Abstract
Dysregulated bile acid (BA)/lipid metabolism and gut bacteria dysbiosis are tightly associated with the development of obesity and non-alcoholic fatty liver disease (NAFLD). The orphan nuclear receptor, Small Heterodimer Partner (SHP/NR0B2), is a key regulator of BA/lipid metabolism, and its gene-regulating function is markedly enhanced by phosphorylation at Thr-58 mediated by a gut hormone, fibroblast growth factor-15/19 (FGF15/19). To investigate the role of this phosphorylation in whole-body energy metabolism, we generated transgenic SHP-T58A knock-in mice. Compared with wild-type (WT) mice, the phosphorylation-defective SHP-T58A mice gained weight more rapidly with decreased energy expenditure and increased lipid/BA levels. This obesity-prone phenotype was associated with the upregulation of lipid/BA synthesis genes and downregulation of lipophagy/β-oxidation genes. Mechanistically, defective SHP phosphorylation selectively impaired its interaction with LRH-1, resulting in de-repression of SHP/LRH-1 target BA/lipid synthesis genes. Remarkably, BA composition and selective gut bacteria which are known to impact obesity, were also altered in these mice. Upon feeding a high-fat diet, fatty liver developed more severely in SHP-T58A mice compared to WT mice. Treatment with antibiotics substantially improved the fatty liver phenotypes in both groups but had greater effects in the T58A mice so that the difference between the groups was largely eliminated. These results demonstrate that defective phosphorylation at a single nuclear receptor residue can impact whole-body energy metabolism by altering BA/lipid metabolism and gut bacteria, promoting complex metabolic disorders like NAFLD. Since posttranslational modifications generally act in gene- and context-specific manners, the FGF15/19-SHP phosphorylation axis may allow more targeted therapy for NAFLD.
Keywords: NAFLD, lipid metabolism, bile acid composition, gut microbiota, FXR, lipophagy
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease and a leading cause of liver transplant and liver-related death (1). NAFLD begins with excess triglyceride (TG) accumulation in the liver partly due to an imbalance in lipid synthesis, degradation via β-oxidation, secretion via very low-density lipoprotein (VLDL), and uptake of adipose-derived fatty acids (2). Defective lipophagy, autophagy-mediated lipid degradation, also promotes NAFLD (3, 4). Despite the striking global increase in NAFLD and its clinical importance, the pathogenesis of NAFLD is not clearly understood.
Bile acids (BAs) facilitate the intestinal absorption of lipid nutrients and also function as signaling molecules that can profoundly impact energy metabolism by modulating the activity of the BA nuclear receptor, Farnesoid-X-Receptor (FXR, NR1H4), and a membrane receptor, Takeda G protein-coupled receptor-5 (TGR5) (5). In the liver, primary BAs, cholic acid (CA), and chenodeoxycholic acid (CDCA) are synthesized and conjugated with taurine or glycine and then are further metabolized by gut bacteria to secondary BAs (6). Emerging evidence indicates that changes in BA composition and gut bacteria populations are tightly linked to the development of obesity and NAFLD (6, 7).
The liver and the intestine communicate with each other to maintain metabolic homeostasis. After a meal, BA-activated FXR regulates postprandial metabolism by inducing numerous genes in enterohepatic tissues, including Small Heterodimer Partner (SHP, NR0B2), and Fibroblast Growth Factor-19 (human FGF19, mouse FGF15) (8). The FXR-induced gut hormone, FGF15/19, is secreted from the ileum in the late fed-state and acts on both the liver and the intestine to ensure a smooth metabolic transition from the fed to the fasted state (8, 9), and SHP mediates postprandial metabolic actions of FGF15/19 (10, 11, 12, 13).
SHP is an unusual orphan nuclear receptor that does not contain a DNA binding domain but generally acts as a repressor by interacting with transcriptional activators involved in diverse biological processes, including LRH-1, SREBP-1/2, AHR, NF-κB, and CREB (10, 11, 14, 15, 16, 17). Previously, we reported that the gene-regulating function of SHP is markedly enhanced via post-translational modifications (PTMs), such as SUMOylation and phosphorylation, in response to BA-induced FGF15/19 signaling (10, 11, 15, 18, 19). Utilizing viral-mediated acute overexpression and downregulation approaches in mice, we showed that SHP mediates the metabolic functions of FGF15/19 in BA/lipid synthesis, cholesterol absorption, and autophagy in the late fed-state when plasma FGF15/19 levels peak (10, 11, 12, 13, 20). However, the long-term in vivo effects of defective SHP phosphorylation in whole-body energy metabolism and related pathologies have not been determined.
To investigate the role of the FGF15/19-SHP phosphorylation axis in obesity and NAFLD development, we generated transgenic SHP-T58A knock-in mice and show that defective phosphorylation at a single nuclear receptor residue can impact whole-body energy metabolism, promoting NAFLD. We further show that the fatty liver-prone phenotypes are associated with selectively altered expression of BA/lipid-regulating genes, and with changes in BA composition and gut bacteria that are known to impact obesity.
Results
SHP-T58A male mice display obesity-prone phenotypes compared with WT mice
To investigate the in vivo role of SHP phosphorylation by FGF15/19 in energy metabolism, mice carrying a T58A (human T55A) mutation in Shp gene were generated by CRISPR/Cas9. Transgenic SHP-T58A knock-in mice were born normally without any gross phenotypic changes and SHP protein levels detected by immunoblotting (Fig. 1A) and immunofluorescence (Fig. 1B) were comparable in both C57BL/6 wild-type (WT) and SHP-T58A mice. As expected, SHP phosphorylation at Thr-58 was absent in the SHP-T58A mice after treatment with FGF19 (Fig. 1C). Consistent with previous reports that FGF15/19-mediated activation of SHP via phosphorylation increases its nuclear localization (10, 11, 15), nuclear levels of SHP were reduced in the phosphorylation-defective SHP-T58A mice (Fig. 1D). When fed normal chow, male SHP-T58A mice gained weight more rapidly than WT mice, while this was not observed in female mice (Fig. 1E), consistent with the known increased susceptibility of male mice to obesity and fatty liver development compared to female mice (21). We, therefore, carried out further studies in male mice.
Figure 1.
SHP-T58A male mice gain weight more rapidly and have an obesity-prone phenotype when maintained on normal chow. A and B, expression of SHP in liver extracts from WT, SHP-T58A mice, and control SHP-KO mice were detected by immunoblotting (IB) (A) and the protein levels and cellular localization of SHP in mouse liver sections were examined by immunofluorescence (IF) (B) (n = 3 mice/group). C and D, WT and SHP-T58A mice were treated with FGF19 (1 mg/kg) for 30 min, and protein levels of phosphorylated SHP-T58 and SHP in the whole liver extracts (C) and protein levels of SHP and LAMIN in the liver nuclear extracts (D) were detected by IB (n= 3 mice/group). E, WT and SHP-T58A (n = 5 mice) were fed a normal chow diet and body weight was determined from 8 to 24 weeks. F, Triglyceride (TG) and cholesterol (CH) levels in the liver, plasma, and stools (n = 3 ∼ 4 mice). G, plasma metabolite levels determined by GC-MS analysis (n = 5 mice). H, images of livers from WT and SHP-T58A mice (left) and the ratio of liver weight/body weight (center) and visceral WAT weights (right). E–H, statistical significance was determined by (E) two-way ANOVA with the FDR test or (F–H) the Student’s t test. The mean ± standard deviation (SD), ∗p < 0.05, ∗∗p < 0.01.
TG and cholesterol levels were elevated in the liver and plasma and reduced in the stools of SHP-T58A mice, compared with WT mice (Fig. 1F). Plasma levels of glycerol and free fatty acids, such as stearate, palmitate, and fumarate, were also elevated in SHP-T58A mice (Fig. 1G). Consistent with these findings, the ratio of liver/body weight, an indicator of fatty liver, was increased and the weight of visceral white adipose tissue was also increased in these mice (Fig. 1H). These results demonstrate that SHP-T58A male mice display obesity-prone phenotypes compared with WT mice when maintained on normal chow.
SHP-T58A mice have decreased metabolic rates and energy expenditure, compared to WT mice
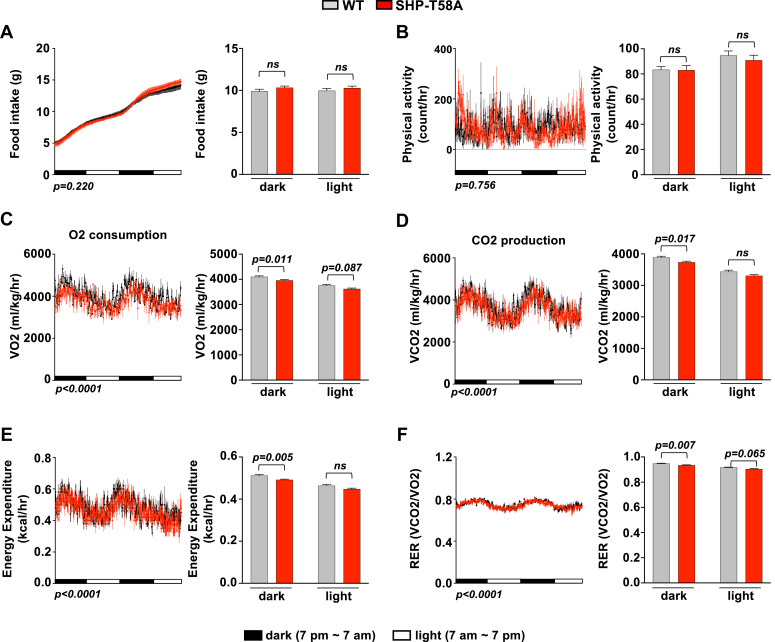
To understand the role of SHP phosphorylation in whole-body energy metabolism, we performed Comprehensive Lab Animal Monitoring System (CLAMS) analyses. Compared to WT mice, O2 consumption, CO2 production, and energy expenditure were decreased in SHP-T58A mice, particularly during the dark cycle, whereas food intake and physical activity were not changed (Fig. 2, A–E). Changes in the respiratory exchange ratio (RER) suggested that preferential utilization of lipids as a fuel source is reduced during the dark cycle, in SHP-T58A mice (Fig. 2F). These results indicate that SHP-T58A mice had reduced energy expenditure and metabolic rates, which contributes to increased weight gain.
Figure 2.
CLAMS metabolic cage studies revealed that SHP-T58A male mice have decreased energy expenditure and metabolic rates, compared to WT mice. A–F, WT and SHP-T58A mutant mice were individually housed for 2 days in CLAMS open-circuit Oxymax chambers. Food intake (A), physical activity (B), O2 consumption (C), CO2 production (D), energy expenditure or heat production (E), and RER (the ratio of CO2 production to O2 consumption) (F), during the dark and light periods. Statistical significance was determined by two-way ANOVA with the FDR test, (n = 5 mice/group). p-values are indicated, and ns, statistically not significant.
Expression of lipid-regulating genes in the liver and intestine is altered in SHP-T58A mice
Because hepatic TG levels were increased and the CLAMS studies indicated that lipids as a fuel source were decreased in SHP-T58A mice, we next examined the expression of lipid-regulating genes in the liver. The mRNA levels of fatty acid synthesis/uptake genes, Fasn, Srebp1, and Cd36 (Fig. 3A), and those of Pemt, a direct SHP target promoting lipogenesis through one-carbon metabolism (15), were increased. In contrast, mRNA levels of genes promoting β-oxidation and lipophagy, autophagy-mediated lipid catabolism, such as Fgf21, Cpt1, Pparα, Jmjd3, Tfeb and Atg7 (3, 22), were decreased in these mice (Fig. 3B).
Figure 3.
Lipid synthesis genes are upregulated and β-oxidation/lipophagy genes are downregulated in SHP-T58A mice. WT or SHP-T58A mice fed a normal chow diet were sacrificed after brief fasting for 4 h. A–D, levels of mRNAs of the indicated genes in the liver and intestine determined by RT-qPCR with WT levels set to 1. E, levels of Mtp mRNA (left) and ApoB48 and ACTIN proteins (right). F, images (left) of small intestines and measured lengths (right). A–F, statistical significance was determined by the Student’s t test. Mean ± SD is plotted (n = 5 mice), ∗p < 0.05, ∗∗p < 0.01, ns, statistically not significant.
Because intestinal lipid absorption is a key determinant of systemic lipid levels, we also examined intestinal lipid-regulating genes. The mRNA levels of Npc1l1, a direct SHP target promoting cholesterol uptake (11), and Cd36, a lipid uptake gene, were increased, whereas those of tight junction genes inhibiting gut permeability, Zo-1 and Occludin, were decreased in SHP-T58A mice (Fig. 3C).
Recently, we showed that gut lipophagy is paradoxically activated after feeding by FGF15/19-SHP-TFEB axis, which contributed to reducing intestinal TG-rich chylomicron levels (13). In intestinal gene expression studies, the mRNA levels of lipophagy genes, Tfeb, Atgl, and Ulk1, were reduced in SHP-T58A mice as were those of Cpt1, whereas those of Dgat1/2 were increased (Fig. 3D). Consistent with these results, expression of a chylomicron assembly gene, Mtp, was increased and protein levels of ApoB48, a marker of TG-rich intestinal chylomicrons (23), were also elevated in these mice (Fig. 3E). Notably, the length of small intestines was also increased (Fig. 3F), which was associated with reduced TG/cholesterol levels in the stools (Fig. 1F). Collectively, these results show that enterohepatic expression of lipid-regulating genes was altered in SHP-T58A mice, contributing to increased systemic TG levels.
Enterohepatic expression of BA metabolism genes is also altered in SHP-T58A mice
Tight regulation of BA metabolism is critical for the maintenance of normal lipid levels (5, 24, 25). We, thus, examined BA levels and enterohepatic expression of BA-related genes. Compared with WT mice, gallbladders were enlarged (Fig. 4A), and the total BA pool levels in the gallbladder, intestine, and liver combined were increased in SHP-T58A mice (Fig. 4B).
Figure 4.
Expression of BA synthesis/transport genes are altered in SHP-T58A mice. A, images of representative gallbladders. B, levels of total BAs in the gallbladder, intestine, and liver combined. C, levels of mRNAs in livers of the indicated genes determined by RT-qPCR with WT levels set to 1 (n = 5 mice). D, protein levels of CYP7A1 in liver extracts detected by IB (left) and quantitation using Image J of CYP7A1 protein bands relative to ACTIN (right, n = 3 mice). E, levels of intestinal mRNAs of the indicated genes determined by RT-qPCR with WT levels set to 1 (n = 5 mice). B–E, statistical significance was determined by the Student’s t test. The mean ± SD are plotted (n = 5 mice). ∗p < 0.05, ∗∗p < 0.01. ns, statistically not significant.
Consistent with these results, mRNA levels of the hepatic BA synthesis genes, Cyp7a1 and Cyp8b1, were elevated (Fig. 4C) as were protein levels of CYP7A1 in the liver extracts (Fig. 4D). Further, mRNA levels of the hepatic BA transporters, Ntcp and Bsep, were also elevated in SHP-T58A mice (Fig. 4C). Notably, the mRNA levels of genes promoting BA recycling, Asbt and Ostβ, were elevated in these mice (Fig. 4E). The mRNA levels of Fgf15, a direct FXR target and a repressor of hepatic BA/lipid synthesis by activating SHP (8, 10, 26), were also elevated (Fig. 4E). These results indicate that BA synthesis/recycling genes were upregulated, and BA pool levels were elevated in SHP-T58A mice.
Mechanistically, impaired interaction of SHP with LRH-1 leads to upregulation of BA/lipid synthesis genes in SHP-T58A mice
We next performed molecular biochemical studies to understand why BA/lipid synthesis genes were upregulated in SHP-T58A mice. Because SHP interacts with and represses LRH-1 (14), a nuclear receptor that activates the BA synthetic genes, Cyp7a1 and Cyp8b1, we first tested whether defective FGF15/19-SHP phosphorylation alters the interaction of SHP with LRH-1 in the SHP-T58A mice.
In CoIP assays, FGF19 treatment increased the interaction of SHP with LRH-1 in WT mice but not in SHP-T58A mice (Fig. 5A). We further examined the functional interaction between SHP and LRH-1 at the Cyp7a1 gene by ChIP assays. Because Osbpl3, which promotes lipogenesis via SREBP-1 processing, is a direct target of LRH-1 (27), we also examined this gene. In re-ChIP assays, SHP association with LRH-bound chromatin at the promoter region of Cyp7a1 and Osbpl3 genes was increased by FGF19 treatment (Fig. 5B), and the occupancy of RNA polymerase II, a positive indicator of transcription (Fig. 5C), was decreased in WT mice, but these changes were not observed in SHP-T58A mice.
Figure 5.
FGF19-mediatedphosphorylation of SHP and its interaction withLRH-1are abolished inSHP-T58Amice. A–C, WT and SHP-T58A mice were fasted overnight and treated with FGF19 (1 mg/kg) for 30 min and livers were collected. A, interaction between SHP and LRH-1 detected by Co-IP. B and C, re-ChIP: Liver chromatin was immunoprecipitated with LRH-1 antibody, eluted, and re-precipitated with SHP or RNA pol II antibody to detect the occupancy at the Cyp7a1 and Osbpl3 genes. D, pre-mRNA levels of indicated genes in mice treated with vehicle or FGF19 for 30 min. E, model: SHP interacts with and represses LRH1 on BA/lipid synthesis genes, Cyp7a1, Cyp8b1, and Osbpl3, which requires phosphorylation at Thr-58. F–H, defective SHP phosphorylation selectively impairs its interaction with SHP-interacting partners in FGF19-treated mice. WT and SHP-T58A mice were fasted overnight and treated with FGF19 for 30 min and livers were collected. F, interaction between SHP and SREBP-2, AHR, or CREB detected by Co-IP. Liver extracts from three mice were pooled and Co-IP assays were done. G and H, mRNA levels of direct SHP target genes involved in BA/lipid/sterol synthesis, autophagy/lipophagy, and BA transport determined by RT-qPCR. B–D, G and H, statistical significance was determined by two-way ANOVA with the FDR test. Mean and SD are plotted (n = 5 mice). ∗p < 0.05, ∗∗p < 0.01. ns, statistically not significant. Or p-values are indicated.
Consistent with these results from ChIP assays, pre-mRNA levels of hepatic genes promoting BA/lipid synthesis, Cyp7a1, Cyp8b1, Osbpl3, and Fasn were reduced by FGF19 treatment in WT mice but not in SHP-T58A mice (Fig. 5D). Collectively, these results suggest that compared to WT mice, impaired interaction of SHP with LRH-1 in the SHP-T58A mice leads to upregulation of direct LRH-1/SHP target genes, promoting BA/lipid synthesis, contributing to increased BA/lipid levels (Model, Fig. 5E).
Defective SHP phosphorylation selectively impairs its interaction with SHP-interacting partners, contributing to gene-selective transcriptional reprogramming
SHP interacts with many transcription factors, including LRH-1, SREBP-1/2, AHR, and CREB, and inhibits their gene-activating function (10, 11, 14, 15, 17, 20). In gene expression studies above, hepatic BA/lipid synthesis genes were upregulated, whereas lipophagy/β-oxidation genes are downregulated in the SHP-T58A mice (Figs. 3 and 4). To further understand this gene-selective alteration, we tested if the lack of SHP phosphorylation, generally or selectively, impairs its interaction with known SHP-interacting proteins, SREBP-2, AHR, and CREB (11, 15, 20).
In CoIP assays utilizing liver extracts from FGF19-treated mice, the SHP interaction with SREBP-2 and AHR, a transcriptional activator of sterol biosynthesis and lipogenesis via one-carbon metabolism, respectively (12, 15), was detected in WT mice, but not in SHP-T58A mice (Fig. 5F). In contrast, the SHP interaction with CREB, a gene activator of hepatic autophagy/lipophagy (20), was still retained in SHP-T58A mice. Consistent with these results, FGF19 treatment decreased mRNA levels of direct SHP/LRH-1 target genes involved in BA/lipid synthesis, such as Cyp7a1 and Osbpl3, in WT mice, but not in SHP-T58A mice (Fig. 5G). Similar effects were observed with direct SHP/AHR and SHP/SREBP-2 target genes, Pemt and Hmgcr, respectively (Fig. 5G). In contrast, FGF19-mediated repression of selected SHP/CREB target autophagy/lipophagy genes, such as Atg5 and Uvrag, was largely retained in SHP-T58A mice (Fig. 5H). FGF19-mediated repression of Ntcp, a direct target of SHP/RAR and a BA uptake transporter (28), was also retained in SHP-T58A mice (Fig. 5H), indicating the gene-selective effects of the SHP phosphorylation.
Collectively, these results suggest that defective phosphorylation in the SHP-T58A mice selectively impaired its interaction with SHP-interacting transcriptional factors, resulting in gene-selective reprogramming. While FGF15/19-mediated repression of LRH-1/SHP target BA/lipid synthesis genes is reversed, the repression of CREB/SHP target lipophagy genes is largely retained in the SHP-T58A mice, which contributes to increased lipid/BA levels.
BA composition and selective gut bacteria levels are also altered in SHP-T58A mice
Some BAs activate and some repress BA receptors, which can impact energy metabolism (24, 29). Since bile in the gallbladder best represents the BAs that are synthesized in the liver and circulated, we next examined the composition of BAs from the gallbladder and intestine combined. LC-MS metabolomic analysis revealed that in SHP-T58A mice, levels of hydrophilic BAs, particularly tauro-α/β-muricholic acid, an antagonist for FXR (6, 30), were reduced, whereas levels of tauro-conjugated CDCA, CA, and LCA, known agonists for FXR, were elevated (Fig. 6, A and B). These results are consistent with the upregulation of the direct FXR targets, intestinal Fgf15 and Ostβ, in SHP-T58A mice (Fig. 4E).
Figure 6.
BA composition and stool levels of selected gut bacteria that are known to impact obesity are altered in SHP-T58A mice. A, composition of BAs in the intestine and gallbladder combined determined by LC-MS/MS analysis, and the relative distribution of bile acids is represented in the stacked bar chart. B, relative levels of individual BAs in WT and SHP-T58A mice. C, WT and SHP-T58A mice were individually housed, and feces were collected. Levels of the indicated bacteria in the feces were measured by qPCR. B and C, statistical significance was determined by the Student’s t test. The mean ± SD are plotted (n = 5 mice/group). p-values are indicated, ns, statistically not significant.
BA composition can be altered by gut bacteria metabolism so that the changed BA composition in SHP-T58A mice suggested that the levels or composition of gut bacteria might be changed (6). We, thus, measured the levels of selected gut bacteria that are known to either promote or counter obesity (31, 32, 33). PCR analysis of 16 different gut bacteria in stool samples revealed that the abundance of Bacteroides was increased, whereas that of Enterococcus, Clostridium, Firmicutes, Actinobacteria, and Proteobacteria were decreased in SHP-T58A mice (Fig. 6C). Levels of some bacteria, such as Lactobacillus, were unchanged. Collectively, these results suggest that BA composition and selected gut bacteria levels are changed in SHP-T58A mice, which may also contribute to the obesity-prone phenotype.
Fatty liver developed more severely in SHP-T58A mice compared to WT mice when fed a high-fat/high-sucrose (HF/HS) diet
Because SHP-T58A mice had altered BAs and gut microbiota and increased BA/lipid levels compared to WT mice even when fed normal chow, we next examined whether the development of obesity and NAFLD in SHP-T58A mice was exaggerated after feeding a HF/HS diet.
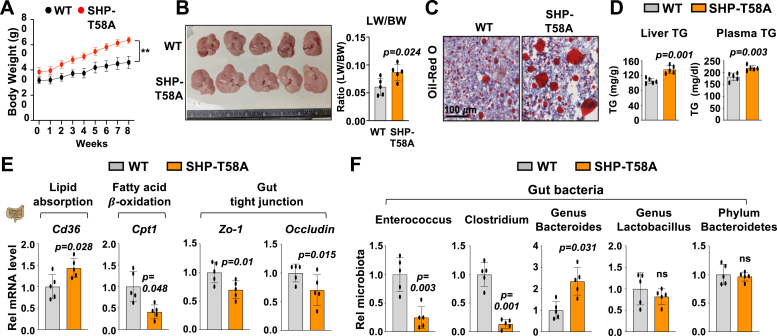
Compared to WT mice, male SHP-T58A mice fed a HF/HS diet gained weight more rapidly (Fig. 7A), had larger livers with increased ratios of liver/body weight (Fig. 7B), and had increased neutral lipids determined by Oli Red O staining (Fig. 7C). Liver and plasma TG levels were also increased (Fig. 7D). In gene expression studies, mRNA levels of an intestinal lipid absorption gene, Cd36, were increased, whereas those of a fatty acid β-oxidation gene, Cpt1, were reduced (Fig. 7E). Levels of mRNA of tight junction genes, such as Zo-1 and Occludin, were also decreased, suggesting gut permeability is increased in these mice. Notably, bacteria with known roles in lipid metabolism, promoting or preventing obesity, including Enterococcus, Clostridium, and Bacteroids (31, 33), were also altered in the SHP-T58A mice (Fig. 7F). These results demonstrate that SHP-T58A mice are more prone to the development of obesity and fatty liver when fed an HF/HS diet.
Figure 7.
Fatty liver is more prominent in SHP-T58A mice fed a high fat/high sucrose diet and selected gut bacteria levels are altered. WT and SHP-T58A male mice (18–24 weeks old) were fed a high-fat/high-sucrose diet (HF/HS) for 8 weeks. A, body weight with HF/HS diet starting at week 0. B, liver images and the ratio of liver weight (LW)/body weight (BW). C, liver sections stained for neutral lipids with Oil red O. D, liver and plasma TG levels. E, levels of mRNAs in intestines of the indicated genes determined by RT-qPCR with WT levels set to 1. F, mice were individually caged for 24 h and feces was collected. Levels of the indicated bacteria in the feces with the WT level set to 1. A, D–F, the mean and standard deviation are plotted. Statistical significance was determined by the Student’s t test (n = 5). ∗∗p < 0.01, p-values are indicated, ns, statistically not significant.
Antibiotic treatment improved the fatty liver in both WT and SHP-T58A mice but had greater effects in the SHP-T58A mice so that the difference between the groups was largely eliminated
To test whether altered gut microbiota in SHP-T58A mice have a role in more severe fatty liver phenotypes, mice fed an HF/HS diet were treated with antibiotics to deplete the gut bacteria. Compared to WT mice, SHP-T58A mice increased body weight more rapidly (Fig. 8A) and had larger livers with increased ratios of liver/body weight (Fig. 8B), and elevated neutral lipids determined by H&E and Oil Red O staining (Fig. 8C). Antibiotic treatment reduced body weights, liver weights, and neutral lipids in both WT and SHP-T58A mice but had greater effects in the SHP-T58A mice, so that difference between the groups were largely abolished (Fig. 8, A–C).
Figure 8.
Antibiotic treatment abolished the diet-induced fatty liver in both SHP-T58A and WT mice but had greater effects in SHP-T58A mice so that the difference between the groups was eliminated. WT and SHP-T58A mice (20–24 weeks old) were fed HF/HS diet for 4 weeks, and 7 days before sacrifice, antibiotics (vancomycin, neomycin, metronidazole; 1 g/l each) (ABs) were added to the drinking water (n = 5 mice/group). A, body weights. B, images of representative livers with the ratio of liver weight (LW)/body weight (BW). C, liver sections stained for neutral lipids with Oil red O and with H&E. D, serum FGF15 levels (n = 3 mice/group). E and F, mRNA levels in the liver and intestine of the indicated genes with WT set to 1 (n = 5 mice/group). G and H, liver and intestinal proteins detected by IB (n = 3 mice/group). A, B, D–F, statistical significance was determined by two-way ANOVA with the FDR test. The mean ± SD are plotted, ∗p < 0.05, ∗∗p < 0.01, ns, statistically not significant, p-values are indicated.
The correlation between circulating FGF15/19 levels in obesity and NAFLD has been controversial and is not clearly understood (34, 35, 36). HF/HS feeding increased plasma FGF15 levels in both WT and SHP-T58A mice, and antibiotic treatment improved the fatty liver in both groups (Fig. 8, A–C) but did not change the FGF15 levels in WT mice, while it modestly, but not significantly, reduced the levels in the SHP-T58A mice (Fig. 8D). These results suggest that FGF15/19 does not have a major role in improving the fatty liver by antibiotics in WT mice but may contribute to the beneficial response in the SHP-T58A mice.
In gene expression studies, the increased expression of BA synthesis genes, Cyp7a1 and Cyp8b1, and lipid transport/synthesis genes, Cd36 and Fasn, in SHP-T58A mice and decreased expression of a β-oxidation gene, Cpt1, were reversed by antibiotic treatment (Fig. 8E). Increased intestinal expression of genes promoting BA recycling, Asbt and Ostβ, and lipid absorption/synthesis, Npc1l1, Cd36, and Dgat2, in SHP-T58A mice were also reversed by antibiotic treatment (Fig. 8F). Protein levels of hepatic CYP7A1 and FASN and intestinal NPC1L1 and ApoB48, the marker of TG-rich intestinal chylomicrons (23), were elevated in SHP-T58A mice, and these differences were blocked by antibiotic treatment (Fig. 8, G and H).
These results from antibiotic experiments indicate that in addition to altered expression of genes regulating lipid/BA metabolism, changes in gut bacteria in the SHP-T58A mice may contribute to the more prominent development of fatty liver.
Discussion
Numerous studies have reported the importance of PTMs in nuclear receptor action, but studies reporting the long-term in vivo effects on energy metabolism and related pathologies are very limited. Utilizing transgenic SHP-T58A knock-in mice, we demonstrate that the phosphorylation status at a single amino acid in SHP can impact whole-body energy metabolism, promoting obesity and NAFLD. Obesity-prone phenotypes in the SHP-T58A mice are partly due to gene-selective reprogramming of BA/lipid-regulating genes but are also associated with changes in BA composition and levels of selective gut bacteria that are known to impact obesity. Remarkably, SHP-T58A mice developed more severe fatty liver than WT mice upon feeding a high-fat diet, and while antibiotic treatment improved fatty liver in both groups, the difference between groups was largely abolished, suggesting that altered gut bacteria may contribute to the more severe fatty liver phenotype in the SHP-T58A mice.
Compared to WT mice, SHP-T58A male mice gained weight more rapidly with increased BA/lipid levels, even when fed normal chow. These obesity-prone phenotypes were associated with altered expression of direct SHP target genes regulating lipid/BA metabolism and lipophagy/lipid degradation in a gene-selective manner. For example, direct SHP target BA synthetic genes, Cyp7a1 and Cyp8b1, lipogenic genes, including Fasn and Osbpl3, were upregulated in the SHP-T58A mice. In contrast, direct SHP target lipophagy/β-oxidation genes were still repressed in these mice. Intriguingly, defective SHP phosphorylation selectively impaired its interaction with known SHP-interacting partners. While the interaction of SHP with LRH-1, SREBP-2, and AHR, key activators of lipid synthesis, was dependent on the SHP phosphorylation, the interaction with CREB, an autophagic activator (20), was not. The selective impairment with SHP-interacting partners can lead to gene-selective transcriptional reprogramming, that is, upregulation of BA/lipid synthesis genes and continued repression of autophagy/lipophagy genes, promoting obesity and NAFLD in SHP-T58A mice.
The intestine plays a crucial role in the maintenance of normal lipid levels. SHP-T58A mice had increased plasma TG/cholesterol levels, whereas levels were reduced in the stool, suggesting that intestinal lipid absorption may be increased. Notably, the lengths of small intestines were longer, and intestinal expression of the lipid absorption genes, Cd36 and Npc1l1, was increased, and protein levels of ApoB48, the intestinal marker of TG-rich chylomicrons (23), were elevated. Expression of the gut lipophagy genes, Tfeb, Atgl, and Ulk1, was also reduced in the SHP-T58A mice, which was consistent with a recent finding that feeding activates gut lipophagy by an FGF15/19-SHP-TFEB axis, reducing postprandial TG levels (13). Obesity-prone SHP-T58A mice also had an elevated BA pool, associated with upregulation of the BA recycling genes, Asbt and Ostα/β. Consistent with these findings, inhibition of enterohepatic recycling of BAs by targeting ASBT, a key ileal BA transporter, prevented NAFLD/NASH in mice (37).
Remarkably, the SHP-T58A mice also had reduced levels of T-α/βMCA, an FXR antagonist, and increased levels of T-CA, T-CDCA, and T-LCA, FXR agonists (5, 6). These changes in BA composition correlated with altered levels of selective gut bacteria that metabolize BAs. The SHP-T58A mice had reduced Clostridium levels and upregulation of the intestinal lipid absorption/synthesis genes, Cd36 and Dgat2, which is consistent with previous studies showing that reduction of Clostridium leads to obesity by upregulation of these genes (33). In contrast, levels of Bacteroides were elevated in the SHP-T58A mice. However, the role of Bacteroides in obesity and metabolic disease is controversial. Levels of Bacteroides were elevated in mice treated with fexaramine, a gut-specific FXR agonist (38), which improved obesity and diabetes, via increased T-LCA levels, adipose tissue browning, and GLP-1 secretion (38, 39). In contrast, the abundance of Bacteroides is positively correlated with BMI in obese individuals (31) and increased in patients with NASH (7). In the present study, T-α/βMCA levels were decreased and Bacteroides levels were elevated in the NAFLD-prone SHP-T58A mice. However, 16S rRNA sequencing and gut bacteria transfer studies in germ-free conditions will be required to establish the role of gut bacteria in NAFLD in these mice.
While SHP is an unequivocal regulator of BA metabolism, its role in energy metabolism is controversial. In contrast to obesity/fatty liver-prone SHP-T58A knock-in mice, mice lacking SHP had a leaner phenotype and were resistant to diet-induced obesity and fatty liver (40), and combined deletion of both SHP/FXR reduced liver TG levels with increased fatty acid utilization (41, 42). These conflicting results could be partly due to gene-selective regulation mediated by SHP-T58 phosphorylation, which was observed in the present study. In contrast, other studies are consistent with the obesity-prone phenotype of SHP-T58A mice. BA-activated FXR reduced hepatic TG levels via a pathway requiring SHP and SREBP-1 (43). We also reported that SHP physiologically represses hepatic lipogenesis and intestinal lipid absorption by inhibiting SREBP-1/2 upon BA-activated FGF15/19 signaling in the late fed state (10, 11, 15). Recently, viral-mediated downregulation of hepatocyte SHP promoted hepatic inflammation and fibrosis in NASH mice (17). The reasons for these conflicting results, with different mechanisms, of the role of SHP in energy metabolism are not clear but likely depend on the experimental models and conditions that are used.
Overall, our study demonstrates that the phosphorylation status of SHP at a single residue can impact systemic energy metabolism, promoting complex multifactorial metabolic disorders like NAFLD. Notably, levels of SHP phosphorylated at Thr-55 (Thr-58 in mice) were dramatically reduced in NAFLD patients (15), implying the potential human relevance of these studies. Since PTMs of nuclear receptors, including SHP, FXR, and LRH-1, generally act in gene- and context-specific manners as shown previously (27, 44, 45, 46, 47) and in the current study, restoring the FGF15/19-SHP phosphorylation may lead to more targeted therapy for NAFLD.
Experimental procedures
Generation of T58A-SHP mice by CRISPR/CAS
A point mutation at the mouse Shp/Nr0b2 locus (GenBank accession number: NM_011850.3) that introduced the mutation T58A (ACC to GCA) was generated by CRISPR/Cas9-mediated genome engineering in a C57BL6 background by Cyagen Biosciences, Inc. Pups were genotyped by PCR using the primer set, Nr0b2(T58A)-F: 5′-AGAAACAGGAACAAGATACTAACCATGAGCT-3′, and Nr0b2(T58A)-R: 5′-TTGAAGAGGATCGTGCCCTTCAG-3’. Heterozygote pups were bred to produce homozygous mice with the T58A mutation (SHP-T58A) and homozygous C57BL/6 (designated WT) mice. The homozygous SHP-T58A and WT mice were bred to establish colonies for the experimental transgenic and control mice, respectively.
CLAMS metabolic cage study
Whole-body energy metabolism of 10- to 14-week-old male WT and SHP-T58A mice was measured by indirect calorimetry in an open-circuit Oxymax chamber of a Comprehensive Lab Animal Monitoring System (CLAMS) (Columbus Instruments), and food intake and physical activity were also measured. Male mice were individually housed in chambers, and food and water were freely available. The chamber was maintained on a 12-h light–dark cycle at 23 °C, and O2 consumption (VO2) and CO2 (VCO2) production were measured continuously. The respiratory exchange ratio (RER) was calculated as VCO2 produced divided by VO2 consumed.
Animal experiments
WT and SHP-T58A mice were housed at 23 °C with 50% humidity under a 12 h/12 h light/dark cycle, and food and water were available ad libitum. For analysis of metabolites, mice were briefly fasted for 4 h and the liver, gallbladder, intestines, blood, and feces were collected. For the generation of diet-induced obese mice with fatty liver, WT, and SHP-T58A mice were fed an HF/HS (42% kcal from fat, 34% high sucrose, 0.2% total cholesterol: Harlan Teklad, TD88137) diet for 8 weeks. To examine the role of gut bacteria in fatty liver development, 20- to 24-week-old male mice were fed an HF/HS diet for 4 weeks and 1 g/L each of vancomycin, neomycin, and metronidazole was added to the drinking water for 7 days before sacrifice. All experiments were approved by the Institutional Animal Care and Use and Biosafety Committees of the University of Illinois, Urbana-Champaign (UIUC), and the University of Wisconsin-Madison.
Histological analyses
Neutral lipids in quick-frozen liver sections were detected by Oil Red O staining. Paraffin-fixed liver and intestinal ileum sections were stained with H&E and imaged with a NanoZoomer Scanner (Hamamatsu). For immunostaining, paraffin-embedded liver sections were incubated with rabbit anti-SHP (ab96605, Abcam, dilution 1:500) overnight at 4 °C followed by the secondary antibody Alexa Fluor 647 goat anti-rabbit IgG (A21245, Life Technologies, dilution 1:200) for 1 h. Nuclei were labeled with DAPI (20,004, Bioquest) for 5 min. Images were taken and processed using the EVOS M5000 Imaging System (Thermo).
Measurement of TG, cholesterol, BA, and plasma metabolites
TG and cholesterol levels were measured using kits (TR0100 and MAK043, respectively, Sigma) and total BA levels in the liver, intestine, plasma, and gallbladder were measured with a kit (MAK309, Sigma). Plasma metabolites were identified by gas chromatography-mass spectrometry (GC-MS) in the Metabolomics Center at UIUC.
Measurement of plasma FGF15 levels
Serum FGF15 levels were determined using FGF15 ELISA Kit (MBS707181, MyBioSource) according to the manufacturer’s protocol.
Measurement of BA composition
Extracts from the intestine and gallbladder combined were adjusted to 0.4 N perchloric acid and centrifuged, and supernatants from the two tissues were combined for analysis. Individual BAs were identified using the 5500 QTRAP LC-MS/MS system (Sciex) in the Metabolomics Center at UIUC.
Measurement of selected gut bacteria levels by qPCR
DNA was isolated from 200 mg of mouse feces using DNA Miniprep Kits (Zymo Research, #D4300). The levels of selected bacteria that are known to impact obesity were determined by qPCR analysis using primer sets designed to target 16S ribosomal RNA genes as previously reported (48).
Co-immunoprecipitation
Mouse livers were finely chopped, washed in ice-cold PBS, resuspended in lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.3% Nonidet P40, 10% glycerol), and homogenized. Then, samples were incubated for 20 min on ice and centrifuged at 16,000g for 10 min. The supernatants were incubated overnight with 1 μg antibodies indicated (SHP: Abcam ab96605, CREB: Abcam ab178322, AHR: Santa Cruz Biotech sc-133088, SREBP2: BD biosciences 557,037, LRH1: Sigma Aldrich ABE2867) and then with 35 μl of 25% protein G agarose. Two h later, agarose beads were washed with the CoIP buffer 3 times and bound proteins were detected by IB.
Chromatin immunoprecipitation
Mouse liver tissue was minced, washed twice with PBS, and incubated with 1% (w/v) formaldehyde for 10 min and then, with 125 mM glycine for 5 min. Chromatin samples were sonicated for 30 min using a QSonica 800R2-110 sonicator at an amplitude setting of 70% with a sonication pulse rate of 15 s on and 45 s off. Chromatin was immunoprecipitated by LRH-1 antibody (Santa Cruz Biotech, sc-393369) and then, immunoprecipitated chromatin was eluted by incubation with 10 mM DTT for 30 min at 37 °C, the eluate was diluted 20-fold and re-precipitated by antibody against SHP (Abcam, ab96605) or RNA polymerase II (Novus biologicals, NB200–598). DNA was isolated and quantified by qPCR using a primer set for detecting SHP and RNA pol II occupancies at Cyp7a1 (49, 50) and Osbpl3 (27).
Immunoblotting
Mouse intestine and liver tissues were washed with ice-cold PBS and homogenized in RIPA buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.1% SDS, 1 mM DTT) containing protease and phosphatase inhibitors, and then, sonicated briefly for 3 to 5 s and centrifuged at 14,000 rpm 4 °C for 10 min. Supernatants were subjected to electrophoresis, transferred to PVDF membranes, blocked in TBS-Tween 20 containing 5% non-fat milk for 1 h at room temperature, and incubated with primary antibodies O/N at 4 °C, SHP (Abcam, ab96605), CYP7A1 (Invitrogen, PA5-100892), FASN (Cell Signaling, #3180), NPC1L1 (LifeSpan BioSciences, LS-B506), and ApoB48 (ThermoFisher, MA5-35458). Antibody for p-SHP-T58 was produced commercially (Abmart, Inc) and utilized in previous studies (11, 12, 15, 18, 19).
Nuclear SHP protein levels
Mouse liver tissues were washed with ice-cold PBS, homogenized in hypotonic buffer (10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 0.2% NP40, 1 mM EDTA, and 5% sucrose), and nuclei were pelleted by cushion buffer (10 mM Tris-HCl, pH 7.5, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, and 10% sucrose) after centrifugation at 5000 rpm for 3 min. Nuclear SHP and LAMIN levels were detected by IB.
RT-qPCR
RNA was isolated from the liver or small intestine using TRIzol (Invitrogen, Thermo Fisher Scientific) or an RNeasy mini kit (Qiagen, 74,104) and quantified by RT-qPCR, using primer sequences as previously reported (3, 11, 13, 15, 22) and normalized to 36B4 mRNA.
Statistical analysis
GraphPad Prism 9 (GraphPad software version 9.2.0) was used for data analysis. Statistical significance was determined by the Student’s two-tailed t test or one- or two-way ANOVA with the false discovery rate post-test for single or multiple comparisons as appropriate. p-values < 0.05 were considered statistically significant.
Data availability
Data will be made available on request.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank H. Eric Xu at Van Andel Research Institute for providing recombinant FGF19. We also thank Lucas Li at Metabolomics Center at the UIUC for his help with the LC-MS/MS and GC-MS analyses.
Author contributions
Y.-C. K., S. S., T. F., and J. K. K. methodology; Y.-C. K., M. Q., X. D., S. S., H. S., X. D., and T. F. investigation; Y.-C. K., M. Q., X. D., S. S., H. S., B. K., T. F., and J. K. K. formal analysis; and Y.-C. K., S. S., B. K., T. F., and J. K. K. writing – original manuscript.
Funding and additional information
This work was supported by an American Heart Association Scientist Development Award (16SDG27570006) to Y. C. Kim, UW-Madison startup grants and an American Cancer Society Institutional Research Grant (MSN228402) to T. F., and by grants from the National Institutes of Health (R01DK062777 and R01DK095842) to J. K. K.
Reviewed by members of the JBC Editorial Board. Edited by Phillip A. Cole
Footnotes
Present addresses for: Young-Chae Kim, Cardiovascular Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD 20892; Sunmi Seok, Cardiovascular Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD20892.
Contributor Information
Ting Fu, Email: ting.fu@wisc.edu.
Jongsook Kim Kemper, Email: jongsook@illinois.edu.
References
- 1.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loomba R., Friedman S.L., Shulman G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell. 2021;184:2537–2564. doi: 10.1016/j.cell.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byun S., Seok S., Kim Y.C., Zhang Y., Yau P., Iwamori N., et al. Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase. Nat. Commun. 2020;11:807. doi: 10.1038/s41467-020-14384-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grefhorst A., van de Peppel I.P., Larsen L.E., Jonker J.W., Holleboom A.G. The role of lipophagy in the development and treatment of non-alcoholic fatty liver disease. Front. Endocrinol. (Lausanne) 2020;11 doi: 10.3389/fendo.2020.601627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang J.Y.L., Ferrell J.M. Bile acid receptors FXR and TGR5 signaling in fatty liver diseases and therapy. Am. J. Physiol. Gastrointest. Liver Physiol. 2020;318:G554–G573. doi: 10.1152/ajpgi.00223.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wahlstrom A., Sayin S.I., Marschall H.U., Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Chu H., Williams B., Schnabl B. Gut microbiota, fatty liver disease, and hepatocellular carcinoma. Liver Res. 2018;2:43–51. doi: 10.1016/j.livres.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kliewer S.A., Mangelsdorf D.J. Bile acids as hormones: the FXR-FGF15/19 pathway. Dig. Dis. 2015;33:327–331. doi: 10.1159/000371670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadaleta R.M., Moschetta A. Metabolic messengers: fibroblast growth factor 15/19. Nat. Metab. 2019;1:588–594. doi: 10.1038/s42255-019-0074-3. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y., Seok S., Zhang Y., Ma J., Kong B., Guo G., et al. Intestinal FGF15/19 physiologically repress hepatic lipogenesis in the late fed-state by activating SHP and DNMT3A. Nat. Commun. 2020;11:5969. doi: 10.1038/s41467-020-19803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y.C., Byun S., Seok S., Guo G., Xu H.E., Kemper B., et al. Small heterodimer partner and fibroblast growth factor 19 inhibit expression of NPC1L1 in mouse intestine and cholesterol absorption. Gastroenterology. 2018;156:1052–1065. doi: 10.1053/j.gastro.2018.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y.C., Byun S., Zhang Y., Seok S., Kemper B., Ma J., et al. Liver ChIP-seq analysis in FGF19-treated mice reveals SHP as a global transcriptional partner of SREBP-2. Genome Biol. 2015;16:268. doi: 10.1186/s13059-015-0835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seok S., Kim Y.C., Zhang Y., Kong B., Guo G., Ma J., et al. Feeding activates FGF15-SHP-TFEB-mediated lipophagy in the gut. EMBO J. 2022;41 doi: 10.15252/embj.2021109997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y., Moore D.D. Dual mechanism for repression of the monomeric orphan receptor liver receptor homologous protein-1 (LRH-1) by the orphan small heterodimer partner (SHP) J. Biol. Chem. 2002;277:2463–2467. doi: 10.1074/jbc.M105161200. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y.C., Seok S., Byun S., Kong B., Zhang Y., Guo G., et al. AhR and SHP regulate phosphatidylcholine and S-adenosylmethionine levels in the one-carbon cycle. Nat. Commun. 2018;9:540. doi: 10.1038/s41467-018-03060-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuk J.M., Shin D.M., Lee H.M., Kim J.J., Kim S.W., Jin H.S., et al. The orphan nuclear receptor SHP acts as a negative regulator in inflammatory signaling triggered by Toll-like receptors. Nat. Immunol. 2011;12:742–751. doi: 10.1038/ni.2064. [DOI] [PubMed] [Google Scholar]
- 17.Zou A., Magee N., Deng F., Lehn S., Zhong C., Zhang Y. Hepatocyte nuclear receptor SHP suppresses inflammation and fibrosis in a mouse model of nonalcoholic steatohepatitis. J. Biol. Chem. 2018;293:8656–8671. doi: 10.1074/jbc.RA117.001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D.H., Kwon S., Byun S., Xiao Z., Park S., Wu S.Y., et al. Critical role of RanBP2-mediated SUMOylation of small heterodimer partner in maintaining bile acid homeostasis. Nat. Commun. 2016;7 doi: 10.1038/ncomms12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seok S., Kanamaluru D., Xiao Z., Ryerson D., Choi S.E., Suino-Powell K., et al. Bile acid signal-induced phosphorylation of small heterodimer partner by protein kinase Czeta is critical for epigenomic regulation of liver metabolic genes. J. Biol. Chem. 2013;288:23252–23263. doi: 10.1074/jbc.M113.452037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byun S., Kim Y.C., Zhang Y., Kong B., Guo G., Sadoshima J., et al. A postprandial FGF19-SHP-LSD1 regulatory axis mediates epigenetic repression of hepatic autophagy. EMBO J. 2017;36:1755–1769. doi: 10.15252/embj.201695500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hart-Unger S., Korach K.S. Estrogens and obesity: is it all in our heads? Cell Metab. 2011;14:435–436. doi: 10.1016/j.cmet.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seok S., Kim Y.C., Byun S., Choi S., Xiao Z., Iwamori N., et al. Fasting-induced JMJD3 histone demethylase epigenetically activates mitochondrial fatty acid beta-oxidation. J. Clin. Invest. 2018;128:3144–3159. doi: 10.1172/JCI97736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussain M.M. Intestinal lipid absorption and lipoprotein formation. Curr. Opin. Lipidol. 2014;25:200–206. doi: 10.1097/MOL.0000000000000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavez-Talavera O., Tailleux A., Lefebvre P., Staels B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology. 2017;152:1679–1694.e1673. doi: 10.1053/j.gastro.2017.01.055. [DOI] [PubMed] [Google Scholar]
- 25.Xue R., Su L., Lai S., Wang Y., Zhao D., Fan J., et al. Bile acid receptors and the gut-liver axis in nonalcoholic fatty liver disease. Cells. 2021;10:2806. doi: 10.3390/cells10112806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C.L., McDonald J.G., et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Stein S., Lemos V., Xu P., Demagny H., Wang X., Ryu D., et al. Impaired SUMOylation of nuclear receptor LRH-1 promotes nonalcoholic fatty liver disease. J. Clin. Invest. 2017;127:583–592. doi: 10.1172/JCI85499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denson L.A., Sturm E., Echevarria W., Zimmerman T.L., Makishima M., Mangelsdorf D.J., et al. The orphan nuclear receptor, shp, mediates bile acid-induced inhibition of the rat bile acid transporter, ntcp. Gastroenterology. 2001;121:140–147. doi: 10.1053/gast.2001.25503. [DOI] [PubMed] [Google Scholar]
- 29.Tripathi A., Debelius J., Brenner D.A., Karin M., Loomba R., Schnabl B., et al. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K., et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Schwiertz A., Taras D., Schafer K., Beijer S., Bos N.A., Donus C., et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Guryn K., Hubert N., Frazier K., Urlass S., Musch M.W., Ojeda P., et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe. 2018;23:458–469.e455. doi: 10.1016/j.chom.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen C., Bell R., Klag K.A., Lee S.H., Soto R., Ghazaryan A., et al. T cell-mediated regulation of the microbiota protects against obesity. Science. 2019;365 doi: 10.1126/science.aat9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozadjieva N., Heppner K.M., Seeley R.J. Targeting FXR and FGF19 to treat metabolic diseases-lessons learned from bariatric surgery. Diabetes. 2018;67:1720–1728. doi: 10.2337/dbi17-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher J.D., Kong B., Pan Y., Zhan L., Sun R., Aa J., et al. The effect of fibroblast growth factor 15 deficiency on the development of high fat diet induced non-alcoholic steatohepatitis. Toxicol. Appl. Pharmacol. 2017;330:1–8. doi: 10.1016/j.taap.2017.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez-Sola G., Uriarte I., Latasa M.U., Fernandez-Barrena M.G., Urtasun R., Elizalde M., et al. Fibroblast growth factor 15/19 (FGF15/19) protects from diet-induced hepatic steatosis: development of an FGF19-based chimeric molecule to promote fatty liver regeneration. Gut. 2017;66:1818–1828. doi: 10.1136/gutjnl-2016-312975. [DOI] [PubMed] [Google Scholar]
- 37.Rao A., Kosters A., Mells J.E., Zhang W., Setchell K.D., Amanso A.M., et al. Inhibition of ileal bile acid uptake protects against nonalcoholic fatty liver disease in high-fat diet-fed mice. Sci. Transl. Med. 2016;8:357ra122. doi: 10.1126/scitranslmed.aaf4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang S., Suh J.M., Reilly S.M., Yu E., Osborn O., Lackey D., et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat. Med. 2015;21:159–165. doi: 10.1038/nm.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pathak P., Xie C., Nichols R.G., Ferrell J.M., Boehme S., Krausz K.W., et al. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology. 2018;68:1574–1588. doi: 10.1002/hep.29857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park Y.J., Kim S.C., Kim J., Anakk S., Lee J.M., Tseng H.T., et al. Dissociation of diabetes and obesity in mice lacking orphan nuclear receptor small heterodimer partner. J. Lipid Res. 2011;52:2234–2244. doi: 10.1194/jlr.M016048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K.H., Choi S., Zhou Y., Kim E.Y., Lee J.M., Saha P.K., et al. Hepatic FXR/SHP axis modulates systemic glucose and fatty acid homeostasis in aged mice. Hepatology. 2017;66:498–509. doi: 10.1002/hep.29199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akinrotimi O., Riessen R., VanDuyne P., Park J.E., Lee Y.K., Wong L.J., et al. Small heterodimer partner deletion prevents hepatic steatosis and when combined with farnesoid X receptor loss protects against type 2 diabetes in mice. Hepatology. 2017;66:1854–1865. doi: 10.1002/hep.29305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe M., Houten S.M., Wang L., Moschetta A., Mangelsdorf D.J., Heyman R.A., et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kemper J.K., Xiao Z., Ponugoti B., Miao J., Fang S., Kanamaluru D., et al. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D.H., Xiao Z., Kwon S., Sun X., Ryerson D., Tkac D., et al. A dysregulated acetyl/SUMO switch of FXR promotes hepatic inflammation in obesity. EMBO J. 2015;34:184–199. doi: 10.15252/embj.201489527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byun S., Kim D.H., Ryerson D., Kim Y.C., Sun H., Kong B., et al. Postprandial FGF19-induced phosphorylation by Src is critical for FXR function in bile acid homeostasis. Nat. Commun. 2018;9:2590. doi: 10.1038/s41467-018-04697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Appelman M.D., van der Veen S.W., van Mil S.W.C. Post-translational modifications of FXR; implications for cholestasis and obesity-related disorders. Front. Endocrinol. (Lausanne) 2021;12 doi: 10.3389/fendo.2021.729828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hermann-Bank M.L., Skovgaard K., Stockmarr A., Larsen N., Molbak L. The Gut Microbiotassay: a high-throughput qPCR approach combinable with next generation sequencing to study gut microbial diversity. BMC Genomics. 2013;14:788. doi: 10.1186/1471-2164-14-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim Y.C., Fang S., Byun S., Seok S., Kemper B., Kemper J.K. FXR-induced lysine-specific histone demethylase, LSD1, reduces hepatic bile acid levels and protects the liver against bile acid toxicity. Hepatology. 2015;62:220–231. doi: 10.1002/hep.27677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y.C., Jung H., Seok S., Zhang Y., Ma J., Li T., et al. MicroRNA-210 promotes bile acid-induced cholestatic liver injury by targeting mixed-lineage leukemia-4 methyltransferase in mice. Hepatology. 2019;71:2118–2134. doi: 10.1002/hep.30966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.