Abstract
All mycobacteria studied to date have an rRNA operon, designated rrnA, located downstream from a single copy of the murA gene, which encodes an enzyme (EC 2.5.1.7) important for peptidoglycan synthesis. The rrnA operon has a promoter, P1(A), located within the coding region of murA, near the 3′ end. Samples of RNA were isolated from Mycobacterium tuberculosis at different stages of the growth cycle and from Mycobacterium smegmatis grown under different conditions. RNase protection assays were used to investigate transcripts of both murA and rrnA. Transcription of murA was found to continue into the 16S rRNA gene, as if murA and rrnA form a hybrid (protein coding-rRNA coding) operon. During the growth of M. tuberculosis, the hybrid operon contributed approximately 2% to total pre-rRNA. Analysis of M. smegmatis RNA revealed that the level of murA RNA depended on the growth rate and that the patterns of expression during the growth cycle were different for murA and rrnA. M. smegmatis has a second rRNA operon, rrnB, located downstream from a single copy of the tyrS gene, encoding tyrosyl-tRNA synthetase. Transcription of tyrS was found to continue into the 16S rRNA gene rrnB. The hybrid tyrS-rrnB operon contributed 0.2 to 0.6% to rrnB transcripts. The pattern of tyrS expression during the growth cycle matched the pattern of rrnB expression, reflecting the essential role of TyrS and rRNA in protein biosynthesis.
All the mycobacteria studied to date have either one or two rrn operons, designated rrnA and rrnB. The rrnA operon, which is present in all mycobacteria, is located downstream from the murA gene, which encodes the enzyme UDP-N-acetylglucosamine 1-carboxyvinyl transferase (EC 2.5.1.7), or MurA (10). This enzyme catalyzes the transfer of the enol ether from phosphoenolpyruvate to the 3′ OH of UDP-N-acetylglucosamine during the early stages of peptidoglycan synthesis. Mycobacterium smegmatis, which is representative of many fast-growing mycobacteria (10), has a second rrn operon (rrnB), located downstream from the tyrS gene, encoding the enzyme tyrosyl-tRNA synthetase (EC 6.1.1.1), or TyrS (8, 15). TyrS is essential for protein biosynthesis; its role is the attachment of l-tyrosine to the ribose moiety of the 3′-terminal adenosine residue of tyrosyl-tRNA.
Each of the known rrnA operons has a promoter, P1(A), which has a transcription start point (tsp) situated within the coding region of murA or no more than 3 nucleotides downstream from the stop codon (10) (Fig. 1). Thus, a region of the murA gene near the 3′ end is transcribed both as part of the murA gene and as part of the rrnA operon, raising the possibility that murA and rrnA are transcribed as a single unit. Similarly, it is possible that tyrS and rrnB are transcribed as a single unit because of the absence of a transcription terminator in the 54-bp region downstream from the 3′ end of the tyrS coding region and the tsp for rrnB (8, 15).
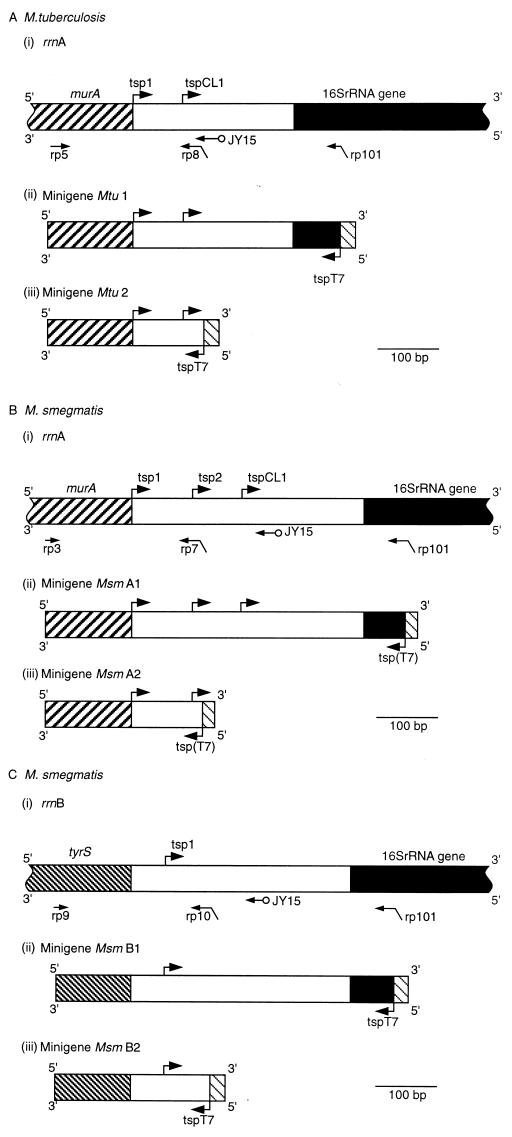
FIG. 1.
Upstream regions of mycobacterial rrn operons and constructs used in RNase protection assays. A horizontal arrow indicates the location of the binding site for the primer specified; a diagonally bent arrow indicates the location of the binding site for the primer specified (horizontal section of the arrow) and the presence of the T7 promoter sequence (angled section); a horizontal arrow with a circle at the end indicates the binding site for primer JY15, used in DNA sequencing reactions. An arrow bent at a right angle denotes the location of a tsp (vertical section) and the direction of transcription (horizontal section). ▨, murA; ▧, tyrS; ■, 16S rRNA gene; ▧, T7 promoter; tspT7 or tsp(T7), tsp for the T7 promoter. Mtu, M. tuberculosis; Msm, M. smegmatis. (A) rrnA operon of M. tuberculosis. (i) Upstream region of the rrnA operon. The scheme shows the locations of murA, tsp’s (arrows bent at right angles labelled tsp1 and tspCL1) for rrnA, binding sites for the primers used to construct the minigenes, and the binding site for primer JY15. (ii) Minigene Mtu 1 constructed by amplification of a section of the upstream region with primers rp5 and rp101. (iii) Minigene Mtu 2 constructed by amplification of a section of the upstream region with primers rp5 and rp8. The same alignment and scale (shown by the bar) are used in panels i to iii. (B) rrnA operon of M. smegmatis. (i) Upstream region of rrnA. The scheme shows the locations of murA, tsp’s (arrows labelled tsp1, tsp2, and tspCL1) for rrnB binding sites for the primers used to construct the minigenes, and the binding site for primer JY15. (ii) Minigene Msm A1 constructed by amplification with primers rp3 and rp101. (iii) Minigene Msm A2 constructed by amplification with primers rp3 and rp7. The same alignment and scale (shown by the bar) are used in panels i to iii. (C) rrnB operon of M. smegmatis. (i) Upstream region of rrnB. The scheme shows the locations of tyrS, the tsp (arrow labelled tsp1) for the rrnB operon, the binding sites for the primers used to construct the minigenes, and the binding site for primer JY15. (ii) Minigene Msm B1 constructed with primers rp9 and rp101. (iii) Minigene Msm B2 constructed with primers rp9 and rp10. The same alignment and scale (shown by the bar) are used in panels i to iii.
In this study, the RNA fraction was isolated from Mycobacterium tuberculosis and M. smegmatis at different stages of growth and from M. smegmatis grown in two different media. RNase protection assays were used to investigate the extents of coordination of the expression of murA with rrnA and of tyrS with rrnB.
MATERIALS AND METHODS
Materials.
A Sequenase (U.S. Biochemicals) sequencing kit was supplied by Cambridge Biosciences. [α-35S]dATP was obtained from Amersham. A GeneClean kit was obtained from Bio 101. Oligonucleotide primers were prepared with an automated DNA synthesizer (model 370A; Applied Biosystems).
Bacterial strains, media, and vector.
M. smegmatis NCTC 8159 (National Collection of Type Cultures) was maintained on Löwenstein-Jensen slopes and grown at 37°C with vigorous shaking in either Lemco broth (3) or Kohn-Harris glucose medium (described below) containing 0.1% Tween 80. Seed cultures of M. smegmatis for inoculation were grown in Lemco broth for 26 h. These cultures were used to inoculate medium at the rate of 20 ml of inoculum/liter. Kohn-Harris glucose medium was based (6) on the medium of Kohn and Harris (13) with glucose (5 g/liter) as the carbon source and trace elements provided at 5 ml/liter as described by Kelly and Clarke (12). M. tuberculosis H37Rv was grown in Dubos medium (7) containing 0.1% Tween 80.
Isolation of RNA.
Cells were collected, resuspended in 1 ml of guanidinium buffer (6 M guanidinium chloride, 0.1% [vol/vol] Tween 80, 10 mM EDTA, 1 mM 2-mercaptoethanol), and kept at −20°C for 15 min. The suspension was added to half the volume of heat-sterilized glass beads (0.15-mm diameter) contained in a 2-ml screw-cap microcentrifuge tube. Mycobacteria were ruptured by three pulses of 1 min each on a Mini-BeadBeater device (Biospec Products); debris and beads were sedimented by centrifugation (10,000 × g for 3 min), and the cleared lysate was retained. The pellet of beads and mycobacterial residue was briefly reextracted on the Mini-BeadBeater (30-s pulse) with 300 μl of fresh guanidinium buffer, and the resulting extract was pooled with the first. The lysate was extracted three times with 2 volumes of chloroform–3-methyl-1-butanol (24:1, vol/vol). The RNA fraction was precipitated by the addition of ethanol and redissolved in an appropriate volume of morpholinepropanesulfonic acid (MOPS) buffer, and residual DNA was removed by digestion with RNase-free DNase. The integrity of the RNA was checked by electrophoresis through formaldehyde gels.
Construction of minigenes.
Regions upstream from the 5′ end of the 16S rRNA coding region of rrnA of M. tuberculosis and rrnB of M. smegmatis were amplified by PCR with the primers shown in Table 1 used to amplify the sequences illustrated in Fig. 1. Appropriate plasmids for PCR amplification for M. tuberculosis and M. smegmatis were described previously (8). The procedures used resulted in a T7 promoter at the 3′ end of the minigene to provide a probe for RNase protection assays.
TABLE 1.
Primers used in this study
| Name | Targeta | Sequence |
|---|---|---|
| JY15 | Invariant motif of rrn leader regions (e.g., 462 to 478; X87943) | 5′CACACTATTGAGTTCTC3′ |
| rp101 | 16S rRNA coding region (e.g., 48 to 66; X58890) | 5′AATTCTAATACGACTCACTATAGGGCGACTTGCATGTGTTAAGC3′ |
| rp5 | M. tuberculosis murA (202 to 219; X87943) | 5′GGGGCCGGCTTGGTGCTG3′ |
| rp8 | M. tuberculosis rrn leader region (435 to 482; X87943) | 5′AATTCTAATACGACTCACTATAGGGGCTTGCTTGTTTCCGCCC3′ |
| rp3 | M. smegmatis murA (202 to 219; X87944) | 5′GGTGCCGGCCTGGTGCTG3′ |
| rp7 | M. smegmatis rrnA leader region (433 to 451; X87944) | 5′AATTCTAATACGACTCACTATAGGGCGCGGCGACTTCATAAAG3′ |
| rp9 | M. smegmatis tyrS (−179 to −762; U09862) | 5′GTCGAAAGCGACGAGTGG3′ |
| rp10 | M. smegmatis rrnB leader region (49 to 66; U09862) | 5′AATTCTAATACGACTCACTATAGGGGGATCCCCCACCTTAGTG3′ |
Data in parentheses show positions and GenBank accession numbers.
Transcription of minigenes.
Radiolabelled transcripts were obtained by use of a Riboprobe kit (Promega, Madison, Wis.) according to the instructions provided. Each reaction mixture contained 50 μCi of [α-32P]CTP; transcription was carried out at 37°C for 1 h, and the reaction was terminated by heating the sample at approximately 90°C for 3 min. DNase was added to the cooled reaction mixture, which was then kept at 37°C for 15 min. The products were purified by electrophoresis through a 6% (wt/vol) polyacrylamide–8 M urea sequencing gel. An appropriate band was excised from the gel, and radiolabelled RNA was recovered after the gel fragment was soaked overnight in elution buffer (0.5 M ammonium acetate, 1 mM EDTA, 0.2% [wt/vol] sodium dodecyl sulfate). The eluate was desalted and then made 0.3 M in sodium acetate, and RNA was precipitated with ethanol.
RNase protection assays.
Hybridizations were carried out with either formamide buffer [80% (vol/vol) formamide, 0.2 M sodium acetate, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.4)] at 50°C for 16 h as described by Sambrook et al. (16) or PES buffer (1 M NaCl, 1 mM EDTA, 25 mM PIPES [pH 6.8]) at 90°C for 70 min (14). Each sample contained radiolabelled probe (1 × 105 to 4 × 105 cpm) and 10 μg of M. tuberculosis RNA or 30 μg of M. smegmatis RNA.
After the hybridization step, samples were treated with RNase A and RNase T1 and then protein was removed by treatment with phenol-chloroform. RNA was precipitated, redissolved, and analyzed by electrophoresis through 6% polyacrylamide–8 M urea gels. The gels were calibrated by use of either HaeIII-digested φX174 DNA molecular size markers (Promega, Madison, Wis.) or the products of sequencing reactions generated by the methods described below. After separation by electrophoresis, radioactive products were located by autoradiography at either approximately 20°C or −70°C with an intensifying screen.
Sequencing of double-stranded DNA.
DNA sequences were determined by the dideoxy chain termination procedure with [α-35S]dATP as described by Ji et al. (11) and with primer JY15.
Measurements of radioactivity.
Quantitative measurements of radioactivity were obtained with a PhosphorImager (model 4005; Molecular Dynamics, Chesham, Buckinghamshire, United Kingdom) and the software supplied with the instrument.
RESULTS
In order to investigate the coordinated expression of murA and rrnA of both M. tuberculosis and M. smegmatis and tyrS and rrnB of M. smegmatis, RNase protection assays were carried out. The constructs used to generate two sets of appropriate radioactive probes are illustrated in Fig. 1. One set was designed to detect the 3′ end of M. tuberculosis murA RNA (Fig. 2A) and the 3′ ends of murA RNA and tyrS RNA of M. smegmatis (Fig. 3A). 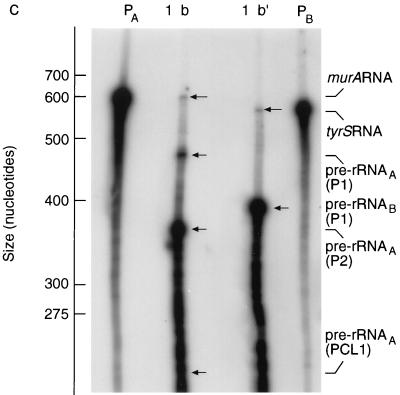 The second set was designed to examine the expression of murA and tyrS and to relate the abundance of their transcripts to the abundance of pre-rRNA.
The second set was designed to examine the expression of murA and tyrS and to relate the abundance of their transcripts to the abundance of pre-rRNA.
FIG. 2.
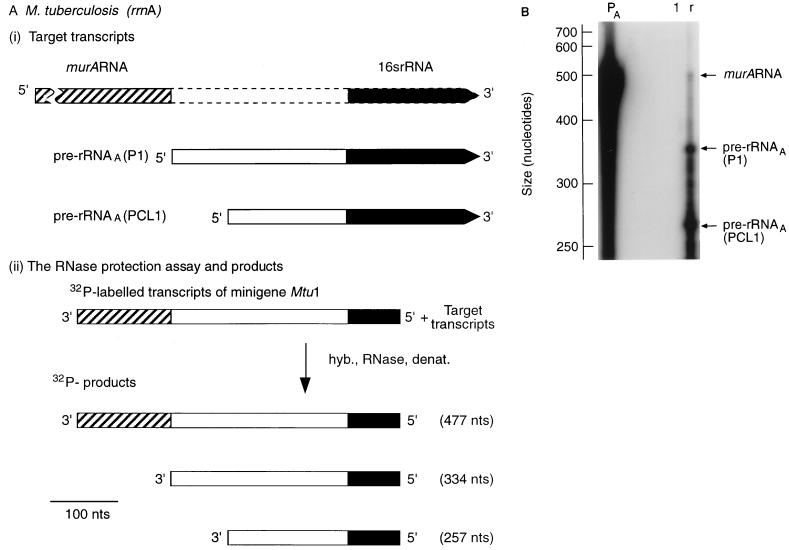
Transcription of M. tuberculosis murA continues into the rRNA coding region of the downstream rrn operon. (A) Design of RNase protection experiments. The scale is shown by the bar. nts, nucleotides. hyb., RNase, denat., respectively, denote the main steps in the RNase protection assay, namely, hybridization, RNase treatment, and denaturation. (i) Target transcripts. The broken lines denote the initial uncertainty in the length of the murA transcript. (ii) RNase protection assay and products. The 32P-labelled probe was obtained by transcribing minigene Mtu 1 (Fig. 1A). The radioactive products (see panel B) were the result of protection of the probe by the target transcripts shown in panel i. (B) Autoradiograph of separated products. The assay described in panel A was carried out, the products were separated, and the sizes of the products were established by use of molecular weight markers as described in Materials and Methods. Lanes: PA, 32P-labelled transcripts of minigene Mtu 1; 1, control sample containing tRNA (10 μg); r, sample (10 μg) of RNA isolated from M. tuberculosis after 5 days of culturing.
FIG. 3.
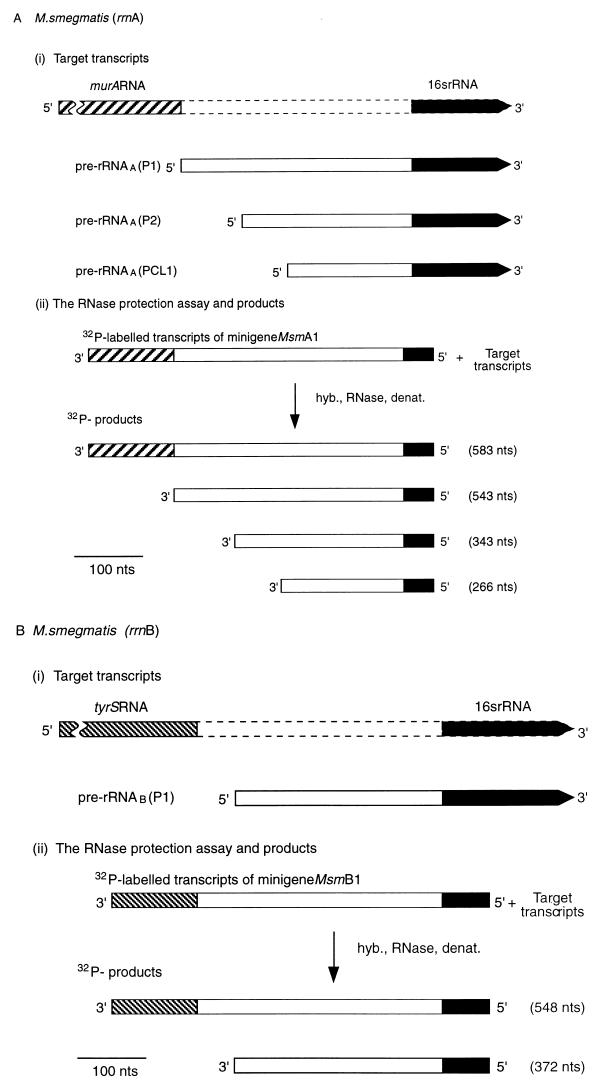
Transcription of M. smegmatis murA and tyrS continues into the rRNA coding regions of the downstream rrn operons. (A and B) Schemes for RNase protection experiments. The scale is shown by the bars. nts, nucleotides. hyb., RNase, denat., respectively, denote the main steps in the RNase protection assay, namely, hybridization, RNase treatment, and denaturation. (A) Expression of murA and rrnA. (i) Target transcripts. The broken lines denote the initial uncertainty in the length of the murA transcript. Pre-rRNAA(P2) is a transcript of rrnA initiated at the P2 promoter and starting from tsp2 (Fig. 1B). (ii) RNase protection assay and products. The 32P-labelled probe was obtained by transcription of minigene Msm A1 (Fig. 1B). The radioactive products (see panel C) were the result of protection of the probe by the target transcripts shown in panel i. (B) Expression of tyrS and rrnB. (i) Target transcripts. The broken lines denote the initial uncertainty in the length of the tyrS transcript. (ii) RNase protection assay and products. The 32P-labelled probe was obtained by transcribing minigene Msm B1 (Fig. 1C). The radioactive products (see panel C) resulted from protection of the probe by the target transcripts shown in panel i. (C) Autoradiograph of products after separation. The assays described in panels A and B were carried out, the products were separated, and the sizes of the products were established by use of molecular weight markers as described in Materials and Methods. Lanes: PA, 32P-labelled transcripts of minigene Msm A1 (Fig. 1B); PB, 32P-labelled transcripts of minigene Msm B1 (Fig. 1C); 1, control sample containing tRNA (30 μg); b, sample (30 μg) of RNA isolated from M. smegmatis after 7 h of culturing and hybridized with PA; b′, sample (30 μg) of RNA isolated from M. smegmatis after 7 h of culturing and hybridized with PB.
The data shown in Fig. 2B (for M. tuberculosis murA) and 3C (for M. smegmatis murA and tyrS) provide evidence for large transcriptional units which extend through the protein coding region and into the 16S rRNA coding region. These transcripts comprise two regions, one coding for protein (MurA or TyrS) and the other coding for rRNA. For this reason, these RNA species are considered to be hybrid transcripts derived from hybrid operons. The hybrid operon identified in M. tuberculosis (Fig. 2) comprises murA at the 5′ end with the rrnA operon located downstream. The hybrid operons identified in M. smegmatis (Fig. 3) comprise murA at the 5′ end with the rrnA operon located downstream and tyrS at the 5′ end with the rrnB operon located downstream.
Previously (10), we described two categories of promoters that are present in M. tuberculosis in particular and in mycobacterial rrn operons in general (Fig. 1) and that are dedicated to pre-rRNA synthesis. One category (P1) is located within the coding region of murA, near the 3′ end; the second category (PCL1) is located within the hypervariable multiple-promoter region which extends from the 3′ end of murA to a conserved sequence motif, CL2, which lies upstream from the 16S rRNA gene and within the leader region of pre-rRNA transcripts (10). PCL1 promoters are associated with another conserved sequence motif, (CL1) (10, 11). A third category of promoter (PmurA), which lies upstream from murA, has now been identified. Previously, we identified a single (P1) promoter of the rrnB operon of M. smegmatis, located between tyrS and the 16S rRNA gene (8, 9). We have now identified a second promoter upstream from tyrS; this promoter is responsible for transcription of the hybrid operon.
M. tuberculosis has a single copy of murA per genome (4). Restriction enzyme digests of M. smegmatis genomic DNA revealed murA and tyrS to be present in single copies per genome (results not shown). Thus, murA, which forms part of a hybrid operon with rrnA, is the sole source of MurA in both M. tuberculosis and M. smegmatis. Similarly, tyrS, which forms part of a hybrid operon with rrnB in M. smegmatis, is the sole source of TyrS in M. smegmatis. We therefore carried out RNase protection studies using the probes described in Fig. 4A, 5A, and 5B in order to study murA RNA and tyrS RNA synthesis under different conditions of mycobacterial growth and to relate their synthesis to pre-rRNA synthesis.
FIG. 4.
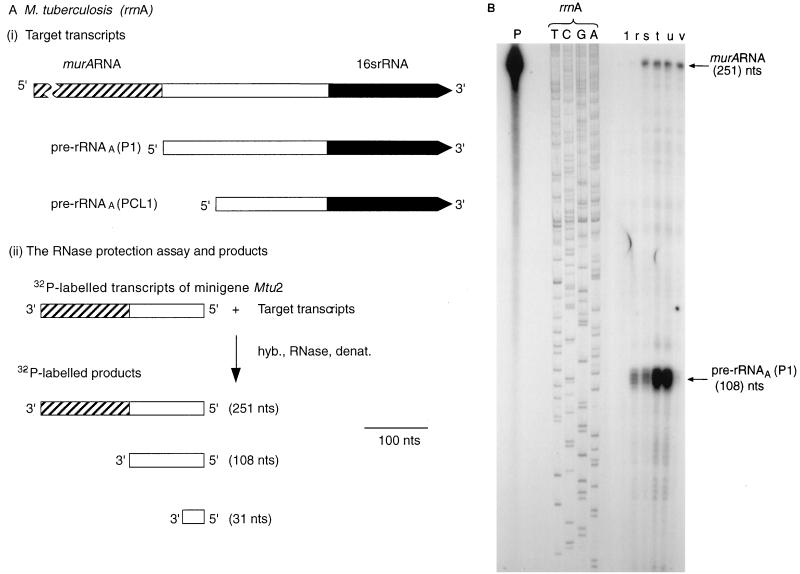
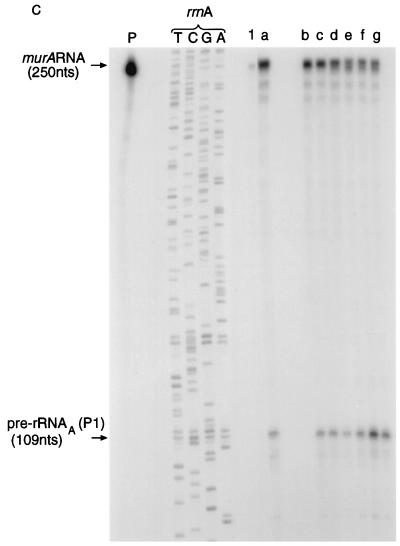
Expression of the murA gene during the growth of M. tuberculosis. (A) Design of RNase protection experiments. The scale is shown by the bar. nts, nucleotides. hyb., RNase, denat., respectively, denote the main steps in the RNase protection assay, namely, hybridization, RNase treatment, and denaturation. (i) Target transcripts. (ii) RNase protection assay and products. The 32P-labelled probe was obtained by transcribing minigene Mtu 2 (Fig. 1A). The radioactive products (see panel B) resulted from protection of the probe by the target transcripts shown in panel i. (B) Autoradiograph of products after separation. The assays described in panel A were carried out, and the products were separated and identified by autoradiography as described in Materials and Methods. The sizes of the products were established by use of the products of sequencing reactions as markers. T, C, G, and A denote reactions carried out with the rrnA operon of M. tuberculosis as a substrate and JY15 as a primer. Lanes: P, 32P-labelled transcripts of minigene Mtu 2; 1, control sample containing tRNA (10 μg). r, s, t, u, and v, respectively, samples (10 μg) of RNA isolated from M. tuberculosis after 5, 10, 13, 22, and 40 days of culturing.
FIG. 5.
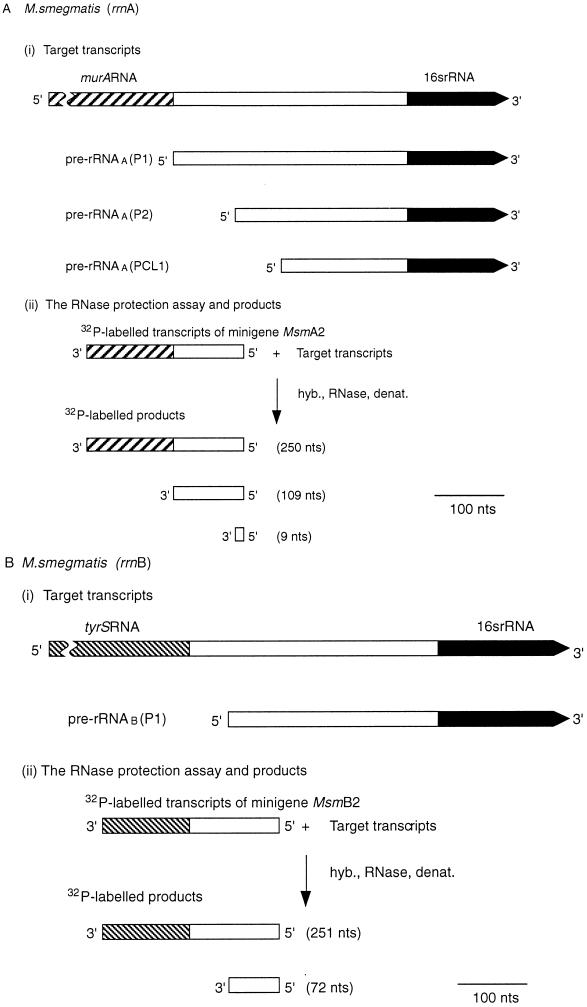
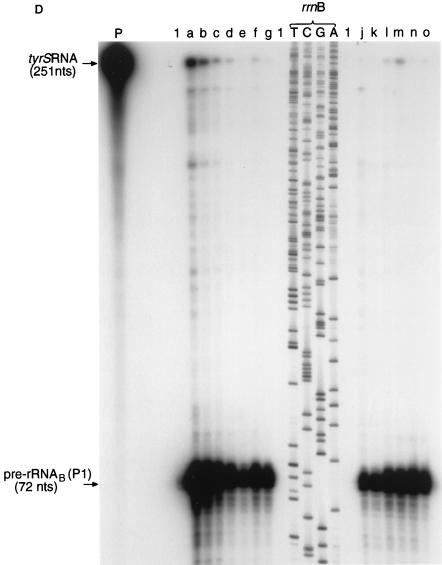
Expression of murA, rrnA, tyrS, and rrnB during the growth of M. smegmatis. (A and B) Schemes for RNase protection experiments. The scale is shown by the bars. nts, nucleotides. hyb., RNase, denat., respectively, denote the main steps in the RNase protection assay, namely, hybridization, RNase treatment, and denaturation. (A) Expression of murA and rrnA. (i) Target transcripts. Pre-rRNAA(P2) is a transcript of rrnA initiated at the P2 promoter and starting from tsp2 (Fig. 1B). (ii) RNase protection assay and products. The 32P-labelled probe was obtained by transcribing minigene Msm A2 (Fig. 1B). The radioactive products (see panel C) resulted from protection of the probe sequences by the target transcripts shown in panel i. (B) Expression of tyrS and rrnB. (i) Target transcripts. (ii) RNase protection assay and products. The 32P-labelled probe was obtained by transcribing minigene Msm B2 (Fig. 1C). The radioactive products (see panel D) were the result of protection of probe sequences by the target transcripts shown in panel i. (C) Autoradiograph of murA RNA- and pre-rRNAA-protected products after separation. The assay described in panel A was carried out, and the products were separated as described in Materials and Methods. The sizes of the protected products were established by use of the products of sequencing reactions as markers. T, C, G, and A denote reactions carried out with the rrnA operon of M. smegmatis as a substrate and JY15 as a primer. Lanes: P, 32P-labelled transcripts of minigene Msm A2; 1, control sample containing tRNA (30 μg); a, b, c, d, e, f, and g, respectively, samples containing RNA (30 μg) isolated from M. smegmatis after 3, 7, 11, 16, 26, 31, and 36 h of culturing in Lemco broth. (D) Autoradiograph of tyrS RNA- and pre-rRNAB-protected products after separation. The assay described in panel B was carried out, and the products were separated as described in Materials and Methods. The sizes of the protected products were established by use of the products of sequencing reactions as markers. T, C, G, and A denote reactions carried out with the rrnB operon of M. smegmatis as a substrate and JY15 as a primer. Lanes: P, 32P-labelled transcripts of minigene Msm B2; 1, control sample containing tRNA (30 μg); a, b, c, d, e, f, and g, respectively, samples containing RNA (30 μg) isolated from M. smegmatis after 3, 7, 11, 16, 26, 31, and 36 h of growth in Lemco broth; j, k, l, m, n, and o, respectively, samples containing RNA (30 μg) isolated from M. smegmatis after 20, 36, 56, 90, 121, and 150 h of growth in Kohn-Harris glucose medium.
The RNA fraction comprises mainly (approximately 83%) rRNA (2) and much lower levels of mRNA. The abundance of a particular mRNA within the RNA fraction represents a steady-state value which reflects both the rate of synthesis and the rate of degradation or processing. Comparative values of radioactivity per assay provide a measure of the relative number of transcripts at the steady state. The abundance of murA RNA within the RNA fraction was related to the abundance of transcripts of rrnA originating from tsp one (tsp1) and designated pre-rRNAA(P1) as shown in Fig. 4 and Table 2 for M. tuberculosis and in Fig. 5A, 5B, and 6A for M. smegmatis.
TABLE 2.
Correlation of the abundance of murA transcripts of M. tuberculosis measured by RNase protection assays with the abundance of pre-rRNAA(P1) transcripts
| Sample | Radioactivitya (counts/assay) of:
|
murA RNA/pre-rRNAA(P1) ratio | |
|---|---|---|---|
| murA RNA | 2.32 × pre-rRNAA(P1)b | ||
| r | 13,000 | 243,000 | 0.03 |
| s | 94,000 | 264,000 | 0.36 |
| t | 118,000 | 986,000 | 0.12 |
| u | 102,000 | 900,000 | 0.11 |
| v | 106,000 | 46,000 | 2.30 |
The radioactivities of the products of the RNase protection experiment shown in Fig. 4B were measured by phosphorimaging (see Material and Methods).
Observed values were multiplied by 2.32, the ratio of the sizes (251 nucleotides and 108 nucleotides, respectively) of the murA RNA- and pre-rRNAA(P1)-protected products. The modified values were used to calculate the molar ratios shown in column 4.
FIG. 6.
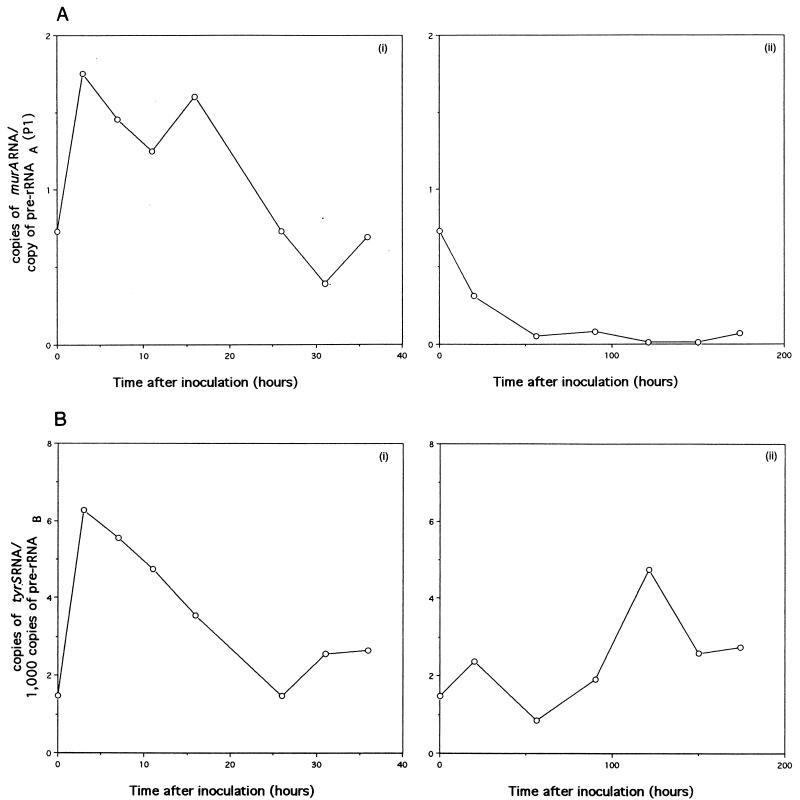
Correlation of murA RNA with pre-rRNAA(P1) synthesis and of tyrS RNA with pre-rRNAB synthesis during the growth of M. smegmatis. The radioactivities of the products shown in Fig. 5C and D were measured by phosphorimaging. (A) Number of copies of murA RNA per copy of pre-rRNAA(P1). The ordinate is equivalent to the radioactivity of murA RNA divided by 2.3 times the radioactivity of pre-rRNAA(P1). The factor 2.3 is the ratio of the sizes (250 nucleotides and 109 nucleotides, respectively) of murA RNA- and pre-rRNAA(P1)-protected products; this factor allows pre-rRNAA(P1) synthesis to be compared with murA RNA synthesis. (i) growth in Lemco broth. (ii) Growth in Kohn-Harris glucose medium. (B) Number of copies of tyrS RNA per 1,000 copies of pre-rRNAB. The ordinate is equivalent to 1,000 times the radioactivity of tyrS RNA divided by 3.5 times the radioactivity of pre-rRNAB. The factor 3.5 is the ratio of the sizes (251 nucleotides and 72 nucleotides, respectively) of tyrS RNA- and pre-rRNAB-protected products; this factor allows pre-rRNAB synthesis to be compared with tyrS RNA synthesis. (i) growth in Lemco broth. (ii) Growth in Kohn-Harris glucose medium.
The results for M. tuberculosis (Fig. 4B and Table 2) show that murA was expressed at all stages of growth, that is, not only during balanced growth but also during the stationary phase. For each assay, the number of murA transcripts was related first to the number of pre-rRNAA(P1) transcripts. The results obtained for samples r, s, t, and u (Table 2) show that there were 50 to 360 copies of murA RNA per 1,000 copies of pre-rRNAA(P1). Previously, we showed (9) that pre-rRNAA(P1) comprises approximately 20% of pre-rRNA. These estimates suggest that, on average, there were 10 to 70 copies of murA RNA for every 1,000 copies of pre-rRNA. The low level of pre-rRNA in the late log phase (sample v), originating from the P1 and PCL1 promoters and reported earlier (9), accounts for the observed high ratio of murA RNA to pre-rRNAA(P1) shown in Table 2.
The data obtained for M. smegmatis (Fig. 5C) show that murA was expressed during the stationary phase as well as during balanced growth. A comparison of the radioactivities of murA RNA- and pre-rRNAA(P1)-protected fragments revealed that more copies of murA RNA per copy of pre-rRNAA(P1) were found during exponential growth in Lemco broth than during the stationary phase (Fig. 6A, panel i) or when growth conditions were less favorable, as in Kohn-Harris glucose medium (Fig. 6A, panel ii).
Previously, it was shown that the contribution of pre-rRNAA(P1) to all transcripts (pre-rRNAA) of rrnA depended on the growth rate; pre-rRNAA(P1) accounted for approximately 1% of pre-rRNAA during early growth (e.g., Table 3, samples a and b) in Lemco broth; this value rose to approximately 15% during the stationary phase (e.g., Table 3, samples e, f, and g) or in Kohn-Harris glucose medium (9). During early growth in Lemco broth, the numbers of murA transcripts were found to exceed the numbers of transcripts of pre-rRNAA(P1) (Fig. 6A, panel i). On the basis of the above-mentioned data, we estimate that, during culturing in Lemco broth, for every 1,000 copies of pre-rRNAA, we detected approximately 15 copies of murA RNA during the early stages of growth and 60 to 100 copies during the stationary phase. However, the overall pattern of murA RNA synthesis does not reflect the pattern of pre-rRNAA synthesis. The burst of pre-rRNAA synthesis [approximately 100-fold that of pre-rRNAA(P1)] that is known to take place during early growth in Lemco broth (9) has no counterpart in murA RNA synthesis. We infer that although the stimuli for murA and rrnA transcription may have features in common, they are not identical.
TABLE 3.
RNA fractions isolated from mycobacteria at representative stages of growth
| Sample | Timea | E640 | Stage of growth |
|---|---|---|---|
| M. smegmatis (Lemco broth, 2 hb) | |||
| i | 0 | 0.034 | Inoculum |
| a | 3 | 0.090 | Balanced growth |
| b | 7 | 0.430 | Balanced growth |
| c | 11 | 1.000 | Early stationary |
| d | 16 | 1.350 | Stationary |
| e | 26 | 1.700 | Stationary |
| f | 31 | 1.860 | Stationary |
| g | 36 | 1.700 | Stationary |
| M. smegmatis (Kohn-Harris glucose medium, 23 hb) | |||
| h | 0 | 0.034 | Inoculum |
| j | 20 | 0.070 | Balanced growth |
| k | 36 | 0.240 | Balanced growth |
| l | 56 | 0.590 | Early stationary |
| m | 90 | 0.870 | Stationary |
| n | 121 | 1.230 | Stationary |
| o | 150 | 1.310 | Stationary |
| M. tuberculosis (Dubos medium, 72 hb) | |||
| i | 0 | 0.025 | Inoculum |
| r | 5 | 0.103 | Balanced growth |
| s | 10 | 0.360 | Balanced growth |
| t | 13 | 0.580 | Early stationary |
| u | 22 | 0.680 | Stationary |
| v | 40 | 1.250 | Stationary |
Reported in hours for M. smegmatis and in days for M. tuberculosis.
Generation time during exponential growth.
The results show that tyrS RNA was detected at all stages of growth of M. smegmatis (Fig. 5D). When the mycobacterium was grown in Lemco broth, tyrS RNA was found to be most abundant during the early stages of growth and least abundant during the early and late stationary phases. When growth took place in Kohn-Harris glucose medium, the abundance of tyrS RNA was scarcely higher than the value found during the stationary phase in Lemco broth. In general, the profiles of tyrS RNA synthesis were found to correlate with the profiles of transcripts (pre-rRNAB) of rrnB (Fig. 5D); for example, the burst in the synthesis of tyrS RNA noted during early growth in Lemco broth was also seen for the synthesis of pre-rRNAB. The abundance of tyrS RNA transcripts was related to the abundance of transcripts of pre-rRNAB (Fig. 6B); ratios within the range of 1 to 6 copies of tyrS RNA for every 1,000 copies of pre-rRNAB were obtained. The highest ratio of tyrS RNA to pre-rRNAB, which was found during early growth in Lemco broth, is thought to reflect the need of the cells for a higher ratio of TyrS per ribosome during a period of rapid protein biosynthesis.
DISCUSSION
Here we have shown that the rrn operons of mycobacteria may form parts of larger hybrid operons in which both protein encoding and rRNA encoding DNAs are transcribed as single units. The starting point for the transcription of the hybrid operon of M. tuberculosis is likely to be between the insertion sequences IS1557, which lies immediately upstream of murA (4), and the 5′ end of the coding region of murA. No similar map is available for M. smegmatis; therefore it is not certain that murA is the first gene of the putative hybrid operon, and it is not known whether it is preceded by other genes. The RNase protection experiments show that transcription in both species continues beyond the 5′ end of the 16S rRNA coding region, suggesting that transcription will continue until an intrinsic terminator downstream from the 5S rRNA gene is reached because of the operation of particular (antitermination) mechanisms that ensure complete transcription of the rrnA operon (5). Initiation of murA transcription from the PmurA promoter ensures that there is one copy of pre-rRNA for each copy of murA RNA derived from that promoter. However, the contribution of the hybrid operon to pre-rRNA synthesis is small (approximately 1 to 4% [Table 2]) because of the presence of additional promoters dedicated to pre-rRNA synthesis and located between the end of the murA gene and the beginning of the 16S rRNA gene (8).
The second rRNA operon of M. smegmatis, rrnB, also may form part of a hybrid operon, in this case including the tyrS gene. In the genome of M. tuberculosis, tyrS is preceded by the Rv1688 gene, which probably codes for 3-methylpurine DNA glycosylase; the two genes appear to be transcribed as a single unit (4), as judged by the size and composition of the sequence separating them. Should a similar arrangement occur in the M. smegmatis genome, then the homologue of the Rv1688 gene would be part of the hybrid operon.
Within the bacterial cell, transcription and translation are coupled so that mRNA is translated as it is being synthesized. If, like mRNA translation, both ribosome assembly and rRNA processing take place as transcription proceeds, then it is likely that only a small proportion of hybrid transcripts will remain intact after transcription is complete. Early pre-rRNA processing events would have the effect of releasing mRNA into the cytosol; a transcriptional terminator also has this effect. Thus, the hybrid operon appears to make economical use of cell resources by using a minimum number of initiation and termination steps.
Our analysis of the pre-rRNA fractions of both M. tuberculosis and M. smegmatis has shown that in normal growth, transcripts originating from promoters dedicated to the expression of rRNA gene sequences represent the majority. The str operon of Escherichia coli provides another example of promoters located within an operon but dedicated to the expression of a particular gene within the operon. This operon (19, 20) comprises four genes in the order rpsL, rpsG, fus, and tufA. In addition to Pstr, the principal promoter for the operon, promoters dedicated to the expression of tufA (encoding elongation factor Tu) are thought to lie within fus (encoding elongation factor G).
Results obtained for E. coli rrn operons help to place our findings in a wider context. The rrnG operon, which does not appear to form part of a hybrid operon, is located downstream from clpB, a gene coding for E. coli heat shock protein F84.1 (18). An intrinsic terminator for clpB is thought to be located 31 bp downstream from the 3′ end of the coding region and 83 bp upstream from the −35 box of promoter P1 of rrnG (17), suggesting that clpB and rrnG are transcribed independently. In contrast, transcripts of the open reading frame upstream from the rrnB operon of E. coli were shown to continue into the 16S rRNA coding region; this finding was the first evidence for a hybrid operon (1). The open reading frame, which is regulated by two promoters (1), encodes glutamate racemase, an enzyme that catalyzes the synthesis of d-glutamate, an essential component of bacterial peptidoglycan.
ACKNOWLEDGMENTS
We thank our colleague Andrew Lane for helpful discussions. We thank Simon A. Cox for help in the preparation of the manuscript.
J. A. G.-y-M. received financial support from COFAA and EDD, IPN, Mexico. This work was supported as part of the European Commission Science Research and Development Programme (contract ERBIC 18CT 9720253).
REFERENCES
- 1.Boros I, Csordás-Tóth E, Kiss A, Török I, Udvardy K, Venetianer P. Identification of two new promoters probably involved in the transcription of a ribosomal RNA gene of Escherichia coli. Biochim Biophys Acta. 1983;739:173–180. doi: 10.1016/0167-4781(83)90027-1. [DOI] [PubMed] [Google Scholar]
- 2.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell growth rate. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1527–1542. [Google Scholar]
- 3.Clarke P H, Meadow P M. Evidence for the occurrence of permeases for tricarboxylic acid intermediates in Pseudomonas aeruginosa. J Gen Microbiol. 1959;20:144–155. doi: 10.1099/00221287-20-1-144. [DOI] [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Draper P. The aliphatic acylamide amidohydrolase of Mycobacterium smegmatis: its inducible nature and relation to acyltransfer to hydroxylamine. J Gen Microbiol. 1967;46:111–123. doi: 10.1099/00221287-46-1-111. [DOI] [PubMed] [Google Scholar]
- 7.Dubos P J, Davis B D. Factors affecting the growth of tubercle bacilli in liquid media. J Exp Med. 1946;83:409–423. [PubMed] [Google Scholar]
- 8.Gonzalez-y-Merchand J A, Colston M J, Cox R A. The rRNA operons of Mycobacterium smegmatis and Mycobacterium tuberculosis: comparison of promoter elements and of neighbouring upstream genes. Microbiology. 1996;142:667–674. doi: 10.1099/13500872-142-3-667. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-y-Merchand J A, Colston M J, Cox R A. Roles of multiple promoters in transcription of ribosomal DNA: effects of growth conditions on precursor rRNA synthesis in mycobacteria. J Bacteriol. 1998;180:5756–5761. doi: 10.1128/jb.180.21.5756-5761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-y-Merchand J A, Garcia M J, Gonzalez-Rico S, Colston M J, Cox R A. Strategies used by pathogenic and nonpathogenic mycobacteria to synthesize rRNA. J Bacteriol. 1997;179:6949–6958. doi: 10.1128/jb.179.22.6949-6958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji Y-E, Colston M J, Cox R A. Nucleotide sequence and secondary structures of precursor 16S rRNA of slow-growing mycobacteria. Microbiology. 1994;140:123–132. doi: 10.1099/13500872-140-1-123. [DOI] [PubMed] [Google Scholar]
- 12.Kelly M, Clarke P H. An inducible amidase produced by a strain of Pseudomonas aeruginosa. J Gen Microbiol. 1962;27:305–316. doi: 10.1099/00221287-27-2-305. [DOI] [PubMed] [Google Scholar]
- 13.Kohn H I, Harris J S. On the mode of action of sulphonamides. I. Action on Escherichia coli. J Pharmacol Exp Ther. 1941;73:343–348. [Google Scholar]
- 14.Mironov V N, Van Montagu M, Inzé D. High throughput RNase protection assay. Nucleic Acids Res. 1995;23:3359–3360. doi: 10.1093/nar/23.16.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Predich M, Doukhan L, Nair G, Smith I. Characterization of RNA polymerase and two sigma factors from Mycobacterium smegmatis. Mol Microbiol. 1995;15:355–366. doi: 10.1111/j.1365-2958.1995.tb02249.x. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 17.Shen W-F, Squires C, Squires C L. Nucleotide sequence of the rrnG ribosomal RNA promoter region of Escherichia coli. Nucleic Acids Res. 1982;10:3303–3313. doi: 10.1093/nar/10.10.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Squires C L, Pedersen S, Ross B M, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zengel J M, Archer R H, Lindahl L. The nucleotide sequence of the Escherichia coli fus gene, coding for elongation factor G. Nucleic Acids Res. 1984;12:2181–2192. doi: 10.1093/nar/12.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zengel J M, Lindahl L. Mapping of two promoters for elongation factor Tu within the structural gene for elongation factor G. Biochim Biophys Acta. 1990;1050:317–322. doi: 10.1016/0167-4781(90)90188-8. [DOI] [PubMed] [Google Scholar]