Abstract
When phosphorylated, the dimeric form of nitrogen regulatory protein C (NtrC) of Salmonella typhimurium forms a larger oligomer(s) that can hydrolyze ATP and hence activate transcription by the ς54-holoenzyme form of RNA polymerase. Studies of Mg-nucleoside triphosphate binding using a filter-binding assay indicated that phosphorylation is not required for nucleotide binding but probably controls nucleotide hydrolysis per se. Studies of binding by isothermal titration calorimetry indicated that the apparent Kd of unphosphorylated NtrC for MgATPγS is 100 μM at 25°C, and studies by filter binding indicated that the concentration of MgATP required for half-maximal binding is 130 μM at 37°C. Filter-binding studies with mutant forms of NtrC defective in ATP hydrolysis implicated two regions of its central domain directly in nucleotide binding and three additional regions in hydrolysis. All five are highly conserved among activators of ς54-holoenzyme. Regions implicated in binding are the Walker A motif and the region around residues G355 to R358, which may interact with the nucleotide base. Regions implicated in nucleotide hydrolysis are residues S207 and E208, which have been proposed to lie in a region analogous to the switch I effector region of p21ras and other purine nucleotide-binding proteins; residue R294, which may be a catalytic residue; and residue D239, which is the conserved aspartate in the putative Walker B motif. D239 appears to play a role in binding the divalent cation essential for nucleotide hydrolysis. Electron paramagnetic resonance analysis of Mn2+ binding indicated that the central domain of NtrC does not bind divalent cation strongly in the absence of nucleotide.
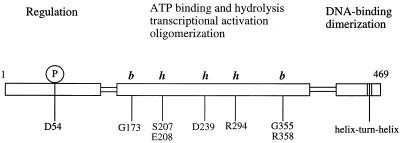
NtrC (nitrogen regulatory protein C) activates transcription by an alternative holoenzyme form of bacterial RNA polymerase that contains ς54 as the ς factor (for a review, see reference 50). NtrC binds to DNA sites that have the properties of eukaryotic transcriptional enhancers and contacts the polymerase by means of DNA loop formation. To activate transcription, NtrC must hydrolyze the β-γ bond of ATP or GTP and couple the energy made available to the process of open complex formation by the polymerase (66). Although both ATPγS and ADP can inhibit the ATPase activity of NtrC, indicating that they bind to the protein (67), neither can substitute for ATP in allowing open complex formation. Nucleotide hydrolysis and transcriptional activation by NtrC are functions of its central domain, a domain of ∼240 residues (Fig. 1). They depend on phosphorylation of an aspartate residue in its amino (N)-terminal regulatory domain, D54, and concomitant formation of large NtrC oligomers (72).
FIG. 1.
Domain structure of NtrC. NtrC is comprised of three functional domains: an amino-terminal regulatory domain (∼120 amino acids) that is phosphorylated at aspartate 54 (D54), a central catalytic domain (∼240 amino acids) that contains determinants for ATP binding and hydrolysis, as well as for oligomerization and transcriptional activation, and a carboxy-terminal domain (∼90 amino acids) that contains a helix-turn-helix DNA-binding motif and dimerization determinants. Residues within the central catalytic domain of NtrC that appear to be required for MgATP binding (this work) are indicated by b, whereas residues that appear to be required for ATP hydrolysis but not binding are indicated by h. Residues G173, S207/E208, D239, R294, and G355/R358 lie in conserved regions 1, 3, 4, 6, and 7, respectively, of Morett and Segovia (36).
The requirement for an activator protein capable of hydrolyzing a nucleoside triphosphate (NTP) appears to be universal for transcription from ς54-dependent promoters (3, 6, 10, 22, 31, 48, 50), and a homologue of the central catalytic domain of NtrC is found in all such activators, a family of over a dozen members (36, 43). Moreover, in the case of the activator NifA, it has been shown that the isolated central domain is sufficient for transcriptional activation both in vivo and in vitro (6, 23). The central domain for activators of ς54-holoenzyme contains a glycine-rich motif (GXXXXGK, where X is any residue) that has been proposed to correspond to the Walker A motif (also known as the phosphate binding or P loop) found in many purine nucleotide-binding proteins (43, 52, 63). This motif is unusual in activators of ς54-holoenzyme in that the conserved lysine is followed by a glutamate or aspartate residue rather than the consensus threonine or serine. In a number of purine nucleotide-binding proteins, the Walker A motif has been shown to form a loop that wraps around the β phosphate group of the bound nucleotide (1, 5, 14, 16, 28, 34, 44, 53, 61, 62). In the case of NtrC, an alteration of the second glycine in this motif, G173, to asparagine results in loss of both ATPase activity and the capacity to activate transcription (67). Another highly conserved region in the central domain for activators of ς54-holoenzyme, BBBD, where B represents a hydrophobic amino acid, has been proposed to correspond to the Walker B motif of purine nucleotide-binding proteins (43, 52, 63). Typically, the highly conserved aspartate residue of the Walker B motif occurs at the carboxy terminus of a β strand (1, 5, 14, 16, 28, 34, 44, 53, 61, 62). For NtrC of Salmonella typhimurium, the putative aspartate residue, D239, is predicted to lie at the end of a β strand (43). Structural analyses of several purine nucleotide-binding proteins indicate that the highly conserved aspartate residue plays a role in coordinating the divalent cation which is essential for hydrolysis of the β-γ bond of ATP or GTP (1, 5, 14, 16, 28, 34, 44, 53, 61, 62). This coordination usually occurs indirectly through an intervening H2O molecule.
Recent secondary structure predictions coupled with the use of recognition algorithms for protein folds indicated that the central domain of activators of ς54-holoenzyme adopts a mononucleotide-binding fold similar to those of the G domains of the bacterial polypeptide elongation factor EF-Tu and the eukaryotic signaling protein p21ras (43). The functional significance of the Walker A and B motifs of such proteins has been underscored by a number of studies showing that key residues within each motif were essential for nucleotide binding and/or hydrolysis (2, 7, 9, 12, 19, 20, 24, 37, 46, 57, 65, 70, 74).
To further our understanding of the mechanism of ATP hydrolysis and transcriptional activation by NtrC, we have investigated the nature of the MgATP-binding site. By testing mutant forms of NtrC previously known to be defective in ATP hydrolysis (42, 67) for their ability to bind ATP, we have identified several regions of the central domain, including the proposed Walker A motif, that are involved in ATP binding and/or hydrolysis. By changing the highly conserved aspartate residue D239 in the putative Walker B motif of NtrC, we have explored the role of this residue in transcriptional activation and in binding and hydrolysis of MgATP. Finally, we have investigated the effect of the oligomerization state of NtrC on its binding of MgATP.
MATERIALS AND METHODS
Materials.
NTPs and dNTPs were purchased from Boehringer Mannheim. For studies of ATP binding by isothermal titration calorimetry (ITC), ATP was dissolved in the appropriate buffer (see below), the pH was adjusted to 7.8 at room temperature with 3 M KOH, and the final ATP concentration was determined spectrophotometrically at 260 nm. MgATP was handled similarly except that the buffer contained 5.1 mM MgCl2. All enzymes used to manipulate DNA were from New England Biolabs or USB Life Research Products and were used as recommended by the manufacturer.
Cloning techniques and mutagenesis.
DNA isolation and cloning were carried out by using standard procedures (4). Site-directed mutagenesis was performed as described elsewhere (29), using a Muta-Gen in vitro phagemid mutagenesis kit from Bio-Rad. Briefly, single-stranded DNA was prepared from phagemid pJES594 (26), a pTZ18U derivative that contains all of the ntrC gene. The oligonucleotides used for mutagenesis were 5′-GCTGTTTCTGGCGGAAATTGGCG-3′ for D239A, 5′-CGCTGTTTCTGTGCGAAATTGGCG-3′ for D239C, and 5′-CGCTGTTTCTGAACGAAATTGGCG-3′ for D239N (mutated nucleotides are indicated by underlining). The DNA sequence of the mutated gene was verified by DNA sequencing using Sequenase (USB Life Research Products). Following mutagenesis, 449-bp AgeI-AscI fragments from the mutated ntrC gene were subcloned into the corresponding region of pJES311 (overexpression plasmid for wild-type NtrC [67]). The resultant plasmids were pJES1063 (D239A), pJES1064 (D239C), and pJES1065 (D239N). DNA fragments encoding the D239 substitutions were similarly subcloned into pJES559, an overexpression plasmid that carries the malE gene directly upstream of ntrC and yields a maltose-binding protein (MBP) fusion to the N terminus of NtrC. This yielded plasmids pJES1089 (D239A), pJES1090 (D239C), and pJES1091 (D239N).
Phenotypic analysis.
Plasmids pJES311 and pJES1063 to 1065 were transformed into Escherichia coli DH5α. Transformants were initially selected on Luria broth plates supplemented with 2 mM glutamine and 100 μg of ampicillin per ml. Individual colonies containing each construct of interest were then screened on nutrient broth plates. Control experiments indicated that transformants expressing NtrCrepressor proteins—i.e. those with a defect only in positive control—could be distinguished from those expressing wild-type NtrC or nonfunctional NtrC proteins because the former failed to grow, whereas the latter grew poorly (wild-type NtrC) or well (nonfunctional NtrC) (32).
Protein purification.
NtrC, NtrCrepressor, NtrC with the D54E and 3Ala (see below) mutations (NtrCD54E,3Ala), NtrC lacking residues 444 to 469 (NtrCΔ444–469), MBP-N terminus, MBP-NtrC, and MBP-NtrCrepressor proteins were overproduced and purified essentially as described elsewhere (references 40 and 42 and references therein). NtrC3Ala (a non-DNA-binding derivative of NtrC that has three alanine substitutions in the helix-turn-helix DNA-binding motif) was purified essentially as described previously (51) and was not frozen before use. The N-terminus (residues 1 to 124) and N-terminusD54E (N-terminal domain carrying the D54E substitution) fragments were a kind gift from D. Kern and D. Wemmer, University of California, Berkeley. The S. typhimurium ς54 protein was purified as described previously (26, 67). Core RNA polymerase was kindly supplied by D. Hager and R. Burgess, University of Wisconsin, Madison.
Transcription assay.
Open complex formation on a supercoiled template (1 nM) was assessed in a single-cycle transcription assay as described elsewhere (40, 51). The template was pJES534 (51), which contains a strong enhancer situated ∼460 bp from the glnA promoter.
ATPase assay.
The release of Pi from [γ-32P]ATP was monitored as described elsewhere (67), with the modifications of Flashner et al. (15), including the omission of polyethylene glycol. The ATP and Mg2+ concentrations were 0.4 and 5.4 mM, respectively. Assays in which Mn2+ was used as the divalent cation were carried out with 2.4 mM MnCl2. This concentration was judged to be optimal from titrations in which MnCl2 was used in place of MgCl2.
ATP binding by filtration.
In a standard filter-binding assay, each NtrC protein was first mixed with ATP binding buffer (50 mM Tris acetate [pH 8.0], 40 mM KCl, 5.4 mM MgCl2, 1.0 mM dithiothreitol [DTT], 0.1 mM EDTA [pH 8.0]) to which a mixture of premixed ATP and [α-32P]ATP was then added; the final volume was 25 μl. The reaction mixture was incubated at 37°C for 20 to 30 min, after which time it was filtered through a polyvinylidene fluoride membrane (1.5-cm diameter) placed on a sintered glass filter, and a vacuum was briefly applied (<2 s) to remove the liquid. The membrane (Immobilon P; 0.45-μm pore size; was prepared as specified by the manufacturer (Millipore). After sample application, the membrane was immediately washed with 1 ml of washing buffer (20 mM Tris Cl [pH 8.0], 10 mM magnesium acetate) and placed directly into a scintillation vial. Scintiverse E scintillant (Fisher) was added to the vial, and the sample was counted in a Beckman LS6800 scintillation counter. In assays that tested the effect of omitting the divalent cation on ATP binding, the assay was performed essentially as described above except that MgCl2 was omitted from the buffer and the EDTA concentration was increased to 1.1 mM. To test the effect of Mn2+ on ATP binding, DTT was omitted from the binding buffer, and MnCl2 (2.4 mM) was used in place of MgCl2 and added just prior to ATP. In competition experiments, standard binding reaction mixtures were incubated for 20 to 30 min at 37°C, challenged with unlabeled competitor ATP (at equimolar concentration to 200-fold excess) for an additional 2 min, and filtered and washed as described above. To test the effect of phosphorylation of NtrC on nucleotide binding, MBP-NtrC was first incubated with binding buffer in the presence of 10 mM carbamoyl phosphate for 10 min at 37°C to allow phosphorylation to occur (33). Then a mixture of the nonhydrolyzable ATP analogue ATPγS and [35S]ATPγS (in place of ATP and [α-32P]ATP, respectively) was added, and incubation was continued for an additional 20 min. The binding mixture was filtered and washed as described above. ATPγS was used because phosphorylated NtrC has ATPase activity; binding of the analogue by unphosphorylated NtrC was also assessed.
To determine the concentration of ATP required for 50% binding by each NtrC protein, 5 μM protein (final dimer concentration) was titrated with various ATP concentrations, typically in the range of 6 μM (∼2 × 104 cpm/pmol) to 1.0 mM (∼200 cpm/pmol). Using Kaleidograph (Adelbeck Software), we fitted data to a standard binding curve, assuming a stoichiometry of one ATP bound per NtrC monomer and no cooperativity: [ATP]bound/[NtrC]total = [ATP]free/(Kd + [ATP]free), where [ATP]free denotes the concentration of free ATP at equilibrium; we report the calculated Kd simply as concentration of ATP required for 50% maximal binding. These values are reported with standard deviations. At 5 mM MgCl2, we assumed that [ATP]total = [MgATP]total. Protein concentrations were determined by the Bradford method (Bio-Rad), with bovine serum albumin as the standard, or were calculated from absorbance at 280 nm in the presence of guanidine hydrochloride, using extinction coefficients of 42,514 M−1 cm−1 for NtrC monomer, 107,345 M−1 cm−1 for MBP-NtrC monomer, 13,514 M−1 cm−1 for the N-terminal domain of NtrC, and 77,149 M−1 cm−1 for MBP-N terminus fusion (4).
ATP binding by ITC.
To achieve the high protein concentrations required (68), we used NtrC3Ala. For reasons that are not understood, this derivative is considerably more soluble than wild-type NtrC and more soluble than MBP-NtrC. Calorimetric experiments were performed in a MCS isothermal titration calorimeter (MicroCal, Inc.) at 25.00°C in a cell volume of 1.348 ml. Binding of MgATP and MgATPγS to NtrC was measured by titrating 4 mM ligand (using a 250-μl injection syringe) into NtrC3Ala (80 to 110 μM dimer) with stirring at 400 rpm. Injections for MgATP were 5 of 2 μl, 10 of 4 μl, and 23 of 8 μl, yielding a final MgATP concentration of 620 μM in the cell. Injections for MgATPγS were 5 of 2 μl, 10 of 4 μl, 20 of 8 μl, 3 of 15 μl, and 1 of 20 μl, yielding a final ligand concentration of 690 μM in the cell. Binding of ATP was measured as described for MgATP by titrating 10 mM ATP into NtrC3Ala (103 μM dimer), yielding a final ligand concentration of 1.5 mM. The buffer composition for the ATP titration was 50 mM Tris-acetate (pH 7.8)–50 mM KCl–0.1 mM EDTA. For titrations of MgATP and MgATPγS, 5.1 mM MgCl2 was present in the buffer to maintain a constant free magnesium concentration and to avoid unwanted heat exchange effects due to the dissociation of the Mg-coordinated nucleotide. For each experiment, the NtrC3Ala protein was dialyzed extensively against the appropriate buffer just before the experiment was performed, and the protein concentration was determined after dialysis by the Bradford method, with bovine serum albumin as the standard. The Bradford method gave the same values for protein concentrations as absorbance at 280 nm in the presence of guanidine hydrochloride (see above). The heat change for dilution of the ligand in the absence of protein was measured for each experiment (control) and was subtracted from the measured heat change of ligand binding to protein (experimental). Data analysis was performed with the Origin program, provided by MicroCal. The equations used for fitting were those for single binding sites and are based on the theory described by Wiseman et al. (68). During all titrations of MgATP and MgATPγS, significantly higher drifts of the baseline for the injection peaks were observed when ligand was titrated into protein than when ligand was titrated into buffer (data not shown). These drifts of the baseline (60% greater drift for MgATP and 40% greater drift for MgATPγS compared to the blank drift, respectively) may have been due to protein precipitation. Centrifugation of samples immediately after ITC revealed that 40 to 50% of the protein had precipitated upon completion of the titration with MgATP, whereas 20% had precipitated after titration with MgATPγS or ATP. In the case of ATP, however, no significant difference in the baseline drift was observed between experimental and control samples. Attempts to study MgNTP binding at 37°C, the temperature used for filter-binding assays, were unsuccessful because more than 80% of the NtrC3Ala protein had precipitated under these conditions.
After titration of MgATP and MgATPγS into buffer (control) and into the NtrC3Ala protein (experimental), samples were analyzed for hydrolysis of nucleotide. They were applied to cellulose polyetheneimine thin-layer plates (Baker-Flex; J. T. Baker, Griesheim, Germany) and were subjected to chromatography in 0.7 M boric acid–0.4 M K2HPO4 as liquid phase. Nucleotides were then visualized by UV shadowing, and their mobilities were compared to those of standards. At the end of the titration with MgATP, approximately 30% of the unbound ATP had been hydrolyzed to ADP. Whether this ATP hydrolysis was due to an intrinsic hydrolysis capacity of unphosphorylated NtrC at concentrations of 100 μM or to contaminating ATPase activities was not determined. For MgATPγS, no hydrolysis was detected by thin-layer chromatography. The NtrC3Ala protein, which is the most soluble form of NtrC, is more difficult to purify away from contaminating ATPases than MBP fusion forms, which allow affinity chromatography, or DNA-binding forms, which bind to heparin matrices.
Divalent cation binding by EPR spectroscopy.
For electron paramagnetic resonance (EPR) experiments, NtrC proteins or their isolated N-terminal domains were dialyzed against buffer (40 mM KCl, 50 mM Tris acetate [pH 8.2]), 5% [vol/vol] glycerol, 1 mM DTT) to eliminate EDTA and then concentrated in Centricon microconcentrators (10- or 3-kDa cutoff for the intact protein or N-terminal domain, respectively; Amicon, Beverly, Mass.). NtrC protein (at a final dimer concentration of 50 to 100 μM) was incubated with MnCl2 in the presence of 40 mM KCl and 50 mM Tris acetate (pH 8.0) for 5 min at 25°C. Fresh stocks of Mn2+ were prepared daily in double-distilled H2O, using AR grade MnCl2 · 4H2O (99.99%) from Aldrich. After incubation, 5-μl samples were transferred to quartz capillaries, and EPR spectra were determined in a Bruker ESP300 EPR spectrometer equipped with a loop-gap resonator (Medical Advances) and a low-noise microwave amplifier (Miteq). To minimize lineshape distortions, the incident microwave power was nonsaturating (2 mW), and a modulation amplitude of 6 G was used, which is small compared to the linewidth of the individual peaks of the EPR spectrum for Mn2+ free in solution (22 G). Plots of the average peak-to-peak height (spectra are first-derivative spectra) of the EPR spectra at a series of Mn2+ concentrations (25 μM to 5 mM) versus Mn2+ concentration were linear. The relative amount of Mn2+ bound to the protein was calculated according to the following formula: [Mn2+]bound/[NtrC]total = ([Mn2+]total − [Mn2+]free)/[NtrC]total, where [Mn2+]free is the concentration of free Mn2+ in solution determined by EPR spectroscopy, and [Mn2+]total and [NtrC]total are the total concentrations of Mn2+ and protein, respectively. Analysis of standard curves, which were determined for each experiment, revealed that the precision for the determination of the free Mn2+ concentration by EPR is approximately 2% and thus is not the main source of experimental uncertainty in the determination of the ratio ([Mn2+]bound/[NtrC]total). The major contributions to experimental uncertainty arose from the pipetting of small volumes of NtrC protein and uncertainties in estimating protein concentration.
RESULTS
MgATP binding by wild-type and mutant NtrC proteins.
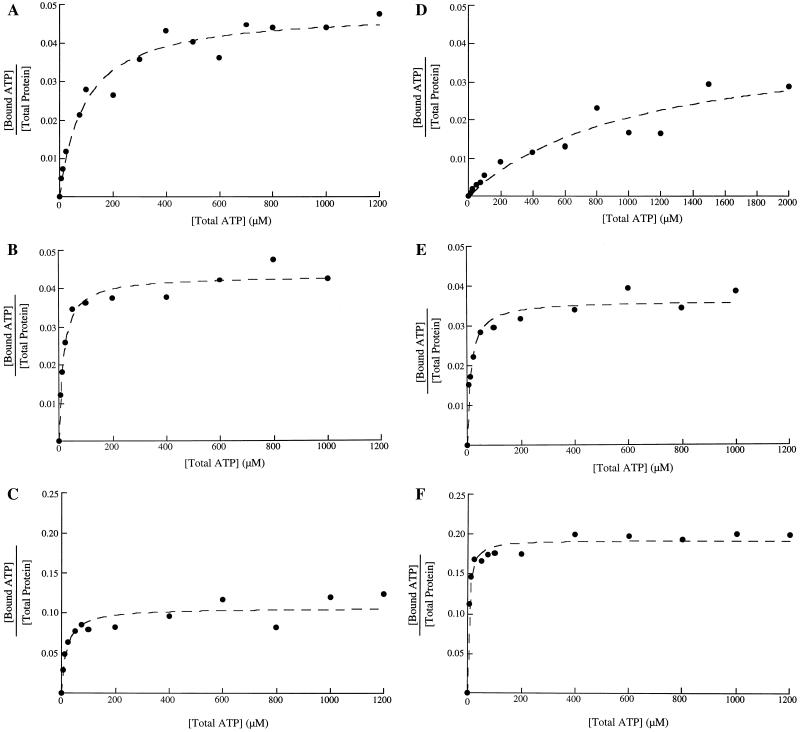
Binding of MgATP by wild-type and mutant forms of NtrC was assessed via a filter-binding assay as described in Materials and Methods. Nucleotide-binding and competition experiments with wild-type NtrC showed that both binding and exchange of ATP occur rapidly, reaching equilibrium in less than 1 min (not shown). Under the experimental conditions used, the maximal level of binding at saturation was typically only a few percent of the total concentration of NtrC monomers (Fig. 2). This observation and the loss of complexes as a function of increased washing of filters (data not shown) were commensurate with the view that NtrC-MgATP complexes are labile. Consequently, we were unable to determine stoichiometries, and we report simply concentrations of MgATP required for half-maximal binding (see Materials and Methods). The concentration of MgATP required for 50% maximal binding by wild-type NtrC was 135 ± 10 μM (Fig. 2A; Table 1), indicating that the affinity of NtrC for MgATP is lower than that of many other MgNTP-binding proteins.
FIG. 2.
Binding of MgATP and ATP to wild-type and D239 mutant forms of NtrC. The monomer concentration of NtrC was 10 μM. The amount of ATP bound to protein was measured by filter binding as described in Materials and Methods and plotted by using Kaleidograph. (A to C) Binding of MgATP to wild-type NtrC, NtrCD239C, and NtrCD239A, respectively; (D to F) binding of ATP to the same proteins. Since less than 1% of the ATP added was bound, [ATP]total = [ATP]free. All of the proteins with substitutions at D239 showed higher maximum binding of nucleotide in the presence or absence of divalent cation than did the wild-type protein (∼12% versus 4%). Given their higher apparent affinities for nucleotide, this result is commensurate with the view that low values for all proteins may be accounted for by the lability of protein-nucleotide complexes.
TABLE 1.
Concentrations of MgATP and ATP required for half-maximal binding by wild-type and mutant forms of NtrC
| Protein | ATPase activitya (%) | Concn for half-maximal binding (μM)b
|
|
|---|---|---|---|
| MgATP | ATP | ||
| Wild type | 100c | 135d ± 10 (6) | 730 ± 120 (3) |
| NtrCG173N | ≤2 | ≥10,000 (3) | ND |
| NtrCS207F | ≤2 | 50 ± 5 (4) | 230 ± 25 (3) |
| NtrCE208Q | 12 | 105 ± 10 (4) | 400 (2) |
| NtrCA216V | 100 | 100 ± 10 (3) | 350 (1) |
| NtrCG219K | 56 | 40 ± 10 (3) | 510 (2) |
| NtrCA220T | 80 | 100 ± 10 (4) | 300 (1) |
| NtrCD239N | ≤2 | 20 ± 5 (3) | 40 ± 15 (3) |
| NtrCD239C | ≤2 | 25 ± 5 (3) | 20 ± 5 (3) |
| NtrCD239A | ≤2 | 30 ± 10 (3) | 10 ± 5 (3) |
| NtrCR294C | ≤2 | 190 ± 35 (3) | 850 (1) |
| NtrCG355V | ≤2 | 800 ± 250 (3) | ND |
| NtrCR358C | ≤2 | ≥10,000 (3) | ND |
| NtrCR358H | ≤2 | ≥10,000 (2) | ND |
From references 67 and 42, with the exception of data for proteins carrying substitutions at position 239. Proteins were phosphorylated.
Binding by unphosphorylated proteins was assessed as described in Materials and Methods, and titration data were plotted to determine concentrations for half-maximal binding (see Materials and Methods). The number of independent titrations carried out for each NtrC protein (usually 10 μM monomer; range from 7 to 20 μM monomer in occasional experiments) is shown in parentheses, and standard deviations are indicated where appropriate. ND, not determined.
100% corresponds to ∼500 pmol/15 min/10 μl at a protein concentration of 1 μM.
Weighted average for two independently purified wild-type NtrC preparations. For the remaining proteins, single preparations were titrated.
Most mutant forms of NtrC that fail specifically in transcriptional activation (42, 67), so-called NtrCrepressor forms or forms with positive control defects, were known to have defects in ATP hydrolysis. To determine which of the ATPase-defective proteins were unable to bind the substrate, and thereby to discriminate between regions of the central domain of NtrC needed for MgATP binding and those involved in hydrolysis per se, we measured the ability of seven such mutant forms to bind MgATP. Proteins NtrCG173N, NtrCR358C, and NtrCR358H have no detectable ability to bind MgATP (concentration of MgATP required for 50% maximal binding of ∼10 mM or higher [Table 1]). NtrCG355V has noticeably reduced affinity (concentration required for 50% maximal binding of 800 ± 250 μM). By contrast, NtrCS207F, NtrCE208Q, and NtrCR294C appear to bind MgATP at least as well as wild-type NtrC. As expected, inactive mutant forms of NtrC that retain the ability to hydrolyze ATP efficiently—NtrCA216V, NtrCG219K, and NtrCA220T—are fully capable of binding MgATP. The apparently increased affinity of binding by NtrCG219K was not accompanied by an increased rate of ATP hydrolysis.
The putative Walker B motif is essential for transcriptional activation.
To investigate the role of the highly conserved aspartate residue within the putative Walker B motif of NtrC (43, 52), we changed this residue to asparagine (D239N), cysteine (D239C), or alanine (D239A). The mutant proteins were expressed either with or without the MBP fused to their N termini. (MBP-fusion forms of NtrC are more soluble than NtrC itself [41].) When plasmids encoding the mutant NtrC proteins were introduced into E. coli DH5α the resulting strains failed to grow on nutrient broth medium unless it was supplemented with glutamine (see Materials and Methods). This phenotype is commensurate with the view that the NtrCD239N, NtrCD239C, and NtrCD239A proteins are NtrCrepressor forms, that is, forms specifically defective in positive control (42, 67): they appear to be folded correctly, at least in their DNA-binding domains. To confirm that the glutamine requirement caused by these mutant proteins in vivo was due to their inability to activate transcription of the glnA gene, which encodes glutamine synthetase, each was purified and tested for the ability to activate transcription from the glnA promoter by ς54-holoenzyme in vitro. All of the mutant proteins and their MBP fusion counterparts failed to activate transcription (data not shown).
The Walker B motif is essential for ATP hydrolysis but not for ATP binding.
NtrC proteins carrying single amino acid substitutions for aspartate 239 had no detectable ATPase activity (Table 1). However, these proteins and their MBP fusion counterparts apparently still bound MgATP (Fig. 2B and C; Table 1). Moreover, their apparent affinities for MgATP were ∼5-fold higher than the affinity of wild-type NtrC. Because the aspartate residue of the Walker B motif has been shown to be involved in coordination of magnesium in the MgATP complexes of several purine nucleotide-binding proteins, the D239 mutant forms of NtrC were tested for the ability to bind ATP in the absence of magnesium. Omission of magnesium resulted in a fivefold decrease in the apparent affinity of wild-type NtrC for ATP (Fig. 2D; Table 1) and a small decrease for the NtrCD239N protein. However, the absence of magnesium resulted in an increase in the affinity of NtrCD239A for ATP (Fig. 2F), whereas there was no change in the affinity of NtrCD239C (Fig. 2E). Despite differences in the effect of omitting magnesium on ATP binding by the D239 mutant proteins, all had a higher apparent affinity for ATP in the absence of Mg2+ than did wild-type NtrC. By contrast to the case for the D239 mutant proteins, the response of the remaining NtrCrepressor proteins to omission of magnesium was similar to that of wild-type NtrC. Thus, the proteins carrying substitutions at position 239 were unique in their ability to bind nucleotide strongly in the absence of Mg2+. To test whether the increased apparent affinity of the three D239-substituted NtrC proteins for nucleotide was due to a reduced ability to undergo nucleotide exchange, these proteins were allowed to bind MgATP (200 μM ATP) under standard conditions and were then challenged with unlabeled ATP as competitor (equimolar to 50-fold excess). Binding was reduced by the expected factor for both the D239 mutant forms and wild-type NtrC, indicating that the former also bind nucleotides reversibly (data not shown).
In cases where coordination of a divalent cation by aspartate is direct, replacement of the aspartate with cysteine can increase the specificity for Mn2+ over Mg2+ due to preferential coordination of Mn2+ by sulfur (reference 49 and references therein). Since Mn2+ can substitute functionally for Mg2+ in transcriptional activation by NtrC (41), we tested its ability to serve as divalent cation for the ATPase activity of the protein and its effect on ATP binding. Although the rate of ATP hydrolysis by the wild-type protein (MBP-NtrC) was the same with Mn2+ as Mg2+, Mn2+ failed to serve as divalent cation for ATP hydrolysis by mutant proteins with substitutions at position 239 (Table 2). The affinity of wild-type MBP-NtrC for ATP was similar in the presence of either divalent cation (and was similar to that of NtrC itself for MgATP), as was the case for MBP-NtrCD239A. However, the affinity of MBP-NtrCD239C for nucleotide was severalfold lower in the presence of Mn2+ than Mg2+. Thus, Mn2+ did not stimulate nucleotide binding or serve as the cation for nucleotide hydrolysis by MBP-NtrCD239C, and hence these experiments provided no evidence that D239 coordinates the divalent cation directly.
TABLE 2.
ATPase activities of D239 mutant forms of NtrC and concentrations of MnATP or MgATP for half-maximal binding
| Protein | Pi releasea (pmol/15 min/10 μl)
|
Concn for half-maximal binding (μM)b
|
||
|---|---|---|---|---|
| Mg2+ | Mn2+ | MnATP | MgATP | |
| MBP-NtrC | 500 | 450 | 110 ± 15 (3) | 180 ± 20 (3) |
| MBP-NtrCD239N | ≤20 | ≤20 | ND | ND |
| MBP-NtrCD239C | ≤20 | ≤20 | 95 (2) | 25 (2) |
| MBP-NtrCD239A | ≤20 | ≤20 | 50 (1) | 35 (1) |
ATPase activity of phosphorylated MBP-NtrC fusion proteins was measured in the presence of MgATP or MnATP as described in Materials and Methods. Values were corrected for the background of Pi, which was 18 pmol/15 min/10 μl.
See footnote b to Table 1.
Binding of divalent cation.
To determine whether residue D239 of NtrC plays a role in binding divalent cation, various forms of the NtrC protein were analyzed for Mn2+ binding by EPR spectroscopy. Although Mg2+, which is EPR silent, is the divalent cation that is normally used to assess the ATPase activity and transcriptional activation capacity of NtrC, the paramagnetic cation Mn2+ can substitute functionally in both cases (Table 2 and reference 41).
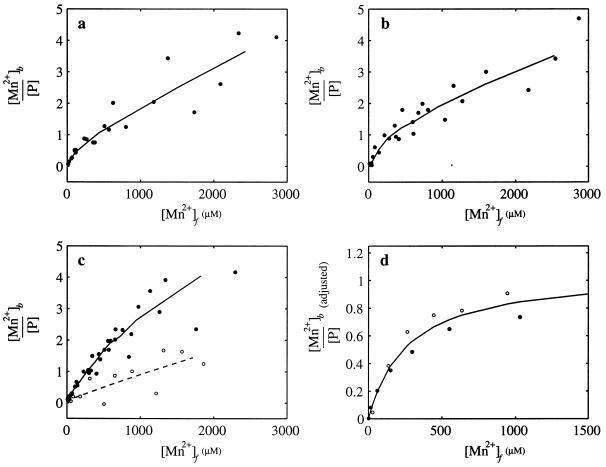
It has been shown previously that the amplitude of the peaks of the Mn2+ EPR spectrum (in the first derivative) are proportional to the concentration of Mn2+ and that binding of Mn2+ to a protein effectively quenches the EPR signal, allowing the level of Mn2+ binding to the protein to be measured (reference 71 and references therein). Standard curves correlating the amplitude of the EPR signal with the concentration of Mn2+ over the appropriate range were used to determine the free Mn2+ concentration in the presence of NtrC proteins. N-terminal MBP fusion forms of NtrC were used because they remained soluble at the high protein concentrations required for EPR analysis. Mn2+ EPR spectra obtained in the presence of MBP-NtrC and MBP-NtrCD239A were used to construct Mn2+-binding curves for these proteins (as described in Materials and Methods) (Fig. 3a and b). The two binding curves were very similar, suggesting that residue D239 is unlikely to be involved in strong Mn2+ binding.
FIG. 3.
Binding of Mn2+ to NtrC proteins and their isolated N-terminal domains (residues 1 to 124). Isotherms for the binding of Mn2+ to MBP-NtrC (178 μM monomer; a), MBP-NtrCD239A (188 μM monomer; b), the N-terminal domain of NtrC (170 μM monomer; c, filled circles), and the N-terminal domain of NtrCD54E (112 μM monomer; c, open circles). The amount of bound Mn2+ was calculated from the difference between the total amount of Mn2+ and the EPR measurements of Mn2+ that remained free in the presence of protein. The lines in panels a to c present three-point smoothing averages of the data. (d) Adjusted isotherms for MBP-NtrC (filled circles) and MBP-NtrCD239A (open circles), corrected by subtracting nonspecific quenching from the smoothed data in panels a and b, respectively. Nonspecific quenching was estimated from the slopes of the isotherms at concentrations of free Mn2+ above 1 mM (see Results). The adjusted isotherms for MBP-NtrC, and MBP-NtrCD239A were very similar, and the solid line represents a least squares fit of the combined data, which yielded a Kd of 260 μM and a stoichiometry of 1.1. Binding reactions and EPR analyses were carried out as described in Materials and Methods. [Mn2+]b is [Mn2+]bound, [P] is [NtrC]total, and [Mn2+]f is [Mn2+]free.
To gauge the affinity of the central domain of NtrC for Mn2+, we measured Mn2+ binding by the isolated N-terminal regulatory domain of the protein, which is known to contain at least one specific Mg2+-binding site (39), and compared it to that of intact NtrC. Surprisingly, we found that the binding curve for the N-terminal domain (Fig. 3c) was very similar to that for full-length NtrC (Fig. 3a), indicating that binding by the regulatory domain accounts for that by the intact protein and hence that the central domain of NtrC does not contain a high-affinity binding site for Mn2+. It is important to note that these experiments were carried out in the absence of ATP because addition of ATP caused a high degree of quenching of the EPR signal under the experimental conditions used. The Mn2+-binding curves for both full-length NtrC and the N-terminal domain were largely hyperbolic in nature. However, some nonspecific Mn2+ binding, which appeared to increase linearly with respect to Mn2+ concentration, was also observed. We showed that this nonspecific binding of Mn2+ to NtrC could be reduced by the addition of 10 mM CaCl2, indicating that it was probably due to nonspecific electrostatic attraction.
To confirm that the hyperbolic component of the binding curve for the N-terminal domain of NtrC was predominantly due to Mn2+ binding at a specific site, we measured binding to N terminusD54E. Previous nuclear magnetic resonance analysis indicated that this mutant form of the N-terminal domain can no longer bind Mg2+ with high affinity (39). The Mn2+-binding curve for N terminusD54E (Fig. 3c) showed loss of the hyperbolic component observed for the wild-type N terminus, with the residual nonspecific binding increasing linearly with respect to Mn2+ concentration. A similar result was observed for full-length NtrC carrying the D54E substitution (not shown).
Nonspecific quenching of the Mn2+ EPR signal by NtrC proteins appeared to be linear, and the average slope was 10−3 for the data points corresponding to concentrations of free Mn2+ between 1 and 3 mM (Fig. 3a and b). Using this average slope, we corrected the binding curves for MBP-NtrC and MBP-NtrCD239A by subtracting the nonspecific quenching contribution (1.0 × 10−3 · [Mn2+]free) to produce the adjusted binding data shown in Fig. 3d. The corrected data should yield the isotherms for strongly bound Mn2+ (Fig. 3D). Fitting the data to a model of n equivalent noninteracting sites, [Mn2+]bound/[NtrC]total = n[Mn2+]free/(Kd + [Mn2+]free), gave a good fit with Kd = 260 μM and n = 1.1. Given the experimental uncertainties, the value for n cannot be considered a precise determination of the stoichiometry of metal binding. However, the data are most consistent with one strong Mn2+-binding site per NtrC monomer, located in the N-terminal domain. Furthermore, it is clear from the adjusted isotherms (Fig. 3d) that MBP-NtrC and MBP-NtrCD239A bound Mn2+ with the same stoichiometry. This conclusion is not affected by the correction for nonspecific quenching since it was the same for the two proteins.
Influence of NtrC oligomerization state on MgATP binding.
To determine whether the dimeric configuration of NtrC is essential for MgATP binding, we tested binding by a largely monomeric mutant form of NtrC, NtrCΔ444–469 (MBP fusion form). NtrCΔ444–469 lacks the helix-turn-helix motif at the end of the C-terminal domain. This motif is directly responsible for specific DNA binding and makes a large contribution to dimerization (27, 40). MBP-NtrCΔ444–469 retains only a weak albeit measurable ability to hydrolyze ATP and activate transcription (40); it appears to retain the ability to bind MgATP, the concentration required for half-maximal binding being 600 ± 80 μM (Table 3). The qualitative nature of the binding curve for MBP-NtrCΔ444–469 (not shown) was similar to that of MBP-NtrC or NtrC (Fig. 2A), suggesting first that the binding observed was, in fact, due to monomer and not residual dimer in the preparation (20% [not shown]) and second that dimerization does not influence MgATP binding in a cooperative manner.
TABLE 3.
Concentrations of MgATP required for half-maximal binding by monomeric, dimeric, and oligomeric forms of NtrC
| Protein | Substrate | Oligomeric state | Concn for half-maximal binding of MgNTP (μM)a |
|---|---|---|---|
| MBP-NtrC | MgATP | Dimer | 180 ± 20 (5) |
| MBP-NtrCΔ444–469 | MgATP | Monomerb | 600 ± 80 (3) |
| MBP-NtrC | MgATPγS | Dimer | 170 (1) |
| P-MBP-NtrC | MgATPγS | >2 dimers | 155 ± 10 (4) |
Binding was assessed, and titration data were plotted to determine concentrations for half-maximal binding (see Materials and Methods). The number of independent titrations carried out for each NtrC protein is shown in parentheses, and standard deviations are indicated where appropriate. Proteins were unphosphorylated unless otherwise indicated (P-MBP-NtrC).
Gel filtration chromatography indicated that there was ≤20% residual dimer in the preparation (not shown).
We have previously shown that the active form of the NtrC protein is a phosphorylated oligomer larger than a tetramer (72). To test the effect of phosphorylation on nucleotide binding, MBP-NtrC (which remains soluble at high protein concentrations under phosphorylating conditions) was phosphorylated and assayed for MgNTP-binding ability. The nonhydrolyzable analog ATPγS (and [35S]ATPγS) was used in place of ATP (and [α-32P]ATP) so that binding could be measured in the absence of hydrolysis by the active, phosphorylated NtrC oligomer. Under nonphosphorylating conditions, the concentration of MgATPγS required for half-maximal binding by MBP-NtrC was 170 μM (Table 3), not significantly different from the concentration of MgATP required for half-maximal binding by MPP-NtrC or NtrC itself. Upon phosphorylation, the concentration of MgATPγS required for half-maximal binding by MBP-NtrC did not change appreciably (155 μM ± 10 μM), providing evidence that phosphorylation and the accompanying higher-order oligomerization of NtrC do not change its affinity for MgNTP. Although we do not know the degree of phosphorylation of NtrC, we know that at a concentration of 10 μM, there is a high percentage of large oligomer (73).
Determination of the binding constant for MgATPγS by ITC.
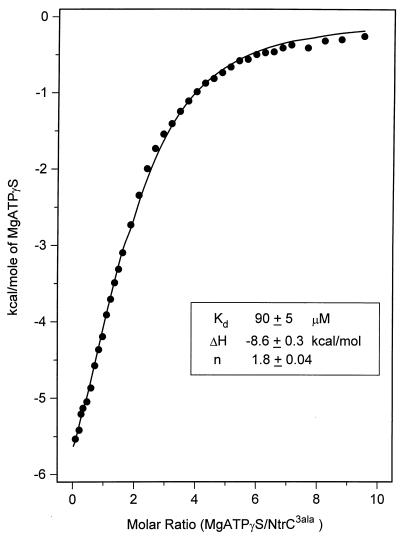
To assess binding of nucleotide by NtrC by a technique other than filter binding, we performed binding studies by ITC. Two independent titrations of MgATPγS at 80 and 95 μM NtrC3Ala (our most soluble form; see Materials and Methods) yielded Kds of 90 ± 5 μM (error of curve fitting [Fig. 4]) and 120 ± 9 μM, respectively, giving an average of 105 μM. Enthalpy values were negative (−9.6 ± 0.6 and −8.6 ± 0.3 kcal/mol, respectively, for the two experiments), indicating that the binding of nucleotide to NtrC was an exothermic process. Binding stoichiometries were 1.8 and 2 mol/mol dimer, respectively, providing evidence of one active site per monomer. The average Kd for the dissociation of MgATPγS from the NtrC3Ala protein complex, 105 μM at 25°C, was of the same order of magnitude as that for dissociation of MgATPγS from the MBP–wild-type NtrC complex, 190 μM at 37°C, as measured by filter binding.
FIG. 4.
Titration calorimetry binding isotherm observed upon injecting MgATPγS into NtrC3Ala protein. The titration was carried out at a dimer concentration of NtrC3Ala of 80 μM as described in Materials and Methods. ●, experimental points; ——, fit according to the equation of Wiseman et al. (68); Kd, dissociation constant; ΔH, enthalpy change; n, stoichiometry factor. Deviations represent the fitting error of this single experiment.
We also performed three independent titrations of MgATP by ITC at concentrations of NtrC3Ala on the order of 100 μM. Although they yielded an average Kd of ∼40 ± 3 μM (data not shown; error reflects standard deviation for the three experiments), the otherwise soluble NtrC3Ala protein showed a strong tendency to precipitate in the presence of Mg2+, and there was significant hydrolysis of MgATP to MgADP (see Materials and Methods). Because the Kds obtained by ITC are a function of heat absorbed or evolved, the observed values may reflect a combination of several different heat change events—binding of MgATP, binding of the product inhibitor MgADP (63), and precipitation of the protein, presumably as a protein-ligand complex(es). Hence, it is likely to be a minimum estimate.
A single titration of ATP in the absence of magnesium yielded a Kd of 570 ± 50 μM (error of curve fitting), value of the same order of magnitude as that obtained by filter binding. The enthalpy change was negative (−8.8 ± 2.1 kcal/mol), and the binding stoichiometry was 1.7 ± 0.4.
The high concentrations of NtrC3Ala needed to detect a calorimetic signal upon binding of the various nucleotides confirm that the Kds reported are of the correct order of magnitude. Had these values been significantly lower, we would have been able to detect a signal at lower concentrations of NtrC.
DISCUSSION
Regions of the central domain of NtrC needed for ATP binding and hydrolysis.
To effect its role as a transcriptional activator, the phosphorylated form of NtrC binds and hydrolyzes MgATP or MgGTP and couples the resultant energy of hydrolysis to the process of open complex formation by ς54-holoenzyme (66). To better understand nucleotide hydrolysis and energy coupling, we have explored the requirements for nucleotide binding. Based on studies by ITC, unphosphorylated dimeric NtrC has a modest affinity for the nonhydrolyzable MgATP analogue MgATPγS, having an apparent Kd of ∼100 μM at 25°C (Fig. 4). The stoichiometry of binding of 1.9 mol of MgATPγS per mol of dimer (Fig. 4) (13) is commensurate with the finding that a largely monomeric mutant form of NtrC retained the ability to bind MgATP in a filter-binding assay (Table 3). Nucleotide binding by the phosphorylated oligomeric form of NtrC, which functions as a transcriptional activator, was not detectably different from that by the unphosphorylated dimeric form (assessed with the nonhydrolyzable analogue MgATPγS in the filter-binding assay). In light of the fact that both ATP hydrolysis and transcriptional activation by NtrC are predicated on the formation of large oligomers, whereas binding of the MgATP substrate is not, one of the functions of higher-order oligomerization appears to be to increase the rate of ATP hydrolysis per se, this in turn being coupled to the formation of open complexes by ς54-holoenzyme.
By virtue of sequence and predicted structural similarities, NtrC appears to belong to a large and diverse group of purine nucleotide-binding proteins that includes the bacterial polypeptide elongation factor EF-Tu, the eukaryotic regulatory protein p21ras, and the F1-ATPase (43, 56). X-ray crystallographic structures of a number of these proteins show that residues within the two highly conserved Walker A and B motifs are directly involved in coordinating the phosphates and the Mg2+ of the MgNTP substrate (1, 5, 14, 16, 28, 34, 44, 53, 54, 61, 62). However, the remaining residues that serve as ligands for nucleotide binding are not sufficiently conserved to be predicted from the primary sequence, nor are additional residues involved in nucleotide hydrolysis. Hence we tested mutant forms of NtrC deficient in MgATP hydrolysis to identify those that were also deficient in nucleotide binding. In this way we were able to identify one region different from the Walker A and B motifs that appears to be essential for nucleotide binding and two regions that are involved in ATP hydrolysis (Fig. 1; Table 4). Like the Walker A and B motifs, all three additional regions are highly conserved among activators of ς54-holoenzyme (36, 42, 43, 67). We also obtained evidence that the conserved aspartate of the putative Walker B motif of NtrC plays a role in coordinating the divalent cation required for nucleotide hydrolysis.
TABLE 4.
Motifs in the central domain of NtrCa
| Binding | Hydrolysis | Hydrolysis | Hydrolysis | Binding |
|---|---|---|---|---|
| G173 | S207/E208 | D239 | R294 | G355/R358 |
| Walker A, binds β phosphate | Switch I effector | Walker B/switch II, coordinates Mg2+ | Catalytic residue? | Binds nucleotide base |
| GESGTG173KE | ES207ELFGHEKGAFTGA | GGTLFLD239EIG | FR294EDLfhRLNV | WPG355NVR358qLEN |
For the central domains of NtrC proteins of S. typhimurium, E. coli, K. pneumoniae, Proteus vulgaris, Rhizobium meliloti, Bradyrhizobium parasponiae, and Agrobacterium tumefaciens (15, 43). The motifs listed are also referred to as conserved regions 1, 3, 4, 6, and 7, respectively, among activators of ς54-holoenzyme (36, 43). Conserved region 6 begins with residue 275 of NtrC and extends through residue 312. For cases in which fewer than four of the five NtrC sequences are identical, the residue is that for the protein from S. typhimurium and is in lowercase. The numbers indicate positions at which amino acid substitutions in the NtrC protein of S. typhimurium were studied in this work.
The two regions found to be required for ATP binding by NtrC were the Walker A motif [(G168ESGTGKE175 [52, 63]) and the region around positions 355 to 358 (W353PGNVRQLEN362), which may interact with the nucleotide base (Table 4). (These regions are referred to as conserved regions 1 and 7 among activators of ς54-holoenzyme [36, 43].) An amino acid substitution in either of these regions can decrease MgATP binding by NtrC by 50- to 100-fold, to our limit of reliable detection (Table 1). Secondary and tertiary structure predictions for the central domain of activators of ς54-holoenzyme led to the postulate that the 355–358 region might be involved in binding to the nucleotide base (43). Given that NtrC—and its homologue NifA—can utilize GTP in place of ATP (6, 30, 67), these proteins are not expected to show great discrimination between purine bases. Interestingly, there are precedents for arginine residues interacting with the adenine ring (28, 53), and replacement of R358 of NtrC with either H or C led to loss of detectable binding of MgATP. It is unlikely that the 355–358 region interacts primarily with the sugar rather than the base because both dATP and ddATP serve well as the nucleotide substrate for NtrC. Hence, as in the case of many other purine nucleotide-binding proteins, the ribose is likely to be largely solvent exposed and not a strong determinant of nucleotide binding (5, 28, 34, 44, 55).
The three positions in the central domain of NtrC that appear to affect ATP hydrolysis without decreasing ATP binding are R294 (in conserved region 6 of activators of ς54-holoenzyme [36, 43]), which may be a catalytic residue, the 207/208 region (in conserved region 3), which appears to correspond to a portion of the switch I effector region, and D239, the conserved aspartate of the putative Walker B motif ([GGT]LFLD239EIG [conserved region 4] [43, 52]) (Table 4). The R294C protein appears to oligomerize normally (73), and hence R294 appears to be required directly for nucleotide hydrolysis. We postulate that R294 may be a catalytic residue because a number of other purine nucleotide-binding proteins have catalytic arginines (8, 11, 35, 58). Replacement of the catalytic arginine in Giα1 with cysteine, the replacement that we studied, greatly impaired GTPase activity without affecting GTP binding (11). Moreover, this replacement resulted in a substantial decrease in the affinity for GDP · A1F4−, an analogue of the transition state for GTP hydrolysis.
The consequences of amino acid substitutions in the region of NtrC between residues 207/208 and 220 (42, 67), which lies between the Walker A and B motifs, in concert with the location of this region in the predicted secondary and tertiary structures for activators of ς54-holoenzyme, led to the postulate that it was analogous to the switch I region of other purine nucleotide-binding proteins (43). Switch I, which undergoes a large conformational change upon nucleotide hydrolysis, can play a critical role in biological output (8, 21, 25, 53, 55, 56, 59, 61, 69). In agreement with the view that positions 207 and 208 lie within the switch I region of NtrC, we have noted that the decrease in ATPase activity caused by the sterically conservative E208Q substitution is not sufficient to account for the profound decrease in transcriptional activation by NtrCE208Q (42). This protein must also be defective in contacting ς54-holoenzyme or in coupling the energy available from ATP hydrolysis to open complex formation. Unfortunately, these are functions of NtrC for which we do not have direct biochemical assays. Similar to E208Q, several amino acid substitutions at positions 216 to 220, just downstream, cause profound defects in transcriptional activation. However, in these cases there is little, if any, decrease in ATP binding or hydrolysis (Table 1) (42, 67). Hence the substitutions at positions 216 to 220 apparently affect only contact between NtrC and polymerase or energy coupling. Strikingly, one substitution for the serine residue that corresponds to S207 of NtrC in the homologous activator DctD impaired the ability of the mutant protein (DctDS212I) to cross-link to ς54-holoenzyme (64).
Site-directed replacement of D239 of NtrC with the uncharged residues N, C, or A resulted in loss of ATPase activity but an apparent increase in the affinity for ATP in the presence or absence of Mg2+ (Table 1). Improved ATP binding may be due to a decrease in negative charge in the vicinity of the nucleotide and hence a decrease in electrostatic repulsion. As appears to be true for NtrC, replacement of the conserved aspartate residue in the Walker B motif of the bacterial rho factor or eIF-4a with an uncharged residue gave rise to an increase in affinity for ATP (12, 46). The loss of ATPase activity by NtrC proteins carrying substitutions at position 239 may result from failure of the proteins to coordinate the divalent cation necessary for ATP hydrolysis or to coordinate it properly (discussed below).
Role of D239 in coordinating divalent cation.
There are several lines of evidence that D239 of NtrC does, in fact, play a role in coordinating the divalent cation needed for ATP hydrolysis. This residue influences both nucleotide binding, as noted above, and the effect of Mg2+ on nucleotide binding. In the absence of Mg2+, the affinity of the wild-type protein for nucleotide decreased ∼5-fold (apparent Kd of ∼500 μM by ITC). The decrease in affinity was also manifested in the filter-binding assay, and there appeared to be a similar decrease for all of the mutant proteins used in this study except those carrying amino acid substitutions at position 239 (Table 1). Omission of Mg2+ had a different effect on nucleotide binding by each mutant protein carrying a substitution at position 239. It decreased the apparent affinity of NtrCD239N slightly, commensurate with the view that this protein retains residual ability to coordinate Mg2+, had no effect on NtrCD239C, which may be explained by the fact that this protein has lost the ability to coordinate Mg2+, and increased the apparent affinity of NtrCD239A, for which there is no obvious explanation. The unusual response of proteins with substitutions at position 239 to omission of Mg2+ provides evidence that D239 interacts with the divalent cation-nucleotide complex (though not necessarily directly; see below). By analogy to the role of the conserved aspartate in the Walker B motif of other purine nucleotide-binding proteins, D239 is likely to play a role in orienting the divalent cation for nucleotide hydrolysis. Changes in F237 and L238, two of the hydrophobic residues that precede D239, presumably in a β strand, result in precipitation of the mutant proteins (45).
Assessment of Mn2+ binding by EPR spectroscopy indicated that the central domain of NtrC does not bind divalent cation strongly in the absence of nucleotide. Although NtrC does have a single strong binding site for Mn2+, it appears to lie in the N-terminal regulatory domain of the protein and to depend on the presence of an aspartate residue at position 54, the site of phosphorylation. Results of EPR spectroscopy were in agreement with previous nuclear magnetic resonance spectroscopic analyses of Mg2+ binding by the N-terminal domain of NtrC (39). The divalent cation bound strongly by this domain is essential for its (self-catalyzed) phosphorylation, and a divalent cation appears to be essential for phosphorylation of homologous regulatory domains (so-called receiver domains) (60).
Comparisons to other activators of ς54-holoenzyme.
Several laboratories have isolated mutant forms of activators of ς54-holoenzyme with lesions in the putative Walker A motif (conserved region 1 [36, 43]); specifically, this was done for the NifA (nitrogen fixation A) and DctD (dicarboxylate transport D) proteins, for the XylR (xylene R) protein, and for the NtrC protein of Klebsiella pneumoniae (3, 10, 17, 48). In all instances the mutant proteins had a defect in transcriptional activation in vivo. A number of the DctD proteins, a XylR protein, and an NtrC protein from K. pneumoniae have also been studied in vitro, and most of these fail in both transcriptional activation and ATP hydrolysis (3, 17, 48). With the exception of the XylR protein, which failed to bind ATP, their ATP-binding properties have not been studied.
Many mutant forms of both DctD and NifA that carry amino acid substitutions in the conserved region corresponding to the 206–220 region of NtrC (conserved region 3 [18, 64]) have been isolated. Of these, many failed to activate transcription in vivo and in vitro (tested only for DctD). As was true of NtrC, decreases in ATP hydrolysis by the mutant forms of DctD were generally not sufficient to account for their defects in transcriptional activation. DctDS212I, which was mentioned above, failed to activate transcription but had little, if any, defect in ATP hydrolysis (64).
Finally, a substitution at position 453 of XylR (R453H), which corresponds precisely to the R358H substitution that we studied in NtrC (conserved region 7), allowed multimerization of the protein in the absence of ATP, in contrast to the case for the wild-type protein (47). It was claimed on the basis of an assay at a single ATP concentration (2.5 mM) that the mutant protein bound ATP as well as its parent (48). Given our discordant finding for the NtrCR358H protein, it is difficult to understand the nucleotide-independent multimerization of the mutant XylR protein, particularly since the G268N substitution in XylR, which corresponds precisely to the G173N substitution in NtrC, prevented both ATP-binding and ATP-dependent multimerization of XylR.
Comparisons to other purine nucleotide-binding proteins.
Perhaps the most interesting comparison to be made between NtrC and other classes of purine nucleotide-binding proteins concerns the role of the conserved aspartate in the Walker B motif. Although the precise functional consequences of altering this residue cannot be predicted (20, 24, 37), loss of ability to catalyze nucleotide hydrolysis and hence loss of biological function, as occurred for NtrC, is a probable outcome (2, 7, 9, 12, 19, 46, 57, 65). Structural studies indicate that the conserved aspartate of the Walker B motif usually coordinates the divalent cation indirectly through an intervening water molecule (5, 16, 34, 38, 44, 59, 61, 69). The cation has a total of six coordinating ligands, two of which are oxygens of the β and γ phosphates of the nucleotide. Given its relatively modest affinity for nucleotide and the small stimulatory effect of divalent cation on nucleotide binding, NtrC may depend on the two ligands of the nucleotide for binding the catalytic metal ion. In the absence of these two liganding groups, the cation apparently cannot bind to the central domain of NtrC with an affinity that can be detected by EPR (Kd ≥ 500 μM).
ACKNOWLEDGMENTS
I.R. and P.P.-W. contributed equally to this work.
We thank Jieli Li for construction of plasmids encoding substitutions for D239 of NtrC and help in purification of proteins.
This work was funded by NIH grants GM38361 and GM51290 to S.K. and Y.-K.S., respectively, and a Searle scholarship to Y.-K.S.
REFERENCES
- 1.Abrahams J P, Leslie A G, Lutter R, Walker J E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 2.Al-Shawi M K, Pasonage D, Senior A E. Directed mutagenesis of the strongly conserved aspartate 242 in the β-subunit of Escherichia coli proton-ATPase. J Biol Chem. 1988;263:19633–19639. [PubMed] [Google Scholar]
- 3.Austin S, Kundrot C, Dixon R. Influence of a mutation in the putative nucleotide binding site of the nitrogen regulatory protein NtrC on its positive control function. Nucleic Acids Res. 1991;19:2281–2287. doi: 10.1093/nar/19.9.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 5.Berchtold H, Reshetnikova L, Reiser C O, Schirmer N K, Sprinzl M, Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- 6.Berger D K, Narberhaus F, Kustu S. The isolated catalytic domain of NifA, a bacterial enhancer-binding protein, activates transcription in vitro: activation is inhibited by NifL. Proc Natl Acad Sci USA. 1994;91:103–107. doi: 10.1073/pnas.91.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black M E, Hruby D. Site-directed mutagenesis of a conserved domain in vaccinia virus thymidine kinase. J Biol Chem. 1992;267:6801–6806. [PubMed] [Google Scholar]
- 8.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 9.Brosh R M, Matson S W. Mutations in motif II of Escherichia coli DNA helicase II render the enzyme nonfunctional in both mismatch repair and excision repair with differential effects on the unwinding reaction. J Bacteriol. 1995;177:5612–5621. doi: 10.1128/jb.177.19.5612-5621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon W, Buck M. Central domain of the positive control protein NifA and its role in transcriptional activation. J Mol Biol. 1992;225:271–286. doi: 10.1016/0022-2836(92)90921-6. [DOI] [PubMed] [Google Scholar]
- 11.Coleman D E, Berghuis A M, Lee E, Linder M E, Gilman A G, Sprang S R. Structures of active conformation of Giα1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 12.Dombroski A J, Brennan C A, Spear P, Platt T. Site-directed alterations in the ATP-binding domain of Rho protein affect its activities as a termination factor. J Biol Chem. 1988;263:18802–18809. [PubMed] [Google Scholar]
- 13.Farez-Vidal M E, Wilson T J, Davidson B E, Howlett G J, Austin S, Dixon R A. Effector-induced self-association and conformational changes in the enhancer-binding protein NtrC. Mol Microbiol. 1996;5:779–788. doi: 10.1046/j.1365-2958.1996.01530.x. [DOI] [PubMed] [Google Scholar]
- 14.Fisher A J, Smith C A, Thoden J B, Smith R, Sutoh K, Holden H M, Rayment I. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.A1F4−. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 15.Flashner Y, Weiss D S, Keener J, Kustu S. Constitutive forms of the enhancer-binding protein NtrC: evidence that essential oligomerization determinants lie in the central activation domain. J Mol Biol. 1995;249:700–713. doi: 10.1006/jmbi.1995.0330. [DOI] [PubMed] [Google Scholar]
- 16.Fry D C, Kuby S A, Mildvan A S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci USA. 1986;83:907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, Wang Y K, Hoover T R. Mutational analysis of the phosphate-binding loop of Rhizobium meliloti DctD, a ς54-dependent activator. J Bacteriol. 1998;180:2792–2795. doi: 10.1128/jb.180.10.2792-2795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González V, Olvera L, Soberón X, Morett E. In vivo studies on the positive control function of NifA: a conserved hydrophobic amino acid patch at the central domain involved in transcriptional activation. Mol Microbiol. 1998;28:55–67. doi: 10.1046/j.1365-2958.1998.00772.x. [DOI] [PubMed] [Google Scholar]
- 19.Gross C H, Shuman S. Mutational analysis of vaccinia virus nucleoside triphosphate phosphohydrolase II, a DExH box RNA helicase. J Virol. 1995;69:4727–4736. doi: 10.1128/jvi.69.8.4727-4736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmack K, Anborgh P, Marcello M, Clark B, Parmeggiani A. Substitution of aspartic acid-80, a residue involved in coordination of magnesium, weakens the GTP binding and strongly enhances the GTPase of the G domain of elongation factor Tu. Biochemistry. 1992;31:7367–7372. doi: 10.1021/bi00147a022. [DOI] [PubMed] [Google Scholar]
- 21.Hilgenfeld R. Regulatory GTPases. Curr Opin Struct Biol. 1995;5:810–817. doi: 10.1016/0959-440x(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 22.Hopper S, Bock A. Effector-mediated stimulation of ATPase activity by the ς54-dependent transcriptional activator FHLA from Escherichia coli. J Bacteriol. 1995;177:2798–2803. doi: 10.1128/jb.177.10.2798-2803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huala E, Ausubel F M. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J Bacteriol. 1989;171:3354–3365. doi: 10.1128/jb.171.6.3354-3365.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John J, Rensland H, Schlichting I, Vetter I, Borasio G D, Goody R, Wittinghofer A. Kinetic and structural analysis of the Mg2+-binding site of the guanine nucleotide-binding protein p21H-ras. J Biol Chem. 1993;268:923–929. [PubMed] [Google Scholar]
- 25.Kim S-H, Privé G G, Milburn M V. Conformational switch and structural basis for oncogenic mutations of Ras proteins. In: Dickey B F, Birnbaumer L, editors. Handbook of experimental pharmacology. 108I. Berlin, Germany: Springer-Verlag; 1993. pp. 177–194. [Google Scholar]
- 26.Klose K E. Ph.D. thesis. Berkeley, California: University of California; 1993. [Google Scholar]
- 27.Klose K E, North A K, Stedman K M, Kustu S. The major dimerization determinants of the nitrogen regulatory protein NtrC from enteric bacteria lie in its carboxy-terminal domain. J Mol Biol. 1994;241:233–245. doi: 10.1006/jmbi.1994.1492. [DOI] [PubMed] [Google Scholar]
- 28.Kull F J, Sablin E P, Lau R, Fletterick R J, Vale R D. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 30.Lee H S, Berger D K, Kustu S. Activity of purified NifA, a transcriptional activator of nitrogen fixation genes. Proc Natl Acad Sci USA. 1993;90:2266–2270. doi: 10.1073/pnas.90.6.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J H, Scholl D, Nixon B T, Hoover T R. Constitutive ATP hydrolysis and transcriptional activation by a stable, truncated form of Rhizobium meliloti DCTD, a ς54-dependent transcriptional activator. J Biol Chem. 1994;269:20401–20409. [PubMed] [Google Scholar]
- 32.Li, J., and S. Kustu. Unpublished data.
- 33.Lukat G, McCleary W, Stock A, Stock J. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci USA. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milburn M V, Tong L, DeVos A M, Brünger A, Yamaizumi Z, Nishimura S, Kim S-H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic Ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 35.Mittal R, Ahmadian M R, Goody R S, Wittinghofer A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science. 1996;273:115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 36.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myles G M, Hearst J E, Sancar A. Site-specific mutagenesis of conserved residues within Walker A and B sequences of Escherichia coli UvrA protein. Biochemistry. 1991;30:3824–3834. doi: 10.1021/bi00230a004. [DOI] [PubMed] [Google Scholar]
- 38.Noel J B, Hamm H E, Sigler P B. The 2.2Å crystal structure of transducin-α complexed with GTPγS. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 39.Nohaile M, Kern D, Wemmer D, Stedman K, Kustu S. Structural and functional analyses of activating amino acid substitutions in the receiver domain of NtrC: evidence for an activating surface. J Mol Biol. 1997;273:299–316. doi: 10.1006/jmbi.1997.1296. [DOI] [PubMed] [Google Scholar]
- 40.North A K, Kustu S. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J Mol Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- 41.North, A. K., and S. Kustu. Unpublished data.
- 42.North A K, Weiss D S, Suzuki H, Flashner Y, Kustu S. Repressor forms of the enhancer-binding protein NtrC: some fail in coupling ATP hydrolysis to open complex formation by ς54-holoenzyme. J Mol Biol. 1996;260:317–331. doi: 10.1006/jmbi.1996.0403. [DOI] [PubMed] [Google Scholar]
- 43.Osuna J, Soberón X, Morett E. A proposed architecture for the central domain of the bacterial enhancer-binding proteins based on secondary structure prediction and fold recognition. Protein Sci. 1997;6:543–555. doi: 10.1002/pro.5560060304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pai E F, Krengel U, Petsko G A, Goody R S, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of h-Ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Passaglia, L., and S. Kustu. Unpublished data.
- 46.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Martin J, de Lorenzo V. ATP binding to the ς54-dependent activator XylR triggers a protein multimerization cycle catalyzed by UAS DNA. Cell. 1996;86:331–339. doi: 10.1016/s0092-8674(00)80104-x. [DOI] [PubMed] [Google Scholar]
- 48.Perez-Martin J, de Lorenzo V. In vitro activities of an N-terminally truncated form of XylR, a ς54-dependent transcriptional activator of Pseudomonas putida. J Mol Biol. 1996;258:575–587. doi: 10.1006/jmbi.1996.0270. [DOI] [PubMed] [Google Scholar]
- 49.Piccirilli J A, Vyle J S, Caruthers M H, Cech T R. Metal ion catalysis in the tetrahymena ribozyme reaction. Nature. 1993;361:85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- 50.Porter S C, North A K, Kustu S. Mechanism of transcriptional activation by NtrC. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 147–158. [Google Scholar]
- 51.Porter S C, North A K, Wedel A B, Kustu S. Oligomerization of NtrC at the glnA enhancer is required for transcriptional activation. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 52.Ronson C W, Astwood P M, Nixon B T, Ausubel F. Deduced products of C4-dicarboxylate transport regulatory genes of Rhizobium leguminosarum are homologous to nitrogen regulatory gene products. Nucleic Acids Res. 1987;15:7921–7934. doi: 10.1093/nar/15.19.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sablin E P, Kull F J, Cooke R, Vale R D, Fletterick R J. Crystal structure of the motor domain of the kinesin-related motor Ncd. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 54.Schindelin H, Kisker C, Schlessman J L, Howard J B, Rees D C. Structure of ADP × AIF4(−) stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 55.Schweins T, Wittinghofer A. GTP-binding proteins—structures, interactions and relationships. Curr Biol. 1994;4:547–550. doi: 10.1016/s0960-9822(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 56.Seefeldt L C, Dean D R. Role of nucleotides in nitrogenase catalysis. Acc Chem Res. 1997;30:260–266. [Google Scholar]
- 57.Senior A E, Al-Shawi M K. Further examination of seventeen mutations in Escherichia coli F1-ATPase β-subunit. J Biol Chem. 1992;267:21471–21478. [PubMed] [Google Scholar]
- 58.Sondek J, Lambright D G, Noel J P, Hamm H E, Sigler P B. GTPase mechanism of G proteins from the 1.7-Å crystal structure of transducin α · GDP · A1F4−. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 59.Sprinzl M. Elongation factor Tu: a regulatory GTPase with an integrated effector. Trends Biochem Sci. 1994;19:245–250. doi: 10.1016/0968-0004(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 60.Stock A M, Martinez-Hackert E, Rasmussen B F, West A H, Stock J B, Ringe D, Petsko G A. Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 61.Story R M, Steitz T A. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 62.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 63.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y-K, Lee J H, Brewer J M, Hoover T R. A conserved region in the ς54-dependent activator DctD is involved in both binding to RNA polymerase and coupling ATP hydrolysis to activation. Mol Microbiol. 1997;26:373–386. doi: 10.1046/j.1365-2958.1997.5851955.x. [DOI] [PubMed] [Google Scholar]
- 65.Washington M T, Rosenberg A H, Griffin K, Studier F W, Patel S S. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J Biol Chem. 1996;271:26825–26834. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- 66.Wedel A, Kustu S. The bacterial enhancer-binding protein NtrC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 67.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. The phosphorylated form of the enhancer-binding protein NtrC has an ATPase activity that is essential for activation of transcription. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 68.Wiseman T, Williston S, Brandts J F, Lin L N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 69.Wittinghofer A. The structure of transducin Gαt: more to view than just Ras. Cell. 1994;76:201–204. doi: 10.1016/0092-8674(94)90327-1. [DOI] [PubMed] [Google Scholar]
- 70.Wittinghofer A, Pai E F. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem Sci. 1991;16:382–387. doi: 10.1016/0968-0004(91)90156-p. [DOI] [PubMed] [Google Scholar]
- 71.Woody A-Y M, Eaton S S, Osumi-Davis P A, Woody R W. Asp537 and Asp812 in bacteriophage T7 RNA polymerase as metal ion-binding sites studied by EPR, flow-dialysis, and transcription. Biochemistry. 1996;35:144–152. doi: 10.1021/bi952037f. [DOI] [PubMed] [Google Scholar]
- 72.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 73.Yan, D., and S. Kustu. Unpublished data.
- 74.Yohda M, Ohta S, Hisabori T, Kagawa Y. Site-directed mutagenesis of a stable adenosine triphosphate synthase. Biochim Biophys Acta. 1988;933:156–164. doi: 10.1016/0005-2728(88)90065-5. [DOI] [PubMed] [Google Scholar]