Abstract
Background and aims
Current prognostic scores of patients with acutely decompensated cirrhosis (AD), particularly those with acute-on-chronic liver failure (ACLF), underestimate the risk of mortality. This is probably because systemic inflammation (SI), the major driver of AD/ACLF, is not reflected in the scores. SI induces metabolic changes, which impair delivery of the necessary energy for the immune reaction. This investigation aimed to identify metabolites associated with short-term (28-day) death and to design metabolomic prognostic models.
Methods
Two prospective multicentre large cohorts from Europe for investigating ACLF and development of ACLF, CANONIC (discovery, n=831) and PREDICT (validation, n=851), were explored by untargeted serum metabolomics to identify and validate metabolites which could allow improved prognostic modelling.
Results
Three prognostic metabolites strongly associated with death were selected to build the models. 4-Hydroxy-3-methoxyphenylglycol sulfate is a norepinephrine derivative, which may be derived from the brainstem response to SI. Additionally, galacturonic acid and hexanoylcarnitine are associated with mitochondrial dysfunction. Model 1 included only these three prognostic metabolites and age. Model 2 was built around 4-hydroxy-3-methoxyphenylglycol sulfate, hexanoylcarnitine, bilirubin, international normalised ratio (INR) and age. In the discovery cohort, both models were more accurate in predicting death within 7, 14 and 28 days after admission compared with MELDNa score (C-index: 0.9267, 0.9002 and 0.8424, and 0.9369, 0.9206 and 0.8529, with model 1 and model 2, respectively). Similar results were found in the validation cohort (C-index: 0.940, 0.834 and 0.791, and 0.947, 0.857 and 0.810, with model 1 and model 2, respectively). Also, in ACLF, model 1 and model 2 outperformed MELDNa 7, 14 and 28 days after admission for prediction of mortality.
Conclusions
Models including metabolites (CLIF-C MET) reflecting SI, mitochondrial dysfunction and sympathetic system activation are better predictors of short-term mortality than scores based only on organ dysfunction (eg, MELDNa), especially in patients with ACLF.
Keywords: cirrhosis, liver failure, liver metabolism
WHAT IS ALREADY KNOWN ON THIS TOPIC.
Model for end-stage liver disease (MELD) score is an excellent predictor of survival.
MELD and MELDNa are used for liver transplant allocation.
Both scores underestimate the risk of death of patients with acutely decompensated cirrhosis (AD), particularly those with acute-on-chronic liver failure (ACLF).
WHAT THIS STUDY ADDS
Identify three prognostic metabolites strongly associated with death.
Two novel models outperforming MELD and MELDNa scores.
Particularly better in patients with AD and ACLF.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
These models may be used for risk stratification in studies.
Ultimately, these models may be implemented and optimise allocation of scarce medical resources such as liver transplantation.
Introduction
Acutely decompensated cirrhosis (AD) is a heterogeneous condition.1 1–3 The association of AD with organ failure(s) defines its maximal form, the acute-on-chronic liver failure (ACLF) syndrome.3 AD occurs in the setting of bursts of systemic inflammation (SI) which are particularly severe in patients developing ACLF,4 5 rendering SI of critical pathogenetic importance in AD.
Prognosis of patients hospitalised with AD relates to clinical features present at hospital admission. Cirrhosis decompensations are precipitated by proinflammatory events, mainly infections and/or acute alcoholic hepatitis, or even potentially from episodes of bacterial translocation from the intestinal microbiota in patients without an identified precipitant.6–8
ACLF is present at admission in 20%–25% of patients with AD3 and is associated with high short-term mortality (22% at 28 days).1 The course of ACLF is highly dynamic since it may improve or deteriorate or follow a steady course within a few days after admission, which also correlates with prognosis.9 Similarly, approximately 20% of all patients hospitalised without ACLF develop the syndrome within 3 months (pre-ACLF) and have a mortality rate comparable to that of patients with ACLF (30% at 28 days).10–12 Therefore, the dynamics of AD also determine short-term prognosis.
Prognostication of patients with AD improved with the introduction of the MELD score in 2001.13 However, MELD (Model for end-strage liver disease) was designed to be independent of subjective clinical features. The modifications of MELD, including sodium to MELD-Na14 and female sex in MELD 3.015 scores, slightly improved the global mortality prediction and has been incorporated to guide transplant allocation (online supplemental tables 1, 2). Despite these improvements, recent studies have shown that MELD-Na may not be sufficiently accurate to predict early prognosis in patients with ACLF.16–18 Even the specific Chronic Liver Failure Consortium ACLF (CLIF-C ACLF) score fails to predict short-term prognosis in a significant number of patients.16
gutjnl-2022-328708supp001.pdf (252.9KB, pdf)
Metabolomics seem to deliver appropriate biomarkers for prognosis, as shown for several diseases also driven by SI and organ dysfunction, including sepsis and COVID-19.19–22 In cirrhosis, a set of metabolites, the so-called ‘ACLF-related metabolomic fingerprint’, strongly correlates with severity of SI and ACLF.23 In addition, survival and the outcome of encephalopathy or kidney failure in decompensated cirrhosis correlate with plasma, cerebrospinal fluid, serum and urinary metabolites,21–24 which may at least partly derive from intestinal microbes.25
Therefore, we aimed to identify metabolites that correlate with short-term mortality and to construct short-term prognostic metabolomic models that would improve prognostication in AD and ACLF.
Patients and methods
The CANONIC and PREDICT cohorts
The study was based on data from the CANONIC and PREDICT study cohorts. The CANONIC study, performed on 1343 consecutive patients from February to September 2011, aimed to assess prevalence, diagnostic criteria, mechanisms, clinical course and outcome of ACLF.3 The PREDICT study, performed from March 2017 to July 2018 in 1271 patients, was initially designed to explore precipitating events and early (3-month) clinical course of AD only in patients hospitalised without ACLF. However, a few months after study onset, we also recruited 202 consecutive patients hospitalised with ACLF as controls.1 6
Clinical and standard laboratory data and biobanking material were obtained in both cohorts at hospital admission and sequentially during hospitalisation in the CANONIC cohort, and at admission and during 3 months after admission in the PREDICT cohort. Follow-up liver transplantation and mortality were recorded in both studies. The current article includes 1682 patients explored by untargeted metabolomics of serum samples obtained at enrolment (831 and 851 patients from the CANONIC and PREDICT studies, respectively). The identification of the death-related metabolomic fingerprint was obtained by using both cohorts and clustering prognostic results hierarchically. The design of the metabolomic models was performed in the CANONIC cohort and validated in the PREDICT cohort. The PREDICT study has been registered in ClinicalTrials.gov as NCT03056612.
Untargeted metabolomics
Untargeted metabolomics was performed in the two cohorts using identical methodology and at the same laboratory, using conditions described before.23 Metabolites were extracted from serum by methanol-assisted protein precipitation and analysed by liquid chromatography coupled to high-resolution mass spectrometry (LC-HRMS) using a combination of two complementary chromatographic methods consisting of reversed-phase chromatography (C18 chromatographic column) and hydrophilic interaction liquid chromatography (HILIC) for the analysis of metabolites (excluding fatty acids and complex lipids). LC-HRMS experiments were conducted on an Ultimate 3000 chromatographic system (Thermo Fisher Scientific, Courtaboeuf, France) coupled to an Exactive or a Q-Exactive high-resolution mass spectrometer (Thermo Fisher Scientific) interfaced with an electrospray ionisation source operating in the positive and negative ion modes for C18 and HILIC separations, respectively. Quality control (QC) samples were obtained by pooling aliquots of each sample and were injected every 10 samples throughout the experiments for further data normalisation/standardisation. In addition, a cocktail of standards, carefully chosen not to interfere with endogenous metabolites, was added to every analysed sample to monitor performances of chromatographic separation and mass spectrometry detection.24 25 Data processing was performed using the Workflow4Metabolomics (W4M) platform.26 Annotation of metabolite features was first performed by comparison of accurate measured masses, chromatographic retention times and MS/MS spectra to an in-house library made from ~1200 authentic pure standards analysed under identical conditions. To ensure accurate and consistent metabolite quantification, chromatographic peaks of the annotated metabolites were integrated for subject, patient and QC samples by using the Trace Finder software (Thermo Fisher Scientific). Peak integrations were then manually reviewed for each sample and the resulting peak areas were normalised with respect to QC samples with the LOESS algorithm to correct for variation resulting from batch-to-batch signal differences.27 Metabolite signals were then filtered according to the following criteria: (1) the correlation between QC dilution factors and areas of chromatographic peaks (relevant metabolites should have coefficients of correlation above 0.7); (2) repeatability (the coefficient of variation obtained on chromatographic peak areas of QC samples should be below 30%); and (iii) ratio of chromatographic peak areas of biological to blank samples above a value of 3. The resulting dataset was expressed in arbitrary units, representing the area of the chromatographic peak for each metabolite. We did not apply batch effect correction between cohorts.
Analysis of data
Variables are described by their absolute value and relative percentage or by their median and IQR and compared between study cohorts through χ2 or Kruskal-Wallis’ hypothesis testing, as appropriate. A log2 transformation of the metabolite levels has been carried out before computing the fold change and corresponding p value when comparing survivors and non-survivors at 28 days in each study cohort. These comparisons have been represented using volcano and Cleveland plots. P values offered in the volcano plots were adjusted by Benjamini & Hochberg methods to control the false discovery rate. How mortality is distributed along time has been evaluated through the estimation of the cumulative incidence function and compared with Gray’s test.
Ability to predict the risk of death has been computed using Harrell’s concordance index (C-index). The C-index allows for the combination of both the event occurrence and the time to such event, ranging from 0 to 1. The estimated C-index for a certain model quantifies its ability to discriminate early events (ie, high-risk patients). Thus, C-index values closer to one indicate the tested model can predict, among two random patients, which one has the larger risk of death, while values closer to 0.5 would correspond to a model discriminating at random. Considering the estimated C-index for individual metabolites in both study cohorts, hierarchical clustering analysis has been performed to discriminate a common death-related metabolomic fingerprint, a cluster of blood metabolites that shares a large ability to predict patients’ risk of death. The identification of the death-related metabolomic fingerprint was performed with a time limit of mortality of 28 days.
The 10 best common and quantifiable individual predictors from this fingerprint have been included in a multivariate Cox regression model, adjusted by age, to assess their independent association with 28-day mortality in the CANONIC study cohort. Working within Fine and Gray’s framework, the models consider death as the primary event and liver transplant as the competing one. Backwards stepwise procedures have been carried out to finalise the model (variables with the largest non-significant p values (p>0.01) were iteratively removed from the model until all variables shared a significant association with the outcome). Two metabolomic models were developed. Model 1 (online supplemental table 1) includes three metabolites independently associated with 28-day mortality, in addition to age as a possible confounder. In model 2 (online supplemental table 1), the same selection procedures were followed adding a small set of four common laboratory measures with well-known prognostic significance (creatinine, bilirubin, international normalised ratio (INR) and white cell count (WCC)). Once models 1 and 2 were found, they were scaled so they ranged from 0 to 100 (with an approximate median of 50), and their predictive ability was estimated through the C-index and compared with the MELDNa score in the whole cohort of patients and in patients with or without ACLF separately. To assess whether the described models performed better, a minimum difference of 0.05 between C-indices was considered clinically relevant when predicting a patient’s outcome. That difference has been estimated and tested by bootstrapping C-index estimates over 100 iterations in a randomly selected subsample representing 70% of the overall sample. The significance level for such differences has been reduced to 1% to account for the multiple comparisons performed. Uncertainty of the resulting C-indices has been assessed by estimating the SD at 28 days by jack-knife. All analyses presented have been conducted on a complete-case basis.
Results
Characteristics of discovery and validation cohorts
There were more patients with ACLF in the CANONIC cohort compared with the PREDICT (table 1), which is related to differences in intended design of both cohorts and explains significant differences in clinical and laboratory characteristics at enrolment. Patients in the CANONIC cohort presented significantly higher prevalence of precipitating events and organ failures. Mortality rates, however, were not significantly different between both cohorts and between the subgroups of patients with ACLF (table 1). The percentage of patients who received liver transplantation within 28 days was very low in both cohorts (1.83% and 3.37%; p value=0.0709), which is consistent with the low indication for this procedure for ACLF in Europe over the past decade (1). The PREDICT cohort served as a validation set for relevant pathophysiological metabolomic pathways, prognostic metabolites and metabolomic models discovered in the CANONIC cohort.
Table 1.
Characteristics of the patients in the CANONIC and PREDICT study cohorts at enrolment
| Characteristic | Discovery set: CANONIC study cohort (n=831) |
Validation set: PREDICT study cohort (n=851) |
P value |
| Age (years) | 57 (50–66) | 59 (52–67) | <0.001 |
| Sex (female, %) | 292 (35.14) | 268 (31.49) | 0.125 |
| Ascites (%) | 526 (63.6) | 602 (70.74) | 0.002 |
| Hepatic encephalopathy (%) | 275 (33.09) | 276 (32.43) | 0.813 |
| Gastrointestinal bleeding (%) | 134 (16.13) | 127 (14.92) | 0.540 |
| Bacterial infection (%) | 194 (23.35) | 279 (32.78) | <0.001 |
| Alcoholic steatohepatitis (%) | 116 (14.36) | 65 (7.64) | <0.001 |
| Sepsis (%) | 127 (15.7) | 108 (12.84) | 0.112 |
| Liver failure (%) | 115 (13.86) | 76 (8.93) | 0.002 |
| Renal failure (%) | 112 (13.48) | 64 (7.52) | 0.000 |
| Brain failure (%) | 52 (6.27) | 33 (3.88) | |
| Coagulation failure (%) | 76 (9.31) | 44 (5.18) | 0.002 |
| Circulatory failure (%) | 33 (4.05) | 15 (1.76) | |
| Respiratory failure (%) | 21 (3) | 6 (0.71) | |
| Presence of ACLF (%) | 181 (21.78) | 109 (12.84) | <0.001 |
| ACLF grade (%) | <0.001 | ||
| ACLF-1 | 97 (11.67) | 74 (8.72) | |
| ACLF-2 | 65 (7.82) | 25 (2.94) | |
| ACLF-3 | 19 (2.29) | 10 (1.18) | |
| Patients in intensive care unit (%) | 44 (5.32) | 78 (9.17) | 0.003 |
| Liver transplant at 28 days (%) | 28 (3.37) | 15 (1.83) | 0.071 |
| Mortality at 28 days (%) | |||
| All patients | 79 (9.51) | 75 (8.81) | 0.683 |
| ACLF patients | 48 (26.52) | 30 (27.52) | 0.852 |
| AD non-ACLF patients | 31 (4.77) | 45 (6.08) | 0.283 |
| Hematocrit (%) | 30.4 (26.4–34.38) | 29.5 (26–34) | 0.024 |
| White cell count (×109/L) | 5.94 (4.14–9.29) | 6.47 (4.4–9.24) | 0.185 |
| International normalised ratio | 1.48 (1.29–1.83) | 1.46 (1.27–1.73) | 0.038 |
| Albumin (g/dL) | 2.9 (2.5–3.3) | 2.8 (2.44–3.3) | 0.171 |
| Aspartate aminotransferase (U/L) | 62 (37.5–101) | 54 (34–87) | 0.001 |
| Alanine aminotransferase (U/L) | 35 (22–55) | 29 (19–44.93) | <0.001 |
| Serum bilirubin (mg/dL) | 3 (1.6–6.88) | 2.67 (1.42–5.8) | 0.003 |
| Serum creatinine (mg/dL) | 0.95 (0.72–1.4) | 0.93 (0.7–1.29) | 0.075 |
ACLF, acute-on-chronic liver failure; AD, acutely decompensated cirrhosis.
Elevated levels of serum metabolites were associated with high 28-day mortality
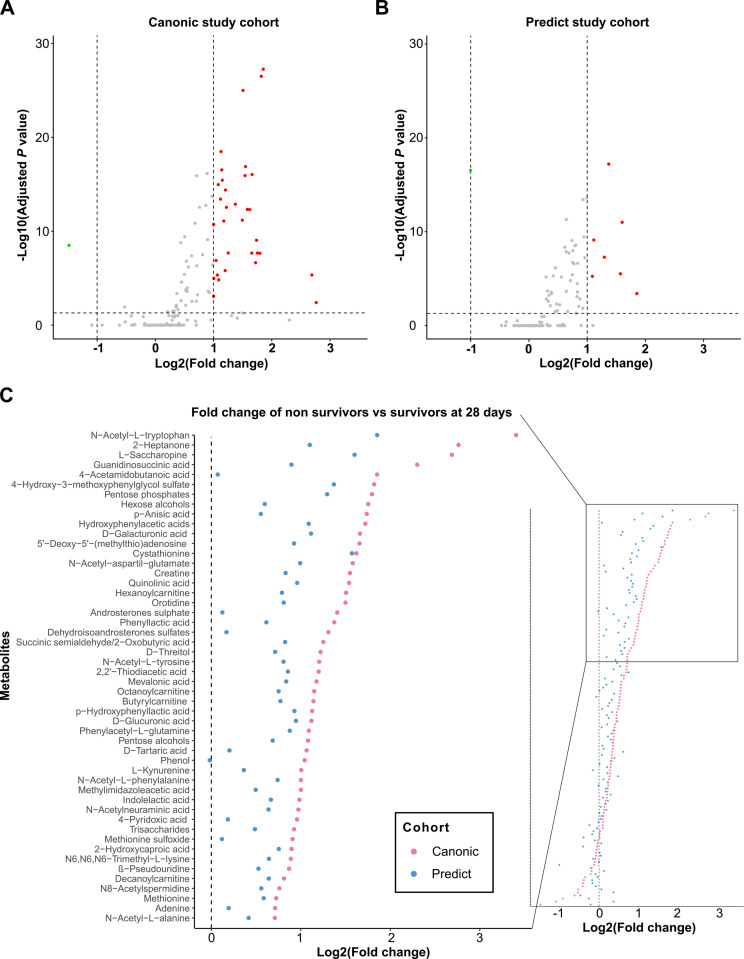
Figure 1A, B shows the volcano plots illustrating the pairwise comparison of each of the 130 annotated metabolites expressed in fold change (FC) between patients who died within 28 days after enrolment versus those who survived in each cohort. There was a clear shift to the right of most metabolites in the two cohorts indicating upregulated metabolites in patients who died versus those who survived. The FC increase was more than twofold for 34 metabolites in the CANONIC cohort (figure 1A) and in seven metabolites in the PREDICT cohort (figure 1B). Figure 1C illustrates the whole set of metabolites in the two cohorts ranked according to the FC between non-survivors and survivors in the CANONIC cohort. The 50 top metabolites in the CANONIC cohort are represented in the zooming inset. The FC of all these metabolites was higher in the CANONIC than in the PREDICT study patients, indicating a more intense metabolomic change in the former group. There was, however, a close relationship between the FC of these metabolites from both cohorts (Pearson’s correlation=0.81; p value <0.0001).
Figure 1.
Mortality at 28 days is associated with a general increase in metabolite levels. (A) Volcano plots showing the results of pairwise comparisons of blood metabolites levels of patients from CANONIC (left) and PREDICT (right) study cohorts relative to mortality at 28 days. The vertical dashed lines indicate the threshold for the twofold abundance difference. The horizontal dashed line indicates the p value=0.05 threshold. (B) Cleveland plots. Overview of the fold change of all detected metabolites between patients who died within 28 days after enrolment versus those who survived in each cohort in both CANONIC and PREDICT (red and blue dots, respectively) study cohorts, using CANONIC cohort as reference. Left inset shows the annotation of the top 50 metabolites.
Of note, we tried to build prediction models using clinical routine parameters, such as interleukin-6 (IL-6) and WCC. When trying to build a clinical model with age, creatinine, bilirubin, INR, WCC and IL-6, a final clinical model was obtained with only age, creatinine, bilirubin and INR. This represents an updated version of MELD together with age that shared the same predictive ability at 28 days as the MELDNA score (0.780 vs 0.785). Therefore, we moved to the metabolites as next.
Identification of a common death-related metabolomic fingerprint in the CANONIC and PREDICT cohorts
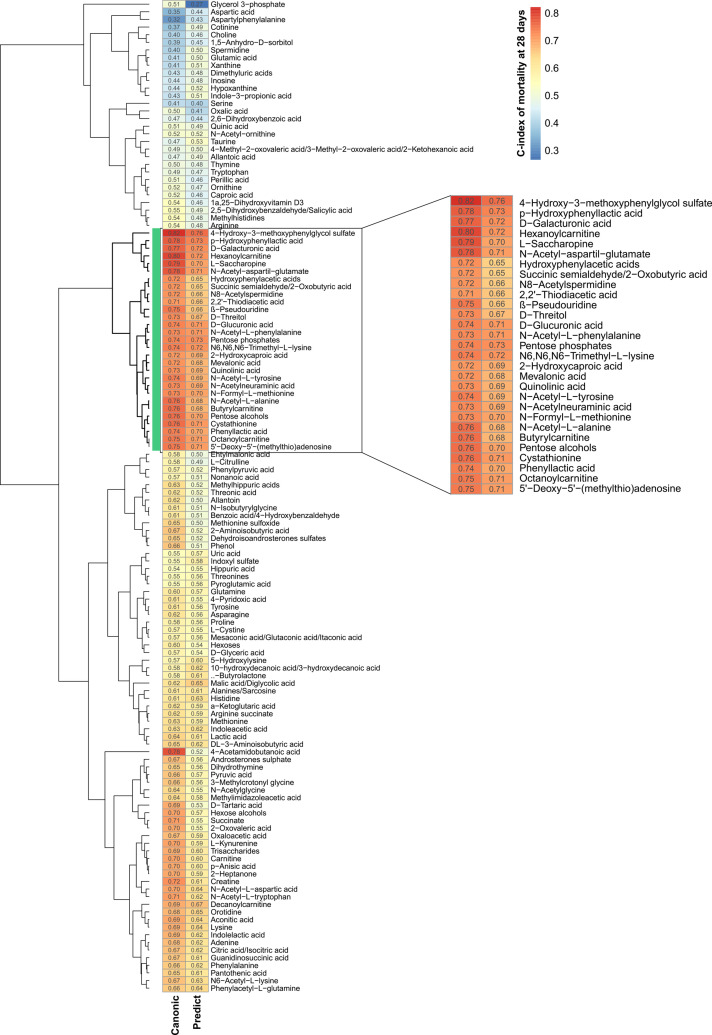
We next investigated the association between the serum metabolome and 28-day mortality. For each of the 130 metabolites and for each cohort, we estimated the Harrel’s concordance index (C-index) assessing the discriminating accuracy of the metabolite levels, expressed in relative units corresponding to chromatographic peak areas, in differentiating prognosis (considering death as the primary event and liver transplant as the competing risk) (online supplemental table 3). We then created a two-column heatmap, one for each cohort, in which each row (one per metabolite) was grouped according to the hierarchical clustering of the observed C-indices (figure 2). A 29-metabolite cluster with the highest C-index values was then shown to be shared by the two cohorts, defining the death-related metabolomic fingerprint of AD.
Figure 2.
Metabolite abilities to predict mortality at 28 days and their similarities between study cohorts. Hierarchical clustering analysis of C-index values obtained assessing mortality at 28 days with each measured metabolite in the CANONIC (left column) and the PREDICT (right column) study cohort. Vertical green bar and inset identify the cluster formed by the 29 metabolites with the highest C-index in both cohorts.
The death-related metabolomic fingerprint of AD largely coincides with the ACLF-related metabolomic fingerprint
In a previous study in the CANONIC cohort,23 we identified a fingerprint of 38 metabolites whose intensity correlated with the severity of SI and discriminated patients with ACLF of any grade from patients with mere AD and healthy controls. Based on this ACLF-related metabolomic fingerprint, we were able to understand the metabolomic hallmark of AD and to propose new pathophysiological mechanisms for the progression of compensated to decompensated cirrhosis and to ACLF.2 23 Importantly, 25 metabolites of the death-related metabolomic fingerprint were among the 38 metabolites of the ACLF-related metabolomic fingerprint (online supplemental table 4), indicating that both pathophysiological mechanisms and prognosis of AD relies on the dysfunctional metabolic pathways.
Metabolomic models (CLIF-C MET) 1 and 2 predict prognosis better than the MELDNa score in AD
We next developed models 1 and 2 (online supplemental table 1) as follows:
CLIF-C MET model 1: [0.02396·Age+0.32981·log2(4-hydroxy-3-methoxyphenylglycol sulfate)+0.45602·log2(hexanoylcarnitine)+0.27226·log2(D-galacturonic acid)–18.1561]/0.0965.
CLIF-C MET model 2: [0.03432·Age+0.34020·log2(4-hydroxy-3-methoxyphenylglycol sulfate) + 0.50724·log2(hexanoylcarnitine)+0.04037·Serum bilirubin+0.34674·INR–14.6517]/0.1218.
The rank of the prognostic estimates was derived from the CANONIC cohort. Eleven out of the top 15 quantifiable prognostic metabolites in the CANONIC cohort (table 2, upper list) were among the top 15 prognostic metabolites in the PREDICT cohort, indicating a high level of concordance between both cohorts. The chromatographic peak area of the top 10 metabolites with the largest prognostic estimates in the CANONIC study plus age were entered to construct model 1. 4-Hydroxy-3-methoxyphenylglycol sulfate, hexanoylcarnitine, D-galacturonic acid and age were the final variables included into the model. Model 2 was designed to assess whether adding clinical prognostic variables to prognostic metabolites could improve prediction of mortality. Therefore, model 2 was constructed using the top 10 prognostic metabolomic estimates in the CANONIC cohort plus age, serum bilirubin, serum creatinine, INR and WCC. At the end, model 2 included 4-hydroxy-3-methoxyphenylglycol sulfate, hexanoylcarnitine, bilirubin, INR and age. The process, however, resulted in a non-significant increase in prognostic prediction with respect to model 1 (table 3).
Table 2.
Fifteen top metabolites from the death-related metabolomic fingerprint and standard laboratory variables in the CANONIC cohort ranked by their predictive ability through C-index estimation and their corresponding position and C-index in the PREDICT cohort
| Variables | Discovery set: CANONIC study cohort | Validation set: PREDICT study cohort | ||
| Rank | C-index | Rank | C-index | |
| Metabolite | ||||
| 4-Hydroxy-3-methoxyphenylglycol sulfate | 1 | 0.822 | 1 | 0.760 |
| Hexanoylcarnitine | 2 | 0.799 | 4 | 0.721 |
| L-Saccharopine | 3 | 0.786 | 12 | 0.704 |
| N-Acetyl-aspartil-glutamate | 4 | 0.782 | 9 | 0.709 |
| p-Hydroxyphenyllactic acid | 5 | 0.777 | 2 | 0.733 |
| D-Galacturonic acid | 6 | 0.766 | 3 | 0.724 |
| N-Acetyl-L-alanine | 7 | 0.759 | 20 | 0.684 |
| Butyrylcarnitine | 8 | 0.759 | 21 | 0.681 |
| Cystathionine | 9 | 0.757 | 8 | 0.711 |
| Octanoylcarnitine | 10 | 0.747 | 10 | 0.707 |
| 5'-Deoxy-5'-(methylthio)adenosine | 11 | 0.747 | 11 | 0.707 |
| β-Pseudouridine | 12 | 0.746 | 26 | 0.664 |
| Phenyllactic acid | 13 | 0.742 | 14 | 0.699 |
| N6,N6,N6-Trimethyl-L-lysine | 14 | 0.742 | 5 | 0.719 |
| N-Acetyl-L-tyrosine | 15 | 0.741 | 18 | 0.687 |
| Standard laboratory variables* | ||||
| INR | 1 | 0.726 | 1 | 0.687 |
| Bilirubin | 2 | 0.695 | 2 | 0.676 |
| Creatinine | 3 | 0.672 | 3 | 0.621 |
| Sodium | 4 | 0.630 | 4 | 0.616 |
*Standard laboratory variables consider the cut-offs presented in the definition of the MELDNa score.
INR, international normalised ratio.
Table 3.
Description of models to predict risk of mortality at several time points and their predictive ability in the CANONIC (discovery set) and the PREDICT (validation set) study cohorts
| Model | C-index | ||||
| At 7 days | At 14 days | At 28 days | At 60 days | At 90 days | |
| Discovery set: CANONIC study cohort (n=831) | |||||
| % of deaths | 2.41% | 5.78% | 9.51% | 15.52% | 18.53% |
| CLIF-C MET 1 | 0.927 | 0.900 | 0.842 | 0.805 | 0.784 |
| CLIF-C MET 2 | 0.937 | 0.921 | 0.853 | 0.808 | 0.784 |
| MELDNa score | 0.810*† | 0.813*† | 0.788*† | 0.779 | 0.772 |
| Validation set: PREDICT study cohort (n=851) | |||||
| % of deaths | 1.18% | 4.58% | 8.81% | 15.28% | 18.21% |
| CLIF-C MET 1 | 0.940 | 0.834 | 0.791 | 0.780 | 0.771 |
| CLIF-C MET 2 | 0.947 | 0.857 | 0.810 | 0.788 | 0.777 |
| MELDNa score | 0.793*† | 0.773† | 0.755† | 0.713*† | 0.700*† |
Set of variables included initially in model 1: log2(4-hydroxy-3-methoxyphenylglycol sulfate, log2(hexanoylcarnitine), log2(L-Saccharopine), log2(N-Acetyl-aspartil-glutamate), log2(p-Hydroxyphenyllactic acid), log2(D-Galacturonic acid), log2(N-Acetyl-L-alanine), log2(butyrylcarnitine), log2(cystathionine), log2(octanoylcarnitine) and age. Model 2 also included creatinine, bilirubin, INR and white cell count.
*P value <0.01 compared with the metabolomic model 1 for differences larger than 0.05.
†P value <0.01 compared with the metabolomic model 2 for differences larger than 0.05.
CLIF-C ACLF, Chronic Liver Failure Consortium ACLF; INR, international normalised ratio.
We then compared the short-term prognostic accuracy of models 1 and 2 with the MELDNa score at days 7, 14, 28 and 90 after enrolment in all patients included in the CANONIC and PREDICT cohorts (table 3) and in the subgroups of patients with or without ACLF separately (table 4). Models 1 and/or 2 were more accurate than the MELDNa score in the CANONIC discovery cohort at days 7, 14 and 28, and these findings were validated in the PREDICT cohort. In patients with or without ACLF, such differences extended up to day 90 after enrolment. The CLIF-C ACLF score also functioned better than the MELDNa score in patients with ACLF, while the CLIF-C AD score performed better than the MELDNa in patients without ACLF during the first 2 weeks and at 90 days.
Table 4.
Description of models to predict risk of mortality at several time points and their predictive ability in patients diagnosed with or without ACLF in the CANONIC (discovery set) and the PREDICT (validation set) study cohorts
| Model | C-index | ||||
| At 7 days | At 14 days | At 28 days | At 60 days | At 90 days | |
| AD-ACLF patients | |||||
| Discovery set: CANONIC study cohort (n=181) | |||||
| % of deaths | 7.73% | 18.23% | 26.52% | 35.91% | 40.33% |
| CLIF-C MET 1 | 0.868 | 0.844 | 0.794 | 0.757 | 0.744 |
| CLIF-C MET 2 | 0.875 | 0.872 | 0.819 | 0.791 | 0.772 |
| CLIF-C ACLF score | 0.757*† | 0.793† | 0.760† | 0.734 | 0.720† |
| MELDNa score | 0.671*†‡ | 0.711*†‡ | 0.696*†‡ | 0.707† | 0.703† |
| Validation set: PREDICT study cohort (n=109) | |||||
| % of deaths | 4.59% | 16.51% | 27.52% | 34.86% | 37.61% |
| CLIF-C MET 1 | 0.914 | 0.724 | 0.717 | 0.714 | 0.714 |
| CLIF-C MET 2 | 0.892 | 0.730 | 0.733 | 0.722 | 0.721 |
| CLIF-C ACLF score | 0.879 | 0.763 | 0.728 | 0.683† | 0.670*† |
| MELDNa score | 0.601*†‡ | 0.592*†‡ | 0.644*†‡ | 0.649*† | 0.643*† |
| AD-no ACLF patients | |||||
| Discovery set: CANONIC study cohort (n=650) | |||||
| % of deaths | 0.92% | 2.31% | 4.77% | 9.85% | 12.46% |
| CLIF-C MET 1 | 0.916 | 0.871 | 0.781 | 0.764 | 0.741 |
| CLIF-C MET 2 | 0.942 | 0.895 | 0.787 | 0.750 | 0.729 |
| CLIF-C AD score | 0.768*† | 0.769*† | 0.752 | 0.733 | 0.736 |
| MELDNa score | 0.715*† | 0.686*†‡ | 0.693*†‡ | 0.723 | 0.723 |
| Validation set: PREDICT study cohort (n=742) | |||||
| % of deaths | 0.67% | 2.83% | 6.06% | 12.40% | 15.36% |
| CLIF-C MET 1 | 0.928 | 0.841 | 0.769 | 0.769 | 0.760 |
| CLIF-C MET 2 | 0.945 | 0.864 | 0.781 | 0.769 | 0.759 |
| CLIF-C AD score | 0.839*† | 0.798† | 0.723 | 0.715 | 0.707 |
| MELDNa score | 0.757*†‡ | 0.726*†‡ | 0.694*† | 0.668*† | 0.660*†‡ |
*P value <0.01 compared to the metabolomic model 1 for differences larger than 0.05.
†P value <0.01 compared to the metabolomic model 2 for differences larger than 0.05.
‡P value <0.01 compared to the CLIF-C ACLF/CLIF-C AD score for differences larger than 0.05.
ACLF, acute-on-chronic liver failure; AD, acutely decompensated cirrhosis; CLIF-C ACLF, Chronic Liver Failure Consortium ACLF.
Table 2 (lower list) compares the individual predictive ability of the four standard laboratory variables of the MELDNa score, estimated through the C-index, in both study cohorts. Variables are ranked according to their corresponding C-index. The individual predictive ability of the standard laboratory variables was lower than that of any of the 15 top prognostic metabolites in both cohorts. The distinct predictive value of the MELD scores and the metabolomic models, therefore, relates to differences in the intrinsic predicting ability of their variables.
Finally, online supplemental figure 1 evaluates how patients with or without ACLF behave along time and the uncertainty their number might introduce. The incidence of mortality and its short-term distribution does not differ between cohorts (p value=0.8499 for patients with AD-ACLF, p value=0.2415 for AD-no ACLF patients). C-index estimates are more imprecise for patients with ACLF in the PREDICT cohort, with larger SD for all models and scores considered, while the opposite is found for AD-no ACLF patients.
Discussion
Patients with pre-ACLF and ACLF, who represent approximately 40% of patients hospitalised for AD, are at high risk of dying (25%) within 28 days after admission. Patients with ACLF are easily identifiable by clinical and standard laboratory features. However, those with pre-ACLF cannot be differentiated from the rest of the patients without ACLF and better prognosis. Patients hospitalised with AD, therefore, require highly sensitive short-term prognostic scoring systems for rapid indication of extreme therapeutic measures, including emergency liver transplantation. The MELDNa score is the universal model used for prediction of prognosis and prioritisation of liver transplantation in patients with AD.12 However, as suggested by recent studies,13–15 and clearly shown by the current investigation, MELD score and its modification (MELDNa score) are imperfect indicators of short-term mortality in patients with AD, pre-ACLF and ACLF.
In this study, we showed that the models CLIF-C MET, based on metabolites and more specifically from glucose, fatty acids and amino acid metabolic pathways, which currently can only be measured by liquid chromatography coupled to mass spectrometry, were significantly better prognosticators than MELD scores and its upgrades predicting short-term mortality in our 1682 patients hospitalised with AD. This finding was supported by the observation that the C-index of each of the top 15 prognostic metabolites identified in the CANONIC study cohort was higher than the C-index of each of the four standard laboratory variables included in the MELDNa score (INR, bilirubin, creatinine and sodium).
To value the superior accuracy of CLIF-C MET over the MELDNa score or of some metabolites versus others in predicting short-term mortality in AD, it is important to understand the mechanisms by which SI impacts on metabolism and survival. This issue, which has been extensively studied in sepsis,26–29 is also increasingly being explored in cirrhosis,20 30 31 with solid data showing that both conditions share similar pathophysiological pathways. Severe SI is among the most common acute stressful situations in humans and leads to a coordinated stress (alarm) response by the central nervous system, endocrine system and metabolism, to fight against the stressor.27–29 The routine inflammation markers (WCC, C-reactive protein (CRP), IL-6) either alone or in combination with (or within) CLIF-C AD and CLIF-C ACLF score performed worse than the new models in the prediction of mortality in patients with and without ACLF.
Most of the 29 metabolites included in the ‘death-related metabolomic fingerprint’ belonged to metabolic pathways related to the alarm response to SI and secondary impairment in mitochondrial function. The top short-term prognostic metabolite identified in our patients was 4-hydroxy-3-methoxyphenylglycol sulfate, which belongs to the phenylalanine-tyrosine pathway and is the terminal metabolite of norepinephrine and, therefore, marks the intensity of the alarm reaction to SI. In order to explain these findings, one should remember that the alarm response is initiated within specific areas in the brainstem and hypothalamus which have no blood-brain barrier and are inflamed by circulating pathogen associated molecular pattern (PAMPs), danger associated molecular pattern (DAMPs) and inflammatory mediators.27 Central nervous nodes (eg, locus coeruleous32) integrate inputs from multiple alarm responsive circuits and releases norepinephrine initiating the metabolic response to SI, which results in a massive release of glucose, fatty acids and amino acids from tissue stores to the periphery and prioritisation of energy metabolism to the immune cells, to cover the high energy demands of the systemic inflammatory reaction.28 29 As discussed above, the top prognostic metabolite identified in our patients was 4-hydroxy-3-methoxyphenylglycol sulfate, a terminal metabolite of norepinephrine closely related to the alarm reaction.
Interestingly, the acylcarnitine hexanoylcarnitine was the second top predictor of the 28-day mortality. Due to the downregulation of mitochondrial metabolism of fatty acids in AD, acylcarnitines accumulate in the cytosol and in blood, and are thus a sensitive marker of mitochondrial dysfunction. In our patients, there was downregulation of mitochondrial metabolism of fatty acids, as indicated by the high circulating levels of acylcarnitines. Inhibition of strategic mitochondrial enzymatic processes (pyruvate dehydrogenase complex and enzymes regulating β-oxidation) by SI likely contributes to the dysregulation of glucose and fatty acid metabolism in AD.33 However, in extreme cases with intense SI, that is, patients with rapid development of severe ACLF, an exaggerated mitochondrial generation of reactive oxygen species and physical mitochondrial damage may play a predominant role.33
Finally, five molecules derived from downregulation of the mitochondrial metabolism of glucose (galacturonic acid, glucuronic acid, pentose-phosphates, pentose alcohols and D-threitol) were components of the death-related metabolomic fingerprint. Among them, galacturonic acid, together with 4-hydroxy-3-methoxyphenylglycol sulfate and hexanoylcarnitine, was the third independent predictor of the 28-day survival identified in our patients. The individual 28-day prognostic accuracy of 4-hydroxy-3-methoxyphenylglycol sulfate and hexanoylcarnitine (C-index 0.822 and 0.799, respectively) exceeded the accuracy of the MELDNa score in the whole CANONIC study cohort (0.788) and particularly in the subgroup of patients with ACLF (0.6). Therefore, similar to sepsis,26 the second most relevant metabolic change associated with SI in our patients indicates a dysregulation of intracellular metabolism of glucose and fatty acids by the immune cells. Instead of being sequentially metabolised by glycolysis in the cytosol and by the Krebs cycle and oxidative phosphorylation in the mitochondria, glucose metabolism in our patients was probably metabolised in the cytosol through glycolysis and the pentose phosphate pathway. This study validates the pathophysiological importance of mitochondrial dysfunction not only in immune cells but probably also in organs inducing dysfunction and/or failure.
Consistent with the above observations, model 1, including age, the plasma levels of 4-hydroxy-3-methoxyphenylglycol sulfate, hexanoylcarnitine and galacturonic acid, showed significantly higher short-term prognostic accuracy than the MELDNa score in the CANONIC and PREDICT cohorts. The accuracy of this model was also remarkably higher than the accuracy of the MELDNa score in patients with ACLF, for whom emergency liver transplantation is specifically indicated.15 34 35 In patients without ACLF, CLIF-C MET models also improved the predictive ability of MELDNa score. While CLIF-C AD score accuracy was considerably worse than the metabolomic model 1, we reported minor differences because of the restrictions we imposed for the comparisons. Defining a minimum difference to be found between C-indices is not common practice, but we believe it gives clinical relevance to the results obtained. It is interesting to note that model 2, which used the two top prognostic metabolites and prognostic standard laboratory tests, did not significantly improve the results obtained by model 1 using only metabolites.
The reduced number of patients with ACLF in the PREDICT cohort did not seem a source of bias in the C-index estimates, as mortality occurrence and distribution along time were not different between cohorts. The uncertainty in such estimates was slightly larger for all models in the validation cohort for this subset of patients, which could be partially explained by its reduced sample size. Even with this increase in the uncertainty, we could still find significant differences among the models’ accuracy.
Our study has one potential limitation. While the CANONIC study was performed in consecutive patients hospitalised with AD, the PREDICT study initially enrolled only patients without ACLF and later incorporated patients hospitalised with ACLF, leading to differences in the prevalence of organ failures between the two cohorts. However, the two cohorts presented a high level of agreement in the composition of metabolomic fingerprints, top prognostic metabolites and superiority of metabolomic models over MELD scores in predicting short-term mortality, indicating that this limitation had no great impact on the results.
On the contrary, our investigation had several major strengths. First, it was performed in the largest series of patients (1682) hospitalised with AD reported to date. Second, it is the first prognostic metabolomic study in cirrhosis whose results were confirmed in a large external validation cohort. Third, our study used a liquid chromatography platform coupled with electrospray ionisation high-resolution mass spectrometry and an analytical pipeline that allowed us to retain high-confidence metabolite annotations, based on a combination of accurate mass, retention time and tandem mass spectrometry characteristics. This approach reduced the dimension of the dataset from hundreds of metabolic features (‘peaks’) into a manageable number of ca. 130 metabolites with interpretable biological and pathophysiological significance in a complex context of severe SI and multiorgan dysfunction/failure. This pragmatic strategy, which in the end led to the identification of a death-related metabolomic fingerprint and specifically three highly sensitive prognostic metabolites closely related to new pathophysiological mechanisms of AD (alarm response and mitochondrial dysfunction associated with SI), paves the way for future implementation of metabolomics in routine clinical practice, as previously described.28 29 Finally, in our study, the metabolomic assessment was performed shortly after the onset of AD in all our patients, when SI and metabolic dysregulation were maximal, as suggested before.30 31 Therefore, we captured the most important metabolic changes promoted by the exogenous (infections or acute alcoholic hepatitis) and endogenous (bacterial translocation) proinflammatory precipitants of AD, and this may also contribute to the higher accuracy of metabolomic scores versus clinical scores based on liver and renal function. For example, while in patients with pre-ACLF, organ function deteriorates days or weeks after the onset of AD, limiting short-term mortality prediction of standard laboratory variables in a significant number of patients, these subjects already show severe SI1 and likely metabolomic derangement at the onset of the process.
In summary, our investigation uncovered four features that may represent major steps forward for identification of prognosis and early management of patients with AD: (1) It identified a death-related metabolomic fingerprint of 29 molecules that correlated with short-term mortality. (2) It identified two top metabolites from this fingerprint (4-hydroxy-3-methoxyphenylglycol sulfate and hexanoylcarnitine), which were individually better predictors of short-term mortality than the MELDNa score, and the galacturonic acid, which was also independently associated with short-term mortality. (3) Such biomarkers are related to the alarm response to SI (4-hydroxy-3-methoxyphenylglycol sulfate) and the secondary impairment in mitochondrial function (hexanoylcarnitine and galacturonic acid). (4) CLIF-C MET models based on these metabolites were better predictors of short-term mortality than MELD scores in patients with AD, and particularly when separating patients with or without ACLF and may improve organ allocation and promote expediting of liver transplantation in these patients. However, prospective validation and cost-effective analysis on the performance of these markers is required before routine implementation in the clinics.
Acknowledgments
The study was supported by the European Foundation for the Study of Chronic Liver Failure (EF CLIF). EF CLIF is a non-profit private organisation. EF CLIF receives unrestricted donations from Cellex Foundation and Grifols and is partner or contributor in several EU Horizon 2020 program projects MICROB-PREDICT (project ID 825694), DECISION (project ID 847949) and IHMCSA (project ID 964590). The funders had no influence on study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Twitter: @tgustot, @Zac_MD, @tony_bruns, @JosPresa3
Correction notice: This article has been corrected since it published Online First. The affiliation has been updated for the author Paolo Caraceni.
Collaborators: PREDICT Study Investigators of the EASL–CLIF Consortium: Jonel Trebicka, Javier Fernandez, Maria Papp, Paolo Caraceni, Wim Laleman, Carmine Gambino, Ilaria Giovo, Frank Erhard Uschner, Christian Jansen, Cesar Jimenez, Rajeshwar Mookerjee, Thierry Gustot, Agustin Albillos, Rafael Bañares, Peter Jarcuska, Christian Steib, Thomas Reiberger, Juan Acevedo, Pietro Gatti, Debbie L. Shawcross, Stefan Zeuzem, Alexander Zipprich, Salvatore Piano, Thomas Berg, Tony Bruns, Karen Vagner Danielsen, Minneke Coenraad, Manuela Merli, Rudolf Stauber, Heinz Zoller, José Presa Ramos, Cristina Solé, Germán Soriano, Andrea de Gottardi, Henning Gronbaek, Faouzi Saliba, Christian Trautwein, Haluk Tarik Kani, Sven Francque, Stephen Ryder, Pierre Nahon, Manuel Romero-Gomez, Hans Van Vlierberghe, Claire Francoz, Michael Manns, Elisabet Garcia-Lopez, Manuel Tufoni, Alex Amoros, Marco Pavesi, Cristina Sanchez, Michael Praktiknjo, Anna Curto, Carla Pitarch, Antonella Putignano, William Bernal, Ferran Aguilar, Joan Clària, Paola Ponzo, Zsuzsanna Vitalis, Giacomo Zaccherini, Boglarka Balogh, Alexander Gerbes, Victor Vargas, Carlo Alessandria, Mauro Bernardi, Pere Ginès, Richard Moreau, Paolo Angeli, Rajiv Jalan, Vicente ArroyoMiriam Maschmeier, David Semela, Laure Elkrief, Ahmed Elsharkawy, Tamas Tornai, Istvan Tornai, Istvan Altorjay, Agnese Antognoli, Maurizio Baldassarre, Martina Gagliardi, Eleonora Bertoli, Sara Mareso, Alessandra Brocca, Daniela Campion, Giorgio Maria Saracco, Martina Rizzo, Jennifer Lehmann, Alessandra Pohlmann, Maximilian J. Brol, Johannes Chang, Robert Schierwagen, Elsa Solà, Nesrine Amari, Miguel Rodriguez, Frederik Nevens, Ana Clemente, Martin Janicko, Daniel Markwardt, Mattias Mandorfer, Christoph Welsch, Tanja M. Welzel, Emanuela Ciraci, Vish Patel, Cristina Ripoll, Adam Herber, Paul Horn, Flemming Bendtsen, Lise Lotte Gluud, Jelte Schaapman, Oliviero Riggio, Florian Rainer, Jörg Tobiasch Moritz, Mónica Mesquita, Edilmar Alvarado-Tapias, Osagie Akpata, Luise Aamann, Didier Samuel, Sylvie Tresson, Pavel Strnad, Roland Amathieu, Macarena Simón-Talero, Francois Smits, Natalie van den Ende, Javier Martinez, Rita Garcia, Harald Rupprechter, Cornelius Engelmann, Osman Cavit Özdogan;
CANONIC Study Investigators of the EASL–CLIF Consortium: Patricia Aguilar Melero, Agustin Albillos, Carlo Alessandria, Paolo Angeli, Vicente Arroyo, Rafael Bañares, Daniel Benten, Mauro Bernardi, Massimo Bocci, Paolo Caraceni, María-Vega Catalina, Jun Liong Chin, Minneke J Coenraad, Audrey Coilly, Mar Concepción, Juan Cordoba, Andrea de Gottardi, Manuel de la Mata, Carme Deulofeu, Marco Domenicali, Livia Dorn, François Durand, Laure Elkrief, Javier Fernandez, Elisabet Garcia, Angelo Gatta, Ludmila Gerber, Alexander Gerbes, Pere Ginès, Henning Grøenbæk, Monica Guevara, Thierry Gustot, AnneKristin Hausen, Corinna Hopf, Rajiv Jalan, Stine Karlsen, Wim Laleman, Ansgar W Lohse, Caterina Maggioli, Daniel Markwardt, Javier Martinez, Alfredo Marzano, P Aiden McCormick, Francisco Mesonero, José Luis Montero Álvarez, Rajeshwar P Mookerjee, Filippo Morando, Richard Moreau, Christophe Moreno, Bernhard Morrell, Christian Mortensen, Frederik Nevens, Marco Pavesi, Markus Peck-Radosavljevic, Gustavo Pereira, Alessandro Risso, Mario Rizzetto, Ezequiel Rodriguez, Antonietta Romano, Faouzi Saliba, Didier Samuel, Tilman Sauerbruch, Macarena Simon-Talero, Pablo Solis-Muñoz, German Soriano, Jan Sperl, Walter Spindelboeck, Rudolf Stauber, Christian Steib, Jonel Trebicka, Dominique Valla, Hans Van Vlierberghe, Len Verbeke, Wolfgang Vogel, Henninge Wege, Tania Welzel, Julia Wendon, Chris Willars, Maria Yago Baenas, Giacomo Zaccherini, Stefan Zeuzem.
Contributors: Study concept and design: EW, JT, RM, JC, VA. Acquisition of data: JMD, TG, WL, AA, CA, MD, PC, SP, FS, SZ, AG, JW, CI, MP, RM, CG, CJ, IG, GZ, MM, AP, RB, JP, LV, JF. Logistics and data management: CS, PS, PIB. Statistical analysis and bioinformatics: CdlP-R, FA, JJL. Metabolomics: FC, FF, CJ. Drafting the manuscript: EW, JT, RM, VA. critical revision of the manuscript: JC, VV, JG, JF, MB, PA, RJ. Guarantor: VA.
Funding: Cellex Foundation
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was performed in accordance with the principles of the Declaration of Helsinki. All study participants gave written informed consent. The study was approved by the local ethics committees (Lead Ethics Committee University of Bonn 311/16) of the participating centres. The PREDICT study has been registered in ClinicalTrials.gov as NCT03056612. Participants gave informed consent to participate in the study before taking part.
References
- 1. Trebicka J, Fernandez J, Papp M, et al. The predict study uncovers three clinical courses of acutely decompensated cirrhosis that have distinct pathophysiology. J Hepatol 2020;73:842–54. 10.1016/j.jhep.2020.06.013 [DOI] [PubMed] [Google Scholar]
- 2. Arroyo V, Angeli P, Moreau R, et al. The systemic inflammation hypothesis: towards a new paradigm of acute decompensation and multiorgan failure in cirrhosis. J Hepatol 2021;74:670–85. 10.1016/j.jhep.2020.11.048 [DOI] [PubMed] [Google Scholar]
- 3. Moreau R, Jalan R, Gines P, et al. Acute-on-chronic liver failure is a distinct syndrome that develops in patients with acute decompensation of cirrhosis. Gastroenterology 2013;144:1426–37. 10.1053/j.gastro.2013.02.042 [DOI] [PubMed] [Google Scholar]
- 4. Clària J, Stauber RE, Coenraad MJ, et al. Systemic inflammation in decompensated cirrhosis: characterization and role in acute-on-chronic liver failure. Hepatology 2016;64:1249–64. 10.1002/hep.28740 [DOI] [PubMed] [Google Scholar]
- 5. Trebicka J, Amoros A, Pitarch C, et al. Addressing profiles of systemic inflammation across the different clinical phenotypes of acutely decompensated cirrhosis. Front Immunol 2019;10:476. 10.3389/fimmu.2019.00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trebicka J, Fernandez J, Papp M, et al. Predict identifies precipitating events associated with the clinical course of acutely decompensated cirrhosis. J Hepatol 2021;74:1097–108. 10.1016/j.jhep.2020.11.019 [DOI] [PubMed] [Google Scholar]
- 7. Trebicka J, Bork P, Krag A, et al. Utilizing the gut microbiome in decompensated cirrhosis and acute-on-chronic liver failure. Nat Rev Gastroenterol Hepatol 2021;18:167–80. 10.1038/s41575-020-00376-3 [DOI] [PubMed] [Google Scholar]
- 8. Fernández J, Clària J, Amorós A, et al. Effects of albumin treatment on systemic and portal hemodynamics and systemic inflammation in patients with decompensated cirrhosis. Gastroenterology 2019;157:149–62. 10.1053/j.gastro.2019.03.021 [DOI] [PubMed] [Google Scholar]
- 9. Gustot T, Fernandez J, Garcia E, et al. Clinical course of acute-on-chronic liver failure syndrome and effects on prognosis. Hepatology 2015;62:243–52. 10.1002/hep.27849 [DOI] [PubMed] [Google Scholar]
- 10. Hernaez R, Solà E, Moreau R, et al. Acute-On-Chronic liver failure: an update. Gut 2017;66:541–53. 10.1136/gutjnl-2016-312670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mezzano G, Juanola A, Cardenas A, et al. Global burden of disease: acute-on-chronic liver failure, a systematic review and meta-analysis. Gut 2022;71:148–55. 10.1136/gutjnl-2020-322161 [DOI] [PubMed] [Google Scholar]
- 12. Schulz M, Trebicka J. Acute-on-chronic liver failure: a global disease. Gut 2022;71:5–6. 10.1136/gutjnl-2020-323973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamath PS, Wiesner RH, Malinchoc M, et al. A model to predict survival in patients with end-stage liver disease. Hepatology 2001;33:464–70. 10.1053/jhep.2001.22172 [DOI] [PubMed] [Google Scholar]
- 14. Kim WR, Biggins SW, Kremers WK, et al. Hyponatremia and mortality among patients on the liver-transplant waiting list. N Engl J Med 2008;359:1018–26. 10.1056/NEJMoa0801209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim WR, Mannalithara A, Heimbach JK, et al. MELD 3.0: the model for end-stage liver disease updated for the modern era. Gastroenterology 2021;161:1887–95. 10.1053/j.gastro.2021.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jalan R, Saliba F, Pavesi M, et al. Development and validation of a prognostic score to predict mortality in patients with acute-on-chronic liver failure. J Hepatol 2014;61:1038–47. 10.1016/j.jhep.2014.06.012 [DOI] [PubMed] [Google Scholar]
- 17. Wu T, Li J, Shao L, et al. Development of diagnostic criteria and a prognostic score for hepatitis B virus-related acute-on-chronic liver failure. Gut 2018;67:2181–91. 10.1136/gutjnl-2017-314641 [DOI] [PubMed] [Google Scholar]
- 18. Sundaram V, Jalan R, Wu T, et al. Factors associated with survival of patients with severe acute-on-chronic liver failure before and after liver transplantation. Gastroenterology 2019;156:1381–91. 10.1053/j.gastro.2018.12.007 [DOI] [PubMed] [Google Scholar]
- 19. Roberts I, Wright Muelas M, Taylor JM, et al. Untargeted metabolomics of COVID-19 patient serum reveals potential prognostic markers of both severity and outcome. Metabolomics 2021;18:6. 10.1007/s11306-021-01859-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Valdés A, Moreno LO, Rello SR, et al. Metabolomics study of COVID-19 patients in four different clinical stages. Sci Rep 2022;12:1650. 10.1038/s41598-022-05667-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Z, Triba MN, Amathieu R, et al. Nuclear magnetic resonance-based serum metabolomic analysis reveals different disease evolution profiles between septic shock survivors and non-survivors. Crit Care 2019;23:169. 10.1186/s13054-019-2456-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eckerle M, Ambroggio L, Puskarich MA, et al. Metabolomics as a driver in advancing precision medicine in sepsis. Pharmacotherapy 2017;37:1023–32. 10.1002/phar.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moreau R, Clària J, Aguilar F, et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J Hepatol 2020;72:688–701. 10.1016/j.jhep.2019.11.009 [DOI] [PubMed] [Google Scholar]
- 24. Boudah S, Olivier M-F, Aros-Calt S, et al. Annotation of the human serum metabolome by coupling three liquid chromatography methods to high-resolution mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2014;966:34–47. 10.1016/j.jchromb.2014.04.025 [DOI] [PubMed] [Google Scholar]
- 25. Imbert A, Rompais M, Selloum M, et al. ProMetIS, deep phenotyping of mouse models by combined proteomics and metabolomics analysis. Sci Data 2021;8:311. 10.1038/s41597-021-01095-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giacomoni F, Le Corguillé G, Monsoor M, et al. Workflow4Metabolomics: a collaborative research infrastructure for computational metabolomics. Bioinformatics 2015;31:1493–5. 10.1093/bioinformatics/btu813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dunn WB, Broadhurst D, Begley P, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 2011;6:1060–83. 10.1038/nprot.2011.335 [DOI] [PubMed] [Google Scholar]
- 28. Bajaj JS, Reddy KR, Tandon P, et al. Association of serum metabolites and gut microbiota at hospital admission with nosocomial infection development in patients with cirrhosis. Liver Transpl 2022;28:1831–40. 10.1002/lt.26552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bajaj JS, Tandon P, O’Leary JG, et al. Admission serum metabolites and thyroxine predict advanced hepatic encephalopathy in a multicenter inpatient cirrhosis cohort. Clin Gastroenterol Hepatol 2022. 10.1016/j.cgh.2022.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Monteiro S, Grandt J, Uschner FE, et al. Differential inflammasome activation predisposes to acute-on-chronic liver failure in human and experimental cirrhosis with and without previous decompensation. Gut 2021;70:379–87. 10.1136/gutjnl-2019-320170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J, Liang X, Jiang J, et al. Pbmc transcriptomics identifies immune-metabolism disorder during the development of HBV-ACLF. Gut 2022;71:163–75. 10.1136/gutjnl-2020-323395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwarz LA, Luo L. Organization of the locus coeruleus-norepinephrine system. Curr Biol 2015;25:R1051–6. 10.1016/j.cub.2015.09.039 [DOI] [PubMed] [Google Scholar]
- 33. Zhang IW, Curto A, López-Vicario C, et al. Mitochondrial dysfunction governs immunometabolism in leukocytes of patients with acute-on-chronic liver failure. J Hepatol 2022;76:93–106. 10.1016/j.jhep.2021.08.009 [DOI] [PubMed] [Google Scholar]
- 34. Arroyo V. Acute-on-chronic liver failure in cirrhosis requires expedited decisions for liver transplantation. Gastroenterology 2019;156:1248–9. 10.1053/j.gastro.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 35. Belli LS, Duvoux C, Artzner T, et al. Liver transplantation for patients with acute-on-chronic liver failure (ACLF) in europe: results of the ELITA/EF-CLIF collaborative study (ECLIS). J Hepatol 2021;75:610–22. 10.1016/j.jhep.2021.03.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-328708supp001.pdf (252.9KB, pdf)
Data Availability Statement
Data are available upon reasonable request.