targets in S. erythraea. (A) Heatmaps of the ChIP-seq signal density at the peak center and TSSs (±5 kb). (B) ChIP-seq signal in S. erythraea ObldDK11Q and ObldD strains. Color-coding of the ChIP samples is as follows: ObldD (blue) and ObldDK11Q (red). (C) ChIP-seq data for the indicated BldD-targeted promoters. Color-coding of the ChIP samples is as follows: ObldD (blue) and ObldDK11Q (red). Plots span ∼2 kb of DNA sequence.
K11 acetylation enhances development and inhibits antibiotic production
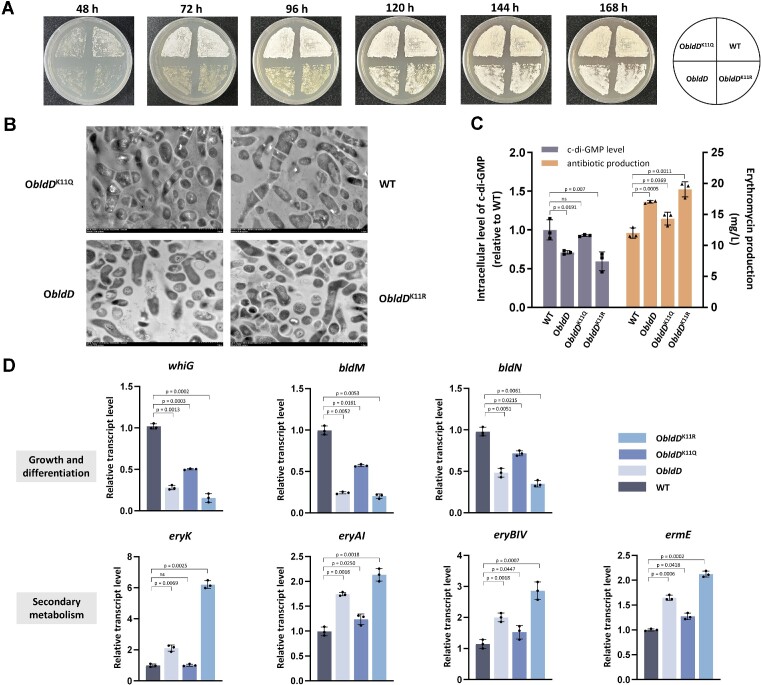
BldD is a repressor of development in actinobacteria (26). To further examine the function of K11 acetylation, we used S. erythraea WT, ObldD, ObldDK11Q and ObldDK11R strains and cultivated on R2YE agar (23) at 30°C. In comparison with WT, ObldD turned white in appearance later, suggesting notably delayed differentiation and sporulation, and the ObldDK11R strain behaved similarly to ObldD (Figure 4A). In contrast, ObldDK11Q displayed accelerated differentiation and sporulation, which was nearly identical to that of the WT strain (Figure 4A). Detailed transmission electron microscopy (TEM) examination further confirmed that K11 acetylation inactivates BldD to release the negative regulatory role of BldD in development (Figure 4B). Since BldD coupled with c-di-GMP-mediated regulation directly controls all the promoters in the biosynthetic gene cluster for the clinical antibiotic erythromycin (30) in addition to its role in regulating morphological differentiation in S. erythraea, the influence of K11 acetylation on antibiotic production was determined by HPLC and bioassays. We found that ObldDK11Q led to an obvious reduction in erythromycin compared with ObldD (Figure 4C and Supplementary Figure S1C), indicating that K11 acetylation abolished the positive effect of BldD overexpression on antibiotic biosynthesis. However, ObldDK11R relieved acetylation inhibition and relatively increased erythromycin production (Figure 4C and Supplementary Figure S1C). Since diguanylate cyclase has emerged as a direct target of BldD (26), the intracellular c-di-GMP level was detected and exhibited an increase in ObldDK11Q and a decrease in ObldDK11R compared with ObldD (Figure 4C), which revealed that K11 acetylation directly influenced the c-di-GMP concentration in vivo. The relationship between BldD acetylation and differentiation/antibiotic production was then verified by real-time RT-PCR monitoring the transcription of selected genes from ChIP-seq. As expected, the repression of the known genes that play important roles in development, including whiG, bldN and bldM, was enhanced in ObldDK11R, while overexpression of BldDK11Q relieved the repression caused by BldD (Figure 4D). Expression of the ery cluster, including eryK, eryAI, eryBIV and ermE, was improved in ObldDK11R and almost completely lost in response to BldD-induced activation in ObldDK11Q (Figure 4D). The growth behaviors and bldD transcription were analyzed and showed that there were no differences in growth behaviors among the S. erythraea WT, ObldD, ObldDK11Q, and ObldDK11R strains (Supplementary Figure S1A), while bldD transcription in the ObldD, ObldDK11Q, and ObldDK11R strains appeared to be relatively synchronized ∼3.5-fold increase compared with that in the WT strain (Supplementary Figure S1B). The results further confirmed that the differences in development and antibiotic production among the detected strains were indeed caused by the acetylation status of BldD at K11. In sum, these observed data fit the results obtained in Figure 3 and revealed that repression of binding with the BldD box in ObldDK11Q indeed accelerated morphological differentiation and inhibited antibiotic production.
Figure 4.
K11 acetylation enhances BldD-mediated development and inhibits antibiotic production. (A) Phenotypic observation of the S. erythraea WT, ObldD, ObldDK11Q and ObldDK11R strains cultivated on R2YE at 30°C. (B) TEM examination of S. erythraea WT, ObldD, ObldDK11Q and ObldDK11R strains grown for 96 h on R2YE at 30°C. (C) Intracellular c-di-GMP levels and antibiotic production in cell extracts of S. erythraea WT, ObldD, ObldDK11Q and ObldDK11R strains grown in TSB medium. (D) The transcription levels of the indicated genes in S. erythraea WT, ObldD, ObldDK11Q and ObldDK11R strains grown in TSB medium till the middle exponential phase. Fold change represents the expression level compared to the WT strain. Error bars show the SDs of three independent experiments.
Nitrogen starvation elevated the intracellular AcP concentration, thereby increasing the acetylation level of BldD at K11
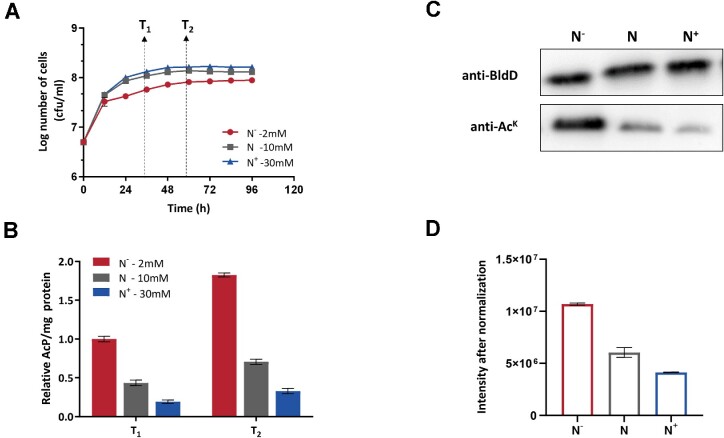
Protein acetylation is considered a consequence of carbon overflow or an environmental carbon-nitrogen imbalance (24,31). We thus considered the possibility that nitrogen starvation might exert a general effect on the in vivo acetylation level of BldD, that is, this nutrient stress appeared to alert the intracellular AcP pool, based on our previous observation that only K11 was identified to be acetylated in vivo and that its acetylation occurred in an AcP-dependent manner (Table 1). To investigate this speculation, the S. erythraea WT strain was cultured in either nitrogen-limited (N−) or nitrogen-rich (N+) media, and the standard nitrogen condition (N) was used as a control (glucose as the carbon source). As shown in Figure 5A, the growth of the WT was positively associated with nitrogen availability, with a significant reduction when cultured in N−, while a promotion was observed with N+. Cells were harvested at the middle exponential (T1) and early stationary phases (T2), and the intracellular AcP was determined, respectively. Both the T1 and T2 phases showed an evidently 5-fold higher intracellular AcP concentration under nitrogen limitation than under nitrogen-rich conditions, and this difference was even more dramatic during the T2 phase (Figure 5B). The acetylation level of BldD protein immunoprecipitated during the T2 phase was then analyzed. As expected, nitrogen limitation led to a markedly higher acetylation level of BldD (Figure 5C), consistent with the higher intracellular AcP level observed in Figure 5B. Accordingly, quantitative mass spectrometry further confirmed that the acetylation level of BldD under nitrogen limitation was nearly 3-fold higher than that under nitrogen excess conditions at K11 (Figure 5D). BldD acetylation at different growth stage was also detected and presented accumulation after entering into stationary phase (Supplementary Figure S15), which is consistent with the AcP level shown in Figure 5B. These results are in line with the model that nitrogen starvation resulted in intracellular AcP accumulation, thus inducing hyperacetylation of K11, which exerted a synergistic regulatory effect on the bldD regulon in S. erythraea.
Figure 5.
Nitrogen starvation increased acetylation of BldD at K11. (A) Growth curves of S. erythraea WT cultured in minimal Evans medium with standard nitrogen (N) and excess/limited nitrogen (N+/N−). ‘T1’ and ‘T2’ denote the middle exponential and early stationary phases, respectively. (B) Quantification of intracellular AcP concentration in the S. erythraea WT strain at N/N+/N− conditions. ‘T1’ and ‘T2’ denote the middle exponential and early stationary phases, respectively. (C) Acetylation level of BldD of the S. erythraea WT strain under N/N+/N− conditions at the T2 phase. Each lane was loaded with equal amount of BldD protein. (D) Acetylation level of BldD at K11 under N/N+/N− conditions in the T2 phase. Areas under the curve (AUCs) of the precursor ion peak were used to evaluate the peptide intensity. Unmodified peptide (AVVVGSYER) was used for protein level normalization. The ratios of the acylated peptides were normalized to the ratios of their corresponding protein levels. Normalized ratios of the peptides were used for analysis.
The cooperation between AcP-induced acetylation and c-di-GMP-induced multimerization is conserved in actinobacteria
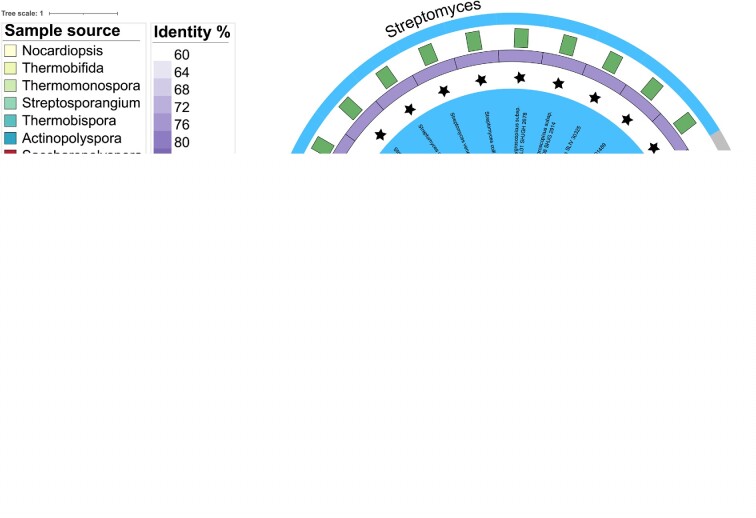
Very few of the Bld regulators are conserved across actinomycetes, but BldD is an exception (26). BldD exists throughout sporulating actinomycetes, and its c-di-GMP binding motif is perfectly conserved in all these homologs. The fact that BldD/c-di-GMP controls key developmental processes throughout sporulating actinomycetes raised the possibility that a similar interaction between AcP-induced acetylation of BldD and c-di-GMP might exist in actinobacteria. To complement these evolutionary insights, BldD homologs from 29 species, including SCO1489 from S. coelicolor, SLIV_30325 from S. lividans, SAVERM_6861 from S. avermitilis, and AMIS62190 from non-Streptomyces actinobacteria Actinoplanes missouriensis (32) as representative actinobacteria, were screened for phylogenetic survey. Indeed, according to the alignment of the S. erythraea BldD sequence with its homologs, it is notable that the K11 was widespread in the BldD sequences across actinobacteria, particularly for streptomyces species (Figure 6). We therefore speculated that K11 acetylation of BldD contributed to the changes in morphological differentiation or antibiotic production and would be comparatively conserved in streptomycetes and some other genera of actinobacteria.
Figure 6.
Phylogenetic tree of the BldD protein within actinomycetes. Sequences of BldD proteins were aligned with MAFFT v7.489 with L-INS-I. Trees were reconstructed from the resulting alignments using FastTree and subsequently visualized with the interactive Tree of Life (iTol) Version 6.5.6. The black star represents species containing the conserved K11 site, and the gray bar represents the aa length of the BldD protein. Left panels show the genus names analyzed in the phylogenetic survey and the identity of the S. erythraea BldD sequence aligned with the corresponding homologs.
DISCUSSION
For all domains of life, the core pathways composed of anabolism and catabolism generate a multitude of different metabolites. The potential chemical reactions in which these metabolites can participate greatly exceed cellular metabolic requirements. Therefore, the cell has evolved diverse mechanisms to control these extrametabolic compounds to limit their toxicity or the damage imparted upon macromolecular structures via nonenzymatic reactions (33). It is well established that bacteria respond to environmental challenges with appropriate regulation of transcriptional and translational programs (34). Compared to time- and energy-intensive transcription and translation processes, posttranslational control over functional proteins can alter cellular physiology rapidly and permit fitness or survival in otherwise suboptimal or lethal conditions through diverse covalent chemical modifications. In addition to enzyme-dependent acetylation, accumulated evidence has shown that nonenzymatic acetylation induced by AcP is also assessed in a few bacteria, such as E. coli (24), B. subtilis (35), and Neisseria gonorrhoeae (36). In the current study, we show that in actinobacteria, AcP is the predominant acetyl donor during carbon/nitrogen imbalance conditions and exerts posttranscriptional regulation on the activity of the master regulator BldD, which exerts a global impact on actinobacterial physiology. Notably, we found that AcP-dependent acetylation of BldD led to loss of function in dimerization as well as interaction with c-di-GMP, which caused weakened BldD–DNA binding and inhibited its transcriptional activity.
The erection of aerial hyphae and sporulation are the most critical developmental transitions in the life cycle of filamentous actinobacteria. Recent advances have illuminated that these developmental transitions are controlled through the regulatory network of specific regulators, in which BldD acts as a top regulator of a developmental hierarchy and a key regulator connecting stress to developmental processes. BldD represses the transcription of genes participating in aerial hyphae and sporulation, such as bldN and whiG (14,15,17). Generally, transcriptional repressors are regulated by a modulatory partner that induces posttranslational modification, dissociation into monomers, and/or regulatory domain cleavage of the oligomer. Our findings elucidated a fine tuning of the molecular regulation mechanism of BldD activity by alternative small molecule metabolites (AcP or c-di-GMP) or the cooperation between both of them, determining when and how repression is relieved to protect the cell metabolic balance.
BldD is a homodimeric protein with each subunit predominantly folded into two independent domains (37), and there is no significant interdomain interaction observed between the two distinct domains in the BldD dimer (38). The N-terminal domain (residues 1–79) belongs to the Xenobiotic Response Element (XRE) family and is responsible for DNA binding through the helix-turn-helix motif (27,38). The C-terminal domain (residues 80–167) is involved in the protein–protein interactions that provide transcriptional regulation in developmental regulation (39). Another important functional role of the C-terminal domain is binding a tetrameric c-di-GMP to the RxD-X8-RxxD motif to drive BldD dimerization and the consequent transcriptional repression of a broad regulon of sporulation genes (15). The highly conserved K11 identified in this study suggests that the N-terminal domain of BldD could be involved in the BldD-c-di-GMP interaction.
According to our results, acetylation on K11 of BldD governed the inactivation of BldD, leading to derepression of the BldD regulon of sporulation genes, thereby accelerating morphological differentiation. Protein acetylation is thought to be a consequence of intracellular AcP accumulation induced by C-N imbalance, including carbon overflow or nitrogen limitation (6,7,24,31,40–43). We propose that defining this signal integration through BldD acetylation to facilitate morphological differentiation might be a safety net to prevent major damage to actinobacteria under extreme nutritional or environmental conditions.
Furthermore, BldD is also a key activator involved in antibiotic production, including actinorhodin, undecylprodigiosin, methylenomycin, and calcium-dependent antibiotics of S. coelicolor (44), erythromycin of S. erythraea (30), daptomycin of S. roseosporus (13), and avermectin of S. avermitilis (14). In this context, it is plausible that repression of BldD acetylation on its regulon of secondary metabolite biosynthesis genes and antibiotic production provided K11 of BldD as a target for the posttranslational modification-metabolic engineering strategy (41). The result obtained from ObldDK11R indeed showed a much higher erythromycin yield. However, it should be noted that the implications of this may differ slightly between the species and need further investigation.
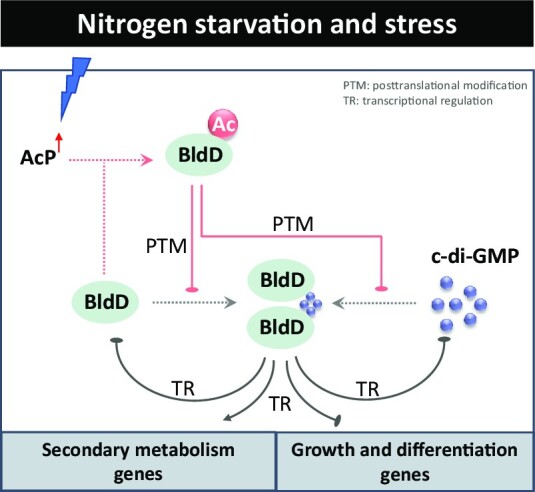
In summary, this study reveals an unusual feature of acetylation assembly responses to nutritional stress and c-di-GMP signals via K11 of BldD, which formed a coherent and conserved regulatory network (Figure 7), drawing new insight into the direct connections from specific regulators to the biological process associated with major morphogenetic events. In this regard, our findings are conducive to a comprehensive understanding of BldD-mediated developmental regulation in actinobacteria.
Figure 7.
Crosstalk of c-di-GMP and AcP integrates signal transduction from nutrient sensing to morphological differentiation and secondary metabolism in streptomycetes and some other genera of actinobacteria.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: Di You and Bang-Ce Ye designed research; Yu Fu and Jin-Long Shen performed research; Yu Fu analyzed data; and Di You, Bin-Cheng Yin and Bang-Ce Ye wrote the paper.
Contributor Information
Yu Fu, Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai 200237, China.
Yu-Qi Dong, Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai 200237, China.
Jin-Long Shen, Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai 200237, China.
Bin-Cheng Yin, Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai 200237, China.
Bang-Ce Ye, Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai 200237, China; Institute of Engineering Biology and Health, Collaborative Innovation Center of Yangtze River Delta Region Green Pharmaceuticals, College of Pharmaceutical Sciences, Zhejiang University of Technology, Hangzhou 310014, Zhejiang, China.
Di You, Laboratory of Biosystems and Microanalysis, State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai 200237, China.
Data Availability
ChIP-seq raw data are available in the GEO database (accession ID: GSE225241), which are accessible with the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE225241.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2020YFA0909100]; National Natural Science Foundation of China [32070066, 31730004]. Funding for open access charge: National Key Research and Development Program of China; National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Procopio R.E., Silva I.R., Martins M.K., Azevedo J.L., Araujo J.M.. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012; 16:466–471. [DOI] [PubMed] [Google Scholar]
- 2. Martin J.F., Liras P.. The balance metabolism safety net: integration of stress signals by interacting transcriptional factors in streptomyces and related actinobacteria. Front. Microbiol. 2019; 10:3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liao C.H., Yao L., Xu Y., Liu W.B., Zhou Y., Ye B.C.. Nitrogen regulator GlnR controls uptake and utilization of non-phosphotransferase-system carbon sources in actinomycetes. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:15630–15635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu Y., You D., Ye B.C.. Nitrogen regulator GlnR directly controls transcription of genes encoding lysine deacetylases in Actinobacteria. Microbiology-Sgm. 2017; 163:1702–1710. [DOI] [PubMed] [Google Scholar]
- 5. Y X., D Y., LL Y., X C., BC Y.. Phosphate regulator PhoP directly and indirectly controls transcription of the erythromycin biosynthesis genes in Saccharopolyspora erythraea. Microb. Cell Fact. 2019; 18:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. You D., Wang M.M., Ye B.C.. Acetyl-CoA synthetases of Saccharopolyspora erythraea are regulated by the nitrogen response regulator GlnR at both transcriptional and post-translational levels. Mol. Microbiol. 2017; 103:845–859. [DOI] [PubMed] [Google Scholar]
- 7. You D., Yin B.C., Li Z.H., Zhou Y., Yu W.B., Zuo P., Ye B.C.. Sirtuin-dependent reversible lysine acetylation of glutamine synthetases reveals an autofeedback loop in nitrogen metabolism. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:6653–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. You D., Zhang B.Q., Ye B.C.. GntR family regulator DasR controls acetate assimilation by directly repressing the acsA gene in Saccharopolyspora erythraea. J. Bacteriol. 2018; 200:e00685-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu Y., Li Y.X., Ye B.C.. Lysine propionylation modulates the transcriptional activity of phosphate regulator PhoP in Saccharopolyspora erythraea. Mol. Microbiol. 2018; 110:648–661. [DOI] [PubMed] [Google Scholar]
- 10. Flardh K., Buttner M.J.. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 2009; 7:36–49. [DOI] [PubMed] [Google Scholar]
- 11. Del Carratore F., Hanko E.K., Breitling R., Takano E.. Biotechnological application of Streptomyces for the production of clinical drugs and other bioactive molecules. Curr. Opin. Biotechnol. 2022; 77:102762. [DOI] [PubMed] [Google Scholar]
- 12. Li J., Wang N., Tang Y., Cai X., Xu Y., Liu R., Wu H., Zhang B.. Developmental regulator BldD directly regulates lincomycin biosynthesis in Streptomyces lincolnensis. Biochem. Biophys. Res. Commun. 2019; 518:548–553. [DOI] [PubMed] [Google Scholar]
- 13. Makitrynskyy R., Tsypik O., Nuzzo D., Paululat T., Zechel D.L., Bechthold A.. Secondary nucleotide messenger c-di-GMP exerts a global control on natural product biosynthesis in streptomycetes. Nucleic Acids Res. 2020; 48:1583–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan H., Lu X., Sun D., Zhuang S., Chen Q., Chen Z., Li J., Wen Y.. BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species. Mol. Microbiol. 2020; 113:123–142. [DOI] [PubMed] [Google Scholar]
- 15. den Hengst C.D., Tran N.T., Bibb M.J., Chandra G., Leskiw B.K., Buttner M.J.. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 2010; 78:361–379. [DOI] [PubMed] [Google Scholar]
- 16. Elliot M.A., Leskiw B.K.. The BldD protein from Streptomyces coelicolor is a DNA-binding protein. J. Bacteriol. 1999; 181:6832–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tschowri N., Schumacher M.A., Schlimpert S., Chinnam N.B., Findlay K.C., Brennan R.G., Buttner M.J.. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell. 2014; 158:1136–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hull T.D., Ryu M.H., Sullivan M.J., Johnson R.C., Klena N.T., Geiger R.M., Gomelsky M., Bennett J.A.. Cyclic Di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces coelicolor. J. Bacteriol. 2012; 194:4642–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tran N.T., Den Hengst C.D., Gomez-Escribano J.P., Buttner M.J.. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J. Bacteriol. 2011; 193:3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. You D., Yao L.L., Huang D., Escalante-Semerena J.C., Ye B.C.. Acetyl coenzyme a synthetase is acetylated on multiple lysine residues by a protein acetyltransferase with a single Gcn5-Type N-Acetyltransferase (GNAT) domain in Saccharopolyspora erythraea. J. Bacteriol. 2014; 196:3169–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu J.Y.X.Y., Xu Z., Zhai L.H., Ye Y., Zhao Y.M., Chu X.H., Tan M.J., Ye B.C.. Protein acylation is a general regulatory mechanism in biosynthetic pathway of Acyl-CoA-derived natural products. Cell Chem. Biol. 2018; 25:984–995. [DOI] [PubMed] [Google Scholar]
- 22. Wilkinson C.J., Hughes-Thomas Z.A., Martin C.J., Bohm I., Mironenko T., Deacon M., Wheatcroft M., Wirtz G., Staunton J., Leadlay P.F.. Increasing the efficiency of heterologous promoters in actinomycetes. J. Mol. Microbiol. Biotechnol. 2002; 4:417–426. [PubMed] [Google Scholar]
- 23. Thompson C.J., Ward J.M., Hopwood D.A.. DNA cloning in Streptomyces: resistance genes from antibiotic-producing species. Nature. 1980; 286:525–527. [DOI] [PubMed] [Google Scholar]
- 24. Weinert B.T., Iesmantavicius V., Wagner S.A., Scholz C., Gummesson B., Beli P., Nystrom T., Choudhary C.. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell. 2013; 51:265–272. [DOI] [PubMed] [Google Scholar]
- 25. Christensen D.G., Xie X., Basisty N., Byrnes J., McSweeney S., Schilling B., Wolfe A.J.. Post-translational protein acetylation: an elegant mechanism for bacteria to dynamically regulate metabolic functions. Front. Microbiol. 2019; 10:1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bush M.J., Tschowri N., Schlimpert S., Flardh K., Buttner M.J.. c-di-GMP signalling and the regulation of developmental transitions in streptomycetes. Nat. Rev. Microbiol. 2015; 13:749–760. [DOI] [PubMed] [Google Scholar]
- 27. Kim I.K., Lee C.J., Kim M.K., Kim J.M., Kim J.H., Yim H.S., Cha S.S., Kang S.O.. Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2). Mol. Microbiol. 2006; 60:1179–1193. [DOI] [PubMed] [Google Scholar]
- 28. Ren Q., Gorovsky M.A.. Histone H2A.Z acetylation modulates an essential charge patch. Mol. Cell. 2001; 7:1329–1335. [DOI] [PubMed] [Google Scholar]
- 29. Schumacher M.A., Zeng W., Findlay K.C., Buttner M.J., Brennan R.G., Tschowri N.. The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex. Nucleic Acids Res. 2017; 45:6923–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chng C., Lum A.M., Vroom J.A., Kao C.M.. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:11346–11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schilling B., Basisty N., Christensen D.G., Sorensen D., Orr J.S., Wolfe A.J., Rao C.V.. Global lysine acetylation in Escherichia coli results from growth conditions that favor acetate fermentation. J. Bacteriol. 2019; 201:e00768-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mouri Y., Konishi K., Fujita A., Tezuka T., Ohnishi Y.. Regulation of sporangium formation by BldD in the rare Actinomycete Actinoplanes missouriensis. J. Bacteriol. 2017; 199:e00840-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dickinson B.C., Chang C.J.. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 2011; 7:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gottesman S. Stress reduction, bacterial style. J. Bacteriol. 2017; 199:e00433-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suzuki S., Kondo N., Yoshida M., Nishiyama M., Kosono S.. Dynamic changes in lysine acetylation and succinylation of the elongation factor Tu in Bacillus subtilis. Microbiology. 2019; 165:65–77. [DOI] [PubMed] [Google Scholar]
- 36. Post D.M.B., Schilling B., Reinders L.M., D'Souza A.K., Ketterer M.R., Kiel S.J., Chande A.T., Apicella M.A., Gibson B.W.. Identification and characterization of AckA-dependent protein acetylation in Neisseria gonorrhoeae. PLoS One. 2017; 12:e0179621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Elliot M., Locke T., Galibois C., Leskiw B.. BldD from Streptomyces coelicolor is a non-essential global regulator that binds its own promoter as a dimer. FEMS Microbiol. Lett. 2003; 225:35–40. [DOI] [PubMed] [Google Scholar]
- 38. Lee C.J., Won H.S., Kim J.M., Lee B.J., Kang S.O.. Molecular domain organization of BldD, an essential transcriptional regulator for developmental process of Streptomyces coelicolor A3(2). Proteins. 2007; 68:344–352. [DOI] [PubMed] [Google Scholar]
- 39. Kim J.M., Won H.S., Kang S.O.. The C-terminal domain of the transcriptional regulator BldD from Streptomyces coelicolor A3(2) constitutes a novel fold of winged-helix domains. Proteins. 2014; 82:1093–1098. [DOI] [PubMed] [Google Scholar]
- 40. Christensen D.G., Orr J.S., Rao C.V., Wolfe A.J.. Increasing growth yield and decreasing acetylation in Escherichia coli by optimizing the carbon-to-magnesium ratio in peptide-based media. Appl. Environ. Microbiol. 2017; 83:e03034-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. You D., Wang M.M., Yin B.C., Ye B.C.. Precursor supply for erythromycin biosynthesis: engineering of propionate assimilation pathway based on propionylation modification. ACS Synth. Biol. 2019; 8:371–380. [DOI] [PubMed] [Google Scholar]
- 42. Zhang B.Q., Bu H.L., You D., Ye B.C.. Acetylation of translation machinery affected protein translation in E. coli. Appl. Microbiol. Biotechnol. 2020; 104:10697–10709. [DOI] [PubMed] [Google Scholar]
- 43. Zhang B.Q., Chen Z.Q., Dong Y.Q., You D., Zhou Y., Ye B.C.. Selective recruitment of stress-responsive mRNAs to ribosomes for translation by acetylated protein S1 during nutrient stress in Escherichia coli. Commun. Biol. 2022; 5:892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elliot M., Damji F., Passantino R., Chater K., Leskiw B.. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J. Bacteriol. 1998; 180:1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq raw data are available in the GEO database (accession ID: GSE225241), which are accessible with the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE225241.