Abstract
Objective
The role of N6-methyladenosine (m6A) in tumour immune microenvironment (TIME) remains understudied. Here, we elucidate function and mechanism of YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) in colorectal cancer (CRC) TIME.
Design
Clinical significance of YTHDF1 was assessed in tissue microarrays (N=408) and TCGA (N=526) cohorts. YTHDF1 function was determined in syngeneic tumours, intestine-specific Ythdf1 knockin mice, and humanised mice. Single-cell RNA-seq (scRNA-seq) was employed to profile TIME. Methylated RNA immunoprecipitation sequencing (MeRIP-seq), RNA sequencing (RNA-seq) and ribosome sequencing (Ribo-seq) were used to identify YTHDF1 direct targets. Vesicle-like nanoparticles (VNPs)-encapsulated YTHDF1-siRNA was used for YTHDF1 silencing in vivo.
Results
YTHDF1 expression negatively correlated with interferon-γ gene signature in TCGA-CRC. Concordantly, YTHDF1 protein negatively correlated with CD8+ T-cell infiltration in independent tissue microarrays cohorts, implying its role in TIME. Genetic depletion of Ythdf1 augmented antitumour immunity in CT26 (MSS-CRC) and MC38 (MSI-H-CRC) syngeneic tumours, while Ythdf1 knockin promoted an immunosuppressive TIME facilitating CRC in azoxymethane-dextran sulphate-sodium or ApcMin/+ models. scRNA-seq identified reduction of myeloid-derived suppressor cells (MDSCs), concomitant with increased cytotoxic T cells in Ythdf1 knockout tumours. Integrated MeRIP-seq, RNA-seq and Ribo-seq revealed p65/Rela as a YTHDF1 target. YTHDF1 promoted p65 translation to upregulate CXCL1, which increased MDSC migration via CXCL1-CXCR2 axis. Increased MSDCs in turn antagonised functional CD8+ T cells in TIME. Importantly, targeting YTHDF1 by CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) or VNPs-siYTHDF1 boosted anti-PD1 efficacy in MSI-H CRC, and overcame anti-PD1 resistance in MSS CRC.
Conclusion
YTHDF1 impairs antitumour immunity via an m6A-p65-CXCL1/CXCR2 axis to promote CRC and serves as a therapeutic target in immune checkpoint blockade therapy.
Keywords: colorectal cancer, immunotherapy, colon carcinogenesis
WHAT IS ALREADY KNOWN ON THIS TOPIC
N6-methyladenosine (m6A) modification plays crucial roles in cancer by regulating RNA splicing, translation and degradation.
YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) is a m6A reader that determines the fate of m6A modified mRNA. However, its potential role in colorectal cancer (CRC) immune microenvironment (TIME) is understudied.
WHAT THIS STUDY ADDS
High YTHDF1 expression is inversely correlated with interferon-γ gene signatures and CD8+ T cell infiltration in multiple cohorts of patient with CRC.
Single-cell sequencing revealed that YTHDF1 contributes to an immunosuppressive TIME with increased infiltration of MDSCs and reduced effector T cells.
YTHDF1 promotes tumour growth via an immunosuppressive TIME in syngeneic mice, intestine-specific Ythdf1 knockin mice, and CD34+ humanised mice of CRC.
Integrative methylated RNA immunoprecipitation sequencing, RNA-seq and Ribo-seq revealed p65/RELA as a YTHDF1 target, leading to increased expression of the cytokine CXCL1, a chemoattractant for MDSCs via CXCL1-CXCR2 axis.
YTHDF1-recuited MDSCs antagonise effector CD8+ and CD4+ T cells in TIME of CRC, thereby promoting tumourigenesis.
Targeting of YTHDF1 by gene knockout or nanoparticle-encapsulated YTHDF1-siRNA synergised with anti-PD1 to suppress the growth of microsatellite instability-high (MSI-H) CRC, and overcomes anti-PD1 resistance in microsatellite stable (MSS) CRC.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Therapeutic targeting of YTHDF1 is a potential strategy to sensitise both MSI-H and MSS CRC to immune checkpoint blockade therapy.
YTHDF1 serves as a prognostic factor in patients with CRC.
Introduction
Colorectal cancer (CRC) is one of the most common cancers worldwide. It is still a deadly cancer with a 5-year survival rate lower than 20% among metastatic patients.1 Immune checkpoint blockade (ICB) has shown beneficial effects to patients with CRC. However, only a minor fraction of patients with microsatellite instability-high (MSI-H) or deficient mismatch repair are responsive to ICB.1 2 Therefore, understanding the molecular mechanisms of immune evasion in CRC and identifying therapeutic strategies that could improve the response to immunotherapies for patients with proficient mismatch repair, microsatellite instability-low, or microsatellite stable (MSS) CRC are crucial.
N6-methyladenosine (m6A) is one of the most abundant RNA modifications, with an estimated 3–5 m6A sites on each mRNA molecule.3 m6A modification is regulated by writers, erasers and readers. m6A writers catalyse m6A formation, consisting of a protein complex of METTL3, METTL14 and WTAP. m6A markers are also actively removed by erasers including FTO and ALKBH5. m6A readers, such as YTH N6-methyladenosine RNA binding protein 1/2/3 (YTHDF1/2/3), YTHDC1/2 and IGF2BP1/2/3 bind to m6A-modified mRNAs to determine the fate of m6A-modified mRNA.3 m6A regulators such as YTHDF1, METTL3 and ALKBH5 have been intensively studied in cancer. Upregulation of YTHDF1 has been reported to be associated with poor prognosis in various cancer types4–6 and YTHDF1 could promote tumourigenesis and cancer metastasis.4–6 However, the role of YTHDF1 in tumour immune microenvironment (TIME) is largely unclear.
Here, we identified that YTHDF1 expression negatively correlated with the interferon gamma (IFN-γ) signature in The Cancer Genome Atlas (TCGA) CRC dataset. Furthermore, we demonstrated for the first time that genetic depletion of Ythdf1 in murine CRC cells of both MSS and MSI-H could induce accumulation of MDSCs and inhibit the infiltration of T cells in syngeneic CRC mice and humanised mice. On the contrary, intestine-specific Ythdf1 knockin drives an immunosuppressive TIME that facilitates spontaneous CRC formation. By integrative analyses of methylated RNA immunoprecipitation sequencing (MeRIP-seq), RNA-seq and Ribo-seq, a YTHDF1-m6A-p65-CXCL1 axis was identified as the mechanism of YTHDF1-induced immunosuppression, leading to the recruitment of MDSCs to suppress effector cells such as functional CD8+ T cells. Moreover, targeting of YTHDF1 by CRISPR or vesicle-like nanoparticles (VNPs)-siRNA enhanced anti-PD1 efficacy in both MSS and MSI-H models, supporting YTHDF1 as a therapeutic target in the immunotherapy of CRC.
Methods
Clinical samples
Cohort I comprised 206 patients with surgically excised CRC tissues, which derived from Prince of Wales Hospital, Hong Kong. Cohort II was Beijing cohort, consisting of 202 patients with CRC. Paraffinised sample of these two cohorts were used to establish the tissue microarrays (TMA). The clinicopathological features of the two cohorts were provided in online supplemental tables S1and S2. Informed consent was obtained for all patients.
gutjnl-2022-328845supp003.pdf (61.7KB, pdf)
Humanised mouse models
Four-to-five week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were exposed to total body gamma irradiation (150–170 cGy/animal) and then intravenously injected with 5×104 human cord blood-derived CD34+ haematopoietic stem cells (STEMCELL) within 24 hours. The percentage of human CD45 positive cells in CD34+ humanised mice was monitored at weeks 8 and 16 by flow cytometry, after CD34+ haematopoietic stem cells transplantation. CD34+ humanised mice with>20% human CD45+ cells in blood were used for establishment of xenografts. All animal studies followed the guidelines approved by the Animal Experimentation Ethics Committee of The Chinese University of Hong Kong.
Murine CRC models with intestinal-specific Ythdf1 knockin
Intestinal-specific Ythdf1 knockin mice (Ythdf1loxp/loxpCDX2-cre) were established. Six to eight weeks old transgenic mice were intraperitoneally injected with tamoxifen (100 mg/kg) to activate Ythdf1 overexpression. For azoxymethane-dextran sulphate-sodium (AOM/DSS) CRC mice model, 7–8 week-old mice were injected intraperitoneally with AOM (10 mg/kg body weight) (#A5486, Sigma-Aldrich), and the mice were given water containing 1.5% dextran sulfate sodium (DSS) (#9011-18-1, MP Biomedicals) for 4 days, 5 days after AOM injection. The regular drinking water was used for the following 2 weeks. DSS treatment was given for an additional 2 cycles, and the mice were sacrificed on day 80. For ApcMin/+ CRC mice model, Ythdf1loxp/loxpCDX2-cre mice were crossbred with ApcMin/+ mice to generate ApcMin/+ Ythdf1loxp/loxpCDX2-cre, all mice were sacrificed after 16 weeks. All animal experiments in this study were approved by the Animal Experimentation Ethics Committee of CUHK and Xiamen University.
TCGA data analysis
Colorectal adenocarcinoma TCGA data (TCGA, PanCancer Atlas) were acquired from cBioPortal (https://www.cbioportal.org/).7 8 Coexpression data performed with Spearman’s correlation in 526 samples was downloaded also from cBioPortal. Genes correlated with YTHDF1 in mRNA expression were subjected to gene set enrichment analysis (GSEA_4.1.0).9 The normalised enrichment scores for IFN-γ signature response was shown. For analysing the association between expression of YTHDF1 and CXCL1 family members, data from stage IV patients were used. High expression tumour was defined with a z-score higher than 0.7, while low expression with a z-score lower than −0.7. The associations were determined with χ2 tests.
Statistical analysis
All measurements were acquired using independent samples rather than collected with repeated measurements. GraphPad Prism V.8 (GraphPad Software; San Diego, California) was used for data analysis, and the data were shown as means ± SD, unless stated otherwise. Two-tailed student’s t-test was used to conduct statistical analysis, unless stated otherwise. A p value<0.05 was regarded as statistically significant.
Additional methods are provided in the online supplemental material.
gutjnl-2022-328845supp001.pdf (206.4KB, pdf)
Results
YTHDF1 is associated with reduced IFN-γ-related gene signatures and poor prognosis in CRC
By analysing copy number variations of 21 known m6A regulators using TCGA data, we identified that YTHDF1 is the top m6A regulator displaying copy number gain or amplification in nearly 80% of CRC tumours (online supplemental figure S1A). Such an increase of copy number is also concordant with upregulation of mRNA expression (online supplemental figure S1A) and protein expression (online supplemental figure S1C), implicating that YTHDF1 functions in promoting CRC. To establish a link between antitumour immunity and m6A regulators, we then performed GSEA to analyse the correlation between m6A regulators with IFN-γ response gene signature. We found that YTHDF1 showed a striking negative correlation (q<0.0001) with the IFN-γ response pathway (online supplemental figure S1B). Of note, IFN-γ response is associated with the induction of antitumour immunity10 11 and responsiveness to ICB therapy.12 13 Consistently, YTHDF1 expression strongly anticorrelated with an 18 IFN-γ-related gene signature (figure 1A), which predicts anti-PD1 responsiveness in multiple cancer types.14 15 Furthermore, expression of CD8A or CD8+ T cell signature16 is negatively correlated with YTHDF1 expression (online supplemental figure S1D). In agreement with TCGA data, IHC staining of TMA from our CRC TMA cohorts showed that high protein expression of YTHDF1 was correlated with low infiltration of CD8+ T cells in cohort I (p<0.001, r=−0.248, n=206) (figure 1B) and cohort II (p<0.0001, r=−0.269, n=202) (figure 1C). These data strongly imply that m6A reader YTHDF1 is associated with impaired antitumour immunity and reduced ICB treatment efficacy.
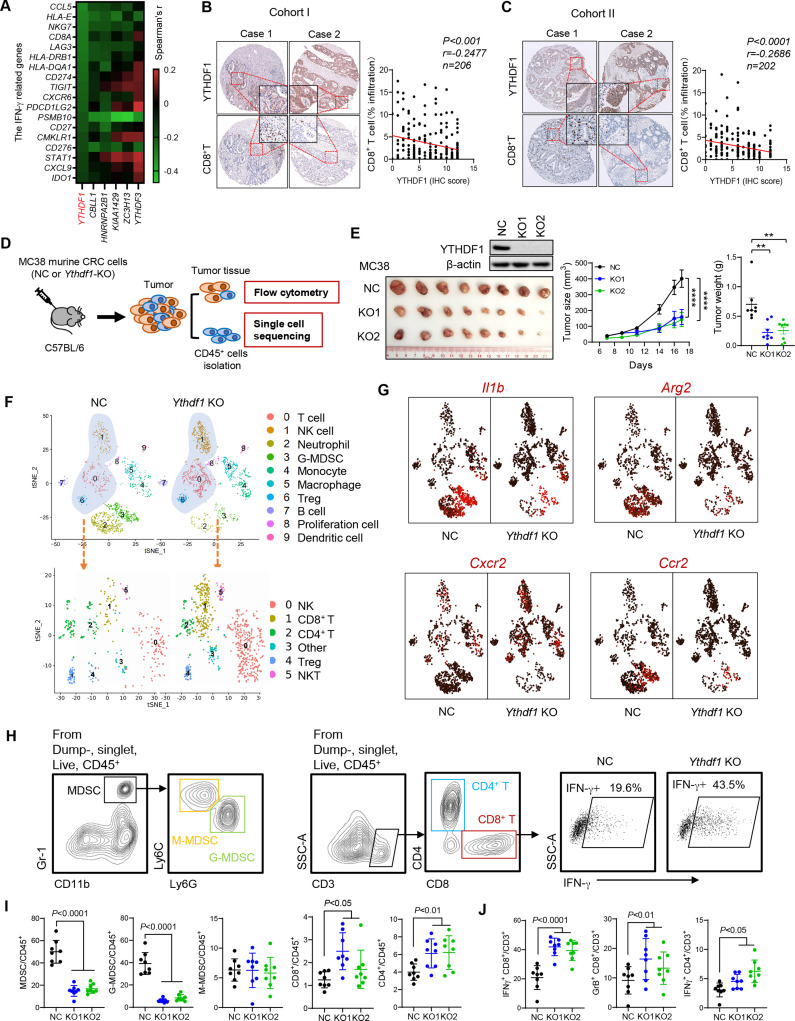
Figure 1.
YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) is associated with an immunosuppressive microenvironment in patients with colorectal cancer (CRC) and its validation by scRNA-seq in immunocompetent mice. (A) Spearman’s correlation in mRNA expression between interferon gamma (IFN-γ)-related genes and m6A regulators in The Cancer Genome Atlas dataset. (B) Correlation between protein levels of YTHDF1 and CD8+ T cell infiltration was determined by immunohistochemistry staining in cohort I (p<0.0001, r=−0.2686, n=202), and (C) validation in cohort II (p<0.001, r=−0.2477, n=206). (D) Experimental design for single-cell analysis of CD45+ cells sorted from the tumours with or without Ythdf1 by FACS, followed by droplet-enabled scRNA-seq. NC: cells with control sgRNA. Ythdf1-KO: cells with CRISPR knockout of Ythdf1. (E) Representative image (left), tumour volume (middle), and weight (right) of MC38 syngeneic tumours with or without Ythdf1 knockout injected in C57BL/6 mice. (F) Upper panel: tSNE plot showing the components of immune cells in MC38 syngeneic tumours with or without Ythdf1 knockout. Each dot represents a single cell. The same cell type was colour coded. Lower panel: subset analysis of T cell and NK cell clusters shown in tSNE projection regions. (G) Feature plots of characteristic markers of all cell types showing expression levels with low expression in dark red to high expression in bright red. (H) Gating strategies for flow cytometry. Identification of myeloid-derived suppressor cells (MDSC), M-MDSC, and G-MDSC (left). Identification of CD8+ T and CD4+ T cells (middle). Representative images showing increase of IFN-γ+ CD8+ T cells in Ythdf1-KO tumours in (D) (Right). (I) Flow cytometry analysis performed with tumours derived from the indicated cells in (D) (**p<0.01; ****p<0.0001) (linear regression (B, C), two tailed t-test (E, H, I), analysis of variance test (E)). NC: cells with control sgRNA. KO1: cells with Ythdf1 sgRNA #1. KO2: cells with Ythdf1 sgRNA #2. GrB: granzyme B.
gutjnl-2022-328845supp002.pdf (11.2MB, pdf)
In agreement with the notion that low CD8+ T cell infiltration is associated with poor prognosis,17 high expression of YTHDF1 (54.3%, 100/184) predicted poor survival of patients with CRC (p<0.01, log-rank test) (online supplemental figure S1E, F). Multivariate Cox regression analysis validated that YTHDF1 was an independent prognostic factor for CRC in cohort II (HR, 1.764; 95% CI 1.058 to 2.939; p<0.05) (online supplemental figure S1F). These findings were further verified in cohort I by multivariate Cox regression analysis (online supplemental figure S1E).
Single-cell transcriptomics reveal YTHDF1-induced immunosuppression
To investigate the role of YTHDF1 in regulating antitumour immunity, we used the CRISPR-Cas9 system to knockout Ythdf1 (Ythdf1-KO) in MC38 murine MSI-H CRC cells and injected the cells to syngeneic C57BL6 mice (figure 1D). We found that both tumour volume and weight were reduced by knockout of Ythdf1 compared with control (NC) (figure 1E). To ask if Ythdf1-KO impact TIME, we isolated CD45+ immune cells from the tumours and performed single cell RNA-seq (scRNA-seq) (NC: 1480 cells; Ythdf1-KO: 1816 cells). Tumours with Ythdf1-KO exhibited strong reduction of granulocytic myeloid-derived suppressor cells (G-MDSCs, cluster 3) and neutrophils compared with NC group (figure 1F and online supplemental figure S2A). In contrast, T cells and NK cells were largely increased in Ythdf1-KO tumours (figure 1F and online supplemental figure S2A). We further reclustered T and NK cells into CD4+ T, CD8+ T, NKT and NK cell subsets, and identified that they were simultaneously increased in Ythdf1-KO tumours compared with controls (figure 1F). We thus speculated that YTHDF1 can suppress the antitumour immunity through inducing MDSC accumulation. Functional markers of MDSCs, Il1b, Arg2, Cxcr2 and Ccr2 were examined, and these genes were mainly enriched in MDSC clusters (cluster 3 and cluster 4), especially from tumours without Ythdf1 knockout (figure 1G). Thus, data from this syngeneic model supports an immunosuppressive function of YTHDF1 in CRC.
Ythdf1 knockout reduces MDSCs but increases cytotoxic T cell infiltration
To validate our findings in scRNA-seq analyses, the composition of tumour-infiltrating immune cells in MC38 syngeneic mice was determined by flow cytometry. We confirmed that Ythdf1 knockout significantly suppressed tumour weight and volume (figure 1C), and flow cytometry revealed that Ythdf1 knockout decreased MDSCs, but increased CD8+ T and CD4+ T cells in tumours (figure 1H, I). Of MDSCs, G-MDSCs were the predominant subset, and knockout of Ythdf1 led to the remarkable reduction of G-MDSCs (figure 1I). In line with the immunosuppressive function of MDSCs, we observed significant increase of functional T cells, including IFN-γ+ CD8+ T cells, Granzyme B+ CD8+ T cells, and IFN-γ+ CD4+ T cells in Ythdf1-KO group (figure 1H, J).
We next performed the experiment in CT26 (MSS-CRC) with Ythdf1-KO to validate the role of YTHDF1 in regulating antitumour immunity. As expected, Ythdf1-KO led to reduced tumour volume and weight (figure 2A, B) in CT26 syngeneic mice, together with reduced functional T cells and accumulation of MDSCs (figure 2C, D). Immunofluorescence staining confirmed the decreased infiltration of MDSCs (CD11b+Gr-1+) in both MC38 and CT26 syngeneic tumours on Ythdf1 knockout (figure 2E and online supplemental figure S2B). Collectively, Ythdf1 depletion in CRC cells reduced MDSCs and increased functional T cell infiltration. These findings are in line with clinical data demonstrating YTHDF1 anticorrelated with CD8+ T cell and IFN-γ-related signatures (figure 1A–C, online supplemental figureS1B, D).
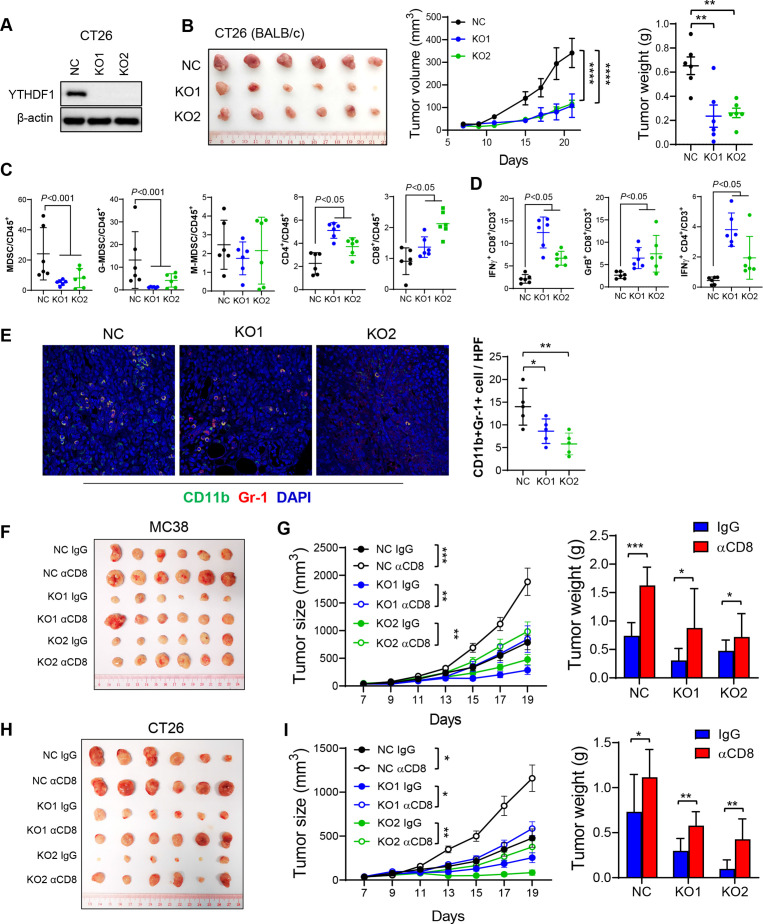
Figure 2.
YTH N6-methyladenosine RNA binding protein 1 (Ythdf1) knockout induces antitumour immunity by reduction of myeloid-derived suppressor cell (MDSC) and increase of functional T cells in syngeneic tumours, an effect reversed by CD8+ T cell depletion. (A) Western blot validated Ythdf1 knockout in CT26 cells. (B) Representative image of CT26 syngeneic tumours with or without Ythdf1 Knockout (left). Knockout of Ythdf1 in CT26 cells inhibits tumour growth (middle) and tumour weight (right) in BALB/c mice. (C) MDSC, G-MDSC, M-MDSC, CD4+ T cells and CD8+ T cells from tumours in (B) were analysed by flow cytometry. (D) Flow cytometry analysis assessing the percentage of T cell functional markers interferon gamma (IFN-γ) and granzyme B (GrB) from tumours in (B). (E) Immunofluorescence identifying MDSCs in subcutaneous tumours from BALB/c injected with the indicated CT26 cells (n=5 each group). (F) MC38 NC or Ythdf1 knockout cells were implanted in C57BL/6 (n=6). Isotype control (IgG) or anti-mouse CD8 antibody (αCD8) were given at 200 µg/mouse on days 4, 7 and 9 post cell injection. Representative images of tumours from each group were shown. (G) Tumour volume (left) and weight (right) from tumours in (F). (H) CT26 NC or Ythdf1 knockout cells were implanted in BALB/c (n=6). Isotype control (IgG) or αCD8 were given at 200 µg/mouse on days 4, 7 and 9 post cell injection. Representative images of tumours from each group were shown. (I) Tumour volume (left) and weight (right) from tumours in (H) (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001) (two tailed t-test (B, C, D, E, G, H), two-way analysis of variance test (D, G, I)). NC: cells with control sgRNA. KO1: cells with YTHDF1 sgRNA #1. KO2: cells with YTHDF1 sgRNA #2.
We asked if the attenuated tumour formation in Ythdf1-KO was dependent on CD8+ T cell antitumour immunity. To address this, we depleted CD8+ T cells with an anti-CD8 antibody in the MC38 syngeneic model. Consistent with our hypothesis, depletion of CD8+ T cells restored growth of Ythdf1-KO tumours (figure 2F, G), demonstrating that tumour-suppressing function of Ythdf1-KO was dependent, at least partially, on CD8+ T cells. This was confirmed in CT26 syngeneic mice showing that anti-CD8 antibody treatment rescued arrested tumour growth in Ythdf1-KO group (figure 2H, I). Depletion of CD8+ T cells by anti-CD8 antibody was confirmed by flow cytometry (online supplemental figure S3A, B). Together, Ythdf1 knockout suppresses CRC growth through the induction of CD8+ T cell-dependent antitumour immunity.
Intestine-specific Ythdf1 knockin promotes colorectal tumourigenesis and suppresses antitumour immunity in mice
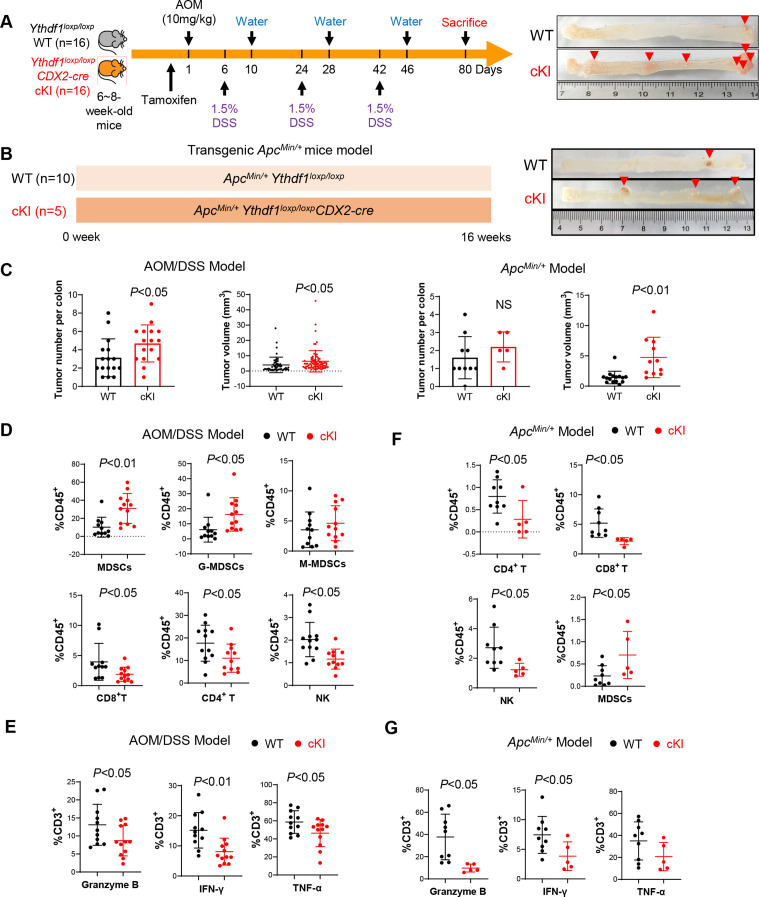
To verify the role of YTHDF1 in spontaneous colorectal tumourigenesis, we generated intestine-specific Ythdf1 knockin mice (Ythdf1loxp/loxpCDX2-cre) and initiated CRC in these mice by AOM/DSS treatment (figure 3A). We found that the overexpression of Ythdf1 resulted in increased colon tumour number and size in the AOM/DSS model (figure 3C). Flow cytometry revealed increased MDSC infiltration together with reduced NK, CD4+ T and CD8+ T cells in colon tumours of Ythdf1 knockin mice compared with wildtype mice (figure 3D). Moreover, we found that Ythdf1 knockin reduced the proportion of functional T cells as identified by granzyme B, INF-γ and TNF-α expression (figure 3E).
Figure 3.
Intestine-specific YTH N6-methyladenosine RNA binding protein 1 (Ythdf1) knockin promotes colorectal tumourigenesis and inhibits antitumour immunity in mice. (A) Scheme for azoxymethane-dextran sulphate-sodium (AOM/DSS) mouse model (left). Representative images of the colon at sacrifice (right). (B) Scheme for ApcMin/+ mouse model (left). Representative images of the colon at sacrifice (right). (C) Tumour number, and tumour size in WT littermates (n=16) and Ythdf1 Ki mice (n=16) mice treated with AOM-DSS (left). Tumour number, and tumour size in ApcMin/+ Ythdf1loxp/loxp (n=10) and ApcMin/+ Ythdf1loxp/loxpCDX2-cre mice (n=5) mice (right). (D) Infiltration of the indicated immune cells in tumours derived from AOM/DSS mice assessed by flow cytometry. (E) Composition of functional T cells in the tumours derived from AOM/DSS mice assessed by flow cytometry. (F) Infiltration of the indicated immune cells in the tumours derived from ApcMin/+ mice assessed by flow cytometry. (G) Composition of functional T cells in the tumours derived from ApcMin/+ mice assessed by flow cytometry. Two tailed t-test (C, E, F, G, H). WT: Ythdf1 wild type; cKI: conditional Ythdf1 knockin.
We next sought to validate these results in ApcMin/+ -driven spontaneous CRC (figure 3B) by establishing ApcMin/+Ythdf1loxp/loxpCDX2-cre mice. Consistently, ApcMin/+ mice with intestine-specific knockin of Ythdf1 developed significantly bigger colon tumours than their wild-type littermates (figure 3C). Analysis of the tumour-infiltrating immune cells demonstrated remarkably decreased infiltration of NK, CD4+ T and CD8+ T cells in tumours with ApcMin/+ with knockin of Ythdf1, together with the induction of MDSCs (figure 3F). Furthermore, Ythdf1 knockin reduced granzyme B+, INF-γ+, or TNF-α+ T cells in ApcMin/+ mice (figure 3G). Collectively, these results support that YTHDF1 fosters an immunosuppressive microenvironment promoting spontaneous CRC.
Targeting YTHDF1 via VNP-siYTHDF1 boosts antitumour immunity in CD34+ humanised mice
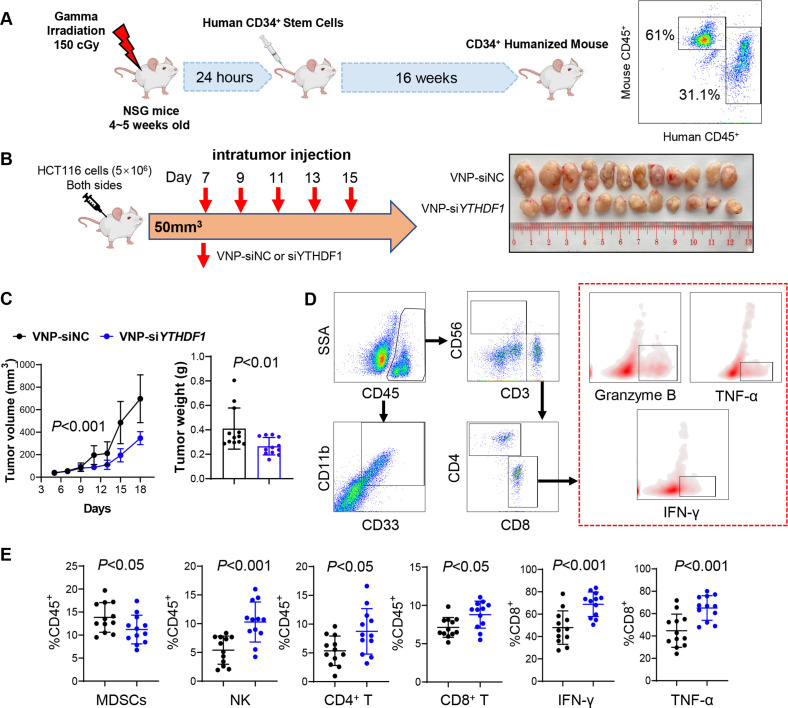
To confirm the role of YTHDF1 in modulating human antitumour immune response, we established the CD34+ humanised mouse model.18 Mice with peripheral blood mononuclear cells (PBMCs) comprising >20% human CD45+ cells were used (figure 4A). To target YTHDF1 in vivo, we developed VNPs19 to carry siRNA against YTHDF1. The humanised NSG mice bearing human CRC HCT116 xenografts were treated with VNP-siNC or -siYTHDF1 after tumours reached 50–100 mm3 (figure 4B). VNP-siYTHDF1 significantly inhibited tumour volume and weight compared with VNP-siNC (figure 4B, C). We also performed flow cytometry to analyse the TIME (figure 4D). VNP-siYTHDF1 decreased MDSC infiltration, but increased CD4+ T cell, CD8+ T cell and NK cell accumulation (figure 4E). In addition, more IFN-γ+, TNF-α+ and granzyme B+ CD8+ T cells were identified in tumours receiving VNP-siYTHDF1 (figure 4E). We also determined the safety of VNP-siYTHDF1 by measuring serum markers of liver (alanine aminotransferase and aspartate transaminase) and kidney function (creatinine and blood urea nitrogen). Mice treated with VNP-siNC or VNP-siYTHDF1 did not demonstrate abnormal liver or kidney function indicators (online supplemental figure S4), showing that VNP treatment is well tolerated. Thus, the targeting of YTHDF1 using VNP-siYTHDF1 is a safe and effective means to enhance antitumour immunity in humanised mice.
Figure 4.
VNP-siYTHDF1 boosted antitumour immunity in CD34+ humanised mice. (A) Workflow for establishment of CD34+ humanised mice (left). Percentage of human CD45+ cells in the CD34+ humanised mice was identified by flow cytometry (right). (B) Design for establishment of HCT116 xenografts and treatment with VNP-siNC or VNP-siYTHDF1 in CD34+ humanised mice (left). Representative image of xenografts in different groups (right). (C) Tumour volume (left) and tumour weight (right) in mice in (B). (D) Gating strategies of immune cells in CD34+ humanised mice. (E) Infiltration of CD4+ T, CD8+ T, NK cells, MDSCs and functional CD8 T cells was assessed by flow cytometry in the tumours from (B) (two tailed t-test (C, E), analysis of variance test (C)). VNP, vesicle-like nanoparticles.
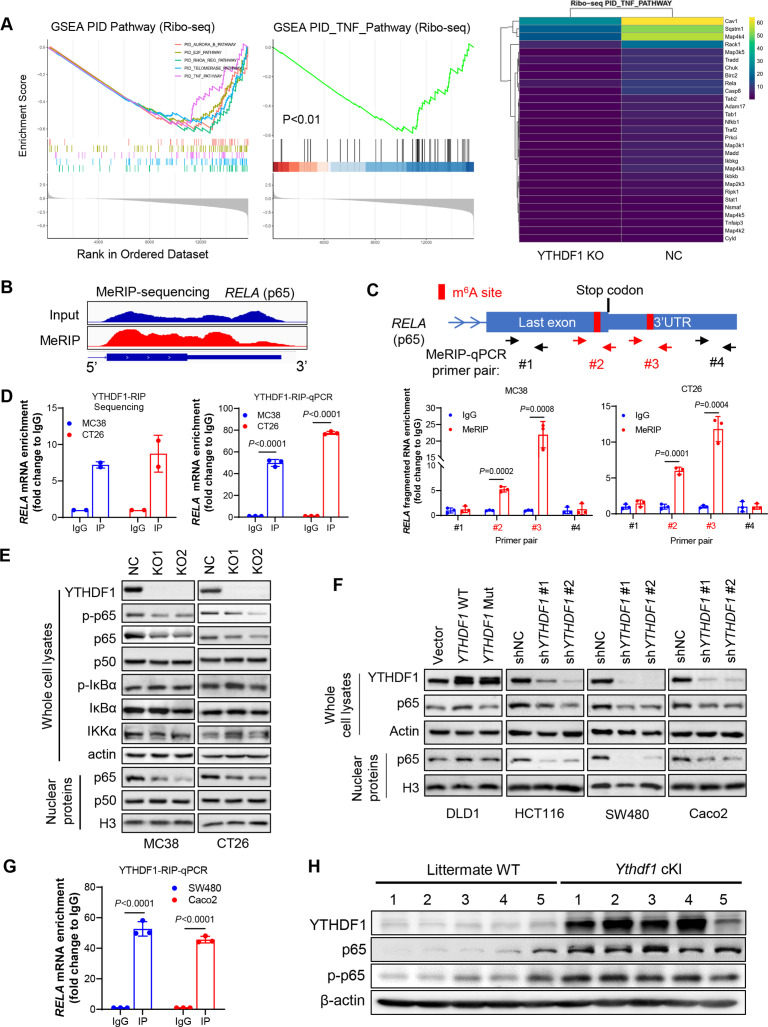
YTHDF1 promotes p65 translation to activate TNF/NF-κB signalling
To identify the molecular mechanism by which YTHDF1 elicits immunosuppression, we performed RNA-seq and Ribo-seq in CRC cells with or without knockout of YTHDF1. By RNA-seq analysis, differentially expressed genes between MC38-NC and MC38-YTHDF1-KO cells were enriched in TNF and NF-κB signalling pathways (online supplemental figure S5A). Consistent result was obtained in another CRC cell line CT26 showing that YTHDF1 regulated TNF/NF-κB signalling pathway (online supplemental figure S5B). In support of these, qPCR validated that Ythdf1-KO reduced mRNA expression of TNF/NF-κB targets (online supplemental figure S6A). Furthermore, Ribo-seq data revealed that loss of YTHDF1 was significantly associated with inactivation of TNF signalling (figure 5A). Accordingly, YTHDF1 knockout reduced ribosome protected fragment abundance of genes involved in TNF signalling (figure 5A). Thus, YTHDF1 could regulate TNF/NF-κB signalling by promoting protein translation. Since YTHDF1 functions as a m6A reader, we next performed m6A immunoprecipitation sequencing (MeRIP-seq) to pinpoint m6A-modified transcripts. By screening m6A peaks in mRNAs involved in TNF signalling, we identified two m6A sites close to the stop codon of p65 mRNA (figure 5B), which was validated by MeRIP-qPCR (figure 5C). Importantly, we identified a direct interaction between YTHDF1 and p65 mRNA by RNA immunoprecipitation (RIP) sequencing and RIP-qPCR with anti-YTHDF1 antibody (figure 5D). Thus, p65 mRNA is a direct target of YTHDF1. We found that knockout of Ythdf1 attenuated p65 protein expression, especially nuclear p65 expression, in both CT26 and MC38 cells, without affecting the expression of other regulators of NF-κB pathways such as IKKα and IκBα (figure 5E). Notably, mRNA expression of p65 was unchanged in Ythdf1-KO cells (online supplemental figure S6B), supporting that YTHDF1 regulates p65 primarily at the level of protein translation, which is in line with the reported YTHDF1 function of facilitating translation of its targets. Consistent results were obtained in human CRC cells showing that overexpression of wild-type YTHDF1, but not dysfunctional mutant,5 6 elevated p65 protein expression; conversely, YTHDF1 knockdown attenuated p65 protein in human CRC cells (figure 5F). RIP-qPCR using anti-YTHDF1 antibody also confirmed a direct interaction between YTHDF1 and p65 mRNA in human CRC cells (figure 5G). We next sought to validate the association of YTHDF1 and p65 in vivo. In Ythdf1 knockin mice, both p65 and phospho-p65 protein were increased in colon tumours (figure 5H). Collectively, YTHDF1 promotes p65 protein expression to activate TNF and NF-κB signalling in vitro and in vivo.
Figure 5.
YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) promotes TNF/NF-kB signalling in CRC through promoting RELA (p65) mRNA translation. (A) Differentially expressed genes between CT26 cells with and without knockout of Ythdf1 were enriched in TNF signalling pathway identifying by Ribo-seq (left). Heatmap of genes on TNF signalling pathway (right). (B) Methylated RNA immunoprecipitation (MeRIP) sequencing on CT26 cells, showing m6A modifications on last exon or 3’UTR of RELA (p65) mRNA. (C) Scheme showing the design of primers for MeRIP-qPCR to validate m6A modifications on p65 mRNA. Potential m6A sites were highlighted in red (upper). MeRIP-qPCR primers are indicated by arrows. MeRIP-qPCR validated m6A modification with primers #2 and #3 (lower). (D) RNA immunoprecipitation sequencing with anti-YTHDF1 antibody (YTHDF1-RIP) showing the enrichment of p65 mRNA compared with IgG control (left). YTHDF1-RIP-qPCR with primers specific to murine p65 mRNA (right). (E) Western blot of key effectors of TNF/NF-κB signalling pathway on knockout of Ythdf1. (F) Western blot analysis performed with indicated human CRC cells overexpressing wild-type (WT) or mutant (mut) YTHDF1, or YTHDF1 knockdown with shRNA. Non-targeted shRNA (shNC) were used as control. (G) YTHDF1-RIP-qPCR with primers specific to human p65 mRNA. (H) Expression of p65 and phospho-p65 was determined by western blot in colon tumours from the intestine-specific Ythdf1 knockin mice and wildtype littermates. cKI: conditional knockin (two tailed t-test (C, D, G)).
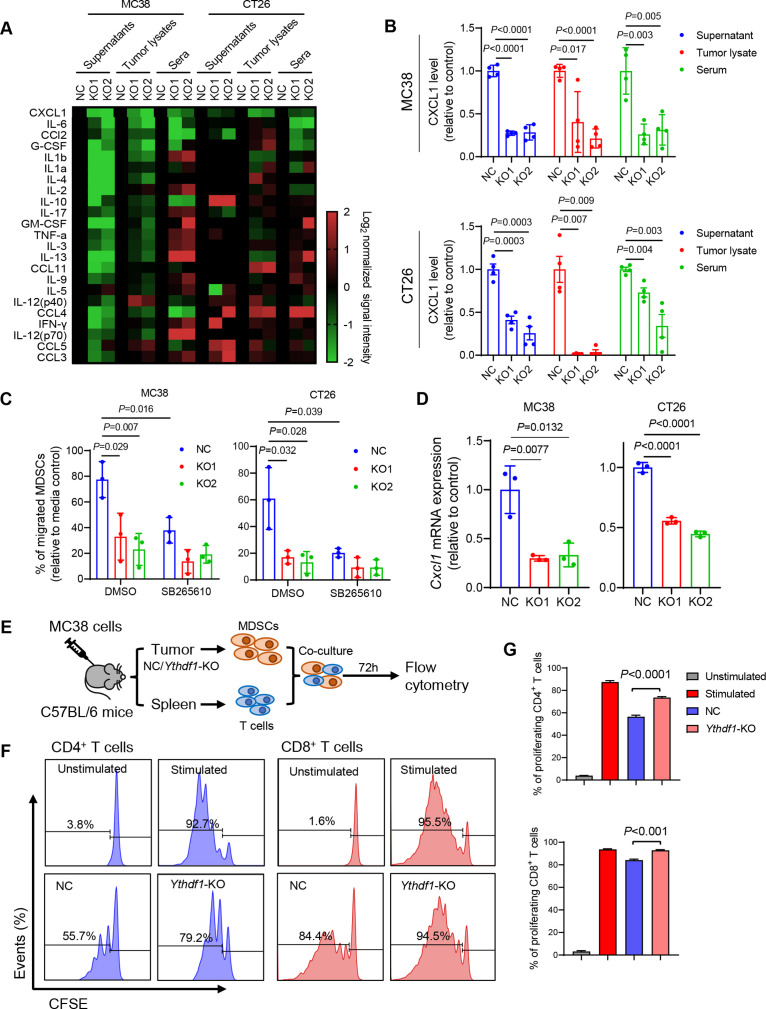
YTHDF1 promotes MDSC migration via p65-CXCL1 axis
To understand the link between YTHDF1-induced p65 and its immunosuppression, we performed a cytokine multiplex immunoassay for the detection of 23 different mouse cytokines in conditioned medium of CRC cells, tumour lysates, and serum from mice bearing syngeneic tumours. Among these cytokines, CXCL1 was consistently reduced by Ythdf1-KO (figure 6A), and this reduction of CXCL1 was confirmed by ELISA assay (figure 6B). CXCL1 has been reported as a transcriptional target of NF-κB signalling,20 21 and it promotes MDSC chemotaxis via interaction with its receptor CXCR2.22–24 Given that Ythdf1-KO reduced MDSC infiltration in CRC TIME, we asked whether YTHDF1 modulates MDSC migration. As such, in vitro MDSC migration assay was conducted. We found that conditioned medium from wild-type CRC cells enhanced MDSC migration, which was impaired by the knockout of Ythdf1 (figure 6C). Blocking CXCL1-CXCR2 interaction by CXCR2 inhibitor SB265610 eliminated the difference between control and Ythdf1-KO culture supernatant in mediating MDSC migration (figure 6C). Thus, YTHDF1 promotes MDSC migration through CXCL1/CXCR2 axis. We next asked whether YTHDF1 could regulate Cxcl1 mRNA expression. As expected, Ythdf1 knockout significantly inhibited Cxcl1 mRNA levels in murine (figure 6D) and human CRC cell lines (online supplemental figure S7A). Conversely, the overexpression of wild-type YTHDF1, but not its dysfunctional mutant, elevated CXCL1 transcription and protein levels in human CRC cells (online supplemental figure S7A). NF-κB activators, such as TNF-α and IL-1β, have been reported to induce CXCL1 expression.25 We thus treated MC38 cells with TNF-α and measured CXCL1 mRNA and secretion. TNF-α stimulated Cxcl1 mRNA and secretion, an effect abrogated by the loss of Ythdf1 (online supplemental figure S7B). To confirm the link between YTHDF1 and CXCL1 in human CRC, we examined the association between YTHDF1 and CXCL1 expression in TCGA CRC cohort. Consistently, YTHDF1-high CRC demonstrated high CXCL1 expression (online supplemental figure S7C). Besides CXCL1, CXCL2 also positively correlated with YTHDF1 (online supplemental figure S7C). In contrast, CXCL5 and CXCL8 expression negatively correlated with YTHDF1 (online supplemental figure S7C). Considering the role of YTHDF1 in promoting MSDC infiltration, we next performed CRC TMA of CD33, a marker for G-MDSCs.26 27 We found that YTHDF1 protein levels positively correlated with the proportion of intratumoural CD33+ cells (online supplemental figure S8). Our findings thus support a YTHDF1-p65-CXCL1/CXCR2 axis mediating MDSC migration in CRC.
Figure 6.
Loss of YTH N6-methyladenosine RNA binding protein 1 (Ythdf1) promotes reduction of myeloid-derived suppressor cells (MDSCs) by decreasing CXCL1 secretion. (A) Multiplex mouse cytokine immunoassay assessing levels of 23 cytokines in cell culture supernatants from MC38 and CT26 cell culture, and tumour lysates and sera from MC38 and CT26 syngeneic tumour models. (B) ELISA validation of levels of CXCL1 in the indicated materials as in (A) (n=4 each group). (C) MDSC migration was assessed in vitro with transwell migration assay using conditional medium derived from the indicated cells. CXCR2 inhibitor SB265610 was used at 5 µM. (D) RT-qPCR determination of Cxcl1 mRNA expression in the indicated cells. (E) Flow chart of T cell suppression assay. (F) Representative images of flow cytometry assessing the proliferation of CFSE-labelled T cells cocultured with MDSCs isolated as (E). (G) Quantification of (F) (two tailed t-test (C, D, E, G)).
We next investigated if YTHDF1 affects MDSC function. CD11b+Gr-1+ MDSCs were isolated from MC38 syngeneic tumours and then cocultured with T cells in vitro (figure 6E). MDSCs isolated from control tumours suppressed T cell proliferation; however, MDSCs from Ythdf1-KO tumours exhibited significantly less suppressive activity against the proliferation of both CD8+ T cells and CD4+ T cells compared with MDSCs from the control (figure 6F, G). In support of this, MDSCs isolated from Ythdf1-KO tumours have reduced expression of MDSC functional markers Nos2, Arg1, and Cd274 compared with Ythdf1-WT tumours (online supplemental figure S9A). Flow cytometry confirmed that iNOS+ MDSCs was attenuated Ythdf1-KO tumours (online supplemental figure S9B). These data validate the reported immunosuppressive function of MDSCs on key effector cells, including CD8+ T cells and CD4+ T cells,28 and support that YTHDF1-expressing CRC recruits functional MDSCs.
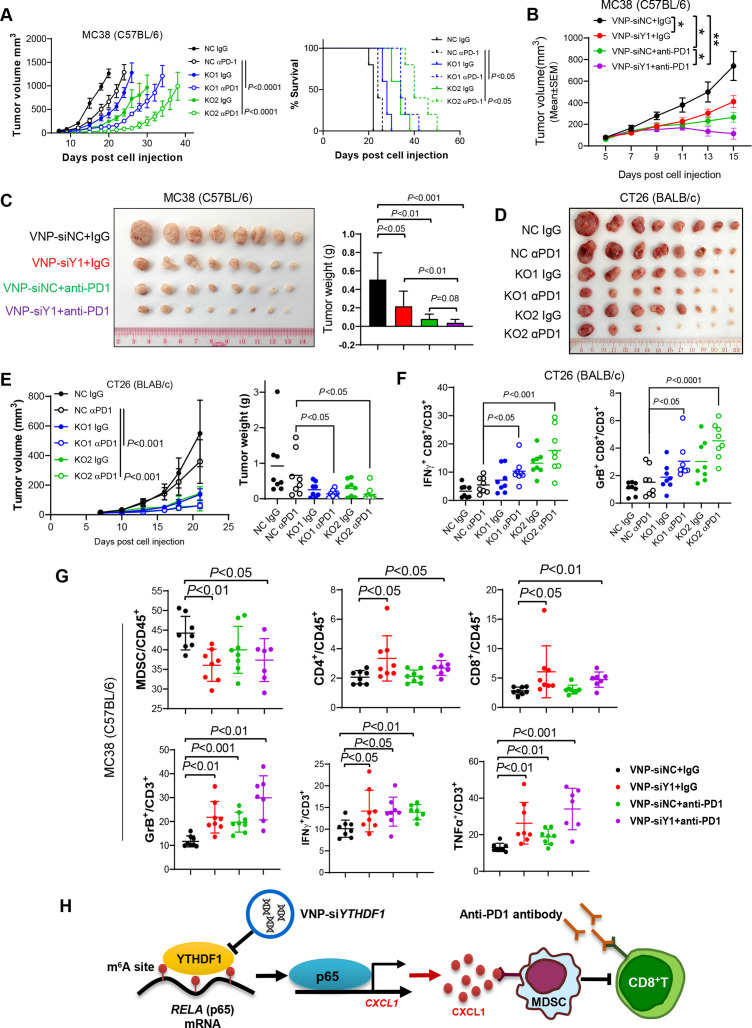
YTHDF1 is a potential therapeutic target for CRC immunotherapy
Considering that reduced infiltration of MDSCs has been reported to correlate with enhanced immunotherapeutic efficacy in various cancer types,22 29 30 we sought to test if targeting YTHDF1 augments anti-PD1 therapy in CRC. As expected, we found that knockout of Ythdf1 boosted efficacy of anti-PD1 in MC38 (MSI-H) syngeneic tumours and prolonged survival of the tumour-bearing mice (figure 7A). We further exploited the VNPs system to deliver specific Ythdf1-siRNA into tumours. When MC38 syngeneic tumours reached 50~100 mm3, we treated the mice with VNP-siYthdf1 (or VNP-siNC) and anti-PD1 treatment (or IgG). VNP-siYthdf1 significantly suppressed MC38 tumour growth compared with VNP-siNC (figure 7B, C). Remarkably, the combination of VNP-siYthdf1 plus anti-PD1 exerted the strongest inhibitory effect against tumour growth (figure 7B, C).
Figure 7.
Targeting YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) augmented anti-PD1 blockade therapy in both microsatellite instability-high (MSI-H) and microsatellite stable colorectal cancer (CRC). (A) MC38 cells with or without Ythdf1 knockout were implanted into C57BL/6 mice and treated with IgG or anti-PD1 (αPD1; n=5 per group). Tumour volume (left) and mice survival (right) were presented. Euthanasia was applied when tumour burden was greater than or equal to 1500 mm3 or when the mouse was moribund. (B) Growth curve of subcutaneous tumours from C57BL/6 injected with MC38 and treated with the indicated treatments. Vesicle-like nanoparticle (VNP)-siRNAs were given intratumorally. And αPD1 was given with intraperitoneal injection. (C) Images and weight of the tumours from (B). (D) The indicated CT26 cells were subcutaneously injected in BALB/c mice. The mice were treated with control or αPD1 (n=8 each group). Tumours removed from the mice at sacrifice were shown. (E) Tumour growth curve (left) and tumour weight (right) in (D). (F) Flow cytometry performed with the tumours in (D). (G) Top panel: flow cytometry assessing the proportion of MDSCs, CD4+ T cells, and CD8+ T cells in total CD45+ cells in tumours in (B). Lower panel: flow cytometry assessing functional CD8+ T cells in tumours in (B). (H) Schematic of the m6A-YTHDF1-p65-CXCL1 axis inducing MDSC accumulation and hence suppressing T cell function in CRC (two tailed t-test (C, D, F, G, H); analysis of variance test (A, C, F); long-rank test (A)). siY1: siYTHDF1. GrB: granzyme B.
We further addressed if targeting YTHDF1 can overcome anti-PD1 resistance in MSS CRC based on syngeneic CT26 (MSS CRC) tumour model. CT26 cells with Ythdf1 knockout were thus injected to the syngeneic mice and treated with anti-PD1. We found that knockout of Ythdf1 significantly enhanced anti-PD1 treatment efficacy in CT26 syngeneic tumours, which otherwise were non-responsive to ICB therapy (figure 7D, E).
Flow cytometry analysis further revealed that combination of Ythdf1 silencing and anti-PD1 remarkably increased tumour-infiltrating functional CD8+ T cells, including IFN-γ+ CD8+ T cells and Granzyme B+ CD8+ T cells in both CT26 and MC38 syngeneic models (figure 7F, G). Furthermore, combinational treatment strongly reduced accumulation of MDSCs, whereas CD4+ T cells and CD8+ T cells were induced (figure 7G). Thus, targeting of YTHDF1 not only potentiates ICB therapeutic efficacy in MSI-H CRC, but also overcomes ICB resistance in MSS CRC by suppressing recruitment of MDSCs and improving functionality of CD8+ T cells.
Discussion
YTHDF1 is a m6A reader is frequently upregulated in CRC, but its role in antitumour immunity is largely unknown. Here, our study demonstrates for the first time that high YTHDF1 expression in CRC drives immunosuppression. Single-cell profiling reveals that YTHDF1 is crucial to the recruitment of MDSCs, which in turn attenuated T cell infiltration and function in CRC. This phenomenon is consistently found in syngeneic tumours, intestine-specific YTHDF1 knockin mice and CD34+ humanised mice. Taken together, our data demonstrate that YTHDF1 promotes an immunosuppressive tumour microenvironment to facilitate CRC tumourigenesis.
A series of in vivo models validated the role of YTHDF1 in recruitment of MDSCs in CRC. In CRC syngeneic models, Ythdf1 knockout inhibited MDSC accumulation. Conversely, intestine-specific Ythdf1 knockin accelerated colorectal tumourigenesis in AOM/DSS and ApcMin/+ models, concomitant with increased intratumoural MDSCs. Moreover, YTHDF1 silencing also impaired infiltration of human MDSCs in CD34+ humanised mice. MDSCs are a highly diverse population of immature myeloid cells, including granulocytic MDSCs and monocytic MDSCs. Both of them, especially G-MDSCs, possess potent immunosuppressive activity.31 In vitro MDSC migration assays directly validated that YTHDF1 plays a crucial role in MDSC chemotaxis. Our results support that YTHDF1 promotes colorectal tumourigenesis via MDSC-mediated immunosuppression.
MDSCs primarily exert their immunosuppressive effect by inhibiting key effector cell proliferation and activation.28 Indeed, we found that Ythdf1 knockout induces infiltration of CD4+ T-cells, CD8+ T cells and NK cells in CRC tumours and CD34+ humanised mice, with increased expression of cytotoxic markers granzyme B, INF-γ and TNF-α whereas intestine-specific Ythdf1 knockin correlated with dampened infiltration and activity status of these effector cells. In vitro studies also demonstrated that MDSCs isolated from Ythdf1-null tumours have impaired capacity to suppress T cell proliferation. Together, our data support that YTHDF1-driven functional MDSC accumulation exerts an inhibitory effect on antitumour immune cells to promote immune evasion in CRC.
We next deciphered the molecular basis of YTHDF1-driven MDSC accumulation in CRC. As YTHDF1 functions as a m6A reader, we performed integrative RNA-seq, Ribo-seq and MeRIP-seq to identify candidate targets of YTHDF1 in CRC. Although several signalling pathways were identified in MSS (CT26) and MSI-H (MC38) CRC, TNF/NF-κB signalling pathway is the top pathway commonly induced by YTHDF1 in both models. We further unravelled p65 subunit of NF-κB as a direct target of YTHDF1. YTHDF1, but not its dysfunctional mutant, binds to m6A-modified p65 mRNA to increase p65 protein levels without affecting its mRNA expression, implying that YTHDF1 promotes p65 translation in m6A-dependent manner. TNF/NF-κB signalling is often activated in tumour cells, and induces expression of proinflammatory genes encoding cytokines and chemokines.32 By cytokine profiling, we found that CXCL1 mRNA expression and secretion were unanimously repressed by genetic inhibition of Ythdf1 in mouse and human CRC cell lines, syngeneic mice and transgenic mice. CXCL1 plays a pivotal role in promoting MDSCs chemotaxis in TIME via interaction with CXCR2.33 In line with this, we found that YTHDF1 mediates the migration of MDSCs via a CXCL1-CXCR2 axis, an effect abolished by an CXCR2 inhibitor. Together, YTHDF1-mediated p65 signalling promotes MDSCs via CXCL1 secretion. Apart from CXCL1, alternative NF-κB downstream targets were also down-regulated by Ythdf1 knockout. It will be of interest to further explore the collateral impact of YTHDF1 on NF-κB-dependent expression of cytokines and chemokines on the immunosuppressive TIME of CRC.
MDSCs have been reported to exert immunosuppressive function on key effector cells, including CD8+ T cells, CD4+ T cells and NK cells.28 MDSCs exert their antagonistic effect on antitumour T cells via a myriad of mechanisms, including depletion of intratumoural arginine via Arginase-1, induction of oxidative stress via iNOS, and secretion of immunosuppressive molecules such as TGF-β.31 Here, we showed that depleting Ythdf1 in cancer cells suppressed intratumoural MDSCs and mitigated the immunosuppressive function of MDSCs on effector cells. MDSCs and T cell coculture studies demonstrated that MDSCs from Ythdf1-KO tumours have impaired capacity to suppress the cell proliferation of both CD8+ T cells and CD4 T+ cells when compared with control tumours. Our findings thus support that YTHDF1 mediates the recruitment of functional MSDCs, which inhibit the function and proliferation of effector T cells in CRC, leading to compromised immune surveillance (figure 7H).
In the clinic, ICI therapy only shows beneficial effects to a small proportion of patients with MMR or MSI-H metastatic CRC, whereas the majority of CRCs with MSS status are largely non-responsive.34 This can be partially attributed to presence of MDSCs and the formation of immunosuppressive tumour microenvironment. Given that knockout of Ythdf1 reduced MDSC infiltration in CRC tissues to revert immune suppression, we thus explored therapeutic implications of targeting YTHDF1 in both MC38 (MSI-H) and CT26 (MSS) syngeneic CRC models. Ythdf1 silencing boosted the anti-PD1 efficacy in MC38 syngeneic model. More importantly, targeting of YTHDF1 reversed anti-PD1 resistance in CT26 (MSS) syngeneic model, highlighting the broad utility of YTHDF1-targeting approach in promoting ICB efficacy not only in MSI-H CRC, but also MSS CRC that otherwise is resistant to ICB therapy.
As there is no currently available drug for YTHDF1, here we developed a nanoparticle-based delivery system35 to deliver YTHDF1-siRNA in vivo. In syngeneic mouse models and humanised mice models of CRC, we observed the robust effect of VNP-siYTHDF1 that remarkably elevated the infiltration of functional T cells and enhanced tumour response to anti-PD1 therapy. Targeting of YTHDF1 thus represents a promising immunotherapeutic target in CRC.
In conclusion, YTHDF1 expression in CRC recruits immunosuppressive MDSCs via activating m6A-p65-CXCL1 axis to inhibit T cells, thereby promoting CRC. Targeting YTHDF1 plus anti-PD1 therapy demonstrates promising antitumour efficacy against CRC, corroborating that YTHDF1 is a potential therapeutic target for CRC.
Footnotes
YB and JZ contributed equally.
Contributors: YB and JZ performed the experiments, analysed the data and drafted the manuscript. HC and CCW commented on the study and revised the paper. CL performed animal experiments. YD and DH performed bioinformatics analyses. HG, DC and YP performed experiments. WK and KFT performed histological staining and evaluation. JY designed and supervised the study and revised the manuscript. JY acts as the guarantor of the study.
Funding: This study was supported by RGC Collaborative Research Fund (C4039-19GF), National Natural Science Foundation of China (81972576), RGC-GRF (14110819, 14111621), RGC Research Impact Fund Hong Kong (R4032-21F), Vice-Chancellor's Discretionary Fund CUHK.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by Clinical Research Ethics Committee of the Chinese University of Hong Kong and Beijing Caner Hospital.
References
- 1. Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA 2021;325:669–85. 10.1001/jama.2021.0106 [DOI] [PubMed] [Google Scholar]
- 2. André T, Shiu K-K, Kim TW, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med 2020;383:2207–18. 10.1056/NEJMoa2017699 [DOI] [PubMed] [Google Scholar]
- 3. Delaunay S, Frye M. Rna modifications regulating cell fate in cancer. Nat Cell Biol 2019;21:552–9. 10.1038/s41556-019-0319-0 [DOI] [PubMed] [Google Scholar]
- 4. Lin X, Chai G, Wu Y, et al. RNA m6a methylation regulates the epithelial mesenchymal transition of cancer cells and translation of snail. Nat Commun 2019;10:2065. 10.1038/s41467-019-09865-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5. Pi J, Wang W, Ji M, et al. YTHDF1 promotes gastric carcinogenesis by controlling translation of FZD7 Cancer Res 2021;81:2651–65. 10.1158/0008-5472.CAN-20-0066 [DOI] [PubMed] [Google Scholar]
- 6. Liu T, Wei Q, Jin J, et al. The M6a reader YTHDF1 promotes ovarian cancer progression via augmenting eIF3c translation. Nucleic Acids Res 2020;48:3816–31. 10.1093/nar/gkaa048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cerami E, Gao J, Dogrusoz U, et al. The cbio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tigano M, Vargas DC, Tremblay-Belzile S, et al. Nuclear sensing of breaks in mitochondrial DNA enhances immune surveillance. Nature 2021;591:477–81. 10.1038/s41586-021-03269-w [DOI] [PubMed] [Google Scholar]
- 11. Sun L, Kees T, Almeida AS, et al. Activating a collaborative innate-adaptive immune response to control metastasis. Cancer Cell 2021;39:1361–74. 10.1016/j.ccell.2021.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao J, Shi LZ, Zhao H, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 2016;167:397–404. 10.1016/j.cell.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mrna profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang W, Green M, Choi JE, et al. Cd8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569:270–4. 10.1038/s41586-019-1170-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018;391:2128–39. 10.1016/S0140-6736(18)30789-X [DOI] [PubMed] [Google Scholar]
- 18. Choi Y, Lee S, Kim K, et al. Studying cancer immunotherapy using patient-derived xenografts (pdxs) in humanized mice. Exp Mol Med 2018;50:99. 10.1038/s12276-018-0115-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rezvantalab S, Drude NI, Moraveji MK, et al. PLGA-based nanoparticles in cancer treatment. Front Pharmacol 2018;9:1260. 10.3389/fphar.2018.01260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scortegagna M, Cataisson C, Martin RJ, et al. Hif-1Alpha regulates epithelial inflammation by cell autonomous NFkappaB activation and paracrine stromal remodeling. Blood 2008;111:3343–54. 10.1182/blood-2007-10-115758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kemp SB, Carpenter ES, Steele NG, et al. Apolipoprotein E promotes immune suppression in pancreatic cancer through NF-κB-mediated production of CXCL1. Cancer Res 2021;81:4305–18. 10.1158/0008-5472.CAN-20-3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Highfill SL, Cui Y, Giles AJ, et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med 2014;6:237ra67. 10.1126/scitranslmed.3007974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang D, Sun H, Wei J, et al. Cxcl1 is critical for premetastatic niche formation and metastasis in colorectal cancer. Cancer Res 2017;77:3655–65. 10.1158/0008-5472.CAN-16-3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Q, Ma C, Duan Y, et al. Gut microbiome directs hepatocytes to recruit mdscs and promote cholangiocarcinoma. Cancer Discov 2021;11:1248–67. 10.1158/2159-8290.CD-20-0304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olivera I, Sanz-Pamplona R, Bolaños E, et al. A therapeutically actionable protumoral axis of cytokines involving IL-8, TNFα, and IL-1β. Cancer Discov 2022;12:2140–57. 10.1158/2159-8290.CD-21-1115 [DOI] [PubMed] [Google Scholar]
- 26. Bronte V, Brandau S, Chen S-H, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 2016;7:12150. 10.1038/ncomms12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cassetta L, Baekkevold ES, Brandau S, et al. Deciphering myeloid-derived suppressor cells: isolation and markers in humans, mice and non-human primates. Cancer Immunol Immunother 2019;68:687–97. 10.1007/s00262-019-02302-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-Derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol 2021;21:485–98. 10.1038/s41577-020-00490-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Steele CW, Karim SA, Leach JDG, et al. CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell 2016;29:832–45. 10.1016/j.ccell.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greene S, Robbins Y, Mydlarz WK, et al. Inhibition of MDSC trafficking with SX-682, a CXCR1/2 inhibitor, enhances NK-cell immunotherapy in head and neck cancer models. Clin Cancer Res 2020;26:1420–31. 10.1158/1078-0432.CCR-19-2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gabrilovich DI, Nagaraj S. Myeloid-Derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009;9:162–74. 10.1038/nri2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer 2013;12:86. 10.1186/1476-4598-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar V, Patel S, Tcyganov E, et al. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol 2016;37:208–20. 10.1016/j.it.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol 2018;36:773–9. 10.1200/JCO.2017.76.9901 [DOI] [PubMed] [Google Scholar]
- 35. Mitchell MJ, Billingsley MM, Haley RM, et al. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov 2021;20:101–24. 10.1038/s41573-020-0090-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-328845supp003.pdf (61.7KB, pdf)
gutjnl-2022-328845supp001.pdf (206.4KB, pdf)
gutjnl-2022-328845supp002.pdf (11.2MB, pdf)
Data Availability Statement
Data are available upon reasonable request.