Abstract
Tumor evolution is the accumulation of various tumor cell behaviors from tumorigenesis to tumor metastasis and is regulated by the tumor microenvironment (TME). However, the mechanism of solid tumor progression has not been completely elucidated, and thus, the development of tumor therapy is still limited. Recently, Tumor chips constructed by culturing tumor cells and stromal cells on microfluidic chips have demonstrated great potential in modeling solid tumors and visualizing tumor cell behaviors to exploit tumor progression. Herein, we review the methods of developing engineered solid tumors on microfluidic chips in terms of tumor types, cell resources and patterns, the extracellular matrix and the components of the TME, and summarize the recent advances of microfluidic chips in demonstrating tumor cell behaviors, including proliferation, epithelial-to-mesenchymal transition, migration, intravasation, extravasation and immune escape of tumor cells. We also outline the combination of tumor organoids and microfluidic chips to elaborate tumor organoid-on-a-chip platforms, as well as the practical limitations that must be overcome.
Keywords: Tumor evolution, Microfluidic chips, Tumor cell behaviors, Solid tumors, Tumor organoids
Graphical abstract
1. Introduction
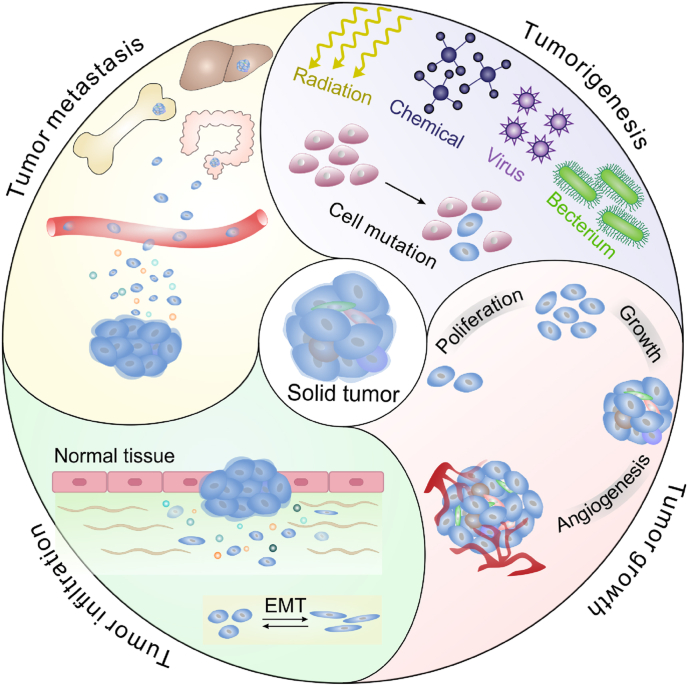
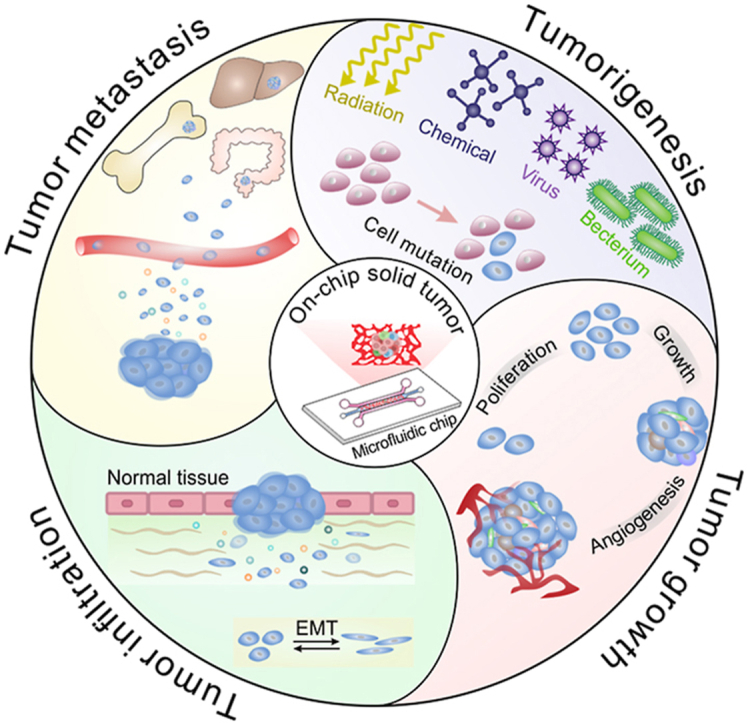
Cancer continues to be a leading cause of death worldwide [1]. Based on the most recent global cancer statistics, nine of the top ten cancers with high mortality are solid malignant tumors [1]. The incidences of various malignant solid tumors increase annually in both developed and developing countries, resulting from more excessive exposure to some risk factors, such as viruses [2], chemicals [3,4], radiation [5] and cigarettes [6]. Solid tumors can arise in almost all human organs and are often considered abnormal organ-like structures mainly composed of tumor cells [7]. The evolution of solid tumors refers to a complicated process from tumorigenesis [8] to tumor growth and infiltration [9] to tumor metastasis [10], which is achieved through a series of dynamic tumor cell behaviors. As tumors evolve, the proliferation speed and invasiveness of tumor cells tend to be accelerated [11,12], and the spread of tumor cells into distant organs or tissues may occur, as well as immune escape and drug resistance [13,14]. The multiple biological behaviors of tumor cells ultimately cause tumor metastasis and the failure of tumor treatment.
To demonstrate the dynamic behaviors of tumor cells and elucidate the mechanism of tumor evolution, researchers have developed many tumor models. Animal models, which are often constructed by subcutaneously implanting tumor tissues or tumor cells in rodents, such as rats and mice, have greatly advanced our understanding of complex tumor progression [[15], [16], [17]]. However, there is a species difference between humans and animals [15,18], and it is difficult to clearly visualize how a single tumor cell behaves over time in tumor evolution for animal models [18,19]. Compared to animal models, cell culture models have an advantage in visualizing cell morphology, are cost-efficient and simple to use [20,21]. However, two-dimensional (2D) cell monocultures fail to replicate the cellular spatial arrangement of solid tumors [22,23], resulting in the inconsistent morphology and behavior of tumor cells compared to those in vivo [24,25]. Traditional three-dimensional (3D) cell culture models (e.g. tumor spheroids) [[26], [27], [28]] can mimic solid tumors in terms of the spatial structure of tumor cells [29]. However, they lack some crucial components of the tumor's physicochemical environment, such as fluid shear force and perfusable vasculatures [30,31].
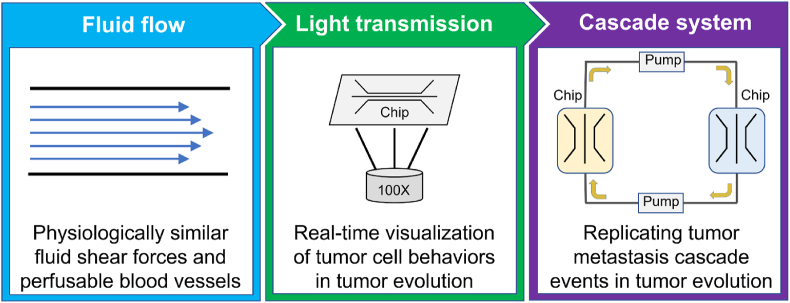
Organ chips have emerged as a novel in vitro research platform in the life sciences and engineering [[32], [33], [34]]. Organ chips focusing on tumor research are referred to as tumor chips. Materials used to fabricate tumor chips mainly include polydimethylsiloxane (PDMS) [35] and polymethyl methacrylate (PMMA) [36]. These materials are optically transparent and can be integrated with microscopy imaging systems (Fig. 1), allowing for easy visualization of a single tumor cell's behavior. The patterns and sizes of tumor chips can be customized, enabling easy perfusion with ultralow-volumes of culture medium [18,19]. Notably, the physiochemical microenvironment of a solid tumor can be modeled on a tumor chip. Currently, tumor chips have been widely applied to visualize the dynamic process of tumor growth, infiltration and metastasis [37], providing new insights into human cancer physiology. This review focuses on human solid tumors and outlines microfluidic chips designed to model tumor cell behaviors in tumor evolution. We first described the methods of modeling human solid tumors on microfluidic chips, and then elaborated on how microfluidic chips are employed to demonstrate tumor cell behaviors. Finally, we evaluated the current challenges and discussed the future opportunities of microfluidic chips in tumor research.
Fig. 1.
Advantages of microfluidic chips in tumor research.
2. Engineered solid tumors
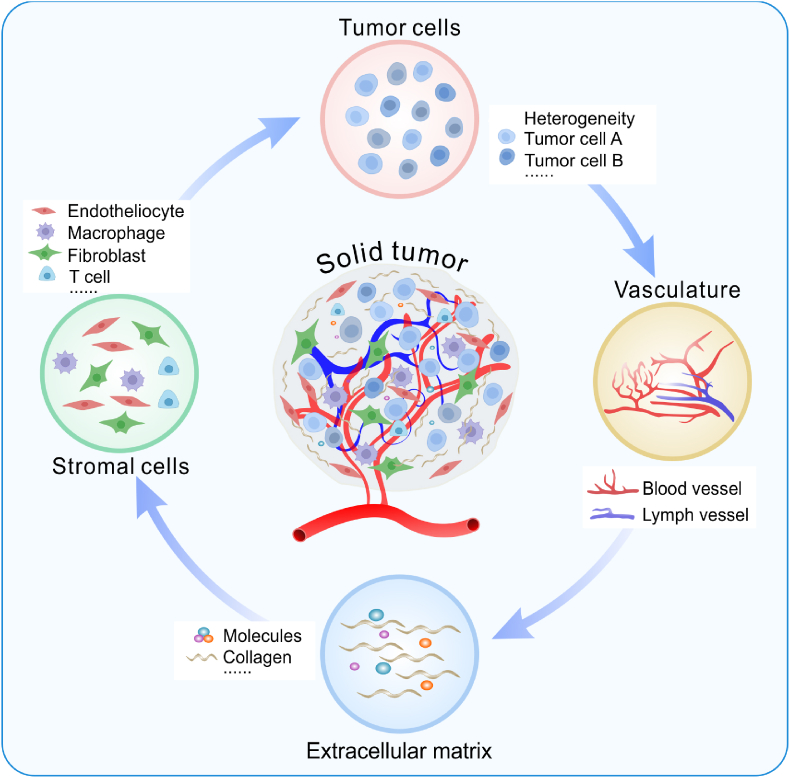
In tumor evolution, the TME influences tumor growth, infiltration, metastasis, drug resistance and recurrence [38,39]. The TME refers to tumor cells and their surrounding microenvironment [40]. In the TME, in addition to heterogeneous tumor cells, there are some stromal cells and acellular components (Fig. 2). Stromal cells in the TME are mainly endothelial cells [41], pericytes [42], fibroblasts [43] and immune cells [44], while the acellular components include blood vessels [45], lymphatic vessels [46], the extracellular matrix (ECM) [47] and some diverse soluble cytokines [48]. To replicate solid tumors in vitro, a physiologically similar TME has been successfully created on microfluidic chips over the past few decades.
Fig. 2.
Schematic of the main components of the TME.
2.1. Construction of on-chip solid tumors
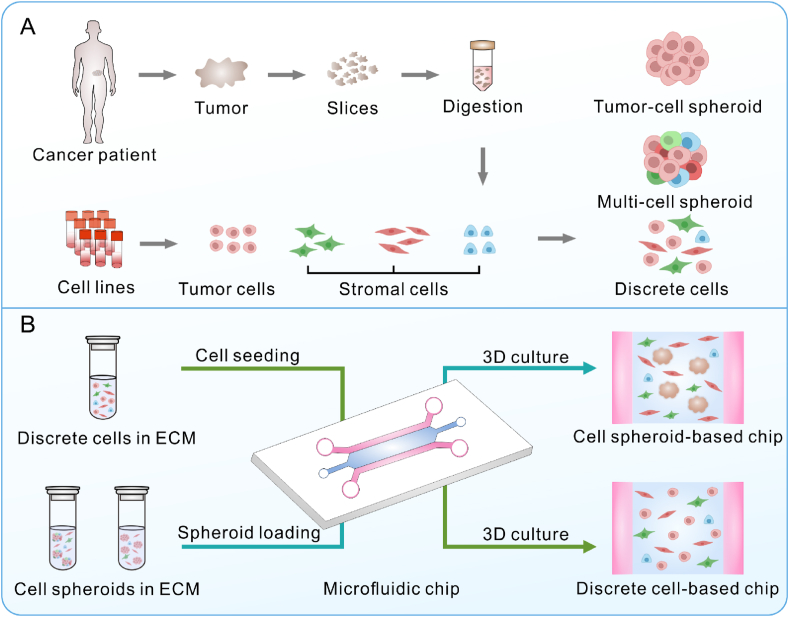
Solid tumors with high incidence, such as lung, breast and liver cancer, are frequently modeled on microfluidic chips [49,50]. Typically, to develop an engineered solid tumor on a chip (Fig. 3), cell lines or primary cells of tumor cells and stromal cells (e.g., endotheliocytes, fibroblasts and macrophages) are obtained from commercial cell banks or isolated from the tumor tissues of cancer patients (Fig. 3A). Discrete tumor cells, stromal cells or multi-cell spheroids of tumor cells and stromal cells are mixed with a biocompatible hydrogel material [51]. The mixture is manually loaded into the tissue culture zone of a microfluidic chip, and the cell culture medium is driven into the chip using gravity pumps, peristaltic pumps or syringe pumps to achieve dynamic long-term culture [52]. During chip culture, the behaviors of tumor cells or stromal cells can be observed or tracked momentarily using a microscope due to the excellent transparency of the chip. In terms of the pattern of the on-chip tumor cells (Fig. 3B), tumor chips can usually be classified into discrete cell-based chips (tumor cells are directly cultured on a chip) [53,54] and cell spheroid-based chips (cell spheroids are often preprepared and cultured on a chip) [[55], [56], [57], [58], [59], [60], [61]].
Fig. 3.
Schematic of developing engineered solid tumors on a microfluidic chip: (A) the cell resource and (B) cell pattern of on-chip tumors.
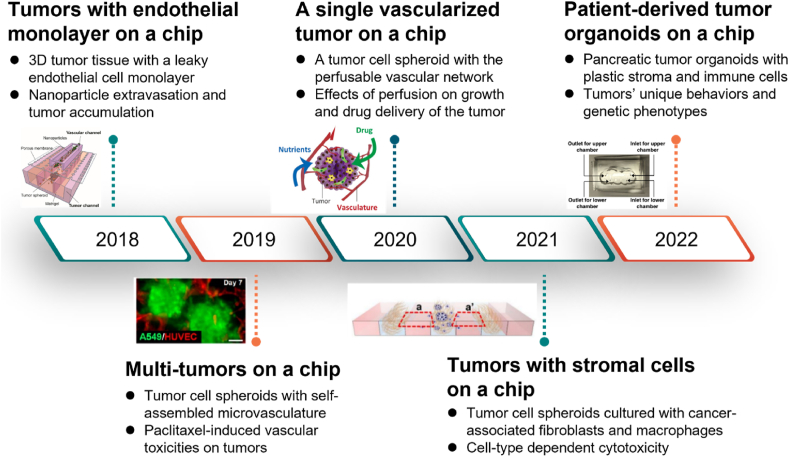
Compared to discrete tumor cell-based chips, cell spheroid-laden tumor chips have an advantage in replicating the spatial architectures, gradients of metabolism and oxygen, and cell distribution of solid tumors [18,62] since the cell spheroids can be prepared with tumor cells and stromal cells, and thus possess a more biomimetic TME. Recently, Yokokawa's group developed a vascularized tumor chip based on cell spheroids of tumor cells and fibroblasts and evaluated the activities of tumor cells after treatment with anticancer drugs on the chip [61]. They found that a vascular network could guarantee long-term perfusion culture of the tumor cell spheroid, enhance the proliferation of tumor cells, and suppress the death of cells in the spheroid [61]. The evolution of typical tumor cell spheroid-based microfluidic chips is shown in Fig. 4.
Fig. 4.
Development of the recent typical cell spheroid-based tumor chips (Reprinted with permission from Ref. [63], Copyright 2018 American Chemical Society; Reprinted with permission from Ref. [64], Copyright 2019 American Chemical Society; Reprinted with permission from Ref. [61], Copyright 2020 Elsevier; Reprinted with permission from Refs. [65,66], Creative Commons By 4.0 (https://creativecommons.org/licenses/by/4.0)).
2.2. Source of tumor cells
The choice of tumor cells for tumor chips varies from the established secondary cancer cell lines to tumor cells extracted from surgically-resected tumor tissues of cancer patients (Fig. 3A). The frequently used tumor cell lines for different tumors on tumor chips are shown in Table 1. Immortalized cancer cells have extensively promoted the understanding of tumor biology [67]. However, it is difficult to comprehend the genetic and epigenetic diversities of different patients using cancer cell lines, and cancer cell lines often exhibit genetic aberrations with increasing passage numbers [68,69]. Patient-derived tumor cells retain native genetic information and tumor heterogeneity, and thus have the potential to replicate patient-dependent tumor variability. It should be noted that the methods of tumor cell dissociation and culture need to be wisely chosen to maintain the parental characteristics of primary tumor cells [70].
Table 1.
A summary of some on-chip engineered solid tumors.
| Tumor type | Cell line | References |
|---|---|---|
| Lung cancer | H1975 cell | [71] |
| SPCA-1 cell | [[72], [73], [74]] | |
| A549 cell | [73,[75], [76], [77], [78]] | |
| NCI–H1437 cell | [79,80] | |
| PC9 cell | [81,82] | |
| Colorectal cancer | HCT-116 cell | [[83], [84], [85], [86]] |
| HT29 cell | [55,[87], [88], [89]] | |
| SW620 cell | [90,91] | |
| Liver cancer | HepG2 cell | [82,83,92,93] |
| C3A cell | [94,95] | |
| Gastric cancer | SGC-7901 cell | [96] |
| Breast cancer | MCF7 cell | [93,[97], [98], [99], [100], [101], [102]] |
| DCIS cell | [57,103] | |
| T47D cell | [58,59] | |
| BT549 cell | [59] | |
| SK-BR-3 cell | [104] | |
| MDA-MB-231 cell | [58,97,102,[104], [105], [106], [107]] | |
| Esophagus cancer | JH-EsoAd1 cell | [108] |
| KYSE-150 cell | [109] | |
| Pancreas cancer | AsPC1 cell | [110] |
| BxPC3 cell | [110,111] | |
| Prostate cancer | PC-3 cell | [[112], [113], [114]] |
| LNCaP cell | [112,114,115] | |
| C4-2 cell | [112] | |
| Cervical cancer | HeLa cell | [56,116] |
| CaSki cell | [117,118] |
2.3. ECM used in tumor replication
The ECM is a 3D acellular network typically composed of collagen, fibronectin, laminin, elastin, polysaccharide, and other proteins [119,120], which provides necessary biochemical and structural support for cellular constituents (Fig. 2). In particular, the structure of the ECM can be remodeled during tumor evolution [121]. The degradation of ECM components, such as collagens and proteoglycans, directly contributes to the migration, invasion and metastasis of tumor cells [122].
To model the ECM of solid tumors, some commercial biomaterials with good biological compatibility and gelatinous capability, such as collagen, Matrigel, GelMA, fibrinogen and Cultrex BME, have been widely employed on tumor chips [123]. Collagen type I is a rich ECM component [120], and it can be obtained from the tendon of the mouse tail [124]. Collagen solution can form hydrogels at a neutral pH [125] (Table 2). Matrigel is a temperature-sensitive hydrogel material containing laminin, type IV collagen, heparan sulfate proteoglycan, nestin and some growth factors, and is extracted from the soluble basement membrane of mice with sarcoma [126]. Matrigel has been proven to promote the adhesion and differentiation of cells, and it has been widely used as the ECM in a variety of 3D cell culture models [127,128]. GelMA is a photosensitive hydrogel biomaterial composed of methacrylate anhydride and gelatin [129]. When mixed with a photoinitiator, GelMA can rapidly crosslink into a 3D hydrogel with a certain strength under blue or ultraviolet light [130]. There are sites in GelMA hydrogel for cell adhesion and matrix metalloproteinase, which can support the proliferation and migration of different cells, such as tumor cells [[131], [132], [133]], endotheliocytes [126,134], cardiomyocytes [135,136] and chondrocytes [114]. Fibrinogen is often sourced from mammals, such as bovines, pigs and goats, and it participates in the process of blood coagulation [137,138]. When mixed with thrombin, fibrinogen can form a 3D hydrogel at room temperature [125]. Bovine fibrinogen is widely used in modeling angiogenesis and self-assembled vascular networks on microfluidic chips due to its good fiber structural support [139,140]. Cultrex BME is another commercially soluble basal membrane material purified from mice with synovial sarcoma [141,142]. Similar to Matrigel [143], it is liquid at 2–8 °C, and polymerizes at 37 °C. In addition to commercial hydrogel biomaterials, decellularized matrix (dECM) prepared in the laboratory after the decellularization of tissues and organs from mammals is also used as the ECM to support cells on microfluidic chips [131,144,145]. The dECM scaffold can preserve the biomolecular profile and composition of the native organ microenvironment where solid tumors arise [146,147]. However, there is still a gap between the reported hydrogel materials and the in vivo ECM of tumors in terms of the stiffness, topography and viscoelastic properties [146,148], and the drawbacks of these materials limit their application. The choice of more biomimetic ECM is still crucial for the construction of on-chip tumors (Table 2).
Table 2.
A summary of the ECM used on tumor chips.
| Category | Gel condition | Concentration range | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| Rat tail collagen type I | At a neutral pH | 1.3–10.0 (mg/mL) | Similar to the ECM of tumors, biodegradability, containing cell adhesive domains, commercially available | Derived from animals, weak stability, monocomponent | [55,[149], [150], [151], [152], [153], [154], [155]] |
| Matrigel | Above room temperature | 2.5%–10% (v/v) | A variety of protein components, biodegradability, commercially available | Derived from animals, undefined compositions, poor mechanical properties | [[156], [157], [158], [159], [160], [161]] |
| Bovine fibrinogen | Mixed with bovine thrombin | 2.5–10.0 (mg/mL) | Promoting the self-assembly of HUVECs, enzymatical degradability, occurring RGD adhesion domains, commercially available | Risks of immunogen and pathogen, monocomponent | [56,140,[162], [163], [164], [165]] |
| GelMA | Exposure to light with the assistance of a photoinitiator | 3%–10% (v/v) | Customized components, easy to pattern, rapid prototyping, good biocompatibility and bioactivity, commercially available | Risk of cell death induced by photoinitiators, lack of microscale pores to accommodate tumor cell migration, difficult to degrade | [133,156,166,167] |
| Cultrex BME | Above room temperature | 2.5%–10% (v/v) | Similar to an early developmental basement membrane, commercially available | Risks of immunogen and pathogen, undefined compositions, poor mechanical properties | [[168], [169], [170]] |
| dECM | Chemical, biological and physical methods | – | Reconstituted native ECM, customized components | Expensive cost, risk of cytotoxicity, require a different protocol to ensure efficient decellularization | [145,155] |
2.4. Stromal cells used in tumor replication
In the TME, the dominant constituent of stromal cells mainly includes fibroblasts, endotheliocytes, pericytes and immune cells. Fibroblasts are often fusiform or spindle-like in shape and can synthesize collagens, fibronectin, laminin and ECM-degrading proteases [171]. The activated fibroblasts associated with tumors are called cancer-associated fibroblasts (CAFs) [172]. Several studies have demonstrated that CAFs can promote tumor evolution and that transforming growth factor-β (TGFβ), platelet-derived growth factor (PDGF) and fibroblast growth factor-2 (FGF2) are crucial mediators of fibroblast activation [171,173]. Endothelial cells and pericytes are crucial for tumor angiogenesis [174,175].
To replicate stromal cells in the TME when developing an engineered tumor on a microfluidic chip, human lung fibroblasts (HLFs) and human umbilical vein endothelial cells (HUVECs) are widely used due to their commercial availability [139]. HLFs are either cultured directly on tumor chips [78,140] or mixed with tumor cells to prepare multicellular spheroids [56,61]. HUVECs derived from the human umbilical cord have the capacity to self-assemble into a vascular network under special culture conditions [139,176,177], and thus, they are frequently employed to construct vascularized tumor chips [60,61,64]. In addition, CAFs [55], pericytes [178], macrophages [179], dendritic cells [180], natural killer cells [181,182] and T cells [183,184] have been loaded on tumor chips to reveal the mechanism of tumor immunity and immunotherapy [185]. However, it remains a challenge to integrate all adaptive stromal cells to build a biomimetic solid tumor on a chip, as it is still difficult to co-culture tumor cells with these primary stromal cells in vitro [177].
2.5. Other components of the TME
There are blood vessels and lymphatic vessels in the TME. Similar to human organs or normal tissues, tumors also need oxygen and nutrients delivered by blood vessels for growth [186]. As early as 1971, Judah Folkman proposed that the cascade events of tumor growth, infiltration and metastasis are angiogenesis dependent [187]. According to this concept of the “angiogenic switch”, tumor progression of solid tumors can be mainly divided into two stages: avascular and vascular growth. At the avascular growth stage, tumors grow slowly, and the tumor diameter is typically within 2 mm [188]. When the oxygen and nutrients from diffusion are no longer sufficient for tumor growth, tumor cells will secrete some biochemical factors, such as hypoxia inducible factor and vascular endothelial growth factor (VEGF), to induce the formation of new blood vessels, namely, tumor angiogenesis [189]. Afterwards, the tumor enters the vascular growth stage and grows quickly, benefiting from the availability of oxygen and nutrients (Fig. 2); that is, in this stage, the tumor spreads into surrounding tissues and even metastasizes to distant sites through the vasculature [188].
Blood vessels not only supply oxygen and nutrients for the growth of solid tumors, but they are also an important pathway for tumor metastasis [190,191]. Lymphatic vessels contribute to the lymphatic invasion and dissemination of tumor cells [192]. The blood vessels of solid tumors are often unorganized and distorted, with variable vessel diameters and abnormal blood flow [193,194]. To form a perfusable vascular network on a microfluidic chip, HUVECs are often mixed with collagen type I or bovine fibrinogen, and then loaded into a chip and dynamically cultured using cell culture medium containing vascular endothelial growth factor (VEGF) [139,140], which promotes the growth of blood vessels. In addition, HFLs were also adopted to support HUVEC morphogenesis and promote angiogenesis through the secretion of some angiogenic growth factors [140]. Yuji Nashimoto et al. presented a tumor-on-a-chip with an engineered tumor vascular network and evaluated tumor activity upon administration of an anticancer drug, thus showing the importance of vascular perfusion in cell proliferation and drug administration [61]. A previous study conducted by our group demonstrated that tumor angiogenesis could promote tumor growth and tumor cell migration in tumor progression [56].
A lymphatic vessel model was developed by culturing lymphatic endothelial cell lines on a microfluidic chip [195]. Immortalized lymphatic endothelial cells were engineered to possess an elongated lifespan with typical cellular morphology, the expression of special protein markers to form a perfusable vessel-like structure, and luminated sprouts were induced by a gradient of lymphangiogenic factors. In another work, a tumor-lymphatic microfluidic model was designed, consisting of a lymphatic endothelial vessel cultured adjacent to breast cancer cells [196]. They found that the vessel growth, permeability, metabolism, hypoxia, and apoptosis of lymphatic endothelial cells were influenced by breast cancer cells. Importantly, these changes in the gene expression of lymphatic endothelial cells were related to the dysfunction of the lymphatic vessels’ barrier. These studies collectively demonstrate the utility of lymphatic vessels in studying tumor evolution.
3. Replication of tumor behaviors
The human body is composed of trillions of cells [197]. When cells mutate and grow abnormally (e.g., unlimited proliferation and loss of contact inhibition), tumors may occur (Fig. 5). It is believed that the formation of solid tumors is similar to the process of human organogenesis built by normal stem cells [198]; that is, cancer stem cells (CSCs) self-renew and differentiate into heterogeneous tumor cells, and then these tumor cells continually proliferate and eventually form a solid tumor.
Fig. 5.
Schematic of the evolution of in vivo solid tumors.
Tumor evolution is the results of a series of dynamic behaviors of tumor cells (Fig. 5). During tumor evolution, tumor behaviors vary with the regulation of the TME. The TME is also constantly changed under the influence of tumor growth and the cytokine secretion from tumor cells. From tumorigenesis to metastasis, the behaviors of solid tumors are the combined effect of tumor cells and their surrounding microenvironment. Scientists and researchers have developed a variety of customized tumor chips with discrete cell patterns and tumor spheroid structures to investigate tumor behaviors in the avascular and vascular stages. The combination of a single-organ chip/multiorgan chip and tumor chips has also emerged, providing a better understanding of the mechanisms of tumor progression.
3.1. Tumor cell proliferation
To study the proliferation of tumor cells affected by the ECM, an automated microfluidic platform was designed to culture breast cancer cell line (T47D cells) with human mammary fibroblasts using three different proteins (i.e., collagen, fibronectin, laminin) [199]. They found that components of the ECM could change the morphology of the T47D cell clusters, the proliferation of T47D cells, and the enzyme expression of several matrix metalloproteinases. In another work, some tumor cell spheroids prepared with human colorectal cancer cells were embedded in collagen hydrogel and indirectly co-cultured with CAFs on a microfluidic chip [55]. Through immunofluorescent staining and analyzing the expression of some proteins, it was concluded that the growth of tumor cell spheroids was positively simulated by CAFs [55]. Similarly, a microfluidic chip with three layers was designed to study the effects of macrophages on the proliferation of tumor cells [58]. Researchers cultured some tumor cell spheroids prepared with different breast cancer cell lines (T47D and MD-MBA-231 cells) on the chip. They found that invasive MD-MBA-231 cells, rather than T47D cells, could polarize monocytes into tumor-associated macrophages (TAMs) and that the presence of TAMs could enhance the proliferation of tumor cells by secreting the cytokine TGF-β1 [58], which was in accordance with the results of an indirect contact culture of breast cancer cells and monocytes on a chip with zigzag signal-blocking channels [200]. Microfluidic chips allow the soluble cell factors secreted from stromal cells to be transported across the microfluidic channel and thus produce local concentration gradients, which can mimic the bidirectional interaction between tumor cells and stromal cells in the TME. Some studies have focused on tumor growth at an early stage. Choi et al. modeled early-stage breast ductal carcinoma in terms of 3D structural organization and the microenvironment of breast cancer on a microfluidic chip with multiple layers by coculturing the cell spheroids of breast cancer cells with human mammary ductal epithelial cells and human mammary fibroblasts [57]. In this study, the enlargement of tumor cell spheroids was observed, and the morphologies of the cell spheroids were not altered by the underlying fibroblasts. The model also examined the contribution of the TME to the progression of breast ductal carcinoma and evaluated the efficacy and toxicity of the drug paclitaxel on tumor growth [57].
Some chemical factors in the TME, such as interferon, interleukin, growth factors, chemokines, tumor necrosis factor (TNF) and transforming growth factor (TGF), are known to mediate the proliferation and chemotaxis of tumor cells [201]. Benefiting from the advantage of microfluidic chips in manipulating trace amounts of liquid, the effects of TNF-α [202,203], IL-1β [204], VEGF [205,206], EGF [101,207], and hypoxic conditions [208,209] on the proliferation of tumor cells have been widely studied. Specifically, a microfluidic tumor slice model was developed to present the behaviors of tumor cells under metabolic starvation gradients [84]. In this study, when tumor cells were exposed to pH gradients and low nutrient conditions, these cells showed multiple changes in the gene expression profile. The cells located further from the media channels upregulated several genes related to stress and survival response and downregulated the genes related to cell proliferation and DNA repair. In summary, the ability of microfluidic chips to directly visualize the biological responses of tumor cells is significant for presenting tumor cell proliferation.
3.2. EMT of tumor cells
Tumor infiltration describes the process by which tumor cells detach from the primary tumor site, cross the structural barriers, including the cell basement membrane and surrounding ECM, and infiltrate into the adjacent tissues of the tumor (Fig. 2) [210]. Tumor infiltration is a prerequisite for the distant metastasis of tumors, and the infiltration degree is regarded as a crucial indicator of malignant tumors. Tumor infiltration is generally achieved through cell migration [211,212]. Two migration patterns of tumor cells have been reported: individual migration and collective migration. For individual migration [213,214], a single tumor cell penetrates the ECM through epithelial-mesenchymal transition (EMT), which is the transition of tumor cells from an epithelial state to a mesenchymal state, facilitating the invasion of tumor cells into surrounding zones or blood vessels [212]. For collective migration, a single tumor cell leads other tumor cells in a manner of narrow lines or clusters, which refers to tumor tissue pushing forward as a whole and displacing the surrounding cells [215].
To in vitro capture the dynamic EMT process of tumor cells, tumor cells are often labeled with green/red fluorescent protein or pre-stained with a cell tracker. The morphology change of human lung cancer cells (A549 cell line) in EMT was observed on a microfluidic chip [216]. On the chip, EMT of A549 cells was induced by TGF-β1 and negligible shear stress. It was clearly observed that A549 cells elongated with typical spindle-shaped morphological changes under a high TGF-β1 concentration, suggesting that the chip is a powerful tool for monitoring EMT induced by a variety of chemical factors. A multichannel microfluidic chip was developed to evaluate the contribution of flow-induced hydrodynamic shear stress on EMT [217]. Morphological changes in A549 cells occurred under fluidic culture conditions. In particular, the dynamic process of EMT in HeLa cells was for the first time presented on a microfluidic chip through the direct-contact coculture of HUVECs and a single tumor spheroid prepared with HeLa cells and HLFs [56]. Microfluidic chips have also been used to detect the expression of EMT-related proteins. EMT is classically characterized by changes in the protein expression of E-cadherin and N-cadherin proteins since E-cadherin is mainly expressed in epithelial cells, whereas N-cadherin is upregulated in mesenchymal cells [218,219]. The soluble active fragments of the E-cadherin and N-cadherin proteins were reported to be biomarkers for cancer diagnosis and prognosis [220]. A microfluidic chip was developed to simultaneously detect the soluble E-cadherin and N-cadherin proteins for EMT monitoring [221]. The chip can accurately detect these proteins at concentrations as low as 10 cells/mL, potentially providing early indications of cancer invasion and metastasis to guide cancer treatment management. More recently, a microfluidic chip integrated with a micromixer was designed to exploit the EMT index based on tumor-derived extracellular vesicles [222]. Using this chip, over 90% of extracellular vesicles expressing the EMT index (i.e., epithelial and mesenchymal biomarkers) can be selectively isolated from plasma samples of breast cancer patients. Compared with healthy people, the EMT index is significantly higher in patients with aggressive breast cancer subtypes. In addition, patients with a high EMT index showed recurrence within 5 years after adjuvant treatment [222].
3.3. Tumor cell migration
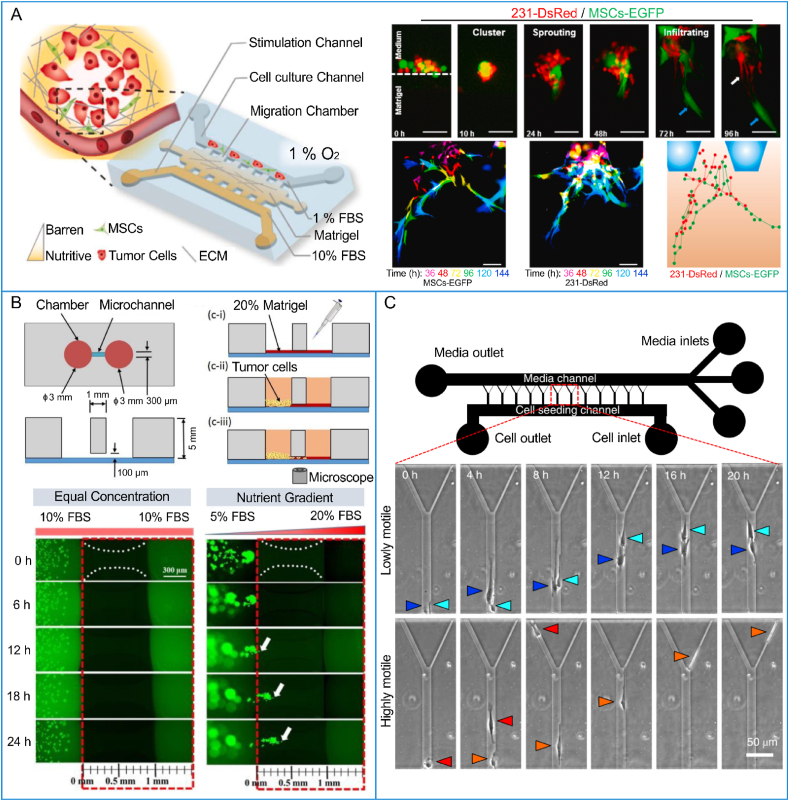
Tumor cell migration refers to the morphological transition and locomotion of tumor cells [223]. The migration capability of tumor cells varies with their type. In one study, breast cancer cells with mild, moderate and severe phenotypes were modeled on a microfluidic chip [105]. It was found that the migration ability of severe breast cancer cells (MDA-MB-231) increased with the secretion of IL-6 by breast cancer cells [105]. Furthermore, IL-6 contributes to the invasiveness of breast cancer cells [106]. Stromal cells in the TME, such as endothelial cells, fibroblasts and macrophages, also regulate the aggressiveness of tumors even at the early stage of tumor evolution [43,224,225]. On a vascularized tumor chip, when HeLa cells were co-cultured with endothelial cells and fibroblasts, they tended to migrate more [56]. By analysis of the paracrine loop between lung cancer cells and fibroblasts on a microfluidic chip, it was found that the cytokines secreted from lung cancer cells stimulated the fibroblasts into myofibroblasts, and that TGF-β1 secreted from myofibroblasts promoted the migration speed of lung cancer cells [226]. Indeed, macrophages and fibroblasts have been proven to produce synergistic effects in accelerating the migration of lung cancer cells [227]. In addition, the migration of MDA-MB-231 cells mediated by mesenchymal stem cells (MSCs) was recently monitored on a microfluidic-based 3D co-culture device [228]. It was observed that MSCs guided cancer cell migration in a “cluster-sprout-infiltrating” mode (Fig. 6A), and the migration of both MDA-MB-231 cells and MSCs was significantly accelerated under nutrient-deficient hypoxic culture conditions. A microfluidic plate containing 96 chambers connected by a microchannel was developed for visualizing and quantifying the migration of HeLa cells [229]. In the experiments, the microchannel was perfused with Matrigel, and HeLa cells migrated along the Matrigel-filled microchannel under presupposed stimulation. By visualizing the process of cell migration and counting the migration speed [229], researchers found that HeLa cells migrated toward the chamber with a high nutrient concentration (high fetal bovine serum in cell culture medium), indicating that tumor cell migration is nutrition dependent (Fig. 6B). Yuan et al. adopted collagen hydrogel as the ECM of breast tumors on a microfluidic chip and presented the migration of MD-MBA-231 cells [58]. They observed that tumor cell migration decreased when the concentration and stiffness of collagen-ECM increased. In addition, it was also found that cell spheroids of T47D and MDA-MB-231 cells could cause monocytes to polarize into tumor-associated macrophages, and the migration ability of T47D and MDA-MB-231 cells was enhanced by these macrophages. The bidirectional crosstalk between tumor cells and macrophages was presented on another microfluidic chip [230]. The invasion of breast cancer cells was promoted by tumor-associated macrophages while the phenotype of macrophages was maintained by cancer cells. Microfluidic chips aimed at generating an oxygen gradient have also been used to reveal the effects of oxygen concentration on tumor cell migration [118,208,231]. Tumor cells showed a migration directionality toward higher oxygen [231].
Fig. 6.
The migration of tumor cells on microfluidic chips: (A) migration of MDA-MB-231 cells (red) mediated by MSCs (green) under hypoxic conditions on a microfluidic device (Reprinted with permission from Ref. [228], Copyright 2022 Elsevier), (B) nutrition-induced migration of HeLa cells (Reprinted with permission from Ref. [229], Copyright 2019 American Chemical Society), and (C) representative time-lapse images of human glioblastoma cells migrating in a microfluidic channel (Reprinted with permission from Ref. [232], Copyright 2021 Spring Nature). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The quantification of the migration of primary glioblastoma cells to predict progression-free survival and recurrence time of glioblastoma patients was conducted on a microfluidic chip (Fig. 6C) [232]. In this study, the widths of the microchannels on the chip ranged from 3 μm to 20 μm. Glioblastoma cells preferentially entered the narrower 3-μm-wide branches of microchannels and were called highly motile cells. Based on live-cell imaging, researchers calculated the percentage of these highly motile cells and analyzed the ability of cells to deform their cytoskeleton and migrate in confining microenvironments. The ability of glioblastoma cell migration was correlated with its aggressiveness and invasiveness [232], suggesting that cell migration and proliferation levels can predict patient-specific clinical outcomes.
3.4. Intravasation of tumor cells
Tumor metastasis is the main cause of cancer mortality in humans. It is a cascade of steps involved in the infiltration of tumor cells into blood vessels and/or lymphatics and the colonization of distant organs/tissues (Fig. 2) [9,10]. The tumor cells that enter blood vessels or lymph vessels are known as circulating tumor cells (CTCs), a few of which can escape the monitoring of host immune cells [52]. After CTCs arrive at a new organ/tissue, they proliferate and form a metastatic tumor [52]. Blood vessels are a crucial barrier to tumor metastasis. A perfusable vascular network was typically constructed on a microfluidic chip based on the spatially controlled 3D co-culture of HUVECs with fibroblasts and pericytes [140]. On the chip, the developed blood vessels possessed the characteristic morphology and biochemical markers compared with in vivo blood vessels and exhibited strong barrier function and long-term stability. This study provides an on-chip angiogenesis method for vascularized organs and tumors. Lee's group presented the process of vasculogenesis, sprouting angiogenesis, and anastomosis in sequential order, which simulated the interconnected perfused vessels from artery to vascularized tissue to vein on a microfluidic chip [139]. The formed perfusable microvascular network connected to the microfluidic channels without appreciable leakage enables the physiological vascular interconnection of multiple tissue constructs on microfluidic chips. The construction of on-chip perfusable blood vessels paves the way for the study of tumor metastasis.
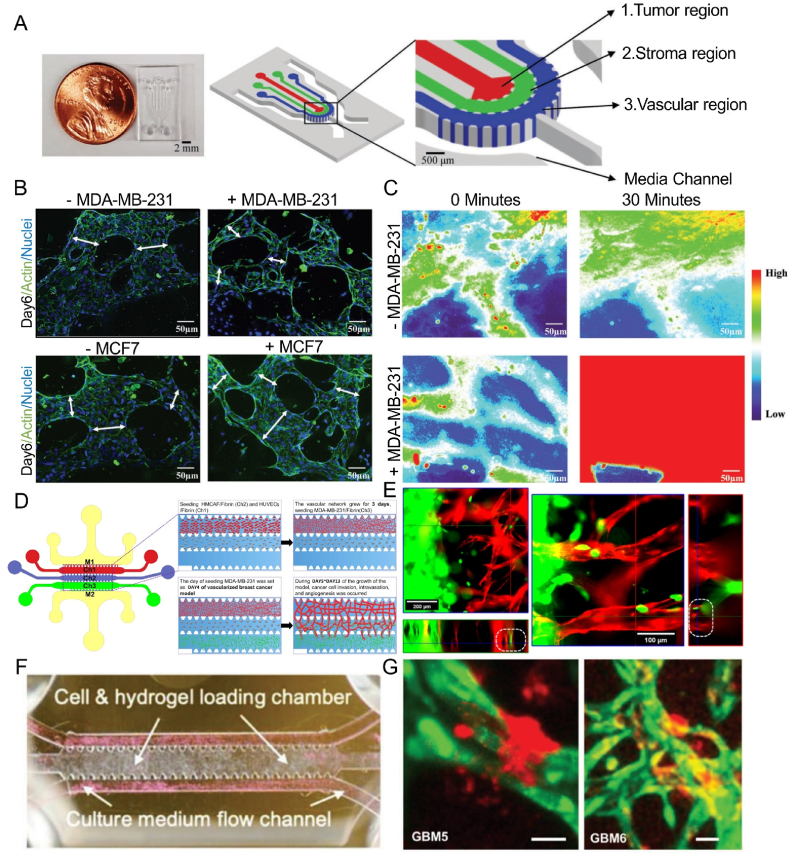
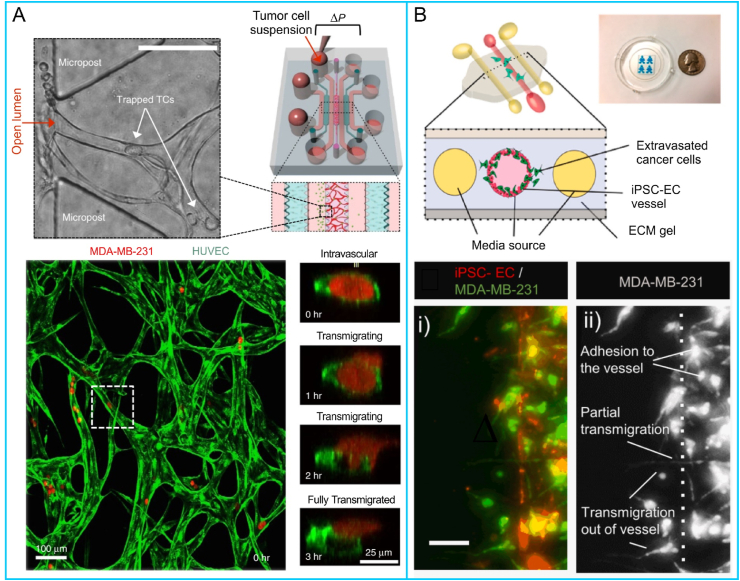
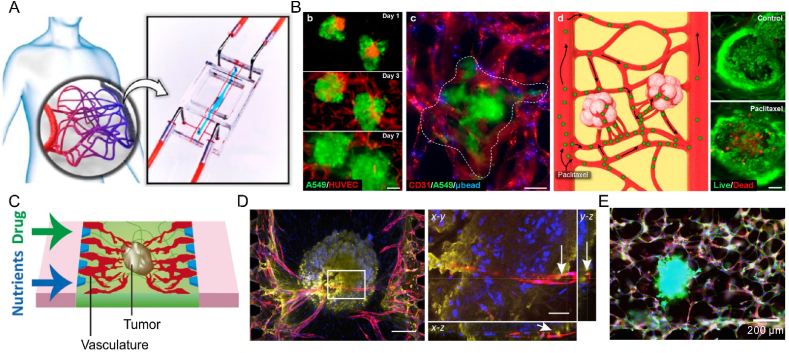
To simultaneously investigate the invasion and intravasation of breast cancer cells, a microfluidic chip composed of three cell-laden hydrogel channels was designed (Fig. 7A) [97]. The intravasation of breast cancer cells into blood vessels following their invasion into the ECM was visualized (Fig. 7B). The presence of the vasculature enhanced the invasion of MDA-MB-231 cells into the stroma. The diameters of blood vessels were significantly reduced after the intravasation of the tumor cells. In contrast, the permeability of blood vessels increased (Fig. 7C), which was consistent with previous in vivo studies. This study provided unique insights into intravasation events of the metastatic cascade. Zhang et al. constructed a self-assembled microvascular network on a microfluidic chip (Fig. 7D) [233], which could be maintained for up to 13 days with the assistance of fibroblasts, and the invasion ability and intravasation behavior of breast cancer cells were observed (Fig. 7E). Several key steps of tumor metastasis in blood vessels were recapitulated on a microfluidic vessel-on-a-chip platform [234]. A cluster of PC3 cells was seeded on the chip to mimic the primary tumor. Cell culture medium was perfused through vessel lumens to induce tumor cell migration. A few PC3 cells gradually shed from the PC3 cell cluster and migrated toward the vessels. After a period of migration, some tumor cells intravasated into the vessel. In another study, the clustered CTCs of breast cancer cells displayed a higher shedding rate and metastasis formation, and tumor metastasis could be restrained through prevention of CTC cluster generation [235]. In addition, a tissue-engineered microvasculature-on-a-chip system was developed (Fig. 7F) [236], and the dynamic microvessel colocalization of brain tumor stem-like cells from glioblastoma patients was demonstrated on a microvasculature chip (Fig. 7G). These platforms provide new ideas for the study of tumor metastasis.
Fig. 7.
Intravasation of tumor cells on microfluidic chips: (A) the real picture and the structure of the chip, (B) fluorescent images of the vascular network and (C) permeability of blood vessels after intravasation of breast cancer cells (Reprinted with permission from Ref. [97], Copyright 2018 Wiley); (D) chip structure and (E) confocal images of breast cancer cells intravasated into blood vessels (Reprinted with permission from Ref. [233], Copyright 2022 AIP Publishing); (F) the cell-laden chip and (G) tumor cells derived from glioblastoma patients in the microvasculature (Reprinted with permission from Ref. [236], Creative Commons By 4.0 (https://creativecommons.org/licenses/by/4.0)).
3.5. Extravasation of tumor cells
Some tumors tend to metastasize to specific sites; for example, breast cancer cells preferentially metastasize to the lung, liver, and bone [237,238]. Studies have shown that tumor metastasis is not entirely determined by the TME of the primary tumor location but is also closely related to the microenvironment of the metastatic site [38,239]. Organ-specific tumor cell extravasation has been modeled on microfluidic chips [53,240]. Jeon et al. developed a microfluidic model with a perfusable microvascular network to analyze the extravasation rate of human breast cancer cells and the permeability of microvasculature after tumor extravasation [53]. They found that a higher extravasation rate of cancer cells was achieved after A3 adenosine receptor was blocked. Chen et al. described a detailed protocol for how to recapitulate the discrete cascades of early metastatic sites of breast cancer on a microfluidic chip [214]. The extravasation events were tracked over 72 h via fluorescence confocal microscopy (Fig. 8A). Compared with most in vivo extravasation assays, microfluidic chips integrated with a high-resolution imaging system allow the visualization of the morphological dynamics of both tumor cells and endothelial cells. An organotypic microfluidic model was developed to investigate the interactions between human breast cancer cells and vasculature generated from induced pluripotent stem cells [240]. On the chip, the dynamic extravasation behavior of breast cancer cells and cancer-vascular interactions were recorded using a microscope (Fig. 8B). In addition, the secretion of IL-6, IL-8 and MMP-3 increased, especially when breast cancer cells were of a highly invasive phenotype. The molecular mechanism of tumor cell extravasation was investigated on microfluidic chips. Gilardi et al. exploited a vascularized microfluidic chip to study the role of the Cdk5/Tln1/FAK axis in the extravasation of breast cancer cells [241]. The integrated results showed that the phosphorylation of FAK serine 732 was a promising target of the metastatic cascade. In addition, the upregulation of cancer-vascular paracrine signaling, such as IL-6, IL-8 and MMP-3, promoted the extravasation of breast cancer cells [240], as well as the early metastatic niche [242].
Fig. 8.
On-chip visualization of the dynamic extravasation process of tumor cells: (A) the extravasation behaviors of breast cancer cells (Reprinted with permission from Ref. [214], Copyright 2017 Spring Nature), and (B) cancer-vascular interactions (Reprinted with permission from Ref. [240], Copyright 2021 Elsevier).
The construction of vascularized tumor-spheroid-on-a-chip models is an alternative approach to study tumor metastasis. Recently, some microfluidic chips with vascularized tumor spheroids have been reported [64]. For example, some multicellular spheroids of lung cancer cells were co-cultured with HUVECs and HFLs on a microfluidic chip to develop a vascularized lung tumor (Fig. 9A and B) [64], a single pre-vascularized multicellular spheroid of breast cancer cells was seeded on a chip to achieve perfusable blood vessels (Fig. 9C and D) [61], and a single multicellular spheroid of HeLa cells and HFLs was on-chip cultured with HUVECs to model early-stage tumor angiogenesis (Fig. 9E) [56]. Jeon and his colleagues adopted a 3D printer to create a cell spheroid culture platform based on a standardized 96-well plate with a user-friendly interface [60]. Based on the culture platform, some events of vasculogenesis, tumor infiltration and tumor angiogenesis were modeled [60]. The approach presented in this study provides a new idea to model human solid tumors as well as the tumor metastasis process. In addition, since solid tumors have a leaky vasculature, the characteristics of the on-chip blood vessels may affect the extravasation process of tumor cells. Therefore, preparation of pre-vascularized tumor spheroids, customization of the structures of microfluidic chips, optimization of the vascular structure and adjustment of fluid flow provide avenues to model the behaviors of tumor metastasis on-chip.
Fig. 9.
Vascularized tumor-spheroid-on-a-chip models: (A) structures of the chip and (B) tumor spheroids of human lung adenocarcinoma cells with a perfusable vascular network on the chip (Reprinted with permission from Ref. [64], Copyright 2019 Elsevier); (C) schematic of the chip and (D) tumor spheroids of human breast cancer cells, HUVECs and fibroblasts with the vasculature (Reprinted with permission from Ref. [61], Copyright 2020 Elsevier); (E) a single vascularized tumor spheroid prepared with human cervix cancer cells and fibroblasts (Reprinted with permission from Ref. [56], Copyright 2021 American Chemical Society).
3.6. Immune escape of tumor cells
Solid tumors often possess an immunosuppressive microenvironment that imposes an overwhelming burden on the immune system. Microfluidic chips are a novel platform to reveal tumor-immune crosstalk and present the behaviors of the immune escape of tumor cells. A microfluidic device mimicking nutrient, pH, proliferation, and necrosis gradients of breast cancer was designed to evaluate NK cell biology [181]. In this study, 1.5 million MCF7 cells/mL were seeded on the device to induce an immunosuppressive environment, and the interaction of NK-92 cells and MCF7 cells was observed. The cytotoxicity of NK cells decreased in the tumor-induced suppressive microenvironment. After NK cells were removed from the device, NK cell exhaustion remained for a period. Checkpoint inhibitors and immunomodulatory agents can alleviate NK cell exhaustion. Pavesi et al. reported a customizable microfluidic platform [243], in which human cancer hepatocytes were cultured as a single cell or tumor cell aggregates under different oxygen levels, as well as human T cells engineered to express tumor-specific T-cell receptors (TCR–T cells). In the presence of inflammatory cytokines, the ability of TCR–T cells to migrate and kill the tumor target and the profile of soluble factors were investigated. They concluded that tumor growth was related to the function of TCR–T cells, oxygen levels and inflammatory environment that affected the function of TCR–T cells. For tracking the behavior of dendritic cells (DCs) toward tumor cells, a microfluidic device mimicking the TME of colorectal cancer and immune systems with advanced microscopy was described [91]. Interferon-α-conditioned DCs tended to migrate toward drug-treated cancer cells, largely guided by the CXCR4/CCL12 axis. Notably, IFN-DCs were found to modify their migration to recognize tumor cells and take up tumor antigens. Overall, microfluidic chips allow crosstalk between tumor cells and immune cells and thus possess the potential to visually reveal the mechanism of immune escape of tumor cells during tumor development.
4. Challenges
Tumor cells become more aggressive, and metastasis often occurs during tumor progression. It remains a challenge to capture the dynamic behaviors of tumor cells from tumorigenesis to tumor metastasis due to the complexity and variability of tumor evolution. Advances in microfluidic technologies and tissue engineering have made tumor chips an attractive visualized platform for demonstrating tumor cell behaviors from tumor growth to infiltration to metastasis. However, some obstacles remain in fully replicating human solid tumors on chips.
Currently, some solid tumors with high mortality, such as lung, breast, colorectal and liver cancer, have been widely modeled on microfluidic chips based on the diverse sources of related tumor cells. Since intrinsic heterogeneity exists in solid tumors [244], tumor cell lines are not optimal for replicating tumor heterogeneity. With the development of the isolation of primary cells from cancer patients, more types of heterogeneous tumors can be expected to be modeled on chips. To mimic the structure of solid tumors, monocellular tumor spheroids (prepared with tumor cells only) and multicellular tumor spheroids (prepared with tumor cells and stromal cells) have been widely generated using different methods [245,246]. To facilitate the study of the cellular behaviors of solid tumors, cells within tumor spheroids should not be restricted to a confined space (e.g., cell spheroids prepared with calcium alginate hydrogel), and the self-assembly of tumor spheroids with multiple cell types is recommended. The size and cell types in tumor spheroids also need to be considered when using them to study different stages during tumor progression.
The tumoral ECM is constantly reshaped during tumor evolution, and its variable structure often affects tumor behaviors such as the migration and infiltration of tumor cells [247]. The ECM is composed of multicomponent proteins; however, the current on-chip ECM is simple in composition and far from the in vivo ECM of tumors. In the future, new matrix biomaterials with good biocompatibility need to be explored. Most solid tumors contain regions of hypoxia because tumor cells are typically farther from the nearest capillary than cells in normal tissues, the blood vessels of solid tumors are abnormal, and hypoxic regions of tumors are likely to have a decreased supply of nutrients such as glucose and essential amino acids, leading to low interstitial pH [39]. Therefore, blood vessels are prominent for on-chip tumors. The engineered vascular network has been achieved on microfluidic chips; however, perfusable blood vessels for an on-chip solid tumor are still a challenge. The construction of the tumor spheroids with inner biomimetic ECM may facilitate the penetration of new blood vessels into the spheroids and the formation of a perfusable vascular network. In addition, the fabrication materials of microfluidic chips remain to be optimized to meet the demands of the hypoxic microenvironment of human solid tumors.
Immune cells play both tumor-promoting and tumor-inhibiting roles in tumor evolution [248]. Despite the dynamics of tumor-immune interactions, such as the tumor killing of tumor-infiltrating lymphocytes [249,250], the cytotoxicity of NK cells [251] and the bidirectional crosstalk between tumor cells and macrophages [230], have been examined on tumor chips [164,252], the incorporation of various types of immune cells on a tumor chip has been a big challenge due to the complexity of human immune system. Long-term maintenance of the function of immune cells on chips still needs further exploration especially when immune cells are cocultured with other cells. In the light of the importance of the immune cells in tumor evolution, if immune components are fully integrated into tumor chips successfully, the promotion on the mechanism discovery of tumor progression and the developments of tumor vaccines and immunotherapy will be greatly expected.
5. Opportunities
Rapid technological advances have facilitated the development of tumor organoids [[253], [254], [255]]. Tumor organoids are spherical, 3D cell clusters cultured on an ultralow attachment surface based on cells derived from patient cancer tissues (Table 3) [256]. Biopsies or surgically resected tumor tissues are cut and digested to obtain cells. The cells are mixed with Matrigel material and cultured in the customized cell culture medium using cell culture plates, allowing the cells to self-assemble into organoids [257]. In recent years, some living biobanks of tumor organoids have been established, such as breast cancer organoids [258] and colorectal cancer organoids [259]. Compared to engineered tumors on microfluidic chips, tumor organoids preserve characteristics of the original tumors in cancer patients in terms of cell phenotype, genetic composition, morphological and mutational signatures [260,261], which is of significance for researchers to dissect tumor biology on a personalized level [[262], [263], [264]], as well as precision medicine for patients [265,266]. Recently, the initiation and growth of breast cancer [267], EMT of breast cancer cells [268] and T-cell-mediated cytotoxicity in breast cancer have been reported based on tumor organoid platforms [269]. Additionally, organoids of colorectal cancer [270], gastric cancer [255] and liver cancer [271] have been developed to analyze tumor genomic evolution, oncogenic pathogens related to tumor origin, and the interactions between cancer genotypes and phenotypes [272].
Table 3.
A summary of in vitro tumor models for tumor evolution.
| Category | Modeling methods | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Monolayer culture | Cells are cultured on cell culture plates/dishes/bottles or glass | Cost-efficient, simple to use, high throughput | Lack of the ECM, the spatial arrangement of tumors and cellular interactions | [18,273] |
| Tumor spheroids | Cells are cultured on the ultralow attachment surfaces to form cell aggregation | Similar to the architecture of solid tumors, it produces gradients of oxygen and nutrition and preserves cellular interactions | Lack of biomimetic mechanical forces, dynamic fluid flow and blood vessels | [28,65,245] |
| Tumor chips | Cells or cell spheroids are mixed with biocompatible hydrogels and cultured on a chip | Easy to control the TME, fluid flow and mechanical forces, dynamic cell-cell and cell-ECM interactions, in situ monitoring tumors, easy to integrate microscopic imaging and sensing platforms | Lack of hypoxic microenvironment and tumor heterogeneity | [72,73,85,274] |
| Tumor organoids | Cells from cancer patients are mixed with Matrigel hydrogel and cultured on the ultralow attachment surfaces | Preserve cell phenotypes, genetic diversity and heterogeneity of the original tumor, produce gradients of oxygen and nutrition | Stochasticity and variability, fail to long-term tumor organoid cultures, absence of immune cells and perfusable blood vessels | [260,275,276,277] |
| Tumor organoid chips | Tumor organoids are cultured on a microfluidic chip | Preserve genetic diversity and heterogeneity of the original tumors, produce gradients of oxygen and nutrition, fluid flow and mechanical forces, have a potential for perfusable blood vessels | Need to customize chip pattern, weak in reliability, robustness and consistency | [253,278,279] |
Despite some advantages of tumor organoids, certain obstacles still exist. It remains a challenge to develop a highly vascularized tumor organoid. Due to the lack of perfusable blood vessels, oxygen and nutrients hardly diffuse into the interior of tumor organoids as they grow larger [280], leading to the loss of cell viability at the inner core of organoids [275]. Some efforts have been made to promote the vascularization of tumor organoids [278,281]. However, a biomimetic perfusable vascular network of tumor organoids has still not been achieved [282]. In addition, there is no fluid flow within tumor organoids. These drawbacks hamper the application of tumor organoids in the modeling of tumor metastasis, as well as precision in drug testing and screening.
The combination of organoid tumor technology and organ chip technology to develop organoid-on-a-chip or organoid chips opens up new frontiers of tumor chips. The synergistic integration of these two technologies can be expected to bridge the enormous gap between engineered and in vivo solid tumors [254,269], thus broadening the deployments of tumor chips in tumor research (Fig. 10A). By culturing tumor organoids in microfluidic chips (Fig. 10B), the lack of perfusable vasculatures and biomimetic fluid flow in tumor organoids may be overcome. Furthermore, some innate immune populations that are preserved in tumor organoids [261] may improve the immune microenvironment of tumor chips and thus promote the application of tumor chips in immunotherapy. The human colon tumor organoid-on-a-chip model mimicking peristalsis was recently developed by culturing human colon tumor organoids on a microfluidic chip [283]. The microfluidic chip was composed of hundreds of semi-open microwells interconnected by a channel for medium flow, which can supply nutrition/oxygen and remove waste, and a surrounding parallel pressure channel providing rhythmic contraction and relaxation. Decreased drug uptake and anti-tumor efficiency of colon tumor cells were observed on the chip. The advancement of the human body-on-chips also promotes the development of tumor organoid chips (Fig. 10C) [284]. However, efforts still need to be made to design more physiologically similar tumor organoid chips.
Fig. 10.
Tumor organoid chips and organ-chip system: (A) microfluidic platform for tumor-organoid culture (Reprinted with permission from Ref. [279], Creative Commons By 4.0 (https://creativecommons.org/licenses/by/4.0)), (B) human pancreatic tumor organoid-on-a-chip model (Reprinted with permission from Ref. [157], Copyright 2020 Wiley), and (C) multi-organ human-body-on-chips (Reprinted with permission from Ref. [284], Copyright 2020 Spring Nature).
6. Summary and prospects
After a decade of development, there have been many reports of microfluidic chips with multicellular tumor spheroids and discrete tumor cells, allowing the visualization of tumor behaviors in tumor evolution, resulting in different implementations and a surge in the understanding of tumor progression. Here, we review the most recent advances in modeling solid tumors on microfluidic chips. This includes the construction of engineered tumors and the on-chip modeling of the TME, such as the ECM, stromal cells, blood vessels and lymph vessels, outlining stages in tumor evolution and discussing certain characteristics of tumor cell behaviors in different stages. We also focus on replicating tumor cell behaviors on microfluidic chips, such as proliferation, EMT, migration, intravasation, extravasation and immune escape. These advancements have led to the creation of tumor chips with biomimetic structure and broad application, enhancing the benefits of demonstrating how a single cell behaves in tumor progression and treatment. Additionally, we described recent advances in tumor-organoid chips.
Tumor chips have seen tremendous progress since the inception of organ-on-a-chip, but many challenges and opportunities still lie ahead. Almost all tumor chips can precisely control the biochemical and physical microenvironments of solid tumors. Few can replicate the tumor heterogeneity and multi-tissue interfaces because complicated dynamic interactions regulate tumor cell behaviors. To encourage the adoption of tumor chips as more versatile and predictive preclinical platforms for efficient and safe drug discovery and personalized medicine, more future research should be directed toward tumor organoid chips. Since tumor organoids preserve genetic and phenotypical stability and tumor heterogeneity, the combination of tumor organoids and microfluidic chips facilitates the capture of tumor-specific genetic alterations and histopathology, visualizing the behaviors of patient-derived tumor cells, and discovering new spatiotemporal dynamics of tumor evolution. Then, more mechanisms of tumor evolution can be uncovered based on tumor organoid chips and new targets for tumor therapy may be developed. In practice, tumor organoid chips can be constructed using the patient-derived tumor tissues and thus, the personalized medication advice for cancer patients can be reached.
We believe that combining organoids and microfluidic chips, along with integrating biosensors and electronic systems into tumor-organoids-on-chip platforms coupled with handled real-time imaging devices, will revolutionize tumor chips. Definitely, the announcement of the Food and Drug Administration (FDA) Modernization Act on alternative in vitro models is of great benefit to translate tumor chips to the clinic and drug development industry [285]. However, standards of organ chips still need to be developed. It is foreseeable that with the advancement of standardization and industrialization of organ chips [286], the application fields of organoid/organ-on-chip models will be greatly extended. In addition, in the future, the integration of tumor chip systems to the health/medical network with the advances made in other fields of science and technology, such as artificial intelligence and big data management strategies [287,288], may accelerate the innovative development and clinical application of tumor chip platforms in revealing the mechanism of tumor evolution and exploiting new strategies for tumor treatment.
Credit author statement
C. L. collected the literature, drew the figures, and wrote the paper. J. H. assisted with the paper writing. Z. S. assisted with the literature collection. B. Q. supervised the study and provided some suggestions. W. D. conceived the review, designed the structure, and revised the paper. All authors discussed the paper and approved the submission.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was partially supported by the China Postdoctoral Science Foundation (2022M713055), the Anhui Provincial Natural Science Foundation (2208085QH256), the National Natural Science Foundation of China (82072018), and the Key Research and Development Program of Anhui Province (2022e07020012).
Contributor Information
Bensheng Qiu, Email: bqiu@ustc.edu.cn.
Weiping Ding, Email: wpdings@ustc.edu.cn.
Data availability
No data was used for the research described in the article.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA-A Cancer. J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Moore P.S., Chang Y.A. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiffman M., Doorbar J., Wentzensen N., de Sanjose S., Fakhry C., Monk B.J., Stanley M.A., Franceschi S. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Prim. 2016;2 doi: 10.1038/Nrdp.2016.86. [DOI] [PubMed] [Google Scholar]
- 4.Poirier M.C. Chemical-induced DNA damage and human cancer risk. Nat. Rev. Cancer. 2004;4:630–637. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- 5.D'Orazio J., Jarrett S., Amaro-Ortiz A., Scott T. UV radiation and the skin. Int. J. Mol. Sci. 2013;14:12222–12248. doi: 10.3390/ijms140612222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hecht S.S. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat. Rev. Cancer. 2004;4:733–744. doi: 10.1038/nrc1272. [DOI] [PubMed] [Google Scholar]
- 7.Fukumura D., Jain R.K. Tumor microenvironment abnormalities: causes, consequences, and strategies to normalize. J. Cell. Biochem. 2007;101:937–949. doi: 10.1002/jcb.21187. [DOI] [PubMed] [Google Scholar]
- 8.Wang M., Zhao J., Zhang L., Wei F., Lian Y., Wu Y., Gong Z., Zhang S., Zhou J., Cao K. Role of tumor microenvironment in tumorigenesis. J. Cancer. 2017;8:761. doi: 10.7150/jca.17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yilmaz M., Christofori G., Lehembre F. Distinct mechanisms of tumor invasion and metastasis. Trends Mol. Med. 2007;13:535–541. doi: 10.1016/j.molmed.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 11.Czekay R.P., Cheon D.J., Samarakoon R., Kutz S.M., Higgins P.J. Cancer-associated fibroblasts: mechanisms of tumor progression and novel therapeutic targets. Cancers. 2022;14:1231. doi: 10.3390/Cancers14051231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vitale I., Shema E., Loi S., Galluzzi L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021;27:212–224. doi: 10.1038/s41591-021-01233-9. [DOI] [PubMed] [Google Scholar]
- 13.Yousefi H., Vatanmakanian M., Mahdiannasser M., Mashouri L., Alahari N.V., Monjezi M.R., Ilbeigi S., Alahari S.K. Understanding the role of integrins in breast cancer invasion, metastasis, angiogenesis, and drug resistance. Oncogene. 2021;40:1043–1063. doi: 10.1038/s41388-020-01588-2. [DOI] [PubMed] [Google Scholar]
- 14.Khalaf K., Hana D., Chou J.T.T., Singh C., Mackiewicz A., Kaczmarek M. Aspects of the tumor microenvironment involved in immune resistance and drug resistance. Front. Immunol. 2021;12 doi: 10.3389/Fimmu.2021.656364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruggeri B.A., Camp F., Miknyoczki S. Animal models of disease: pre-clinical animal models of cancer and their applications and utility in drug discovery. Biochem. Pharmacol. 2014;87:150–161. doi: 10.1016/j.bcp.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Mironova N., Shklyaeva O., Andreeva E., Popova N., Kaledin V., Nikolin V., Vlassov V., Zenkova M. Animal model of drug-resistant tumor progression. Ann. Ny. Acad. Sci. 2006;1091:490–500. doi: 10.1196/annals.1378.090. [DOI] [PubMed] [Google Scholar]
- 17.Gould S.E., Junttila M.R., de Sauvage F.J. Translational value of mouse models in oncology drug development. Nat. Med. 2015;21:431–439. doi: 10.1038/nm.3853. [DOI] [PubMed] [Google Scholar]
- 18.Hachey S.J., Hughes C.C.W. Applications of tumor chip technology. Lab Chip. 2018;18:2893–2912. doi: 10.1039/c8lc00330k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sontheimer-Phelps A., Hassell B.A., Ingber D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer. 2019;19:65–81. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- 20.Duval K., Grover H., Han L.H., Mou Y., Pegoraro A.F., Fredberg J., Chen Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology. 2017;32:266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 21.Lee S.I., Ko Y., Park J.B. Evaluation of the osteogenic differentiation of gingiva-derived stem cells grown on culture plates or in stem cell spheroids: comparison of two- and three-dimensional cultures. Exp. Ther. Med. 2017;14:2434–2438. doi: 10.3892/etm.2017.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nunes A.S., Barros A.S., Costa E.C., Moreira A.F., Correia I.J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019;116:206–226. doi: 10.1002/bit.26845. [DOI] [PubMed] [Google Scholar]
- 23.Imamura Y., Mukohara T., Shimono Y., Funakoshi Y., Chayahara N., Toyoda M., Kiyota N., Takao S., Kono S., Nakatsura T., Minami H. Comparison of 2D-and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015;33:1837–1843. doi: 10.3892/or.2015.3767. [DOI] [PubMed] [Google Scholar]
- 24.Stock K., Estrada M.F., Vidic S., Gjerde K., Rudisch A., Santo V.E., Barbier M., Blom S., Arundkar S.C., Selvam I., Osswald A., Stein Y., Gruenewald S., Brito C., van Weerden W., Rotter V., Boghaert E., Oren M., Sommergruber W., Chong Y., de Hoogt R., Graeser R. Capturing tumor complexity in vitro: comparative analysis of 2D and 3D tumor models for drug discovery. Sci. Rep-UK. 2016;6:1–15. doi: 10.1038/Srep28951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benien P., Swami A. 3D tumor models: history, advances and future perspectives. Future Oncol. 2014;10:1311–1327. doi: 10.2217/fon.13.274. [DOI] [PubMed] [Google Scholar]
- 26.Edmondson R., Broglie J.J., Adcock A.F., Yang L.J. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarockyte G., Dapkute D., Karabanovas V., Daugmaudis J.V., Ivanauskas F., Rotomskis R. 3D cellular spheroids as tools for understanding carboxylated quantum dot behavior in tumors. Bba-Gen Subjects. 2018;1862:914–923. doi: 10.1016/j.bbagen.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 28.Han S.J., Kwon S., Kim K.S. Challenges of applying multicellular tumor spheroids in preclinical phase. Cancer Cell Int. 2021;21:1–19. doi: 10.1186/s12935-021-01853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravi M., Paramesh V., Kaviya S.R., Anuradha E., Solomon F.D.P. 3D cell culture systems: advantages and applications. J. Cell. Physiol. 2015;230:16–26. doi: 10.1002/jcp.24683. [DOI] [PubMed] [Google Scholar]
- 30.Guan P.P., Yu X., Guo J.J., Wang Y., Wang T., Li J.Y., Konstantopoulos K., Wang Z.Y., Wang P. By activating matrix metalloproteinase-7, shear stress promotes chondrosarcoma cell motility, invasion and lung colonization. Oncotarget. 2015;6:9140–9159. doi: 10.18632/oncotarget.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polacheck W.J., Charest J.L., Kamm R.D. Interstitial flow influences direction of tumor cell migration through competing mechanisms. P. Natl. Acad. Sci. USA. 2011;108:11115–11120. doi: 10.1073/pnas.1103581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan J., Li Z., Guo J., Liu S., Guo J. Organ-on-a-chip: a new tool for in vitro research. Biosens. Bioelectron. 2022;216 doi: 10.1016/j.bios.2022.114626. [DOI] [PubMed] [Google Scholar]
- 33.Ma C., Peng Y., Li H., Chen W. Organ-on-a-chip: a new paradigm for drug development. Trends Pharmacol. Sci. 2021;42:119–133. doi: 10.1016/j.tips.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B., Korolj A., Lai B.F.L., Radisic M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018;3:257–278. doi: 10.1038/s41578-018-0034-7. [DOI] [Google Scholar]
- 35.Fujii T. PDMS-based microfluidic devices for biomedical applications. Microelectron. Eng. 2002;61–2:907–914. doi: 10.1016/S0167-9317(02)00494-X. [DOI] [Google Scholar]
- 36.Romoli L., Tantussi G., Dini G. Experimental approach to the laser machining of PMMA substrates for the fabrication of microfluidic devices. Opt Laser. Eng. 2011;49:419–427. doi: 10.1016/j.optlaseng.2010.11.013. [DOI] [Google Scholar]
- 37.Zhang B.Y., Radisic M. Organ-on-a-chip devices advance to market. Lab Chip. 2017;17:2395–2420. doi: 10.1039/c6lc01554a. [DOI] [PubMed] [Google Scholar]
- 38.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tredan O., Galmarini C.M., Patel K., Tannock I.F. Drug resistance and the solid tumor microenvironment. J. Natl. Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 40.Di Martino J.S., Akhter T., Bravo-Cordero J.J. Remodeling the ECM: implications for metastasis and tumor dormancy. Cancers. 2021;13:4916. doi: 10.3390/Cancers13194916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang D., Guo P., He T., Powell C.A. Role of endothelial cells in tumor microenvironment. Clin. Transl. Med. 2021;11 doi: 10.1002/ctm2.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeiro A.L., Okamoto O.K. Combined effects of pericytes in the tumor microenvironment. Stem Cell. Int. 2015;2015 doi: 10.1155/2015/868475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalluri R., Zeisberg M. Fibroblasts in cancer. Nat. Rev. Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 44.Gajewski T.F., Schreiber H., Fu Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immun. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forster J.C., Harriss-Phillips W.M., Douglass M.J., Bezak E. A review of the development of tumor vasculature and its effects on the tumor microenvironment. Hypoxia. 2017;5:21–32. doi: 10.2147/Hp.S133231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaahtomeri K., Alitalo K. Lymphatic vessels in tumor dissemination versus immunotherapy. Cancer Res. 2020;80:3463–3465. doi: 10.1158/0008-5472.CAN-20-0156. [DOI] [PubMed] [Google Scholar]
- 47.Brassart-Pasco S., Brezillon S., Brassart B., Ramont L., Oudart J.B., Monboisse J.C. Tumor microenvironment: extracellular matrix alterations influence tumor progression. Front. Oncol. 2020;10:397. doi: 10.3389/Fonc.2020.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bezel P., Valaperti A., Steiner U., Scholtze D., Wieser S., Vonow-Eisenring M., Widmer A., Kowalski B., Kohler M., Franzen D.P. Evaluation of cytokines in the tumor microenvironment of lung cancer using bronchoalveolar lavage fluid analysis. Cancer Immunol. Immunother. 2021;70:1867–1876. doi: 10.1007/s00262-020-02798-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Del Piccolo N., Shirure V.S., Bi Y., Goedegebuure S.P., Gholami S., Hughes C.C.W., Fields R.C., George S.C. Tumor-on-chip modeling of organ-specific cancer and metastasis. Adv. Drug Deliv. Rev. 2021;175 doi: 10.1016/j.addr.2021.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Radhakrishnan J., Varadaraj S., Dash S.K., Sharma A., Verma R.S. Organotypic cancer tissue models for drug screening: 3D constructs, bioprinting and microfluidic chips. Drug Discov. Today. 2020;25:879–890. doi: 10.1016/j.drudis.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Terrell J.A., Jones C.G., Kabandana G.K.M., Chen C. From cells-on-a-chip to organs-on-a-chip: scaffolding materials for 3D cell culture in microfluidics. J. Mater. Chem. B. 2020;8:6667–6685. doi: 10.1039/D0TB00718H. [DOI] [PubMed] [Google Scholar]
- 52.Castro-Giner F., Aceto N. Tracking cancer progression: from circulating tumor cells to metastasis. Genome Med. 2020;12:31. doi: 10.1186/S13073-020-00728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeon J.S., Bersini S., Gilardi M., Dubini G., Charest J.L., Moretti M., Kamm R.D. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. P. Natl. Acad. Sci. USA. 2015;112:214–219. doi: 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sobrino A., Phan D.T., Datta R., Wang X., Hachey S.J., Romero-López M., Gratton E., Lee A.P., George S.C., Hughes C.C. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep-UK. 2016;6:1–11. doi: 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeong S.-Y., Lee J.-H., Shin Y., Chung S., Kuh H.-J. Co-culture of tumor spheroids and fibroblasts in a collagen matrix-incorporated microfluidic chip mimics reciprocal activation in solid tumor microenvironment. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C., Li S., Du K., Li P., Qiu B., Ding W. On-chip replication of extremely early-stage tumor behavior. ACS Appl. Mater. Interfaces. 2021;13:19768–19777. doi: 10.1021/acsami.1c03740. [DOI] [PubMed] [Google Scholar]
- 57.Choi Y., Hyun E., Seo J., Blundell C., Kim H.C., Lee E., Lee S.H., Moon A., Moon W.K., Huh D. A microengineered pathophysiological model of early-stage breast cancer. Lab Chip. 2015;15:3350–3357. doi: 10.1039/c5lc00514k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuan T.Y., Gao D., Li S.F., Jiang Y.Y. Co-culture of tumor spheroids and monocytes in a collagen matrix-embedded microfluidic device to study the migration of breast cancer cells. Chin. Chem. Lett. 2019;30:331–336. doi: 10.1016/j.cclet.2018.07.013. [DOI] [Google Scholar]
- 59.Chen Y.L., Gao D., Wang Y.W., Lin S., Jiang Y.Y. A novel 3D breast-cancer-on-chip platform for therapeutic evaluation of drug delivery systems. Anal. Chim. Acta. 2018;1036:97–106. doi: 10.1016/j.aca.2018.06.038. [DOI] [PubMed] [Google Scholar]
- 60.Ko J., Ahn J., Kim S., Lee Y., Lee J., Park D., Jeon N.L. Tumor spheroid-on-a-chip: a standardized microfluidic culture platform for investigating tumor angiogenesis. Lab Chip. 2019;19:2822–2833. doi: 10.1039/C9LC00140A. [DOI] [PubMed] [Google Scholar]
- 61.Nashimoto Y., Okada R., Hanada S., Arima Y., Nishiyama K., Miura T., Yokokawa R. Vascularized cancer on a chip: the effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials. 2020;229 doi: 10.1016/j.biomaterials.2019.119547. [DOI] [PubMed] [Google Scholar]
- 62.Kunz-Schughart L.A., Kreutz M., Knuechel R. Multicellular spheroids: a three-dimensional in vitro culture system to study tumour biology. Int. J. Exp. Pathol. 1998;79:1–23. doi: 10.1046/j.1365-2613.1998.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H.F., Ran R., Liu Y., Hui Y., Zeng B.J., Chen D., Weitz D.A., Zhao C.X. Tumor-vasculature-on-a-chip for investigating nanoparticle extravasation and tumor accumulation. ACS Nano. 2018;12:11600–11609. doi: 10.1021/acsnano.8b06846. [DOI] [PubMed] [Google Scholar]
- 64.Paek J., Park S.E., Lu Q.Z., Park K.T., Cho M., Oh J.M., Kwon K.W., Yi Y.S., Song J.W., Edelstein H.I., Ishibashi J., Yang W.L., Myerson J.W., Kiseleva R.Y., Aprelev P., Hood E.D., Stambolian D., Seale P., Muzykantov V.R., Huh D. Microphysiological engineering of self-assembled and perfusable microvascular beds for the production of vascularized three-dimensional human microtissues. ACS Nano. 2019;13:7627–7643. doi: 10.1021/acsnano.9b00686. [DOI] [PubMed] [Google Scholar]
- 65.Jang S.D., Song J., Kim H.A., Im C.N., Khawar I.A., Park J.K., Kuh H.J. Anti-cancer activity profiling of chemotherapeutic agents in 3D co-cultures of pancreatic tumor spheroids with cancer-associated fibroblasts and macrophages. Cancers. 2021;13:5955. doi: 10.3390/Cancers13235955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haque M.R., Wessel C.R., Leary D.D., Wang C.Y., Bhushan A., Bishehsari F. Patient-derived pancreatic cancer-on-a-chip recapitulates the tumor microenvironment. Microsyst. Nanoeng. 2022;8:1–13. doi: 10.1038/S41378-022-00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gillet J.P., Calcagno A.M., Varma S., Marino M., Green L.J., Vora M.I., Patel C., Orina J.N., Eliseeva T.A., Singal V., Padmanabhan R., Davidson B., Ganapathi R., Sood A.K., Rueda B.R., Ambudkar S.V., Gottesman M.M. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. P. Natl. Acad. Sci. USA. 2011;108:18708–18713. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gazdar A.F., Gao B.N., Minna J.D. Lung cancer cell lines: useless artifacts or invaluable tools for medical science? Lung Cancer. 2010;68:309–318. doi: 10.1016/j.lungcan.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirk R. Genetics: personalized medicine and tumour heterogeneity. Nat. Rev. Clin. Oncol. 2012;9:250. doi: 10.1038/nrclinonc.2012.46. [DOI] [PubMed] [Google Scholar]
- 70.Mitra A., Mishra L., Li S.L. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol. 2013;31:347–354. doi: 10.1016/j.tibtech.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hassell B.A., Goyal G., Lee E., Sontheimer-Phelps A., Levy O., Chen C.S., Ingber D.E. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 2017;21:508–516. doi: 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 72.Xu Z.Y., Gao Y.H., Hao Y.Y., Li E.C., Wang Y., Zhang J.N., Wang W.X., Gao Z.C., Wang Q. Application of a microfluidic chip-based 3D co-culture to test drug sensitivity for individualized treatment of lung cancer. Biomaterials. 2013;34:4109–4117. doi: 10.1016/j.biomaterials.2013.02.045. [DOI] [PubMed] [Google Scholar]
- 73.Xu Z.Y., Li E.C., Guo Z., Yu R.F., Hao H.L., Xu Y.T., Sun Z., Li X.C., Lyu J.X., Wang Q. Design and construction of a multi-organ microfluidic chip mimicking the in vivo microenvironment of lung cancer metastasis. ACS Appl. Mater. Interfaces. 2016;8:25840–25847. doi: 10.1021/acsami.6b08746. [DOI] [PubMed] [Google Scholar]
- 74.Meng Q., He Z., Zhang L., Zhao L., Li E., Zhang Q., Zhang X., Yang D., Zou L., Gao Z. Development of a double-layer microfluidic chip with flow medium for chemotherapy resistance analysis of lung cancer. Electrophoresis. 2011;32:3446–3453. doi: 10.1002/elps.201100086. [DOI] [PubMed] [Google Scholar]
- 75.Zuchowska A., Jastrzebska E., Chudy M., Dybko A., Brzozka Z. 3D lung spheroid cultures for evaluation of photodynamic therapy (PDT) procedures in microfluidic Lab-on-a-Chip system. Anal. Chim. Acta. 2017;990:110–120. doi: 10.1016/j.aca.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 76.Zheng Y., Wu Z., Lin J.-M., Lin L. Imitation of drug metabolism in cell co-culture microcapsule model using a microfluidic chip platform coupled to mass spectrometry. Chin. Chem. Lett. 2020;31:451–454. doi: 10.1016/j.cclet.2019.07.036. [DOI] [Google Scholar]
- 77.Dhiman N., Shagaghi N., Bhave M., Sumer H., Kingshott P., Rath S.N. Selective cytotoxicity of a novel Trp-rich peptide against lung tumor spheroids encapsulated inside a 3d microfluidic device. Adv. Biosyst. 2020;4 doi: 10.1002/adbi.201900285. [DOI] [PubMed] [Google Scholar]
- 78.Kim D., Hwang K.S., Seo E.U., Seo S., Lee B.C., Choi N., Choi J., Kim H.N. Vascularized lung cancer model for evaluating the promoted transport of anticancer drugs and immune cells in an engineered tumor microenvironment. Adv. Healthc. Mater. 2022;11 doi: 10.1002/adhm.202102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu Z.H., Aref A.R., Cohoon T.J., Barbie T.U., Imamura Y., Yang S.H., Moody S.E., Shen R.R., Schinzel A.C., Thai T.C., Reibel J.B., Tamayo P., Godfrey J.T., Qian Z.R., Page A.N., Maciag K., Chan E.M., Silkworth W., Labowsky M.T., Rozhansky L., Mesirov J.P., Gillanders W.E., Ogino S., Hacohen N., Gaudet S., Eck M.J., Engelman J.A., Corcoran R.B., Wong K.K., Hahn W.C., Barbie D.A. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov. 2014;4:452–465. doi: 10.1158/2159-8290.CD-13-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khalid M.A.U., Kim Y.S., Ali M., Lee B.G., Cho Y.J., Choi K.H. A lung cancer-on-chip platform with integrated biosensors for physiological monitoring and toxicity assessment. Biochem. Eng. J. 2020;155 doi: 10.1016/J.Bej.2019.107469. [DOI] [Google Scholar]
- 81.Yeo T., Tan S.J., Lim C.L., Lau D.P.X., Chua Y.W., Krisna S.S., Iyer G., Tan G.S., Lim T.K.H., Tan D.S.W., Lim W.T., Lim C.T. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci. Rep-UK. 2016;6:1–12. doi: 10.1038/Srep22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu W.W., Song J., Du X.H., Zhou Y., Li Y., Li R., Lyu L., He Y.T., Hao J.X., Ben J., Wang W., Shi H.B., Wang Q. AKR1B10 (Aldo-keto reductase family 1 B10) promotes brain metastasis of lung cancer cells in a multi-organ microfluidic chip model. Acta Biomater. 2019;91:195–208. doi: 10.1016/j.actbio.2019.04.053. [DOI] [PubMed] [Google Scholar]
- 83.Skardal A., Devarasetty M., Forsythe S., Atala A., Soker S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol. Bioeng. 2016;113:2020–2032. doi: 10.1002/bit.25950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ayuso J.M., Virumbrales-Munoz M., McMinn P.H., Rehman S., Gomez I., Karim M.R., Trusttchel R., Wisinski K.B., Beebe D.J., Skala M.C. Tumor-on-a-chip: a microfluidic model to study cell response to environmental gradients. Lab Chip. 2019;19:3461–3471. doi: 10.1039/C9LC00270G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao L., Liu Y., Liu Y., Zhang M., Zhang X. Microfluidic control of tumor and stromal cell spheroids pairing and merging for three-dimensional metastasis study. Anal. Chem. 2020;92:7638–7645. doi: 10.1021/acs.analchem.0c00408. [DOI] [PubMed] [Google Scholar]
- 86.Aeby E.A., Misun P.M., Hierlemann A., Frey O. Microfluidic hydrogel hanging-drop network for long-term culturing of 3D microtissues and simultaneous high-resolution imaging. Adv. Biosyst. 2018;2 doi: 10.1002/adbi.201800054. [DOI] [Google Scholar]
- 87.Khot M.I., Levenstein M.A., de Boer G.N., Armstrong G., Maisey T., Svavarsdottir H.S., Andrew H., Perry S.L., Kapur N., Jayne D.G. Characterising a PDMS based 3D cell culturing microfluidic platform for screening chemotherapeutic drug cytotoxic activity. Sci. Rep-UK. 2020;10:1–13. doi: 10.1038/s41598-020-72952-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nair A.L., Mesch L., Schulz I., Becker H., Raible J., Kiessling H., Werner S., Rothbauer U., Schmees C., Busche M. Parallelizable microfluidic platform to model and assess in vitro cellular barriers: technology and application to study the interaction of 3D tumor spheroids with cellular barriers. Biosensors. 2021;11:314. doi: 10.3390/bios11090314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee B.-E., Kim D.-K., Lee H., Yoon S., Park S.-H., Lee S., Yoo J. Recapitulation of first pass metabolism using 3D printed microfluidic chip and organoid. Cells. 2021;10:3301. doi: 10.3390/cells10123301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Petreus T., Cadogan E., Hughes G., Smith A., Pilla Reddy V., Lau A., O'Connor M.J., Critchlow S., Ashford M., Oplustil O'Connor L. Tumour-on-chip microfluidic platform for assessment of drug pharmacokinetics and treatment response. Commun. Biol. 2021;4:1001. doi: 10.1038/s42003-021-02526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parlato S., De Ninno A., Molfetta R., Toschi E., Salerno D., Mencattini A., Romagnoli G., Fragale A., Roccazzello L., Buoncervello M. 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Sci. Rep-UK. 2017;7:1093. doi: 10.1038/s41598-017-01013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deng J., Zhang X., Chen Z., Luo Y., Lu Y., Liu T., Wu Z., Jin Y., Zhao W., Lin B. A cell lines derived microfluidic liver model for investigation of hepatotoxicity induced by drug-drug interaction. Biomicrofluidics. 2019;13 doi: 10.1063/1.5070088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shen S., Zhang X., Zhang F., Wang D., Long D., Niu Y. Three-gradient constructions in a flow-rate insensitive microfluidic system for drug screening towards personalized treatment. Talanta. 2020;208 doi: 10.1016/j.talanta.2019.120477. [DOI] [PubMed] [Google Scholar]
- 94.Ozkan A., Ghousifam N., Hoopes P.J., Yankeelov T.E., Rylander M.N. In vitro vascularized liver and tumor tissue microenvironments on a chip for dynamic determination of nanoparticle transport and toxicity. Biotechnol. Bioeng. 2019;116:1201–1219. doi: 10.1002/bit.26919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ozkan A., Stolley D.L., Cressman E.N., McMillin M., DeMorrow S., Yankeelov T.E., Rylander M.N. Tumor microenvironment alters chemoresistance of hepatocellular carcinoma through CYP3A4 metabolic activity. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.662135. 1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Z.Z., Li W.M., Zhang Y., Yu M., Shan L.F., Yuan D.Z., Liu F.R., Fang J. Establishment of a gastric cancer subline with high metastatic potential using a novel microfluidic system. Sci. Rep-UK. 2016;6 doi: 10.1038/srep38376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nagaraju S., Truong D., Mouneimne G., Nikkhah M. Microfluidic tumor-vascular model to study breast cancer cell invasion and intravasation. Adv. Healthc. Mater. 2018;7 doi: 10.1002/Adhm.201701257. [DOI] [PubMed] [Google Scholar]
- 98.Lin D.G., Li P.W., Feng J., Lin Z., Chen X., Yang N., Wang L.H., Liu D.Y. Screening therapeutic agents specific to breast cancer stem cells using a microfluidic single-cell clone-forming inhibition assay. Small. 2020;16 doi: 10.1002/Smll.201901001. [DOI] [PubMed] [Google Scholar]
- 99.Wu L.Y., Di Carlo D., Lee L.P. Microfluidic self-assembly of tumor spheroids for anticancer drug discovery. Biomed. Microdev. 2008;10:197–202. doi: 10.1007/s10544-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 100.Huang X., Li L., Tu Q., Wang J., Liu W., Wang X., Ren L., Wang J. On-chip cell migration assay for quantifying the effect of ethanol on MCF-7 human breast cancer cells. Microfluid. Nanofluid. 2011;10:1333–1341. doi: 10.1007/s10404-011-0766-9. [DOI] [Google Scholar]
- 101.Liu T., Li C., Li H., Zeng S., Qin J., Lin B. A microfluidic device for characterizing the invasion of cancer cells in 3-D matrix. Electrophoresis. 2009;30:4285–4291. doi: 10.1002/elps.200900289. [DOI] [PubMed] [Google Scholar]
- 102.Rodoplu D., Matahum J.S., Hsu C.-H. A microfluidic hanging drop-based spheroid co-culture platform for probing tumor angiogenesis. Lab Chip. 2022;22:1275–1285. doi: 10.1039/D1LC01177D. [DOI] [PubMed] [Google Scholar]
- 103.Moon H.R., Ospina-Munoz N., Noe-Kim V., Yang Y., Elzey B.D., Konieczny S.F., Han B. Subtype-specific characterization of breast cancer invasion using a microfluidic tumor platform. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cognart H.A., Viovy J.L., Villard C. Fluid shear stress coupled with narrow constrictions induce cell type-dependent morphological and molecular changes in SK-BR-3 and MDA-MB-231 cells. Sci. Rep-UK. 2020;10:1–14. doi: 10.1038/S41598-020-63316-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mi S., Du Z., Xu Y., Wu Z., Qian X., Zhang M., Sun W.J.S.r. Microfluidic co-culture system for cancer migratory analysis and anti-metastatic drugs screening. Sci. Rep-UK. 2016;6:1–11. doi: 10.1038/srep35544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Du Z., Mi S., Yi X., Xu Y., Sun W. Microfluidic system for modelling 3D tumour invasion into surrounding stroma and drug screening. Biofabrication. 2018;10 doi: 10.1088/1758-5090/aac70c. [DOI] [PubMed] [Google Scholar]
- 107.Morales X., Cortes-Dominguez I., Ortiz-de-Solorzano C. Modeling the mechanobiology of cancer cell migration using 3D biomimetic hydrogels. Gels. 2021;7:17. doi: 10.3390/Gels7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Calibasi Kocal G., Güven S., Foygel K., Goldman A., Chen P., Sengupta S., Paulmurugan R., Baskin Y., Demirci U. Dynamic microenvironment induces phenotypic plasticity of esophageal cancer cells under flow. Sci. Rep-UK. 2016;6:1–11. doi: 10.1038/srep38221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nan L., Yang Z., Lyu H., Lau K.Y.Y., Shum H.C. A microfluidic system for one-chip harvesting of single-cell-laden hydrogels in culture medium. Adv. Biosyst. 2019;3 doi: 10.1002/adbi.201900076. [DOI] [PubMed] [Google Scholar]
- 110.Eduati F., Utharala R., Madhavan D., Neumann U.P., Longerich T., Cramer T., Saez-Rodriguez J., Merten C.A. A microfluidics platform for combinatorial drug screening on cancer biopsies. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-04919-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beer M., Kuppalu N., Stefanini M., Becker H., Schulz I., Manoli S., Schuette J., Schmees C., Casazza A., Stelzle M. A novel microfluidic 3D platform for culturing pancreatic ductal adenocarcinoma cells: comparison with in vitro cultures and in vivo xenografts. Sci. Rep-UK. 2017;7:1–12. doi: 10.1038/s41598-017-01256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kirby B.J., Jodari M., Loftus M.S., Gakhar G., Pratt E.D., Chanel-Vos C., Gleghorn J.P., Santana S.M., Liu H., Smith J.P. Functional characterization of circulating tumor cells with a prostate-cancer-specific microfluidic device. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hsiao A.Y., Torisawa Y.-s., Tung Y.-C., Sud S., Taichman R.S., Pienta K.J., Takayama S. Microfluidic system for formation of PC-3 prostate cancer co-culture spheroids. Biomaterials. 2009;30:3020–3027. doi: 10.1016/j.biomaterials.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Luo D., Liu N., Chen Y., Peng Y., Yue T., Cao S., Liu Y. Microfluidic assessment of drug effects on physical properties of androgen sensitive and non-sensitive prostate cancer cells. Micromach. Basel. 2021;12:532. doi: 10.3390/mi12050532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huang C., Liu H., Bander N.H., Kirby B.J. Enrichment of prostate cancer cells from blood cells with a hybrid dielectrophoresis and immunocapture microfluidic system. Biomed. Microdev. 2013;15:941–948. doi: 10.1007/s10544-013-9784-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nath B., Raza A., Sethi V., Dalal A., Ghosh S.S., Biswas G. Understanding flow dynamics, viability and metastatic potency of cervical cancer (HeLa) cells through constricted microchannel. Sci. Rep-UK. 2018;8:1–10. doi: 10.1038/s41598-018-35646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin L., Lin X., Lin L., Feng Q., Kitamori T., Lin J.-M., Sun J. Integrated microfluidic platform with multiple functions to probe tumor–endothelial cell interaction. Anal. Chem. 2017;89:10037–10044. doi: 10.1021/acs.analchem.7b02593. [DOI] [PubMed] [Google Scholar]
- 118.Lin X., Chen Q., Liu W., Zhang J., Wang S., Lin Z., Lin J.-M. Oxygen-induced cell migration and on-line monitoring biomarkers modulation of cervical cancers on a microfluidic system. Sci. Rep-UK. 2015;5:1–7. doi: 10.1038/srep09643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frantz C., Stewart K.M., Weaver V.M. The extracellular matrix at a glance. J. Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Theocharis A.D., Skandalis S.S., Gialeli C., Karamanos N.K. Extracellular matrix structure. Adv. Drug Deliv. Rev. 2016;97:4–27. doi: 10.1016/j.addr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 121.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hynes R.O. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.117600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu H., Wang Y., Cui K., Guo Y., Zhang X., Qin J. Advances in hydrogels in organoids and organs-on-a-chip. Adv. Mater. 2019;31 doi: 10.1002/adma.201902042. [DOI] [PubMed] [Google Scholar]
- 124.Brand D.D., Latham K.A., Rosloniec E.F. Collagen-induced arthritis. Nat. Protoc. 2007;2:1269–1275. doi: 10.1038/nprot.2007.173. [DOI] [PubMed] [Google Scholar]
- 125.Huh D., Kim H.J., Fraser J.P., Shea D.E., Khan M., Bahinski A., Hamilton G.A., Ingber D.E. Microfabrication of human organs-on-chips. Nat. Protoc. 2013;8:2135–2157. doi: 10.1038/nprot.2013.137. [DOI] [PubMed] [Google Scholar]
- 126.Aisenbrey E.A., Murphy W.L. Synthetic alternatives to matrigel. Nat. Rev. Mater. 2020;5:539–551. doi: 10.1038/s41578-020-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Osaki T., Uzel S.G.M., Kamm R.D. On-chip 3D neuromuscular model for drug screening and precision medicine in neuromuscular disease. Nat. Protoc. 2020;15:421–449. doi: 10.1038/s41596-019-0248-1. [DOI] [PubMed] [Google Scholar]
- 128.Zhang B., Montgomery M., Chamberlain M.D., Ogawa S., Korolj A., Pahnke A., Wells L.A., Massé S., Kim J., Reis L. Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis. Nat. Mater. 2016;15:669–678. doi: 10.1038/nmat4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yue K., Trujillo-de Santiago G., Alvarez M.M., Tamayol A., Annabi N., Khademhosseini A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials. 2015;73:254–271. doi: 10.1016/j.biomaterials.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yin J., Yan M., Wang Y., Fu J., Suo H. 3D bioprinting of low-concentration cell-laden gelatin methacrylate (GelMA) bioinks with a two-step cross-linking strategy. ACS Appl. Mater. Interfaces. 2018;10:6849–6857. doi: 10.1021/acsami.7b16059. [DOI] [PubMed] [Google Scholar]
- 131.Lu S., Cuzzucoli F., Jiang J., Liang L.G., Wang Y., Kong M., Zhao X., Cui W., Li J., Wang S. Development of a biomimetic liver tumor-on-a-chip model based on decellularized liver matrix for toxicity testing. Lab Chip. 2018;18:3379–3392. doi: 10.1039/c8lc00852c. [DOI] [PubMed] [Google Scholar]
- 132.Duan J., Cao Y., Shen Z., Cheng Y., Ma Z., Wang L., Zhang Y., An Y., Sang S. 3D bioprinted GelMA/PEGDA hybrid scaffold for establishing an in vitro model of melanoma. J. Microbiol. 2022;32:531–540. doi: 10.4014/jmb.2111.11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cao X., Ashfaq R., Cheng F., Maharjan S., Li J., Ying G., Hassan S., Xiao H., Yue K., Zhang Y.S. A tumor-on-a-chip system with bioprinted blood and lymphatic vessel pair. Adv. Funct. Mater. 2019;29 doi: 10.1002/adfm.201807173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Deng J., Wei W., Chen Z., Lin B., Zhao W., Luo Y., Zhang X. Engineered liver-on-a-chip platform to mimic liver functions and its biomedical applications: a review. Micromach. Basel. 2019;10:676. doi: 10.3390/mi10100676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Annabi N., Selimovic S., Acevedo Cox J.P., Ribas J., Afshar Bakooshli M., Heintze D., Weiss A.S., Cropek D., Khademhosseini A. Hydrogel-coated microfluidic channels for cardiomyocyte culture. Lab Chip. 2013;13:3569–3577. doi: 10.1039/c3lc50252j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sun L., Chen Z., Xu D., Zhao Y. Electroconductive and anisotropic structural color hydrogels for visual heart-on-a-chip construction. Adv. Sci. 2022;9 doi: 10.1002/advs.202105777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Blombäck B., Hessel B., Hogg D., Therkildsen L.J.N. A two-step fibrinogen–fibrin transition in blood coagulation. Nature. 1978;275:501–505. doi: 10.1038/275501a0. [DOI] [PubMed] [Google Scholar]
- 138.Guan Y., Zhang K., Xu F., Guo R., Fang A., Sun B., Meng X., Liu Y., Bai M. An integrated platform for fibrinogen quantification on a microfluidic paper-based analytical device. Lab Chip. 2020;20:2724–2734. doi: 10.1039/D0LC00439A. [DOI] [PubMed] [Google Scholar]
- 139.Wang X., Phan D.T., Sobrino A., George S.C., Hughes C.C., Lee A.P. Engineering anastomosis between living capillary networks and endothelial cell-lined microfluidic channels. Lab Chip. 2016;16:282–290. doi: 10.1039/C5LC01050K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim S., Lee H., Chunga M., Jeon N.L. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip. 2013;13:4891. doi: 10.1039/C3LC41320A. 4891. [DOI] [PubMed] [Google Scholar]
- 141.Yamada Y., Hozumi K., Aso A., Hotta A., Toma K., Katagiri F., Kikkawa Y., Nomizu M. Laminin active peptide/agarose matrices as multifunctional biomaterials for tissue engineering. Biomaterials. 2012;33:4118–4125. doi: 10.1016/j.biomaterials.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 142.Sasser A.K., Mundy B.L., Smith K.M., Studebaker A.W., Axel A.E., Haidet A.M., Fernandez S.A., Hall B.M. Human bone marrow stromal cells enhance breast cancer cell growth rates in a cell line-dependent manner when evaluated in 3D tumor environments. Canc. Lett. 2007;254:255–264. doi: 10.1016/j.canlet.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 143.Mizutani T., Clevers H. In: Intestinal Stem Cells: Methods and Protocols. Ordóñez-Morán P., editor. Humana; New York: 2020. Primary intestinal epithelial organoid culture; pp. 185–200. [DOI] [PubMed] [Google Scholar]
- 144.McCrary M.W., Bousalis D., Mobini S., Song Y.H., Schmidt C.E.J.A.b. Decellularized tissues as platforms for in vitro modeling of healthy and diseased tissues. Acta Biomater. 2020;111:1–19. doi: 10.1016/j.actbio.2020.05.031. [DOI] [PubMed] [Google Scholar]
- 145.Yi H.-G., Jeong Y.H., Kim Y., Choi Y.-J., Moon H.E., Park S.H., Kang K.S., Bae M., Jang J., Youn H., Paek S.H., Cho D.-W. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019;3:509–519. doi: 10.1038/s41551-019-0363-x. [DOI] [PubMed] [Google Scholar]
- 146.Ferreira L.P., Gaspar V.M., Mano J.F. Decellularized extracellular matrix for bioengineering physiomimetic 3D in vitro tumor models. Trends Biotechnol. 2020;38:1397–1414. doi: 10.1016/j.tibtech.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 147.Marques-Magalhães Â., Cruz T., Costa Â.M., Estêvão D., Rios E., Canão P.A., Velho S., Carneiro F., Oliveira M.J., Cardoso A.P.J.C. Decellularized colorectal cancer matrices as bioactive scaffolds for studying tumor-stroma interactions. Cancers. 2022;14:359. doi: 10.3390/cancers14020359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ramadan Q., Zourob M.J.B. Organ-on-a-chip engineering: toward bridging the gap between lab and industry. Biomicrofluidics. 2020;14 doi: 10.1063/5.0011583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen D., Yin J., Yang Z., Qin W., Huo J., Huang J., Sun J., Piao W. Construction and application of hepatocyte model based on microfluidic chip technique in evaluating emodin. Nutrients. 2022;14 doi: 10.3390/nu14132768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nguyen D.-H.T., Lee E., Alimperti S., Norgard R.J., Wong A., Lee J.J.-K., Eyckmans J., Stanger B.Z., Chen C.S. A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hou Y., Ai X., Zhao L., Gao Z., Wang Y., Lu Y., Tu P., Jiang Y. An integrated biomimetic array chip for high-throughput co-culture of liver and tumor microtissues for advanced anticancer bioactivity screening. Lab Chip. 2020;20:2482–2494. doi: 10.1039/D0LC00288G. [DOI] [PubMed] [Google Scholar]
- 152.Saha B., Mathur T., Tronolone J.J., Chokshi M., Lokhande G.K., Selahi A., Gaharwar A.K., Afshar-Kharghan V., Sood A.K., Bao G., Jain A. Human tumor microenvironment chip evaluates the consequences of platelet extravasation and combinatorial antitumor-antiplatelet therapy in ovarian cancer. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abg5283. eabg5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miller C.P., Tsuchida C., Zheng Y., Himmelfarb J., Akilesh S. A 3D human renal cell carcinoma-on-a-chip for the study of tumor angiogenesis. Neoplasia. 2018;20:610–620. doi: 10.1016/j.neo.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hayn A., Fischer T., Mierke C.T. Inhomogeneities in 3D collagen matrices impact matrix mechanics and cancer cell migration. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.593879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Park S., Kim T.H., Kim S.H., You S., Jung Y. Three-dimensional vascularized lung cancer-on-a-chip with lung extracellular matrix hydrogels for in vitro screening. Cancers. 2021;13 doi: 10.3390/cancers13163930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen Z., Wang F., Zhang J., Sun X., Yan Y., Wang Y., Ouyang J., Zhang J., Honore T., Ge J., Gu Z. Study on development of composite hydrogels with tunable structures and properties for tumor-on-a-chip research. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.611796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lai B.F.L., Lu R.X.Z., Hu Y., Davenport Huyer L., Dou W., Wang E.Y., Radulovich N., Tsao M.S., Sun Y., Radisic M. Recapitulating pancreatic tumor microenvironment through synergistic use of patient organoids and organ-on-a-chip vasculature. Adv. Funct. Mater. 2020;30 doi: 10.1002/adfm.202000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhuang J., Zhang J., Wu M., Zhang Y. A dynamic 3D tumor spheroid chip enables more accurate nanomedicine uptake evaluation. Adv. Sci. 2019;6 doi: 10.1002/advs.201901462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Huang C.-H., Lei K.F. Quantitative study of tumor angiogenesis in three-dimensional matrigel barrier using electric impedance measurement technique. Sens. Actuat. B-Chem. 2022;370 doi: 10.1016/j.snb.2022.132458. [DOI] [Google Scholar]
- 160.Carvalho M.R., Barata D., Teixeira L.M., Giselbrecht S., Reis R.L., Oliveira J.M., Truckenmüller R., Habibovic P. Colorectal tumor-on-a-chip system: a 3D tool for precision onco-nanomedicine. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aaw1317. eaaw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pan Y., Hu N., Wei X., Gong L., Zhang B., Wan H., Wang P. 3D cell-based biosensor for cell viability and drug assessment by 3D electric cell/matrigel-substrate impedance sensing. Biosens. Bioelectron. 2019;130:344–351. doi: 10.1016/j.bios.2018.09.046. [DOI] [PubMed] [Google Scholar]
- 162.Alonzo L.F., Moya M.L., Shirure V.S., George S.C. Microfluidic device to control interstitial flow-mediated homotypic and heterotypic cellular communication. Lab Chip. 2015;15:3521–3529. doi: 10.1039/c5lc00507h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ko J., Lee Y., Lee S., Lee S.-R., Jeon N.L. Human ocular angiogenesis-inspired vascular models on an injection-molded microfluidic chip. Adv. Healthc. Mater. 2019;8 doi: 10.1002/adhm.201900328. [DOI] [PubMed] [Google Scholar]
- 164.Bi Y., Shirure V.S., Liu R., Cunningham C., Ding L., Meacham J.M., Goedegebuure S.P., George S.C., Fields R.C. Tumor-on-a-chip platform to interrogate the role of macrophages in tumor progression. Integr. Biol-Uk. 2020;12:221–232. doi: 10.1093/intbio/zyaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hachey S.J., Movsesyan S., Nguyen Q.H., Burton-Sojo G., Tankazyan A., Wu J., Hoang T., Zhao D., Wang S., Hatch M.M., Celaya E., Gomez S., Chen G.T., Davis R.T., Nee K., Pervolarakis N., Lawson D.A., Kessenbrock K., Lee A.P., Lowengrub J., Waterman M.L., Hughes C.C.W. An in vitro vascularized micro-tumor model of human colorectal cancer recapitulates in vivo responses to standard-of-care therapy. Lab Chip. 2021;21:1333–1351. doi: 10.1039/d0lc01216e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Silvani G., Basirun C., Wu H., Mehner C., Poole K., Bradbury P., Chou J. A 3D-bioprinted vascularized glioblastoma-on-a-chip for studying the impact of simulated microgravity as a novel pre-clinical approach in brain tumor therapy. Adv. Therapeut. 2021;4 doi: 10.1002/adtp.202100106. [DOI] [Google Scholar]
- 167.Hu X., Zhao S., Luo Z., Zuo Y., Wang F., Zhu J., Chen L., Yang D., Zheng Y., Zheng Y., Cheng Y., Zhou F., Yang Y. On-chip hydrogel arrays individually encapsulating acoustic formed multicellular aggregates for high throughput drug testing. Lab Chip. 2020;20:2228–2236. doi: 10.1039/d0lc00255k. [DOI] [PubMed] [Google Scholar]
- 168.Zhang Q., Liu T., Qin J. A microfluidic-based device for study of transendothelial invasion of tumor aggregates in realtime. Lab Chip. 2012;12:2837–2842. doi: 10.1039/c2lc00030j. [DOI] [PubMed] [Google Scholar]
- 169.Lyu Z., Park J., Kim K.-M., Jin H.-J., Wu H., Rajadas J., Kim D.-H., Steinberg G.K., Lee W. A neurovascular-unit-on-a-chip for the evaluation of the restorative potential of stem cell therapies for ischaemic stroke. Nat. Biomed. Eng. 2021;5:847–863. doi: 10.1038/s41551-021-00744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Strelez C., Chilakala S., Ghaffarian K., Lau R., Spiller E., Ung N., Hixon D., Yoon A.Y., Sun R.X., Lenz H.-J., Katz J.E., Mumenthaler S.M. Human colorectal cancer-on-chip model to study the microenvironmental influence on early metastatic spread. iScience. 2021;24 doi: 10.1016/j.isci.2021.102509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kalluri R. The biology and function of fibroblasts in cancer. Nat. Rev. Canc. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 172.Ishii G., Ochiai A., Neri S. Phenotypic and functional heterogeneity of cancer-associated fibroblast within the tumor microenvironment. Adv. Drug Deliv. Rev. 2016;99:186–196. doi: 10.1016/j.addr.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 173.Bhowmick N.A., Neilson E.G., Moses H.L. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wei C.L., Tang M., Xu Z.L., Yang L., Lv Y.G. Role of endothelial cells in the regulation of mechanical microenvironment on tumor progression. Acta Mech. Sin. 2021;37:218–228. doi: 10.1007/s10409-021-01056-4. [DOI] [Google Scholar]
- 175.Raza A., Franklin M.J., Dudek A.Z. Pericytes and vessel maturation during tumor angiogenesis and metastasis. Am. J. Hematol. 2010;85:593–598. doi: 10.1002/ajh.21745. [DOI] [PubMed] [Google Scholar]
- 176.Baudin B., Bruneel A., Bosselut N., Vaubourdolle M.J.N.p. A protocol for isolation and culture of human umbilical vein endothelial cells. Nat. Protoc. 2007;2:481–485. doi: 10.1038/nprot.2007.54. [DOI] [PubMed] [Google Scholar]
- 177.Ahn J., Sei Y.J., Jeon N.L., Kim Y. Tumor microenvironment on a chip: the progress and future perspective. Bioengineering. 2017;4:64. doi: 10.3390/bioengineering4030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ruppen J., Wildhaber F.D., Strub C., Hall S.R., Schmid R.A., Geiser T., Guenat O.T. Towards personalized medicine: chemosensitivity assays of patient lung cancer cell spheroids in a perfused microfluidic platform. Lab Chip. 2015;15:3076–3085. doi: 10.1039/C5LC00454C. [DOI] [PubMed] [Google Scholar]
- 179.Elitas M., Brower K., Lu Y., Chen J.J., Fan R. A microchip platform for interrogating tumor–macrophage paracrine signaling at the single-cell level. Lab Chip. 2014;14:3582–3588. doi: 10.1039/c4lc00676c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rosa P.M., Gopalakrishnan N., Ibrahim H., Haug M., Halaas Ø. The intercell dynamics of T cells and dendritic cells in a lymph node-on-a-chip flow device. Lab Chip. 2016;16:3728–3740. doi: 10.1039/C6LC00702C. [DOI] [PubMed] [Google Scholar]
- 181.Ayuso J.M., Rehman S., Virumbrales-Munoz M., McMinn P.H., Geiger P., Fitzgerald C., Heaster T., Skala M.C., Beebe D.J. Microfluidic tumor-on-a-chip model to evaluate the role of tumor environmental stress on NK cell exhaustion. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abc2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Marzagalli M., Pelizzoni G., Fedi A., Vitale C., Fontana F., Bruno S., Poggi A., Dondero A., Aiello M., Castriconi R., Bottino C., Scaglione S. A multi-organ-on-chip to recapitulate the infiltration and the cytotoxic activity of circulating NK cells in 3D matrix-based tumor model. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.945149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Aung A., Kumar V., Theprungsirikul J., Davey S.K., Varghese S. An engineered tumor-on-a-chip device with breast cancer–immune cell interactions for assessing t-cell recruitment. Cancer Res. 2020;80:263–275. doi: 10.1158/0008-5472.CAN-19-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lee J., Kim S.-E., Moon D., Doh J. A multilayered blood vessel/tumor tissue chip to investigate T cell infiltration into solid tumor tissues. Lab Chip. 2021;21:2142–2152. doi: 10.1039/D1LC00182E. [DOI] [PubMed] [Google Scholar]
- 185.Parlato S., Grisanti G., Sinibaldi G., Peruzzi G., Casciola C.M., Gabriele L. Tumor-on-a-chip platforms to study cancer-immune system crosstalk in the era of immunotherapy. Lab Chip. 2021;21:234–253. doi: 10.1039/d0lc00799d. [DOI] [PubMed] [Google Scholar]
- 186.Yang S., Chen C., Qiu Y., Xu C., Yao J. Paying attention to tumor blood vessels: cancer phototherapy assisted with nano delivery strategies. Biomaterials. 2021;268 doi: 10.1016/j.biomaterials.2020.120562. [DOI] [PubMed] [Google Scholar]
- 187.Lugano R., Ramachandran M., Dimberg A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020;77:1745–1770. doi: 10.1007/s00018-019-03351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chaplain M. Avascular growth, angiogenesis and vascular growth in solid tumours: the mathematical modelling of the stages of tumour development. Math. Comput. Model. 1996;23:47–87. doi: 10.1016/0895-7177(96)00019-2. [DOI] [Google Scholar]
- 189.Roy H., Bhardwaj S., Ylä-Herttuala S. Biology of vascular endothelial growth factors. FEBS Lett. 2006;580:2879–2887. doi: 10.1016/j.febslet.2006.03.087. [DOI] [PubMed] [Google Scholar]
- 190.Dewhirst M.W., Secomb T.W. Transport of drugs from blood vessels to tumour tissue. Nat. Rev. Cancer. 2017;17:738–750. doi: 10.1038/nrc.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Swartz M.A., Lund A.W. Lymphatic and interstitial flow in the tumour microenvironment: linking mechanobiology with immunity. Nat. Rev. Cancer. 2012;12:210–219. doi: 10.1038/nrc3186. [DOI] [PubMed] [Google Scholar]
- 192.Alitalo A., Detmar M. Interaction of tumor cells and lymphatic vessels in cancer progression. Oncogene. 2012;31:4499–4508. doi: 10.1038/onc.2011.602. [DOI] [PubMed] [Google Scholar]
- 193.Jain R.K. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 194.Jain R.K., Stylianopoulos T. Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol. 2010;7:653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Frenkel N., Poghosyan S., Alarcon C.R., Garcia S.B., Queiroz K., van den Bent L., Laoukili J., Rinkes I.B., Vulto P., Kranenburg O. Long-lived human lymphatic endothelial cells to study lymphatic biology and Lymphatic Vessel/Tumor Coculture in a 3D Microfluidic Model. ACS Biomater. Sci. Eng. 2021;7:3030–3042. doi: 10.1021/acsbiomaterials.0c01378. [DOI] [PubMed] [Google Scholar]
- 196.Ayuso J.M., Gong M.M., Skala M.C., Harari P.M., Beebe D.J. Human tumor-lymphatic microfluidic model reveals differential conditioning of lymphatic vessels by breast cancer cells. Adv. Healthc. Mater. 2020;9 doi: 10.1002/adhm.201900925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sherwood L. Cengage learning; 2015. Human Physiology: from Cells to Systems. [Google Scholar]
- 198.Al-Hajj M., Clarke M.F. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 199.Montanez-Sauri S.I., Sung K.E., Berthier E., Beebe D.J. Enabling screening in 3D microenvironments: probing matrix and stromal effects on the morphology and proliferation of T47D breast carcinoma cells. Integr. Biol-Uk. 2013;5:631–640. doi: 10.1039/c3ib20225a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fang G.C., Lu H.X., Es H.A., Wang D.J., Liu Y., Warkiani M.E., Lin G.G., Jin D.Y. Unidirectional intercellular communication on a microfluidic chip. Biosens. Bioelectron. 2021;175 doi: 10.1016/J.Bios.2020.112833. [DOI] [PubMed] [Google Scholar]
- 201.Xu H., Cheng C., Le W.D. Recent research advances of the biomimetic tumor microenvironment and regulatory factors on microfluidic devices: a systematic review. Electrophoresis. 2022;43:839–847. doi: 10.1002/elps.202100360. [DOI] [PubMed] [Google Scholar]
- 202.Valero A., Merino F., Wolbers F., Luttge R., Vermes I., Andersson H., van den Berg A. Apoptotic cell death dynamics of HL60 cells studied using a microfluidic cell trap device. Lab Chip. 2005;5:49–55. doi: 10.1039/b415813j. [DOI] [PubMed] [Google Scholar]
- 203.Hyvärinen T., Hagman S., Ristola M., Sukki L., Veijula K., Kreutzer J., Kallio P., Narkilahti S. Co-stimulation with IL-1β and TNF-α induces an inflammatory reactive astrocyte phenotype with neurosupportive characteristics in a human pluripotent stem cell model system. Sci. Rep-UK. 2019;9 doi: 10.1038/s41598-019-53414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chernyavska M., Hermans C.K., Chan C., Baumann N., Rösner T., Leusen J.H., Valerius T., Verdurmen W.P. Evaluation of immunotherapies improving macrophage anti-tumor response using a microfluidic model. Organs-on-a-Chip. 2022;4 doi: 10.1016/j.ooc.2022.100019. [DOI] [Google Scholar]
- 205.Sarkar S., Peng C.-C., Tung Y.-C. Comparison of VEGF-A secretion from tumor cells under cellular stresses in conventional monolayer culture and microfluidic three-dimensional spheroid models. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sharafeldin M., Chen T., Ozkaya G.U., Choudhary D., Molinolo A.A., Gutkind J.S., Rusling J.F. Detecting cancer metastasis and accompanying protein biomarkers at single cell levels using a 3D-printed microfluidic immunoarray. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Unachukwu U.J., Sauane M., Vazquez M., Redenti S. Microfluidic generated EGF-gradients induce chemokinesis of transplantable retinal progenitor cells via the JAK/STAT and PI3kinase signaling pathways. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sun W., Chen Y., Wang Y., Luo P., Zhang M., Zhang H., Hu P.J.A. Interaction study of cancer cells and fibroblasts on a spatially confined oxygen gradient microfluidic chip to investigate the tumor microenvironment. Analyst. 2018;143:5431–5437. doi: 10.1039/C8AN01216D. [DOI] [PubMed] [Google Scholar]
- 209.Morshed A., Dutta P. Hypoxic behavior in cells under controlled microfluidic environment. Bba-Gen Subjects. 2017;1861:759–771. doi: 10.1016/j.bbagen.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 210.Mignatti P., Rifkin D.B. Biology and biochemistry of proteinases in tumor invasion. Physiol. Rev. 1993;73:161–195. doi: 10.1152/physrev.1993.73.1.161. [DOI] [PubMed] [Google Scholar]
- 211.Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr. Opin. Cell Biol. 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 212.Thiery J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 213.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 214.Chen M.B., Whisler J.A., Fröse J., Yu C., Shin Y., Kamm R.D. On-chip human microvasculature assay for visualization and quantification of tumor cell extravasation dynamics. Nat. Protoc. 2017;12:865–880. doi: 10.1038/nprot.2017.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Friedl P., Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 216.Kim S.H., Hwang S.M., Lee J.M., Kang J.H., Chung I.Y., Chung B.G. Epithelial-to-mesenchymal transition of human lung alveolar epithelial cells in a microfluidic gradient device. Electrophoresis. 2013;34:441–447. doi: 10.1002/elps.201200386. [DOI] [PubMed] [Google Scholar]
- 217.Mani V., Lyu Z.L., Kumar V., Ercal B., Chen H., Malhotra S.V., Demirci U. Epithelial-to-mesenchymal transition (EMT) and drug response in dynamic bioengineered lung cancer microenvironment. Adv. Biosyst. 2019;3 doi: 10.1002/Adbi.201800223. [DOI] [PubMed] [Google Scholar]
- 218.Cano A., Pérez-Moreno M.A., Rodrigo I., Locascio A., Blanco M.J., del Barrio M.G., Portillo F., Nieto M.A. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 219.Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- 220.De Wever O., Derycke L., Hendrix A., De Meerleer G., Godeau F., Depypere H., Bracke M. Soluble cadherins as cancer biomarkers, Clin. Exp. Metastas. 2007;24:685–697. doi: 10.1007/s10585-007-9104-8. [DOI] [PubMed] [Google Scholar]
- 221.Zhang Z., Wang J., Shanmugasundaram K.B., Yeo B.L.D., Moeller A., Wuethrich A., Lin L.L., Trau M. Tracking drug-induced epithelial-mesenchymal transition in breast cancer by a microfluidic surface-enhanced Raman spectroscopy Immunoassay. Small. 2020;16 doi: 10.1002/Smll.201905614. [DOI] [PubMed] [Google Scholar]
- 222.Gwak H., Park S., Kim J., Lee J.D., Kim I.S., Kim S.I., Hyun K.A., Jung H.I. Microfluidic chip for rapid and selective isolation of tumor-derived extracellular vesicles for early diagnosis and metastatic risk evaluation of breast cancer. Biosens. Bioelectron. 2021;192 doi: 10.1016/J.Bios.2021.113495. [DOI] [PubMed] [Google Scholar]
- 223.Zhang D., Sheng Y.Q., Piano N., Jakuszeit T., Cozens E.J., Dong L.Q., Buell A.K., Pollet A., Lei I.M., Wang W.Y., Terentjev E., Huang Y.Y.S. Cancer cell migration on straight, wavy, loop and grid microfibre patterns. Biofabrication. 2022;14 doi: 10.1088/1758-5090/Ac48e6. [DOI] [PubMed] [Google Scholar]
- 224.Rizvi I., Gurkan U.A., Tasoglu S., Alagic N., Celli J.P., Mensah L.B., Mai Z., Demirci U., Hasan T. Flow induces epithelial-mesenchymal transition, cellular heterogeneity and biomarker modulation in 3D ovarian cancer nodules. P. Natl. Acad. Sci. USA. 2013;110:E1974–E1983. doi: 10.1073/pnas.1216989110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Takahashi Y., Akishima-Fukasawa Y., Kobayashi N., Sano T., Kosuge T., Nimura Y., Kanai Y., Hiraoka N. Prognostic value of tumor architecture, tumor-associated vascular characteristics, and expression of angiogenic molecules in pancreatic endocrine tumors. Clin. Cancer Res. 2007;13:187–196. doi: 10.1158/1078-0432.CCR-06-1408. [DOI] [PubMed] [Google Scholar]
- 226.Hsu T.H., Xiao J.L., Tsao Y.W., Kao Y.L., Huang S.H., Liao W.Y., Lee C.H. Analysis of the paracrine loop between cancer cells and fibroblasts using a microfluidic chip. Lab Chip. 2011;11:1808–1814. doi: 10.1039/c1lc20090a. [DOI] [PubMed] [Google Scholar]
- 227.Hsu T.H., Kao Y.L., Lin W.L., Xiao J.L., Kuo P.L., Wu C.W., Liao W.Y., Lee C.H. The migration speed of cancer cells influenced by macrophages and myofibroblasts co-cultured in a microfluidic chip. Integr. Biol-Uk. 2012;4:177–182. doi: 10.1039/c2ib00112h. [DOI] [PubMed] [Google Scholar]
- 228.An F., Hou Z., Wang X., Wang Z., Jing C., Sui R., Wu Y., Ma Y., Chang C., Liu S., Li M., Sun L., Gao Z., Zhang W., Lang Z., Zhao J., Qu Y., Xu L., Luo Y., Yan J., Wang Y., Xu J., Liu Q. A microfluidic demonstration of “cluster-sprout-infiltrating” mode for hypoxic mesenchymal stem cell guided cancer cell migration. Biomaterials. 2022;290 doi: 10.1016/j.biomaterials.2022.121848. [DOI] [PubMed] [Google Scholar]
- 229.Goh A., Yeh C.C., Lei K.F. Visualization and quantification of 3d tumor cell migration under extracellular stimulation. ACS Appl. Bio Mater. 2020;3:1506–1513. doi: 10.1021/acsabm.9b01134. [DOI] [PubMed] [Google Scholar]
- 230.Mi S., Liu Z., Du Z., Yi X., Sun W. Three-dimensional microfluidic tumor–macrophage system for breast cancer cell invasion. Biotechnol. Bioeng. 2019;116:1731–1741. doi: 10.1002/bit.26961. [DOI] [PubMed] [Google Scholar]
- 231.Nam H., Funamoto K., Jeon J.S.J.B. Cancer cell migration and cancer drug screening in oxygen tension gradient chip. Biomicrofluidics. 2020;14 doi: 10.1063/5.0011216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wong B.S., Shah S.R., Yankaskas C.L., Bajpai V.K., Wu P.-H., Chin D., Ifemembi B., ReFaey K., Schiapparelli P., Zheng X., Martin S.S., Fan C.-M., Quiñones-Hinojosa A., Konstantopoulos K. A microfluidic cell-migration assay for the prediction of progression-free survival and recurrence time of patients with glioblastoma. Nat. Biomed. Eng. 2021;5:26–40. doi: 10.1038/s41551-020-00621-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhang K., Du Z., Yuan T., Huang J., Zhao X., Mi S. Long-term cultured microvascular networks on chip for tumor vascularization research and drug testing. Biomicrofluidics. 2022;16 doi: 10.1063/5.0090027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wu Y., Zhou Y., Paul R., Qin X., Islam K., Liu Y. Adaptable microfluidic vessel-on-a-chip platform for investigating tumor metastatic transport in bloodstream. Anal. Chem. 2022;94:12159–12166. doi: 10.1021/acs.analchem.2c02556. [DOI] [PubMed] [Google Scholar]
- 235.Donato C., Kunz L., Castro-Giner F., Paasinen-Sohns A., Strittmatter K., Szczerba B.M., Scherrer R., Di Maggio N., Heusermann W., Biehlmaier O.J.C.r. Hypoxia triggers the intravasation of clustered circulating tumor cells. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Xiao Y., Kim D., Dura B., Zhang K., Yan R., Li H., Han E., Ip J., Zou P., Liu J., Chen A.T., Vortmeyer A.O., Zhou J., Fan R. Ex vivo dynamics of human glioblastoma cells in a microvasculature-on-a-chip system correlates with tumor heterogeneity and subtypes. Adv. Sci. 2019;6 doi: 10.1002/advs.201801531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Langley R.R., Fidler I.J. The seed and soil hypothesis revisited-The role of tumor-stroma interactions in metastasis to different organs. Int. J. Cancer. 2011;128:2527–2535. doi: 10.1002/ijc.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Qu W.L., Li S.H., Zhang M., Qiao Q. Pattern and prognosis of distant metastases in nasopharyngeal carcinoma: a large-population retrospective analysis. Cancer Med-US. 2020;9:6147–6158. doi: 10.1002/cam4.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Steeg P.S. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 240.Humayun M., Ayuso J.M., Brenneke R.A., Virumbrales-Munoz M., Lugo-Cintron K., Kerr S., Ponik S.M., Beebe D.J. Elucidating cancer-vascular paracrine signaling using a human organotypic breast cancer cell extravasation model. Biomaterials. 2021;270 doi: 10.1016/j.biomaterials.2020.120640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gilardi M., Bersini S., Valtorta S., Proietto M., Crippa M., Boussommier-Calleja A., Labelle M., Moresco R.M., Vanoni M., Kamm R.D., Moretti M. The driving role of the Cdk5/Tln1/FAKS732 axis in cancer cell extravasation dissected by human vascularized microfluidic models. Biomaterials. 2021;276 doi: 10.1016/j.biomaterials.2021.120975. [DOI] [PubMed] [Google Scholar]
- 242.Crippa M., Bersini S., Gilardi M., Arrigoni C., Gamba S., Falanga A., Candrian C., Dubini G., Vanoni M., Moretti M. A microphysiological early metastatic niche on a chip reveals how heterotypic cell interactions and inhibition of integrin subunit beta(3) impact breast cancer cell extravasation. Lab Chip. 2021;21:1185. doi: 10.1039/d1lc90024b. 1185. [DOI] [PubMed] [Google Scholar]
- 243.Pavesi A., Tan A.T., Koh S., Chia A., Colombo M., Antonecchia E., Miccolis C., Ceccarello E., Adriani G., Raimondi M.T. A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors. JCI Insight. 2017;2 doi: 10.1172/jci.insight.89762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Alizadeh A.A., Aranda V., Bardelli A., Blanpain C., Bock C., Borowski C., Caldas C., Califano A., Doherty M., Elsner M., Esteller M., Fitzgerald R., Korbel J.O., Lichter P., Mason C.E., Navin N., Pe'er D., Polyak K., Roberts C.W.M., Siu L., Snyder A., Stower H., Swanton C., Verhaak R.G.W., Zenklusen J.C., Zuber J., Zucman-Rossi J. Toward understanding and exploiting tumor heterogeneity. Nat. Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Shen H.L., Cai S.X., Wu C.X., Yang W.G., Yu H.B., Liu L.Q. Recent advances in three-dimensional multicellular spheroid culture and future development. Micromachines-Basel. 2021;12:96. doi: 10.3390/Mi12010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Nath S., Devi G.R. Three-dimensional culture systems in cancer research: focus on tumor spheroid model. Pharmacol. Therapeut. 2016;163:94–108. doi: 10.1016/j.pharmthera.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Cui J., Dean D., Hornicek F.J., Chen Z., Duan Z. The role of extracelluar matrix in osteosarcoma progression and metastasis. J. Exp. Clin. Cancer Res. 2020;39:178. doi: 10.1186/s13046-020-01685-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Christofides A., Strauss L., Yeo A., Cao C., Charest A., Boussiotis V.A. The complex role of tumor-infiltrating macrophages. Nat. Immun. 2022;23:1148–1156. doi: 10.1038/s41590-022-01267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Moore N., Doty D., Zielstorff M., Kariv I., Moy L.Y., Gimbel A., Chevillet J.R., Lowry N., Santos J., Mott V., Kratchman L., Lau T., Addona G., Chen H., Borenstein J.T. A multiplexed microfluidic system for evaluation of dynamics of immune–tumor interactions. Lab Chip. 2018;18:1844–1858. doi: 10.1039/C8LC00256H. [DOI] [PubMed] [Google Scholar]
- 250.Camargo C.P., Muhuri A.K., Alapan Y., Sestito L.F., Khosla M., Manspeaker M.P., Smith A.S., Paulos C.M., Thomas S.N. Adhesion analysis via a tumor vasculature-like microfluidic device identifies CD8(+) T cells with enhanced tumor homing to improve cell therapy. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Ayuso J.M., Truttschel R., Gong M.M., Humayun M., Virumbrales-Munoz M., Vitek R., Felder M., Gillies S.D., Sondel P., Wisinski K.B., Patankar M., Beebe D.J., Skala M.C. Evaluating natural killer cell cytotoxicity against solid tumors using a microfluidic model. OncoImmunology. 2019;8 doi: 10.1080/2162402X.2018.1553477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Liu Y., Yang Q., Cao L., Xu F. Analysis of leukocyte behaviors on microfluidic chips. Adv. Healthc. Mater. 2019;8 doi: 10.1002/adhm.201801406. [DOI] [PubMed] [Google Scholar]
- 253.Park S.E., Georgescu A., Huh D. Organoids-on-a-chip. Science. 2019;364:960–965. doi: 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tuveson D., Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 255.Zhang H., Schaefer A., Wang Y., Hodge R.G., Blake D.R., Diehl J.N., Papageorge A.G., Stachler M.D., Liao J., Zhou J., Wu Z., Akarca F.G., de Klerk L.K., Derks S., Pierobon M., Hoadley K.A., Wang T.C., Church G., Wong K.K., Petricoin E.F., Cox A.D., Lowy D.R., Der C.J., Bass A.J. Gain-of-function RHOA mutations promote focal adhesion kinase activation and dependency in diffuse gastric cancer. Cancer Discov. 2020;10:288–305. doi: 10.1158/2159-8290.CD-19-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Broutier L., Mastrogiovanni G., Verstegen M.M.A., Francies H.E., Gavarró L.M., Bradshaw C.R., Allen G.E., Arnes-Benito R., Sidorova O., Gaspersz M.P., Georgakopoulos N., Koo B.-K., Dietmann S., Davies S.E., Praseedom R.K., Lieshout R., Ijzermans J.N.M., Wigmore S.J., Saeb-Parsy K., Garnett M.J., van der Laan L.J.W., Huch M. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017;23:1424–1435. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Dekkers J.F., van Vliet E.J., Sachs N., Rosenbluth J.M., Kopper O., Rebel H.G., Wehrens E.J., Piani C., Visvader J.E., Verissimo C.S., Boj S.F., Brugge J.S., Clevers H., Rios A.C. Long-term culture, genetic manipulation and xenotransplantation of human normal and breast cancer organoids. Nat. Protoc. 2021;16:1936–1965. doi: 10.1038/s41596-020-00474-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Sachs N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A.V., Wind K., Gracanin A., Begthel H., Korving J., van Boxtel R., Duarte A.A., Lelieveld D., van Hoeck A., Ernst R.F., Blokzijl F., Nijman I.J., Hoogstraat M., van de Ven M., Egan D.A., Zinzalla V., Moll J., Boj S.F., Voest E.E., Wessels L., van Diest P.J., Rottenberg S., Vries R.G.J., Cuppen E., Clevers H. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 259.Fujii M., Shimokawa M., Date S., Takano A., Matano M., Nanki K., Ohta Y., Toshimitsu K., Nakazato Y., Kawasaki K., Uraoka T., Watanabe T., Kanai T., Sato T. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 260.LeSavage B.L., Suhar R.A., Broguiere N., Lutolf M.P., Heilshorn S.C. Next-generation cancer organoids. Nat. Mater. 2022;21:143–159. doi: 10.1038/s41563-021-01057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Neal J.T., Li X., Zhu J., Giangarra V., Grzeskowiak C.L., Ju J., Liu I.H., Chiou S.-H., Salahudeen A.A., Smith A.R., Deutsch B.C., Liao L., Zemek A.J., Zhao F., Karlsson K., Schultz L.M., Metzner T.J., Nadauld L.D., Tseng Y.-Y., Alkhairy S., Oh C., Keskula P., Mendoza-Villanueva D., De La Vega F.M., Kunz P.L., Liao J.C., Leppert J.T., Sunwoo J.B., Sabatti C., Boehm J.S., Hahn W.C., Zheng G.X.Y., Davis M.M., Kuo C.J. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Osuna de la Peña D., Trabulo S.M.D., Collin E., Liu Y., Sharma S., Tatari M., Behrens D., Erkan M., Lawlor R.T., Scarpa A., Heeschen C., Mata A., Loessner D. Bioengineered 3D models of human pancreatic cancer recapitulate in vivo tumour biology. Nat. Commun. 2021;12:5623. doi: 10.1038/s41467-021-25921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lo Y.-H., Karlsson K., Kuo C.J. Applications of organoids for cancer biology and precision medicine. Nat. Canc. 2020;1:761–773. doi: 10.1038/s43018-020-0102-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yuki K., Cheng N., Nakano M., Kuo C.J. Organoid models of tumor immunology. Trends Immunol. 2020;41:652–664. doi: 10.1016/j.it.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Xia X., Li F., He J., Aji R., Gao D. Organoid technology in cancer precision medicine. Canc. Lett. 2019;457:20–27. doi: 10.1016/j.canlet.2019.04.039. [DOI] [PubMed] [Google Scholar]
- 266.Driehuis E., van Hoeck A., Moore K., Kolders S., Francies H.E., Gulersonmez M.C., Stigter E.C.A., Burgering B., Geurts V., Gracanin A., Bounova G., Morsink F.H., Vries R., Boj S., van Es J., Offerhaus G.J.A., Kranenburg O., Garnett M.J., Wessels L., Cuppen E., Brosens L.A.A., Clevers H. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. P. Natl. Acad. Sci. USA. 2019;116:26580–26590. doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Prince E., Cruickshank J., Ba-Alawi W., Hodgson K., Haight J., Tobin C., Wakeman A., Avoulov A., Topolskaia V., Elliott M.J., McGuigan A.P., Berman H.K., Haibe-Kains B., Cescon D.W., Kumacheva E. Biomimetic hydrogel supports initiation and growth of patient-derived breast tumor organoids. Nat. Commun. 2022;13:1–12. doi: 10.1038/S41467-022-28788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zhao N., Powell R.T., Yuan X.Y., Bae G., Roarty K.P., Stossi F., Strempfl M., Toneff M.J., Johnson H.L., Mani S.A., Jones P., Stephan C.C., Rosen J.M. Morphological screening of mesenchymal mammary tumor organoids to identify drugs that reverse epithelial-mesenchymal transition. Nat. Commun. 2021;12:1–13. doi: 10.1038/S41467-021-24545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zhou Z., Van der Jeught K., Fang Y., Yu T., Li Y., Ao Z., Liu S., Zhang L., Yang Y., Eyvani H., Cox M.L., Wang X., He X., Ji G., Schneider B.P., Guo F., Wan J., Zhang X., Lu X. An organoid-based screen for epigenetic inhibitors that stimulate antigen presentation and potentiate T-cell-mediated cytotoxicity. Nat. Biomed. Eng. 2021;5:1320–1335. doi: 10.1038/s41551-021-00805-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Kawasaki K., Fujii M., Sugimoto S., Ishikawa K., Matano M., Ohta Y., Toshimitsu K., Takahashi S., Hosoe N., Sekine S., Kanai T., Sato T. Chromosome engineering of human colon-derived organoids to develop a model of traditional serrated adenoma. Gastroenterology. 2020;158:638–651. doi: 10.1053/j.gastro.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 271.Artegiani B., van Voorthuijsen L., Lindeboom R.G.H., Seinstra D., Heo I., Tapia P., Lopez-Iglesias C., Postrach D., Dayton T., Oka R., Hu H.L., van Boxtel R., van Es J.H., Offerhaus J., Peters P.J., van Rheenen J., Vermeulen M., Clevers H. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell. 2019;24:927–943. doi: 10.1016/j.stem.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 272.Nanki K., Toshimitsu K., Takano A., Fujii M., Shimokawa M., Ohta Y., Matano M., Seino T., Nishikori S., Ishikawa K., Kawasaki K., Togasaki K., Takahashi S., Sukawa Y., Ishida H., Sugimoto S., Kawakubo H., Kim J., Kitagawa Y., Sekine S., Koo B.K., Kanai T., Sato T. Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell. 2018;174:856–869. doi: 10.1016/j.cell.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 273.Bhadriraju K., Chen C.S. Engineering cellular microenvironments to improve cell-based drug testing. Drug Discov. Today. 2002;7:612–620. doi: 10.1016/S1359-6446(02)02273-0. [DOI] [PubMed] [Google Scholar]
- 274.Ingber D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022;23:467–491. doi: 10.1038/s41576-022-00466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Podaza E., Kuo H.-H., Nguyen J., Elemento O., Martin M.L. Next generation patient derived tumor organoids. Transl. Res. 2022;250:84–97. doi: 10.1016/j.trsl.2022.08.003. [DOI] [PubMed] [Google Scholar]
- 276.Ooft S.N., Weeber F., Dijkstra K.K., McLean C.M., Kaing S., van Werkhoven E., Schipper L., Hoes L., Vis D.J., van de Haar J.J.S.t.m. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- 277.Huang L., Holtzinger A., Jagan I., BeGora M., Lohse I., Ngai N., Nostro C., Wang R., Muthuswamy L.B., Crawford H.C. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell–and patient-derived tumor organoids. Nat. Med. 2015;21:1364–1371. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Rajasekar S., Lin D.S.Y., Abdul L., Liu A., Sotra A., Zhang F., Zhang B. IFlowPlate-A customized 384-well plate for the culture of perfusable vascularized colon organoids. Adv. Mater. 2020;32 doi: 10.1002/adma.202002974. [DOI] [PubMed] [Google Scholar]
- 279.Schuster B., Junkin M., Kashaf S.S., Romero-Calvo I., Kirby K., Matthews J., Weber C.R., Rzhetsky A., White K.P., Tay S. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat. Commun. 2020;11:5271. doi: 10.1038/s41467-020-19058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Grebenyuk S., Ranga A. Engineering organoid vascularization. Front. Bioeng. Biotechnol. 2019;7:39. doi: 10.3389/fbioe.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wörsdörfer P., Dalda N., Kern A., Krüger S., Wagner N., Kwok C.K., Henke E., Ergün S. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci. Rep-UK. 2019;9 doi: 10.1038/s41598-019-52204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Shi Y., Sun L., Wang M., Liu J., Zhong S., Li R., Li P., Guo L., Fang A., Chen R., Ge W.P., Wu Q., Wang X. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Fang G., Lu H., Al-Nakashli R., Chapman R., Zhang Y., Ju L.A., Lin G., Stenzel M.H., Jin D. Enabling peristalsis of human colon tumor organoids on microfluidic chips. Biofabrication. 2021;14 doi: 10.1088/1758-5090/ac2ef9. [DOI] [PubMed] [Google Scholar]
- 284.Novak R., Ingram M., Marquez S., Das D., Delahanty A., Herland A., Maoz B.M., Jeanty S.S.F., Somayaji M.R., Burt M., Calamari E., Chalkiadaki A., Cho A., Choe Y., Chou D.B., Cronce M., Dauth S., Divic T., Fernandez-Alcon J., Ferrante T., Ferrier J., FitzGerald E.A., Fleming R., Jalili-Firoozinezhad S., Grevesse T., Goss J.A., Hamkins-Indik T., Henry O., Hinojosa C., Huffstater T., Jang K.-J., Kujala V., Leng L., Mannix R., Milton Y., Nawroth J., Nestor B.A., Ng C.F., O'Connor B., Park T.-E., Sanchez H., Sliz J., Sontheimer-Phelps A., Swenor B., Thompson G., Touloumes G.J., Tranchemontagne Z., Wen N., Yadid M., Bahinski A., Hamilton G.A., Levner D., Levy O., Przekwas A., Prantil-Baun R., Parker K.K., Ingber D.E. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng. 2020;4:407–420. doi: 10.1038/s41551-019-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Stewart A., Denoyer D., Gao X., Toh Y.-C. The FDA modernisation act 2.0: bringing non-animal technologies to the regulatory table. Drug Discov. Today. 2023;28 doi: 10.1016/j.drudis.2023.103496. [DOI] [PubMed] [Google Scholar]
- 286.Piergiovanni M., Leite S.B., Corvi R., Whelan M. Standardisation needs for organ on chip devices. Lab Chip. 2021;21:2857–2868. doi: 10.1039/D1LC00241D. [DOI] [PubMed] [Google Scholar]
- 287.Fetah K.L., DiPardo B.J., Kongadzem E.-M., Tomlinson J.S., Elzagheid A., Elmusrati M., Khademhosseini A., Ashammakhi N. Cancer modeling-on-a-chip with future artificial intelligence integration. Small. 2019;15 doi: 10.1002/smll.201901985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zare Harofte S., Soltani M., Siavashy S., Raahemifar K. Recent advances of utilizing artificial intelligence in lab on a chip for diagnosis and treatment. Small. 2022;18 doi: 10.1002/smll.202203169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.