Abstract
Objective
To explore the interplay between dietary modifications, microbiome composition and host metabolic responses in a dietary intervention setting of a personalised postprandial-targeting (PPT) diet versus a Mediterranean (MED) diet in pre-diabetes.
Design
In a 6-month dietary intervention, adults with pre-diabetes were randomly assigned to follow an MED or PPT diet (based on a machine-learning algorithm for predicting postprandial glucose responses). Data collected at baseline and 6 months from 200 participants who completed the intervention included: dietary data from self-recorded logging using a smartphone application, gut microbiome data from shotgun metagenomics sequencing of faecal samples, and clinical data from continuous glucose monitoring, blood biomarkers and anthropometrics.
Results
PPT diet induced more prominent changes to the gut microbiome composition, compared with MED diet, consistent with overall greater dietary modifications observed. Particularly, microbiome alpha-diversity increased significantly in PPT (p=0.007) but not in MED arm (p=0.18). Post hoc analysis of changes in multiple dietary features, including food-categories, nutrients and PPT-adherence score across the cohort, demonstrated significant associations between specific dietary changes and species-level changes in microbiome composition. Furthermore, using causal mediation analysis we detect nine microbial species that partially mediate the association between specific dietary changes and clinical outcomes, including three species (from Bacteroidales, Lachnospiraceae, Oscillospirales orders) that mediate the association between PPT-adherence score and clinical outcomes of hemoglobin A1c (HbA1c), high-density lipoprotein cholesterol (HDL-C) and triglycerides. Finally, using machine-learning models trained on dietary changes and baseline clinical data, we predict personalised metabolic responses to dietary modifications and assess features importance for clinical improvement in cardiometabolic markers of blood lipids, glycaemic control and body weight.
Conclusions
Our findings support the role of gut microbiome in modulating the effects of dietary modifications on cardiometabolic outcomes, and advance the concept of precision nutrition strategies for reducing comorbidities in pre-diabetes.
Trial registration number
Keywords: diet, diabetes mellitus, nutrition
WHAT IS ALREADY KNOWN ON THIS TOPIC
Previously reported clinical results from an randomised controlled trial in pre-diabetes demonstrated beneficial effects of a personalised postprandial-targeting (PPT) diet on glycaemic and other cardiometabolic outcomes, beyond the established benefits of a Mediterranean-style diet.
Gut microbiome is intimately linked to host diet and cardiometabolic health, but current knowledge about gut microbiome at the intersection between diet and human health mostly relies on observational studies.
WHAT THIS STUDY ADDS
PPT diet prompted greater dietary modifications and increased microbiome diversity and richness.
A causal mediation analysis demonstrates a partial mediatory role for nine microbial species in the association between specific dietary changes and clinical outcomes.
Machine-learning models trained on dietary changes and baseline clinical data predict personalised metabolic responses to dietary modifications in several cardiometabolic markers of blood lipids, glycaemic control and body weight.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our findings suggest concrete microbiota species targets, which may form the basis for future mechanistic experiments on their role in human diet and health, as well as potential therapeutic directions to be evaluated in preclinical and intervention studies to improve cardiometabolic health in pre-diabetes.
Introduction
Diet is a major contributor to cardiometabolic health and plays a fundamental role in the prevention, management and even reversal of many chronic diseases, including but not limited to diabetes, cardiovascular disease (CVD) and fatty liver disease.1 2 Increasing evidence suggests complex nutrient-disease relationships with numerous factors affecting the relative contribution of a specific nutrient to disease risk, as has been suggested for example for saturated fatty acids (SFAs) and CVD.3 This comprehension has recently led international health organisations to shift the focus of nutritional recommendations in dietary guidelines away from macronutrient composition towards the quality of foods and dietary patterns.4 However, there is high interpersonal variability in metabolic responses to dietary interventions even when evaluating nutritional intake by means of specific foods or dietary patterns rather than macronutrient composition.5–7 This suggests that while dietary guidelines are based on population averages, a deeper understanding of the factors that drive the variation between individuals in metabolic response to dietary modifications may inform more effective and precise dietary recommendations to individuals in clinical practice.8
The gut microbiota, which consists of trillions of bacterial microorganisms, has a central role in human health and disease.9 10 Specifically, its role in cardiometabolic health has been studied extensively in recent years, including in diabetes,11 12 obesity13 14 and CVD.15 16 Gut microbiome is also intimately linked in a bidirectional relationship with the host diet. On the one hand, gut microbiome significantly affects host metabolism and how one responds to food.5 6 17 On the other hand, gut microbial composition is responsive to the host diet which may exert downstream effects on human health.18–20 Additionally, since the gut microbiome is highly individualised in its nature, it is reasonable to hypothesise that it has a significant role in the highly variable effects that specific dietary interventions have on individual cardiometabolic outcomes. However, despite the recognition of strong links between microbiomes with both cardiometabolic health and the host diet, most studies that explored the interplay between these three elements in humans were observational,21 22 and only few formally tested it in interventional settings.23–26
We recently reported the clinical results of a randomised controlled trial (RCT) in adults with pre-diabetes that demonstrated beneficial effects of a personalised postprandial-targeting (PPT) diet, which is based on a machine-learning algorithm for predicting personal postprandial glucose responses (PPGR), on primary glycaemic outcomes and other secondary metabolic outcomes, beyond the established metabolic benefits of a Mediterranean-style diet.27 In the current work, we perform ad hoc analyses aimed at evaluating other exploratory outcomes of the 6-month dietary intervention, including high-resolution assessment of the dietary changes and microbiome compositional changes observed on both arms. We further explore associations between changes in multiple dietary features, microbiome species and metabolic readouts across the cohort (regardless of arm assignment) and evaluate the role of microbiota species in mediating the effects of dietary changes on metabolic outcomes. Finally, we devise machine-learning models trained on dietary changes and baseline clinical data, to predict personalised metabolic responses to dietary modifications and assess features importance for clinical improvement in cardiometabolic markers of blood lipids, glycaemic control and body weight in this RCT setting.
Methods
Recruitment to the study
This randomised clinical trial (ClinicalTrials.gov ID: NCT03222791) was carried out at the Weizmann Institute of Science, Israel, between January 2017 and March 2020. All details regarding recruitment to the study were previously published in the primary publication of this RCT.27 In short, inclusion criteria included: (1) fasting plasma glucose (FPG) levels between 100 and 125 mg/dL (5.6 and 6.9 mmol/L), (2) HbA1c level between 5.7 and 6.5% (39 and 48 mmol/mol), (3) age of 18–65 years and (4) capability to work with a smartphone app on a daily basis (for dietary intake logging). Key exclusion criteria were any use of diabetes or weight loss medications, use of antibiotics in the 3 months before enrollment, diagnosed chronic diseases or chronic use of medications that affect glucose/energy metabolism or HbA1c. Volunteers from Israel who self-reportedly declared themselves as having pre-diabetes on the trial website were invited for a screening visit to determine eligibility according to measured FPG and HbA1c at the trial’s central laboratory. Eligible participants were then invited for a ‘profiling’ visit at the trial’s site (Weizmann Institute of Science) during which they were informed in detail of all study procedures and requirements, provided a stool sample for baseline microbiome analysis (required for algorithm predictions), and were connected to continuous glucose monitoring (CGM) sensors (FreeStyle Libre, Abbott Laboratories) for a run-in period of 2–4 weeks before the start of the intervention. All participants signed an informed consent prior to participation.
Study design, randomisation and interventions
Design and interventions of this RCT, as well as primary and secondary clinical outcomes of the trial, were previously reported in detail.27 The current report is focused on exploratory outcomes of this trial, including high-resolution assessment of the 6-month dietary and microbiome compositional changes observed in the 200 participants who completed the intervention (baseline characteristics summarised in online supplemental table 1), and investigation of the gut microbiota’s role at the intersection between diet and human health (figure 1A). Briefly, the study was a biphasic, randomised, controlled, single-blind dietary intervention. Phase 1 included a 6-month intervention that compared two diets targeting glycaemic control, while phase two included a 6-month follow-up period. The two dietary interventions included (1) an MED diet and (2) an algorithm-based personalised PPT diet aimed at lowering PPGRs with real-time feedback through a smartphone application (app). After completion of the run-in period, participants were randomly assigned in a 1:1 ratio to an MED or a PPT diet, while ensuring minimal differences between the groups in six prognostic baseline characteristics: sex, age, weight, body mass index (BMI), HbA1c and FPG. Participants and medical lab examiners were blinded to arm assignment, while the investigators and dietitians were not. Dietary guidance was provided by certified dietitians, with identical number and length of sessions and other monitoring and mentoring tools to ensure equal support and intervention intensity in both arms.
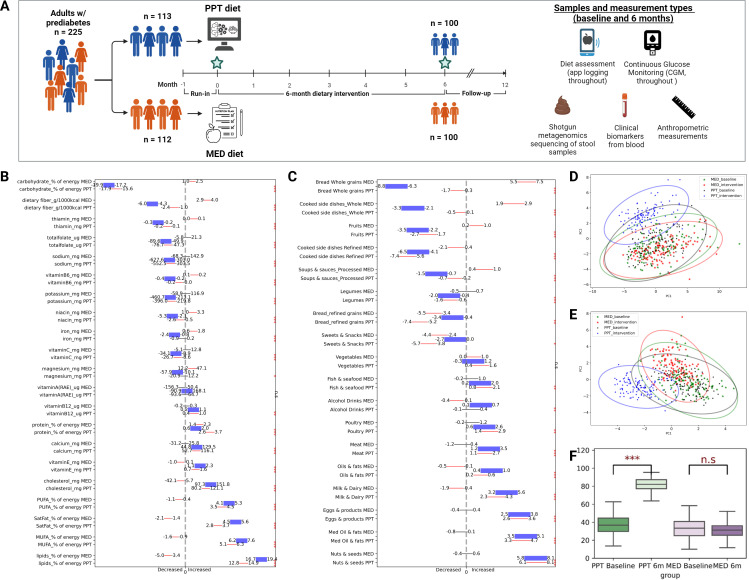
Figure 1.
PPT intervention induces greater changes in multiple dietary features compared to MED intervention. (A) Study design scheme and total sample count by category. Artwork created with ‘BioRender.com’. (B) CIs of 6 months changes in nutrients consumption. For each nutrient on the y-axis, the upper and lower thin lines indicate CIs of change within the MED and PPT arms, respectively. CIs length is normalised by the mean change in the respective dietary feature across the cohort. The thick purple line indicates CI of between group change difference (PPT change minus MED change). Red and black lines denote significant (p<0.05) and non-significant changes within arms, respectively (one-sided t-test). Asterisks on the right side of the panel denote between-group significant differences (two-sided t-test, *p<0.05, **p<0.01, ***p<0.001. n.s, non significant). (C) As in B but for 6 months changes in food categories consumption (presented as per cent of total energy intake). (D, E) Principal component analysis (PCA) of dietary consumption by arm and study phase (baseline vs intervention) evaluated based on nutrients (D) or food categories (E). (F) Box plots showing the PPT adherence score by arm and study phase (baseline vs intervention). Bars on top denote significance level for difference within arms (***p<0.001. n.s, non significant). PPT, personalised postprandial-targeting.
gutjnl-2022-329201supp001.xlsx (122.6KB, xlsx)
Dietary recommendations for both groups were administered as menus, with meals selected from a meal bank generated for this study. The selection of meals for the menus relied on the diet principles in each group, as previously described in detail.27 In short, menus were designed in a way that will allow diversity, guarantee safety (a balanced diet) and suit personal taste. Participants also received, on inquiring, recommendations or discouragement to consume any other desired food or meal outside their menus, depending on the principles of the diet arm they were assigned to (for PPT, based on the algorithm; for MED, based on dietitian judgement). Meals were self prepared by participants at home. By design, no total calorie restriction was advised and menus were designed with a daily caloric target that was personally set to each participant to match their estimated energy expenditure.
The design of the two diets in this RCT was previously described in detail.27 In short, the MED diet was based on standard-of-care recommendations as commonly advised by certified dietitians in Israel.28 Recommended foods on the MED diet included whole-wheat bread and grains, legumes, low-fat dairy products, fish, poultry, olive oil, fruits and vegetables. Discouraged foods included commercial bakery goods, sweets and pastries, fried foods and snacks, fatty and processed meat, and high-fat dairy products. Menus in this diet arm were designed with the following diet composition: 45%–65% of energy intake from carbohydrates, 15%–20% from protein and <35% from fat, with <10% from saturated fat. The PPT diet was based on personal predicted glucose responses according to a previously published algorithm that integrates clinical and gut microbiome features to predict PPGR to meals, and that was adjusted for use in a clinical setting.5 27 In short, among the features used by the algorithm to predict PPGR to meals were anthropometrics, blood tests (FPG, HbA1c% and haemoglobin), lifestyle features derived from questionnaires, and microbiome features: abundances of species estimated by MetaPhlAn2 and functional modules estimated with mapping to KEGG database. Additionally, macronutrient and micronutrient composition of the meal was used and features extracted from a person’s CGM (percentiles of blood glucose). Since no events around the meal were used for prediction, trained predictors could predict response for any profiled participant to any given meal. By design, macronutrient composition was not predetermined for the PPT diet.
Dietary and adherence assessment
As part of the study design, participants in both arms were asked to record their full dietary intake in real time every day for the full study period using a designated smartphone app (‘Personalised Nutrition Project’) that we developed. Each food item within every meal was logged along with its weight or portion units by selecting it from a database of >7000 foods with full nutritional values based on the Israeli Ministry of Health database that we further expanded with additional items from certified sources. Adherence to the prescribed diets was evaluated during the intervention by the self-recorded dietary intake in the logging app and by monthly follow-up questionnaires.
In the current work, we conducted a high-resolution assessment of the actual dietary changes observed, by extracting dietary data from the extensive self-reported meal logging (mean±SD of 747±299 meals (range 26–1618) and 242 000±97 000 kcal (range 12 084–521 330) per person). Analyses include 21 macronutrients and micronutrients and 18 food categories. For macronutrients (carbohydrates, protein, total dietary fat, saturated fatty acids (hereafter ‘SatFat’), monounsaturated fatty acids (hereafter ‘MUFA’) and polyunsaturated fatty acids (hereafter ‘PUFA’)), values were computed as proportion (%) of kcal intake per day. Total dietary fibre was evaluated as gram per 1000 kcal consumed per day, as accepted. For micronutrients (cholesterol, calcium, magnesium, iron, potassium, sodium, vitamin A (RAE), vitamin B1 (thiamin), vitamin B3 (niacin), vitamin B6 (pyridoxine), vitamin B9 (total folate) vitamin B12, vitamin C, vitamin E), values were computed as absolute amount consumed (mg or µg) per day, as accepted. Micronutrients with counts close to 0 or that were not correctly annotated across the food database were not included in the analysis (eg, vitamin K, vitamin B7 (biotin), iodine, trans fatty acids). Food categories were determined by assigning all food items recorded by participants on the mobile app (>10 counts) into 18 common food categories (based on botanical and nutritional properties). Consumption of food categories was evaluated as proportion (%) from energy intake per day. Dietary changes in all features were assessed as the delta between mean daily consumption over the 6-month intervention and mean daily consumption over the run-in (profiling) phase.
To encourage dietary adherence and self-monitoring during the intervention, we originally devised composite adherence scores on a scale of 0–100 (from worst to best) for both MED and PPT diets, based on their respective dietary principles. These diet scores were delivered to participants during the intervention, aiming to motivate and convey to them their compliance to their assigned diets in terms of meal composition, as previously described in detail.27 Here, we took advantage of the thorough dietary data collected from participants’ logging and our ability to run algorithm PPGR predictions for every profiled participant, to compute, as a post hoc analysis (ie, retrospectively), the PPT adherence score across the entire cohort (ie, regardless of arm assignment). In short, the PPT adherence score was devised to indicate how well one sticks to algorithm-based recommendations. As such, the calculation of the PPT adherence score was based on our meal recommendation scoring system, which stands at the core of the PPT approach (meal scores 1–5, best to worst, as previously described in detail).27 Each meal recommendation score was assigned with an adherence score as follows: meal score 1=adherence score 100; meal score 2=80; meal score 3=50; meal score 4=25; meal score 5=0. These meal adherence scores were averaged calorie-wise (meal energy trimmed to be within 100–500 kcal interval) in order to proportionally account for the contribution of large versus small meals to the overall adherence score. For example, if a person logged three meals: 600 kcal of meal score 2, 1000 kcal of meal score 5 and 80 kcal of meal score 1, he received a PPT adherence score of: (500×80+500×0+100×100)/(500+500+100)=45. The PPT adherence scores were computed per day and averaged over the run-in and intervention phases. If too few (100 by default) calories were logged (overall), we did not compute an adherence score for that day.
Blood, CGM and anthropometric measurements
Blood samples were obtained at baseline and 6 months, at the trial site (Weizmann Institute of Science) or at the central medical laboratory of the trial (AMC Medical Center Laboratory). All blood specimens were processed and biochemical assays performed by one technician at the central laboratory, who was not aware of arm assignment or any other characteristics of participants. All blood biochemical assays were previously described in detail.27
To obtain the most informative view of glucose levels possible, participants in both arms were continuously connected to CGM sensors throughout the run-in and the entire intervention period (15 727±4430 glucose measurements per person), with sensors replaced every 2 weeks and participants being blinded to glucose tracings. The CGM measurements were used to calculate measures of glycaemic control, including daily time of glucose levels above 140 mg/dL (7.8 mmol/L) (hereafter ‘time>140’) and mean CGM glucose levels.
Anthropometric measurements were taken at the trial site (Weizmann Institute of Science) during the monthly follow-up meetings with dietitians. Body weight, and body composition measures (‘BMI’; basal metabolic rate (hereafter ‘BMR’); body fat percentage) were measured using a BC-418MA Segmental Body Composition Analyser; Tanita. Hips and waist circumference were measured in each participant by their respective dietitian using a standard measuring tape. Blood pressure and heart rate were measured using an automated blood pressure monitor (M6 model, Omron, Hoofddorp, the Netherlands).
Stool collection, microbiome sequencing and processing
Stool samples for microbiome analysis (at baseline and 6 months time points) were collected by participants at home using an OMNIgene GUT (OMR-200; DNA Genotek) stool collection kit. Participants were asked to collect the stool sample as close as possible to their scheduled visit to the trial site and store it in a home freezer (−18°C) until then. When arrived at the trial site, samples were transferred to −20°C pending DNA extraction. The stool samples were processed to extract and sequence bacterial DNA, as previously described in detail.27 A total of 382 samples from 191 participants (94 from PPT and 97 from MED arm) were included in the analysis. The relative abundance (RA) of bacterial species was obtained from metagenomic sequencing via an expanded microbial genome reference recently published by our lab29 with default parameters. After filtering by species present in at least 20% of samples we obtained 320 species, and out of these, 296 that were present in both baseline and 6-month samples were included in the analysis. Changes in microbiome features were calculated as follows: log(RAt6)−log(RAt0).
Statistical analysis
All statistical analyses were performed in Python V.3.7. To assess dietary changes within and between arms we used one-sided and two-sided t-test, respectively. To assess microbiome changes within and between arms we used one-sided t-test and Mann-Whitney U test, respectively. All multiple hypothesis testing were controlled for false discovery rate (FDR) on the level of 0.1
Assessing the mediatory effect of microbiome species at the intersection between dietary changes and clinical outcomes, we applied causal mediation analysis as previously described,30 where changes in dietary features were treated as independent variables (X), changes in clinical outcomes as dependent variables (Y) and changes in microbiome species abundances as potential mediators (M). A total of 456 potential mediation paths (based on correlation matrices of diet features change vs clinical biomarkers change and diet features change vs microbiome species change) were applied into a mediation analysis pipeline using the Python package ‘pingouin.mediation_analysis’. Age, gender and BMI change were treated as covariates to correct for their possible effect as confounders that distort mediator-outcome associations. Significance of the mediation effects was determined based on bootstrapping and a p<0.05 for the indirect effect (average causal mediation effect; ACME).
For predicting 6-month changes (delta) in various clinical parameters of blood lipids, glycaemic and anthropometric measures, we devised gradient-boosted decision trees (GBDT) using the LightGBM Python-package (Sklearn._version_‘1.0.2’) with default parameters except for: feature_fraction=0.6 and bagging_fraction=0.6. Fivefold cross-validation was generated six times for each predictor with a different random state, and the score from each cross-validation was used to estimate the predictor’s accuracy and calculate SE. The models were trained with different combination sets of dietary and personal clinical input features (47 and 31, respectively). Dietary features included PPT score, 27 nutrients and 19 food categories. Personal clinical features included age, gender and all blood biomarkers, CGM and anthropometric measures collected in the study. For each clinical readout, four different models were trained with the following sets of input features: (1) baseline levels of the respective clinical feature that we wish to predict its outcome (‘Basic’ model); (2) baseline levels of all personal clinical features measured in this trial, plus age and gender (‘Personalised’ model); (3) as model 1 plus dietary changes (PPT score, nutrients and food categories features) (‘Basic+Diet’ model) and (4) as model 2 plus dietary changes (PPT score, nutrients and food categories features) (‘Personalised+Diet’ model). To study the relative contribution of different features to the final prediction in the ‘Personalised+Diet’ models, the ‘SHAP V.0.41.0’ library of Python was used.
Results
PPT intervention induce greater changes in multiple dietary features compared with MED intervention
As we previously described in the primary publication of this RCT,27 the study design included full dietary records logged by the study participants on a mobile application during the run-in phase (baseline) and throughout the entire 6-month intervention (figure 1A). This allowed us to extract extensive dietary data and assess in high resolution the actual changes in dietary intake adopted by participants during the intervention compared with baseline, including changes in 21 nutrients (macronutrients and micronutrients) and in 18 food categories (see the Methods section) (figure 1B, C). To evaluate the overall pattern of dietary modifications that characterised the two diet arms (MED vs PPT), we conducted a principal component analysis (PCA) using either nutrients or food categories as features. In accordance with the distinct dietary approaches that the two arms were based on (an algorithm-based PPGR-targeting approach vs a Mediterranean-style dietary pattern approach), the PCA analysis demonstrated, as expected, distinct 6 months shifts in dietary intake between arms, both by means of nutrients and food categories intake (figure 1D, E). Notably, since the Mediterranean diet is considered the standard-of-care in Israel and generally represents the local typical diet, the overall dietary shift that the MED arm exhibited was smaller than that of the PPT arm (figure 1D, E).
Following the high-level assessment of dietary changes, we also assessed specific 6-month changes in nutrients intake. Indeed, we found that the two arms differed significantly in most of the nutrients evaluated. In terms of macronutrients, the PPT arm significantly decreased carbohydrate intake compared with baseline (95% CI −17.9% to −15.6% kcal) while MED arm slightly increased carbohydrate intake (95% CI 1.0% to 2.5% kcal). Intake of total dietary fat and the different fat subtypes (‘SatFat’, ‘MUFA’ and ‘PUFA’) were all significantly increased within the PPT arm (95% CI 12.8% to 14.9%; 2.8% to 3.7%; 5.1% to 6.3%; 3.5% to 4.5% kcal, respectively) and decreased in MED arm (95% CI −5% to −3.4%; −2.1% to −1.4%; −1.6% to −0.9%; −1.1% to −0.4% kcal, respectively). Protein intake was slightly, though significantly, increased in both PPT and MED arms (95% CI 2.6% to 3.7% and 1.4% to 2.3% kcal, respectively). Total dietary fibre intake was significantly decreased within the PPT arm (95% CI −2.4 to −1.0 g/1000kcal) and increased in MED (95% CI 2.9 to 4.0 g/1000 kcal) (figure 1B). Notably, the overall low-carbohydrate and high-fat pattern of the PPT intervention was expected as previously described,27 since meal carbohydrate content constitutes an important component in PPGR prediction. However, we found greater variation in change of these (and other) dietary features within the PPT arm as compared with MED (online supplemental table 2), supporting the personalised design of the PPT approach. In terms of micronutrients, among the 14 micronutrients assessed, the most substantial changes within the PPT arm included increases in intake of cholesterol, vitamin E, calcium, vitamin B12 (95% CI 80.2 to 121.1 mg; 0.7 to 1.6 mg; 52.7 to 116.1 mg; 0.4 to 1.0 µg, respectively) and decreases in thiamin (vitamin B1) total folate (vitamin B9), sodium and potassium (95% CI −0.2 to −0.1 mg; −76.7 to −47.3 µg; −552.5 to −303.5 mg; −396.0 to −219.8 mg, respectively). In the MED arm most substantial changes included increase in intake of iron, magnesium, niacin (vitamin B3) and vitamin B6 (95% CI 0.6 to 1.8 mg; 12.2 to 47.1 mg; 1.0 to 3.3 mg; 0.1 to 0.2 mg, respectively) and a decrease in intake of cholesterol (95% CI −42.1 to −5.7 mg) (figure 1B).
To gain more insight about the actual dietary changes adopted by participants during the intervention in terms of dietary patterns and food choices, we assessed participants’ consumption of 18 food categories (evaluated as proportion of total energy intake per day) during the intervention as compared with baseline. Importantly, this analysis demonstrated significant 6 months changes within the PPT arm for 17 out of 18 food categories evaluated, while the MED arm had significant changes in 11 categories (figure 1C). Among the most substantial changes within the PPT arm were increases in ‘Nuts and Seeds’ (95% CI 6.1% to 8.1% kcal); ‘Med Oil and Fats’ (3.3% to 4.7% kcal); ‘Eggs and products’ (2.6% to 3.6% kcal); ‘Milk and Dairy’ (2.3% to 4.3% kcal), and decreases in ‘Refined-grains cooked side dishes’ (−7.4% to −5.6% kcal) and ‘Refined-grains Bread’ (−7.4% to −5.2% kcal). In MED arm most substantial changes included increases in ‘whole-grains bread’ (5.5% to 7.5% kcal) and ‘whole-grains cooked side dishes’ (95% CI 1.9% to 2.9% kcal). Notably, both PPT and MED arms increased intake of ‘vegetables’ and decreased intake of ‘refined-grains bread’ and ‘sweets and snacks’ (figure 1C), indicating that both interventions were overall promoting healthy dietary modifications in that respect.
Finally, to assess the effects of the PPT-based dietary approach on microbiome and clinical outcomes, we devised a ‘PPT-adherence score’ ranging from 0 to 100, that indicates the adherence level of participants to PPT-based recommendations, (see the Methods section). Importantly, as a post hoc analysis for this RCT, we were able to compute this score across the entire cohort (regardless of arm assignment). As expected, the PPT adherence score significantly increased within the PPT arm during the intervention, while for the MED arm there was no significant difference between adherence to PPT-based recommendations at baseline and during the intervention (figure 1E).
PPT intervention increases microbiome diversity and richness and exerts specific microbiome species changes that associate with clinical outcomes
To examine the effects of the dietary intervention on gut microbiome composition, we used shotgun metagenomic sequencing data from faecal samples collected from participants at baseline and at 6 months, and assessed high-level features of microbiome diversity and richness as well as the RA of observed taxa. Consistent with the overall greater dietary shifts that characterised the PPT intervention as compared with MED, the overall gut microbiome diversity and richness were both increased more in the PPT arm (one-sided t-test, p=0.007 and p=0.00017, respectively), than in MED arm (one-sided t-test, p=0.18 and p=0.04, respectively) (figure 2A, B). However, the change difference between arms in these high-level features of the microbiome was not statistically significant (MW test, p=0.55 and 0.17, respectively).
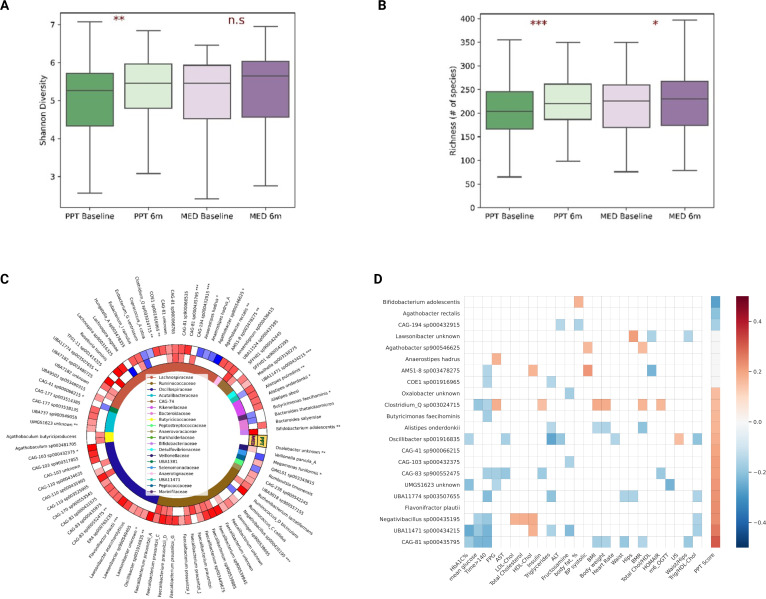
Figure 2.
PPT intervention increases microbiome diversity and richness and exerts specific microbiome species changes that are associated with clinical outcomes. (A) Boxplots showing microbiome diversity (Shannon's diversity index) by arm and time point (baseline vs 6 months). Asterisks on top denote significance level for difference within arms (one-sided t-test). n.s, non significant; *p<0.05, **p<0.01, ***p<0.001. (B) Same as in A but for microbiome richness (# of species). (C) Heatmap of species-level microbiome taxa significantly changed within arms compared to baseline (p<0.05, one-tailed t-test, FDR corrected). Red and blue cells denote enrichment and reduction, respectively. White cells denote no significant change. Asterisks next to species names denote significant differences between arms in the respective species (MW-test, *p<0.05, **p<0.01, ***p<0.001). Species are grouped based on taxonomy hierarchy, with family-level taxonomy represented by colours in the inner circle and in the legend in the centre. (D) Heatmap showing significant associations (p<0.05) between 6 months changes in microbiome species (those distinctly changed between arms) and 6 months changes in clinical readouts or PPT adherence score across the cohort. ALT, alanine transaminase; AST, aspartate aminotransferase; BMI, body mass index; BMR, basal metabolic rate; BP, blood pressure; FDR, false discovery rate; FPG, fasting plasma glucose; HbA1c, hemoglobin A1c; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; OGTT, oral glucose tolerance test; PPT, personalised postprandial-targeting.
To better understand the specific compositional changes that occurred following the dietary interventions, we also assessed changes in bacteria RA at the species level, and identified 69 and 33 bacterial species that changed significantly within the PPT and MED arms, respectively (one-sided t-test, p<0.05, FDR corrected) (figure 2C). Notably, 24 of these identified changes in microbiome species within arms were significantly distinct between arms (MW test, p<0.05; figure 2C and online supplemental table 3) and in some cases are in agreement with findings from other studies that reported links between these species and specific dietary patterns or components. For example, Bifidobacterium adolescentis, a bacterial species consistently linked in the literature to various dietary fibres intake,31–33 was significantly enriched in the MED arm following the intervention, but not in the PPT arm (figure 2C), which is inline with the increase in dietary fibre consumption by MED arm participants in this trial (figure 1A). On the other hand, Flavonifractor plautii, a bacterium previously reported to participate in the metabolism of the flavonoid ‘catechin’ in the gut,34 was significantly enriched in the PPT arm following the intervention but not in the MED arm (figure 2C). Catechins are flavonoids (a type of polyphenols) naturally occurring in certain plant-based foods and are well recognised for their antioxidative properties in vitro and potential to exhibit health benefits in vivo.35 36 Notably, the consumption of some catechin-rich foods including dark chocolate, walnuts, almonds and cashews,36 37 was increased significantly more in PPT arm than in MED arm (online supplemental table 4), which may indicate the underlying mechanism for the enrichment in F. plautii species among PPT dieters. Notably, the RA of F. plautii was also positively associated with almonds consumption across the cohort at baseline. Interestingly, oral administration of F. plautii was recently reported by Mikami et al to attenuate obesity-induced chronic inflammation in mice,38 suggesting a potential therapeutic effect for this species in cardiometabolic health.
Next, in an attempt to explore how microbiome changes are linked to clinical outcomes and whether these are attributable to PPT adherence, we first examined which of the 24 microbiome species that distinctly changed between arms were significantly associated with clinical outcomes or PPT-adherence score across the entire cohort. We identified 22 species with significant associations (p<0.05, figure 2D), out of which 17 were PPT adherence-associated species (14 positively and 3 negatively associated with PPT score). Notably, most of these PPT adherence-associated species were also significantly associated with multiple clinical readouts, suggesting that the beneficial effects of PPT adherence on metabolic readouts may be partially related to these specific bacterial changes (figure 2D). For example, change in the bacterial species Alistipes onderdonkii was positively associated with PPT adherence and negatively with change in clinical readouts of triglycerides, body weight, BMI and BMR. Notably, this species was recently suggested to suppress proliferation of pancreatic primary cancer cells.39
Changes in specific gut microbiome species partially mediate the effects of dietary modifications on clinical outcomes
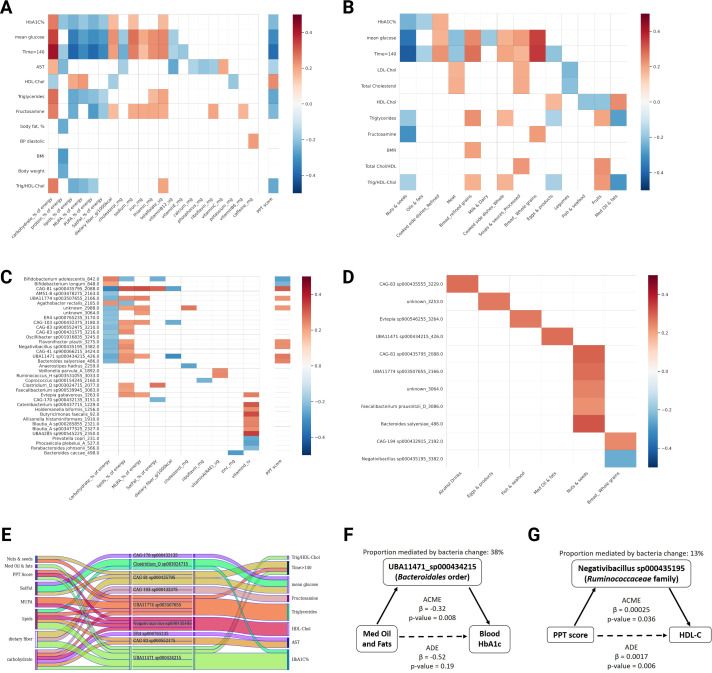
To infer whether the microbiome compositional changes causally mediated the effects of dietary changes on host clinical outcomes, we applied causal mediation analysis as previously described,30 where changes in dietary features were treated as independent variables (X), changes in clinical outcomes as dependent variables (Y) and changes in microbiome species abundances as potential mediators (M). First, we constructed correlation matrices across the entire cohort (regardless of arm assignment) of changes in dietary features (nutrients or food categories) against changes in clinical markers or against changes in microbiome species abundance, while correcting for multiple testing (FDR at 0.1). We detected multiple significant correlations (diet vs clinical, figure 3A, B; diet vs microbiome, figure 3C, D), comprising a total of 384 potential mediation paths (dietary features that significantly correlate with both a clinical outcome and a bacterial species change), that we then applied into a mediation analysis pipeline (see the Methods section). We found 30 significant mediation effects (indirect effect, also termed ACME; p<0.05), comprising 9 bacterial species that partially mediate the effect of different dietary changes on clinical outcomes (figure 3E and online supplemental table 5). For example, change in the bacterial species UBA11471 sp000434215 (from Bacteroidales order) was found to partially mediate the effect of change in ‘Med Oil and Fats’ consumption on HbA1c outcome (proportion mediated by bacteria change, 38% figure 3F).
Figure 3.
Changes in specific gut microbiome species partially mediate the effect of dietary changes on clinical outcomes. (A) Heatmap showing significant associations (p<0.05, FDR corrected) between 6 months changes in clinical readouts and 6 months changes in nutrient consumption or PPT adherence score across the cohort. (B) Heatmap showing significant associations (p<0.05, FDR corrected) between 6 months changes in clinical readouts and 6 months changes in food categories consumption across the cohort. (C) As in A but for microbiome species versus nutrients or PPT adherence score. (D) As in B but for microbiome species versus food categories. (E) Alluvial plot showing significant mediatory effects of microbiome species (middle) in the association between dietary changes (left) and clinical outcomes (right). (F, G) Two examples of mediation paths with assessment of the proportional mediatory effect of microbiome species. ACME, average causal mediation effect; ADE, average direct effect; AST, aspartate aminotransferase; BMI, body mass index; BMR, basal metabolic rate; BP, blood pressure; FDR, false discovery rate; FPG, fasting plasma glucose; HbA1c, hemoglobin A1c; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; MUFA, monounsaturated fatty acids; PPT, personalised postprandial-targeting.
To evaluate whether PPT adherence exerts its beneficial effects on metabolic outcomes through modulation of specific microbiome species, we also estimated the causal mediation effect of microbiome species on metabolic outcomes, using 72 potential mediation paths, based on significantly associated clinical outcomes and microbiome species changes with change in PPT score (figure 3A, B). We found three bacterial species (from Bacteroidales, Lachnospiraceae and Oscillospirales orders) that had a significant partial mediation effect on the association of PPT adherence with clinical outcomes of HbA1c, HDL-cholesterol and triglycerides (figure 3E and online supplemental table 5). For example, change in the bacterial species ‘Negativibacillus sp000435195’ (from the Ruminococcaceae family) was found to partially mediate the effect of PPT adherence on HDL-C outcome (proportion mediated by bacteria change, 13%) (figure 3G and online supplemental table 5). Notably, all three bacterial species found to significantly mediate the association between PPT adherence and clinical readouts also came up as significant mediators of the association between other dietary features and corresponding clinical outcomes (figure 3E), suggesting that these species had a profound effect in mediating the association between dietary modifications and clinical outcomes in this trial.
Machine-learning models trained on dietary changes and baseline clinical data predict clinical outcomes
Lastly, as part of the objective to explore the interplay between dietary modifications, microbiome composition and clinical outcomes, we sought to look beyond the mean effects of the dietary intervention on clinical readouts and to infer what drives personalised metabolic responses to dietary modifications. Traditionally, dietary guidelines are based on population average response to dietary components or patterns. However, in different research settings as well as in clinical practice, high interpersonal differences in metabolic response to similar dietary modifications are often observed.5–7 40 To examine the variation in metabolic response to similar dietary modifications in this RCT setting, we ranked the entire cohort by changes in major dietary features (PPT score and macronutrients) and compared it to ranked changes in major clinical readouts. Indeed, we found large variation in clinical response to similar dietary changes at the individual level, in both arms (figure 4A, online supplemental table 6). Specifically, subjects that adopted similar dietary modifications, in terms of PPT adherence and macronutrients composition, exhibited in some cases vastly different clinical results in terms of glycaemic, blood lipids or BMI outcomes (figure 4A).
Figure 4.
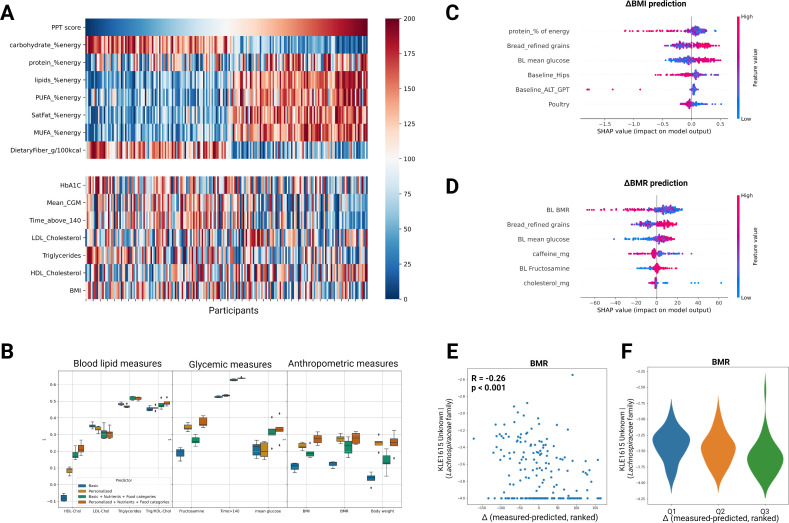
Machine learning models trained on dietary changes and baseline clinical data predict clinical outcomes. (A) Heatmap showing the interindividual variation in clinical response to similar dietary modifications across the cohort. Participants (columns) are ordered by the 6 months change in PPT adherence score (top row). Ranked changes in major dietary features and clinical outcomes (rows) are presented in the upper and lower panels, respectively. (B) Box-plots showing the prediction accuracy (R) of different clinical outcomes for models trained with different sets of input features using GBDT (LightGBM, see Methods). Error bars in each box plot are based on repeated full cross validation. (C) Top six features contributing to BMI prediction (‘Personalised+Diet’ model), using the SHAP analysis. (D) As in C but for the ΔBMR prediction model. (E) Scatter plot of the correlation between baseline levels of one bacterial species from the Lachnospiraceae family (‘KLE1615_Unknown’) and the difference (Δ) between measured and predicted change in BMR outcome. (F) As in E but presented as violin plots for three quantiles of difference (Δ) from prediction (participants with zero levels of this bacterial species at baseline were filtered out). ALT, alanine transaminase; BMI, body mass index; BMR, basal metabolic rate; BP, blood pressure; CGM, continuous glucose monitoring; FDR, false discovery rate; FPG, fasting plasma glucose; GBDT, gradient-boosted decision trees; HbA1c, hemoglobin A1c; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; MUFA, monounsaturated fatty acids; PPT, personalised postprandial-targeting; PUFA, polyunsaturated fatty acid.
Next, in an attempt to explore which dietary changes and personal features modulate metabolic responses, we devised GBDT models to predict the 6 months change (Δ) in various clinical parameters of blood lipids, glycaemic and anthropometric measures. To infer the role of dietary versus personal clinical data in predicting clinical outcomes, we compared for each clinical readout four different models, trained with different sets of input features as follows: (1) baseline levels of the clinical feature that we wish to predict its outcome (hereafter ‘Basic’ model); (2) baseline levels of all personal clinical features measured in this trial (blood biomarkers, CGM and anthropometric measures), plus age and gender (hereafter ‘Personalised’ model); (3) as model 1 plus dietary changes (nutrients and food categories features) (hereafter ‘Basic+Diet’ model) and (4) as model 2 plus dietary changes (nutrients and food categories features) (hereafter ‘Personalised+Diet’ model). Overall, for 9 out of 21 clinical outcomes tested, a diet-based model (‘Basic+Diet’ or ‘Personalised+Diet’ model) yielded a significant prediction accuracy. Among these, for eight clinical outcomes the ‘Personalised+Diet’ model yielded the best correlation between measured and predicted outcome as compared with the other three models (HDL-C, R=0.2, p=0.004; Trig/HDL-C, R=0.52, p<0.001; fructosamine, R=0.33, p<0.001; time>140, R=0.64, p<0.001; mean CGM glucose, R=0.37, p<0.001; BMI, R=0.25, p<0.001; BMR, R=0.23, p=0.001; body-weight, R=0.2, p=0.005) (figure 4B and online supplemental figures S1). To infer which specific dietary and personal clinical features are the main drivers of the 6 months change in these clinical outcomes, we used a feature attribution analysis (SHapley Additive exPlanations, SHAP)41 and computed the individual-level contribution of each dietary and clinical feature in the model (termed SHAP values) to prediction accuracy (figure 4C, D and online supplemental figures S2). Interestingly, for both ΔBMI and ΔBMR prediction models, baseline ‘mean CGM glucose’ was among the top personal clinical features and change in ‘Refined-grains Bread’ intake was among the top dietary features that attributed to prediction accuracy (figure 4C, D).
gutjnl-2022-329201supp002.pdf (362.6KB, pdf)
Lastly, we sought to infer whether prediction accuracy per person also relies on the RA of specific microbiome species at baseline. To address this, we ranked the cohort by the difference (Δ) between measured and predicted change in the respective clinical parameter and then regressed this ranked difference against all baseline microbiome species. We identified five significant correlations (p<0.05, FDR corrected) composed of four bacterial species (from Lachnospiraceae, Ruminococcaceae and Coriobacteriaceae families) that correlated with the ranked difference between measured and predicted change in the predictors of BMR, time>140, mean CGM glucose and low-density lipoprotein cholesterol (LDL-C) (figure 4E, F and online supplemental table 7). For example, for ΔBMR predictor (‘Personalised+Diet’ model) we found a significant negative correlation between the baseline levels of a bacterial species from the Lachnospiraceae family (KLE1615 Unknown) and the difference (Δ) between measured and predicted change in this outcome (R=−0.26, p=0.0002), suggesting that the higher this bacteria was at baseline the better was the actual clinical outcome compared with prediction (figure 4E, F). Interstingly, this species was recently reported to be positively correlated with nuts consumption and with urinary urolithins (secondary polyphenol metabolites derived from the gut microbial action on ellagitannins-rich foods such as pomegranate, berries and nuts) in a dietary intervention study in overweight and obese subjects.23 Importantly, urolithin production was negatively associated with serum hs-CRP, triglycerides, body fat mass, body weight, BMI and urinary carnitine in that study, suggesting overall a beneficial role for that bacterial species in weight loss. To further validate the link between ‘KLE1615 Unknown’ species (from the Lachnospiraceae family) and diet or metabolic measures, we tested for associations of this species with clinical markers and dietary data in another independent and larger cohort (n=7000) of healthy adults, from an ongoing observational study conducted in our lab (the ‘10K’ cohort).42 Interestingly, we found that this species was negatively associated with several metabolic markers, including BMI (R=−0.08, p=6.4×10−11), weight (R=−0.1, p=8.2×10−17), hips (R=−0.07, p=2.2×10−9), waist (r=−0.1, p=8×10−20), triglycerides (R=−0.08, p=4.8×10−11), and positively associated with HDL-C (R=0.08, p=7.4×10−11), vitamin B12 status (R=0.05, p=2.8×10−4) and with dietary consumption of brazil nuts (R=0.04, p=5×10−4), pinenuts (R=0.04, p=1.2×10−3) and walnuts (R=0.05, p=1.8×10−5) (data not shown). Together, the findings from these different cohorts suggest a beneficial role for that species at the intersection between diet and body weight.
Discussion
In the current study, we conducted ad hoc analyses aimed at exploring the interplay between dietary modifications, microbiome composition and cardiometabolic health outcomes in an RCT setting of a 6-month dietary intervention comparing a PPT diet versus MED diet in 200 adults with pre-diabetes. We demonstrate that the PPT diet prompted greater changes in gut microbiota composition, consistent with overall greater dietary modifications observed, as compared with the MED intervention. Specifically, at the species level, we demonstrate that the PPT diet induced significant changes to 2-fold more species than the MED diet (69 vs 33 species, respectively, figure 2C), some of which are inline with findings from other studies that indicated links between microbiome species and specific dietary elements, while others, to the best of our knowledge, are first to be recognised. A notable example for a novel finding from this work is of the newly-classified bacterial species ‘UBA11471_sp000434215’ (from Bacteroidales order), which differentially changed between arms (enriched on PPT and decreased on MED, figure 2C and online supplemental table 3). Indeed, this species is largely uncharacterised but was recently reported to be a diet-associated taxa that is highly associated with complex foods, rather than specific nutrients.43
Beyond the comparison of dietary and microbiome compositional changes between arms, our study provides a comprehensive view of the associations between changes in multiple dietary features (including PPT adherence), microbiome species and metabolic readouts across the entire cohort. Notably, as a post hoc analysis for this RCT, we were able to compute a PPT-adherence score across the entire cohort, thus achieving greater statistical power in assessing the effects of a PPT diet approach on microbiome composition and host clinical outcomes. Furthermore, using causal mediation analysis, we demonstrate the mediatory role of nine microbial species in the association between specific dietary changes and clinical outcomes, including 3 species (from Bacteroidales, Lachnospiraceae and Oscillospirales orders) that mediate the association between adherence to PPT-based recommendations and clinical outcomes of HbA1c, HDL-cholesterol and triglycerides (figure 3E). Interestingly enough, a subanalysis that we conducted at a greater, food-level resolution, also indicated a partial mediatory role for the latter three species in the effect of ‘wholemeal bread’ and ‘almonds’ consumption on the clinical outcome of ‘time>140’ (online supplemental figure S3 and online supplemental table 8). Importantly, two of these species were also found to be similarly associated with clinical markers and dietary consumption in an independent and larger cohort from an ongoing observational study conducted in our lab (n=7000; the ‘10K’ cohort).42 Specifically, the two bacterial species UBA11774 sp003507655 (from Lachnospiraceae family) and UBA11471 sp000434215 (from Bacteroidales order) were reported here to partially mediate the association between PPT-adherence and clinical improvements in triglycerides and HbA1c, respectively. Notably, in the 10K cohort, these two species were also positively associated with blood HDL-C and with dietary intake of nuts, vegetable oils and animal protein-rich foods, and negatively associated with triglycerides and body-related measures and with dietary intake of wheat-based and other carbohydrate-rich foods (online supplemental table 9). On the other hand, the species CAG-81 sp000435795 (from Lachnospiraceae family), reported here to mediate the effects of dietary modifications on glycaemic control outcomes (‘time>140’ and ‘mean CGM glucose’) seems to have inverse associations in the 10K cohort and in few other studies. In the 10K cohort, this species is positively associated with various metabolic markers, including triglycerides, ALT, blood glucose, HbA1c and BMI (online supplemental table 9). Other studies suggested a positive association of this species with a Fatty Liver Index44 and with TMAO levels in urine.45 This controversial finding suggests that specific microbes may function differently in different ecological environments or metabolic contexts, and requires further investigation in future studies.
Traditionally, nutrition research is focused on populations’ average response to diets as the leading approach for drawing global dietary recommendations. Here, we look beyond the mean effects of the dietary intervention on clinical readouts and try to infer what drives personalised metabolic responses to dietary modifications, which may inform more effective and precise dietary recommendations to individuals in clinical practice. Using machine-learning models trained on dietary changes and baseline clinical data of the entire cohort, we predict personalised clinical outcomes in several cardiometabolic markers of blood lipids, glycaemic control and body weight. Importantly, SHAP analyses of these predictors demonstrated the top dietary and clinical features attributing to prediction accuracy, including for example baseline glucose levels and change in consumption of refined-grains bread as most predictive for improvement in BMI and BMR outcomes (figure 4C, D). This important finding suggests that the consideration of these clinical and dietary features may be specifically important in clinical practice for achieving treatment goals of weight loss in individuals with pre-diabetes. Lastly, we identify that for our predictors of BMR, time>140, mean CGM glucose and LDL-C the prediction accuracy per person partially relies on the baseline levels of specific bacterial species (online supplemental table 7). A notable example is of the bacterial species ‘KLE1615 Unknown’ (from Lachnospiraceae family), for which high baseline levels were indicative for greater measured improvement in BMR outcome than predicted by our ‘Personalised+Diet’ prediction model (figure 4E, F).
Our study has several limitations. First, as opposed to observational studies which typically use large cohorts, the data from this RCT is based on a relatively small sample size (n=200). As such, it has a limited statistical power to detect associations between diet, microbiota and clinical features, and specifically to devise prediction models. To partially overcome this limitation, we analysed the cohort as a whole (regardless of arm assignment) to gain more insight about the causal relationships between dietary features, microbiome taxa and cardiometabolic markers. Nevertheless, since the trial was not originally designed to answer these exploratory research questions then results should be interpreted with caution. Second, with respect to the angle of microbiome, the current work focuses solely on microbiome compositional aspects. Broader analyses of other omics layers, such microbiome functional changes and metabolomics, were beyond the scope for the current paper, but are of great interest for future work as these may potentially improve our mechanistic understanding of the microbiome’s role at the intersection between diet and host clinical outcomes.
Together, while the research on gut microbiota at the intersection between diet and human health is mostly based on observational studies, our findings from this 6-month dietary intervention support a causal role for diet in shaping the gut microbiome composition and, for gut microbiota in turn, in modulating diet’s impact on host cardiometabolic markers. Our findings suggest concrete microbiota species targets, which may form the basis for future mechanistic experiments on their role in human diet and health, as well as potential therapeutic directions to be evaluated in preclinical and intervention studies to improve cardiometabolic health through precision nutrition strategies.
Footnotes
Twitter: @OrlyBenYacov
Contributors: OB-Y and AG conceived and conducted the analyses, and equally contributed to the study. OB-Y led the clinical trial, interpreted biological results and wrote the manuscript. AG directed all computational aspects of data analysis. MR contributed to dietary data mapping and analyses. SS was the medical lead of the study. AW and ML-P led specimens processing and sequencing procedures. ES conceived and directed the RCT and all analyses and is the guarantor of this paper. All authors critically revised the manuscript for important intellectual content.
Funding: The funding for the original RCT study was provided jointly by the companies Janssen Pharmaceuticals and DayTwo Company. Janssen is one of the investors of DayTwo.
Competing interests: ES is a regular paid consultant for DayTwo.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Citation: Artwork in figures was created with 'BioRender.com'
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. Shotgun metagenomic sequencing, dietary data and other clinical deidentified participant data are available on reasonable request from ES, by email: Eran.Segal@weizmann.ac.il.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Institutional review board (IRB) of the WeizmannInstitute of Science (protocol #398-1). Participants gave informed consent to participate in the study before taking part.
References
- 1. Sattar N, Gill JMR, Alazawi W. Improving prevention strategies for cardiometabolic disease. Nat Med 2020;26:320–5. 10.1038/s41591-020-0786-7 [DOI] [PubMed] [Google Scholar]
- 2. Mozaffarian D. Dietary and policy priorities for cardiovascular disease, diabetes, and obesity. Circulation 2016;133:187–225. 10.1161/CIRCULATIONAHA.115.018585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siri-Tarino PW, Krauss RM. Diet, lipids, and cardiovascular disease. Curr Opin Lipidol 2016;27:323–8. 10.1097/MOL.0000000000000310 [DOI] [PubMed] [Google Scholar]
- 4. Millen BE, Abrams S, Adams-Campbell L, et al. The 2015 dietary guidelines Advisory Committee scientific report: development and major conclusions. Adv Nutr 2016;7:438–44. 10.3945/an.116.012120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeevi D, Korem T, Zmora N, et al. Personalized nutrition by prediction of glycemic responses. Cell 2015;163:1079–94. 10.1016/j.cell.2015.11.001 [DOI] [PubMed] [Google Scholar]
- 6. Korem T, Zeevi D, Zmora N, et al. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab 2017;25:1243–53. 10.1016/j.cmet.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 7. Asnicar F, Berry SE, Valdes AM, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med 2021;27:321–32. 10.1038/s41591-020-01183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bashiardes S, Godneva A, Elinav E, et al. Towards utilization of the human genome and microbiome for personalized nutrition. Curr Opin Biotechnol 2018;51:57–63. 10.1016/j.copbio.2017.11.013 [DOI] [PubMed] [Google Scholar]
- 9. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 2016;375:2369–79. 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- 10. Cani PD. Human gut microbiome: hopes, threats and promises. Gut 2018;67:1716–25. 10.1136/gutjnl-2018-316723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490:55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 12. Wu H, Tremaroli V, Schmidt C, et al. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metabolism 2020;32:379–390. 10.1016/j.cmet.2020.06.011 [DOI] [PubMed] [Google Scholar]
- 13. Thingholm LB, Rühlemann MC, Koch M, et al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe 2019;26:252–64. 10.1016/j.chom.2019.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee CJ, Sears CL, Maruthur N. Gut microbiome and its role in obesity and insulin resistance. Ann N Y Acad Sci 2020;1461:37–52. 10.1111/nyas.14107 [DOI] [PubMed] [Google Scholar]
- 15. Tang WHW, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013;368:1575–84. 10.1056/NEJMoa1109400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Talmor-Barkan Y, Bar N, Shaul AA, et al. Metabolomic and microbiome profiling reveals personalized risk factors for coronary artery disease. Nat Med 2022;28:295–302. 10.1038/s41591-022-01686-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sonnenburg JL, Bäckhed F. Diet–microbiota interactions as moderators of human metabolism. Nature 2016;535:56–64. 10.1038/nature18846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smits SA, Leach J, Sonnenburg ED, et al. Seasonal cycling in the gut microbiome of the hadza hunter-gatherers of Tanzania. Science 2017;357:802–6. 10.1126/science.aan4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jardon KM, Canfora EE, Goossens GH, et al. Dietary macronutrients and the gut microbiome: a precision nutrition approach to improve cardiometabolic health. Gut 2022;71:1214–26. 10.1136/gutjnl-2020-323715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang DD, Nguyen LH, Li Y, et al. The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat Med 2021;27:333–43. 10.1038/s41591-020-01223-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meslier V, Laiola M, Roager HM, et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020;69:1258–68. 10.1136/gutjnl-2019-320438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rinott E, Meir AY, Tsaban G, et al. The effects of the green-mediterranean diet on cardiometabolic health are linked to gut microbiome modifications: a randomized controlled trial. Genome Med 2022;14:29. 10.1186/s13073-022-01015-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ghosh TS, Rampelli S, Jeffery IB, et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: the NU-AGE 1-year dietary intervention across five European countries. Gut 2020;69:1218–28. 10.1136/gutjnl-2019-319654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fragiadakis GK, Wastyk HC, Robinson JL, et al. Long-term dietary intervention reveals resilience of the gut microbiota despite changes in diet and weight. Am J Clin Nutr 2020;111:1127–36. 10.1093/ajcn/nqaa046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ben-Yacov O, Godneva A, Rein M, et al. Personalized postprandial glucose response-targeting diet versus Mediterranean diet for glycemic control in prediabetes. Diabetes Care 2021;44:1980–91. 10.2337/dc21-0162 [DOI] [PubMed] [Google Scholar]
- 28. Abu-Saad K, Endevelt R, Goldsmith R, et al. Adaptation and predictive utility of a Mediterranean diet screener score. Clin Nutr 2019;38:2928–35. 10.1016/j.clnu.2018.12.034 [DOI] [PubMed] [Google Scholar]
- 29. Leviatan S, Shoer S, Rothschild D, et al. An expanded reference map of the human gut microbiome reveals hundreds of previously unknown species. Nat Commun 2022;13:3863. 10.1038/s41467-022-31502-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol 1986;51:1173–82. 10.1037//0022-3514.51.6.1173 [DOI] [PubMed] [Google Scholar]
- 31. Cox SR, Lindsay JO, Fromentin S, et al. Effects of low FODMAP diet on symptoms, fecal microbiome, and markers of inflammation in patients with quiescent inflammatory bowel disease in a randomized trial. Gastroenterology 2020;158:176–188. 10.1053/j.gastro.2019.09.024 [DOI] [PubMed] [Google Scholar]
- 32. Ramirez-Farias C, Slezak K, Fuller Z, et al. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 2009;101:541–50. 10.1017/S0007114508019880 [DOI] [PubMed] [Google Scholar]
- 33. Birkeland E, Gharagozlian S, Birkeland KI, et al. Correction to: prebiotic effect of inulin‑type fructans on faecal microbiota and short‑chain fatty acids in type 2 diabetes: a randomised controlled trial. Eur J Nutr 2020;59:3339–40. 10.1007/s00394-020-02314-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ozdal T, Sela DA, Xiao J, et al. The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 2016;8:78. 10.3390/nu8020078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Isemura M. Catechin in human health and disease. Molecules 2019;24:528. 10.3390/molecules24030528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vinson JA, Cai Y. Nuts, especially walnuts, have both antioxidant quantity and efficacy and exhibit significant potential health benefits. Food Funct 2012;3:134–40. 10.1039/c2fo10152a [DOI] [PubMed] [Google Scholar]
- 37. Fan F-Y, Sang L-X, Jiang M. Catechins and their therapeutic benefits to inflammatory bowel disease. Molecules 2017;22:484. 10.3390/molecules22030484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mikami A, Ogita T, Namai F, et al. Oral administration of Flavonifractor plautii attenuates inflammatory responses in obese adipose tissue. Mol Biol Rep 2020;47:6717–25. 10.1007/s11033-020-05727-6 [DOI] [PubMed] [Google Scholar]
- 39. Lee K, Oh HJ, Kang M-S, et al. Metagenomic analysis of gut microbiome reveals a dynamic change in Alistipes onderdonkii in the preclinical model of pancreatic cancer, suppressing its proliferation. Appl Microbiol Biotechnol 2021;105:8343–58. 10.1007/s00253-021-11617-z [DOI] [PubMed] [Google Scholar]
- 40. Zeisel SH. A conceptual framework for studying and investing in precision nutrition. Front Genet 2019;10:200. 10.3389/fgene.2019.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lundberg SM, Erion G, Chen H, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell 2020;2:56–67. 10.1038/s42256-019-0138-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shilo S, Bar N, Keshet A, et al. 10 K: a large-scale prospective longitudinal study in Israel. Eur J Epidemiol 2021;36:1187–94. 10.1007/s10654-021-00753-5 [DOI] [PubMed] [Google Scholar]
- 43. Troci A, Rausch P, Waschina S, et al. Long-term dietary effects on human gut microbiota composition employing shotgun metagenomics data analysis. Mol Nutr Food Res 2022:e2101098. 10.1002/mnfr.202101098 [DOI] [PubMed] [Google Scholar]
- 44. Ruuskanen MO, Åberg F, Männistö V, et al. Links between gut microbiome composition and fatty liver disease in a large population sample. Gut Microbes 2021;13:1–22. 10.1080/19490976.2021.1888673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burton KJ, Krüger R, Scherz V, et al. Trimethylamine-N-Oxide postprandial response in plasma and urine is lower after fermented compared to non-fermented dairy consumption in healthy adults. Nutrients 2020;12:234. 10.3390/nu12010234 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-329201supp001.xlsx (122.6KB, xlsx)
gutjnl-2022-329201supp002.pdf (362.6KB, pdf)
Data Availability Statement
Data are available on reasonable request. Shotgun metagenomic sequencing, dietary data and other clinical deidentified participant data are available on reasonable request from ES, by email: Eran.Segal@weizmann.ac.il.